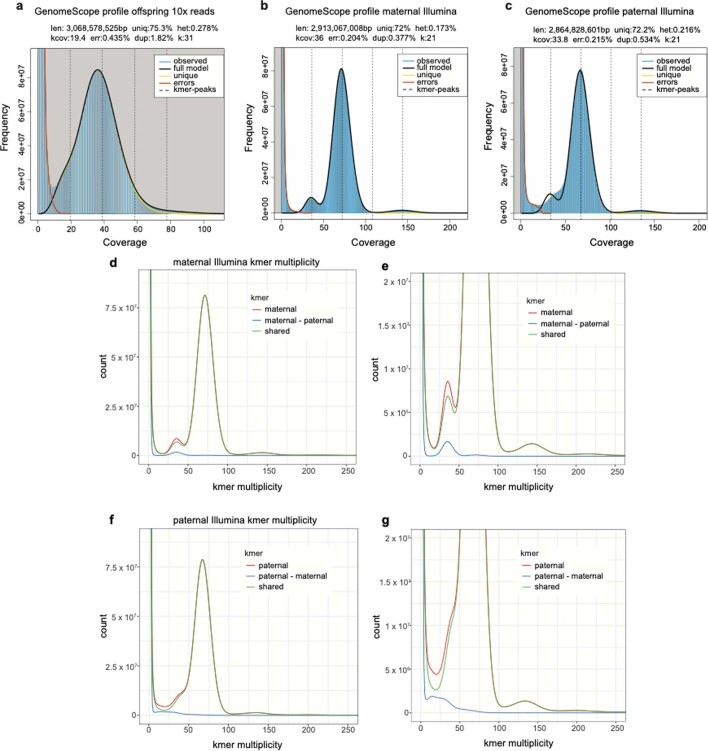
Extended Data Fig. 1. GenomeScope analyses.
a, GenomeScope (v.1.0) profile for 31-mers collected from the F1 10X linked-reads using Meryl (https://github.com/marbl/meryl) (following GEM (gel-bead in emulsion) barcode trimming). Heterozygosity estimated at a maximum of 0.287%. Read error rate estimated at a maximum of 0.435%. Genome haploid length estimated at a maximum of 3,068,578,525 bp, repeat length estimated at a maximum of 757,852,942 bp and unique length estimated at a maximum of 2,310,725,582 bp. b, c, Genomescope profiles of the maternal (b) and paternal (c) 21-mers collected from the raw Illumina data. The observed paternal data do not fit GenomeScope’s robust model (black line) for a diploid organism and exhibit higher overall heterozygosity than the maternal data (0.216% compared to 0.173%). This supports a premise that the father’s sequencing reads contain a level of chimerism, whereas the mother’s reads contain negligible representation of alternative alleles, at most. Further analysis of the parental Illumina data shows that the k-mer multiplicity distribution varies greatly between the maternal and paternal sets. d–g, The maternal k-mers (d, e (e shows a magnified version of d)) show clear distributions with a distinct haploid peak at half coverage (around 35×), whereas the paternal k-mers (f, g (g shows a magnified version of f)) show an irregular distribution with no clearly defined haploid peak. This provides further evidence that the paternal data exhibit a level of chimerism.