Abstract
Substantial changes in the composition of the vaginal microbiome occur following the end of pregnancy. To identify potential drivers of microbiome changes in individual women during the pregnancy to postpartum transition, we evaluated vaginal samples from 48 pregnant women during their first and third trimesters and postpartum. We determined the species composition of vaginal communities and the vaginal fluid levels of compounds involved in mediating changes in host physiology and the immune system at each time point. We used linear mixed-effects models to characterize associations. Consistent with previous reports, but with a larger sample size, a US population, and variations in the dominant bacteria, the vaginal microbiome was found to be more diverse during the postpartum period. There was a lower abundance of Lactobacillus and significantly higher proportions of Streptococcus anginosus and Prevotella bivia. Moreover, we uniquely demonstrated that postpartum vaginal secretions were also altered postpartum. There were elevated levels of hyaluronan and Hsp70 and decreased levels of the D- and L-lactic acid isomers. We posit that these variations are consequences of alterations in the vagina after delivery that profoundly alter the host environment and, thus, lead to changes in the capability of different bacterial species to survive and proliferate.
Supplementary Information
The online version contains supplementary material available at 10.1007/s43032-020-00438-6.
Keywords: Pregnancy, Postpartum, Vagina, Vaginal microbiome
Introduction
During a healthy pregnancy, the vaginal microbiome is characterized by low bacterial species diversity [1, 2] and is typically dominated by one of several different species of Lactobacillus [3]. A few cross-sectional [4] and longitudinal studies [5–7] have compared the vaginal microbiome during pregnancy and in the postpartum period. These studies confirmed that considerable changes in vaginal community composition occur immediately following pregnancy. During this transition, there is an overall decrease in Lactobacillus [4, 5, 7] with a concomitant increase in diverse taxa including but not limited to Clostridia (Peptoniphilus and Anaerococcus), Bacteroidia, Prevotella, Veillonella, Porphyromonas, and Megasphaera [4–7]. As expected, the proliferation of diverse taxa during the postpartum period is paralleled by increases in ⍺-diversity [4–7]. Doyle et al. obtained vaginal samples from a cohort of Malawi women anywhere from 5 to 583 days post-delivery and demonstrated that Lactobacillus spp. were present in less than a third of the women [8]. Furthermore, postpartum vaginal communities resembled those of community state types (CST) III and IV, as defined by Ravel et al. [8, 9]. CST-III consists of communities in which Lactobacillus iners is dominant. In contrast, CST-IV vaginal communities are heterogeneous in composition with low proportions of lactobacilli and elevated levels of an assortment of anaerobic bacteria. They also noted that the abundances of L. iners increased with time throughout the postpartum stage [8]. In comparison to other Lactobacillus-dominant communities, those dominated by L. iners tend to be more diverse and less stable [10–12] as they often transition to communities resembling CST-IV [13]. The observed shift in bacterial community composition during the postpartum period can occur as early as the onset of labor [7, 14] and may persist for up to 1 year [5].
Alterations in vaginal secretions would also be anticipated to accompany changes in the vaginal microbiome in the postpartum period. Specifically, a decrease in the levels of compounds produced chiefly by lactobacilli and increases in those associated with an elevated level of physiological stress or degradation of the extracellular matrix would be expected. Currently, only limited information exists on host-associated changes in the vaginal microbiome in individual women during pregnancy and postpartum. In the present study, we determined the species composition of vaginal communities and the levels of vaginal fluid compounds involved in host physiology and immune system modifications in pregnant women within the first and third trimesters and approximately 1 month postpartum.
Materials and Methods
Study Participants and Sample Collection
Our prospective study included pregnant women seen for outpatient obstetrics services at Weill Cornell Medicine in New York City. Exclusion criteria included inability to give informed consent, signs or symptoms of a gynecological disorder or infection at the time of examination, multifetal gestation, presence of an autoimmune or endocrine problem, antibiotic use in the previous 4 weeks, or vaginal bleeding. In total, 48 women were enrolled and provided vaginal samples during their first (≤ 12 weeks) and third (28–38 weeks) trimesters, and 28–45 days postpartum. All of the women in this study had vaginal deliveries. Of those 48 women, 32 provided samples for each time point. Samples from the posterior vagina were obtained with a cotton swab, vigorously shaken into a sterile tube containing 1 mL of sterile phosphate-buffered saline (PBS) and centrifuged, and aliquots of the supernatant were stored at ‑ 80 °C until analyzed. The study was approved by the Institutional Review Board at Weill Cornell Medicine. All subjects gave informed, written consent.
Compound Measurements
Vaginal levels of the D- and L-lactic acid isomers were quantitated by colorimetric assays using the EnzyChrom D-lactic acid and L-lactic acid kits (BioAssay Systems, Haywood, CA). The levels of matrix metalloproteinase-8 (MMP-8) (R&D Systems, Minneapolis, MN), extracellular matrix metalloproteinase inducer (EMMPRIN) (R&D Systems), neutrophil gelatinase-associated lipocalin (NGAL) (R&D Systems), hyaluronan (R&D Systems), α-amylase (human pancreatic α-amylase, Abcam, Cambridge, UK), the stress-inducible 70-kDa heat shock protein (HSPA1A) commonly known as Hsp70 (R&D Systems), and p62 (Enzo Life Sciences, Farmingdale, NY) were determined using commercially available ELISA kits. Hsp70 and p62 were measured from the supernatant of lysed epithelial cells, while all of the other compounds were measured directly from vaginal secretions. To obtain the epithelial cell lysate, vaginal secretions were centrifuged, and the epithelial cell pellet was immediately lysed in a detergent containing a protease inhibitor cocktail (Sigma, St. Louis, MO). The supernatant from the lysate was stored in aliquots at ‑ 80 °C. The total protein levels in samples were measured using a colorimetric assay (Thermo-Fisher Scientific, Waltham, MA). Concentrations were determined from a standard curve that was generated in parallel to test samples and converted to μmoles per mL, units per mL, or pg per mL. When applicable, measurements were normalized by the total amount of protein in the sample and reported in pg, ng, or units per μg of total protein. All assays were performed by staff blinded to all clinical information. These compounds were chosen for analysis since each had previously been shown to be associated with alterations in the composition of the vaginal microbiota or with immunological properties of vaginal epithelial cells.
Determining Vaginal Community Composition
The species composition of vaginal microbial communities was determined by classifying partial 16S rRNA gene sequences as previously described [15]. In brief, total genomic DNA was extracted from 250 μL of vaginal swabs stored in 1X PBS using chemical and mechanical lysis and purified using QIAamp DNA mini kits (Qiagen). Genomic DNA concentrations were determined using the Quant-iT™ PicoGreen™ dsDNA assay kit (Invitrogen). For amplicon sequencing, the V1-V3 16S rRNA gene regions were amplified using a two-step PCR protocol, first amplifying the gene region using universal primers 27F and 534R, and then adding sample barcodes and sequence adapters. Amplicons were sequenced using the Illumina MiSeq at the University of Idaho. Forward and reverse reads were paired using FLASH [16], processed through DADA2 v 1.12.1 [17] to identify distinct sequences, and the sequences were classified into genus and species level using SPINGO [18].
A total of 358 taxa were identified, and these data were used to analyze the rank abundance of taxa and measure ⍺-diversity. For subsequent analyses, including visualizing community composition, we created a smaller dataset of taxon abundance for 57 taxa. Fifty-six of the 358 taxa were present at a minimum of 1% in at least two individuals. The remaining taxa (302) were categorized as other. The corresponding relative abundance data are reported in Table S1. Samples that did not have at least 3000 sequence reads were discarded from further analysis. This resulted in 47, 45, and 34 samples from the first trimester, third trimester, and postpartum stages, respectively. Thirty-two women had all three time points, 14 women had two time points, and two women had only one time point.
Bioinformatics and Statistical Analyses
All analyses were performed using R v 3.6.0 [19]. In addition to the base packages, the following packages were used for organizing data, performing analyses, and to produce figures: cluster [20], e1071 [21], FSA [22], ggplot2 [23], ggpubr [24], lme4 [25], lmerTest [26], multcomp [27], plyr [28], tidyr [29], and vegan [30]. To determine what kinds of communities were present in this cohort, we performed complete-linkage hierarchical clustering on alt-Gower distances computed from taxon relative abundance data. We then used silhouette information to identify ten clusters, numbered 1 through 10. Alpha diversity was measured using the Shannon and Simpson diversity indices.
Linear mixed-effects models were used to model the means of the response variables between pregnancy stages to evaluate significant differences. From this point forward, stage refers to the first and third trimesters and the postpartum period. In these models, subject was included as a random variable to account for any within-subject variation across stages. The various response variables included bacterial species abundances, measures of ⍺-diversity, and compounds measured in vaginal fluids. Statistical comparisons were performed by testing general linear hypotheses and multiple comparisons of the means using Tukey’s test. If the linear mixed-effects model resulted in a singular fit, we used a Kruskal-Wallis rank sum test to evaluate significant differences in the means of the response variables between stages. A significant Kruskal-Wallis test was followed with post hoc analysis of multiple comparisons using Dunn’s test with Bonferroni adjustment. To test associations with changes in Shannon and Simpson diversity, we created a base mixed-effects model. We included stage and vaginal compounds that differed significantly by stage as constant variables (fixed effects). To account for differences in the subject across stages, we treated the subject as a random variable. Using these base models, we performed stepwise linear regression to determine which variables to include in the final models. A summary of each final model was generated to determine associations. The significance threshold was set to p < 0.05.
Results
The characteristics of the 48 women enrolled in the study are shown in Table 1. Their mean age was 33.4 years, and the mean body mass index was 22.3 kg/m2. All delivered healthy infants at term at a mean gestational age of 39.5 weeks. This was the first pregnancy for 15 (31.3%) of the women, while 33 (68.8%) experienced a prior delivery. The majority of subjects were white. We found that all subjects had a vaginal pH < 4.5 (data not shown).
Table 1.
Characteristics of study participants
| Characteristic | No. of women or % | Mean | SD |
|---|---|---|---|
| Age at delivery (year) | 48 | 33.4 | 3.6 |
| Body mass index (kg/m2) | 44 | 22.3 | 2.6 |
| Weeks of gestation | 48 | 39.5 | 1.0 |
| Race | 48 | ||
| White | 56.3% | ||
| Black | 2.1% | ||
| Asian | 12.5% | ||
| Hispanic | 4.2% | ||
| Other or mixed | 25.0% | ||
| Gravida | 48 | 2.2 | 1.1 |
| Primiparous | 15 | ||
| Prior term births | 23 | ||
| Prior preterm births | 2 | ||
| Prior spontaneous miscarriages | 11 |
SD, standard deviation
Comparison of the Vaginal Microbiome from Pregnancy to Postpartum
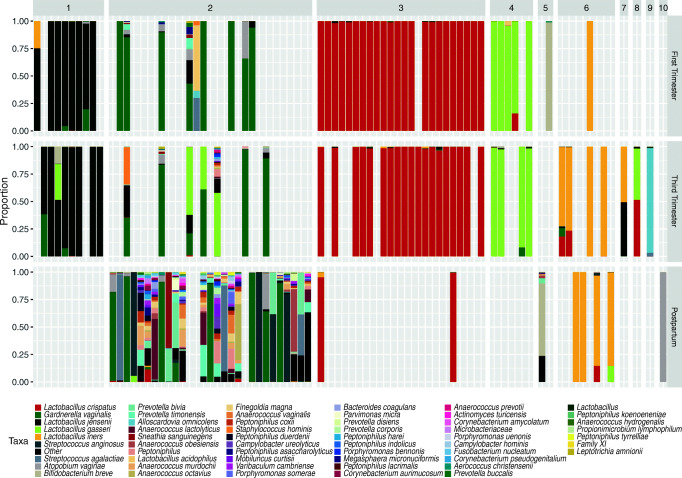
The relative proportions of 57 taxa identified in each stage of pregnancy are shown in Fig. S1. Vaginal communities during the first and third trimesters were mostly dominated by Lactobacillus species, whereas postpartum communities had low proportions of lactobacilli and were more diverse. To get a better sense of the kinds of communities that were present at each stage, we grouped relative abundance data based on similarities and differences using complete-linkage hierarchical clustering. In total, we identified ten clusters of community types. Figure 1 shows a stacked bar chart of samples separated by cluster and pregnancy stage. Vaginal communities in the first and third trimesters tended to be very similar. Roughly, 79% of the first trimester samples and 78% of the third trimester samples had vaginal communities in which Lactobacillus spp. were dominant. In contrast, postpartum vaginal communities were much more heterogeneous in composition. Approximately 77% of the postpartum vaginal communities had low proportions of Lactobacillus spp. and a diverse array of bacteria, including G. vaginalis, Prevotella spp., Streptococcus spp., and numerous others. At each stage, there appeared to be no difference in the composition of vaginal communities between primiparous and multiparous women (data not shown).
Fig. 1.
Relative proportions of bacteria in the vaginal communities of 48 pregnant women separated by stage and cluster. The stacked bars represent the proportions of bacterial taxa within one sample. Bars are separated by the pregnancy stage in which the sample was collected (right heading), and the cluster the sample was grouped into (top heading). Taxa colors are indicated in the legend below the figure. “Other” represents the sum of all bacterial taxa that were not present at 1% or more in at least two women
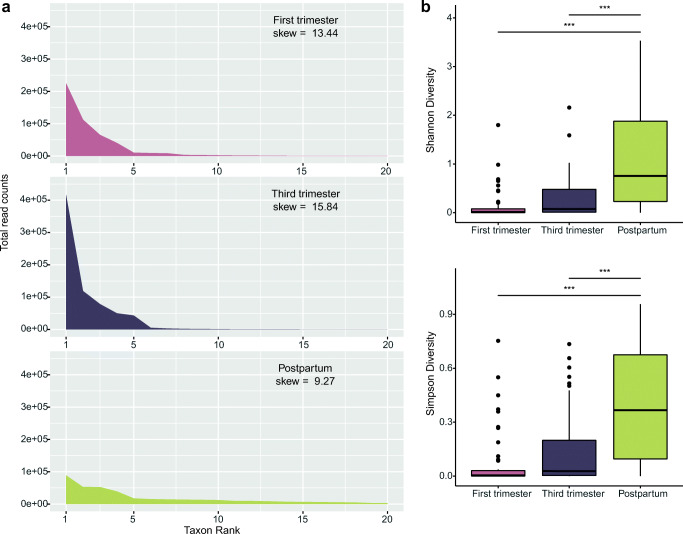
To visualize the distribution of taxa and compare diversity between pregnancy stages, we prepared rank abundance profiles for the top 20 taxa in each stage and measured the skewness in these data (Fig. 2a). Skewness measures the degree to which a sample distribution varies from normal. Compositional microbiome data typically have a long-tailed distribution and are positively skewed. This results from communities having few species that are highly abundant and many that are present at low abundances. Third trimester samples had the highest skew (15.84) followed by first trimester (13.44) and postpartum samples (9.27). This suggests postpartum samples had a distribution of taxa that were more even than the first and third trimester samples.
Fig. 2.
Rank abundance and ⍺-diversity profiles of samples collected during the first trimester, third trimester, and postpartum. Panel a shows the counts of the top-ranked taxa among first trimester, third trimester, and postpartum samples. Panel b shows the distribution of Shannon and Simpson diversity measures for each stage. The thick band within the box represents the median for the data, and the lower and upper boxes represent the 25th and 75th quartiles, respectively. The upper and lower whiskers represent the 95% confidence interval for the median. Individual dots above represent outliers that were not removed. Statistical significance is indicated above the lines (“***” = p < 0.001)
Next, we measured ⍺-diversity using the Shannon and Simpson diversity indices and used linear mixed-effects models to determine whether ⍺-diversity differed between stages (Fig. 2b). The Shannon diversity increased with each stage. Likewise, there were 44 species found in first trimester samples, 112 in third trimester samples, and 225 in postpartum samples. Measures of Simpson diversity followed a similar trend. The increase in ⍺-diversity from early to late pregnancy was not significant. However, ⍺-diversity was significantly higher in postpartum vaginal communities when compared to those of the first (p < 0.001; Shannon and Simpson diversity) and third trimesters (p < 0.001; Shannon and Simpson diversity).
Changes in Numerically Abundant Taxa from Pregnancy to Postpartum
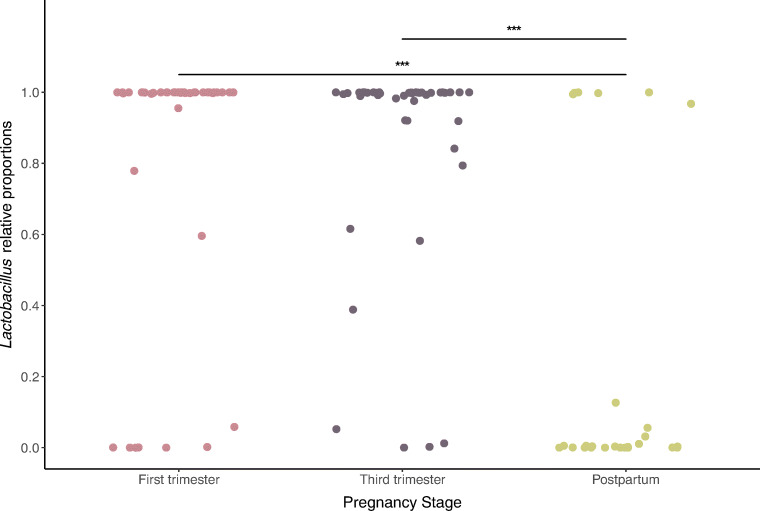
To quantify the change in Lactobacillus during pregnancy and afterward, we used linear mixed-effects models to model the means of Lactobacillus relative proportions between stages and controlled for within-subject variation. The distribution of the relative abundances of Lactobacillus by stage is shown in Fig. 3. There was no change in the mean relative abundance of Lactobacillus over gestation. However, we observed a sharp decrease in the abundances of lactobacilli among vaginal communities sampled during the postpartum stage in comparison to those sampled during the first and third trimesters (p < 0.01). Next, we evaluated whether there were significant shifts in individual lactobacilli and other top-ranked taxa in the transition from pregnancy to the postpartum period (Table 2, Table S2). We found postpartum vaginal communities to have significantly lower abundances of L. crispatus, L. jensenii, and L gasseri, but not L. iners. Instead, vaginal communities sampled during the postpartum period had markedly higher proportions of Streptococcus anginosus and Prevotella bivia. Streptococcus agalactiae was present at higher abundances in a few postpartum samples, but overall, these differences were not significant. Likewise, there was relatively no change in G. vaginalis, Atopobium vaginae, or Bifidobacterium breve across stages.
Fig. 3.
Differences in the relative abundances of Lactobacillus in the first and third trimesters, and postpartum. Each dot is one sample from a woman. Statistical significance is indicated above the lines (“***” = p < 0.001)
Table 2.
Mean relative proportions for the most abundant taxa in each stage of pregnancy. The following ten taxa below were identified by taking the top ten ranked taxa for each stage, collapsing them together, and keeping only those greater than 1% in all samples
| Mean relative proportions | p values for comparison of the meansb | |||||
|---|---|---|---|---|---|---|
| Taxa | First Trimester (Na = 47) |
Third Trimester (N = 45) |
Postpartum (N = 34) |
p valuec | First - Postpartumd | Third - Postpartumd |
| Lactobacillus crispatus | 49.1 | 44.0 | 6.7 | 0.00 | *** | *** |
| Gardnerella vaginalis | 15.0 | 10.0 | 18.6 | 0.29 | ||
| Lactobacillus jensenii | 16.2 | 17.4 | 0.0 | 0.00 | ** | *** |
| Lactobacillus gasseri | 10.2 | 13.9 | 0.6 | 0.01 | · | ** |
| Lactobacillus iners | 2.7 | 8.9 | 11.0 | 0.21 | ||
| Streptococcus anginosus | 0.0 | 0.0 | 10.1 | 0.00 | *** | *** |
| Streptococcus agalactiae | 0.6 | 0.1 | 6.2 | 0.11 | ||
| Atopobium vaginae | 1.2 | 0.3 | 4.6 | 0.35 | ||
| Bifidobacterium breve | 2.1 | 0.4 | 2.0 | 0.69 | ||
| Prevotella bivia | 0.0 | 0.0 | 4.6 | 0.00 | *** | *** |
aN = the number of samples/observations in each category
bThe asterisks correspond to the following levels of significance for the p values: ‘.’, p < 0.1; *, p < 0.05; **, p < 0.01, ***, p < 0.001
cThe p values in this column result from running linear mixed-effects models or Kruskal-Wallis tests to test for significant differences between the means of each stage; p values in italics are significant
dThe p values in this column result from evaluating multiple comparisons between group means using Tukey’s test for linear models or the Dunn’s test with a Bonferroni adjustment
Next, we compared the number of women that transitioned to and from a community that was dominated by Lactobacillus spp. Therefore, we identified the dominant bacterial species in the vaginal communities at each stage sampled (Table S3). Here, dominance was defined as having a relative proportion higher than 50%. A community was labeled as “mixed” if it contained a more even representation of taxa and was not dominated by any one species. Among the three stages, Lactobacillus was dominant in 35, 38, and six communities of the first trimester, third trimester, and postpartum samples, respectively. To assess transitions to and from a Lactobacillus-dominant community, we only considered the 32 women for which samples were available for each stage. Transitions from the third trimester to postpartum occurred more frequently than from first to third trimester (Table 3). Roughly, a third (N = 10) of the women transitioned in community composition between the first and third trimesters, whereas 88% (N = 28) transitioned between the third trimester and postpartum. From early to late pregnancy, transitions to or between communities dominated by Lactobacillus were more common. Despite the high prevalence of Lactobacillus during gestation, most of the transitions that occurred between the third trimester and postpartum were from a community that was dominated by Lactobacillus to one that was not. Notably, there were no transitions in the opposite direction.
Table 3.
Transitions between dominant bacteria among 32 women with samples from all three time points
| Transitiona | First ➝ Third | Third ➝ Postpartum |
|---|---|---|
| LB ➝ LB | 4 | 4 |
| LB ➝ non LB | 1 | 22 |
| non LB ➝ LB | 4 | 0 |
| non LB ➝ non LB | 1 | 2 |
| Total # of transitions | 10 | 28 |
| % transitionsb | 31 | 88 |
aTransitions are defined as LB for communities dominated by species of Lactobacillus or non LB for communities not dominated by species of Lactobacillus
b% transitions = (total number of transitions/number of women) × 100
Analysis of Vaginal Compounds
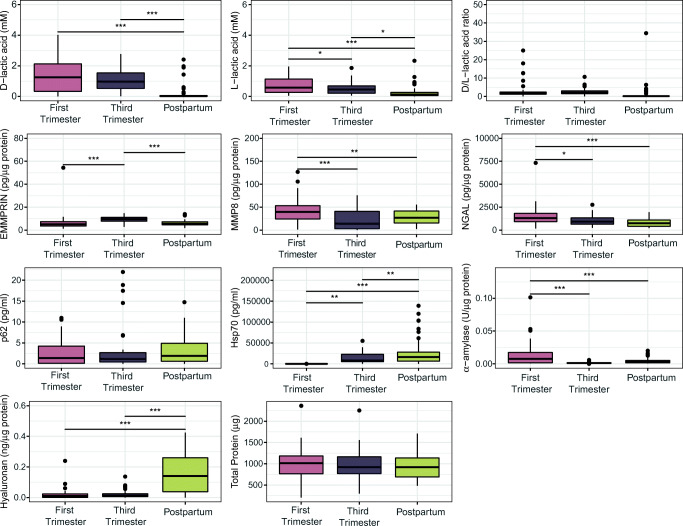
We sought to determine if vaginal biomarkers involved in host physiology and immunity were associated with the changes observed during the postpartum period. To do this, we assessed differences in compounds that were measured in vaginal fluids and epithelial cells using linear mixed-effects models (Fig. 4). In comparison to both early and late pregnancies, postpartum samples were characterized by elevated levels of hyaluronan (p < 0.001) and Hsp70 (p < 0.01) and lower levels of D (p < 0.001) and L-lactic acid (p < 0.001, p < 0.05, respectively). Next, we regressed ⍺-diversity measures against these four compounds with subject as a random effect. Using stepwise linear regression, we selected only those that were significantly associated with an increase in ⍺-diversity observed in the transition from pregnancy to the postpartum stage (Table S4). We found that an increase in ⍺-diversity was negatively associated with levels of L-lactic acid (p < 0.05, Shannon diversity; p < 0.001, Simpson diversity), but positively associated with hyaluronan (p < 0.001, for both), and the postpartum stage (p < 0.01, Shannon diversity).
Fig. 4.
Boxplots showing the concentrations of vaginal biomarkers grouped by stage in pregnancy. The thick band within the box represents the median for the data, and the lower and upper boxes represent the 25th and 75th quartiles, respectively. The upper and lower whiskers represent the 95% confidence interval for the median. Individual dots above represent outliers that were not removed. Statistical significance is indicated above the lines as follows: “*”, p < 0.05; “**”, p < 0.01; “***” = p < 0.001
Discussion
We demonstrated that in the transition from pregnancy to the postpartum period, there was a sharp decline in the abundances of three Lactobacillus spp., namely L. crispatus, L. gasseri, and L. jensenii. This coincided with an increase in the relative abundances of other diverse bacterial taxa, while the levels of L. iners remained largely unchanged. These findings are consistent with previous studies that found the composition of vaginal communities to change considerably from pregnancy to the postpartum stage [5–7]. The majority of the women in our study that had communities dominated by vaginal lactobacilli during late pregnancy transitioned to communities with low proportions of lactobacilli during the postpartum stage. Similar results were reported in a study that surveyed European women during their gestation to 6 weeks after birth. MacIntyre et al. reported that 60% of the postpartum communities shifted to CST-IV from a community previously dominated by Lactobacillus. In that study, the transition in community composition following pregnancy was shown in a small number of samples (N = 15). To our knowledge, the present study is the first to be performed on pregnant women in the USA, contained a larger cohort (N = 32), and differed in the dominant bacterial species observed. In addition to the bacterial changes, we were able to uniquely identify an increase in the levels of hyaluronan and Hsp70 and a decrease in the levels of D- and L-lactic acid in the pregnancy to postpartum transition. These observations strongly reinforce that Lactobacillus-dominant communities, while relatively stable during pregnancy [31], and the levels of compounds in vaginal secretions, are rapidly altered following delivery.
The abundance of Lactobacillus in the vagina has been positively associated with the level of vaginal estrogen (reviewed in [32, 33]). Thus, the decrease in the abundance of Lactobacillus may well be the result of reduced levels of estrogen post-delivery (reviewed in [34]). During pregnancy, the placenta produces two forms of estrogen, namely estradiol and estriol, and this causes a sharp increase in estrogen levels with estradiol and estriol increasing two to three orders of magnitude, respectively (reviewed in [34]). This is paralleled by higher abundances of Lactobacillus during pregnancy (reviewed in [35]). Once the placenta is removed after birth, estrogen levels fall precipitously (reviewed in [34]). However, it is unclear exactly how much the overall levels of estrogen change. One study reported serum estradiol levels were roughly 7 ng/mL at 36 weeks’ gestation [36] while another study determined this level to be 20 ng/mL during the third trimester [37]. During the postpartum period, serum estradiol levels were reported as 20 pg/mL at week 1 and increased slightly to above 40 pg/mL after 6 months [38]. If the decline in estrogen levels is as severe as indicated and if there is a lag time for estrogen levels to rebound after birth, this could lead to lower abundances of lactobacilli in vaginal communities for quite some time post-delivery [5, 6]. A limitation of our study is that we did not directly measure estrogen levels to evaluate how this might contribute to changes in vaginal community composition observed in our cohort.
In addition to the decline in estrogen levels, normal physiological changes that occur during labor and delivery could influence the dramatic shift in vaginal community composition. Immediately before and during parturition, innate immune cells are activated to initiate an inflammatory process that promotes uterine contractions, dilation of the cervix, and rupturing of fetal membranes [39]. In our study, vaginal levels of hyaluronan and Hsp70 were significantly elevated in postpartum samples, while both D- and L-lactic acid were substantially lower. Hyaluronan is a high molecular weight glycosaminoglycan that is present in the extracellular matrix [40]. Prior to and during labor, hyaluronan levels increase corresponding to a softening and dilation of the cervix in preparation for delivery [41]. The increased hyaluronan level observed in postpartum samples most likely reflects the consequences of ongoing cervical remodeling after delivery. Although not measured in the current study, we expect the detected increase was in the level of low molecular weight hyaluronan fragments [42]. Elevations in Hsp70 could indicate the occurrence of functional changes in the genital tract as a result of cervical remodeling. Given that both Hsp70 and hyaluronan fragments activate pro-inflammatory immune responses [43–45], the altered environmental conditions would be expected to induce a stress response in the affected cells. This will modify the properties of vaginal epithelial and immune cells leading to changes in the composition of vaginal fluid. Moreover, a decrease in the levels of D- and L-lactic acid reduce their ability to provide multiple protective mechanisms in the vagina including the suppression of pathogens [46–48], promotion of DNA repair in vaginal epithelial cells [49], and enhancement of innate and adaptive immunity [50]. Although we did not measure changes in vaginal pH, it is probable that the lower levels of lactic acid postpartum reduced vaginal acidification leading to a decrease in inhibition of non-lactobacilli proliferation. In our study, the concentrations of L-lactic acid and hyaluronan also correlated with a decrease or increase in ⍺-diversity, respectively. We anticipate this association is simply the byproduct of changes in host physiology during and after parturition. Nonetheless, the combination of higher levels of Hsp70 and hyaluronan and lower levels of lactic acid creates an environment that contributes to the persistence of diverse bacterial communities in the vagina post-delivery.
Avershina et al. determined that changes in vaginal community composition can occur as soon as labor begins [14]. Due to all postpartum samples being obtained approximately 1 month after delivery, we are unable to comment on possible changes that may have occurred immediately after birth or at later periods. However, we have confirmed a marked shift from a Lactobacillus-dominated vaginal microbiota to one in which lactobacilli are depleted and replaced by a diverse number of other bacteria that occur during the pregnancy to postpartum transition. Furthermore, these variations in the vaginal microbiota are consequences of alterations in nutrient availability, vaginal pH, and local immune parameters that change the ability of different bacterial species to survive and proliferate.
Supplementary Information
Relative proportions of bacteria in the vaginal communities of 48 pregnant women separated by stage. The stacked bars represent the proportions of bacterial taxa within one sample. Bars are separated by the pregnancy stage in which the sample was collected (top headings). Taxa colors are indicated in the legend below the figure. “Other” represents the sum of all bacterial taxa that were not present at 1% in at least two women. (PDF 40 kb)
(XLSX 153 kb)
(DOCX 39 kb)
Acknowledgments
We would like to thank Ann Marie Bongiovanni for maintaining recording and managing all clinical and laboratory data associated with this project and the staff of the Genomics Resources and the Computational Resources Cores of the Institute for Bioinformatics and Evolutionary Studies (IBEST) for technical support.
Code Availability
Sample data in addition to custom R scripts used for the analyses in this manuscript are available on GitHub at https://github.com/kenettanunn/Pregnant_Women_Study.
Authors’ Contributions
SSW and LJF conceptualized and designed the study. AB collected all patient samples and provided and interpreted all clinical data. DN and EM performed the initial sample preparations and all the vaginal fluid analyses. GMS and KG participated in experimental protocol design and data collection. KLN performed data collection, data analysis, and interpretation and wrote the original version of the manuscript with feedback from LJF. All authors contributed to the final version of the manuscript.
Funding
This study was supported by funds provided to SSW by the Department of Obstetrics and Gynecology at Weill Cornell Medicine and by a grant (P20 RR016448) to LJF from the National Institutes of Health. KLN was supported by a fellowship from the Bioinformatics and Computational Biology Graduate Program from the University of Idaho.
Data Availability
The sequence data analyzed in this study along with the corresponding metadata are available in the NCBI Sequence Read Archive under BioProject PRJNA614593 (“Study of the pregnant and postpartum period vaginal microbiome;” http://www.ncbi.nlm.nih.gov/bioproject/614593).
Compliance with Ethical Standards
Conflict of Interest
The authors declare that they have no conflicts of interest.
Ethics Approval
The study was approved by the Institutional Review Board at Weill Cornell Medicine.
Consent to Participate
All subjects provided informed written consent.
Consent for Publication
All authors agreed with the final version of the manuscript and the order of authors.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Aagaard K, Riehle K, Ma J, Segata N, Mistretta TA, Coarfa C, Raza S, Rosenbaum S, van den Veyver I, Milosavljevic A, Gevers D, Huttenhower C, Petrosino J, Versalovic J. A metagenomic approach to characterization of the vaginal microbiome signature in pregnancy. PLoS One. 2012;7(6):e36466. doi: 10.1371/journal.pone.0036466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Freitas AC, Chaban B, Bocking A, et al. The vaginal microbiome of pregnant women is less rich and diverse, with lower prevalence of Mollicutes, compared to non-pregnant women. Sci Rep. 2017;7(1):9212. doi: 10.1038/s41598-017-07790-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hernández-Rodríguez C, Romero-González R, Albani-Campanario M, et al. Vaginal microbiota of healthy pregnant Mexican women is constituted by four Lactobacillus species and several vaginosis-associated bacteria. Infect Dis Obstet Gynecol. 2011;851485. [DOI] [PMC free article] [PubMed]
- 4.Huang Y-E, Wang Y, He Y, Ji Y, Wang LP, Sheng HF, Zhang M, Huang QT, Zhang DJ, Wu JJ, Zhong M, Zhou HW. Homogeneity of the vaginal microbiome at the cervix, posterior fornix, and Vaginal Canal in pregnant Chinese women. Microb Ecol. 2015;69(2):407–414. doi: 10.1007/s00248-014-0487-1. [DOI] [PubMed] [Google Scholar]
- 5.DiGiulio DB, Callahan BJ, McMurdie PJ, et al. Temporal and spatial variation of the human microbiota during pregnancy. Proc Natl Acad Sci U S A. 2015;112(35):11060–11065. doi: 10.1073/pnas.1502875112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MacIntyre DA, Chandiramani M, Lee YS, et al. The vaginal microbiome during pregnancy and the postpartum period in a European population. Sci Rep. 2015;5:e36466–e36469. doi: 10.1038/srep08988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bisanz JE, Enos MK, PrayGod G, Seney S, Macklaim JM, Chilton S, Willner D, Knight R, Fusch C, Fusch G, Gloor GB, Burton JP, Reid G. Microbiota at multiple body sites during pregnancy in a rural Tanzanian population and effects of Moringa-supplemented probiotic yogurt. App Environ Microbiol. 2015;81(15):4965–4975. doi: 10.1128/AEM.00780-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doyle R, Gondwe A, Fan Y-M, et al. A Lactobacillus-deficient vaginal microbiota dominates postpartum women in rural Malawi. App Environ Microbiol. 2018;84(6):4680–4612. doi: 10.1128/AEM.02150-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SSK, McCulle SL, Karlebach S, Gorle R, Russell J, Tacket CO, Brotman RM, Davis CC, Ault K, Peralta L, Forney LJ. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petrova MI, Reid G, Vaneechoutte M, Lebeer S. Lactobacillus iners: friend or foe? Trends Microbiol. 2017;25(3):182–191. doi: 10.1016/j.tim.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 11.Kindinger LM, Bennett PR, Lee YS, Marchesi JR, Smith A, Cacciatore S, Holmes E, Nicholson JK, Teoh TG, MacIntyre DA. The interaction between vaginal microbiota, cervical length, and vaginal progesterone treatment for preterm birth risk. Microbiome. 2017;5(1):6. doi: 10.1186/s40168-016-0223-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goltsman DSA, Sun CL, Proctor DM, DiGiulio DB, Robaczewska A, Thomas BC, Shaw GM, Stevenson DK, Holmes SP, Banfield JF, Relman DA. Metagenomic analysis with strain-level resolution reveals fine-scale variation in the human pregnancy microbiome. Genome Res. 2018;28(10):1467–1480. doi: 10.1101/gr.236000.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gajer P, Brotman RM, Bai G, et al. Temporal Dynamics of the Human Vaginal Microbiota. Sci Transl Med. 2012;4(12):132–152. doi: 10.1126/scitranslmed.3003605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Avershina E, Slangsvold S, Simpson MR, Storrø O, Johnsen R, Øien T, Rudi K. Diversity of vaginal microbiota increases by the time of labor onset. Sci Rep. 2017;7(1):17558. doi: 10.1038/s41598-017-17972-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan S, Cohen DB, Ravel J, Abdo Z, Forney LJ. Evaluation of methods for the extraction and purification of DNA from the human microbiome. PLoS One. 2012;7(3):e33865. doi: 10.1371/journal.pone.0033865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27(21):2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Callahan BJ, McMurdie PJ, Rosen MJ, et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13(7):58. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allard G, Ryan FJ, Jeffery IB, Claesson MJ. SPINGO: a rapid species-classifier for microbial amplicon sequences. BMC Bioinformatics. 2015;16:324. doi: 10.1186/s12859-015-0747-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Team RC. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/. 2019: 1–2673.
- 20.Maechler M, Rousseeuw P, Struyf A, et al. Cluster: Cluster Analysis Basics and Extensions 2019.
- 21.Meyer D, Dimitriadou E, Hornik K, et al. e1071: Misc Functions of the Department of Statistics, Probability Theory Group (Formerly: E1071), TU Wienhttps://CRAN.R-project.org/package=e1071. 2019.
- 22.Ogle DH, Wheeler P, Dinno A. Fisheries Stock Analysishttps://github.com/droglenc/FSA. 2019.
- 23.Wickham H. ggplot2: elegant graphics for data analysis. New York: Springer-Verlag; 2016. [Google Scholar]
- 24.Kassambara A. ggpubr: ‘ggplot2’ based publication ready. Plots. 2019:1–126.
- 25.Bates D, Mächler M, Bolker B, et al. Fitting linear mixed-effects models using lme4. J Stat Soft. 2015;67:1–48. doi: 10.18637/jss.v067.i01. [DOI] [Google Scholar]
- 26.Kuznetsova A, Brockhoff PB, Christensen RHB. lmerTestPackage: tests in linear mixed effects models. J Stat Soft. 2017;82:1–26. doi: 10.18637/jss.v082.i13. [DOI] [Google Scholar]
- 27.Hothorn T, Bretz F, Westfall P. Simultaneous inference in general parametric models. Biom J. 2008;50:346–363. doi: 10.1002/bimj.200810425. [DOI] [PubMed] [Google Scholar]
- 28.Wickham H. The split-apply-combine strategy for data analysis. J Stat Soft. 2011;40:1–29. [Google Scholar]
- 29.Wickham H, Henry L. tidyr: Tidy Messy Datahttps://CRAN.R-project.org/package=tidyr. 2019.
- 30.Oksanen J, Blanchet FG, Friendly M, et al. vegan: Community Ecology Package. R package version 2.5–6. 2019:1–295.
- 31.Romero R, Hassan SS, Gajer P, et al. The composition and stability of the vaginal microbiota of normal pregnant women is different from that of non-pregnant women. 2014;2(1):4. [DOI] [PMC free article] [PubMed]
- 32.Hickey RJ, Zhou X, Pierson JD, Ravel J, Forney LJ. Understanding vaginal microbiome complexity from an ecological perspective. Transl Res. 2012;160(4):267–282. doi: 10.1016/j.trsl.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nunn KL, Forney LJ. Unraveling the dynamics of the human vaginal microbiome. Yale J Biol Med. 2016;89(3):331–337. [PMC free article] [PubMed] [Google Scholar]
- 34.Hendrick V, Altshuler LL, Suri R. Review article hormonal changes in the postpartum and implications for postpartum depression. Psychosomatics. 1998;39(2):93–101. doi: 10.1016/S0033-3182(98)71355-6. [DOI] [PubMed] [Google Scholar]
- 35.Amabebe E, Anumba DOC. The vaginal microenvironment: the physiologic role of lactobacilli. Front Med. 2018;5:181. doi: 10.3389/fmed.2018.00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Glynn LM, Davis EP, Sandman CA, Goldberg WA. Gestational hormone profiles predict human maternal behavior at 1-year postpartum. Horm Behav. 2016;85:19–25. doi: 10.1016/j.yhbeh.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schock H, Zeleniuch-Jacquotte A, Lundin E, Grankvist K, Lakso HÅ, Idahl A, Lehtinen M, Surcel HM, Fortner RT. Hormone concentrations throughout uncomplicated pregnancies: a longitudinal study. BMC Pregnancy Childbirth. 2016;16(1):146. doi: 10.1186/s12884-016-0937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Groer ME, Jevitt C, Ji M. Immune changes and dysphoric moods across the postpartum. Am J Reprod Immunol. 2015;73(3):193–198. doi: 10.1111/aji.12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gomez-Lopez N, StLouis D, Lehr MA, Sanchez-Rodriguez EN, Arenas-Hernandez M. Immune cells in term and preterm labor. Cell Mol Immunol. 2014;11(6):571–581. doi: 10.1038/cmi.2014.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fouladi-Nashta AA, Raheem KA, Marei WF, Ghafari F, Hartshorne GM. Regulation and roles of the hyaluronan system in mammalian reproduction. Reproduction. 2017;153(2):R43–R58. doi: 10.1530/REP-16-0240. [DOI] [PubMed] [Google Scholar]
- 41.Marandy EE, Kanayama N, Kobayashi H, et al. The role of hyaluronic acid as a mediator and regulator of cervical ripening. Hum Reprod. 1997;12(5):1080–1088. doi: 10.1093/humrep/12.5.1080. [DOI] [PubMed] [Google Scholar]
- 42.Taylor KR, Trowbridge JM, Rudisill JA, et al. Hyaluronan Fragments Stimulate Endothelial Recognition of Injury through TLR4*. The Journal of Biological Chemistry. 279:17079–84. [DOI] [PubMed]
- 43.Campisi J, Leem TH, Fleshner M. Stress-induced extracellular Hsp72 is a functionally significant danger signal to the immune system. Cell Stress Chaperones. 2003;8(3):272–286. doi: 10.1379/1466-1268(2003)008<0272:SEHIAF>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garantziotis S, Savani RC. Hyaluronan biology: a complex balancing act of structure, function, location and context. Matrix Biol. 2019;78-79:1–10. doi: 10.1016/j.matbio.2019.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hong Y, Kim YK, Kim GB, Nam GH, Kim SA, Park Y, Yang Y, Kim IS. Degradation of tumour stromal hyaluronan by small extracellular vesicle-PH20 stimulates CD103+ dendritic cells and in combination with PD-L1 blockade boosts anti-tumour immunity. J Extracell Vesicles. 2019;8(1):1670893. doi: 10.1080/20013078.2019.1670893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O'Hanlon DE, Moench TR, Cone RA. In vaginal fluid, bacteria associated with bacterial vaginosis can be suppressed with lactic acid but not hydrogen peroxide. BMC Infect Dis. 2011;11:200. doi: 10.1186/1471-2334-11-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O'Hanlon DE, Moench TR, Cone RA. Vaginal pH and microbicidal lactic acid when Lactobacilli dominate the microbiota. PLoS One. 2013;8(11):e80074–e80078. doi: 10.1371/journal.pone.0080074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aldunate M, Tyssen D, Johnson A, Zakir T, Sonza S, Moench T, Cone R, Tachedjian G. Vaginal concentrations of lactic acid potently inactivate HIV. J Antimicrob Chemother. 2013;68(9):2015–2025. doi: 10.1093/jac/dkt156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wagner W, Ciszewski WM, Kania KD. L- and D-lactate enhance DNA repair and modulate the resistance of cervical carcinoma cells to anticancer drugs via histone deacetylase inhibition and hydroxycarboxylic acid receptor 1 activation. Cell Commun Signal. 2015;13:36. doi: 10.1186/s12964-015-0114-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Witkin SS. The vaginal microbiome, vaginal anti-microbial defense mechanisms and the clinical challenge of reducing infection-related preterm birth. BJOG. 2015;122(2):213–219. doi: 10.1111/1471-0528.13115. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Relative proportions of bacteria in the vaginal communities of 48 pregnant women separated by stage. The stacked bars represent the proportions of bacterial taxa within one sample. Bars are separated by the pregnancy stage in which the sample was collected (top headings). Taxa colors are indicated in the legend below the figure. “Other” represents the sum of all bacterial taxa that were not present at 1% in at least two women. (PDF 40 kb)
(XLSX 153 kb)
(DOCX 39 kb)
Data Availability Statement
The sequence data analyzed in this study along with the corresponding metadata are available in the NCBI Sequence Read Archive under BioProject PRJNA614593 (“Study of the pregnant and postpartum period vaginal microbiome;” http://www.ncbi.nlm.nih.gov/bioproject/614593).