Abstract
Acute respiratory distress syndrome (ARDS), an inflammatory condition with high mortality rates, is common in severe COVID-19, whose risk is reduced by metformin rather than other anti-diabetic medications. Given evidence of inflammasome assembly in post-mortem COVID-19 lungs, we asked whether and how metformin inhibits inflammasome activation and exerts its anti-inflammatory effect. We show that metformin inhibited NLRP3 inflammasome activation and interleukin (IL)-1β production in cultured and alveolar macrophages along with inflammasome-independent IL-6 secretion, thus attenuating lipopolysaccharide (LPS)- and SARS-CoV-2-induced ARDS. Metformin blocked LPS-induced ATP-dependent synthesis of the NLRP3 ligand mtDNA independently of AMP-activated protein kinase (AMPK) or NF-κB. Myeloid-specific ablation of LPS-induced cytidine monophosphate kinase 2 (CMPK2), which is rate limiting for mtDNA synthesis, reduced ARDS severity without a direct effect on IL-6. Thus, inhibition of ATP and mtDNA synthesis is sufficient for ARDS amelioration.
Keywords: ARDS, CMPK2, COVID-19, IL-1β, IL-6, metformin, mitochondrial DNA, NLRP3 inflammasome, SARS-CoV-2, inflammation
Graphical abstract
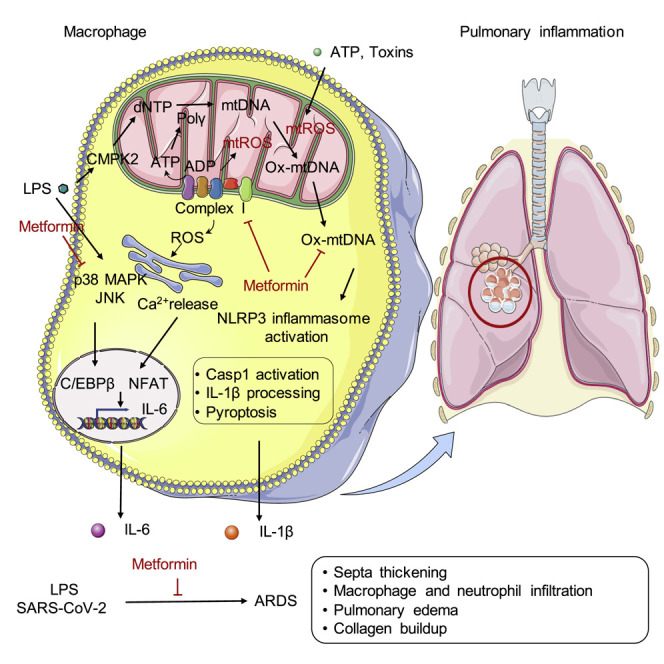
The exact mechanism by which metformin exerts its anti-inflammatory effects is still not known. Xian et al. shows that metformin’s inhibition of ETCCI blocks ATP-dependent mtDNA synthesis, cytoplasmic ox-mtDNA generation and NLRP3 inflammasome activation in macrophages, independent of AMPK and NF-κB. By limiting IL-1β production, metformin blunts pulmonary inflammation.
Introduction
Acute respiratory distress syndrome (ARDS) is a severe pulmonary inflammatory reaction triggered by respiratory viruses and bacteria and even physical trauma (Han and Mallampalli, 2015). Recently, ARDS has emerged as a leading cause of death in ICU hospitalized COVID-19 patients (Tay et al., 2020), although its COVID-19 related features are described as somewhat different and milder than “typical” ARDS (Li and Ma, 2020). ARDS onset depends on innate immune cells, monocytes, macrophages, and neutrophils, which propagate uncontrolled inflammation and tissue injury by secreting interleukin (IL)-1β, IL-18, IL-6, and tumor necrosis factor (TNF) (Han and Mallampalli, 2015). Bacterially triggered ARDS is initiated on Toll like receptor (TLR) engagement, but in sterile or virally induced ARDS, the inflammatory response is thought to be initiated by the NLRP3 inflammasome, which is highly expressed and activated in myeloid cells that have been primed on TLR engagement by viral RNA or proteins, such as the SARS-CoV spike protein (Dolinay et al., 2012; Dosch et al., 2009). Following priming, the NLRP3 inflammasome, composed of the enzyme caspase-1 (Casp1), the sensor NLRP3, the mitotic kinase NEK7, and the scaffold protein ASC (Sharif et al., 2019), is activated by damage associated molecular patterns (DAMPs), e.g., ATP and uric acid (UA) microcrystals, released by injured epithelial cells (Tschopp and Schroder, 2010), or the SARS-CoV pore-forming protein viroporin 3a (Chen et al., 2019). Injured epithelial cells also release IL-1α, which primes airway macrophages via the same adaptors used by TLRs, MyD88, and TRIF (Cohen, 2014). Macrophage priming stimulates DNA polymerase γ (POLγ)-mediated mitochondrial (mt) DNA synthesis, whereas ATP, UA microcrystals, and other DAMPs elicit mitochondrial damage and reactive oxygen species (ROS) production (Zhong et al., 2018; Zhou et al., 2011). Mitochondrial ROS react with newly synthesized mtDNA to generate oxidized (Ox) mtDNA, which binds to cytoplasmic NLRP3 and triggers inflammasome assembly and activation (Shimada et al., 2012; Zhong et al., 2018). Circulating mtDNA is suggested to be an inflammatory mediator that causes distant organ damage (Grazioli and Pugin, 2018) and an early indicator of severe illness and mortality in ARDS (Han and Mallampalli, 2015) and COVID-19 (Scozzi et al., 2020). Other factors that amplify ARDS (Day et al., 2009) and COVID-19 (Gao et al., 2021; Richardson et al., 2020; Singh and Khan, 2020; Verma et al., 2020; Yang et al., 2020a) severity and mortality are old age and co-morbidities like type 2 diabetes mellitus (T2DM) and obesity.
Several commonly used drugs, including dexamethasone, statins, and metformin, are found to reduce COVID-19 severity and mortality (Bramante et al., 2021; Horby. et al., 2021; Zhang et al., 2020). While dexamethasone is currently used in COVID-19 treatment, statins and metformin are only described to have a prophylactic effect. In particular, early studies from China have noted that long-term metformin usage decreased hospital mortality by 4-fold relative to other anti-diabetic drugs (Li et al., 2020; Luo et al., 2020) and reduced COVID-19-associated heart failure and inflammation (Cheng et al., 2020). In a US-based study, metformin reduces the odds ratio of COVID-19 death in African Americans with T2D by 10-fold (Crouse et al., 2021). Extended metformin usage ameliorates other age-related pathologies and extends lifespan and health span in model organisms (Barzilai et al., 2016), effects that are unrelated to glycemic control (Valencia et al., 2017) but may be due to its anti-inflammatory properties (Barzilai et al., 2016; Marcucci et al., 2020; Pollak, 2017). Prolonged use of metformin, but not other anti-diabetic drugs, also correlates with decreased age-related dementia and neurodegenerative disease incidence in elderly T2DM patients (Campbell et al., 2018; Markowicz-Piasecka et al., 2017; Shi et al., 2019). Metformin’s mode of action remains debatable, but most researchers agree that it inhibits oxidative phosphorylation and reduces ATP production through interaction with respiratory complex I (ETCCI) or other mitochondrial (mt) electron transport chain components (Bridges et al., 2014; Pollak, 2017). By increasing the ADP:ATP ratio, metformin activates AMPK and inhibits the nutrient sensor mTORC1, reducing hepatic glucose production (Duca et al., 2015) and enhancing autophagy, a homeostatic process that limits cell and tissue damage (Salminen and Kaarniranta, 2012). Autophagy curtails inflammation in part by eliminating damaged mitochondria and limiting NLRP3 inflammasome activation (Sanchez-Lopez et al., 2019; Zhong et al., 2016). Sustained metformin administration reduces macrophage NLRP3 inflammasome activation in diabetic individuals and in non-myeloid cells via an AMP-activated protein kinase (AMPK)-dependent and poorly defined mechanism (Lee et al., 2013; Li et al., 2016; Yang et al., 2019). Downstream mediators connecting AMPK to the inflammasome and their role in the response to metformin have not been investigated. AMPK has also been suggested to inhibit IKK-mediated NF-κB activation in vascular endothelial cells and cardiomyocytes but the underlying mechanism and its relevance to NLRP3 inflammasome priming remain obscure (Hattori et al., 2006; Vaez et al., 2016). Metformin has also been shown to inhibit NF-κB and cytokine induction in glia independently of AMPK (Labuzek et al., 2010) and to ameliorate air-pollution-induced thrombosis and IL-6 production by alveolar macrophages through ETCCI inhibition (Soberanes et al., 2019). Here, we show that metformin treatment of non-diabetic mice blunted the severity of LPS-induced ARDS when given either before or after LPS challenge. Although metformin did not affect expression of TNF or many other cytokines and chemokines, it inhibits IL-6 and IL-1β secretion. Attenuation of IL-6 secretion was likely to be due to abrogated JNK and p38 MAPK activation and reduced recruitment of C/EBPβ and NFATc1-4 to the IL-6 promoter. Diminished IL-1β production, however, was caused by inhibition of NLRP3 inflammasome activation, correlating with reduced Ox-mtDNA generation as a result of metformin-induced inhibition of mtATP production and LPS-induced ATP-dependent mtDNA synthesis. Myeloid-specific ablation of cytidine monophosphate kinase 2 (CMPK2), the LPS-inducible rate-limiting enzyme required for activation of the salvage pathway of mtDNA synthesis (Zhong et al., 2018), also reduced LPS-induced ARDS severity with little effect on IL-6 production. Neither NF-κB nor AMPK was involved in the ARDS protective activity of metformin. Of note, metformin also attenuated pulmonary inflammation in SARS-CoV2-infected human (h) ACE2 transgenic (Tg) mice. We suggest that a myeloid cell targeted version of metformin, a safe and inexpensive drug, can be used for the treatment of ARDS and other severe inflammatory conditions.
Results
Metformin has a minor effect on inflammatory cytokine mRNAs
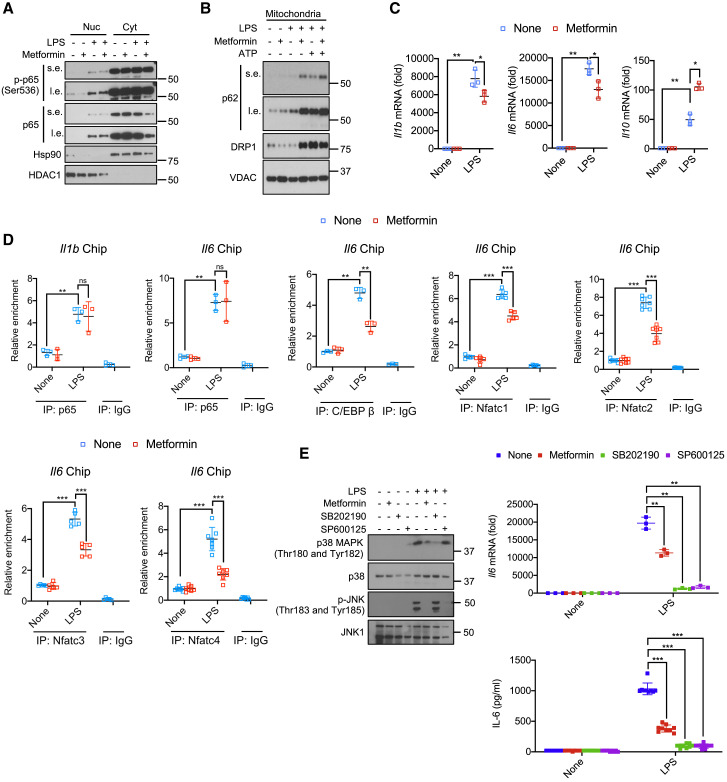
To understand the basis for metformin’s anti-inflammatory activity, we examined its effect on macrophage NF-κB activation and cytokine gene induction in macrophages. Mouse bone-marrow-derived macrophages (BMDMs) were incubated overnight with or without metformin (0.5 mM) and stimulated with LPS minus or plus ATP. In contrast to previous reports, metformin did not interfere with LPS-induced p65/RelA nuclear translocation or phosphorylation (Figures 1 A and S1A). Metformin also did not affect recruitment of the autophagy chaperon p62, encoded by the NF-κB-induced Sqstm1 gene, to mitochondria after LPS+ATP stimulation and enhanced it in LPS-primed cells that were not ATP exposed (Figures 1B and S1B). Metformin had similar effects on recruitment of the mitochondrial fission dynamin GTPase DRP1, enhancing it in LPS primed cells and not affecting it in LPS+ATP challenged macrophages (Figures 1B and S1B). Enhanced p62 and DRP1 recruitment may be the consequence of reduced ATP production and partial mitochondrial membrane depolarization in metformin-treated cells (see below). Metformin did not inhibit STAT1 or STAT3 phosphorylation (Figures S1C and S1D). Despite having no effect on NF-κB or STAT3, metformin led to a 30%–50% decline in Il1b and Il6 mRNA induction by LPS (Figure 1C). AMPKα1 silencing enhanced Il1b and Il6 mRNA accumulation in LPS-treated macrophages, but both transcripts still declined after metformin treatment (Figure S1E). As reported (Kelly et al., 2015), metformin potentiated Il10 mRNA induction (Figure 1C) but had little effect on other cytokine and chemokine mRNAs, many of which are NF-κB responsive, including Tnf, Il12, Il23, Ccl2, Ccl3, Ccl4, and Ccl5 (Figure S1F). Metformin did not inhibit induction of Ifnb and Ifng mRNAs, either (Figure S1F). IL-10 induction has been proposed to contribute to metformin’s anti-inflammatory activity (Kelly et al., 2015; Park et al., 2019). However, despite enhanced Il1b and Il6 mRNA induction in LPS-stimulated Il10 −/− BMDM, metformin still inhibited their expression (Figure S1G). Metformin-enhanced Il10 mRNA expression was AMPK-dependent (Figure S1H), but neither AMPK nor IL-10 was needed for the inhibitory effect on Il1b mRNA (Figures S1E and S1G).
Figure 1.
Metformin attenuates Il1b and Il6 mRNA induction but does not inhibit NF-κB activation
(A) BMDM pretreated -/+ metformin (0.5 mM, 16 h) were harvested 30 min after LPS (100 ng/mL) addition. Nuclear (Nuc) and cytosolic (Cyt) fractions were analyzed for the indicated proteins. s.e., short exposure; l.e., long exposure. One representative immunoblot (IB) out of 2.
(B) IB analysis of p62, DRP1, and VDAC in mitochondria from BMDM pretreated -/+ metformin, primed with LPS, and challenged with ATP (4 mM, 1 h).
(C) Q-PCR quantitation of Il1b, Il6 and Il10 mRNAs before or after LPS stimulation -/+ metformin pre-treatment (n = 3).
(D) p65, C/EBPβ or NFATc1-4 recruitments to the Il1 and Il6 promoters in BMDM before or after LPS stimulation was analyzed by ChIP-qPCR assay (n = 3–7).
(E) IB analysis of p38 MAPK and JNK phosphorylation (left) and quantitation of Il6 mRNA (top) and IL-6 secretion (bottom) from BMDM pretreated with metformin, SB202190 (10 μM) or SP600125 (40 μM) for 16 h and stimulated with LPS for 30 min.
All IB show one representative out of 3. Results in (C–E) are averages ±SD. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. Two-sided unpaired t test. See also Figure S1.
Metformin inhibited IL-6 secretion by LPS-stimulated BMDM, but TNF release was unaffected (Figures S1I and S1J). A similar effect was seen in human peripheral blood mononuclear cell (PBMC)-derived macrophages (Figure S1K). Inhibition of IL-6 production correlated with blunted p38 or JNK MAP kinase activation and decreased nuclear translocation of NFAT family members and C/EBPβ (Figures S1L–S1N), transcription factors that mediate Il6 gene induction (Akira et al., 1990; Luo and Zheng, 2016; Soberanes et al., 2019; Trautwein et al., 1993). Metformin also inhibited C/EBPβ and NFATc1-4 recruitment to the Il6 promoter but had no effect on p65/RelA recruitment to either the Il6 or Il1b promoter (Figure 1D). In concordance with the above results, JNK (SP600125) and p38 (SB202190) inhibitors blocked Il6 mRNA induction and IL-6 secretion (Figures 1E and S1O). Taken together, these results show that apart from a modest reduction in Il1b and Il6 mRNA induction, metformin has very little effect on expression of inflammatory cytokines and chemokines. Moreover, reduced Il6 mRNA correlates with inhibition of C/EBPβ and NFATc1-4 but not with NF-κB or STAT3, whose activation is not affected by metformin.
Metformin inhibits NLRP3 inflammasome activation
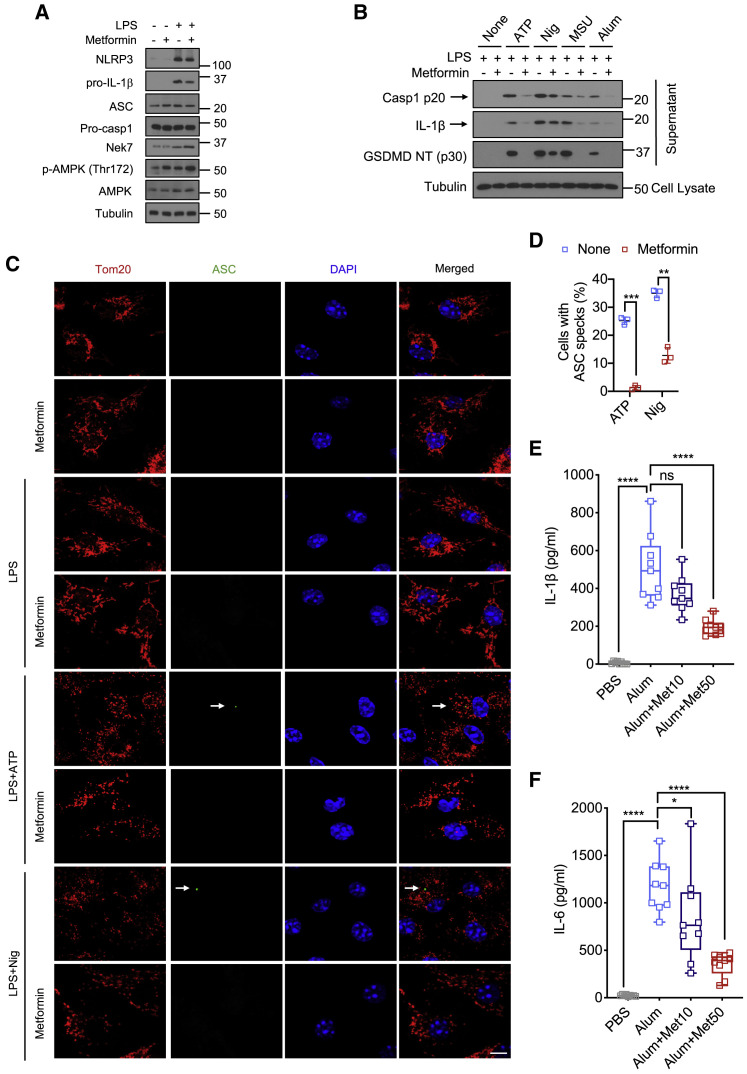
As IL-1β production is mainly regulated post-translationally, we examined effects of metformin on pro-IL-1β processing and NLRP3 inflammasome activation. Metformin modestly decreased pro-IL-1β protein and had no effect on expression of the inflammasome components NLRP3, which was induced by LPS stimulation, ASC, pro-Casp1, and NEK7 (Figures 2 A and S2A). As expected, metformin increased AMPK phosphorylation. Importantly, metformin inhibited Casp1 activation and pro-IL-1β processing to mature IL-1β in LPS-primed macrophages incubated with either ATP, nigericin (Nig), alum, or monosodium urate (MSU) crystals, although the effect on nigericin-challenged macrophages was relatively modest (Figures 2B and S2B–S2D). Metformin also inhibited gasdermin D (GSDMD) cleavage (Figures 2B and S2B) and abrogated Casp1 p20 and IL-1β secretion from human macrophages and LPS-primed mouse microglia stimulated with different NLRP3 inflammasome activators (Figures S2E–S2H). To validate NLRP3 inflammasome inhibition, we assessed metformin’s effect on ASC speck formation, an indicator of inflammasome assembly (Dick et al., 2016). Consistent with its effect on Casp1, metformin inhibited nigericin- and ATP-induced ASC speck formation, although inhibition of nigericin-induced specks was incomplete (Figures 2C and 2D), correlating with residual inflammasome activation in nigericin stimulated cells. In concert with its ability to inhibit GSDMD processing and block production of its pore forming N-terminal fragment (Liu et al., 2016), metformin also reduced LDH release (Figure S2I).
Figure 2.
Metformin inhibits NLRP3 inflammasome activation
(A) IB analysis of NLRP3, pro-IL-1β, ASC, pro-caspase-1, NEK7, phosphorylated AMPK, and total AMPK in BMDM pre-treated -/+ metformin (0.5 mM, 16 h) before or after 4 h of LPS (100 ng/mL) stimulation. Tubulin shows loading control.
(B) IB analysis of cleaved caspase-1 (Casp1 p20), mature IL-1β and cleaved GSDMD (GSDMD NT [p30]) in culture supernatants, and tubulin in lysates of BMDM that were pretreated -/+ metformin, primed with LPS, and challenged with inflammasome activators (Nig-nigericin, 10 μM, 1 h; MSU, 600 μg/mL, 6 h).
(C) Representative fluorescent microscopy images of BMDM co-stained for Tom20 and ASC before or after LPS (100 ng/mL, 4 h) priming followed by ATP (4 mM, 1 h) or nigericin (10 μM, 1 h), -/+ metformin (0.5 mM, 16 h) pre-treatment. DAPI stains nuclei. Arrows indicate ASC specks. Scale bars, 10 μm (n = 3).
(D) Percentages of cells shown in (C) with ASC specks. n = 150 cells per group from 3 independent experiments, magnification × 100.
(E and F) Peritoneal IL-1β (E) and IL-6 (F) in mice treated with different doses of metformin (0, 10, and 50 mg/kg) 30 min prior to i.p. alum (700 mg) injection (n = 9 mice per group).
All blots show one representative IB out of 3. Results in (D) are averages ± SD and in (E) averages ±SEM.∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; ns, not significant. Two- sided unpaired t test. See also Figure S2.
To determine whether short-term metformin treatment inhibits NLRP3-dependent inflammation in vivo we used the alum-induced peritonitis model (Guarda et al., 2011). Mice were given different doses of metformin 30 min prior to intraperitoneal (i.p.) alum challenge. Metformin treatment led to dose-dependent decline in IL-1β and IL-6 production (Figures 2E and 2F), but as seen in vitro had no effect on TNF production (Figure S2J). Thus, IL-1β production by metformin-treated macrophages correlates with inhibition of NLRP3 inflammasome activation.
Metformin inhibits mtDNA synthesis and Ox-mtDNA generation
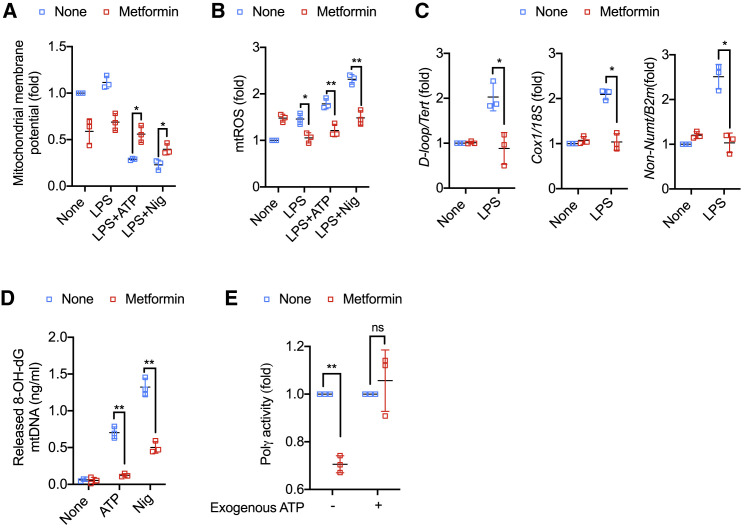
Metformin stimulates mitophagy by activating AMPK (Boyle et al., 2018), and mitophagy limits NLRP3 inflammasome activation (Sanchez-Lopez et al., 2019; Zhong et al., 2016). As expected, inhibition of mitophagy by p62 or ATG7 ablation enhanced Casp1 activation and IL-1β secretion (Figures S3A–S3D). However, metformin still inhibited Casp1 activation and IL-1β secretion in p62- and ATG7-deficient macrophages (Figures S3A–S3D), indicating that metformin’s effect is mitophagy independent. Most NLRP3 inflammasome activators induce mitochondrial depolarization, measured by a drop in membrane potential, and stimulate mtROS production (Tschopp and Schroder, 2010; Zhong et al., 2016; Zhou et al., 2011). Accordingly, treatment of LPS-primed BMDM with ATP or nigericin decreased mitochondrial membrane potential and increased mtROS, and metformin treatment blunted both responses (Figures 3A and 3B). Of note, metformin treatment decreased mitochondrial membrane potential on its own in non-stimulated macrophages, possibly due to defective ETCCI-mediated electron transfer from NADH to coenzyme Q. To determine whether inhibition of mtROS production by metformin contributes to inhibition of NLRP3 inflammasome activation, we used two compounds that stimulate ROS production, 2,3-dimethoxy-1,4-naphtoquinone (DMNQ) and mitoParaquat. Although both compounds increased mtROS in metformin-treated and ATP-stimulated macrophages, neither one restored Casp1 activation and IL-1β secretion (Figure S3E–S3G). In addition to mtROS, metformin strongly inhibited LPS-stimulated mtDNA synthesis (Figure 3C) and blocked generation of cytoplasmic Ox-mtDNA in LPS-primed macrophages challenged with either ATP or nigericin (Figure 3D). Inhibition of cytoplasmic Ox-mtDNA or mtDNA synthesis was independent of mtROS, as neither parameter was restored by DMNQ or mitoParaquat-treated macrophages (Figures S3H and S3I). Inhibition of LPS-induced mtDNA synthesis by metformin was confirmed by ethynyl deoxyuridine (EdU) labeling (Figures S3J and S3K). Metformin had no effect on expression of CMPK2 (Figures S3L–S3N), an LPS-inducible nucleotide kinase that converts dCMP to dCDP and is needed for stimulation of mtDNA synthesis (Zhong et al., 2018). Looking for another cause of blunted LPS-stimulated mtDNA synthesis, we examined effects of metformin on POLγ expression and activity. While having no effect on POLγ expression (Figures S3O and S3P), metformin treatment decreased POLγ activity measured in lysates of LPS-stimulated macrophages with an assay mix containing all 4 dNTP precursors but lacking exogenous ATP (Figure 3E). Addition of ATP at 10 mM fully restored POLγ activity, suggesting that the apparent decline in enzyme activity was due to the low ATP content of metformin treated LPS-stimulated cells (Figures 3E and S3Q). ATP is needed for initiation complex formation and POLγ movement along its DNA template (Burgers and Kornberg, 1982; Graziewicz et al., 2006).
Figure 3.
Metformin inhibits mtDNA synthesis and cytosolic Ox-mtDNA release
(A) Mitochondrial membrane potential of untreated and LPS (100 ng/mL, 4 h)-primed BMDM challenged with ATP (4 mM, 1 h) or nigericin (10 μM, 1 h), -/+ metformin (0.5 mM, 16 h) pretreatment, was measured by TMRM staining.
(B) Relative mtROS amounts measured by MitoSOX staining of BMDM treated as above.
(C) Relative total mtDNA amounts in LPS-primed BMDM -/+ metformin pre-treatment. Shown are the ratio of D-loop mtDNA to Tert nuclear (n) DNA, Cox1 mtDNA to 18S nDNA or mtDNA that is not inserted into nuclear DNA (non-NUMT) to B2m nDNA.
(D) 8-OH-dG amounts in cytosolic mtDNA of LPS-primed BMDM stimulated with ATP or nigericin (Nig) -/+ metformin pre-treatment were measured by ELISA.
(E) Lysates of LPS-primed BMDM -/+ metformin pretreatment, were divided into two aliquots. One aliquot was used to measure POLγ activity without exogenous ATP and the other was supplemented with 10 mM ATP prior to measuring POLγ activity.
Results are averages ± SD (n = 3). ∗p < 0.05; ∗∗p < 0.01; ns, not significant. Two-sided unpaired t test. See also Figure S3.
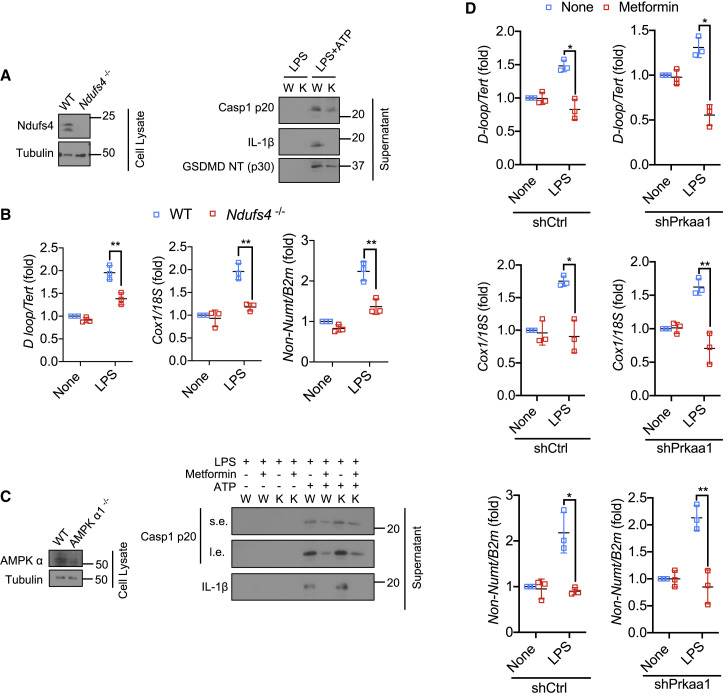
Metformin inhibits ATP production by targeting ETCCI (Wheaton et al., 2014). To validate the role of ETCCI in NLRP3 inflammasome activation, we used Ndufs4 −/− BMDM, missing a subunit needed for ETCCI assembly and function (McElroy et al., 2020; Yang et al., 2020b). Ndufs4 −/− BMDM exhibited defective NLRP3 inflammasome activation and GSDMD cleavage (Figures 4 A and S4A), and metformin did not further reduce IL-1β and Casp1 p20 release in these cells (Figures S4B and S4C). Ndufs4 ablation also interfered with LPS-induced mtDNA synthesis (Figure 4B). A recently developed ETCCI inhibitor, IACS-010759 (Molina et al., 2018), which is chemically unrelated and more potent than metformin, also inhibited IL-1β production and Casp1 and GSMDM cleavage in LPS-primed macrophages stimulated with NLRP3 inflammasome activators (Figures S4D–S4F). As found with metformin, the response to nigericin was not as strongly inhibited as the response to the other activators. IACS-010759 treatment also caused modest inhibition of IL-6 secretion (Figure S4G) but had no effect on TNF (Figure S4H). IACS-010759 also inhibited mtDNA synthesis, but unlike metformin and like the potent ETCCI inhibitor rotenone (Wheaton et al., 2014), it enhanced ROS production (Figures S4I and S4J). These results further confirm that inhibition of mtROS production is not required for metformin inhibition of NLRP3 inflammasome activation. Next, we examined the role of AMPK activation in inhibition of NLRP3 inflammasome activation. AMPKα1 ablation had no effect on inhibition of Casp1 activation or IL-1β processing and secretion by metformin in LPS-primed BMDM stimulated with ATP (Figures 4C and S4K). Importantly, AMPKα1 silencing had no effect on LPS-stimulated mtDNA synthesis and its inhibition by metformin (Figure 4D). Congruently, AMPKα1 deficiency had no effect on POLγ expression or activity in lysates of metformin-treated LPS-stimulated BMDM (Figures S4L–S4N). Collectively, these results suggest that inhibition of NLRP3 inflammasome activation is due to inhibition of new mtDNA synthesis, which is needed for generation of the NLRP3 ligand Ox-mtDNA (Zhong et al., 2018).
Figure 4.
Metformin inhibits NLRP3 inflammasome activation via ETCCI, independently of AMPK
(A) IB analysis of Ndufs4 (left), Casp1 p20, mature IL-1β, and cleaved GSDMD (right) in culture supernatants and cell lysates of LPS (100 ng/mL, 4 h)-primed wild type (W) or Ndufs4−/− (K) BMDM stimulated −/+ ATP (4 mM, 1 h).
(B) Relative total mtDNA amounts in LPS (200 ng/mL, 4 h)-primed wild type (WT) or Ndufs4−/− BMDM. The ratio of D-loop mtDNA to Tert nDNA, Cox1 mtDNA to 18S nDNA, or mtDNA that is not inserted into nuclear DNA (non-NUMT) to B2m nDNA is shown.
(C) IB analysis of AMPKα (left), Casp1 p20 and mature IL-1β (right) in culture supernatants and lysates of LPS-primed wild type (W) or Ampkα1−/− (K) BMDM stimulated -/+ ATP.
(D) Relative total mtDNA amounts in LPS-primed shCtrl- and shPrkaa1-transfected BMDM. The ratio of D-loop mtDNA to Tert nDNA, Cox1 mtDNA to 18S nDNA, or mtDNA that is not inserted into nuclear DNA (non-NUMT) to B2m nDNA is shown.
All IB show one representative out of 3. Results are averages ± SD (n = 3). ∗p < 0.05; ∗∗p < 0.01. Two-sided unpaired t test. See also Figure S4.
Metformin inhibits LPS-induced ARDS
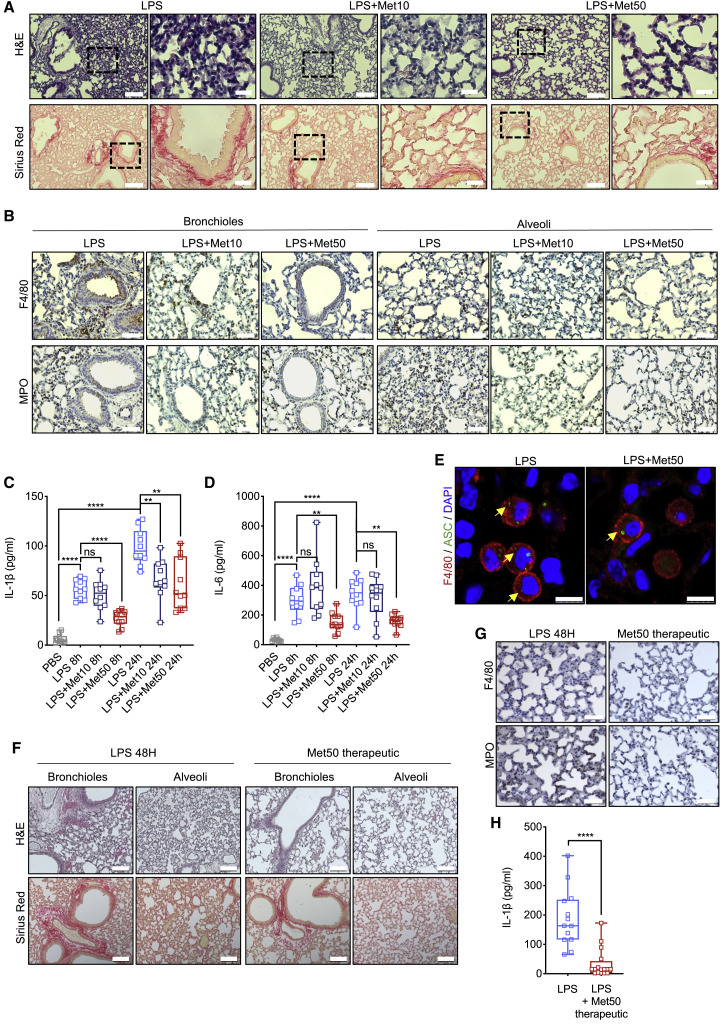
ARDS is a common cause of respiratory failure in critically ill patients, with a mortality rate of 30%–40% (Matthay et al., 2019). We examined whether short-term metformin treatment prevented or ameliorated ARDS in mice i.p. injected with a single bolus of 5 mg/kg LPS followed by collection of bronchioalveolar lavage fluid (BALF) and lung tissue after 8 and 24 h (Figure S5A). As described (Menezes et al., 2005), LPS administration damaged the pulmonary vascular endothelium and led to interstitial edema accompanied by innate immune cell recruitment into the septa and alveolar space with evident alveolar walls thickening and secondary collagen deposition (Figure S5B). Initially, mice were given vehicle or two different doses of metformin for three consecutive days with the last dose administered 30 min prior to LPS challenge (Figure S5A). Whereas at 10 mg/kg metformin had a modest protective effect, at 50 mg/kg it led to strong protection. Histological assessment of lung sections showed that at 50 mg/kg metformin prevented septa thickening and inhibited infiltration of inflammatory cells into the bronchioles, vascular bed, and lung parenchyma, and abrogated collagen buildup (Figures 5 A and S5C). At that dose metformin strongly inhibited macrophage and neutrophil infiltration (Figures 5B, S5D, and S5E), and this was accompanied by decreased IL-1β and IL-6 in BALF, with no effect on TNF (Figures 5C, 5D, and S5F). Consistently, metformin limited ASC speck formation (Figures 5E and S5G), and C/EBPβ and NFAT activation (Figures S5H–S5J), and reduced mortality of mice challenged with a higher LPS dose (Figure S5K).
Figure 5.
Metformin inhibits LPS-induced ARDS
(A) H&E and Sirius red staining of lung tissue from mice that were left untreated or pretreated with 10 or 50 mg/kg metformin and challenged with 5 mg/kg LPS 24 h prior tissue collection. Scale bar, 100 μm and 20 μm. n = 10 mice per group. 10–12 images per mouse were analyzed.
(B) Lung sections from above mice were stained with F4/80 and MPO antibodies. Scale bar, 50 μm. n = 4 mice per group. 10–12 images per mouse were evaluated.
(C and D) IL-1β (C) and IL-6 (D) concentrations in BALF from above mice were measured by ELISA. n = 10 mice per group.
(E) F4/80 and ASC immunostaining of paraffin-embedded lung tissue from above mice. DAPI stains nuclei. Scale bar, 7.5 μm. n = 4 mice per group.
(F) H&E and Sirius red staining of lung tissue from mice that were left untreated or treated with 50 mg/kg metformin daily starting 30 min after 5 mg/kg LPS challenge. Tissue was collected 48 h after LPS administration. Scale bar, 100 μm; n = 5 mice per group. 10–12 images per mouse were evaluated.
(G) F4/80 and MPO staining in lung sections from mice in (F). Scale bar, 50 μm. n = 5 mice per group. 10–12 images per mouse were evaluated.
(H) IL-1β concentration in BALF from mice in (F) were measured by ELISA. n = 5 mice per group.
∗∗p < 0.01; ∗∗∗∗p < 0.001; ns, not significant. Unpaired t test; Mann-Whitney test. See also Figure S5.
Next, we tested whether metformin administration after LPS challenge ameliorates LPS- induced ARDS. Metformin was given by i.p. injection at 50 mg/kg at 30 min after LPS administration with a second dose given after 24 h. Mice were evaluated 24 h after the last metformin dose (Figure S5L). Therapeutic administration of metformin was as effective as the prophylactic treatment, reducing septa thickening, infiltration of monocytic cells into the bronchioles, vascular bed and lung parenchyma, and abrogating collagen buildup (Figure 5F). Therapeutic metformin also inhibited macrophage and neutrophil infiltration (Figures 5G), and decreased IL-1β in BALF (Figure 5H). We also examined the preventive efficacy of oral metformin dissolved in drinking water, starting 48 h prior to LPS challenge (Figure S5M). Oral metformin was as protective as injected metformin (Figures S5N–S5P). We compared the plasma concentrations of metformin given via the two routes. Although oral metformin resulted in higher plasma concentrations than i.p. metformin at the time of LPS administration (942.57 ± 658.76 ng/mL versus 439.57 ± 389.02 ng/mL), at 24 h after LPS challenge the circulating concentrations of metformin were equally low, regardless of its administration route (Figure S5Q). Although the mice were left on oral metformin until the end of the experiment, soon after LPS injection their water intake decreased (Figure S5R), thus explaining why at 24 h after LPS challenge metformin plasma concentrations were equally low in both treatment groups. We conclude that short-term metformin treatment can prevent and ameliorate LPS-induced ARDS.
Myeloid-specific Cmpk2 ablation prevents LPS-induced ARDS but only affects IL-1β
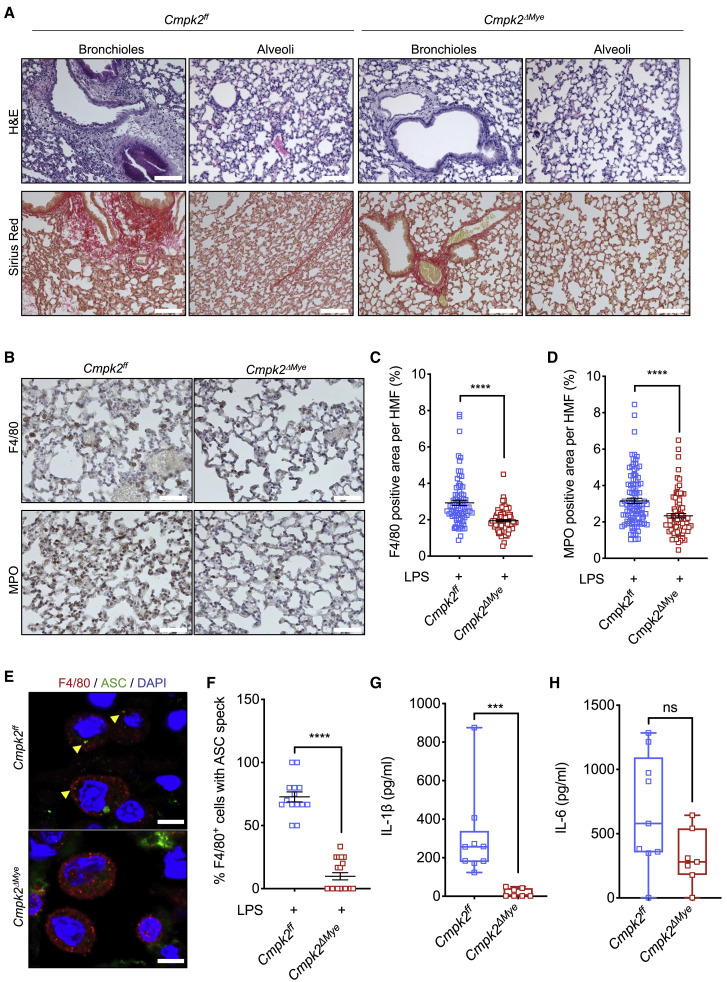
To test whether inhibition of LPS-induced mtDNA synthesis reduces pulmonary inflammation and IL-1β production in vivo, we generated mice with myeloid-specific ablation of CMPK2 (Cmpk2 ΔMye mice). As expected (Zhong et al., 2018), CMPK2-deficient BMDM (Cmpk2 Δ/Δ), exhibited defective LPS-stimulated mtDNA synthesis, Casp1 activation, and IL-1β secretion (Figures S6A–S6E). Consistently, mice lacking CMPK2 in the myeloid compartment were protected from LPS-induced ARDS like metformin-treated animals (Figure 6 A), exhibiting reduced macrophage and neutrophil infiltration (Figures 6B–6D). Cmpk2 ΔMye mice also exhibited fewer alveolar macrophages with ASC specks, indicating reduced NLRP3 inflammasome activation (Figures 6E and 6F). CMPK2 ablation also led to near complete blockade of IL-1β release but had a small and insignificant effect on C/EBPβ activation and IL-6 secretion (Figures 6G and 6H and S6F and S6G). Metformin treatment of Cmpk2 ΔMye mice did not result in noticeable improvement of lung pathology or reduced IL-1β secretion, but it did inhibit C/EBPβ activation and IL-6 secretion (Figures S6G–S6I). These results confirm that metformin affects IL-1β and IL-6 secretion through entirely different mechanisms, with only IL-1β being dependent on LPS-induced mtDNA synthesis, which is blocked by CMPK2 ablation. Moreover, these results demonstrate that myeloid-specific inhibition of TLR-induced mtDNA synthesis is sufficient for conferring protection from LPS-induced ARDS.
Figure 6.
Myeloid-specific Cmpk2 ablation recapitulates metformin protective effects in LPS-induced ARDS
(A) H&E and Sirius red staining of lung tissue from Cmpk2ff and Cmpk2ΔMye mice challenged with 5 mg/kg LPS for 24 h. Scale bar, 100 μm. n = 7–9 mice per group. 10–12 images per mouse were evaluated.
(B) F4/80 and MPO staining in lung sections from mice in (A). Scale bar, 50 μm. n = 5 mice per group. 10–12 images per mouse were evaluated.
(C and D) Area (in %) occupied by F4/80 (C), and MPO (D) staining, in lung sections from (B). Averages ± SEM; n = 5 mice per group. 10–12 HMF per group evaluated.
(E) F4/80 and ASC immunostaining of lung tissue from mice in (A). DAPI stains nuclei. Scale bar, 7.5 μm. n = 4–5 mice per group.
(F) Percentage of F4/80+ cells with ASC specks in lung sections from mice in (E). Cells were quantified in 15 HMF from 4–5 mice per group. Averages ± SEM.
(G and H) IL-1β (G) and IL-6 (H) concentrations in BALF from above mice were measured by ELISA. n = 7–9 mice per group.
∗∗∗p < 0.005; ∗∗∗∗p < 0.001; ns, not significant. Unpaired t test; Mann-Whitney test. See also Figure S6.
Metformin attenuates SARS-CoV-2-induced pulmonary inflammation
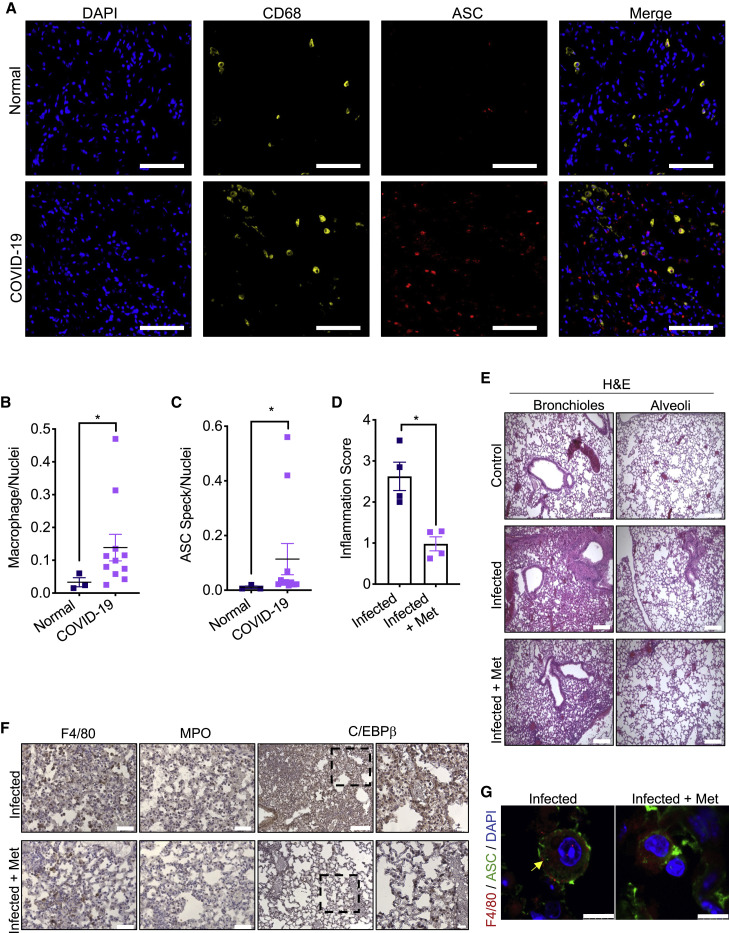
Consistent with a recent report of NLRP3 inflammasome activation in COVID-19 PBMCs (Rodrigues et al., 2021), we observed enhanced macrophage recruitment and ASC speck formation in COVID-19 post-mortem lungs (Figures 7 A–7C). To determine whether metformin treatment reduces SARS-CoV-2 elicited pulmonary inflammation, we used SARS-CoV-2 infectable hACE2 Tg mice (Bao et al., 2020; Winkler et al., 2020). The mice were i.p. injected with 50 mg/kg metformin or vehicle, starting 2 days before intranasal inoculation with 104 PFU of SARS-CoV-2 USA-WA1/2020 strain, and lung tissue was collected for analysis 6 days post-infection. As in LPS-induced ARDS, metformin treatment reduced immune cell infiltration and alveolar wall thickening and C/EBPβ activation in SARS-CoV-2 infected mice (Figures 7D–7F and S7A–S7C). Although at day 6 post-infection, only few macrophages with ASC specks were observed in SARS-CoV-2-infected lungs, almost none were found in metformin treated lungs (Figures 7G and S7D). Metformin treatment also slowed down the infection-associated loss of body weight at day 2 post-inoculation (Figure S7E); however, the mechanism underlying this effect is currently unknown.
Figure 7.
Metformin attenuates ARDS caused by SARS-CoV-2 infection
(A) Representative images from healthy (n = 3) and COVID-19 post-mortem (n = 11) tissue stained for CD68 (yellow) and ASC (red) taken at 20x.
(B and C) Quantification of macrophages (B) (CD68+) and cells with ASC specks (C) normalized to nuclei in above lung sections. ∗p < 0.05. Unpaired t test; Mann-Whitney test.
(D) Lung inflammation score in SARS-CoV-2-infected hACE2 Tg mice infected as indicated in STAR Methods. The scores were averaged and are shown as average ± SEM n = 4 mice. ∗p < 0.05. All groups were compared to each other using ANOVA Kruskal-Wallis test.
(E) H&E staining of lung tissue from hACE2 Tg mice treated as in (D). Scale bar, 200 μm. n = 4 mice per group. 10–12 images per mouse were evaluated.
(F) Lung sections from mice in (E) were stained with F4/80, MPO and C/EBPβ antibodies. Scale bar, 100 μm for F4/80, MPO, and C/EBPβ, and 50 μm for highlighted areas. n = 4 mice per group. 10–12 images per mouse were evaluated.
(G) F4/80 and ASC immunostaining in lungs from mice in (E). Nuclei were counterstained with DAPI and the images were captured by fluorescent confocal microscopy focusing on areas containing alveolar macrophages. Scale bar, 7.5 μm. n = 4 mice per group. All images are representative for each group. See also Figure S7.
Discussion
ARDS is associated with high mortality rates and is a serious and often fatal complication of COVID-19, although its manifestation in the latter differs from “typical” ARDS (Fan et al., 2020; Han and Mallampalli, 2015; Li and Ma, 2020; Wu et al., 2020). Mortality and ARDS risk in SARS-CoV-2 infected individuals are increased by old age and comorbidities such as T2D, obesity and hypertension (Gao et al., 2021; Guan et al., 2020; Richardson et al., 2020; Singh and Khan, 2020; Valencia et al., 2017), but are substantially reduced by long-term metformin usage (Bramante et al., 2021; Crouse et al., 2021; Li et al., 2020; Luo et al., 2020). This protective effect is independent of glycemic control and is related to metformin’s anti-inflammatory properties (Barzilai et al., 2016; Pollak, 2017; Valencia et al., 2017). Nonetheless, whether short-term metformin treatment of non-diabetic subjects can prevent acute inflammatory responses, including ARDS, was heretofore unknown. We now show that abrupt metformin-based intervention prevents and ameliorates LPS-induced ARDS in mice, accompanied with abrogated in situ NLRP3 inflammasome activation and IL-1β and IL-6 secretion. The mechanisms by which metformin inhibited IL-6 and IL-1β secretion were distinct. IL-6 production was mainly reduced transcriptionally, whereas inhibition of IL-1β secretion was primarily due to post-translational inhibition of NLRP3 inflammasome activation and defective pro-IL-1β processing. Consistent with previous studies (Soberanes et al., 2019), inhibition of IL-6 expression correlated with blunted NFAT and C/EBPβ (aka NF-IL6) nuclear translocation and recruitment to the Il6 promoter region, demonstrated by our ChIP experiments. This was most likely due to defective JNK and p38 activation, as inhibitors of these kinases blocked IL-6 mRNA induction and protein secretion. Nonetheless, inhibition of both IL-6 and IL-1β was mediated by metformin’s well-established inhibitory effect on ETCCI (Wheaton et al., 2014). Of note, metformin did not inhibit NF-κB activation and did not block induction of numerous cytokine mRNAs, including IFNβ and IFNγ.
While confirming metformin’s ability to diminish ETCCI-dependent and C/EBPβ and NFAT-mediated IL-6 production in alveolar macrophages (Soberanes et al., 2019), we showed that metformin is an effective inhibitor of NLRP3 inflammasome activation in vitro and in vivo and have identified its mode of action. The principle molecular target for metformin was ETCCI, whose inhibition reduced ATP production (Bridges et al., 2014; Wheaton et al., 2014). Consistent with previous publications showing that LPS induces a glycolytic switch and increases mitochondrial membrane potential (Ip et al., 2017; Mills et al., 2016), metformin elicited a larger drop in ATP concentration in LPS-primed than in non-stimulated macrophages. Genetic ablation of ETCCI subunit Ndufs4 also inhibited NLRP3 inflammasome activation and LPS-induced mtDNA synthesis, and in Ndusf4 −/− macrophages metformin did not lead to further inhibition of Casp1 activation and IL-1β secretion. Newly replicated single stranded mtDNA is highly susceptible to oxidative damage leading to Ox-mtDNA generation (Zhong et al., 2018). Fragments of Ox-mtDNA entering the cytoplasm bind NLRP3 and trigger inflammasome assembly and activation (Shimada et al., 2012; Zhong et al., 2018). Although metformin attenuated mtROS accumulation in macrophages treated with the NLRP3 activators ATP and nigericin, this effect was not important for inhibition of NLRP3 inflammasome activation which was not reversed by treatment of metformin-incubated macrophages with the mtROS generators mitoParaquat and DMNQ. Moreover, another ETCCI inhibitor IACS-010759 also blocked NLRP3 inflammasome activation and IL-1β secretion, but like the ETCCI inhibitor rotenone (Wheaton et al., 2014), it enhanced mtROS production. We therefore conclude that the main mechanism by which metformin prevents NLRP3 inflammasome activation is inhibition of TLR-induced mtDNA synthesis, which blocks NLRP3 inflammasome activation in vitro and in vivo and protects mice from LPS-induced ARDS. Treatment of Cmpk2 ΔMye mice with metformin did not result in any further decrease in IL-1β secretion although it inhibited IL-6, whose production was unaffected by CMPK2 ablation. Disruption of cytoplasmic accumulation of fragmented mtDNA and inhibition of GSDMD cleavage may prevent the circulatory release of mtDNA, thus attenuating distant organ damage (Grazioli and Pugin, 2018), and making further contributing to the protective effects of metformin and myeloid-specific CMPK2 ablation. In contrast to previous studies showing metformin-induced NF-κB inhibition in vascular endothelial cells and cardiomyocytes (Hattori et al., 2006; Vaez et al., 2016), no inhibition of NF-κB or NF-κB-dependent cytokine mRNA induction took place in mouse or human macrophages. Although we don’t rule out that metformin may manifest its ARDS protective effects in endothelial cells, it should be pointed out that NF-κB inhibition in such cells enhances TNF-induced apoptosis (Liu et al., 1996), resulting in vascular leakage. We also found no role for metformin-induced AMPK activation in inhibition of IL-1β and IL-6 production. AMPK-induced autophagy is speculated to be responsible for inhibition of NLRP3 inflammasome by metformin in non-myeloid cell types (Lee et al., 2013; Liu et al., 2016; Yang et al., 2019). However, AMPKα1 ablation or autophagy blockade by p62 or ATG7 ablation did not diminish metformin’s ability to inhibit NLRP3 inflammasome activation and IL-1β secretion.
Inhibition of IL-6 and IL-1β production by lung macrophages is likely to account for the ARDS blocking activity of metformin. The NLRP3 inflammasome and its products IL-1β and IL-18 play critical roles in ARDS pathogenesis (Dolinay et al., 2012; Ganter et al., 2008; Kolb et al., 2001), and recent studies implicate them along with IL-6 in acute airway inflammation triggered by SARS-CoV-2 and its relatives SARS-CoV-1 and MERS-CoV (Freeman and Swartz, 2020). Congruently, NLRP3 inflammasome activation is detected in COVID-19 PBMC (Rodrigues et al., 2021) and post-mortem lungs and our results demonstrate a protective effect of metformin in SARS-CoV-2 infected hACE2 Tg mice. A study of IL-1 receptor antagonist (IL-IRA, anakinra) in a small COVID-19 cohort reveals a rapid decline in inflammation and fever, followed by clinical improvement (Cauchois et al., 2020). Similar results, including reduced mortality or need for mechanical ventilation, are seen in another COVID-19 anakinra study (Huet et al., 2020), and a recent clinical trial has established the COVID-19 ameliorating effect of IL-6 receptor blockade (Gupta et al., 2021). Although metformin is not a specific NLRP3 inflammasome inhibitor, no specific NLRP3 antagonist has advanced to the clinic. Recently, however, FDA-approved BTK inhibitors have been found to inhibit NLRP3 inflammasome activation (Ito et al., 2015) and reduce inflammation in COVID-19 (Roschewski et al., 2020). Another FDA-approved drug that may reduce IL-1β secretion is disulfiram, which is used to fight alcohol addiction. Disulfiram does not inhibit GSDMD cleavage, but it blocks formation of the pore formed by its N-terminal fragment (Hu et al., 2020).
Inhibition of LPS-induced ARDS by metformin is extensive, reducing macrophage infiltration and collagen deposition. The dose of metformin required for these effects, 50 mg/kg per bolus, is equivalent to its maximal human daily dosage, 3 g/day orally in a 60 kg person (Wilcock and Bailey, 1994). Although oral metformin is just as effective as metformin injection, it should be noted that macrophages barely express the metformin and organic cation transporter OCT1, which mediates metformin uptake into hepatocyte, where it inhibits gluconeogenesis (Nies et al., 2009). Thus, the efficacy of acute metformin-based intervention in different ARDS etiologies may be improved by directed delivery into lung macrophages using nanoparticles or liposomes.
Given the important role of IL-1β and IL-6 in tumor development and progression (Greten and Grivennikov, 2019; Grivennikov et al., 2010), it is likely that inhibition of cytokine production by tumor associated macrophages contributes to metformin’s anti-cancer activity (Pollak, 2012). Supporting this hypothesis, a large clinical study of the IL-1β blocking antibody canakinumab in cardiovascular disease reveals a reduction in lung cancer incidence and mortality (Ridker et al., 2017). Given the widespread involvement of IL-1 in multiple inflammatory pathologies (Dinarello, 2011), we propose that metformin should be evaluated for therapeutic efficacy in such diseases. Given its low cost and high safety profile, metformin represents a viable alternative to expensive biological IL-1 blockers, such as anakinra and canakinumab.
Limitations of study
When using animal models there is always a concern regarding human relevance. We sought a mechanistic explanation to observational studies in which metformin for glycemic control was found to reduce ARDS and COVID-19 risk. It is unclear whether short-term metformin treatment fully mimics long-term drug treatment. It is also unclear whether LPS-induced ARDS in mice mimics bacterial-induced human ARDS, as LPS is only one component of gram negative bacterial and absent in gram positive bacteria. To model COVID-19, we used hACE2 transgenic mice. Although these mice succumb to SARS-CoV-2 infection, their lungs were not as heavily infiltrated with inflammatory cells as COVID-19 patients’ lungs. Moreover, hACE2 transgenic mice can only be followed for 1 week after SARS-CoV-2 infection and are unsuitable for studying prolonged COVID-19. It also remains to be seen whether COVID-19 patients who benefited from metformin use, show reduced ASC speck formation in alveolar macrophages.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| IL-1β | Cell Signaling | Cat#12426; N/A |
| Phospho-AMPK (Thr172) | Cell Signaling | Cat#2535; N/A |
| VDAC | Cell Signaling | Cat#4161; RRID:AB_10557420 |
| Cleaved Gasdermin D (Asp275) (E7H9G) | Cell Signaling | Cat#36425; RRID:AB_ 2799099 |
| Phospho-JNK (Thr183/Tyr185) | Cell Signaling | Cat#9251; RRID:AB_331659 |
| NEK7 (C34C3) | Cell Signaling | Cat#3057; RRID:AB_2150676 |
| Phospho-p38 MAPK (Thr180/Tyr182) | Cell Signaling | Cat#9211; RRID:AB_331641 |
| Atg7 | Cell Signaling | Cat#8558; RRID:AB_10831194 |
| Ndufs4 | Proteintech | Cat#15849-1-AP; N/A |
| Polγ | Abcam | Cat#128899; RRID:AB_1114530 |
| AMPK | Santa Cruz Biotechnology | Cat#25792; RRID:AB_2169546 |
| DRP1 | Santa Cruz Biotechnology | Cat#32898; RRID:AB_2093533 |
| p38 | Santa Cruz Biotechnology | Cat#728; N/A |
| JNK1/3 | Santa Cruz Biotechnology | Cat#474; RRID:AB_632384 |
| NFATc1 (IB) | Santa Cruz Biotechnology | Cat#7294; RRID:AB_2152503 |
| NFATc2 (IB) | Santa Cruz Biotechnology | Cat#7296; RRID:AB_628012 |
| NFATc3 ((IB) | Santa Cruz Biotechnology | Cat#8405; RRID:AB_628014 |
| NFATc4 (IB and ChIP) | Santa Cruz Biotechnology | Cat#271597; RRID:AB_10714946 |
| C/EBPβ (IB) | Santa Cruz Biotechnology | Cat#150; RRID:AB_2260363 |
| HDAC1 | Santa Cruz Biotechnology | Cat#7872; RRID:AB_2279709 |
| NF-κB p65 | Cell Signaling Technology | Cat# 8242; RRID:AB_10859369 |
| NFATc1 (ChIP) | Thermo Fisher Scientific | Cat# MA3-024; RRID:AB_2236037 |
| NFATc2 (ChIP) | Thermo Fisher Scientific | Cat#MA1-025; RRID:AB_2152763 |
| NFATc3 (ChIP) | Proteintech | Cat#18222-1-AP; RRID:AB_2152773 |
| C/EBPβ (ChIP) | Thermo Fisher Scientific | Cat# PA5-27244; RRID:AB_2544720 |
| Tubulin | Sigma-Aldrich | Cat#T9026; N/A |
| Caspase-1 | Adipogen | Cat#AG-20B-0042-C100; N/A |
| ASC | Adipogen | Cat#AG-25b-0006-C100; N/A |
| NLRP3 | Adipogen | Cat#AG-20B-0014-C100; N/A |
| p62 | ProGen | Cat#GP62-C; RRID:AB_2687531 |
| Cmpk2 | Novus | Cat#80653; RRID:AB_11037163 |
| Ndufs4 | Proteintech | Cat#15849-1-AP; RRID:AB_2878191 |
| F4/80 | Invitrogen | Cat#MF48000; RRID:AB_1500089 |
| Myeloperoxidase | Abcam | Cat#ab9535; RRID:AB_307322 |
| Anti-ATP Synthase, beta chain, clone 4.3E8.D1 antibody | Millipore | Cat#MAB3494; RRID:AB_177597 |
| Anti-rabbit IgG, HRP-linked Antibody | Cell Signaling | Cat#7074; RRID:AB_2099233 |
| Anti-mouse IgG, HRP-linked Antibody | Cell Signaling | Cat#7076; RRID:AB_330924 |
| Mouse IL-1beta /IL-1F2 Antibody | R&D Systems | Ca#MAB401; RRID:AB_2124620 |
| Mouse IL-1beta /IL-1F2 Biotinylated Antibody | R&D Systems | Ca#BAF401; RRID: AB_356450 |
| Anti-Mouse/Rat TNF alpha antibody | eBioscience | Cat#14-7423; RRID:AB_468492 |
| Anti-Mouse/Rat TNF alpha Biotin antibody | eBioscience | Cat#13-7341; RRID:AB_466951 |
| IL-6 Monoclonal Antibody (MP5-20F3) | eBioscience | Cat#14-7061-81; RRID:AB_468422 |
| IL-6 Monoclonal Antibody (MP5-32C11), Biotin | eBioscience | Cat#13-7062-81; RRID:AB_466910 |
| Donkey anti-rabbit IgG (H+L) secondary antibody, Alexa Fluor 488 | Invitrogen | Cat#A-21206; RRID:AB_2535792 |
| Donkey anti-rabbit IgG (H+L) secondary antibody, Alexa Fluor 594 | Invitrogen | Cat#A-21207; RRID:AB_141637 |
| Donkey anti-mouse IgG (H+L) secondary antibody, Alexa Fluor 488 | Invitrogen | Cat#A-21202; RRID:AB_141607 |
| Donkey anti-mouse IgG (H+L) secondary antibody, Alexa Fluor 594 | Invitrogen | Cat#A-21203; RRID:AB_141633 |
| Donkey anti-rat IgG (H+L) secondary antibody, Alexa Fluor 594 | Invitrogen | Cat#A-21209; RRID:AB_2535795 |
| Bacterial and virus strains | ||
| SARS-CoV-2 USA-WA1/2020 strain. GenBank (Accession number MN985325) | BEI Resources (NR-52281) | N/A |
| Biological samples | ||
| PBMC isolated from fresh peripheral blood of healthy donors (StemExpress, Inc) | StemExpress, Inc | N/A |
| Lung tissue from autopsy of patients with PCR confirmed SARS-CoV-2 deceased due to COVID-related pneumonia and patients deceased due to non-pulmonary-related causes. | Dr. Warren G. Tourtellotte. | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Recombinant Mouse IL-1β | R&D Systems | Cat#401-ML |
| Recombinant Mouse TNFα | eBioscience | Cat#14-8321 |
| Recombinant Mouse IL-6 | eBioscience | Cat#14-8061-62 |
| Recombinant human GM-CSF | R&D Systems | Cat# 7954-GM/CF |
| Recombinant Human IFN-gamma Protein | R&D Systems | Cat#285-IF-100 |
| SARS-CoV-2 Spike S1+S2 ECD | ABclonal | Cat# RP01260LQ |
| SARS-CoV-2 Spike S1 | ABclonal | Cat# RP01262 |
| Recombinant Human coronavirus SARS-CoV-2 Spike Glycoprotein S1 (Fc Chimera) |
Abcam | Cat# ab272105 |
| Ultrapure LPS, E. coli 0111:B4 | Invivogen | Cat#tlrl-3pelps |
| Lipopolysaccharides from Escherichia coli O26:B6 | Sigma-Aldrich | Cat#L2654 |
| Adenosine 5¢-triphosphate disodium salt solution | Sigma-Aldrich | Cat#A6559 |
| Nigericin | Invivogen | Cat#tlrl-..nig |
| Monosodium urate (MSU) | Enzo Life Science | Cat#ALX-400-047-M002 |
| Imject™ Alum Adjuvant | Thermo Scientific | Cat#77161 |
| Metformin | Selleckchem | Cat#S1950 |
| Metformin | Tocris | Cat#2864 |
| IACS-010759 | Selleckchem | Cat#S8731 |
| Metformin-(dimethyl-d6) hydrochloride | Sigma-Aldrich | Cat#53183 |
| SP600125 | Selleckchem | Cat#S1460 |
| SB202190 | Selleckchem | Cat#S1077 |
| MitoParaquat | Selleckchem | Cat#S8978 |
| DMNQ | Selleckchem | Cat#S6563 |
| Complete™, Mini Protease Inhibitor Cocktail | Roche | Cat#11836153001 |
| EDTA | Versene solution | Cat#9314 |
| Phosphatase Inhibitor Cocktail 2 | Sigma-Aldrich | Cat#P5726 |
| Clarity Western ECL Substrate | Biorad | Cat#1705061 |
| DAPI (4’6-Diamidino-2-Phenylindole, Dihydrochloride | Invitrogen | Cat#D1306 |
| Caspase I FLICA dye (660-YVAD-FMK) | Immunochemistry technologies, LLC | Cat#9122 |
| FAM-FLICA® Caspase-1 (YVAD) Assay Kit | Immunochemistry technologies, LLC | Cat#97 |
| Tetramethylrhodamine, Methyl Ester, Perchlorate (TMRM) | Invitrogen | Cat#T668 |
| Ghost Dye™ Red 780 | Tonbo | Cat# 13-0865-T100 |
| MitoSOX™ Red mitochondrial superoxide indicator | Invitrogen | Cat#M36008 |
| Bovine Serum Albumin | Gemini | Cat#700-100 |
| CD-1 (ICR) MSE00PLK2 | BioIVT | Cat#MSEOOPLK2 |
| Triton X-100 | Sigma-Aldrich | Cat#T8787 |
| Normal horse serum | Vectorlabs | Cat#S-2000 |
| Dimethyl Sulfoxide (DMSO) | Sigma-Aldrich | Cat#D2650-100 |
| Protein Assay Dye reagent concentrate | Biorad | Cat#5000006 |
| Polybrene | Sigma-Aldrich | Cat#TR-1003 |
| FluorSave™ | Millipore | Cat#345789 |
| Hematoxyline 560 | Leica | Cat#3801570 |
| Alcoholic Eosine Y 515 | Leica | Cat#3801615 |
| Direct Red 80 | Sigma-Aldrich | Cat#365548 |
| Picric acid solution | Ricca | Cat#5860-16 |
| Ficoll-Paque PREMIUM | GE Healthcare | Cat#17-5442-02 |
| Critical commercial assays | ||
| Mitochondria isolation kit | ThermoFisher Scientific | Cat#89874 |
| NE-PER™ Nuclear and cytoplasmic extraction reagents | ThermoFisher Scientific | Cat#78835 |
| Click-iT™ EdU Cell Proliferation Kit for Imaging | ThermoFisher Scientific |
Cat#C10337 |
| AllPrep DNA/RNA Mini kit | QIAGEN | Cat#80204 |
| Lipofectamine™ 3000 Transfection Reagent | Invitrogen | Cat#L3000001 |
| RNeasy Plus Mini kit | QIAGEN | Cat# 74134 |
| SuperScript™ VILO™ cDNA Synthesis Kit | Invitrogen | Cat#11754050 |
| SsoAdvancedTM Universal SYBR Green Supermix | Biorad | Cat#172-5274 |
| Power SYBR® Green RNA-to-Ct™ 1-Step Kit | Applied Biosystems | Cat# 4391178 |
| LDH-Glo™ Cytotoxicity Assay | Promega | Cat#J2380 |
| CellTiter-Glo Luminescent Assay | Promega | Cat#G924B |
| Human DNA Polymerase Gamma Assay | ProFoldin | Cat#DPG100K |
| 8-hydroxy 2 deoxyguanosine ELISA Kit | Abcam | Cat#ab201734 |
| MILLIPLEX MAP Human Cytokine/Chemokine Magnetic Bead Panel - Premixed 41 Plex - Immunology Multiplex Assay | EMD Millipore | Cat#HCYTMAG-60K-PX41 |
| Sep Mate 15 tubes | Stem Cell technologies | Catalog # 85415 |
| EasySep Human CD14 positive selection cocktail II | Stem Cell technologies | Cat#17858 |
| Waters XBridge Amide column (2.1 × 150 mm, 3.5 μm) | Waters | Cat#186004861 |
| Experimental models: Cell lines | ||
| HEK293T | ATCC | ATCC® CRL-3216™; RRID:CVCL_0063 |
| L929 | ATCC | ATCC® CCL-1™; RRID:CVCL_0462 |
| Immortalized BMDM | Dr. Kate Fitzgerald | (N/A) |
| Vero E6 | ATCC | ATCC Cat# CRL-1586, RRID:CVCL_0574 |
| Experimental models: Organisms/strains | ||
| C57BL/6N female and male (in vitro) | In house breeding | N/A |
| C57BL/6N male (in vivo) | Charles River Laboratories | N/A |
| Il10−/− female and male (in vitro) | In house breeding | N/A |
| Ampkα1−/− female and male (in vitro) | Dr. R. Liu-Bryan | N/A |
| p62ff and p62ΔMye female and male (in vitro) | In house breeding | N/A |
| Cmpk2ff and Cmpk2ΔMye male (in vivo) | In house breeding | N/A |
| Ndufs4−/− female and male (in vitro) | Dr. J. D. Rabinowitz | N/A |
| Oligonucleotides | ||
| Primers for mouse Hprt1 Forward: CTGGTGAAAAGGACCTCTCG Reverse: TGAAGTACTCATTATAGTCAAG GGCA |
Integrated DNA Technologies | N/A |
| Primers for mouse Il1b Forward: AGTTGACGGACCCCAAAAG Reverse: AGCTGGATGCTCTCATCAGG |
Integrated DNA Technologies | N/A |
| Primers for mouse Il6 Forward: CCAGGTAGCTATGGTACTCCA Reverse: GCTACCAAACTGGCTATAATC |
Integrated DNA Technologies | N/A |
| Primers for mouse Tnf Forward: CCCTCACACTCAGATCATCTT Reverse: GCTACGACGTGGGCTACAG |
Integrated DNA Technologies | N/A |
| Primers for mouse Il10Forward: CAG AGCCACATGCTCCTAGAReverse: TGTCCAGCTGGTCCTTTGTT |
Integrated DNA Technologies | N/A |
| Primers for mouse Ifn-β Froward: GCCTTTGCCATCCAAGAGATGC Reverse: ACACTGTCTGCTGGTGGAGTTC |
Integrated DNA Technologies | N/A |
| Primers for mouse Ifn-γFroward: CAGCAACAGCAAGGCGAAAAAGG Reverse: TTTCCGCTTCCTGAGGCTGGAT |
Integrated DNA Technologies | N/A |
| Primers for mouse Il12a Forward: ACGAGAGTTGCCTGGCTACTAG Reverse: CCTCATAGATGCTACCAA GGCAC |
Integrated DNA Technologies | N/AN/A |
| Primers for mouse Il12bFroward: TTGAACTGGCGTTGGAAGCACG Reverse: CCACCTGTGAGTTCTTC AAAGGC |
Integrated DNA Technologies | N/A |
| Primers for mouse Il23 Froward: CATGCTAGCCTGGAACGCACAT Reverse: ACTGGCTGTTGTCCTTGAGTCC |
Integrated DNA Technologies | N/A |
| Primers for mouse Ccl2Froward: GCTACAAGAGGATCACCAGCAG Reverse: GTCTGGACCCATTCCTTCTTGG |
Integrated DNA Technologies | N/A |
| Primers for mouse Ccl3 Froward: ACTGCCTGCTGCTTCTCCTACA Reverse: ATGACACCTGGCTGGGAG CAAA |
Integrated DNA Technologies | N/A |
| Primers for mouse Ccl4 Froward: ACCCTCCCACTTCCTGCTGTTT Reverse: CTGTCTGCCTCTTTTGGTCAGG |
Integrated DNA Technologies | N/A |
| Primers for mouse Ccl5 Froward: CCTGCTGCTTTGCCTACCTCTC Reverse: ACACACTTGGCGGTTCCTTCGA |
Integrated DNA Technologies | N/A |
| Primers for mouse Cmpk2 Forward: GCCCCAGATATAGCATTCCC Reverse: GTTCATCCTGTTCCTGCTCC |
Integrated DNA Technologies | N/A |
| Primers for mouse D-loopForward: AATCTACCATCCTCCGTGAAACC Reverse: TCAGTTTAGCTACCCCCAA GTTTAA |
Integrated DNA Technologies | N/A |
| Primers for mouse 18S Forward: TAGAGGGACAAGTGGCGTTC Reverse: CGCTGAGCCAGTCAGTGT |
Integrated DNA Technologies | N/A |
| Primers for mouse TertForward: CTAGCTCATGTGTCAAGACCCTCTT Reverse: GCCAGCACGTTTCTCTCGTT |
Integrated DNA Technologies | N/A |
| Primers for mouse Cox Forward: GCCCCAGATATAGCATTCCC Reverse: GTTCATCCTGTTCCTGCTCC |
Integrated DNA Technologies | N/A |
| Primers for mouse Non-Numt Forward: CTAGAAACCCCGAAACCAAA Reverse: CCAGCTATCACCAAGCTCGT |
Integrated DNA Technologies | N/A |
| Primers for mouse B2m Forward: ATGGGAAGCCGAACATACTG Reverse: CAGTCTCAGTGGG GGTGAAT |
Integrated DNA Technologies | N/A |
| Primers for human IL1BForward: ATGATGGCTTATTACAGTGGCAA Reverse: GTCGGAGATTCGTAGCTGGA |
Integrated DNA Technologies | N/A |
| Primers for human IL6Forward: ACTCACCTCTTCAGAACGAATTG Reverse: CCATCTTTGGAAGGTTCA GGTTG |
Integrated DNA Technologies | N/A |
| Primers for human 18SForward: GCTTAATTTGACTCAACACGGGA Reverse: AGCTATCAATCTGTCAATCC TGTC |
Integrated DNA Technologies | N/A |
| Primers for mouse Il1b (NF-κB p65 ChIP assay) Forward: AACGGAGGAGCCGTTGATATG Reverse: AGAGCATCTTCCTAATGC |
Integrated DNA Technologies | N/A |
| Primers for mouse Il6 (NF-κB p65 ChIP assay) Forward: AGCTCATTCTGCTCTG Reverse: AGATTGCACAATGTGACGTCG |
Integrated DNA Technologies | N/A |
| Primers for mouse Il6 (C/EBPβ ChIP assay) Forward: CCCCTTCCTAGTTGTGATTC Reverse: AAATCTTTGTTGGAGGGTGG |
Integrated DNA Technologies | N/A |
| Primers for mouse Il6 (Nfatc1 ChIP assay) Forward: CCCACCCTCCAACAAAGATT Reverse: GCTCCAGAGCAGAATGAGCTA |
Integrated DNA Technologies | N/A |
| Primers for mouse Il6 (Nfatc2 ChIP assay) Forward: TCTGGAGACAGGTGGACAGAA Reverse: CCCCTCCAATGCTCAAGTCT |
Integrated DNA Technologies | N/A |
| Primers for mouse Il6 (Nfatc3 ChIP assay) Forward: GCATTCTCCCCAGTGGTCTC Reverse: AGACATGCTCAAGTGCTGAGTC |
Integrated DNA Technologies | N/A |
| Primers for mouse Il6 (Nfatc4 ChIP assay) Forward: AGATATCTGGAGACAGGT GGACAReverse: TCTTCTAGGCTGGGT CAAACT |
Integrated DNA Technologies | N/A |
| Recombinant DNA | ||
| VSV-G | Zhong et al., 2016 | N/A |
| pLV-CMVΔ8.9 | Zhong et al., 2016 | N/A |
| shAtg7: 5¢-CCGGCCAGCTCTGAAC TCAATAATACTCGAGTATTATTGAGT TCAGAGCTGGTTTTTG-3¢ |
Sigma shRNA Mission librarySigma shRNA Mission library | N/A |
| shPrkaa1:5¢-CCGGCCTGAATCG AAATGTGTTCTTCTCGAGAAGAA CACATTTCGATTCAGGTTTTT-3¢ |
Sigma shRNA Mission library | N/A |
| Software and algorithms | ||
| Prism 8.0 | GraphPad Software | N/A |
| ImageJ | ImageJ | N/A |
| FloJo | FloJo | N/A |
| XPONENT | Luminex | N/A |
| MasterPlex Reader fit | Hitachi Solutions America | N/A |
| Other | ||
| DMEM | GIBCO | Cat#11995-065 |
| RPMI 1640 | GIBCO | Cat# 11875085 |
| MEM non-essential amino acids | Hyclone | Cat#SH30050.03 |
| Fetal Bovine Serum (FBS) | GIBCO | Cat#10437-028 (lot.1913181) |
| Penicillin-streptomycin | GIBCO | Cat#15140-122 |
| Penicillin-streptomycin | Corning | Cat#20-002-CL |
| L-glutamine | Corning | Cat#25-005-Cl |
Resource availability
Lead contact
Lead Contact, Dr. Michael Karin (karinoffice@ucsd.edu).
Materials availability
Further information and request for resources and reagents should be directed to the Lead Contact, Dr. Michael Karin (karinoffice@ucsd.edu) or Dr. Elsa Sanchez-Lopez (esl023@health.ucsd.edu).
Data and code availability
All data supporting the findings of this study are available within the paper and its supplementary materials. This study did not generate any unique datasets or code.
Experimental model and subject details
Cell lines and primary culture
Macrophage culture and stimulation
Femurs and tibia bones from C57BL/6, p62 f/f and p62 ΔMye (Zhong et al., 2016), Cmpk2 f/f and Cmpk2 ΔMye (generated in house), Ndsuf4 −/− (kindly provided by Dr. Joshua D. Rabinowitz and Dr. Lifeng Yang), Ampkα1 −/− (kindly provided by Dr. Ru Liu-Bryan), and Il10 −/− mice collected at 8-10 weeks of age (regardless the gender) were used to generate bone-marrow-derived macrophage (BMDM) as described (Hornung et al., 2008). Macrophages were cultured in high glucose DMEM supplemented with 10% FBS, 20% L929-cell conditioned medium, and 100 U/mL penicillin-streptomycin for 7-10 days. Immortalized mouse BMDM (iBMDM) were obtained from Dr. Kate Fitzgerald and grown in DMEM supplemented with 10% FBS and 100 U/mL penicillin-streptomycin.
Primary microglia was isolated from BL6 mice as described (Saura et al., 2003). Briefly, confluent mixed glial cultures were subjected to mild trypsinization (0.05-0.12%) in the presence of 0.2-0.5 mM EDTA and 0.5-0.8 mM Ca2+ to detach an intact layer of astrocytes, leaving attached microglia.
PBMC were isolated from fresh peripheral blood of healthy donors (StemExpress, Inc) by density gradient centrifugation using Ficoll-Paque PLUS (GE healthcare). PBMC layer was collected and CD14+ monocytes were purified by magnetic selection (StemCell Technology) according to the manufacturer’s protocol. CD14+ monocytes, with a final cell density of 106 cells/mL, were cultured in RPMI-1640 medium with L-glutamine and 25 mM HEPES (Hyclone), supplementation includes 100 μg/mL streptomycin, 100 U/mL penicillin (Thermo Fisher Scientific), 1X MEM non-essential amino acids (Hyclone), 1X (55uM) 2-Mercaptoethanol (Thermo fisher Scientific) and 10% heat-inactivated fetal bovine serum (Hyclone) and 50 ng/mL human recombinant GM-CSF (R&D systems 215-GM-010). Medium containing GM-CSF was refreshed every 2–3 days by adding half of the initial volume. After 6 days, macrophages were harvested with Versene solution (Irvine Scientific cat. 9314) for 30 min (37°C, 5% CO2) and collected by cell scrapping. Cells were resuspended in RPMI-1640 media with 2 mM L-glutamine (Thermo Fisher) and supplemented with HEPES (25 mM, Hyclone), 1X penicillin/streptomycin (Thermo Fisher Scientific) and 1% heat-inactivated fetal bovine serum (Hyclone). 2.5x104 macrophages were seeded as triplicate wells in 96-well plate in a volume of 150uL per well. After 4 h period of adaptation (37°C, 5% CO2), the indicated dose of metformin (Tocris Bioscience) was added overnight for 16 h. Macrophages were then stimulated with 20 ng/mL of LPS for 3 h followed by addition of 2 mM ATP (Invivogen) for 30 min. Supernatants were collected and stored at -80°C. Cells were rinsed with PBS and processed for Caspase 1 activity by flow cytometry.
All macrophages were cultured at 37°C with 5% CO2. NLRP3 inflammasome activation was induced after 4 h priming with ultrapure LPS (100 ng/mL) by challenge with the NLRP3 activators ATP (4 mM) and nigericin (10 μM) for 1 h, unless otherwise indicated, and monosodium urate (MSU) crystals (400 μg/mL) and alum (250 μg/mL) for 6 h.
Vero cells culture
African green monkey kidney epithelial cells (Vero-E6, CRL-1586) were obtained from the American Type Culture Collection (ATCC, Bethesda, MD) and maintained in Dulbecco’s modified Eagle medium (DMEM) supplemented with 5% (v/v) fetal bovine serum (VWR) and 100 units/mL penicillin-streptomycin (Corning).
Animal studies
Nine weeks-old C57BL/6 male mice purchased from Charles River Laboratories were maintained at a UCSD animal facility until alum-induced peritonitis and LPS-induced ARDS models were performed. All mouse studies were conducted in accordance with UCSD and NIH guidelines and regulations for the housing and treatment of laboratory animals using protocols approved by the UCSD Institutional Animal Care and Use Committee.
Myeloid specific Cmpk2 deficient (Cmpk2 ΔMye) mice were generated from C57BL/6N- Cmpk2tm1c(KOMP)Wtsi/H mice (MRC Harwell Institute), that harbor a targeted Cmpk2 floxed allele that can be inactivated by Cre-mediated recombination. Cmpk2 floxed homozygous (Cmpk2 f/f) mice were crossed with transgenic mice that expressed Cre recombinase under control of the lysozyme M (LysM) promoter to generate Cmpk2 ΔMye.
LPS-induced ARDS
Ten- to twelve- weeks-old male C57BL/6 mice, Cmpk2 f/f and Cmpk2 ΔMye were subjected to LPS-induced ARDS as described (Yu et al., 2018). Briefly, 5 mg/kg LPS or vehicle was i.p. injected and the mice were euthanized 8, 24 or 48 h later. Metformin’s prophylactic effect was evaluated by i.p. injections at 10 or 50 mg/kg for three consecutive days with the last dose administered 30 min prior LPS challenge. Oral metformin prophylactic effect was evaluated by providing 400 mg/kg metformin in the drinking water ad libitum starting 2 days prior to LPS challenge and until the endpoint. Metformin’s therapeutic effect was studied by i.p. injection at 50 mg/kg starting 30 min after LPS injection with a second dose given after 24 h. In all cases, at the experimental endpoint the lungs were inflated with cold PBS, excised and fixed in 10% formalin for histological evaluation.
Mice were also subjected to metformin pretreatment as above and injected with a lethal LPS dose (30 mg/kg). Survival was monitored up to 72 h and survival rate was determined by Kaplan Meyer analysis.
Alum-induced peritonitis
Ten- to twelve-weeks old male C57BL/6 mice were i.p. injected with alum (700 mg) or PBS. The mice were pretreated with metformin at 10 or 50 mg/kg 30 min prior to alum challenge. Peritoneal lavage was collected to measure cytokine amounts by ELISA.
SARS-CoV-2 infection in hACE2 mice
All the in vitro and in vivo experiments with infectious SARS-CoV-2 have been conducted under appropriated biosafety level (BSL) 3 and animal BSL3 (ABSL3) laboratories, respectively, at The Scripps Research Institute (TSRI). Experiments have been approved by the TSRI Institutional Biosafety (IBC) and Animal Care and Use (IACUC) committees. K18 human-ACE2 B6 background male mice were subjected to intranasal infection with 100 μl volume containing 10,000 PFU of SARS-CoV-2 USA-WA1/2020 strain (Gen Bank: MN985325.1) following gaseous sedation in an isoflurane chamber. Mice were pretreated with vehicle or 50 mg/kg metformin daily starting 2 days before infection. Some mice were mock infected, treated with vehicle and used as healthy controls for scoring purposes. Body weight and general health status was monitored daily during the course of the experiment. At day 5 post-infection mice were euthanized by overdose of isoflurane and lung were inflated with 10% formalin for histopathological assessment.
Method details
shRNA lentiviral silencing
ATG7 and AMPKα1 shRNA silencing were done by lentiviral transduction of iBMDM or primary BMDM as described (Sanchez-Lopez et al., 2019). Sequences of specific shRNAs used in this study were obtained from the MISSION shRNA Library (Sigma). Viral particles were generated in HEK293T cells using VSV-G and pLV- CMVΔ8.9 plasmids and specific shRNAs. Supernatants were collected 48 h after transfection, filtrated through a 0.45-micron pore filter and added to iBMDM or primary BMDM. To increase infection efficiency, 8 μg/mL of polybrene were added. Virus containing medium was washed after 6 h and the cells were cultured with fresh medium. Infected cells were expanded and selected with puromycin at 72 h post-transduction.
Generation of SARS-CoV-2 USA-WA1/2020 viral stock
SARS-CoV-2 USA-WA1/2020 natural isolate was obtained from BEI Resources (NR- 52281) and amplified on Vero E6 cells. Briefly, Vero-E6 cells were plated in T75 flasks with complete DMEM containing 10% FBS, 1X PenStrep, 2 mM L- Glutamine ON at 37 °C 5% CO2 . The media in the flask was removed and SARS-CoV-2 strain USA-WA1/2020 (BEI Resources NR-52281) diluted (final volume 2 ml) in complete DMEM containing 2% FBS and added to the flask (MOI of 0.5) and was allowed to adsorb for 60 min at 37 °C 5% CO2 . After adsorption, the inoculum was removed and 15 mL of complete DMEM containing 2% FBS was added to the flask. Infection was allowed to proceed at 37 °C 5% CO2 for 72 h. Supernatant was harvested and clarified at 1,000 xg for 5 min.
Clarified supernatant was filtered through a 0.22 μM filter and stored at −80 °C. To determine virus titers Vero E6 cells (106 cells/well, 6-well plate format, triplicates) were infected with ten-fold serial dilutions of viral samples for 1 h at 37°C. After viral adsorption, cells were overlaid with post-infection media containing 1% low melting agarose and incubated at 37°C. At 72 h pi, cells were fixed ON with 10% formaldehyde solution. The agarose plug was removed, and the fixed monolayer stained with 0.4% crystal violet in 20% methanol.
Protein immunoblotting
Mitochondria were isolated using Mitochondria Isolation kit. Nuclear and cytosolic fractions were obtained using NE-PER Nuclear and Cytoplasmic Extraction Reagents. Whole cell lysates were prepared in RIPA buffer (25 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS) containing a protease inhibitor cocktail (Roche, 11836153001) and a phosphatase inhibitor cocktail (Sigma-Aldrich, P5726). Protein concentrations were determined using BCA Protein Assay Kit (Pierce, 23225). Equal amounts of protein were separated by SDS–PAGE and transferred onto PVDF membranes, blocked in 5% BSA in 1X TBST for 1 h and incubated with indicated antibodies overnight. Secondary antibodies were added for 1 h and detection was done using Clarity Western ECL Substrate (Biorad).
ELISA (Enzyme-linked immunosorbent assay)
Paired antibodies (capture and detection) and standard recombinant mouse IL-1β (R&D Systems), and TNF and IL-6 (eBioscience) were used to determine mouse cytokine concentrations according to manufacturer’s instructions.
Human 41-plex Luminex kit (EMD Millipore) was used to determine human cytokine and chemokine release by human macrophages according to the manufacturer’s protocol. Samples were captured in the MAGPIX (Luminex XMAP Technology) using the XPONENT software. Raw data were analyzed with MasterPlex software using best fit curve method for each analyte and concentrations were calculated (pg/mL).
Caspase-1 ZYAD FLICA staining
Stimulated macrophages were rinsed with PBS, and Caspase 1 activity detected with 1:60 dilution Caspase 1 FLICA dye (660-YVAD-FMK, Immunochemistry technologies, LLC) per manufacturer’s instructions alongside GhostRED Live dead marker (Tonbo) for 30 min at 37°C. After staining, the cells were PBS rinsed, detached with Versene for 30 min on ice, resuspended in PBS and fluorescence intensity measured by flow cytometry on FACS CANTO II (BD biosciences). Data was analyzed in FloJo software after live singlet gating and Median florescent intensity (MFI) of each channel exported to excel and graphed in Prism.
RNA isolation and quantitative real-time PCR (QPCR)
RNA was extracted using AllPrep DNA/RNA Mini kit, and cDNA was synthesized using SuperScript VILO cDNA Synthesis Kit. mRNA expression was determined by QPCR in a CFX96 thermal cycler (Biorad) as described (Zhong et al., 2016). Data are presented in arbitrary units and calculated by the 2ˆ(–ΔΔCT) method. Primer sequences were obtained from PrimerBank site (https://pga.mgh.harvard.edu/primerbank/) http://mouseprimerdepot.nci.nih.govand provided by Integrated DNA technologies.
Chromatin immunoprecipitation assays
Cells were fixed using 0.75% formaldehyde for 10 min followed by 0.125 M glycine to stop the crosslinking reaction. After 5 min at room temperature, ice-cold PBS was added to the flasks and cells were scraped on ice, washed three times with PBS and then lysed for 10 min in A buffer (50 mM Tris, pH 8.0, 2 mM EDTA, 0.1% Nonidet P-40 and 10% glycerol) supplemented with containing a protease inhibitor cocktail (Roche, 11836153001) and a phosphatase inhibitor cocktail (Sigma-Aldrich, P5726). Nuclei were pelleted at 1,000 xg in a cold microfuge and resuspended in B buffer (50 mM Tris, pH 8.0, 5 mM EDTA and 1% SDS plus inhibitors). Chromatin was then sheared by sonication (six rounds of 15 s pulses each, at 50% of the maximum potency of ultrasonic homogenizer), centrifuged to pellet debris and finally 10x diluted in dilution buffer (50 mM Tris, pH 8.0, 0.5% Nonidet P-40, 0.2 M NaCl and 5 mM EDTA). Nuclear extracts were immunoprecipitated with respective antibodies. The co-immunoprecipitated material was then subjected to qPCR analysis using specific primers.
Immunofluorescence and confocal microscopy
BMDM were fixed in 4% paraformaldehyde (PFA), permeabilized in 0.02% Triton X-100, and blocked in 1x PBS supplemented with 2% BSA and 5% normal horse serum. Primary antibodies were incubated in blocking buffer at 4°C overnight. Secondary Alexa antibodies from Life Technologies or Jackson Immuno Research Laboratories were added for 1 h. Nuclei were counterstained with DAPI. Samples were imaged through a Leica SP5 confocal microscope.
To image mtDNA synthesis, BMDM were stimulated with LPS (1 μg/mL) for 4 h in the presence of 10 μM EdU. Cells were then washed twice with sterile PBS and fixed in 4% PFA for 15 min followed by permeabilization with 0.02% Triton X-100 for 10 min. EdU staining was performed using a Click-iT EdU Microplate Assay Kit (Thermo Fisher Scientific). In brief, BMDM were postfixed in EdU fixative for 5 min, and equal volumes of the EdU reaction cocktail, which was prepared immediately before use, were added to each chamber and left overnight. Primary antibody against the mitochondrial marker Atp5b was included in the EdU reaction cocktail. The next day, the BMDM were washed three times with PBS followed by incubation with secondary Alexa antibodies from Life Technologies or Jackson Immuno Research Laboratories. DAPI was used for nuclear counterstaining.
Histological evaluation and immunohistochemistry
Lungs were fixed in 10% formalin for 24 h and embedded in paraffin, sectioned and stained with hematoxylin and eosin (H&E) to evaluate gross morphology and lung damage and inflammation; and with Sirius red to determine collagen buildup. Ten to twelve H&E-stained sections from each animal were scored for the degree of lung inflammation and damage by two blinded mouse lung pathology experts as described (Jupelli et al., 2013), and the scores were averaged. Briefly, the degree of lung inflammation was assigned an arbitrary score of 0 (normal=no inflammation, no airway thickening, no edema), 1 (minimal cellular infiltration, and minimal edema), 2 (mild-moderate cellular infiltration, plus mild airway thickening and mild edema), 3 (severe cellular infiltration, plus diffuse airway thickening and severe edema) in 10-12 fields per mouse.
Lung sections were subjected to incubation with antibodies specific for F4/80 (macrophage marker), myeloperoxidase (MPO, neutrophil marker), C/EBPβ, NFATc2 and NFATc3. Stained areas were quantified using ImageJ software.
NLRP3 inflammasome activation in the lung was detected by immunofluorescence co-staining with F4/80 and ASC antibodies. Briefly, lung sections were hydrated and boiled at 96°C in citrate buffer 1x for 20 min. Sections were blocked with 2% BSA/PBS1x containing 1% normal horse serum for 1 h at room temperature. F4/80 and ASC antibodies were incubated in blocking solution overnight at 4°C. After washing, Alexa secondary antibodies were added, and the sections were incubated in 2% BSA/PBS1x for 1 h at room temperature. Sections were washed, incubated with DAPI, and mounted using FluoSave. Sections were imaged through a Leica SP5 confocal microscope. The percentage of F4/80+ cells with ASC specks were quantified in 18-26 high magnification fields from 4-5 mice per group.
LC-MS/MS for measuring metformin concentration in plasma
Metformin has high apparent volume of distribution and the protein binding is negligible. Therefore, simple protein precipitation was utilized for plasma metformin extraction. Pooled mice plasma (CD-1 (ICR) MSE00PLK2, BioIVT) was selected as blank matrix for standards and quality control samples. Sample preparation procedure was adopted from literatures with minor modifications. In brief, to 20 μL of blank pooled plasma or mouse plasma samples, 20 μL of 50% methanol-water or standard solutions were added followed by addition of 360 μL of ice-cold acetonitrile solution containing IS (111 nM metformin-D6). The samples mix were vortexed for 2 min then sonicated for 1 min and centrifuged at 6100 g for 10 min. Supernatants were transferred into a new set 96- well plate, and 10 μL were injected for LC-MS/MS analysis. A 2 μg/mL metformin stock solution was prepared in 50% methanol-water. The solution was then serially diluted with 50% methanol-water to obtain standard working solutions over a concentration range of 10–2000 ng/mL. The quality control (QC) samples were prepared in three concentrations: 10, 100, and 500 ng/mL.
LC-MS/MS system was performed using an Agilent 1290 Infinity HPLC and HiP ALS (Autosampler) coupled with AB Sciex 6500 Q-Trap mass spectrometer. All data were acquired using Analyst (AB SCIEX v1.7.1). Metformin from mice plasma samples were separated on a Waters XBridge Amide column (2.1 × 150 mm, 3.5 μm) at 40°C. The mobile phase A was 20 mM ammonium acetate buffer in water and 100% acetonitrile as mobile phase B. A total 7 min gradient run with a flow rate of 0.8 mL/min was implemented. The chromatographic runs started with 95% mobile phase B for 2 min, then gradually increased mobile phase A from 5% to 95% in 1.5 min, kept the phase A at 95% for 1 min and reduced to 5% in 1.5 min, finally equilibrate the column at 95% phase B for 2 min. The mass spectrometer was operated in a positive mode using scheduled multiple reaction morning (sMRM). The ion source parameters were as follow: curtain gas (CUR) at 37, collision gas (CAD) at high, ionspray voltage (IS) at 5 kV, temperature (TEM) at 500, ion source gas 1 (GS1) at 60, ion source gas 2 (GS2) at 50, MRM detection window at 120 s, target scan time (per sMRM expt) at 0.15 s, the retention time (RT) for both metformin and metformin-D6 were at 3.26 min, Q1 were 130.2 and 136.2 for metformin and metformin-D6 respectively, Q3 was 60.2, declustering potential (DP) at 100, collision energy (CE) at 13, entrance potential (EP) at 10, and collision cell exit potential (CXP) at 17.
Immunofluorescence and analysis of lung samples from human autopsies
Tissues were obtained from autopsy of 11 patients with PCR confirmed SARS-CoV-2 and death due to COVID-related pneumonia and diffuse alveolar damage. Lung tissue from 3 patients with death due to non-pulmonary-related causes and no evidence of pneumonia or other pulmonary histopathological abnormalities were used as control. Tissues were fixed in 10% buffered formalin and paraffin embedded (FFPE). Immunofluorescence staining was performed on the Ventana Discovery Ultra instrument (Roche). Tissue sections were deparaffinized and antigen retrieval was performed using citrate buffer (pH = 8.0) at 72 oC (CC1; Roche). Tissue sections were first probed with rabbit anti-ASC (AG-25B-0006, AdipoGen) followed by anti-Rabbit HRP (760-4311, Roche) and fluorescent visualization using tyramide signal amplification (Red610, Roche). Residual peroxidase was blocked (760-4840, Roche), followed by application of mouse anti-CD68 (KP-1, Roche), OmniMap anti-mouse HRP (Roche) and visualization using Discovery Rhod6G (Roche). Representative immunofluorescence images were acquired using a Leica DMi8 microscope and for analysis the slides were scanned using a Zeiss AxiosScan Z1. Staining and imaging were performed by the Cedars-Sinai Biobank and Translational Research Core. ASC speck quantification was performed using open source QuPath software (version 0.2.3) (Bankhead et al., 2017).
Mitochondrial function
Mitochondrial membrane potential was measured using TMRM according to manufacturer’s instructions. Briefly, BMDM were primed or not with LPS for 4 h, and further stimulated with ATP or nigericin for 30 min. Cells were incubated with 200 nM TMRM for 30 min at 37°C. After washing twice, fluorescence intensity was determined per manufacturer’s instructions using a FilterMax F5 multimode plate reader (Molecular Devices). Mitochondrial reactive oxygen species (mtROS) were measured using MitoSOX (Invitrogen) as described (Sanchez-Lopez et al., 2019). Metformin-pretreated BMDM were primed with LPS for 4 h and stimulated with NLRP3 inflammasome activators for 30 min. Cells were loaded with 4 μM MitoSOX for 20 min. After washing with PBS twice, cells were resuspended in PBS and counted. Equal numbers of cells from different treatment groups were then plated onto 96-well plates for fluorescence reading to minimize variation among treatment groups.
Fluorescence intensity was determined at 510/580 nm using a FilterMax F5 plate reader (Molecular Devices). Cellular ATP was measured after 4 h LPS stimulation using CellTiter-Glo Luminescent Assay (Promega) as described (Ip et al., 2017).
Measurement of total mtDNA
Macrophages were primed with LPS (200 ng/mL) for 4 h, with or without 0.5 mM metformin pre-treatment for overnight. Total DNA was isolated using Allprep DNA/RNA Mini Kit (catalog 80204, QIAGEN) according to manufacturer’s instructions. mtDNA was quantified by qPCR using primers specific for the mitochondrial D-loop region, cytochrome c oxidase (Cox1) or a specific region of mtDNA that is not inserted into nuclear DNA (non-NUMT) (Zhong et al., 2018). Nuclear DNA encoding Tert, 18S ribosomal RNA and B2m was used for normalization.
Cellular fractionation and measurement of cytosolic mtDNA
Metformin-treated BMDM were primed with LPS and stimulated with NLRP3 inflammasome activators. Cellular fractionation was performed using Mitochondrial Isolation kit (89874, ThermoScientific) according to manufacturer’s instructions. Cytosolic mtDNA was analyzed as described (Nakahira et al., 2011). For the measurement of Ox-mtDNA, mtDNA was first purified using Allprep DNA/RNA mini kit (QIAGEN) from the cytosolic fraction of BMDM as described above. The 8-OH-dG content of the mtDNA was then quantified using an 8-OHdG quantification kit (ab201734), per manufacturer’s instructions.
POLγ activity assay
Macrophages were pre-treated with 0.5 mM metformin overnight, followed by LPS (200 ng/mL) priming for 4 h. Whole cell lysates were prepared in RIPA lysis buffer and POLγ activity was measured using DNA Polymerase Gamma Assay Kit (ProFoldin, DPG100K) per manufacturer’s instruction.
Quantification and statistical analysis
Data are shown as average ± SD or average ± SEM, as indicated. Statistical significance was determined using two-tailed Student’s t test, and p values lower than 0.05 were considered statistically significant. For mouse data analysis, Mann-Whitney test was used when comparing two groups and ANOVA Kruskal-Wallis test when all groups were compared to each other. GraphPad Prism 8 was used for statistical analysis and graphing.
Acknowledgments
We thank eBioscience, Cell Signaling Technologies, Santa Cruz Technologies, Thermo Fisher, and Promega for gifts of reagents, and the UCSD TTSR supported by an NCI Cancer Center Support Grant (CCSG P30CA23100). We also thank T.N. Ngo for technical assistance; J.D. Rabinowitz and L. Yang for Ndsuf4 −/− mouse bones; and R. Liu-Bryan for Ampkα1 −/− mice. M.A. (R01AI072726) and W.G.T. (R01NS095894) were supported by the NIH. E.S.-L. was supported by NIAMS (K01AR077111) and Resource-based Center for the study of the joint microenvironment in rheumatology UCSD (P30AR073761). Research was supported by NIH grant awards to M.K., M.A. and W.G.T. (U54CA260591), and M.K. (R01A1043477, P42ES010337), who is an American Cancer Research Society Professor and holds the Ben and Wanda Hildyard Chair for Mitochondrial and Metabolic Diseases.
Author contributions
M.K. conceived the project with H.X. and E.S.-L., who designed and performed most of the experiments. Y.L. and A.R.-N. helped with experiments and analyses. R.G. performed IHC analysis. T.R.C. and M.A. blindly scored lung damage in LPS- and SARS-CoV-2-induced ARDS. W.G.T. and Y.Z. stained and quantified human healthy and COVID-19 lung samples. G.R.A.-M. and G.L. performed human macrophage experiments. W.C. measured metformin in mouse plasma. S.K. assisted with IHC and BMDM cultures. D.T. and M.L. performed microglia experiments. J.C.T. and J.T. performed SARS-CoV2 infection in hACE2 Tg mice. M.K., E.S.-L., M.A., P.S., S.A.L., J.C.T., and J.T. provided advice. M.K. supervised the project and wrote the manuscript with E.S.-L., H.X., and input from all co-authors.
Declaration of interests
M.K. is a founder of Elgia Pharmaceuticals and receives research support from Gossamer Bio, Jansen Pharmaceuticals and Merck. G.R.A.-M., G.L., and P.S. are employees of Jansen Pharmaceuticals. The University of California San Diego is in the process of applying for a patent covering the generation and use of novel anti-inflammatory therapy for ARDS listing H.X., E.S.-L., and M.K. as inventors. All other authors declare no competing interests.
Published: June 8, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.immuni.2021.05.004.
Supplemental information
References
- Akira S., Isshiki H., Sugita T., Tanabe O., Kinoshita S., Nishio Y., Nakajima T., Hirano T., Kishimoto T. A nuclear factor for IL-6 expression (NF-IL6) is a member of a C/EBP family. EMBO J. 1990;9:1897–1906. doi: 10.1002/j.1460-2075.1990.tb08316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankhead P., Loughrey M.B., Fernández J.A., Dombrowski Y., McArt D.G., Dunne P.D., McQuaid S., Gray R.T., Murray L.J., Coleman H.G., et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017;7:16878. doi: 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L., Deng W., Huang B., Gao H., Liu J., Ren L., Wei Q., Yu P., Xu Y., Qi F., et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583:830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- Barzilai N., Crandall J.P., Kritchevsky S.B., Espeland M.A. Metformin as a Tool to Target Aging. Cell Metab. 2016;23:1060–1065. doi: 10.1016/j.cmet.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle K.A., Van Wickle J., Hill R.B., Marchese A., Kalyanaraman B., Dwinell M.B. Mitochondria-targeted drugs stimulate mitophagy and abrogate colon cancer cell proliferation. J. Biol. Chem. 2018;293:14891–14904. doi: 10.1074/jbc.RA117.001469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramante C.T., Ingraham N.E., Murray T.A., Marmor S., Hovertsen S., Gronski J., McNeil C., Feng R., Guzman G., Abdelwahab N., et al. Metformin and risk of mortality in patients hospitalised with COVID-19: a retrospective cohort analysis. Lancet Healthy Longev. 2021;2:e34–e41. doi: 10.1016/S2666-7568(20)30033-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges H.R., Jones A.J., Pollak M.N., Hirst J. Effects of metformin and other biguanides on oxidative phosphorylation in mitochondria. Biochem. J. 2014;462:475–487. doi: 10.1042/BJ20140620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgers P.M., Kornberg A. ATP activation of DNA polymerase III holoenzyme of Escherichia coli. I. ATP-dependent formation of an initiation complex with a primed template. J. Biol. Chem. 1982;257:11468–11473. [PubMed] [Google Scholar]
- Campbell J.M., Stephenson M.D., de Courten B., Chapman I., Bellman S.M., Aromataris E. Metformin Use Associated with Reduced Risk of Dementia in Patients with Diabetes: A Systematic Review and Meta-Analysis. J. Alzheimers Dis. 2018;65:1225–1236. doi: 10.3233/JAD-180263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauchois R., Koubi M., Delarbre D., Manet C., Carvelli J., Blasco V.B., Jean R., Fouche L., Bornet C., Pauly V., et al. Early IL-1 receptor blockade in severe inflammatory respiratory failure complicating COVID-19. Proc. Natl. Acad. Sci. USA. 2020;117:18951–18953. doi: 10.1073/pnas.2009017117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I.Y., Moriyama M., Chang M.F., Ichinohe T. Severe Acute Respiratory Syndrome Coronavirus Viroporin 3a Activates the NLRP3 Inflammasome. Front. Microbiol. 2019;10:50. doi: 10.3389/fmicb.2019.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X., Liu Y.M., Li H., Zhang X., Lei F., Qin J.J., Chen Z., Deng K.Q., Lin L., Chen M.M., et al. Metformin Is Associated with Higher Incidence of Acidosis, but Not Mortality, in Individuals with COVID-19 and Pre-existing Type 2 Diabetes. Cell Metab. 2020;32:537–547. doi: 10.1016/j.cmet.2020.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P. The TLR and IL-1 signalling network at a glance. J. Cell Sci. 2014;127:2383–2390. doi: 10.1242/jcs.149831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouse A.B., Grimes T., Li P., Might M., Ovalle F., Shalev A. Metformin Use Is Associated With Reduced Mortality in a Diverse Population With COVID-19 and Diabetes. Front. Endocrinol. (Lausanne) 2021;11:600439. doi: 10.3389/fendo.2020.600439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day C.W., Baric R., Cai S.X., Frieman M., Kumaki Y., Morrey J.D., Smee D.F., Barnard D.L. A new mouse-adapted strain of SARS-CoV as a lethal model for evaluating antiviral agents in vitro and in vivo. Virology. 2009;395:210–222. doi: 10.1016/j.virol.2009.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick I.E., Joshi-Mukherjee R., Yang W., Yue D.T. Arrhythmogenesis in Timothy Syndrome is associated with defects in Ca(2+)-dependent inactivation. Nat. Commun. 2016;7:10370. doi: 10.1038/ncomms10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinarello C.A. Interleukin-1 in the pathogenesis and treatment of inflammatory diseases. Blood. 2011;117:3720–3732. doi: 10.1182/blood-2010-07-273417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinay T., Kim Y.S., Howrylak J., Hunninghake G.M., An C.H., Fredenburgh L., Massaro A.F., Rogers A., Gazourian L., Nakahira K., et al. Inflammasome-regulated cytokines are critical mediators of acute lung injury. Am. J. Respir. Crit. Care Med. 2012;185:1225–1234. doi: 10.1164/rccm.201201-0003OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosch S.F., Mahajan S.D., Collins A.R. SARS coronavirus spike protein-induced innate immune response occurs via activation of the NF-kappaB pathway in human monocyte macrophages in vitro. Virus Res. 2009;142:19–27. doi: 10.1016/j.virusres.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duca F.A., Côté C.D., Rasmussen B.A., Zadeh-Tahmasebi M., Rutter G.A., Filippi B.M., Lam T.K. Metformin activates a duodenal Ampk-dependent pathway to lower hepatic glucose production in rats. Nat. Med. 2015;21:506–511. doi: 10.1038/nm.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan E., Beitler J.R., Brochard L., Calfee C.S., Ferguson N.D., Slutsky A.S., Brodie D. COVID-19-associated acute respiratory distress syndrome: is a different approach to management warranted? Lancet Respir. Med. 2020;8:816–821. doi: 10.1016/S2213-2600(20)30304-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman T.L., Swartz T.H. Targeting the NLRP3 Inflammasome in Severe COVID-19. Front. Immunol. 2020;11:1518. doi: 10.3389/fimmu.2020.01518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganter M.T., Roux J., Miyazawa B., Howard M., Frank J.A., Su G., Sheppard D., Violette S.M., Weinreb P.H., Horan G.S., et al. Interleukin-1beta causes acute lung injury via alphavbeta5 and alphavbeta6 integrin-dependent mechanisms. Circ. Res. 2008;102:804–812. doi: 10.1161/CIRCRESAHA.107.161067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F., Zheng K.I., Wang X.B., Yan H.D., Sun Q.F., Pan K.H., Wang T.Y., Chen Y.P., George J., Zheng M.H. Metabolic associated fatty liver disease increases coronavirus disease 2019 disease severity in nondiabetic patients. J. Gastroenterol. Hepatol. 2021;36:204–207. doi: 10.1111/jgh.15112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziewicz M.A., Longley M.J., Copeland W.C. DNA polymerase gamma in mitochondrial DNA replication and repair. Chem. Rev. 2006;106:383–405. doi: 10.1021/cr040463d. [DOI] [PubMed] [Google Scholar]
- Grazioli S., Pugin J. Mitochondrial Damage-Associated Molecular Patterns: From Inflammatory Signaling to Human Diseases. Front. Immunol. 2018;9:832. doi: 10.3389/fimmu.2018.00832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greten F.R., Grivennikov S.I. Inflammation and Cancer: Triggers, Mechanisms, and Consequences. Immunity. 2019;51:27–41. doi: 10.1016/j.immuni.2019.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov S.I., Greten F.R., Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horby P., Lim W.S., Emberson J.R., Mafham M., Bell J.L., Linsell L., Staplin N., Brightling C., Ustianowski A., et al. RECOVERY Collaborative Group Dexamethasone in Hospitalized Patients with Covid-19. N Engl J Med. 2021;384:693–704. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.J., Liang W.H., Zhao Y., Liang H.R., Chen Z.S., Li Y.M., Liu X.Q., Chen R.C., Tang C.L., Wang T., et al. China Medical Treatment Expert Group for COVID-19 Comorbidity and its impact on 1590 patients with COVID-19 in China: a nationwide analysis. Eur. Respir. J. 2020;55:55. doi: 10.1183/13993003.00547-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarda G., Braun M., Staehli F., Tardivel A., Mattmann C., Förster I., Farlik M., Decker T., Du Pasquier R.A., Romero P., Tschopp J. Type I interferon inhibits interleukin-1 production and inflammasome activation. Immunity. 2011;34:213–223. doi: 10.1016/j.immuni.2011.02.006. [DOI] [PubMed] [Google Scholar]
- Gupta S., Wang W., Hayek S.S., Chan L., Mathews K.S., Melamed M.L., Brenner S.K., Leonberg-Yoo A., Schenck E.J., Radbel J., et al. STOP-COVID Investigators Association Between Early Treatment With Tocilizumab and Mortality Among Critically Ill Patients With COVID-19. JAMA Intern. Med. 2021;181:41–51. doi: 10.1001/jamainternmed.2020.6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Mallampalli R.K. The acute respiratory distress syndrome: from mechanism to translation. J. Immunol. 2015;194:855–860. doi: 10.4049/jimmunol.1402513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori K., Inoue M., Inoue T., Arai H., Tamura H.O. A novel sulfotransferase abundantly expressed in the dauer larvae of Caenorhabditis elegans. J. Biochem. 2006;139:355–362. doi: 10.1093/jb/mvj041. [DOI] [PubMed] [Google Scholar]
- Hornung V., Bauernfeind F., Halle A., Samstad E.O., Kono H., Rock K.L., Fitzgerald K.A., Latz E. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat. Immunol. 2008;9:847–856. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J.J., Liu X., Xia S., Zhang Z., Zhang Y., Zhao J., Ruan J., Luo X., Lou X., Bai Y., et al. FDA-approved disulfiram inhibits pyroptosis by blocking gasdermin D pore formation. Nat. Immunol. 2020;21:736–745. doi: 10.1038/s41590-020-0669-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet T., Beaussier H., Voisin O., Jouveshomme S., Dauriat G., Lazareth I., Sacco E., Naccache J.M., Bézie Y., Laplanche S., et al. Anakinra for severe forms of COVID-19: a cohort study. Lancet Rheumatol. 2020;2:e393–e400. doi: 10.1016/S2665-9913(20)30164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip W.K.E., Hoshi N., Shouval D.S., Snapper S., Medzhitov R. Anti-inflammatory effect of IL-10 mediated by metabolic reprogramming of macrophages. Science. 2017;356:513–519. doi: 10.1126/science.aal3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M., Shichita T., Okada M., Komine R., Noguchi Y., Yoshimura A., Morita R. Bruton’s tyrosine kinase is essential for NLRP3 inflammasome activation and contributes to ischaemic brain injury. Nat. Commun. 2015;6:7360. doi: 10.1038/ncomms8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jupelli M., Shimada K., Chiba N., Slepenkin A., Alsabeh R., Jones H.D., Peterson E., Chen S., Arditi M., Crother T.R. Chlamydia pneumoniae infection in mice induces chronic lung inflammation, iBALT formation, and fibrosis. PLoS ONE. 2013;8:e77447. doi: 10.1371/journal.pone.0077447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly B., Tannahill G.M., Murphy M.P., O’Neill L.A. Metformin Inhibits the Production of Reactive Oxygen Species from NADH:Ubiquinone Oxidoreductase to Limit Induction of Interleukin-1β (IL-1β) and Boosts Interleukin-10 (IL-10) in Lipopolysaccharide (LPS)-activated Macrophages. J. Biol. Chem. 2015;290:20348–20359. doi: 10.1074/jbc.M115.662114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb M., Margetts P.J., Anthony D.C., Pitossi F., Gauldie J. Transient expression of IL-1beta induces acute lung injury and chronic repair leading to pulmonary fibrosis. J. Clin. Invest. 2001;107:1529–1536. doi: 10.1172/JCI12568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labuzek K., Liber S., Gabryel B., Adamczyk J., Okopień B. Metformin increases phagocytosis and acidifies lysosomal/endosomal compartments in AMPK-dependent manner in rat primary microglia. Naunyn Schmiedebergs Arch. Pharmacol. 2010;381:171–186. doi: 10.1007/s00210-009-0477-x. [DOI] [PubMed] [Google Scholar]
- Lee H.M., Kim J.J., Kim H.J., Shong M., Ku B.J., Jo E.K. Upregulated NLRP3 inflammasome activation in patients with type 2 diabetes. Diabetes. 2013;62:194–204. doi: 10.2337/db12-0420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Ma X. Acute respiratory failure in COVID-19: is it “typical” ARDS? Crit. Care. 2020;24:198. doi: 10.1186/s13054-020-02911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A., Zhang S., Li J., Liu K., Huang F., Liu B. Metformin and resveratrol inhibit Drp1-mediated mitochondrial fission and prevent ER stress-associated NLRP3 inflammasome activation in the adipose tissue of diabetic mice. Mol. Cell. Endocrinol. 2016;434:36–47. doi: 10.1016/j.mce.2016.06.008. [DOI] [PubMed] [Google Scholar]
- Li J., Wei Q., Li W.X., McCowen K.C., Xiong W., Liu J., Jiang W., Marin T., Thomas R.L., He M., et al. Metformin use in diabetes prior to hospitalization: Effects on mortality in COVID-19. Endocr. Pract. 2020;26:1166–1172. doi: 10.4158/EP-2020-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z.G., Hsu H., Goeddel D.V., Karin M. Dissection of TNF receptor 1 effector functions: JNK activation is not linked to apoptosis while NF-kappaB activation prevents cell death. Cell. 1996;87:565–576. doi: 10.1016/s0092-8674(00)81375-6. [DOI] [PubMed] [Google Scholar]
- Liu X., Zhang Z., Ruan J., Pan Y., Magupalli V.G., Wu H., Lieberman J. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature. 2016;535:153–158. doi: 10.1038/nature18629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Zheng S.G. Hall of Fame among Pro-inflammatory Cytokines: Interleukin-6 Gene and Its Transcriptional Regulation Mechanisms. Front. Immunol. 2016;7:604. doi: 10.3389/fimmu.2016.00604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo P., Qiu L., Liu Y., Liu X.L., Zheng J.L., Xue H.Y., Liu W.H., Liu D., Li J. Metformin Treatment Was Associated with Decreased Mortality in COVID-19 Patients with Diabetes in a Retrospective Analysis. Am. J. Trop. Med. Hyg. 2020;103:69–72. doi: 10.4269/ajtmh.20-0375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci F., Romeo E., Caserta C.A., Rumio C., Lefoulon F. Context-Dependent Pharmacological Effects of Metformin on the Immune System. Trends Pharmacol. Sci. 2020;41:162–171. doi: 10.1016/j.tips.2020.01.003. [DOI] [PubMed] [Google Scholar]
- Markowicz-Piasecka M., Sikora J., Szydlowska A., Skupien A., Mikiciuk-Olasik E., Huttunen K.M. ). Metformin - a Future Therapy for Neurodegenerative Diseases: Theme: Drug Discovery, Development and Delivery in Alzheimer’s Disease Guest Editor: Davide Brambilla. Pharm Res. 2017;34:2614–2627. doi: 10.1007/s11095-017-2199-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthay M.A., Zemans R.L., Zimmerman G.A., Arabi Y.M., Beitler J.R., Mercat A., Herridge M., Randolph A.G., Calfee C.S. Acute respiratory distress syndrome. Nat. Rev. Dis. Primers. 2019;5:18. doi: 10.1038/s41572-019-0069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElroy G.S., Reczek C.R., Reyfman P.A., Mithal D.S., Horbinski C.M., Chandel N.S. NAD+ Regeneration Rescues Lifespan, but Not Ataxia, in a Mouse Model of Brain Mitochondrial Complex I Dysfunction. Cell Metab. 2020;32:301–308.e306. doi: 10.1016/j.cmet.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menezes S.L., Bozza P.T., Neto H.C., Laranjeira A.P., Negri E.M., Capelozzi V.L., Zin W.A., Rocco P.R. Pulmonary and extrapulmonary acute lung injury: inflammatory and ultrastructural analyses. J Appl Physiol. 2005;98:1777–1783. doi: 10.1152/japplphysiol.01182.2004. [DOI] [PubMed] [Google Scholar]
- Mills E.L., Kelly B., Logan A., Costa A.S.H., Varma M., Bryant C.E., Tourlomousis P., Dabritz J.H.M., Gottlieb E., Latorre I., et al. Succinate Dehydrogenase Supports Metabolic Repurposing of Mitochondria to Drive Inflammatory Macrophages. Cell. 2016;167:457–470.e413. doi: 10.1016/j.cell.2016.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina J.R., Sun Y., Protopopova M., Gera S., Bandi M., Bristow C., McAfoos T., Morlacchi P., Ackroyd J., Agip A.A., et al. An inhibitor of oxidative phosphorylation exploits cancer vulnerability. Nat. Med. 2018;24:1036–1046. doi: 10.1038/s41591-018-0052-4. [DOI] [PubMed] [Google Scholar]
- Nakahira K., Haspel J.A., Rathinam V.A., Lee S.J., Dolinay T., Lam H.C., Englert J.A., Rabinovitch M., Cernadas M., Kim H.P., et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat. Immunol. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nies A.T., Koepsell H., Winter S., Burk O., Klein K., Kerb R., Zanger U.M., Keppler D., Schwab M., Schaeffeler E. Expression of organic cation transporters OCT1 (SLC22A1) and OCT3 (SLC22A3) is affected by genetic factors and cholestasis in human liver. Hepatology. 2009;50:1227–1240. doi: 10.1002/hep.23103. [DOI] [PubMed] [Google Scholar]
- Park M.J., Moon S.J., Baek J.A., Lee E.J., Jung K.A., Kim E.K., Kim D.S., Lee J.H., Kwok S.K., Min J.K., et al. Metformin Augments Anti-Inflammatory and Chondroprotective Properties of Mesenchymal Stem Cells in Experimental Osteoarthritis. J. Immunol. 2019;203:127–136. doi: 10.4049/jimmunol.1800006. [DOI] [PubMed] [Google Scholar]
- Pollak M.N. Investigating metformin for cancer prevention and treatment: the end of the beginning. Cancer Discov. 2012;2:778–790. doi: 10.1158/2159-8290.CD-12-0263. [DOI] [PubMed] [Google Scholar]
- Pollak M. The effects of metformin on gut microbiota and the immune system as research frontiers. Diabetologia. 2017;60:1662–1667. doi: 10.1007/s00125-017-4352-x. [DOI] [PubMed] [Google Scholar]
- Richardson S., Hirsch J.S., Narasimhan M., Crawford J.M., McGinn T., Davidson K.W., Barnaby D.P., Becker L.B., Chelico J.D., Cohen S.L., et al. the Northwell COVID-19 Research Consortium Presenting Characteristics, Comorbidities, and Outcomes Among 5700 Patients Hospitalized With COVID-19 in the New York City Area. JAMA. 2020;323:2052–2059. doi: 10.1001/jama.2020.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridker P.M., MacFadyen J.G., Thuren T., Everett B.M., Libby P., Glynn R.J., CANTOS Trial Group Effect of interleukin-1β inhibition with canakinumab on incident lung cancer in patients with atherosclerosis: exploratory results from a randomised, double-blind, placebo-controlled trial. Lancet. 2017;390:1833–1842. doi: 10.1016/S0140-6736(17)32247-X. [DOI] [PubMed] [Google Scholar]
- Rodrigues T.S., de Sá K.S.G., Ishimoto A.Y., Becerra A., Oliveira S., Almeida L., Gonçalves A.V., Perucello D.B., Andrade W.A., Castro R., et al. Inflammasomes are activated in response to SARS-CoV-2 infection and are associated with COVID-19 severity in patients. J. Exp. Med. 2021;218:218. doi: 10.1084/jem.20201707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roschewski M., Lionakis M.S., Sharman J.P., Roswarski J., Goy A., Monticelli M.A., Roshon M., Wrzesinski S.H., Desai J.V., Zarakas M.A., et al. Inhibition of Bruton tyrosine kinase in patients with severe COVID-19. Sci. Immunol. 2020;5:e20201707. doi: 10.1126/sciimmunol.abd0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen A., Kaarniranta K. AMP-activated protein kinase (AMPK) controls the aging process via an integrated signaling network. Ageing Res. Rev. 2012;11:230–241. doi: 10.1016/j.arr.2011.12.005. [DOI] [PubMed] [Google Scholar]
- Sanchez-Lopez E., Zhong Z., Stubelius A., Sweeney S.R., Booshehri L.M., Antonucci L., Liu-Bryan R., Lodi A., Terkeltaub R., Lacal J.C., et al. Choline Uptake and Metabolism Modulate Macrophage IL-1beta and IL-18 Production. Cell Metab. 2019;29:1350–1362.e1357. doi: 10.1016/j.cmet.2019.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saura J., Tusell J.M., Serratosa J. High-yield isolation of murine microglia by mild trypsinization. Glia. 2003;44:183–189. doi: 10.1002/glia.10274. [DOI] [PubMed] [Google Scholar]
- Scozzi D., Cano M., Ma L., Zhou D., Zhu J.H., O’Halloran J.A., Goss C., Rauseo A.M., Liu Z., Peritore V., et al. Circulating Mitochondrial DNA is an Early Indicator of Severe Illness and Mortality from COVID-19. bioRxiv. 2020 doi: 10.1172/jci.insight.143299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif H., Wang L., Wang W.L., Magupalli V.G., Andreeva L., Qiao Q., Hauenstein A.V., Wu Z., Núñez G., Mao Y., Wu H. Structural mechanism for NEK7-licensed activation of NLRP3 inflammasome. Nature. 2019;570:338–343. doi: 10.1038/s41586-019-1295-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q., Liu S., Fonseca V.A., Thethi T.K., Shi L. Effect of metformin on neurodegenerative disease among elderly adult US veterans with type 2 diabetes mellitus. BMJ Open. 2019;9:e024954. doi: 10.1136/bmjopen-2018-024954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada K., Crother T.R., Karlin J., Dagvadorj J., Chiba N., Chen S., Ramanujan V.K., Wolf A.J., Vergnes L., Ojcius D.M., et al. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity. 2012;36:401–414. doi: 10.1016/j.immuni.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S., Khan A. Clinical Characteristics and Outcomes of Coronavirus Disease 2019 Among Patients With Preexisting Liver Disease in the United States: A Multicenter Research Network Study. Gastroenterology. 2020;159:768–771.e763. doi: 10.1053/j.gastro.2020.04.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soberanes S., Misharin A.V., Jairaman A., Morales-Nebreda L., McQuattie-Pimentel A.C., Cho T., Hamanaka R.B., Meliton A.Y., Reyfman P.A., Walter J.M., et al. Metformin Targets Mitochondrial Electron Transport to Reduce Air-Pollution-Induced Thrombosis. Cell Metab. 2019;29:335–347.e335. doi: 10.1016/j.cmet.2018.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay M.Z., Poh C.M., Rénia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautwein C., Caelles C., van der Geer P., Hunter T., Karin M., Chojkier M. Transactivation by NF-IL6/LAP is enhanced by phosphorylation of its activation domain. Nature. 1993;364:544–547. doi: 10.1038/364544a0. [DOI] [PubMed] [Google Scholar]
- Tschopp J., Schroder K. NLRP3 inflammasome activation: The convergence of multiple signalling pathways on ROS production? Nat. Rev. Immunol. 2010;10:210–215. doi: 10.1038/nri2725. [DOI] [PubMed] [Google Scholar]
- Vaez H., Najafi M., Rameshrad M., Toutounchi N.S., Garjani M., Barar J., Garjani A. AMPK activation by metformin inhibits local innate immune responses in the isolated rat heart by suppression of TLR 4-related pathway. Int. Immunopharmacol. 2016;40:501–507. doi: 10.1016/j.intimp.2016.10.002. [DOI] [PubMed] [Google Scholar]
- Valencia W.M., Palacio A., Tamariz L., Florez H. Metformin and ageing: improving ageing outcomes beyond glycaemic control. Diabetologia. 2017;60:1630–1638. doi: 10.1007/s00125-017-4349-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma N., Duseja A., Singh V. Impact of Pre-existing Chronic Liver Disease on the Outcome of Patients with COVID-19 Disease. Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.05.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheaton W.W., Weinberg S.E., Hamanaka R.B., Soberanes S., Sullivan L.B., Anso E., Glasauer A., Dufour E., Mutlu G.M., Budigner G.S., Chandel N.S. Metformin inhibits mitochondrial complex I of cancer cells to reduce tumorigenesis. eLife. 2014;3:e02242. doi: 10.7554/eLife.02242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcock C., Bailey C.J. Accumulation of metformin by tissues of the normal and diabetic mouse. Xenobiotica. 1994;24:49–57. doi: 10.3109/00498259409043220. [DOI] [PubMed] [Google Scholar]
- Winkler E.S., Bailey A.L., Kafai N.M., Nair S., McCune B.T., Yu J., Fox J.M., Chen R.E., Earnest J.T., Keeler S.P., et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 2020;21:1327–1335. doi: 10.1038/s41590-020-0778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Chen X., Cai Y., Xia J., Zhou X., Xu S., Huang H., Zhang L., Zhou X., Du C., et al. Risk Factors Associated With Acute Respiratory Distress Syndrome and Death in Patients With Coronavirus Disease 2019 Pneumonia in Wuhan, China. JAMA Intern Med. 2020 doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Qin Y., Wang Y., Meng S., Xian H., Che H., Lv J., Li Y., Yu Y., Bai Y., Wang L. Metformin Inhibits the NLRP3 Inflammasome via AMPK/mTOR-dependent Effects in Diabetic Cardiomyopathy. Int. J. Biol. Sci. 2019;15:1010–1019. doi: 10.7150/ijbs.29680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Zheng Y., Gou X., Pu K., Chen Z., Guo Q., Ji R., Wang H., Wang Y., Zhou Y. Prevalence of comorbidities and its effects in patients infected with SARS-CoV-2: a systematic review and meta-analysis. Int. J. Infect. Dis. 2020;94:91–95. doi: 10.1016/j.ijid.2020.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Garcia Canaveras J.C., Chen Z., Wang L., Liang L., Jang C., Mayr J.A., Zhang Z., Ghergurovich J.M., Zhan L., et al. Serine Catabolism Feeds NADH when Respiration Is Impaired. Cell Metab. 2020;31:809–821.e806. doi: 10.1016/j.cmet.2020.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L.L., Zhu M., Huang Y., Zhao Y.M., Wen J.J., Yang X.J., Wu P. Metformin relieves acute respiratory distress syndrome by reducing miR-138 expression. Eur. Rev. Med. Pharmacol. Sci. 2018;22:5355–5363. doi: 10.26355/eurrev_201808_15737. [DOI] [PubMed] [Google Scholar]
- Zhang X.J., Qin J.J., Cheng X., Shen L., Zhao Y.C., Yuan Y., Lei F., Chen M.M., Yang H., Bai L., et al. ). In-Hospital Use of Statins Is Associated with a Reduced Risk of Mortality among Individuals with COVID-19. Cell Metab. 2020;32:176–187.e174. doi: 10.1016/j.cmet.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z., Umemura A., Sanchez-Lopez E., Liang S., Shalapour S., Wong J., He F., Boassa D., Perkins G., Ali S.R., et al. NF-κB Restricts Inflammasome Activation via Elimination of Damaged Mitochondria. Cell. 2016;164:896–910. doi: 10.1016/j.cell.2015.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z., Liang S., Sanchez-Lopez E., He F., Shalapour S., Lin X.J., Wong J., Ding S., Seki E., Schnabl B., et al. New mitochondrial DNA synthesis enables NLRP3 inflammasome activation. Nature. 2018;560:198–203. doi: 10.1038/s41586-018-0372-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R., Yazdi A.S., Menu P., Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the paper and its supplementary materials. This study did not generate any unique datasets or code.