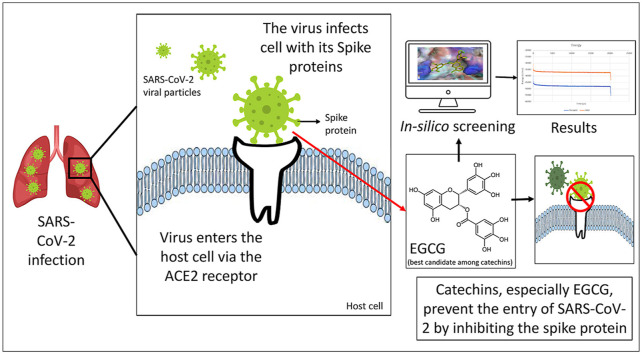
Abstract
Background
The global pandemic caused by a RNA virus capable of infecting humans and animals, has resulted in millions of deaths worldwide. Severe acute respiratory syndrome corona virus 2 (SARS-CoV-2) infects the lungs, and the gastrointestinal tract to some extent. Rapid structural mutations have increased the virulence and infectivity of the virus drastically. One such mutated strain known as the UK variant has caused many deaths in the United Kingdom.
Hypothesis
Among several indigenous natural ingredients used for prevention and cure of many diseases, the catechins have been reported for their antiviral activity, even against SARS-CoV-2. Characteristic mutations present on the spike protein have presented the newer strain its enhanced infectivity. The spike protein helps the virus bind to ACE2 receptor of the host cell and hence is a drug target. Catechins have been reported for their entry-inhibitory activity against several viruses.
Method
In this study, we performed molecular docking of different catechins with the wild and mutant variants of the spike protein of SARS-CoV-2. The stability of the best docked complexes was validated using molecular dynamics simulation.
Results
The in-silico studies show that the catechins form favourable interactions with the spike protein and can potentially impair its function. Epigallocatechin gallate (EGCG) showed the best binding among the catechins against both the strains. Both the protein-ligand complexes were stable throughout the simulation time frame.
Conclusion
The outcomes should encourage further exploration of the antiviral activity of EGCG against SARS-CoV-2 and its variants.
Keywords: SARS-CoV-2, UK variant, Spike protein, Catechins, Epigallocatechin gallate
Graphical abstract
Summary
The UK variant of SARS-CoV-2 has caused a sudden increase in the number of COVID-19 cases. Characteristic point mutations present on the spike protein have made this new strain far more infectious than the wild strain. There are very few reports that have validated the efficiency of vaccines and approved drugs on the mutated strain. Phytoconstituents, specifically catechins have gathered a lot of attention owing to their immunomodulatory and antiviral activity against several viruses. In this manuscript, we screened several natural catechins against the spike proteins of the wild strain of SARS-CoV-2 and VUI 202012/01, commonly known as the UK variant. We used in-silico tools such as molecular docking and molecular dynamics (MD) simulation to study the binding affinities of catechins towards the active binding sites of spike proteins of both the strains. It was found that all the catechins show better binding affinities towards the spike proteins then their native ligands. Hence, catechins can preferentially bind to the protein and form complexes. Among the catechins screened in this study, EGCG showed the highest binding affinity. The complex was formed with several hydrogen bonds, van der Waals interactions, and other hydrophobic bonds. The stability of this complex was validated using MD simulation of 20 ns. The complex of EGCG with both the wild and spike protein was observed to be stable throughout the simulation. These results show that the catechins, especially EGCG, can be developed as entry-inhibitors of SARS-CoV-2 and its variants. Their in-silico activity however needs to be validated in in vitro and in vivo models. Nevertheless, these molecules offer a promising role in the treatment of viral infections like COVID-19. Another drawback of these molecules are their pharmacokinetic properties and poor bioavailability. These need to be improved by developing drug-conjugates and suitable dosage forms.
1. Introduction
The emergence of a positive-sense virus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), since December 2019 has severely affected global health and economics. Its human-to-human transmission through saliva droplets and mucosal discharges substantially contributed to a global pandemic with over 139 million cases and close to 3 million deaths reported as of mid-April 2021 [1]. SARS-CoV-2 is a single stranded RNA virus which has glycosylated spike proteins covering its surface [2]. These proteins facilitate viral entry by binding to angiotensin converting enzyme 2 (ACE2) receptors of the host cell [3]. Viral RNA is released and translated upon entry of the virus into the cell. Further, viral RNA is replicated and more proteins are translated and synthesized. Assembly and packaging of these proteins trigger the release of viral particles [4].
Owing to the extraordinary efforts of scientists and pharmaceutical companies, approval of 9 vaccines worldwide and clinical trials of more than 50 vaccines have been achieved for this outbreak [5]. However, the viral mutations of SARS-CoV-2 are raising concerns [6]. Out of these, three variants named as B.1.1.7 lineage or E484K or VUI 202012/01, first detected in the United Kingdom (UK), B.1.351 lineage or 501Y.V2 or 20H/501Y.V2 detected in South Africa, and P.1 detected in Brazil, have been found to spread more rapidly than other variants [[7], [8], [9], [10], [11]]. The UK variant specifically has been reported with more than 160,000 cases in England, Scotland, and Wales [12]. Its affinity to bind human cells has been reported to be increased partly because of mutations in spike proteins and other functional locations on the genome. The 69–70 and 144 deletions in the N-terminal domain, N501Y in the receptor binding domain, A570D in the sub-domain-1, D614G and P681H near the host processing furin cleavage site, T716I, S982A and D1118H in the S2 domain are the key mutations on the spike protein (14). The UK variant of the coronavirus is reported to have a 70% increased rate of infection and it eventually led to a strict lockdown in the UK [13]. The total number of new cases in UK for the month of December 2020 and January 2021 increased multiple folds as compared to previous months. More than 68,000 cases were reported on January 10, 2021 while on 10th October, there were roughly 15,000 cases [14]. The effectiveness of approved vaccines against the UK as well as other variants is currently being evaluated. One report suggests that Novavax, a vaccine developed by US biotechnology, has 85.6% efficiency against the UK variant, 60% against the South African variant, and 95.6% against the original strain [15].
Catechins are plant-based, polyphenolic, bioactive compounds found in tea, cocoa, red wine, fruits like apples, grapes, cherries, berries. They are flavonoids classified as flavanols and include catechin (C), catechin gallate (CG), epicatechin (EC), gallocatechin (GC), epigallocatechin (EGC), epicatechin gallate (ECG), gallocatechin gallate (GCG), and epigallocatechin gallate (EGCG). Catechins and their derivatives can neutralize free radicals, and this antioxidant property has been found beneficial to conditions such as obesity, diabetes, infections, cancer, cardiovascular and neurodegenerative diseases [16]. These molecules have shown to exhibit strong immunomodulatory, anti-inflammatory, anti-proliferative, and anti-cancer properties [[17], [18], [19]]. Several catechins have been reported to be effective against viral infections including HIV, Hepatitis B and C, influenza, chikungunya, adenovirus, Ebola, and gastroenteritis coronavirus [[20], [21], [22], [23], [24], [25]] and recently against the SARS-CoV-2 [26,27]. There are several reports that show the entry inhibiting activity of catechins in viruses like HIV, HSV, Influenza, etc. [[28], [29], [30]]. A recent study by Joseph et al. highlights the role of catechins as entry-inhibitors against SARS-CoV-1, Middle East Respiratory Syndrome-CoV, and SARS-CoV-2 [31]. Catechins from green tea bind to S1 domain of the spike protein and block its interaction with ACE2 receptor, thus preventing the viral infection. Another study suggests that catechins bind to the amino acids near the receptor binding domain (RBD) of the spike protein and cause fluctuations of alpha helices and beta strands of the RBD-ACE2 complex, thereby hindering the formation of the RBD-ACE2 complex [32].
Considering the increasing number of cases due to SARS-CoV-2 and its new variants, apart from following social distancing protocols, there is a need for developing safe and effective strategies. Although the process of vaccination has already begun in many countries, it will take several months before it is fully completed. The immunomodulatory and entry-inhibiting activity of the catechins offer an effective approach to boost immunity and decrease an individual's chances of getting infected. Herein, we have used computational tools to screen several bioactive catechins against the spike protein of the wild and VUI 202012/01 strain of SARS-CoV-2. We employed tools like molecular docking and molecular dynamics (MD) simulation to predict the behaviour of catechins with both the protein structures. We compared the docking scores of the catechins to identify the catechin that showed the best in-silico activity against the wild strain of SARS-CoV-2. We further performed similar studies with the spike protein of mutated strain to see if the activity is reproduced in presence of mutations. We also identified the most potential active biomolecule against mutant strain using in-silico tools.
2. Methods
2.1. Procuring of structures
The crystallographic 3D structure of the spike protein was procured from the RCSB Protein Data Bank [33].
The prefusion SARS-CoV-2 spike glycoprotein is available on the website by the PDB ID 6VSB [34]. The complete structure contains three chains (A, B, C) of the macromolecule, seventeen chains of oligosaccharides (D-T), and three chains of ligands (A, B, C). The structures of the catechins were obtained from the PubChem database. The IDs of C, CG, EC, GC, EGC, ECG, GCG, and EGCG are 9064, 6419835, 72276, 65084, 72277, 107905, 5276890, and 65064, respectively.
2.2. Mutation in spike protein
The UK variant of SARS-CoV-2 contains multiple point mutations on the spike glycoprotein. The mutations were produced using Mutagenesis wizard of the PyMol Viewer. Each of the amino acid in the wild structure was changed to the mutated amino acid as reported in the literature. Mutations lead to change in the overall stability and energy of the protein. These changes in the energy of the protein due to point mutations were calculated using the DynaMut web server [35].
2.3. Protein and ligand preparation
The structures of wild protein, mutated protein and the catechins were prepared and minimized using the UCSF Chimera visualizer.
A protocol that we previously used in one of our studies for the preparation and minimization of ligands and receptors was employed [36]. The native ligands and non-standard residues were removed. The protein was then minimized with 200 steepest descent steps and 20 conjugate gradient steps of size 0.02 Å each. Dunbrack rotamer library was used to add hydrogen atoms and charges [37]. The resultant structures of receptors and the ligands were saved as pdb files.
2.4. Binding site analysis
The reported structure of wild spike glycoprotein has several hydrophobic cavities and hence a number of binding sites are located on the surface of the protein.
Out of the available binding sites, the active binding site was located using the native ligand and the ligand interactions mentioned in the primary citation of the protein 6VSB [34]. Further, a docking grid was generated using a site mapping tool AutoGridFR [38].
2.5. Molecular docking
After the protein structures and ligands were minimized and the active binding site was located, the ligands were docked to the protein using PyRx, an open-source virtual screening software [39]. The ligands and structures of wild protein and mutated protein were imported as pdbqt files. The conversion of pdb files to corresponding pdbqt files was performed using OpenBabel [40]. The docking grid was set using docking coordinates produced by AutoGridFR. PyRx uses the AutoDock Vina algorithm to perform the molecular docking and evaluate the binding affinities. Top 8 binding poses were generated for all the ligands against both the wild protein and the mutated protein each. The binding poses and the interactions of the best pose were visualized using the Discovery Studio Visualizer.
2.6. MD simulations
MD simulations were performed using Molecular Operating Environment (MOE) software on a Windows 10 system.
Two docked complexes, EGCG and wild spike glycoprotein complex and the EGCG and mutated spike glycoprotein, were simulated. The first step was to add a solvent box around the protein-ligand complex to represent a biological system. A water box of appropriate length to soak entire complex was added using the Water Soak module of MOE. The complexes were then protonated at a pH of 7.4 at a temperature of 298 K. All the atoms of the complex were typed with the AMBER99 forcefield. Next, the complex was minimized to get a stable conformation. Finally, the MD simulation was performed. The complex was heated at a temperature of 310 K from an initial temperature of 300 K for a period of 100 ps in a NVT ensemble at a pressure of 101 kPA. The complex was then simulated for a period of 20 ns at a temperature of 310 K at same pressure in NVT ensemble. Lastly, the complex was cooled down to a temperature of 300 K for a time period of 100 ps. The trajectory generated was analysed by plotting the energy changes through the timesteps.
2.7. Pharmacokinetic properties analysis
Pharmacokinetic properties like absorption, distribution, metabolism, excretion and toxicity were evaluated using the pkCSM web server [41].
The predicted values were compared with previously reported experimental values from literature.
3. Results and discussion
3.1. Locating and analysing the binding site
Proteins usually have several hydrophobic cavities in their surface.
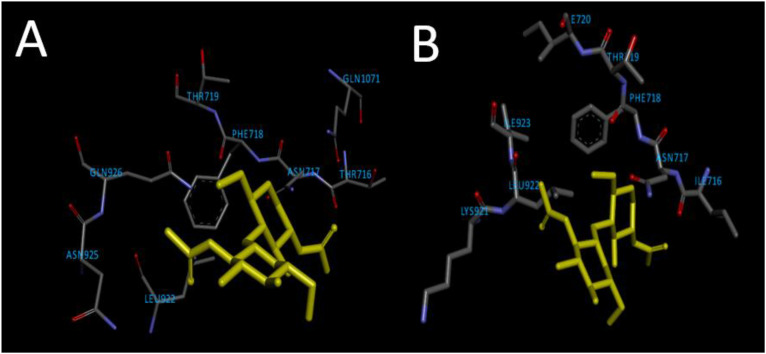
Some or all of these cavities can be potential targets for small molecules like drugs to bind. However, it is important for the drugs to bind to the active binding site to produce their desired therapeutic activity. Hence, the identification of active sites is the important step prior to molecular docking. The primary citation reports the interaction of the native ligand 2-acetamido-2-deoxy-beta-d-glucopyranose (NAG) with the A chain of the spike glycoprotein. The amino acids GLN1071 and ASN717 form hydrogen bonds with the oxygen atom of NAG [34]. This active binding site predicted by AutoGridFR (Fig. 1 ) consists of the amino acids THR719, PHE718, ASN717, GLN1071, THR716, LYS1073, LEU894, PHE1075, LYS1073, GLU1072, TYR1110, and PHE1109 with an active site score of 66.64. Out of all the mutations reported in the UK variant of SARS-CoV-2, only the THR716 amino acid underwent a mutation to ILE716 while the others remained unaltered. After producing the mutation, the binding site was revaluated and it received an active binding score of 63.68, which was close to that of the wild protein. The mutation did not alter the stability, volume and conformation of the active binding site. A binding grid was generated with the centroid coordinates of 224.59,225.80 and 174.19 with a cell length of 20 Å across all three dimensions.
Fig. 1.
Amino acid composition of the active binding site on spike protein of wild strain (A) and that of mutated strain (B), along with the native ligand NAG.
3.2. Mutagenesis analysis
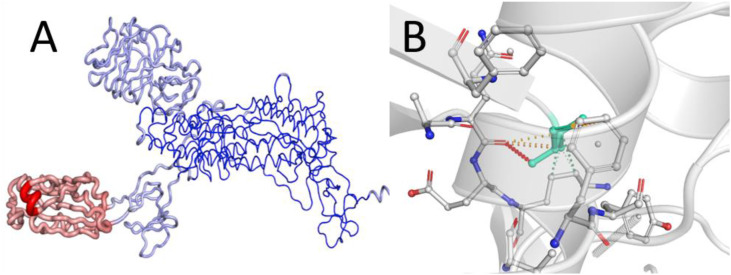
The mutations observed on the UK variant of the SARS-CoV-2 are point mutations at several locations of the A chain of the protein. A change in the amino acid brings about changes in the conformational energy, vibrational entropy and a change in overall Gibbs energy of the protein. The amino acid mutations present in the active binding site have a direct impact on binding of small molecules to that site. The other mutations also bring a change in the protein energy and the amino acid confirmations but have a comparatively lower impact on the targetability of the binding site as these changes do not take place on the active binding site. Nevertheless, these mutations are important from the overall protein structure point of view. The only amino acid residue present in the active binding site was THR716 which mutated to ILE716. This mutation increased the protein energy and thus decreased the stability. However, this change was very small considering the overall protein energy. The significant change that these mutations produce was accounted to the conformation of the active binding site and the types of interactions allowed with the ligand. Considering all the mutations on the protein, the overall stability was negligibly affected. The change in conformation altered the interactions which may alter the binding mode as well as the docking score. Fig. 2 represents the change in orientation of the protein change as well as the binding site. The changes in protein energies are represented in Table 1 .
Fig. 2.
Change in the 3D orientation of the (A) spike protein (B) and the active binding site.
Table 1.
Analysis of change in energy (in kJ/mol) due to point mutations.
| Mutation | Δ (Stability) | Δ (ΔG) | Δ (Vibrational Entropy) |
|---|---|---|---|
| N501Y | 4.406 | 0.427 | −4.008 |
| A570D | −0.027 | 0.03 | 0.033 |
| D614G | −0.098 | −0.257 | 0.123 |
| T716I | 0.205 | 0.76 | −0.256 |
| S982A | −0.034 | 0.467 | 0.043 |
| D1118H | 0.063 | 0.712 | −0.078 |
3.3. Molecular docking
Molecular docking is a technique that explains how two or more molecules when placed in a restricted system under a defined forcefield interact with each other. This technique gives the most stable conformation of the ligand and the protein in the active site. The energy of interaction of the protein-drug complex, i.e., the binding energy is reflected in the docking score. There are several methods to calculate the free binding energies.
In mathematical terms, the binding energy is calculated empirically as
It is the sum of the fractions of energy changes by formation of hydrogen bonds, electrostatic interactions, hydrophobic interactions, van der Waals forces and entropy changes in the protein ligand docked complex [42]. The individual energy terms (ΔG) are multiplied by respective weighing coefficients (cj) to account for their contribution to the overall binding energy. Each of the participating energy term further can be calculated using complex mathematical equations.
A docking score (also sometimes called as binding score) is a software specific scoring function derived from the binding energy, that helps us rank different protein-ligand complexes based on energy calculations. The absolute values of docking scores may vary across softwares, but the relative values of complexes docked on the same platform are analogous. A higher (more negative) docking score indicates a more stable interaction between the drug and the protein. Further, the technique gives the types of interactions between the molecules, which help to identify the key moieties and amino acids participating in the interactions. The docking study gives the relative binding affinities of the ligand towards the protein in comparison with the native ligand and aids in screening drug candidates. In this study, we used molecular docking to screen the activity of the catechins against the spike protein of both the wild and mutated strains of SARS-CoV-2. Table 2 represents the docking scores of the catechins.
Table 2.
Docking scores of catechins against spike protein of wild and mutated strains.
| Catechin | Docking scores |
|
|---|---|---|
| Wild | Mutated | |
| NAG | −4.8 | −5.0 |
| Catechin | −5.7 | −5.7 |
| Catechin Gallate | −5.8 | −6.5 |
| Epicatechin | −5.7 | −6.0 |
| Gallocatechin | −5.8 | −5.9 |
| Epigallocatechin | −5.9 | −5.7 |
| Epicatechin Gallate | −6.1 | −6.1 |
| Gallocatechin Gallate | −5.9 | −6.3 |
| Epigallocatechin Gallate | −6.3 | −6.4 |
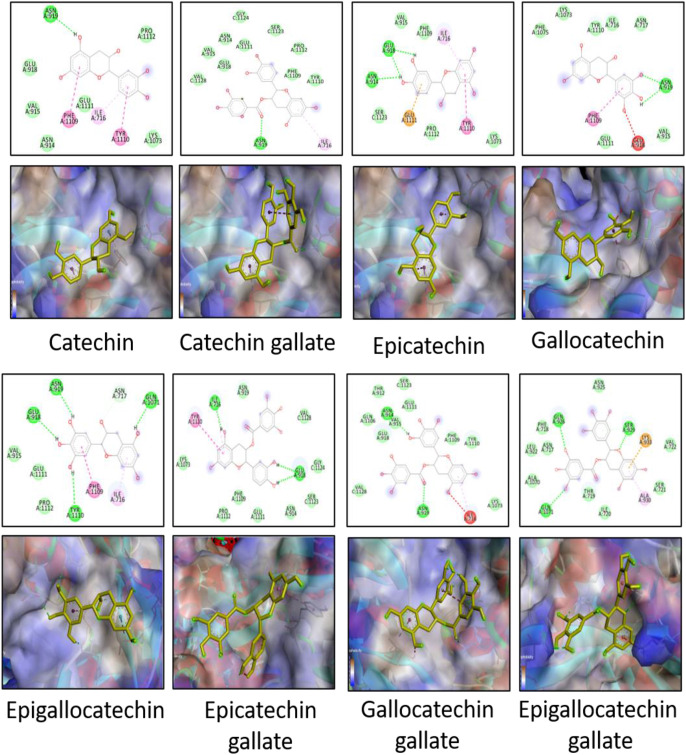
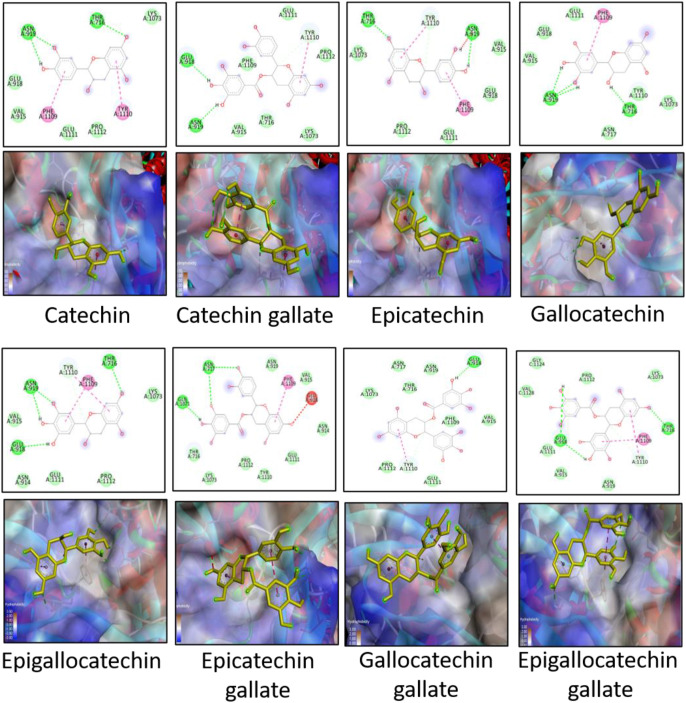
The docking scores of all the catechins were better than that of NAG, indicating that the catechins have a higher affinity to bind to the spike proteins than the native ligand. Thus, these molecules can selectively bind to the active site over NAG. The interactions between catechins and the amino acids of the active binding site form a stronger complex than that with NAG. These interactions are represented in Fig. 3, Fig. 4 . EGCG had better docking scores among the others, although the difference was not significant. This is in agreement with previous reports [31,43]. The same trend is followed for wild and mutated strains. The difference between the docking scores for the two strains for almost all the catechins is small. Hence, the scores of the catechins are not largely affected by the mutation of THR716 to ILE716 and the overall change in the conformation of the binding site. The interaction diagrams reveal the formation of several types of interactions including hydrogen bonding, van der Waals interactions, pi-anion and pi-cation bonds, pi-alkyl bonds, pi-pi and pi-signa bonds. Some key amino acids observed in majority of interactions are ASN919, THR716 (in wild strain), ILE716 (in mutated strain), TYR1110, PHE1109, GLN1071 and GLU918. THR716 residue of the wild strain is prominently involved in hydrogen bonding with the hydroxyl groups on catechin backbone or forms weak van der Waals interactions. On the other hand, ILE716 residue of the mutated strain majorly forms pi-alkyl bonds with the aromatic ring of the catechin backbone, although there are instances of formation of hydrogen bonds as well as van der Waals interactions. As the structures become larger, the number of functionalities in the molecule also increase. These moieties take part in various interactions and the docking score is increased. However, larger structures also produce steric hindrance and may form unfavourable interactions. EGCG has more several favourable interactions while ECG and GCG, although structurally similar to EGCG, show some unfavourable interactions which could explain their slightly lesser docking scores. From the docking scores, there were two conclusions. Firstly, all the catechins can potentially act as entry-inhibitors as they had good binding efficacy. This activity is irrelevant of the type of virus strain. Secondly, EGCG exhibited the best docking score among the other catechins and hence, is a suitable candidate to be further subjected to MD simulation.
Fig. 3.
2-D and 3-D images of docking interactions between catechins and the spike protein of wild strain. Hydrogen bonding is depicted by green dotted lines, van der Waals interaction is depicted by green circles, pi-anion or pi-cation interactions are depicted by orange dotted lines, pi-alkyl bond is shown by pink dotted lines, pi-sigma or pi-pi bond is depicted by purple dotted lines, pi-sulphur bond is depicted by yellow dotted lines, interaction with halogen is depicted by cyan dotted line and certain unfavourable interactions are depicted by red dotted line.
Fig. 4.
2-D and 3-D images of docking interactions between catechins and the spike protein of mutated strain. Hydrogen bonding is depicted by green dotted lines, van der Waals interaction is depicted by green circles, pi-anion or pi-cation interactions are depicted by orange dotted lines, pi-alkyl bond is shown by pink dotted lines, pi-sigma or pi-pi bond is depicted by purple dotted lines, pi-sulphur bond is depicted by yellow dotted lines, interaction with halogen is depicted by cyan dotted line and certain unfavourable interactions are depicted by red dotted line.
3.4. MD simulation
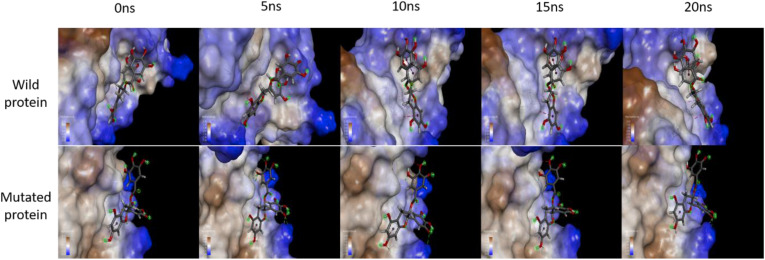
Biomolecules in their natural state, unlike represented in a molecular docking study, are flexible and not rigid. Their conformations and their binding modes keep on altering in response to the energy changes in the complex itself, the surrounding molecules and the physiological conditions as a function of time. Hence, the docked complexes obtained through molecular docking need to be validated for their long-term stability. MD simulation employ the Newtonian physics to estimate the behaviour of atoms and bonds in a predefined environment. On subjecting a molecule or a complex to MD simulation, we can study its energy and conformational changes and comment on its feasibility in a particular system. The docked complexes of EGCG with the spike proteins of wild and mutated strains were thus simulated in a solvent system to understand the changes over a time period. After performing the MD simulations, a frame was captured every 5 ns (Fig. 5 ) and the major interactions observed are reported in Table 3, Table 4 . As seen in the frames of ligand interaction at the active site, the interaction between EGCG and the wild protein with TYR1110 (pi-pi T-shaped), TYR1110 (pi-alkyl) and PRO1112 (alkyl) remained constant throughout the simulation. In case of wild protein, the ligand conformer was further stabilised because of pi-pi stacked interaction between the aromatic rings of EGCG, which might aid a prolonged interaction with the protein, thus inhibiting the viral entry.
Fig. 5.
Frames from MD simulation of EGCG with spike protein of SARS-CoV-2 obtained after every 5 ns during the analysis of the trajectory run for 20 ns, showing interactions of the ligand with both wild and mutated protein. For wild protein, dark purple dashed lines show the pi-pi T-shaped interaction; light pink dashed lines depict the pi-alkyl and alkyl interactions; green colour dashed lines depict the conventional hydrogen bonding; the magenta dashed lines depict the pi-pi stacked interaction in the ligand itself, stabilising the conformation further. For mutated protein, light pink dashed lines depict pi-alkyl interaction; green lines depict conventional hydrogen bonding; grey lines depict carbon-hydrogen bonding; and red lines depict unfavourable donor-donor interaction.
Table 3.
List of major interactions observed between EGCG and wild protein.
| Time frame | Interaction involved | Amino acid involved | Distance |
|---|---|---|---|
| 0 ns |
|
|
5.45 Å 5.42 Å 5.20 Å 2.15 Å |
| 5 ns |
|
|
5.42 Å 5.16 Å 2.15 Å |
| 10 ns |
|
|
5.46 Å 5.39 Å 5.23 Å 4.42 Å |
| 15 ns |
|
|
5.42 Å 5.27 Å 5.08 Å 4.20 Å |
| 20 ns |
|
|
5.52 Å 4.96 Å 4.33 Å |
Table 4.
List of major interactions observed between EGCG and mutated protein.
| Time frame | Interaction involved | Amino acid involved | Distance |
|---|---|---|---|
| 0 ns |
|
|
4.81 Å 2.07 Å 2.27 Å |
| 5 ns |
|
|
4.35 Å 2.73 Å 2.62 Å 2.68 Å 2.76 Å 3.08 Å 2.43 Å |
| 10 ns |
|
|
4.59 Å 2.94 Å 2.86 Å 2.22 Å 2.83 Å 2.22 Å |
| 15 ns |
|
|
4.66 Å 2.65 Å 2.24 Å 2.89 Å |
| 20 ns |
|
|
4.34 Å 2.72 Å 2.59 Å 2.81 Å 2.8 Å |
In case of mutated protein, the interaction with ALA930 (pi-alkyl), ALA930 (carbon-hydrogen) remained constant throughout, with many conventional hydrogen bonds forming and breaking. Since the interacting amino acids changed as compared to wild protein, it can be concluded that the mutations have changed in the orientation of the active site, but the ligand shows stable binding without leaving the binding site throughout the 20 ns simulation.
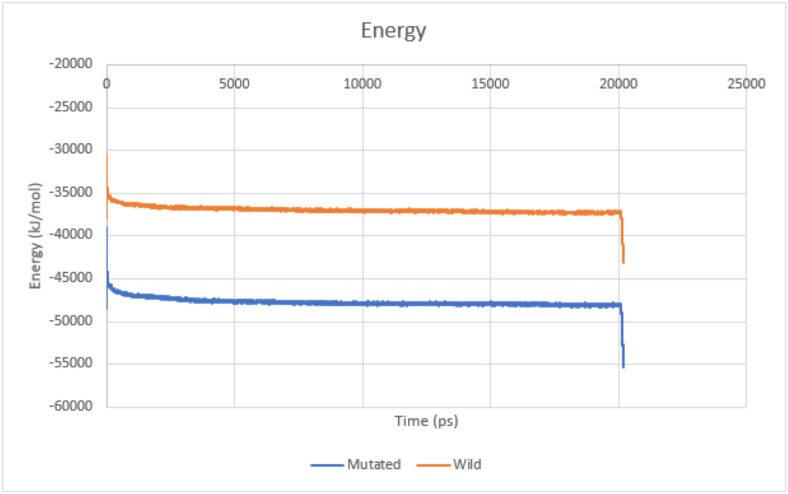
The total energy plots with energy on Y-axis and time on X-axis are shown in Fig. 6 , and as observed, the mutated protein had a relatively lower energy as compared to the wild strain. However, there were no major fluctuations in both the simulations, indicating that the protein-ligand complexes are stable throughout the simulation process.
Fig. 6.
Total energy plot for protein-ligand complex of both wild and mutated strains.
3.5. Pharmacokinetic properties
Pharmacokinetics studies the movement of drug into the body after the medication is administered. It provides the mathematical evaluation of the concentration of drugs and their actions in the body. The study of pharmacokinetic properties is essential to assess the safety and efficacy of drugs and design appropriate regimens. Pharmacokinetic properties of EGCG predicted by the pkCSM webserver showed almost similar results as those from previously reported experimental evaluation (Table 5 ). The negative value of water solubility indicates poor solubility of EGCG, which justifies its poor oral bioavailability. Huang et al. used a human colorectal adenocarcinoma monolayer cell line (Caco 2) to study the intestinal absorption of EGCG [44]. Even though the Caco 2 permeability values were different for computational and previous experimental studies, both these values justify the poor intestinal absorption of less than 20%. A study by Chu et al. suggests that the EGCG absorption occurs in the gastrointestinal tract, but it undergoes decomposition in gastric pH and is unstable for absorption [45]. Lambert et al. had proposed that the poor intestinal absorption could be attributed to the colonic microflora which break EGCG down into phenolic acid. Moreover, absorption into cells occurs only by passive diffusion, acellular and transcellular diffusion due to the absence of any receptor on intestinal epithelial cells [46]. A smaller fraction of unbound drug (0.31, 0.36) suggests high protein binding, which conveys the low bioavailability of EGCG. Intravenous administration of EGCG could avoid the metabolism and encounter with gastric conditions to ensure free form reaching tissues and ensuring its action. The clearance of EGCG is relatively faster as predicted from computational tools as well as from previously reported experimental observations. The study by Lambert et al. suggests that after absorption, it reaches the liver and is prone to methylation by catechol-O-methyltransferase. This methylation further facilitates sulfation/glucuronidation of the methylated product leading to increased clearance by effectively eliminating the methylation product from the body [46]. Another report by Lipinski et al. suggests that the unabsorbed EGCG is degraded into ring-fission metabolites by intestinal microflora, and metabolites of liver degradation are secreted into bile and excreted through faeces and urine [47]. Low values of BBB permeability for EGCG implies less CNS toxicity and side-effects. The maximum tolerance of EGCG is much higher than many antiviral drugs owing to its natural origin and low acute as well as chronic toxicity values. Hence, the formulations involving EGCG can support high dosages to ensure the required bioavailability. Other approaches that have been successfully studied to improve the oral bioavailability of EGCG include structure modification and nano delivery through nanoparticles, nanoliposomes, double emulsions, and nanoemulsions [48].
Table 5.
Comparison of computationally predicted and experimentally observed values.
| Property of EGCG | Computationally predicted values | Previously reported experimental values from literature | Reference |
|---|---|---|---|
| Water solubility (log mol/L) | −2.89 | −2.29 | [49] |
| Caco 2 permeability (log 10-6 cm/s) | −0.89 | −0.37 | [44] |
| Volume of distribution at steady state (log L/kg) | 0.052 | 0.11 | [50] |
| Fraction Unbound (Fu) | 0.31 | 0.36 | [45] |
| Blood Brain Barrier permeability (log BB) | −2.23 | −1.54 | [51] |
| Total clearance (log mL/min/kg) | 0.51 | 1.49 | [52] |
| Max. tolerated dose (log mg/kg/day) | 0.45 | 0.68 | [53] |
| Oral rat acute toxicity (mol/kg) | 2.63 | 2.24 | [54] |
| Oral rat chronic toxicity (log mg/kg bw/day) | 4.07 | 3.30 | [55] |
4. Conclusion
The COVID-19 global pandemic has proved that the fields of science and medicine are much equipped than before to deal with a disease outbreak, but there is a need for more preparedness. The development of safer and widely available strategies to overcome the increasing number of cases is highly anticipated. With the new variants emerging in several parts of the world, the health and immunity of individuals are further at stake. The cost and availability of currently approved drugs makes it difficult for everyone to have access to them. Natural phytoconstituents hold several health benefits and also possess enormous potential to be developed as drug candidates. Phytoconstituents offer a promising alternative to the treatment of viral infections like COVID-19, by exhibiting their wide-spectrum activity. One of the promising class of phytoconstituents is catechins. In this study, we screened several natural catechins for their antiviral activity as entry-inhibitors against the spike protein of the wild strain, and that of the UK variant of the SARS-CoV-2 using computational tools. It was found that all the catechins had a good affinity towards the spike proteins, even higher than the native ligand. Out of all the catechins, EGCG had the best affinity. The EGCG-spike protein complex was observed to be stable when simulated using MD simulation. These results should encourage development of EGCG and other catechins as drug candidates. The pharmacokinetic properties need to be modified to improve the drug-likeliness of these phytoconstituents. Further in vitro and in vivo studies are required to validate these in-silico findings.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author contributions
SM performed the docking studies, MD simulations studies and wrote a part of the manuscript. NG performed the docking studies, analysed and interpreted its results and wrote a part of the manuscript. MS performed the MD simulations studies, analysed and interpreted the results and wrote a part of the manuscript. SM, NG and MS collectively made the figures, tables and supporting sections of the manuscript. NG and MS revised the manuscript. VP planned the manuscript, mentored other authors, as well as administered and supervised the entire work. VP also facilitated all the correspondence of the manuscript.
Declaration of competing interest
We confirm that there are no conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome.
Acknowledgements
Molecular graphics and analyses were performed with UCSF Chimera, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from NIH P41-GM103311. Computational results were obtained by using Dassault Systèmes BIOVIA software programs. BIOVIA Discovery Studio was used to perform the calculations and to generate the graphical results. We are thankful to Dr Sreeranjini Pulakkat for taking efforts to revise the manuscript.
References
- 1.Coronavirus Disease (reportCOVID-19) Situation Reports, (n.d.). https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports (accessed March 26, 2021).
- 2.Mercurio I., Tragni V., Busto F., De Grassi A., Pierri C.L. Protein structure analysis of the interactions between SARS-CoV-2 spike protein and the human ACE2 receptor: from conformational changes to novel neutralizing antibodies, Cell. Mol. Life Sci. 2021;78:1501–1522. doi: 10.1007/s00018-020-03580-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xiong X., Qu K., Ciazynska K.A., Hosmillo M., Carter A.P., Ebrahimi S., Ke Z., Scheres S.H.W., Bergamaschi L., Grice G.L., Zhang Y., Nathan J.A., Baker S., James L.C., Baxendale H.E., Goodfellow I., Doffinger R., Briggs J.A.G. A thermostable, closed SARS-CoV-2 spike protein trimer. Nat. Struct. Mol. Biol. 2020;27:934–941. doi: 10.1038/s41594-020-0478-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.V’kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.COVID-19 Vaccines, (n.d.). https://www.who.int/emergencies/diseases/novel-coronavirus-2019/covid-19-vaccines (accessed March 26, 2021).
- 6.Kupferschmidt K. New mutations raise specter of ‘immune escape. Science. 2021;371:329–330. doi: 10.1126/science.371.6527.329. [DOI] [PubMed] [Google Scholar]
- 7.Burki T. Understanding variants of SARS-CoV-2. Lancet. 2021;397:462. doi: 10.1016/S0140-6736(21)00298-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mahase E. Covid-19: what have we learnt about the new variant in the UK? BMJ. 2020;371:m4944. doi: 10.1136/bmj.m4944. [DOI] [PubMed] [Google Scholar]
- 9.Toovey O.T.R., Harvey K.N., Bird P.W., Tang J.W.-T.W.-T. Introduction of Brazilian SARS-CoV-2 484K.V2 related variants into the UK. J. Infect. 2021 doi: 10.1016/j.jinf.2021.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang J.W., Toovey O.T.R., Harvey K.N., Hui D.D.S. Introduction of the South African SARS-CoV-2 variant 501Y.V2 into the UK. J. Infect. 2021 doi: 10.1016/j.jinf.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kupferschmidt K. Fast-spreading U.K. virus variant raises alarms. Science. 2021;371:9–10. doi: 10.1126/science.371.6524.9. [DOI] [PubMed] [Google Scholar]
- 12.Variants: Distribution of Cases Data, GOV.UK. (n.d.). https://www.gov.uk/government/publications/covid-19-variants-genomically-confirmed-case-numbers/variants-distribution-of-cases-data (accessed March 27, 2021).
- 13.Kirby T. New variant of SARS-CoV-2 in UK causes surge of COVID-19. Lancet Respiratory Med. 2021;9:e20–e21. doi: 10.1016/S2213-2600(21)00005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Official Uk Coronavirus Dashboard https://coronavirus.data.gov.uk n.d.
- 15.Mahase E. Covid-19: Novavax vaccine efficacy is 86% against UK variant and 60% against South African variant. BMJ. 2021;372:n296. doi: 10.1136/bmj.n296. [DOI] [PubMed] [Google Scholar]
- 16.Higdon J.V., Frei B. Tea catechins and polyphenols: health effects, metabolism, and antioxidant functions. Crit. Rev. Food Sci. Nutr. 2003;43:89–143. doi: 10.1080/10408690390826464. [DOI] [PubMed] [Google Scholar]
- 17.Chacko S.M., Thambi P.T., Kuttan R., Nishigaki I. Beneficial effects of green tea: a literature review. Chin. Med. 2010;5:13. doi: 10.1186/1749-8546-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Musial C., Kuban-Jankowska A., Gorska-Ponikowska M. Beneficial properties of green tea catechins. Int. J. Mol. Sci. 2020;21:1744. doi: 10.3390/ijms21051744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yahfoufi N., Alsadi N., Jambi M., Matar C. The immunomodulatory and anti-inflammatory role of polyphenols. Nutrients. 2018;10:1618. doi: 10.3390/nu10111618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tran J. Green tea: a potential alternative anti-infectious agent catechins and viral infections. Adv. Anthropol. 2013:198. doi: 10.4236/aa.2013.34028. 03. [DOI] [Google Scholar]
- 21.Weber J.M., Ruzindana-Umunyana A., Imbeault L., Sircar S. Inhibition of adenovirus infection and adenain by green tea catechins. Antivir. Res. 2003;58:167–173. doi: 10.1016/s0166-3542(02)00212-7. [DOI] [PubMed] [Google Scholar]
- 22.Catherine DeSoto M. Regional differences in use of immune-modulating catechins should be investigated regarding COVID-19, Brain, Behavior, and Immunity. 2020. 89. 526-527. [DOI] [PMC free article] [PubMed]
- 23.Liang W., He L., Ning P., Lin J., Li H., Lin Z., Kang K., Zhang Y. (+)-Catechin inhibition of transmissible gastroenteritis coronavirus in swine testicular cells is involved its antioxidation. Res. Vet. Sci. 2015;103:28–33. doi: 10.1016/j.rvsc.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferreira P.G., Ferraz A.C., Figueiredo J.E., Lima C.F., Rodrigues V.G., Taranto A.G., Ferreira J.M.S., Brandão G.C., Vieira-Filho S.A., Duarte L.P., de Brito Magalhães C.L., de Magalhães J.C. Detection of the antiviral activity of epicatechin isolated from Salacia crassifolia (Celastraceae) against Mayaro virus based on protein C homology modelling and virtual screening. Arch. Virol. 2018;163:1567–1576. doi: 10.1007/s00705-018-3774-1. [DOI] [PubMed] [Google Scholar]
- 25.Patrick Reid St, Shurtleff A.C., Costantino J.A., Tritsch S.R., Retterer C., Spurgers K.B., Bavari S. HSPA5 is an essential host factor for Ebola virus infection. Antivir. Res. 2014;109:171–174. doi: 10.1016/j.antiviral.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 26.Mhatre S., Srivastava T., Naik S., Patravale V. Antiviral activity of green tea and black tea polyphenols in prophylaxis and treatment of COVID-19: a review. Phytomedicine. 2020 doi: 10.1016/j.phymed.2020.153286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Storozhuk M. COVID-19: could green tea catechins reduce the risks? MedRxiv. 2020 doi: 10.1101/2020.10.23.20218479. 2020.10.23.20218479. [DOI] [Google Scholar]
- 28.Song J.-M., Lee K.-H., Seong B.-L. Antiviral effect of catechins in green tea on influenza virus. Antivir. Res. 2005;68:66–74. doi: 10.1016/j.antiviral.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 29.Yamaguchi K., Honda M., Ikigai H., Hara Y., Shimamura T. Inhibitory effects of (−)-epigallocatechin gallate on the life cycle of human immunodeficiency virus type 1 (HIV-1) Antivir. Res. 2002;53:19–34. doi: 10.1016/S0166-3542(01)00189-9. [DOI] [PubMed] [Google Scholar]
- 30.Ciesek S., von Hahn T., Colpitts C.C., Schang L.M., Friesland M., Steinmann J., Manns M.P., Ott M., Wedemeyer H., Meuleman P., Pietschmann T., Steinmann E. The green tea polyphenol, epigallocatechin-3-gallate, inhibits hepatitis C virus entry. Hepatology. 2011;54:1947–1955. doi: 10.1002/hep.24610. [DOI] [PubMed] [Google Scholar]
- 31.Joseph J., Ajay A., Das V.R.A., Raj V.S. Green tea and Spirulina extracts inhibit SARS, MERS, and SARS-2 spike pseudotyped virus entry in vitro. BioRxiv. 2020 doi: 10.1101/2020.06.20.162701. K. T. 2020.06.20.162701. [DOI] [Google Scholar]
- 32.Jena A.B., Kanungo N., Nayak V., Chainy G.B.N., Dandapat J. Catechin and curcumin interact with S protein of SARS-CoV2 and ACE2 of human cell membrane: insights from computational studies. Sci. Rep. 2021;11:2043. doi: 10.1038/s41598-021-81462-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodrigues C.H., Pires D.E., Ascher D.B. DynaMut: predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 2018;46:W350–W355. doi: 10.1093/nar/gky300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mhatre S., Naik S., Patravale V. A molecular docking study of EGCG and theaflavin digallate with the druggable targets of SARS-CoV-2. Comput. Biol. Med. 2021;129 doi: 10.1016/j.compbiomed.2020.104137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shapovalov M.V., Dunbrack R.L. A smoothed backbone-dependent rotamer library for proteins derived from adaptive kernel density estimates and regressions. Structure. 2011;19:844–858. doi: 10.1016/j.str.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ravindranath P.A., Sanner M.F. AutoSite: an automated approach for pseudo-ligands prediction—from ligand-binding sites identification to predicting key ligand atoms. Bioinformatics. 2016;32:3142–3149. doi: 10.1093/bioinformatics/btw367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dallakyan S., Olson A.J. Small-molecule library screening by docking with PyRx. Methods Mol. Biol. 2015;1263:243–250. doi: 10.1007/978-1-4939-2269-7_19. [DOI] [PubMed] [Google Scholar]
- 40.O'Boyle N.M., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R. Open Babel: an open chemical toolbox. J. Cheminf. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pires D.E.V., Blundell T.L., Ascher D.B. pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J. Med. Chem. 2015;58:4066–4072. doi: 10.1021/acs.jmedchem.5b00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Azevedo W.F., Dias R. Computational methods for calculation of ligand-binding affinity. Curr. Drug Targets. 2008;9:1031–1039. doi: 10.2174/138945008786949405. [DOI] [PubMed] [Google Scholar]
- 43.Liu J., Bodnar B.H., Meng F., Khan A., Wang X., Luo G., Saribas S., Wang T., Lohani S.C., Wang P., Wei Z., Luo J., Zhou L., Wu J., Li Q., Hu W., Ho W. Epigallocatechin gallate from green tea effectively blocks infection of SARS-CoV-2 and new variants by inhibiting spike binding to ACE2 receptor. BioRxiv. 2021 doi: 10.1101/2021.03.17.435637. 2021.03.17.435637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang T.-W., Ho Y.-C., Tsai T.-N., Tseng C.-L., Lin C., Mi F.-L. Enhancement of the permeability and activities of epigallocatechin gallate by quaternary ammonium chitosan/fucoidan nanoparticles. Carbohydr. Polym. 2020;242 doi: 10.1016/j.carbpol.2020.116312. [DOI] [PubMed] [Google Scholar]
- 45.Chu K.O., Pang C.C.P. 2018. Pharmacokinetics and Disposition of Green Tea Catechins, Pharmacokinetics and Adverse Effects of Drugs - Mechanisms and Risks Factors. [DOI] [Google Scholar]
- 46.Lambert J.D., Lee M.-J., Lu H., Meng X., Hong J.J.J., Seril D.N., Sturgill M.G., Yang C.S. Epigallocatechin-3-gallate is absorbed but extensively glucuronidated following oral administration to mice. J. Nutr. 2003;133:4172–4177. doi: 10.1093/jn/133.12.4172. [DOI] [PubMed] [Google Scholar]
- 47.Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 48.Dai W., Ruan C., Zhang Y., Wang J., Han J., Shao Z., Sun Y., Liang J. Bioavailability enhancement of EGCG by structural modification and nano-delivery: a review. J. Funct. Foods. 2020;65 doi: 10.1016/j.jff.2019.103732. [DOI] [Google Scholar]
- 49.Moon Y.-H., Lee J.-H., Ahn J.-S., Nam S.-H., Oh D.-K., Park D.-H., Chung H.-J., Kang S., Day D.F., Kim D. Synthesis, structure analyses, and characterization of novel epigallocatechin gallate (EGCG) glycosides using the glucansucrase from leuconostoc mesenteroides B-1299CB. J. Agric. Food Chem. 2006;54:1230–1237. doi: 10.1021/jf052359i. [DOI] [PubMed] [Google Scholar]
- 50.Zhu M., Chen Y., Li R.C. Pharmacokinetics and system linearity of tea catechins in rat. Xenobiotica. 2001;31:51–60. doi: 10.1080/00498250010024988. [DOI] [PubMed] [Google Scholar]
- 51.Pervin M., Unno K., Nakagawa A., Takahashi Y., Iguchi K., Yamamoto H., Hoshino M., Hara A., Takagaki A., Nanjo F., Minami A., Imai S., Nakamura Y. Blood brain barrier permeability of (-)-epigallocatechin gallate, its proliferation-enhancing activity of human neuroblastoma SH-SY5Y cells, and its preventive effect on age-related cognitive dysfunction in mice. Biochem Biophys Rep. 2017;9:180–186. doi: 10.1016/j.bbrep.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen L., Lee M.-J., Li H., Yang C.S. Absorption, distribution, and elimination of tea polyphenols in rats. Drug Metab. Dispos. 1997;25:1045–1050. [PubMed] [Google Scholar]
- 53.Ramachandran B., Jayavelu S., Murhekar K., Rajkumar T. Repeated dose studies with pure Epigallocatechin-3-gallate demonstrated dose and route dependant hepatotoxicity with associated dyslipidemia. Toxicol Rep. 2016;3:336–345. doi: 10.1016/j.toxrep.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee S., Song P.H., Lee Y.J., Ku S.-K., Song C.-H. vol. 2019. 2019. Acute and Subchronic Oral Toxicity of Fermented Green Tea with Aquilariae Lignum in Rodents, Evidence-Based Complementary and Alternative Medicine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chengelis C.P., Kirkpatrick J.B., Regan K.S., Radovsky A.E., Beck M.J., Morita O., Tamaki Y., Suzuki H. 28-Day oral (gavage) toxicity studies of green tea catechins prepared for beverages in rats. Food Chem. Toxicol. 2008;46:978–989. doi: 10.1016/j.fct.2007.10.027. [DOI] [PubMed] [Google Scholar]