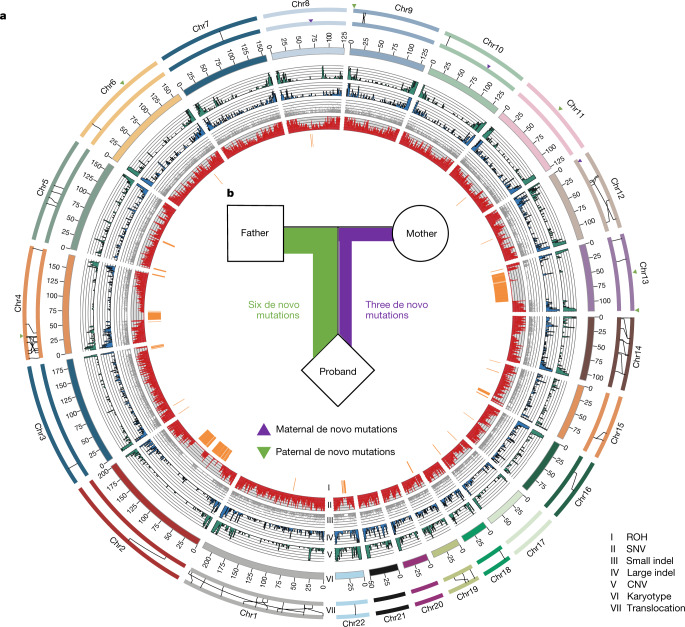
Fig. 1. Distribution of SNVs, small indels and SVs in a diploid marmoset genome.
a, Heterozygosity landscape patterns between the two haploid marmoset genomes. Tracks from inside out (I–VI): distribution of runs of homozygosity (ROH) (>1 Mb), SNV density (window size, 500 kb; range, 0–0.85%), small indel (<50 bp) distribution (y axis, indel length), large indel density (≥50 bp; window size, 1 Mb; count, 0–9), CNV density (window size, 1 Mb; count, 0–9) and karyotype. The links in the outermost circles denote differences in translocation events between maternal (inner) and paternal (outer) assemblies (VII). Triangles indicate locations of the de novo mutations in parental alleles. b, Schematic showing the proportion of parental sources of the de novo mutations.