Graphical abstract
Keywords: Molecular dynamics simulation, RNA-dependent RNA polymerase, SARS-CoV, SARS-CoV-2
Abstract
One of the promising drug targets against COVID-19 is an RNA-dependent RNA polymerase (RdRp) of SARS-CoV-2. The tertiary structures of the SARS-CoV-2 and SARS-CoV RdRps are almost the same. However, the RNA-synthesizing activity of the SARS-CoV RdRp is higher than that of the SARS-CoV-2 RdRp. We performed molecular dynamics simulations and found differences in their dynamic properties. In the SARS-CoV RdRp, motifs A–G, which form the active site, are up to 63% closer to each other. We also observed cooperative domain motion in the SARS-CoV RdRp. Such dynamic differences may cause the activity differences between the two RdRps.
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a novel coronavirus and the causative virus of coronavirus disease 2019 (COVID-19) [1]. The World Health Organization (WHO) declared a pandemic in 2020 due to COVID-19. Currently, therapeutic drugs and vaccines against this disease are being developed in the world. One of the drug targets is an RNA-dependent RNA polymerase (RdRp) which replicates RNA of SARS-CoV-2 [2], [3], [4]. The growth of the virus is expected to be suppressed by inhibition of RNA replications with the therapeutic drugs.
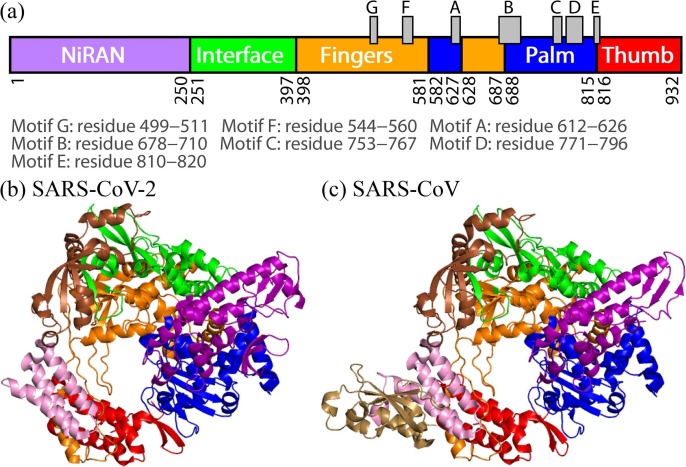
RdRp of SARS-CoV-2 is a complex of viral nonstructural proteins (nsps) such as nsp7, nsp8, and nsp12 [5], [6], [7], [8], [9], [10], [11]. The catalytic core of RdRp for RNA replications is nsp12, which shows little activity by itself [6], [7]. Other subunits including nsp7 and nsp8 assist nsp12 as cofactors [7]. There are mainly three domains in nsp12, which are nidovirus RdRp-associated nucleotidyltransferase (NiRAN) domain (residues 1–250), interface domain (residues 251–397), and conserved polymerase domain (residues 398–932) [12]. The polymerase domain is composed of three domains: fingers, palm, and thumb domains. In the polymerase domain, seven motifs A–G form active site of RdRp. The residues that compose the domains and motifs are shown in Fig. 1 a.
Fig. 1.
(a) Diagram of the domains in the SARS-CoV-2 nsp12 with motifs A–G. Tertiary structures of (b) SARS-CoV-2 RdRp and (c) SARS-CoV RdRp determined by using cryo-EM. NiRAN domain, interface domain, fingers domain, palm domain, and thumb domain are drawn by purple, green, orange, blue, and red, respectively, corresponding to the colors shown in Fig. 1a. The nsp7 and two nsp8s (nsp8-1 and nsp8-2) cofactors are also drawn by pink, brown, and sand.
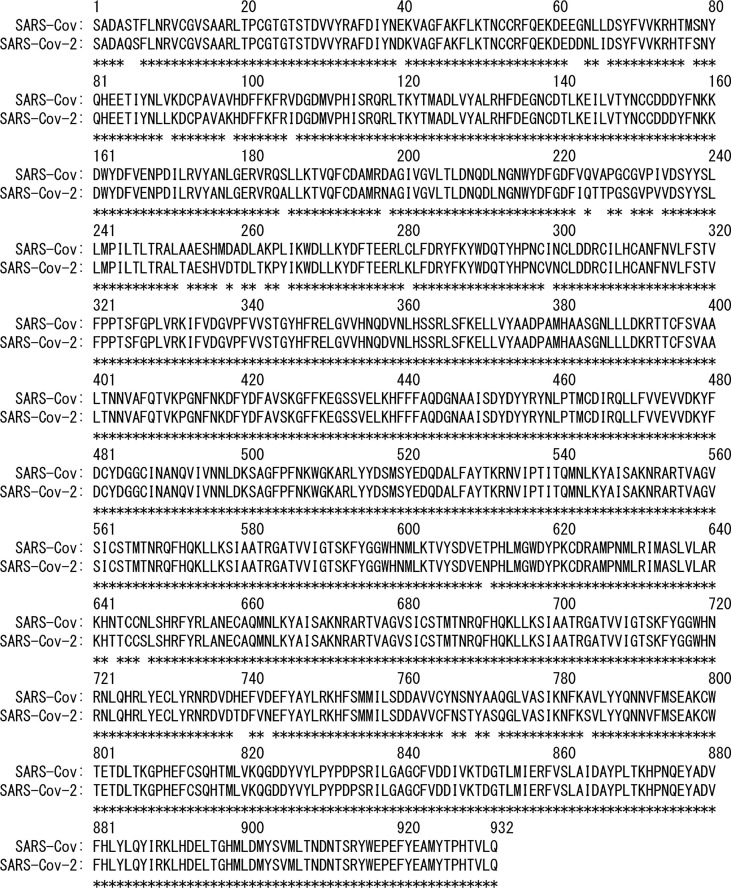
Recently, the tertiary structure of RdRp that consists of nsp7, nsp8, and nsp12 was determined by using cryogenic electron microscopy (cryo-EM), as shown in Fig. 1b (PDB entry: 7bv2) [8]. Not only this structure but also other structures were reported. For example, RdRp (nsp7, nsp8, and nsp12) bound to an RNA duplex [13] and that with an RNA template and nsp13 helicases [14] were determined. The structure of SARS-CoV-2 RdRp is almost the same as that of SARS-CoV RdRp which is also determined by using cryo-EM (Fig. 1c, PDB entry: 6nur) [7]. Both SARS-CoV-2 and SARS-CoV belong to the Coronaviridae family [15], and nsp12s of SARS-CoV-2 and SARS-CoV show more than 96% sequence identity [9], [12]. Amino-acid sequences of both nsp12 are shown in Fig. 2 . Although their tertiary structures and amino-acid sequences are almost the same, it was reported that the polymerase activities of RdRps are different [16]. The activity of the SARS-CoV RdRp is more than three times higher than that of the SARS-CoV-2 RdRp. It was also shown that by replacing only nsp12 of the SARS-CoV-2 RdRp with that of the SARS-CoV RdRp, the activity of the SARS-CoV-2 RdRp increases more than twice. Therefore, it is important to clarify the cause of this difference for understanding the RdRp function.
Fig. 2.
Amino-acid sequences of SARS-CoV-2 and SARS-CoV nsp12s. Asterisks indicate conserved amino-acid residues.
It is expected that the difference in activity between the two RdRps is due to a difference in their dynamic properties. This is because there is no difference in the static properties between the SARS-CoV-2 and SARS-CoV RdRps, such as their tertiary structures. In fact, it was experimentally shown that there is a difference between melting temperatures of the two nsp12s [16]. However, there is no study on the difference in dynamic properties between the two RdRps at the atomic level. To investigate the difference in the dynamic properties at the atomic level, we perform all-atom molecular dynamics (MD) simulations for the SARS-CoV-2 and SARS-CoV RdRps in explicit water. Since the determined tertiary structure for the SARS-CoV-2 RdRp include two Mg2+ ions that are essential as catalysts for RNA synthesis, we also investigate whether the dynamic properties change with and without these ions. In the PDB structure for the SARS-CoV RdRp, Mg2+ ions are not included. Therefore, our MD simulations are performed without these ions for the SARS-CoV RdRp.
2. Results
Movies 1, 2, and 3 show MD simulations for the SARS-CoV-2 RdRp with Mg2+ ions, the SARS-CoV-2 RdRp without Mg2+ ions, and the SARS-CoV RdRp, respectively. We analyzed these MD simulations. The obtained results for each system are compared below.
2.1. Comparison of ability of secondary structure formation
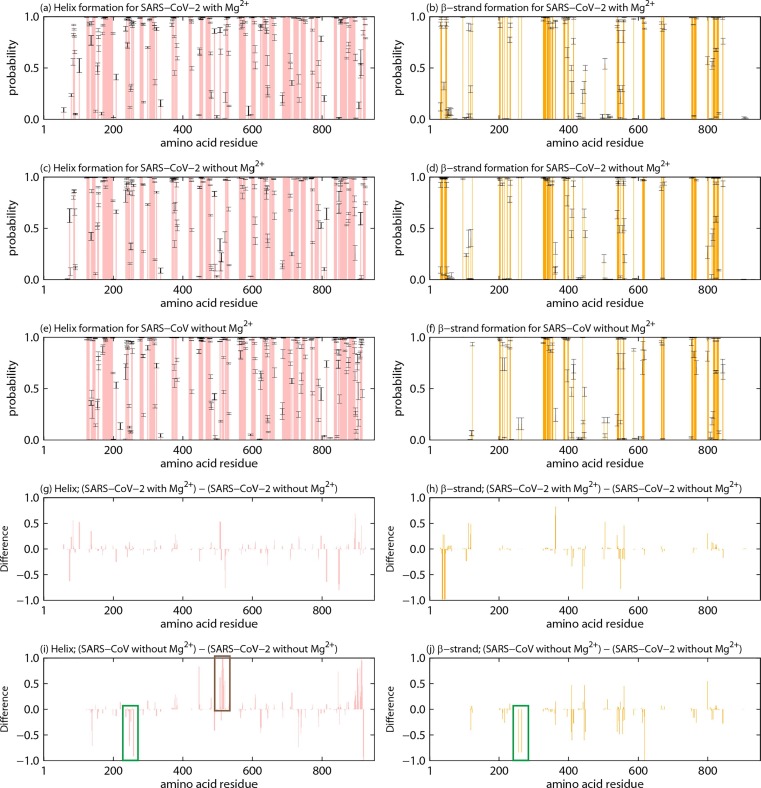
There is no difference between the secondary structures of the SARS-CoV and SARS-CoV-2 RdRps determined by using cryo-EM. However, it has not been clarified whether there is a difference in the ability of secondary structure formation. To investigate the ability of secondary structure formation of RdRps, probabilities of secondary structure formation of residues in nsp12 were calculated. Fig. 3 a, Fig. 3c, and Fig. 3e show the probability of helix formation for the SARS-CoV-2 nsp12 with Mg2+, that for the SARS-CoV-2 nsp12 without Mg2+, and that for the SARS-CoV nsp12, respectively. The probabilities of β-strand formation are presented in Fig. 3b, 3d, and 3f. Here, the define secondary structure of proteins (DSSP) criteria [17] were employed to define the secondary structures. Errors were estimated by the bootstrap method [18] (please see the Methods section for more details). There seems to be no difference between these probabilities for both helix formation and β-strand formation. To clarify the difference, we calculated the difference (subtraction) for each formation probability. The difference in helix formation between nsp12s are shown in Fig. 3g and 3i. The difference in β-strand formation is also presented in Fig. 3h and 3j. As seen in Fig. 3g and 3h, consecutive residues with significantly different probabilities of secondary structure formation were not found. In other words, the secondary structures of nsp12 did not change in the presence and absence of Mg2+. Conversely, large differences were seen between nsp12s of SARS-CoV-2 and SARS-CoV from Fig. 3i and 3j. The helix and β-strand structures were broken at residues in the vicinity of residue 260 in the SARS-CoV nsp12, as shown in the green rectangles in these figures. Residues in the vicinity of residue 515 in the SARS-CoV nsp12 formed more helix structures than the SARS-CoV-2 nsp12, as shown in the brown rectangle.
Fig. 3.
Probabilities of (a) helix and (b) β-strand formation of residues in the SARS-CoV-2 nsp12 with Mg2+. Probabilities of (c) helix and (d) β-strand formation for the SARS-CoV-2 nsp12 without Mg2+. Probabilities of (e) helix and (f) β-strand formation for the SARS-CoV nsp12. The difference between nsp12 of SARS-CoV-2 with Mg2+ and that without Mg2+ in probabilities of (g) helix formation and (h) β-strand formation. The difference between nsp12 of SARS-CoV and that of SARS-CoV-2 without Mg2 in probabilities of (i) helix formation and (j) β-strand formation. The green and brown rectangles show consecutive residues that have large differences.
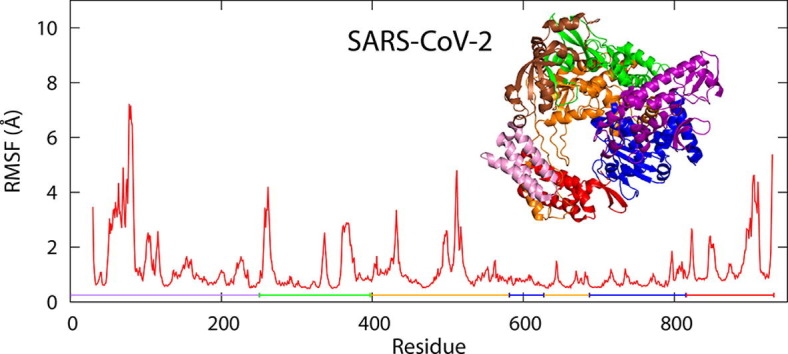
2.2. Comparison of fluctuations
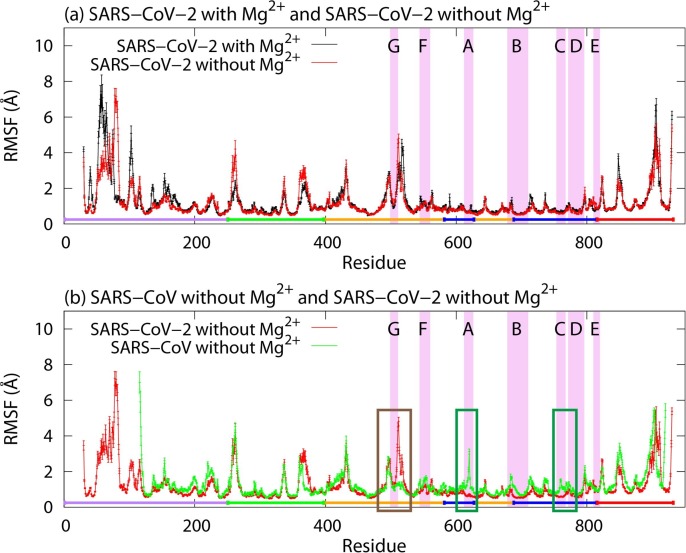
To see the difference between fluctuations of nsp12s, the root-mean-square fluctuation (RMSF) was calculated. Here, RMSF of residue i is defined by , where 〈 〉 represents time average and is the coordinate vector of the Cα atom of residue i. Fig. 4 a and Fig. 4b show RMSF of nsp12 of SARS-CoV-2 with Mg2+ and that of SARS-CoV, respectively. For comparison, RMSF of the SARS-CoV-2 nsp12 without Mg2+ is also shown in both figures. Except for the N-terminal and C-terminal regions, residues in the interface domain have large fluctuations in common to SARS-CoV-2 and SARS-CoV.
Fig. 4.
(a) RMSFs of the SARS-CoV-2 nsp12s with and without Mg2+. (b) RMSFs of nsp12s of SARS-CoV and SARS-CoV-2 without Mg2+. Purple line, green line, orange line, blue line, and red line represent the NiRAN domain, interface domain, fingers domain, palm domain, and thumb domain, respectively. The regions highlighted in violet show residues in motif A–G, and the uppercase letters are the labels of these motifs. The brown and green rectangles show residues that have large differences in RMSF.
Although there was a slight difference, residues before and after motif G fluctuated largely in both SARS-CoV-2 systems, as seen in Fig. 4a. Other residues in the SARS-CoV-2 nsp12s with and without Mg2+ have almost the same fluctuations, except for the N-terminal regions. Conversely, RMSF for the SARS-CoV nsp12 was different from that for the SARS-CoV-2 nsp12 in residues near residue 515, those near residue 620, and those near residue 760, as shown in the brown and green rectangles in Fig. 4b. Large fluctuations at residues near residue 515 seen in the SARS-CoV-2 nsp12 were suppressed in the SARS-CoV nsp12. A part of these residues composes motif G. Residues near residue 620 and residue 760 in the SARS-CoV nsp12 had large fluctuations. These residues are included in motifs A and C, and the two Mg2+ ions are located near these motifs.
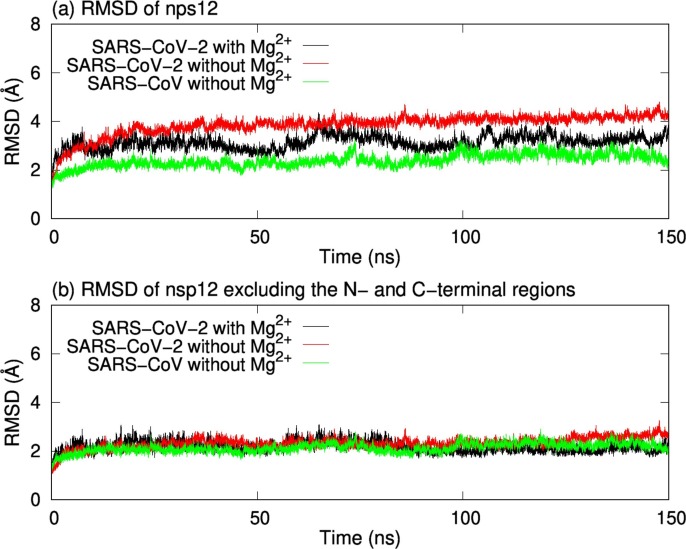
We also calculated the root-mean-square deviations (RMSDs) of nsp12s. Here, RMSD is defined by , where is the coordinate vector of the Cα atom of residue i in the initial structure, N is the number of the Cα atoms, and a rigid superposition is performed to minimize RMSD. Fig. 5 a shows the time series of RMSDs of nsp12s. The SARS-CoV-2 nsp12 without Mg2+ and that with Mg2+ had the largest RMSD and the second largest RMSD, respectively. However, there was no significant difference in fluctuations of RMSD among the three systems. To eliminate the effects of large fluctuations of the N- and C-terminal regions, the time series of RMSDs were calculated by using residues 130–890 in nsp12s, as shown in Fig. 5b. These residues are commonly shown in PDB structures of the SARS-CoV-2 nsp12 and the SARS-CoV nsp12 (the N-terminal region up to residue 116 is missing in PDB structure of the SARS-CoV nsp12). In Fig. 5b, the time series of RMSDs of these residues are almost the same in all systems.
Fig. 5.
(a) Time series of RMSDs of nsp12s. (b) Time series of RMSDs calculated by using residues 130–890 in nsp12s.
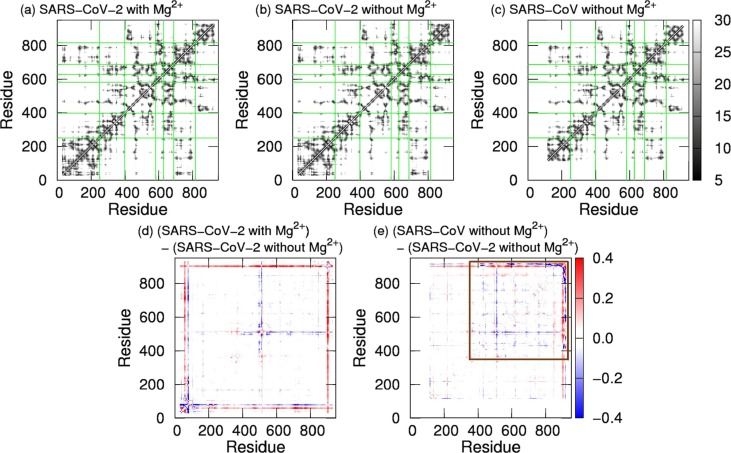
2.3. Difference between the tertiary structures
Distances between Cα atoms in nsp12s were calculated to see difference between the tertiary structures of the SARS-CoV-2 and SARS-CoV nsp12s. The distances were averaged over the production run. The average distances between Cα atoms are shown in Fig. 6 a–6c. From these figures, the following are common to the three systems. The NiRAN is spatially close to the palm domain, and the interface domain is close to the fingers domain. The thumb domain is in contact with only parts of the fingers and palm domains. To clarify the differences between the average distances for two systems, the following ratio of difference was calculated: , where is the average distance between Cα atoms of residues i and j in one system and is that in the other system. The ratios calculated for pairs of two systems are presented in Fig. 6d and 6e. As shown in Fig. 6d, there is not much difference between two SARS-CoV-2 systems, except for residues in the vicinity of residue 520 and the N- and C-terminal regions. The difference at residue 520, shown by the blue lines, indicates that residues near residue 520 in the SARS-CoV-2 nsp12 with Mg2+ are closer to the fingers, palm, and thumb domains than those in the SARS-CoV-2 nsp12 without Mg2+. Such differences between residue 520 in the SARS-CoV-2 nsp12 with and that without Mg2+ are also seen in Fig. 3g and 4a. Differences between the SARS-CoV-2 and SARS-CoV nsp12s are found in the region shown by the brown square in Fig. 6e. The bule lines (or blue mesh) are seen at residues near residues 430, 520, 560, 620, 690, 760, and 800. This indicates that distances between all motifs in the SARS-CoV nsp12 are shorter than those in the SARS-CoV-2 nsp12. This is because residues near residues 520, 560, 620, 690, 760, and 800 correspond to residues that compose motifs A–G. The distance between motifs F and G in the SARS-CoV nsp12 was up to 63% shorter in comparison with the SARS-CoV-2 nsp12 without Mg2+.
Fig. 6.
The average distances between Cα atoms for (a) the SARS-CoV-2 nsp12 with Mg2+, (b) the SARS-CoV-2 nsp12 without Mg2+, and (c) the SARS-CoV nsp12. (d) The ratios calculated from differences between the average distances for the SARS-CoV-2 nsp12 with Mg2+ and those for the SARS-CoV-2 nsp12 without Mg2+. (e) The ratios calculated from differences between the average distances for SARS-CoV and those for SARS-CoV-2 without Mg2. The brown square shows residues that have large differences.
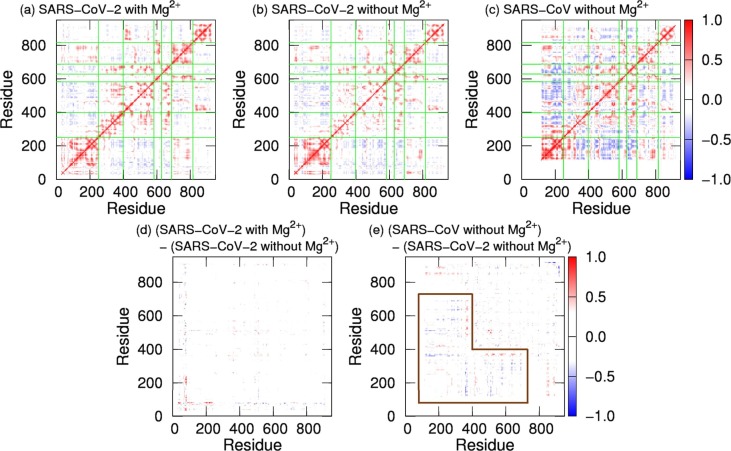
2.4. Correlation of domain motions
To investigate correlation between domain motions, the dynamic cross-correlation (DCC) was calculated. The DCC analysis is a useful method for analyzing domain motions [19]. DCC between residues i and j is given by , where . DCC for the SARS-CoV-2 and SARS-CoV nsp12s are presented in Fig. 7 a–7c. Here, red and blue show a positive correlation and a negative correlation, respectively. A positive (negative) correlation between residues i and j indicates that these residues move in the same (opposite) direction. In all systems, correlations are positive between most residues within the domains. However, the interface domain of the SARS-CoV nsp12 has both positive and negative correlations, and the border between these correlations is at residue 330. Residues in the interface domain before and after residue 330 have positive correlations with the NiRAN domain and the fingers domain, respectively.
Fig. 7.
DCC for (a) the SARS-CoV-2 nsp12 with Mg2+, (b) that without Mg2+, and (c) the SARS-CoV nsp12. (d) Differences between DCCs for the SARS-CoV-2 nsp12 with Mg2+ and those for the SARS-CoV-2 nsp12 without Mg2+. (e) Differences between DCCs for the SARS-CoV nsp12 and those for the SARS-CoV-2 nsp12 without Mg2+. The green lines show the borders between the domains in nsp12. Residues that have large differences are surrounded by the brown lines.
The differences (subtractions) between DCCs are shown in Fig. 7d and 7e. As seen in Fig. 7d, there is no difference between the SARS-CoV-2 nsp12s with and without Mg2+. The difference between the SARS-CoV-2 and SARS-CoV nsp12s is large in the NiRAN and interface domains, as shown by the region surrounded by the brown line. In the SARS-CoV nsp12, the NiRAN and interface domains before residue 330 have a strong negative correlation with the fingers domain. That is, the region before residue 330 has cooperative motion with the fingers domain so that they move closer to and away from each other.
3. Discussion
Using MD simulation, we investigated the secondary structures, fluctuations of structures, tertiary structures, and domain motions for the SARS-CoV-2 and SARS-CoV nsp12s. In the SARS-CoV-2 nsp12, these results without Mg2+ are the same as those with Mg2+. That is, the presence or absence of Mg2+ did not affect the results. Therefore, these structural and dynamical properties for the SARS-CoV nsp12 without Mg2+ are expected to be similar to those with Mg2+. We remark that the Mg2+ ion is essential as catalysts for RNA synthesis and is bound to the SARS-CoV RdRp in the active form [20]. However, Mg2+ is not included in the PDB structure (PDB entry: 6nur) [7].
Although there is no difference between the tertiary structures of the SARS-CoV and SARS-CoV-2 nsp12s determined by using cryo-EM, we found several differences in dynamic properties between the SARS-CoV and SARS-CoV-2 nsp12s. The secondary structures of residues in the vicinity of residue 260 of the SARS-CoV nsp12 tend to be broken in comparison with those of the SARS-CoV-2 nsp12 (Fig. 3i and 3j). There are several substitutions in the residues in the vicinity of residue 260 (see Fig. 2), and it is considered that such differences in residues affect the stabilities of secondary structures. Fluctuations of residues near residue 515 of the SARS-CoV nsp12 are suppressed as shown in Fig. 4b. This is because these residues form the helix structure as shown in Fig. 3i. Conversely, residues composing motifs A and C have larger fluctuations. Motifs A and C play an important role in the RdRp activity, and there is a possibility that their activity is enhanced by large fluctuations of these motifs. Fluctuations of the N-terminal residues before residue 100 are larger than those of other residues in the SARS-CoV-2 nsp12. In the SARS-CoV nsp12, the structure of the N-terminal residues before residue 116 has not been determined. This implies that fluctuations of the N-terminal residues are also large in the SARS-CoV nsp12. Moreover, in the SARS-CoV-2 nsp12, the increase in RMSD over time is larger than in the SARS-CoV nsp12, as shown in Fig. 5a. This is because the structure of the N-terminal residues, which are missing in the PDB structure of the SARS-CoV nsp12, is readily changed. In fact, the time series of RMSD excluding the N-terminal residues of the SARS-CoV-2 nsp12 agrees well with that of the SARS-CoV nsp12, as seen in Fig. 5b.
Distances between all motifs in the SARS-CoV nsp12 are shorter than those in the SARS-CoV-2 nsp12 as seen in Fig. 6e. This also may enhance the SARS-CoV RdRp activity. In the SARS-CoV nsp12, furthermore, the NiRAN and fingers domains have cooperative motion so that they move closer to and away from each other. The NiRAN domain is an important domain for the RdRp activity since removal of this domain reduces the RdRp activity [21]. Cooperative motion of the NiRAN domain with the core (fingers) domain of RdRp may increase the RdRp activity.
4. Conclusions
SARS-CoV-2 and SARS-CoV belong to the Coronaviridae family, and their nsp12s show more than 96% sequence identity. The tertiary structures of nsp12s determined by using cryo-EM are also almost the same. However, it was reported that by replacing only nsp12 of the SARS-CoV-2 RdRp with that of the SARS-CoV RdRp, the activity of the SARS-CoV-2 RdRp increases more than twice [16]. It is expected that there is a difference in the dynamic property between the SARS-CoV-2 and SARS-CoV nsp12s since there is no difference in the static properties between them. To investigate the difference in the dynamic properties, MD simulations of the SARS-CoV-2 and SARS-CoV RdRps were performed. We also investigated whether the dynamic properties of the SARS-CoV-2 nsp12 change with or without Mg2+ ions.
In our MD simulations, dynamic properties of the SARS-CoV-2 nsp12 did not change with or without the Mg2+ ions. Conversely, the dynamic properties of the SARS-CoV nsp12 are different from those of the SARS-CoV-2 nsp12. The secondary structures near residue 260 of the SARS-CoV nsp12 are broken, but the helix structure near residue 515 of the SARS-CoV nsp12 formed. In the SARS-CoV nsp12, fluctuations near residues composing motifs A and C are large. The tertiary structures of the core domains are stable in both nsp12s. Motifs A–G are closer to each other in the SARS-CoV nsp12. Regarding the domain motion, the NiRAN and fingers domains move cooperatively in the SARS-CoV nsp12. Such differences may be the cause of the differences in the activities between the two nsp12s. It may also make a difference in the binding ability of inhibitor candidates [22].
As for the difference between the two nsp12s, it was experimentally reported that the melting temperatures of these are different. Understanding the difference in the melting process is important for clarifying the difference in the activity between the two nsp12s. The generalized-ensemble algorithms [23], [24], [25], [26] such as the replica-permutation method [27], [28], [29], [30] are useful for studying the melting process [31], [32]. In the future, we will perform replica-permutation MD simulations for the RdRps of SARS-CoV-2 and SARS-CoV.
5. Methods
We employed the crystal structure of SARS-CoV RdRp (PDB entry: 6nur) [7] and SARS-CoV-2 RdRp (PDB entry: 7bv2) [8] as initial structures. Since one of the nsp8 of SARS-CoV-2 RdRp is missing in this crystal structure, the nsp8 corresponding to the chain D in the apo RdRp (PDB entry: 7bv1) [8] was added. The non-terminal missing residues in the crystal structure were complemented using the modeller plugin [33] in UCSF Chimera [34]. Hydrogen atoms were added using the LEaP and reduce plugin [35] in AMBER. To make the initial structure of SARS-CoV-2 RdRp without Mg2+ ions, the two Mg2+ ions were simply removed from that with Mg2+ ions. 14, 19, and 15 Na+ ions were added to neutralize the systems for SARS-CoV RdRp, SARS-CoV-2 RdRp without Mg2+, and SARS-CoV-2 RdRp with Mg2+, respectively. Explicit water molecules were also included. The whole system consisted of about 250,000 atoms (250,002, 250,012, and 250,013 atoms for SARS-CoV RdRp, SARS-CoV-2 RdRp without Mg2+, and SARS-CoV-2 RdRp with Mg2+, respectively). The entire system was immersed in a cubic box with the side length of 136.718 Å.
All MD simulations were performed using the Generalized-Ensemble Molecular Biophysics (GEMB) program, which was developed by one of the authors (H. O.). This program has been applied to several protein and peptide systems [36], [37], [38], [39], [40]. We applied the AMBER parm14SB force field [41] to the proteins and ions. We used the TIP3P rigid body model [42] for the water by adopting the symplectic [43] quaternion scheme [44], [45]. The electrostatic potential was calculated using the particle mesh Ewald (PME) method [46]. The cut-off distance was 12 Å for the Lennard–Jones (LJ) potential. Reversible multiple time-step MD techniques were applied. The time step was taken to be Δt = 0.5 fs for the bonding interactions of the protein atoms, Δt = 2.0 fs for the LJ interactions and the real part of the PME calculation of the protein atoms and those between the protein atoms and solvent molecules, and Δt = 4.0 fs for the LJ interaction and the real part of the PME calculation between the solvent molecules and the reciprocal part of the PME calculation of all the atoms. Because the symplectic rigid body algorithm was used for the water molecules, Δt can be taken as long as 4.0 fs [45]. MD simulations in the isothermal–isobaric ensemble were performed for 150 ns at 310 K and 0.1 MPa. The temperature was controlled using the Nosé–Hoover thermostat [47], [48], [49], and the pressure was controlled using the Andersen barostat [50]. The first 10 ns of the simulations was regarded as the equilibration, and the following 140 ns of the simulations was used for the analysis. Errors for physical quantities were estimated by the bootstrap method [18]. For the bootstrap analysis, the production run for 140 ns was divided into 28 bins every 5 ns. Bootstrap cycles were 106 cycles.
CRediT authorship contribution statement
Satoru G. Itoh: Conceptualization, Methodology, Formal analysis, Investigation, Data curation, Writing - original draft, Writing - review & editing, Visualization. Shoichi Tanimoto: Methodology, Validation, Writing - review & editing. Hisashi Okumura: Conceptualization, Methodology, Software, Validation, Formal analysis, Resources, Writing - review & editing, Visualization, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work used supercomputers at the Research Center for Computational Science, Okazaki Research Facilities, National Institutes of Natural Sciences, Japan, and computational resources of the TSUBAME3.0 provided by Tokyo Institute of Technology through the HPCI System Research Project (Project ID: hp200142).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cplett.2021.138819.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., Penzar D., Perlman S., Poon L.L.M., Samborskiy D.V., Sidorov I.A., Sola I., Ziebuhr J. Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. Nat. Microbiol. 2020;5:536. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Hüttenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., Mac Kain A., Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.-P., Liu Y., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O’Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu-Ozturk D., Wang H.-Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d’Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., García-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. Nature. 2020;583:459. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X., Zheng M., Chen L., Li H. Acta Pharmaceutica Sinica B. 2020;10:766. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanders J.M., Monogue M.L., Jodlowski T.Z., Cutrell J.B. JAMA. 2020;323:1824. doi: 10.1001/jama.2020.6019. [DOI] [PubMed] [Google Scholar]
- 5.Romano M., Ruggiero A., Squeglia F., Maga G., Berisio R. Cells. 2020;9:1267. doi: 10.3390/cells9051267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Subissi L., Posthuma C.C., Collet A., Zevenhoven-Dobbe J.C., Gorbalenya A.E., Decroly E., Snijder E.J., Canard B., Imbert I. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E3900. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kirchdoerfer R.N., Ward A.B. Nat. Commun. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yin W., Mao C., Luan X., Shen D.-D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M., Chang S., Xie Y.-C., Tian G., Jiang H.-W., Tao S.-C., Shen J., Jiang Y., Jiang H., Xu Y., Zhang S., Zhang Y., Xu H.E. Science. 2020;368:1499. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., Ge J., Zheng L., Zhang Y., Wang H., Zhu Y., Zhu C., Hu T., Hua T., Zhang B., Yang X., Li J., Yang H., Liu Z., Xu W., Guddat L.W., Wang Q., Lou Z., Rao Z. Science. 2020;368:779. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Venkataraman S., Prasad B.V.L.S., Selvarajan R. Viruses. 2018;10:76. doi: 10.3390/v10020076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y., Anirudhan V., Du R., Cui Q., Rong L. J. Med. Virol. 2021;93:300. doi: 10.1002/jmv.26264. [DOI] [PubMed] [Google Scholar]
- 12.Zhang W.-F., Stephen P., Thériault J.-F., Wang R., Lin S.-X. J. Phys. Chem. Lett. 2020;11:4430. doi: 10.1021/acs.jpclett.0c00571. [DOI] [PubMed] [Google Scholar]
- 13.Hillen H.S., Kokic G., Farnung L., Dienemann C., Tegunov D., Cramer P. Nature. 2020;584:154. doi: 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- 14.Chen J., Malone B., Llewellyn E., Grasso M., Shelton P.M.M., Olinares P.D.B., Maruthi K., Eng E.T., Vatandaslar H., Chait B.T., Kapoor T.M., Darst S.A., Campbell E.A. Cell. 2020;182:1560. doi: 10.1016/j.cell.2020.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. N. Engl. J. Med. 2020;382:727. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng Q., Peng R., Yuan B., Zhao J., Wang M., Wang X., Wang Q., Sun Y., Fan Z., Qi J., Gao G.F., Shi Y. Cell. Rep. 2020;31 doi: 10.1016/j.celrep.2020.107774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kabsch W., Sander C. Biopolymers. 1983;22:2577. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 18.Efron B. Ann. Statist. 1979;7:1. [Google Scholar]
- 19.McCammon J.A. Rep. Prog. Phys. 1984;47:1. [Google Scholar]
- 20.te Velthuis A.J.W., Arnold J.J., Cameron C.E., van den Worm S.H.E., Snijder E.J. Nucl. Acids Res. 2009;38:203. doi: 10.1093/nar/gkp904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lehmann K.C., Gulyaeva A., Zevenhoven-Dobbe J.C., Janssen G.M.C., Ruben M., Overkleeft H.S., van Veelen P.A., Samborskiy D.V., Kravchenko A.A., Leontovich A.M., Sidorov I.A., Snijder E.J., Posthuma C.C., Gorbalenya A.E. Nucl. Acids Res. 2015;43:8416. doi: 10.1093/nar/gkv838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ribaudo G., Ongaro A., Oselladore E., Zagotto G., Memo M., Gianoncelli A. J. Biomol. Struct. Dyn. 2020 doi: 10.1080/07391102.2020.1822209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Itoh S.G., Okumura H., Okamoto Y. J. Chem. Phys. 2010;132 doi: 10.1063/1.3372767. [DOI] [PubMed] [Google Scholar]
- 24.Itoh S.G., Okumura H. J. Comput. Chem. 2013;34:622. doi: 10.1002/jcc.23167. [DOI] [PubMed] [Google Scholar]
- 25.Hukushima K., Nemoto K. J. Phys. Soc. Jpn. 1996;65:1604. [Google Scholar]
- 26.Mitsutake A., Sugita Y., Okamoto Y. Biopolymers. 2001;60:96. doi: 10.1002/1097-0282(2001)60:2<96::AID-BIP1007>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 27.Itoh S.G., Okumura H. J. Comput. Chem. 2013;34:2493. doi: 10.1002/jcc.23402. [DOI] [PubMed] [Google Scholar]
- 28.Itoh S.G., Okumura H. J. Chem. Theory. Comput. 2013;9:570. doi: 10.1021/ct3007919. [DOI] [PubMed] [Google Scholar]
- 29.Yamauchi M., Okumura H. J. Chem. Phys. 2017;147 doi: 10.1063/1.4996431. [DOI] [PubMed] [Google Scholar]
- 30.Yamauchi M., Okumura H. J. Comput. Chem. 2019;40:2694. doi: 10.1002/jcc.26030. [DOI] [PubMed] [Google Scholar]
- 31.Itoh S.G., Okumura H. J. Phys. Chem. B. 2014;118:11428. doi: 10.1021/jp505984e. [DOI] [PubMed] [Google Scholar]
- 32.Itoh S.G., Okumura H. J. Phys. Chem. B. 2016;120:6555. doi: 10.1021/acs.jpcb.6b03828. [DOI] [PubMed] [Google Scholar]
- 33.Sali A., Blundell T.L. J. Mol. Biol. 1993;234:779. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 34.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. J. Comput. Chem. 2004;25:1605. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 35.Word J.M., Lovell S.C., Richardson J.S., Richardson D.C. J. Mol. Biol. 1999;285:1735. doi: 10.1006/jmbi.1998.2401. [DOI] [PubMed] [Google Scholar]
- 36.Okumura H. Proteins. 2012;80:2397. doi: 10.1002/prot.24125. [DOI] [PubMed] [Google Scholar]
- 37.Okumura H., Itoh S.G. Phys. Chem. Chem. Phys. 2013;15:13852. doi: 10.1039/c3cp44443k. [DOI] [PubMed] [Google Scholar]
- 38.Chiang H.L., Chen C.J., Okumura H., Hu C.K. J. Comput. Chem. 2014;35:1430. doi: 10.1002/jcc.23633. [DOI] [PubMed] [Google Scholar]
- 39.Okumura H., Itoh S.G. J. Am. Chem. Soc. 2014;136:10549. doi: 10.1021/ja502749f. [DOI] [PubMed] [Google Scholar]
- 40.Okumura H., Itoh S.G. Sci. Rep. 2016;106:38422. doi: 10.1038/srep38422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maier J.A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K.E., Simmerling C. J. Chem. Theory. Comput. 2015;11:3696. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. J. Chem. Phys. 1983;79:926. [Google Scholar]
- 43.Yoshida H. Phys. Lett. A. 1990;150:262. [Google Scholar]
- 44.Miller T.F., Eleftheriou M., Pattnaik P., Ndirango A., Newns D., Martyna G.J. J. Chem. Phys. 2002;116:8649. [Google Scholar]
- 45.Okumura H., Itoh S.G., Okamoto Y. J. Chem. Phys. 2007;126 doi: 10.1063/1.2434972. [DOI] [PubMed] [Google Scholar]
- 46.Essmann U., Perera L., Berkowitz M.L. J. Chem. Phys. 1995;103:8577. [Google Scholar]
- 47.Nosé S. Mol. Phys. 1984;52:255. [Google Scholar]
- 48.Nosé S. J. Chem. Phys. 1984;81:511. [Google Scholar]
- 49.Hoover W.G. Phys. Rev. A. 1985;31:1695. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 50.Andersen H.C. J. Chem. Phys. 1980;72:2384. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.