Abstract
Recently, the regulatory role of epigenetic modifications in the occurrence and development of malignant tumors has attracted extensive attention. RNA m6A methylation is the most abundant RNA modification in eukaryotic cells and regulates RNA transcription, processing, splicing, degradation, and translation. As important biomarkers, miRNAs play a crucial role in the diagnosis and treatment of diseases as well as in the development of anti-tumor drugs. Recently, increasing evidence has shown that m6A modification plays a vital role in regulating miRNA biosynthesis. We, herein, have reviewed the enzyme system involved in m6A methylation and the crosstalk between m6A modification and miRNAs in cancer. In addition, we have discussed the potential clinical applications and possible development directions of this field in the future.
Subject terms: Biomarkers, Cancer
Facts
N6-methyladenosine regulates miRNA biosynthesis.
miRNAs affect m6A levels by targeting m6A regulatory proteins.
The interactions between m6A modification and miRNAs influence cancer progression.
Open questions
How does m6A inhibit the biological functions of miRNAs?
Are there other m6A readers that recognize m6A modifications to perform different biological functions?
Could the combination of m6A and miRNAs effectively treat cancer, especially drug-resistant cancer?
Introduction
Recently, the regulatory role of epigenetic modifications in the occurrence and development of malignant tumors has attracted much attention. RNA epigenetics is an important way to regulate gene expression at the RNA level. The reversibility of epigenetics provides a scientific basis for early intervention and treatment of diseases. To date, more than 170 RNA modifications have been identified, and methylation modification is known to be the most common1. Studies have found that the RNA methylation modifications usually include N1-methyladenosine (m1A), 5-methylcytosine (m5C), 5-hydroxymethylcytosine (5hmC), N6-methyladenosine (m6A), and 7-methylguanine (m7G)2. Among them, m6A modification is the most abundant and important mRNA modification in mammals, and accounts for ~50% of the total methylated ribonucleotides3. Sequencing analysis showed that m6A modification was mainly concentrated in the common motif RRACH (R = G/A, H = A/C/U). In the RRACH motif, m6A modification is preferentially concentrated near the 3′-UTR, followed by the CDs and 5′-UTR regions4,5. In the 3′-UTRs enriched with m6A, about 67% of UTRs also contain ncRNAs, such as the binding sites of miRNAs, suggesting that m6A and ncRNAs may jointly regulate target mRNAs through cooperation or competition4. m6A modification is involved in the regulation of RNA metabolism, including translation, splicing, and degradation, and plays a crucial regulatory role in the development and disease onset and progression6.
m6A modification exists not only in mRNA but also in non-coding RNAs, such as miRNAs. miRNAs are noncoding single-stranded small RNA molecules with a length of ~21–23 nucleotides. They are widely found in eukaryotes and play an important role in post-transcriptional regulation of gene expression through translation inhibition and mRNA cleavage7,8. As an important biomarker, miRNA has a very important guiding significance in the diagnosis and treatment of diseases and development of antitumor drugs. m6A has been shown to be abnormally expressed in many tumors and plays a vital role in the regulation of a series of malignant biological behaviors such as cell proliferation, invasion, and metastasis. With the wide application of high-throughput sequencing technology and bioinformatics analysis in scientific research, increasing number of m6A-modified miRNAs have been discovered. Subsequently, researchers have also analyzed the interaction between m6A RNA methylation and miRNAs in the context of cancer. We, in this article, have reviewed the research progress regarding the interactions between m6A modification and miRNA in tumors; our aim was to provide a theoretical basis for a deeper understanding of the occurrence and development of malignant tumors as well as that regarding the search for tumor prediction biomarkers and therapeutic targets.
m6A enzyme system
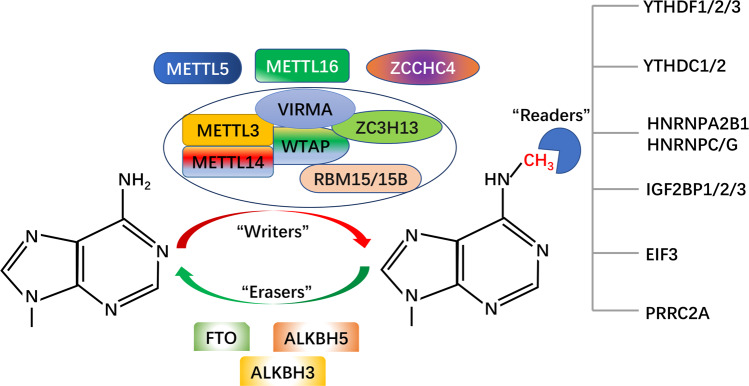
m6A refers to the methylation modification at the sixth N position of adenosine, and has been proven to be reversible. There are three main types of enzymes involved in m6A modification: methylated transferase, demethylated transferase, and methylated recognition protein (Fig. 1).
Fig. 1. Dynamic reversible process and molecular composition of m6A.
m6A modifications are catalyzed by m6A writers, erasers, and readers. The sixth N position of adenylate (A) can be methylated by m6A writers to form N6-methyladenosine (m6A). Writers include METTL3, METTL14, WTAP, RBM15/15B, VIRMA, and ZC3H13. METTL5, METTL16, and ZCCHC4 are independent m6A methyltransferases. Erasers are proteins with demethylase activity, and include FTO, ALKBH5, and ALKBH3. Readers are proteins that perform a biological function by recognizing m6A modifications, and include YTH family, HNRNP family, EIF3, IGF2BPs, and PRRC2A.
m6A methylated transferase
m6A methylation is catalyzed by the methyltransferase complex (MTC) known as “m6A writers”. MTC is composed of m6A-METTL complex (MAC) and m6A-METTL associated complex (MACOM)9. MAC consists of methyl transferase 3 (METTL3) and methyl transferase 14 (METTL14), which can form stable heterodimers. METTL3 is the first discovered methyltransferase with a catalytic subunit, and serves as the main catalytic core10,11. METTL14 can recognize substrates and promote binding to RNA as well as increase the activity of METTL3 methyltransferase12,13. MACOM consists of Wilms tumor-1 associated protein (WTAP), CCCH type 13 zinc finger protein (ZC3H13), m6A methyltransferase-associated virus (VIRMA), and RNA-binding protein 15/15b (RBM15/15b). WTAP itself does not have methyltransferase activity, but it can directly bind to METTL3 to ensure the proper localization of the METTL3–METTL14 heterodimer, thus improving the activity of methyltransferase14. ZC3H13 can promote nuclear localization of the complexes15. VIRMA recruits core catalytic components to mediate preferred methylation in the 3′-UTR16. RBM15/15b can bind complexes and recruit them to specific sites on RNA to promote methylation17. In addition to the above methyltransferase in the form of a complex, independent methyl transferases METTL16, METTL5, and zinc finger CCHC type containing 4 (ZCCHC4) have also been identified. METTL16 was observed to catalyze m6A methylation of U6-like hairpins in U6 snRNA and methionine adenosine transferase (MAT2A) as well as to regulate the variable splicing of MAT2A, thereby maintaining SAM’s homeostasis18. ZCCHC4 mediates m6A methylation of 28S rRNA, which plays an important role in the onset and development of tumors19. A recent study showed that METTL5 was able to mediate the methylation of 18S rRNA m6A and maintain the stability of cell metabolism by forming a heterodimer with TRMT11220. In summary, we, from the literature, have identified that m6A methyltransferase exists mainly in two forms. One is in the form of a methyltransferase complex, in which different methyltransferases perform their respective functions and eventually lead to m6A methylation modification of RNA. The other is independent of the presence of methyltransferase, independent of the form of the complex, but independent of methylation. Further research on “m6A writers” may provide novel biomarkers for tumor diagnosis as well as new directions for the discovery of tumor therapeutic targets.
m6A demethylated transferase
m6A methylation is dynamically reversible and can be reversed by demethylases called “m6A Erasers”. Erasers mainly include FTO and ALKBH5, which belong to the ALKB family, with Fe (II) and alpha-ketone glutaric acid-dependent way of catalyzing m6A demethylation9,21. FTO is the first to be found to methylation enzyme; it can catalyze m6A oxidation of N6-hydroxymethyladenosine (hm6A) and N6-formyladenosine (f6A)22. Studies have found that FTO can also demethylate m6Am in mRNAs and snRNAs, and m1A in tRNAs23–25. ALKBH5 is the second discovered demethylase, and can directly reverse m6A to adenosine26. Recently, ALKBH3, as a demethylase, was found to be capable of removing the m6A modification of tRNAs27. The discovery of m6A dimethyl transferase is of great significance to the researchers analyzing m6A and can provide great help for reverse treatment. However, at present, our understanding of m6A dimethyl transferase is still in the exploratory stage and needs further analysis.
m6A regulatory proteins
m6A modified RNAs require a class of m6A binding proteins that can specifically recognize and mediate their specific biological functions; these proteins known as “m6A readers”9. Readers are mainly divided into proteins containing the YTH domain and proteins without this domain. The YTH domain can selectively bind to the m6A site of RNA. The proteins containing this YTH domain include YTHDF1, YTHDF2, YTHDF3, YTHDC1, and YTHDC228. YTHDF1-3 specifically recognize the mRNA containing m6A in the cytoplasm and promote mRNA translation and degradation29–31. YTHDC1-2 mainly recognize m6A-binding mRNA in the nucleus and promote mRNA degradation32,33. Heterogeneous nuclear ribonucleoproteins A2/B1 (HNRNPA2B1) and C (HNRNPC) of the nuclear heterogeneous ribonucleoprotein family are two rich nuclear RNA-binding proteins involved in the processing of precursor mRNA. HNRNPA2B1 binds to m6A modification sites on RNA to regulate RNA splicing and miRNA maturation34. HNRNPC and HNRNPG can regulate the alternative splicing of transcripts containing m6A modifications by recognizing and binding to m6A dependent structural switches35,36. Eukaryotic initiation factor 3 can directly bind to the 5′-UTR of m6A-modified mRNA, initiating mRNA translation37. The insulin-like growth factor-2 binding protein family (IGF2BPs, including IGF2BP1/2/3) promotes mRNA translation that is more stable in an m6A-dependent manner38. A new m6A reader, PRRC2A, was recently found to be dependent on m6A to stabilize mRNA expression39. Overall, these results indicate that numerous m6A readers have been found thus far, which shows that potentially numerous m6A readers with wide targeting abilities exist, thus offering a broad research space. Since m6A modification relies on “Readers” to perform its biological function; the same m6A modification may have opposite biological effects when combined with different “Readers”. Therefore, promoting or blocking the binding of m6A RNA to “Readers” may provide a new approach for cancer therapy in the future.
Mutual regulation between m6A and miRNAs
m6A regulates the biosynthesis of miRNAs
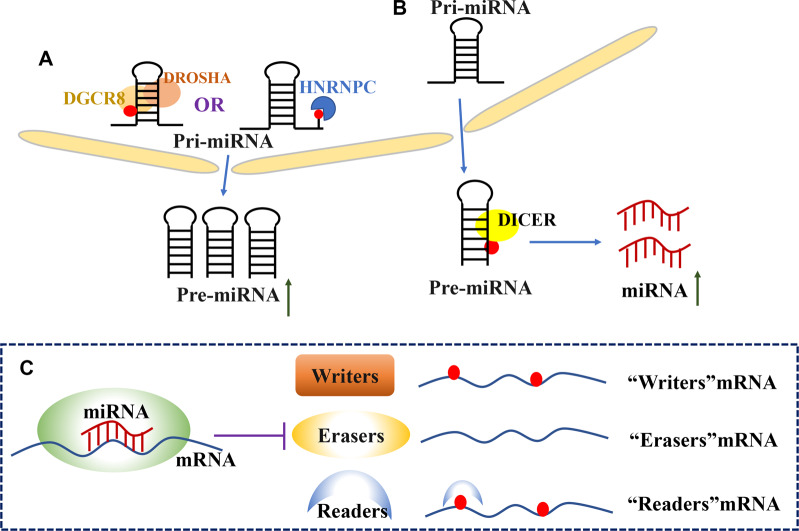
The classical miRNA synthesis route is as follows: first, miRNAs in the cell nucleus are transcribed into pri-miRNAs under the action of RNA polymerase II, and pri-miRNAs form pre-miRNAs under the action of a micro-processing complex composed of endonuclease Drosha and a cofactor, double-stranded RNA binding protein DGCR8. Pre-miRNAs are transported to the cytoplasm and then spliced by the endoribonuclease Dicer to form double-stranded microRNAs7,8. It has been found that m6A modification can regulate miRNA biosynthesis by participating in the processing of pri-miRNAs or splicing of pre-miRNAs (Fig. 2).
Fig. 2. Mutual regulation between m6A and miRNAs.
A In the nucleus, the intranuclear enzyme Drosha and DGCR8 cleave primary microRNA (pri-miRNA) to precursor microRNA (pre-miRNA). m6A modifications helps to recruit DGCR8 to target pri- miRNA and promote the formation of pre-miRNA. Meanwhile, HNRNPC can directly recognize the m6A sites. B In the cytoplasm, Dicer cleaves pre-miRNAs transported to the cytoplasm into mature miRNAs. C miRNAs regulate the expression of m6A regulatory proteins. miRNAs regulate the expression of mRNAs encoding for writers, erasers, and readers.
Alarcon et al. first discovered the regulation of methylase METTL3 in miRNA biosynthesis in 2015. Studies have shown that METTL3 induces m6A modification in pri-miRNAs and enhances DGCR8 recognition and binding to its substrate, thus promoting the processing of pri-miRNAs and miRNA maturation40. Recently, in addition to its interaction with DGCR8, METTL3 has been shown to promote the synthesis of mature miRNAs by increasing the splicing of pre-miRNAs by Dicer41. METTL14 can promote the processing of pri-miR-375 by regulating the m6A modification through interaction with DGCR842. Subsequently, Berulava et al. found that the levels of pri-miRNAs were not altered by knocking down the methyltransferase FTO, however, homeostatic levels of some potential methylated miRNAs were reduced43. Nonetheless, the mechanism by which FTO negatively regulates the stability of miRNAs is still unclear and needs to be further investigated. The interactions between the demethylase ALKBH5 and dead box helicase family member DDX3 regulate the m6A level in mRNAs. DDX3 interacts with AGO2 to regulate the demethylation of miRNAs; however, whether there is a direct interaction between ALKBH5 and AGO2 needs further verification44. In addition, the NF-κB activator protein NKAP, which is recognized by m6A, promotes the processing of pri-miR-25 by interacting with DGCR8 to recognize and bind to the m6A site on pri-miR-2545. HNRNPC can directly recognize and bind to the pri-miR-21 site modified by m6A to promote the expression of miR-2146. HNRNPA2B1 is of particular interest as it has been shown to promote the processing of some of the pri-miRNAs by recruiting DGCR8. In addition, HNRNPA2B1 has been found to inhibit the shearing of some of the pri-miRNAs; however, the specific mechanism underlying this inhibition remains unclear34,47.
miRNAs regulate m6A modifications
In the classical miRNA synthesis pathway, mature miRNAs bind to the 3′-UTR of their target mRNA through complementary base pairing, which leads to the degradation of target mRNA or inhibition of protein translation. m6A modifications can also be used as target genes of miRNAs (Fig. 2).
A study found that miR-33a can inhibit the proliferation and migration of cancer cells by directly binding to the 3′-UTR of METTL3, thus reducing the expression of METTL348. Subsequently, He et al. found that miR-4429 can target and downregulate METTL3 to inhibit the stabilization of SEC62 induced by m6A. The results showed that METTL3 can be used as a target gene by miRNAs and that it can regulate the mRNA stability of downstream genes through m6A modification49. Moreover, a study found that in hepatocellular carcinoma (HCC) cells, miR-145 regulated the level of m6A by targeting the 3′-UTR of YTHDF2 mRNA50. Recent studies have found that YTHDF1 can act as a target of miR-346 to regulate the progression of glioma51.
In conclusion, on one hand, m6A modifications can promote the processing of pri-miRNAs through the interaction with RNA-binding protein DGCR8 or promote the maturation of miRNAs by increasing the splicing of pre-miRNAs by Dicer and positively regulating the synthesis and expression of miRNAs. On the other hand, m6A also plays a negative regulatory role in miRNA synthesis. For example, HNRNPA2B1 can inhibit the over-splicing of some of the pri-miRNAs; nonetheless, the specific inhibitory mechanism remains to be elucidated. In addition, miRNAs can directly target m6A-related proteins, resulting in the translation inhibition of m6A-related protein-encoding genes, thus playing a negative regulatory role.
Interaction between m6A and miRNAs in cancer
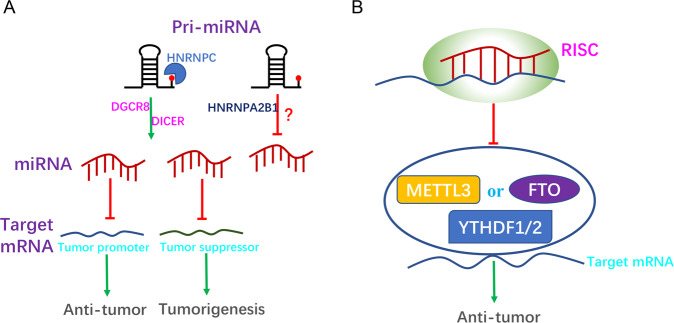
In recent years, it has been found that m6A exists in miRNAs targeting oncogenes or tumor suppressors, and is involved in the onset and development of cancer by affecting the biogenesis or stability of miRNAs (Table 1 and Fig. 3).
Table 1.
M6A regulate miRNAs in different cancers.
| m6A Regulator | Cancer | miRNAs | Biological function | Ref. |
|---|---|---|---|---|
| METTL3 | bladder | miR-221/222 | Promote cells proliferation in bladder | 52 |
| METTL3 | colorectal | miR-1246 | Promote cells migration and invasion in CRC | 53 |
| METTL3 | pancreatic | miR-25-3p | Promote cells proliferation migration and invasion in pancreatic cancer | 45 |
| METTL3 | NSCLC | miR-143-3p | Promote cells invasion in NSCLC | 41 |
| Mettl14 | breast | miR-146a-5p | Promote cells migration and invasion in breast cancer | 55 |
| Mettl14 | hepatoma | miR-126 | Suppress cells migration in HCC | 54 |
| Mettl14 | colorectal | miR-375 | Suppress cells proliferation invasion and migration in CRC | 42 |
| ALKBH5 | ovarian | miR-7 | Promote cells proliferation, invasion, and autophagy in ovarian cancer | 57 |
| NKAP | pancreatic | miR-25-3p | Promote cells proliferation migration and invasion in pancreatic cancer | 45 |
| HNRNPA2B1 | NSCLC | miR-106b-5p | Promote cells proliferation in NSCLC | 58 |
| HNRNPC | glioblastoma | miR-21 | Promote cells migration and invasion in glioblastoma | 46 |
Fig. 3. Interaction between m6A and miRNAs in cancer.
A m6A modifications play different roles in multiple cancers by influencing the maturation of miRNAs. m6A modification promotes miRNA maturation by recruiting DGCR8 to promote pri-miRNA processing or increasing pre-miRNA splicing by Dicer in different cancers. Furthermore, m6A modification can promote or inhibit the occurrence and development of tumors by affecting the maturation of miRNAs. HNRNPA2B1 has also been found to inhibit the shearing of some of the pri-miRNAs; however, the specific mechanism underlying this inhibition remains unclear in breast cancer. B miRNAs affect cancer progression by regulating m6A modification of mRNAs (METTL3, FTO, YTHDF1, and YTHDF2).
Previous studies have shown that METTL3 promotes the processing of pri-miRNAs by recruiting DGCR8 to identify substrates40. As of 2019, this mechanism has been confirmed in various human cancers. As reported by Han et al., METTL3 expression was found to be upregulated in bladder cancer tissues and cell lines. Furthermore, METTL3 promoted the maturation of miR-221/222 in bladder cancer cells by recruiting DGCR8. The expression of miR-221/222 target gene PTEN was downregulated, which promoted the proliferation of bladder cancer cells. The study showed that METTL3 may act as a new prognostic marker for the treatment of bladder cancer52. Subsequently, Peng et al. found a similar mechanism involving METTL3 in colorectal cancer (CRC). METTL3 promoted the biogenesis of miR-1246 by interacting with DGCR8, which in turn promoted the migration and invasion of CRC cells. miR-1246 was found to reverse the inhibition of MAPK signaling pathways by downregulating the tumor suppressor gene SPRED2, which enhanced the metastatic ability of CRC53. Moreover, another study found that cigarette condensate can induce DNA hypomethylation in METTL3 promoter, which promotes the expression of METTL3, resulting in m6A modification of pri-miR-25, followed by enhancing the processing of pri-miR-25 in NKAP-dependent manner. Mature miR-25-3p promoted the proliferation, migration, and invasion of pancreatic cancer cells by inhibiting PHLPP2 and activating the oncogenic AKT-p70S6K signaling pathway45. METTL3 has also been shown to promote miRNA biogenesis by increasing the splicing of pre-miRNAs by Dicer. Recently, a study found that miR-143-3p was highly expressed in lung cancer patients with brain metastasis, which depended on METTL3 promoting Dicer splicing pre-miR-143 into mature miR-143 in lung cancer cells, followed by downregulation of VASH1-dependent degradation of VEGF, which promoted angiogenesis in lung cancer patients with brain metastasis. This was the first study to reveal the role of m6A in pre-RNA processing, thus providing a new therapeutic target for patients with non-small cell lung cancer (NSCLC) brain metastases41. In summary, the results show that METTL3 promotes the occurrence and development of tumors by enhancing the synthesis of tumor-promoting factor miRNAs in bladder, colorectal, pancreatic, and lung cancers. METTL14 plays a similar role in cancer as that of METTL3; it promotes DGCR8 to recognize and bind m6A-modified pri-miRNAs, which further mediates miRNA maturation. Studies have shown that METTL14 promotes pri-miR-126 maturation through the recruitment of DGCR8-dependent m6A in HCC. Deletion of METTL14 in HCC cells reduces m6A levels and the expression of miR-126, leading to the migration and invasion of cancer cells54. Another study found that METTL14 inhibited the progression of CRC by promoting the maturation of miR-375 in an m6A-dependent manner. It not only inhibited the proliferation of cancer cells through miR-375/YAP1, but also inhibited their migration and invasion via the miR-375/SP1 pathway42. Yi et al. observed abnormal expression of METTL14 in breast cancer tissues and cells, and further found that overexpression of METTL14 promoted the migratory and invasive abilities of breast cancer cells by promoting the expression of miR-146a-5p55. In summary, the results indicate that METTL14 can inhibit or promote the occurrence of tumors by regulating the synthesis of miRNAs. In HCC and CRC, METTL14 inhibits the occurrence of tumors by promoting the synthesis of tumor-suppressor miRNAs, whereas in breast cancer, METTL14 promotes the occurrence and development of tumors by enhancing the synthesis of tumor-promoting miRNAs.
The role of the demethylase enzyme FTO in regulating miRNAs in cancer has not been reported till date. However, Shen et al. found that FTO plays an important role as a downstream target gene of miR-1266 in colorectal cancer and that miR-1266 directly targets the 3′-UTR of FTO to inhibit the proliferation of colorectal cancer cells56. Another demethylase, ALKBH5, promoted the maturation of miR-7 in a hub-dependent manner and upregulated the mRNA levels of Bcl2 in m6A-dependent manner, thus promoting the proliferation, migration, and autophagy of ovarian cancer cells57.
Furthermore, the m6A reader participates in miRNA biogenesis. HNRNPA2B1 has been shown to promote miRNA synthesis by recruiting DGCR8 to recognize the m6A marker site of pri-miRNAs34. Consistent with this finding, the overexpressed oncogene LINC01234 in NSCLC promotes maturation from pri-miR-106b to miR-106b-5p by interacting with hnRNPA2B1 to recruit DGCR8, resulting in a decrease in cryptochrome 2 as well as promotion of c-MYC expression. Interestingly, activated c-MYC binds to the promoter of LINC01234 and increases its transcription, thus forming a positive feedback loop that leads to cell proliferation and tumor growth58. However, the role of HNRNPA2B1 in the regulation of miRNAs appears to be complex in breast cancer. Genome-wide miRNA sequencing was performed post the upregulation of HNRNPA2B1 in endocrine-resistant breast cancer cells. Sequencing results showed that miR-1266-5p, miR-1268a, and miR-671-3p were upregulated in the HNRNPA2B1 group, while miR-29a-3p, miR-29b-3p, and miR-222 were downregulated in this group. However, the biological processes by which HNRNPA2B1 inhibits miRNAs are not yet clear47. In summary, these results suggest that HNRNPA2B1 may act as an oncogenic factor in NSCLC; however, its specific role in breast cancer remains unclear and further research is needed in this direction. In glioblastoma, HNRNPC can directly recognize and bind to pri-miR-21 sites that have been modified with m6A and promote the expression of miR-21. Silencing HNRNPC in glioblastoma cells reduced the level of miR-21, thereby increasing the expression of PDCD4, inhibiting the activation of Akt and p70S6K, and inhibiting cell migration and invasion46.
m6A modifications have also been found to play an important role in cancer progression by acting as targets for miRNAs (Table 2 and Fig. 3). In NSCLC, the expression level of METTL3 was higher in lung cancer than in normal tissues. Moreover, miR-33a inhibited the migration and proliferation of lung cancer cells by directly targeting the 3′-UTR of METTL3 and decreasing its mRNA levels48. In addition, in NSCLC, miR-600 induced the mitochondrial apoptosis signaling pathway by downregulating METTL3 expression to promote apoptosis and inhibit proliferation and invasion of lung cancer cells59. In breast cancer, let-7g, as a tumor suppressor, was found to downregulate the expression of METTL3 by targeting the 3′-UTR of METTL3 mRNA, while hepatitis B virus X-interacting protein (HBXIP) could increase the expression of METTL3 by inhibiting the function of let-7g. Interestingly, METTL3 upregulated HBXIP in an m6A dependent manner, forming a positive feedback loop of HBXIP/let-7g/METTL3/HBXIP, and in turn, accelerating the proliferation and invasion of breast cancer cells60. In gastric cancer, miR-4429 was observed to target and reduce the mRNA expression of METTL3, inhibiting the stability of SEC62 induced by m6A, thereby inhibiting the proliferation and inducing apoptosis of gastric cancer cells49. In hepatoblastoma (HB) cells, knockout of METTL3 notably inhibited the proliferation, migration, and invasion of cells. Furthermore, miR-186 suppressed the progression of HB by directly targeting the 3′-UTR of METTL3 and activating the Wnt/β-catenin signaling pathway61. Collectively, these findings indicate that METTL3 can play a tumor-promoting role in NSCLC, gastric cancer, hepatoblastoma, and breast cancer.
Table 2.
MiRNAs regulate m6A key proteins in different cancers.
| miRNAs | Cancer | mRNAs | Biological function | Ref. |
|---|---|---|---|---|
| miR-33a | NSCLC | METTL3 | Suppress cells proliferation in NSCLC | 48 |
| Let-7g | breast | METTL3 | Suppress cells proliferation in breast cancer | 60 |
| miR-600 | NSCLC | METTL3 | Suppress cells proliferation migration and promote apoptosis in NSCLC | 59 |
| miR-4429 | gastric | METTL3 | Suppress cells proliferation induce apoptosis in gastric cancer | 49 |
| miR-186 | hepatoma | METTL3 | Suppress cells proliferation invasion and migration in hepatoma | 61 |
| miR-1266 | colorectal | FTO | Suppress cells proliferation in colorectal cancer | 56 |
| miR-346 | glioblastoma | YTHDF1 | Suppress cells proliferation and tumor growth in glioblastoma | 51 |
| miR-145 | hepatoma | YTHDF2 | Suppress cells proliferation in hepatoma | 50 |
| miR-493-3p | prostate | YTHDF2 | Suppress cells proliferation and migration in prostate cancer | 62 |
| miR-744-5p | ovarian | HNRNPC | Promote cells apoptosis in ovarian cancer | 63 |
miRNAs can participate in cancer progression by targeting the mRNA of m6A regulatory proteins. Studies have shown that miR-493-3p can directly target YTHDF2 and downregulate its expression, and reduce the level of m6A to inhibit the proliferation and invasion of prostate cancer cells62. In addition, YTHDF2 was found to be a downstream target of miR-145, which is involved in the regulation of m6A levels as well as promotion of the proliferation of HCC cells50. In glioblastoma, miR-346 targeting the 3′-UTR of YTHDF1 reduced the mRNA level of YTHDF1, inhibiting cell proliferation and tumor growth51. In ovarian cancer cells, HNRNPC was found to be a target of miR-744-5p, which is known to induce apoptosis. Furthermore, silencing HNRNPC could inhibit the mRNA levels of miR-21 and decrease the phosphorylation of the Akt ser473 site, thus promoting the apoptosis of ovarian cancer cells63. As mentioned above, in a variety of cancers, m6A modification is regulated by different m6A regulatory proteins by facilitating miRNA biosynthesis, and miRNAs, in turn, regulate the biological functions of m6A regulatory proteins (Fig. 4).
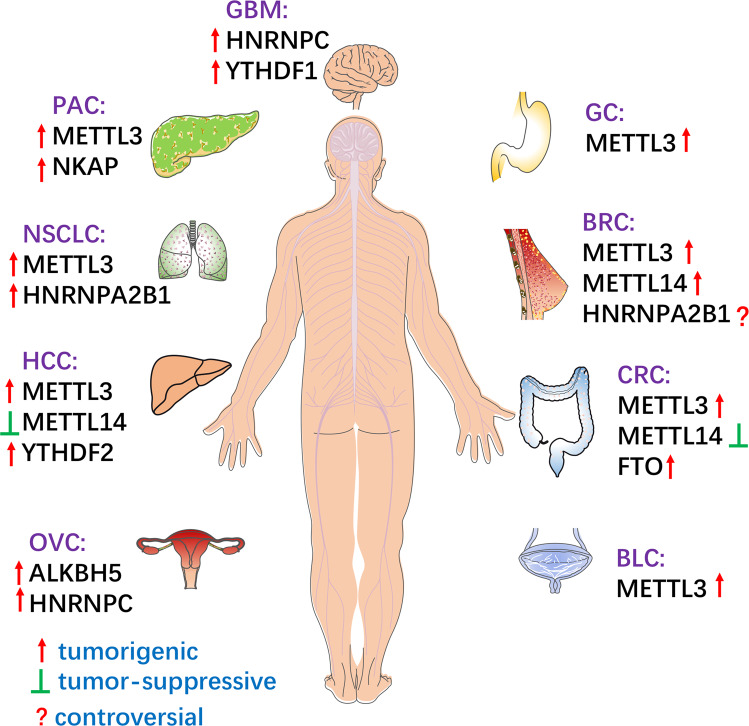
Fig. 4. m6A modifications in different human cancers.
m6A modifications participate in tumorigenesis and metastasis in different cancers. BLC, bladder cancer; CRC, colorectal cancer; BRC, breast cancer; GBM, glioblastoma; GC, gastric cancer; PAC, pancreatic cancer; NSCLC, non-small cell lung cancer; HCC, hepatocellular carcinoma; OVC, ovarian cancer.
Conclusion
In conclusion, m6A methylation modification and miRNAs play an important role in the regulation of tumor development. On one hand, m6A modification controls tumorigenesis by affecting miRNA maturation, while on the other hand, m6A related proteins can be used as downstream targets of miRNAs to participate in the development of tumors. Therefore, it is important to elucidate the role of miRNAs and m6A modifications in the regulation of their interrelationship in tumor therapy for all cancer types. However, current studies mostly focus on the abnormal expression of m6A modifications. Furthermore, the molecular mechanism and downstream signaling pathways involved in the differential transcriptome regulation of m6A modification-related proteins between tumor and normal cells require further analysis. In addition, the current studies on the regulatory mechanism of m6A are mostly limited to in vitro analyses, while only few relevant in vivo studies have been carried out. At the same time, due to the complexity and diversity of m6A modifications and their wide presence in many aspects of life activities, it is still a huge challenge to find translational applications of results obtained from research pertaining to m6A modifications. More research is required in the future to further understand the underlying mechanisms as well as to elucidate the regulatory relationship between m6A modification and miRNAs in tumors. With the newer insights, analysis of m6A methylation modification may lead to novel future directions for the effective management of a variety of diseases. Alone or in combination with miRNAs, it may have a strong therapeutic potential for the treatment of various types of cancer, especially for those with drug resistance.
Author contributions
X. H.: design and conception, manuscript writing, creation of figure and table, and final approval of the manuscript. J. G.: search of literature, manuscript writing, financial support, and final approval of the manuscript. Z. F.: design and conception, manuscript revising, financial support, and final approval of the manuscript. All authors have read and approved the final version of the manuscript.
Funding
This work was supported by the National Sciences Foundation of China (81625005 to Z.P.F.), CAMS Innovation Fund for Medical Sciences (2019-I2M-5-031 to Z.P.F.), and the Program for “Hundred-Thousand- Ten Thousand” Talents in Beijing (2018A16 to Z.P.F.), Grant Key Program of Jiang Xi Province Science and Technology Department of China (20171BBH80012 to J.G.).
Conflict of interest
The authors declare no competing interests.
Footnotes
Edited by G. Calin
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Alderman MH, 3rd, Xiao AZ. N(6)-methyladenine in eukaryotes. Cell Mol. Life Sci. 2019;76:2957–2966. doi: 10.1007/s00018-019-03146-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li J, et al. The role of mRNA m6A methylation in the nervous system. Cell Biosci. 2019;9:66. doi: 10.1186/s13578-019-0330-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wei CM, Gershowitz A, Moss B. Methylated nucleotides block 5’ terminus of HeLa cell messenger RNA. Cell. 1975;4:379–386. doi: 10.1016/0092-8674(75)90158-0. [DOI] [PubMed] [Google Scholar]
- 4.Meyer KD, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yue Y, Liu J, He C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 2015;29:1343–1355. doi: 10.1101/gad.262766.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu ZM, Huo FC, Pei DS. Function and evolution of RNA N6-methyladenosine modification. Int. J. Biol. Sci. 16, 1929–1940 (2020). [DOI] [PMC free article] [PubMed]
- 7.Annese T, Tamma R, De Giorgis M, Ribatti D. microRNAs biogenesis, functions and role in tumor angiogenesis. Front. Oncol. 2020;10:581007. doi: 10.3389/fonc.2020.581007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Achkar NP, Cambiagno DA, Manavella PA. miRNA biogenesis: a dynamic pathway. Trends Plant Sci. 2016;21:1034–1044. doi: 10.1016/j.tplants.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 9.Lence T, Paolantoni C, Worpenberg L, Roignant JY. Mechanistic insights into m6A RNA enzymes. Biochim. Biophys. Acta Gene Regul. Mech. 2019;1862:222–229. doi: 10.1016/j.bbagrm.2018.10.014. [DOI] [PubMed] [Google Scholar]
- 10.Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 3, 1233–1247 (1997). [PMC free article] [PubMed]
- 11.Wang P, Doxtader KA, Nam Y. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell. 2016;63:306–317. doi: 10.1016/j.molcel.2016.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, et al. Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex. Nature. 2016;534:575–578. doi: 10.1038/nature18298. [DOI] [PubMed] [Google Scholar]
- 13.Liu J, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ping XL, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wen J, et al. Zc3h13 regulates nuclear RNA m6A methylation and mouse embryonic stem cell self-renewal. Mol. Cell. 2018;69:1028–1038. doi: 10.1016/j.molcel.2018.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang Z, et al. VIRMA mediates preferential m6A mRNA methylation in 3’UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 4, 10 (2018). [DOI] [PMC free article] [PubMed]
- 17.Patil DP, et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:369–373. doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pendleton KE, et al. The U6 snRNA m6A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell. 2017;169:824–835. doi: 10.1016/j.cell.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma H, et al. N6-Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat. Chem. Biol. 2019;15:88–94. doi: 10.1038/s41589-018-0184-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tran N, Ernst FGM, Hawley BR, Zorbas C, Ulryck N, Hackert P, et al. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res.47, 7719–7733 (2019). [DOI] [PMC free article] [PubMed]
- 21.Zhao W, et al. Epigenetic regulation of m6A modifications in human cancer. Mol. Ther. Nucleic Acids. 2020;19:405–412. doi: 10.1016/j.omtn.2019.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu Y, Jia G, Pang X, Wang RN, Wang X, Li CJ, et al. FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nat. Commun. 4, 1798 (2013). [DOI] [PMC free article] [PubMed]
- 23.Mauer J, et al. Reversible methylation of m6Am in the 5’ cap controls mRNA stability. Nature. 2017;541:371–375. doi: 10.1038/nature21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mauer J, et al. FTO controls reversible m6Am RNA methylation during snRNA biogenesis. Nat. Chem. Biol. 2019;15:340–347. doi: 10.1038/s41589-019-0231-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei J, et al. Differential m6A, m6Am, and m1A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol. Cell. 2018;71:973–985.e5. doi: 10.1016/j.molcel.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng G, et al. Sprouts of RNA epigenetics: the discovery of mammalian RNA demethylases. RNA Biol. 2013;10:915–918. doi: 10.4161/rna.24711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ueda Y, et al. AlkB homolog 3-mediated tRNA demethylation promotes protein synthesis in cancer cells. Sci. Rep. 2017;7:42271. doi: 10.1038/srep42271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Theler D, Dominguez C, Blatter M, Boudet J, Allain FH. Solution structure of the YTH domain in complex with N6-methyladenosine RNA: a reader of methylated RNA. Nucleic Acids Res. 2014;42:13911–13919. doi: 10.1093/nar/gku1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi H, et al. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017;27:315–328. doi: 10.1038/cr.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roundtree IA, et al. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. Elife. 2017;6:e31311. doi: 10.7554/eLife.31311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsu PJ, et al. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017;27:1115–1127. doi: 10.1038/cr.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alarcón CR, et al. HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell. 2015;162:1299–1308. doi: 10.1016/j.cell.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu N, et al. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017;45:6051–6063. doi: 10.1093/nar/gkx141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T.N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature518, 560–564 (2015). [DOI] [PMC free article] [PubMed]
- 37.Meyer KD, et al. 5’ UTR m(6)A promotes cap-independent translation. Cell. 2015;163:999–1010. doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang H, et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018;20:285–295. doi: 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu R, et al. A novel m6A reader Prrc2a controls oligodendroglial specification and myelination. Cell Res. 2019;29:23–41. doi: 10.1038/s41422-018-0113-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alarcón CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519:482–485. doi: 10.1038/nature14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang H, Deng Q, Lv Z, Ling Y, Hou X, Chen Z, et al. N6-methyladenosine induced miR-143-3p promotes the brain metastasis of lung cancer via regulation of VASH1. Mol. Cancer18, 181(2019) [DOI] [PMC free article] [PubMed] [Retracted]
- 42.Chen X, et al. METTL14 Suppresses CRC progression via regulating N6-methyladenosine-dependent primary miR-375 processing. Mol. Ther. 2020;28:599–612. doi: 10.1016/j.ymthe.2019.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Berulava T, Rahmann S, Rademacher K, Klein-Hitpass L, Horsthemke B. N6-adenosine methylation in MiRNAs. PLoS ONE. 2015;10:e0118438. doi: 10.1371/journal.pone.0118438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shah A, et al. The DEAD-Box RNA helicase DDX3 interacts with m6A RNA demethylase ALKBH5. Stem Cells Int. 2017;2017:8596135. doi: 10.1155/2017/8596135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang J, Bai R, Li M, Ye H, Wu C, Wang C, et al. Excessive miR-25-3p maturation via N6-methyladenosine stimulated by cigarette smoke promotes pancreatic cancer progression. Nat. Commun. 10, 1858 (2019). [DOI] [PMC free article] [PubMed]
- 46.Park YM, Hwang SJ, Masuda K, Choi KM, Jeong MR, Nam DH, et al. Heterogeneous nuclear ribonucleoprotein C1/C2 controls the metastatic potential of glioblastoma by regulating PDCD4. Mol. Cell Biol. 32, 4237–4244 (2012). [DOI] [PMC free article] [PubMed]
- 47.Klinge CM, Piell KM, Tooley CS, Rouchka EC. HNRNPA2/B1 is upregulated in endocrine-resistant LCC9 breast cancer cells and alters the miRNA transcriptome when overexpressed in MCF-7 cells. Sci. Rep. 2019;9:9430. doi: 10.1038/s41598-019-45636-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Du M, Zhang Y, Mao Y, Mou J, Zhao J, Xue Q, et al. MiR-33a suppresses proliferation of NSCLC cells via targeting METTL3 mRNA. Biochem. Biophys. Res. Commun.482, 582–589 (2017) [DOI] [PubMed]
- 49.He H, Wu W, Sun Z, Chai L. MiR-4429 prevented gastric cancer progression through targeting METTL3 to inhibit m6A-caused stabilization of SEC62. Biochem. Biophys. Res. Commun. 2019;517:581–587. doi: 10.1016/j.bbrc.2019.07.058. [DOI] [PubMed] [Google Scholar]
- 50.Yang Z, et al. MicroRNA-145 modulates N6-methyladenosine levels by targeting the 3’-untranslated mRNA region of the N6-methyladenosine binding YTH domain family 2 protein. J. Biol. Chem. 2017;292:3614–3623. doi: 10.1074/jbc.M116.749689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu C, Yuan B, He T, Ding B, Li S. Prognostic values of YTHDF1 regulated negatively by mir-3436 in glioma. J. Cell Mol. Med. 10.1111/jcmm.15382.(2020) [DOI] [PMC free article] [PubMed]
- 52.Han J, et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol. Cancer. 2019;18:110. doi: 10.1186/s12943-019-1036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peng W, et al. Upregulated METTL3 promotes metastasis of colorectal Cancer via miR-1246/SPRED2/MAPK signaling pathway. J. Exp. Clin. Cancer Res. 2019;38:393. doi: 10.1186/s13046-019-1408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ma JZ, et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N6 -methyladenosine-dependent primary MicroRNA processing. Hepatology. 2017;65:529–543. doi: 10.1002/hep.28885. [DOI] [PubMed] [Google Scholar]
- 55.Yi D, et al. METTL14 promotes the migration and invasion of breast cancer cells by modulating N6-methyladenosine and hsa-miR-146a-5p expression. Oncol. Rep. 2020;43:1375–1386. doi: 10.3892/or.2020.7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen XP, Ling X, Lu H, Zhou CX, Zhang JK, Yu Q. Low expression of microRNA-1266 promotes colorectal cancer progression via targeting FTO. Eur. Rev. Med. Pharmacol. Sci. 22, 8220–8226(2018). [DOI] [PubMed]
- 57.Zhu H, et al. ALKBH5 inhibited autophagy of epithelial ovarian cancer through miR-7 and BCL-2. J. Exp. Clin. Cancer Res. 2019;38:163. doi: 10.1186/s13046-019-1159-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen Z, et al. Integrative analysis of NSCLC identifies LINC01234 as an oncogenic lncRNA that interacts with HNRNPA2B1 and regulates miR-106b biogenesis. Mol. Ther. 2020;28:1479–1493. doi: 10.1016/j.ymthe.2020.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wei W, Huo B, Shi X. miR-600 inhibits lung cancer via downregulating the expression of METTL3. Cancer Manag Res. 2019;11:1177–1187. doi: 10.2147/CMAR.S181058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cai X, Wang X, Cao C, Gao Y, Zhang S, Yang Z, et al. HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7g. Cancer Lett. 415, 11–19 (2018). [DOI] [PubMed]
- 61.Cui X, et al. Cross talk between RNA N6-methyladenosine methyltransferase-like 3 and miR-186 regulates hepatoblastoma progression through Wnt/β-catenin signalling pathway. Cell Prolif. 2020;53:e12768. doi: 10.1111/cpr.12768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li J, et al. Downregulation of N6-methyladenosine binding YTHDF2 protein mediated by miR-493-3p suppresses prostate cancer by elevating N6-methyladenosine levels. Oncotarget. 2017;9:3752–3764. doi: 10.18632/oncotarget.23365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kleemann M, et al. MiR-744-5p inducing cell death by directly targeting HNRNPC and NFIX in ovarian cancer cells. Sci. Rep. 2018;8:9020. doi: 10.1038/s41598-018-27438-6. [DOI] [PMC free article] [PubMed] [Google Scholar]