Abstract
Cyclic GMP-AMP synthase (cGAS) recognizes cytosolic foreign or damaged DNA to activate the innate immune response to infection, inflammatory diseases, and cancer. In contrast, cGAS reactivity against self-DNA in the nucleus is suppressed by chromatin tethering. We report a 3.3-angstrom-resolution cryo-electron microscopy structure of cGAS in complex with the nucleosome core particle. The structure reveals that cGAS employs two conserved arginines to anchor to the nucleosome acidic patch. The nucleosome binding interface exclusively occupies the strong dsDNA binding surface on cGAS and sterically prevents cGAS from oligomerizing into the functionally active 2:2 cGAS-dsDNA state. These findings provide a structural basis for how cGAS maintains an inhibited state in the nucleus and further exemplify the role of the nucleosome in regulating diverse nuclear protein functions.
A fundamental host-defense mechanism is to detect and respond to nucleic acids from bacterial and viral pathogens and damaged cellular DNA (1, 2). In the mammalian innate immune system, cyclic GMP–AMP synthase (cGAS) and stimulator of interferon genes (STING) mediate a major pathway in response to cytosolic DNA fragments that derive from pathogens or from cellular DNA damage caused by inflammatory diseases and cancer (3). In the cytoplasm, cGAS is activated by recognizing and binding double-stranded DNA (dsDNA) in a sequence-independent manner, resulting in the synthesis of a second messenger, 2’–3’ cyclic GMP-AMP (cGAMP) that features both noncanonical (2’–5’) and canonical (3’–5’) phosphodiester linkages (4, 5). STING, an ER membrane protein, binds cGAMP and triggers a signaling cascade to activate inflammatory responses, including induction of type I interferons (6–8). While it was discovered as a cytosolic dsDNA sensor, cGAS has since been shown to be enriched in the nucleus (9–13). Remarkably, despite being surrounded with endogenous genomic DNA, nuclear cGAS activity is suppressed through tethering to chromatin (9), and the nucleosome, the fundamental unit of the genome, has been shown to competitively suppress the enzymatic activity of cGAS (12). Such suppression is critical to prevent unnecessary triggering of immune responses.
To understand the molecular mechanism through which nucleosome-bound cGAS maintains its resting, inhibited state, we used cryo-electron microscopy (cryo-EM) to solve the structure of the mouse cGAS catalytic domain bound to the nucleosome core particle (NCP) (Fig. 1 and movie S1). Mouse cGAS is composed of an unstructured N terminus (amino acid residues 1–147) and a highly structured, bilobate C terminus (amino acid residues 148–507) (Fig. 1A) (4). The positively charged N terminus plays a role in enhancing cGAS-DNA phase transition to promote cGAMP production (14). The C terminus comprises the nucleotidyltransferase (NTase) catalytic domain and the Mab21 dsDNA recognition domain, where an α-helical spine (residues 148–183) bridges the two cGAS C-terminal lobes (15–21). Because the disordered N-terminal region of cGAS is not required for nuclear tethering (9), we expressed and purified the catalytic domain of mouse cGAS, referred to as cGAS hereafter, for structural and enzymatic studies. We assembled the cGAS-NCP complex by mixing cGAS with recombinant human NCP in a 3:1 ratio and isolated the resulting 2:1 cGAS-NCP complex by gel filtration chromatography (fig. S1). The purified cGAS-NCP complex was used to prepare specimens for cryo-EM single particle analysis. The complex was stable during grid preparation allowing us to collect images in a native state without crosslinking.
Fig. 1. Overall structure of nucleosome-bound cGAS.
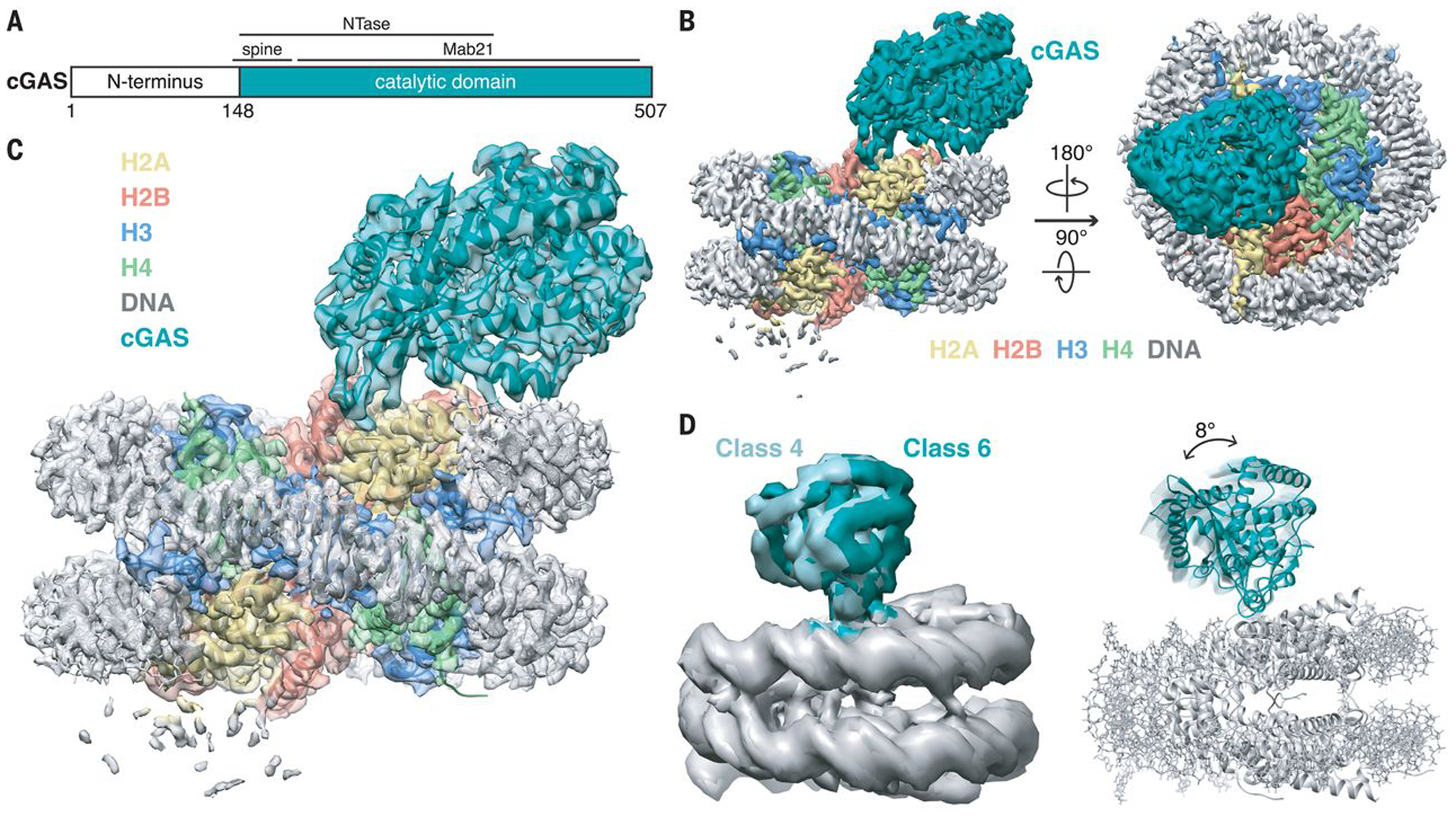
(A) Schematic of the mouse cGAS primary structure colored by domains. (B) Side and top views of the composite cryo-EM density map of cGAS-NCP complex. cGAS, H2A, H2B, H3, H4, and DNA are colored teal, yellow, red, blue, green, and gray, respectively. (C) Transparent composite cryo-EM density map overlaid onto atomic model of cGAS-NCP complex. (D) Overlay of 3D subclasses (left) and cartoon representations that depict the hinge motion of cGAS relative the nucleosome (right) created by morphing between cGAS structures docked into class 4 and class 6 maps.
A total of 2,100 movies were recorded on a Talos Arctica 200 kV microscope equipped with a Gatan K3 direct-electron detector. After iterative rounds of 2D classification, initial 3D classification of 433,445 particles revealed a heterogeneous mixture of 2:1 (Class 4; 27%) and 1:1 (Class 1; 21%) cGAS-NCP complexes (fig. S2). Independent 3D refinements of the 2:1 and 1:1 classes gave initial EM density maps at 4.35 Å resolution for both stoichiometries. Through Bayesian particle polishing, iterative contrast transfer function (CTF) refinement, and 3D re-classification, the resolution of the 1:1 complex map was improved to 3.41 Å with 70,790 particles, and the resolution of the 2:1 complex map was improved to 3.30 Å with 45,587 particles. To maximize the overall resolution at the cGAS-NCP binding interface, we combined all 116,377 particles from the 1:1 and 2:1 cGAS-NCP reconstructions and refined the final map to an overall resolution of 3.26 Å. In this reconstruction, high-resolution structural features are visible throughout the nucleosome and into the nucleosome-interacting face of cGAS (Fig. 1, B and C, and figs. S2–S4). In contrast, regions of the reconstructed map corresponding to parts of cGAS that are farther from the nucleosome interface are less well resolved, suggesting conformational heterogeneity or structural dynamics. Multibody refinement using cGAS and nucleosome masks revealed a rigid body rotation of cGAS hinging from the nucleosome interface (fig. S2 and movie S2). To improve cGAS visualization, we performed an additional 3D classification restricted to local angular searches. This 3D classification recapitulated our earlier observations of a mixture of 1:1 and 2:1 complexes, and further revealed several distinct cGAS conformations relative to the nucleosome (e.g. classes 3, 4, and 6) (Fig. 1D and fig. S2). Subsequent refinement of the most populated class (class 6, 38%) with a mask enveloping cGAS resulted in the 3.9 Å resolution 1:1 cGASmask-NCP reconstruction with clear density for all regions of cGAS and noticeable blurring of nucleosome density (figs. S2 and S3).
To generate an atomic model of the 1:1 cGAS-NCP complex, we docked a high-resolution crystal structure of the apo mouse cGAS catalytic domain (15) and a cryo-EM structure of the NCP (22) into the 3.26 Å resolution 1:1 cGAS-NCP map, and carried out iterative manual model building and real-space refinement. Due to the low signal-to-noise in the map regions corresponding to cGAS residues far away from the nucleosome interface, reference model restraints were applied during refinement to preserve model quality. Additionally, to minimize over-refinement into the full-map, the resultant model was refined against one sharpened, masked half-map, leading to a final model that showed excellent correlation to both the sharpened, masked full map and the other sharpened, masked half-map (Fig. 1C, fig. S3, table S1, and movie S1). The model also correlated well with the 3.9 Å map reconstructed using a cGAS mask. Therefore, we prepared a composite map from the 3.26 Å sharpened map and the 3.9 Å cGAS masked map to best represent our 1:1 complex (Fig. 1C and fig. S4). Through docking and real-space refinement, a structure was solved using the 3.30 Å resolution 2:1 cGAS-NCP map that is nearly identical to the 1:1 complex (figs. S3 and S5, and table S1).
The high-resolution complex structure clearly reveals how cGAS binds the nucleosome. An intricate set of interactions are formed between two cGAS arginines (R222 and R241) and four acidic-patch residues (E61, E64, D90, and E92) in histone H2A (Fig. 2, A–D). cGAS R222 and R241 reside in spatially adjacent loops with conformations stabilized by an inter-loop hydrogen bonding network and direct side chain interactions with histones. R222 forms three intermolecular hydrogen bonds with H2A E61 and E64 as well as two intramolecular hydrogen bonds with backbone atoms of cGAS K240 and R241 in the adjacent loop. cGAS K240 in turn also forms a hydrogen bond with E224. R241, in addition to interacting with R222, inserts its side chain into the cavity surrounded by H2A E61, D90, and E92 and is completely encapsulated with four intermolecular hydrogen bonds. In addition to this major contact site, the backbone of cGAS K315 is hydrogen bonded with the sidechain of H2A R71 (Fig. 2, A and B). Together, these interactions create two pivot points that provide a structural basis for the observed hinge motions of cGAS relative to the nucleosome surface (Fig. 1D).
Fig. 2. Interactions between cGAS and the nucleosome core particle.
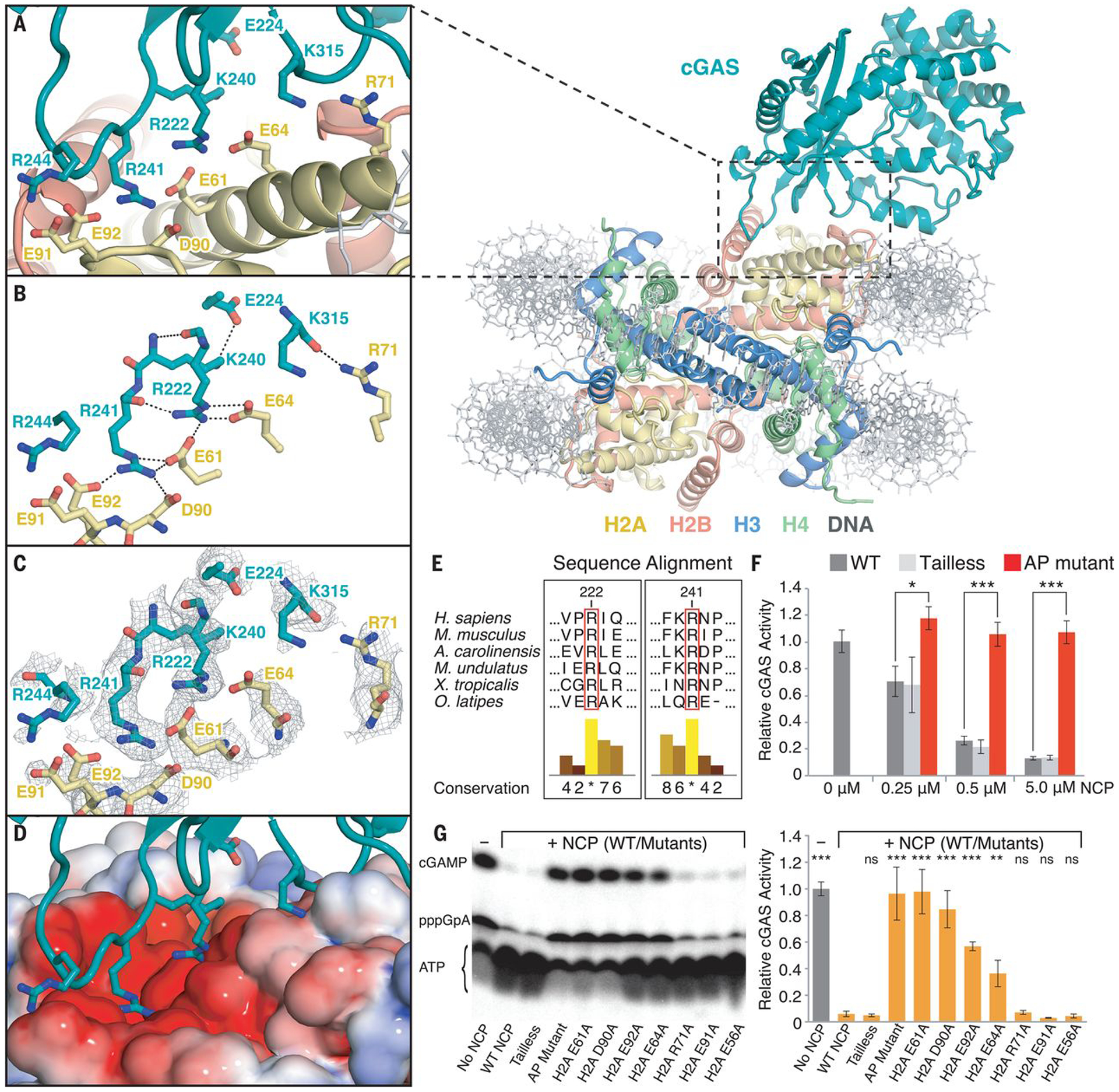
(A) Close-up view of the binding interface of the cGAS-NCP complex. (B) Close-up view of the hydrogen-bond network between cGAS residues (teal) and histone H2A residues (yellow). (C) Close-up view of the binding interface with composite combined focused map overlaid. (D) Identical view of cGAS interacting with the nucleosome acidic patch as highlighted with electrostatic potential surface, where positive is shown in blue and negative is shown in red. (E) Conservation of the arginine anchors, R222 and R241, of cGAS across vertebrates. Numerical and histogram representations of the conservation score are shown, where asterisks denote complete identity conservation (http://www.jalview.org/). (F) Quantification of nucleosome-dependent inhibition of cGAS activity. (G) Quantification of the inhibitory effect of individual nucleosome acidic-patch residues on cGAS activity (0.5 μM cGAS: 5 μM dsDNA) in the presence of 5 μM NCP mutants. ATP, pppGpA, and cGAMP are substrate, intermediate, and product, respectively. Average relative cGAS activities from three replicates are plotted with standard deviations represented as error bars (mean ± SD), where ns, *, ** and *** denote p-values >0.05, <0.05, <0.01 and <0.001, respectively, for statistical differences between WT and mutant NCPs.
This structure provides molecular mechanisms to explain biochemical and cellular data that identified a role for nuclear tethering in cGAS inactivation (9). For example, all cGAS mutations that showed no or mild effects in changing nuclear localization and tethering, including K335E, K395M/K399M, ΔH378–C393, K382A, E211A/D213A, R244E, and loop242–247 (IPRGNP to SGSGSG) (9), based on the structure are not expected to perturb the cGAS-NCP interactions (fig. S6). While cGAS R244 is positioned adjacent to H2A E91, neither side chain is well resolved in our reconstructions, thus providing no evidence of a stable interaction, which is consistent with the mild effects of R244E mutation in perturbing nuclear tethering (fig. S4). In contrast, the R222E and R241E mutants, which charge-reverse the two arginines of cGAS that are key for anchoring to the nucleosome, exhibit the most deleterious effects in abolishing nuclear tethering. Moreover, the K240E mutation also abolishes nuclear tethering of cGAS, suggesting that the observed intramolecular interactions with R222 and E224 are important in stabilizing the local conformation of the anchoring residues of cGAS for NCP binding. The K240E mutation could also have a repulsive effect by sandwiching an additional negatively charged side chain between H2A E61 and E64 on the acidic patch below and cGAS E224 above. Despite cGAS being one of the most rapidly evolved genes with substantial sequence variations (23, 24), both R222 and R241 are completely conserved in cGAS across all vertebrates (Fig. 2E) (9). While R222 also participates in dsDNA binding to activate cGAMP synthesis, the R241E mutation does not impair DNA-dependent cGAMP synthesis by cGAS (17, 18). The lack of a functional role for R241 in cGAS enzymatic activity suggests that the evolutionary conservation of R241 may largely be to preserve interactions with the nucleosome acidic patch. This interaction ensures that cGAS is suppressed when encountering endogenous chromatinized DNA either in the nucleus or in the cytosol during processes such as mitosis and apoptosis. Sequence and structural alignments between cGAS and nuclear Mab21L1/L2 proteins further suggest that the arginine anchors may be evolutionarily conserved in other nuclear Mab21 family proteins that potentially interact with the nucleosome for their functions (fig. S7) (25, 26).
Beyond these mutational analyses of cGAS localization and function, it was reported that the nucleosome can competitively suppress DNA-dependent cGAS activity (12). Moreover, mutating acidic-patch residues within H2A-H2B abolishes binding of histone dimers to cGAS in pulldown assays (12). To verify that cGAS binding to the acidic patch results in the observed competitive suppression of DNA-dependent activity, we carried out cGAS enzymatic assays with titration of nucleosome as a competitor (Fig. 2F and fig. S8). We clearly observed that the nucleosome outcompeted dsDNA and exhibited a dose-dependent inhibition of cGAMP synthesis; in contrast, comprehensive mutation of the acidic-patch (H2A E61A, E64S, N68A, D72S, N89A, D90A, E91S) completely abolishes the ability of the nucleosome to suppress cGAS activity. To further dissect the role of individual acidic patch residues in cGAS inactivation, we performed thorough site-directed mutagenesis of H2A paired with enzymatic and binding assays (Fig. 2G and fig. S9). Single-point mutations of H2A E61A and D90A, the two residues located at the core of the interactions with cGAS R222 and R241, completely abolish the ability of the nucleosome to bind cGAS and suppress its enzymatic activity. Mutant nucleosomes containing H2A E92A and H2A E64A exhibited weaker binding affinities than the wild type, resulting in partial suppressions of cGAS activity. In contrast, nucleosomes assembled with H2A R71A and H2A E91A showed cGAS inhibition comparable to both wild type and negative control of H2A E56A mutated nucleosomes. Altogether, these results not only strengthen the critical role of the nucleosome acidic patch in interacting with cGAS R222 and R241, but also suggest that the observed H2A R71-cGAS K315 interaction is dispensable for the nucleosome-mediated cGAS inactivation. It is well known that histone tails play an important role in nucleosome dynamics and recruitment of proteins to chromatin loci (27). Hence, we further examined whether histone tails may also contribute to the regulation of cGAS. However, we observed no changes in the inhibitory function or binding of the nucleosome upon deleting N-terminal tails from all four histones, suggesting that in the absence of any histone modifications, the nucleosome acidic patch plays the dominant role in sequestering cGAS in its resting, inhibited nuclear state.
How does the nucleosome inhibit cGAS activity? To activate enzymatic synthesis of cGAMP, cGAS needs to oligomerize into the functionally active 2:2 cGAS-DNA state, where each cGAS interacts with two dsDNA helices through two distinct DNA binding sites (site A and site B) (Fig. 3A) (17, 18). Site A contains ten residues (K151, R158, K160, R161, K162, S165, K180, K184, K372, and K395), which not only contribute to the majority of buried surface area between cGAS and dsDNA but also form the site that induces bending of the spine helix to allosterically transform the catalytic pocket to an active conformation. In contrast, site B, which consists of only six residues (R222, K240, K315, K323, K335, and R342), has a higher binding affinity for dsDNA than site A (17). An additional dsDNA binding site C was identified in human cGAS, which is critical for DNA-induced activity and liquid-phase condensation of human cGAS but is much less conserved in mouse cGAS (residues 247–291 and 408–421) (21). The nucleosome exclusively occupies site B on the cGAS surface through binding to the H2A-H2B dimer and proximity to nucleosomal DNA while leaving sites A and C completely exposed (Fig. 3B). Indeed, overlaying the DNA-bound cGAS structure (17) onto the cGAS-NCP structure shows that dsDNA can still be recognized by cGAS at site A without colliding with the nucleosome, even accounting for the observed cGAS conformational heterogeneity (Fig. 3C and fig. S10). However, the nucleosome sterically prevents cGAS from oligomerizing into its functionally active 2:2 cGAS-DNA state. Furthermore, the nucleosome-bound cGAS adopts an overall conformation that is essentially identical to the DNA-free inactive state with a root mean square deviation (RMSD) of 0.7 Å over all backbone atoms, but is distinct from the active conformation (an RMSD of 2.3 Å over all backbone atoms) where DNA binding alters the N-terminal spine helix, repositions the activation loop, and opens the entrance to the catalytic pocket (Fig. 3D) (15–20). Together, these findings show how the nucleosome uses its histone surface, rather than DNA, to recognize and maintain cGAS in the resting, inhibited state.
Fig. 3. Mechanism of nucleosome-dependent inhibition of cGAS activity.
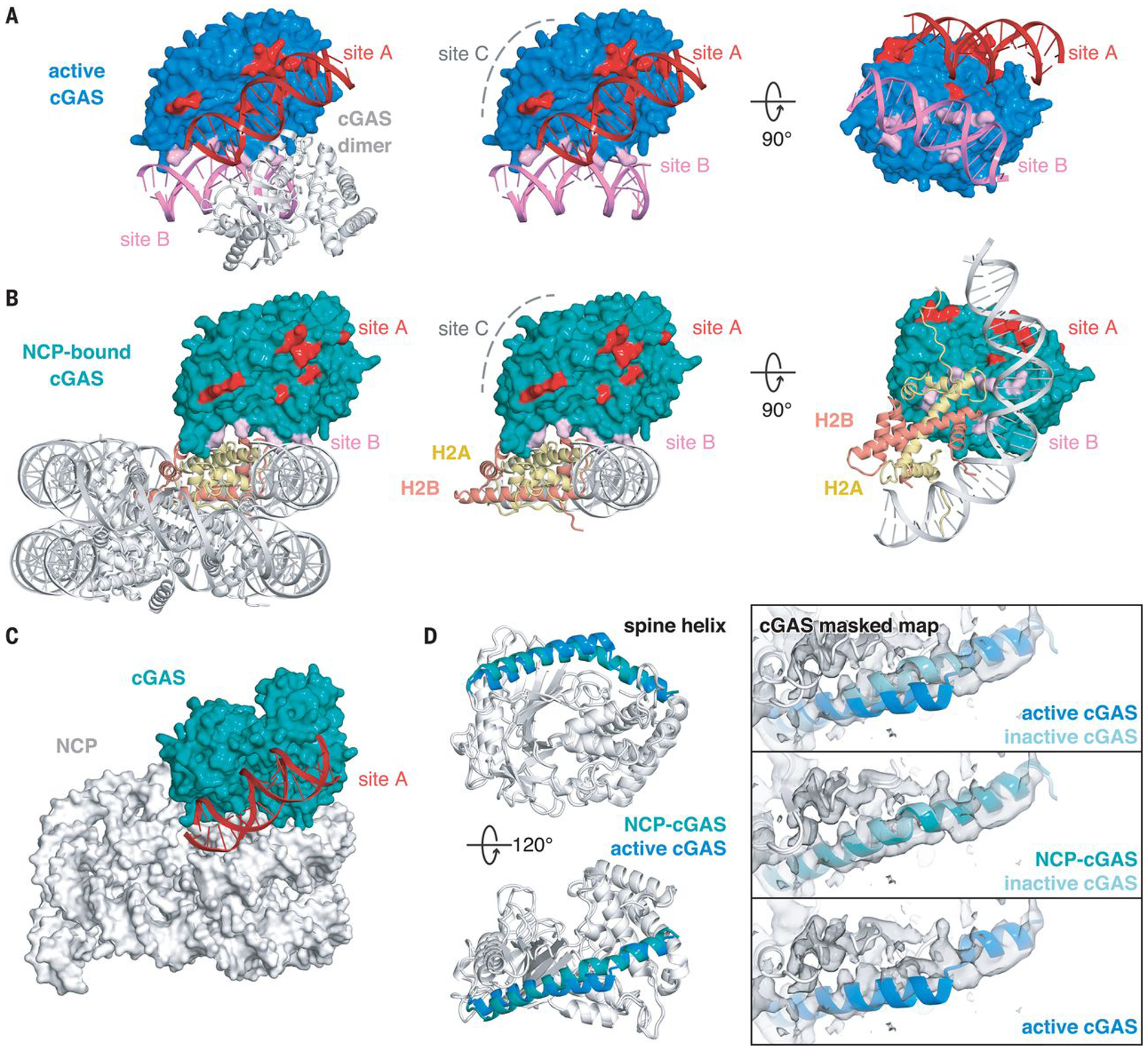
(A) Structure of the functionally active 2:2 cGAS-DNA complex (PDBID: 4LEZ). DNA binding sites A, B, and C are highlighted in red, pink, and gray. Middle and right panels depict side and bottom views of cGAS-DNA interactions for one cGAS monomer. (B) Structure of the cGAS-NCP complex with cGAS DNA-binding sites highlighted in red, pink, and gray. cGAS-interacting H2A and H2B chains are highlighted in yellow and red. Middle and right panels show side and bottom views of cGAS-NCP interactions. (C) Overlay of active cGAS-site A DNA structure onto NCP-cGAS, where dsDNA is shown in red. (D) Side and top views of overlay between NCP-bound and active cGAS structures with the spine helix highlighted in teal and blue, respectively. Right panels show overlays of spine helices onto the 3.9 Å resolution 1:1 cGASmask-NCP map. Inactive, apo-cGAS obtained from PDBID: 4K8V
It has become increasingly clear that the nucleosome plays essential roles in regulating nuclear protein functions, and the acidic patch has emerged as a key hot-spot for interactions (28, 29). Arginines termed arginine anchors, bind to the acidic patch in canonical and variant locations. cGAS employs one canonical arginine anchor (R241) that interacts with H2A E61, D90, and E92 in a nearly identical conformation as that observed for other nucleosome binding proteins, including RCC1, Sir3, and Ring1B (30–32) (Fig. 4). An additional variant-type arginine anchor in cGAS (R222) interacts with similar H2A residues (E61 and E64) as other nucleosome binding proteins (33), but exhibits a distinct conformation (Fig. 4). Overall, our structure establishes the molecular mechanism governing nucleosome-mediated inhibition of nuclear cGAS and further expands the paradigm of nucleosome acidic patch recognition by proteins involved in diverse nuclear processes.
Fig. 4. Arginine anchors bind the nucleosome acidic patch.
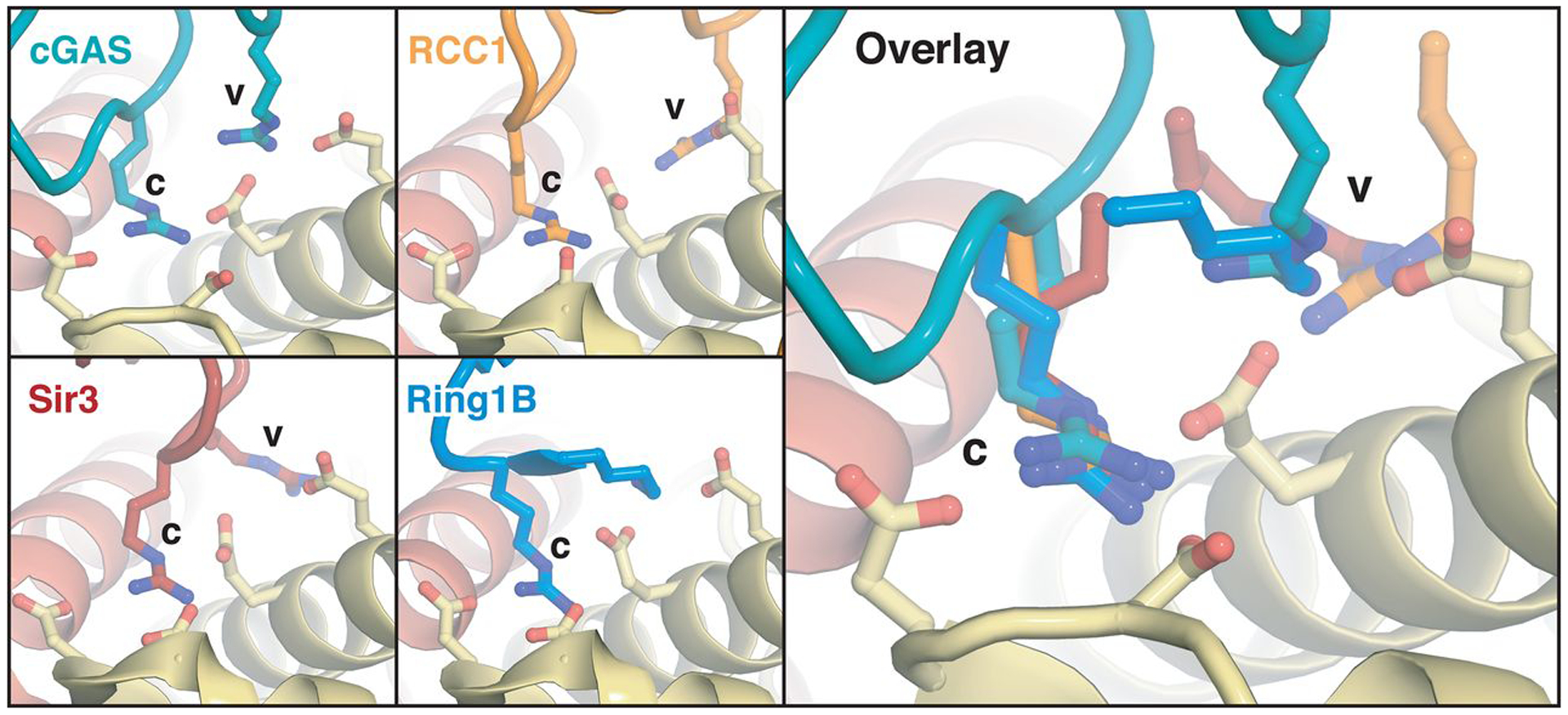
Identical views of canonical (c) and variant (v) arginine anchors employed by cGAS (c: R241, v: R222) and nucleosome binding proteins RCC1 (PDBID: 3MVD), Sir3 (PDBID: 3TU4), and Ring1B (PDBID: 4R8P). Parts of Ring1B structure not shown to allow visualization of arginine anchor.
Supplementary Material
Acknowledgments:
Single particle cryo-EM data were collected at the University of North Carolina at Chapel Hill, School of Medicine, Cryo-Electron Microscopy Facility, which is partially supported by NIH P30CA016086 and is part of the Molecular Microscopy Consortium of the University of North Carolina at Chapel Hill, Duke University, and the NIEHS. We thank M. Kesimer (University of North Carolina at Chapel Hill) for SEC-MALS assistance. We thank members of the Zhang and McGinty labs for critical comments.
Funding:
This work is supported by start-up funds and Jefferson Pilot Fellowship from the University of North Carolina at Chapel Hill to Q.Z., NIH 1R35GM133498 and Searle Scholars and Pew-Stewart Scholars awards to R.K.M, NIH R21CA234979 to P.L., and F99CA253730 to C.J.S.
Footnotes
Publisher's Disclaimer: This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
Competing interests: The authors declare no competing interests.
Data and materials availability: Cryo-EM maps have been deposited in the Electron Microscopy Data Bank under accession codes EMD-22408 (the 1:1 cGAS-nucleosome complex), EMD-22409 (the 2:1 cGAS-nucleosome complex), and EMD-22413 (the 1:1 cGASmask-nucleosome complex). Atomic coordinates of the 1:1 and 2:1 cGAS-nucleosome complexes have been deposited in the Protein Data Bank with accession codes 7JO9 and 7JOA, respectively. All materials in this study are available upon request.
References and Notes:
- 1.Barbalat R, Ewald SE, Mouchess ML, Barton GM, Nucleic acid recognition by the innate immune system. Ann. Rev. Immunol 29, 185–214 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Wu J, Chen ZJ, Innate immune sensing and signaling of cytosolic nucleic acids. Ann. Rev. Immunol 32, 461–488 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Ablasser A, Chen ZJ, cGAS in action: Expanding roles in immunity and inflammation. Science 363, eaat8657 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Sun L, Wu J, Du F, Chen X, Chen ZJ, Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu J et al. , Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339, 826–830 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ishikawa H, Barber GN, STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhong B et al. , The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29, 538–550 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Burdette DL et al. , STING is a direct innate immune sensor of cyclic di-GMP. Nature 478, 515–518 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Volkman HE, Cambier S, Gray EE, Stetson DB, Tight nuclear tethering of cGAS is essential for preventing autoreactivity. eLife 8, e47491 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu H et al. , Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature 563, 131–136 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Gentili M et al. , The N-Terminal domain of cGAS determines preferential association with centromeric DNA and innate immune activation in the nucleus. Cell Rep. 26, 2377–2393 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zierhut C et al. , The cytoplasmic DNA sensor cGAS promotes mitotic cell death. Cell 178, 302–315 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang H et al. , Chromatin-bound cGAS is an inhibitor of DNA repair and hence accelerates genome destabilization and cell death. EMBO J. 38, e102718 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du M, Chen ZJ, DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science 361, 704–709 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao P et al. , Cyclic [G(2’,5’)pA(3’,5’)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell 153, 1094–1107 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Civril F et al. , Structural mechanism of cytosolic DNA sensing by cGAS. Nature 498, 332–337 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X et al. , Cyclic GMP-AMP synthase is activated by double-stranded DNA-induced oligomerization. Immunity 39, 1019–1031 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X et al. , The cytosolic DNA sensor cGAS forms an oligomeric complex with DNA and undergoes switch-like conformational changes in the activation loop. Cell Rep. 6, 421–430 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andreeva L et al. , cGAS senses long and HMGB/TFAM-bound U-turn DNA by forming protein-DNA ladders. Nature 549, 394–398 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Zhou W et al. , Structure of the human cGAS-DNA complex reveals enhanced control of immune surveillance. Cell 174, 300–311 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie W et al. , Human cGAS catalytic domain has an additional DNA-binding interface that enhances enzymatic activity and liquid-phase condensation. Proc. Natl. Acad. Sci. USA 116, 11946–11955 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bilokapic S, Strauss M, Halic M, Structural rearrangements of the histone octamer translocate DNA. Nat. Commun 9, 1330 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu X et al. , Molecular evolutionary and structural analysis of the cytosolic DNA sensor cGAS and STING. Nucleic Acids Res. 42, 8243–8257 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hancks DC, Hartley MK, Hagan C, Clark NL, Elde NC, Overlapping patterns of rapid evolution in the nucleic acid sensors cGAS and OAS1 suggest a common mechanism of pathogen antagonism and escape. PLoS Genet. 11, e1005203 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mariani M et al. , Two murine and human homologs of mab-21, a cell fate determination gene involved in Caenorhabditis elegans neural development. Hum. Mol. Genet 8, 2397–2406 (1999). [DOI] [PubMed] [Google Scholar]
- 26.de Oliveira Mann CC, Kiefersauer R, Witte G, Hopfner KP, Structural and biochemical characterization of the cell fate determining nucleotidyltransferase fold protein MAB21L1. Sci. Rep 6, 27498 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fierz B, Poirier MG, Biophysics of chromatin dynamics. Ann. Rev. Biophys 48, 321–345 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Kalashnikova AA, Porter-Goff ME, Muthurajan UM, Luger K, Hansen JC, The role of the nucleosome acidic patch in modulating higher order chromatin structure. J. R. Soc., Interface 10, 20121022 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGinty RK, Tan S, Recognition of the nucleosome by chromatin factors and enzymes. Curr. Opin. Struct. Biol 37, 54–61 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Makde RD, England JR, Yennawar HP, Tan S, Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature 467, 562–566 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Armache KJ, Garlick JD, Canzio D, Narlikar GJ, Kingston RE, Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 A resolution. Science 334, 977–982 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGinty RK, Henrici RC, Tan S, Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature 514, 591–596 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anderson CJ et al. , Structural basis for recognition of ubiquitylated nucleosome by Dot1L methyltransferase. Cell Rep. 26, 1681–1690 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skrajna A et al. , Comprehensive nucleosome interactome screen establishes fundamental principles of nucleosome binding. Nucleic Acids Res. (2020). doi: 10.1093/nar/gkaa544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glaeser RM, Typke D, Tiemeijer PC, Pulokas J, Cheng A, Precise beam-tilt alignment and collimation are required to minimize the phase error associated with coma in high-resolution cryo-EM. J. Struct. Biol 174, 1–10 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Herzik MA Jr., Wu M, Lander GC, High-resolution structure determination of sub-100 kDa complexes using conventional cryo-EM. Nat. Commun 10, 1032 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mastronarde DN, Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Zivanov J, Nakane T, Scheres SHW, Estimation of high-order aberrations and anisotropic magnification from cryo-EM data sets in RELION-3.1. IUCrJ 7, 253–267 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rohou A, Grigorieff N, CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tan YZ et al. , Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat. Methods 14, 793–796 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Afonine PV et al. , Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D: Struct. Biol 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen VB et al. , MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D: Biol. Crystallogr 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barad BA et al. , EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat. Methods 12, 943–946 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dechassa ML et al. , Structure and Scm3-mediated assembly of budding yeast centromeric nucleosomes. Nat. Commun 2, 313 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


