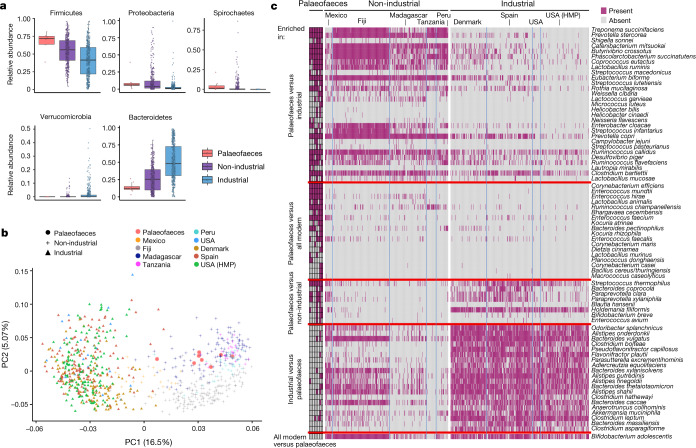
Fig. 1. Phylum, family and species compositions of the palaeofaeces samples are similar to the gut microbiomes of present-day non-industrial individuals.
a, Differentially abundant phyla (one-tailed Wilcoxon rank-sum test with FDR correction) as identified by MetaPhlAn220 (palaeofaeces, n = 8; non-industrial, n = 370; industrial, n = 418). Data are presented as box plots (middle line, median; lower hinge, first quartile; upper hinge, third quartile; lower whisker, the smallest value at most 1.5× the interquartile range from the hinge; upper whisker, the largest value no further than 1.5× the interquartile range from the hinge; data beyond the whiskers are outlying points). b, Principal component analysis of the species composition as identified by MetaPhlAn220. HMP, Human Microbiome Project. c, Presence–absence heat map (fuchsia, present; grey, absent) for differentially enriched species (two-tailed Fisher’s test, FDR correction). Species without fully specified species names are not shown (a complete list is included in Supplementary Table 3).