Abstract
We conducted a genome-wide association study of 1320 centenarians from the New England Centenarian Study (median age = 104 years) and 2899 unrelated controls using >9 M genetic variants imputed to the HRC panel of ~65,000 haplotypes. The genetic variants with the most significant associations were correlated to 4131 proteins that were profiled in the serum of a subset of 224 study participants using a SOMAscan array. The genetic associations were replicated in a genome-wide association study of 480 centenarians and ~800 controls of Ashkenazi Jewish descent. The proteomic associations were replicated in a proteomic scan of approximately 1000 Ashkenazi Jewish participants from a third cohort. The analysis replicated a protein signature associated with APOE genotypes and confirmed strong overexpression of BIRC2 (p < 5E−16) and under-expression of APOB in carriers of the APOE2 allele (p < 0.05). The analysis also discovered and replicated associations between longevity variants and slower changes of protein biomarkers of aging, including a novel protein signature of rs2184061 (CDKN2A/CDKN2B in chromosome 9) that suggests a genetic regulation of GDF15. The analyses showed that longevity variants correlate with proteome signatures that could be manipulated to discover healthy-aging targets.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11357-021-00376-4.
Keywords: SOMAscan array, Genetic variants, Molecular aging rate, Extreme human longevity
Introduction
Over the last decade, several studies have provided evidence that many centenarians and their offspring delay or escape aging-related diseases such as cardiovascular and Alzheimer’s disease and that more than 90% of people living to 100 are functionally independent at the mean age of 92 years and thus markedly delay disability [1, 2]. Many who live to 105 years and older, thus approaching the limit of human lifespan, compress towards the end of their very long lives both the age of onset of these diseases and disability [3]. While a variety of studies of centenarians have provided evidence for the compression of morbidity and disability [4], the genetic, molecular, and environmental determinants of this phenomenon remain elusive and the identification of the modifiable factors that allow centenarians to live long and healthy lives is still an open problem [5].
Evidence that extreme human longevity is heritable is solid: extreme longevity clusters in families [6], and siblings of centenarians have a much better chance of living to extremely old ages compared to their generation [7, 8]. Genome-wide association studies (GWASs) of centenarians have identified genetic variants that associate with extreme human longevity [5, 9–13], including the well-replicated association of APOE [14], FOXO3 [15], and CDKN2A/CDKN2B [16]. Most of the GWASs of extreme human longevity have focused on common genetic variants but the growing size of centenarian studies and the availability of large reference panels that support the accurate imputation of rare genetic variants have created the opportunity to examine the role of both common and rare genetic variants in extreme human longevity.
The discovery of genetic variants that correlate with extreme human longevity can help inform the biological mechanisms involved in this process. However, genetic variants often do not provide directly actionable targets. The functional annotation of these genetic variants through gene expression (eQTLs), or protein expression (pQTLs), provides a link to associated gene products and could point to therapeutic targets [17]. Evidence for an underlying genetic influence on drug-related mechanisms appears to substantially support drug development [18], and therefore, there is interest in identifying biomarker signatures of longevity-associated variants as possible targets for healthy-aging therapeutics.
Among multiple options for molecular biomarkers, for example, tissue-specific gene and protein expression, the serum proteome has emerged as a rich and easily accessible source of biomarkers. The SomaLogic’s aptamer technology provides a highly reproducible means of measuring thousands of serum proteins without the challenges posed by mass spectrometry [19, 20]. Several studies have shown that many serum proteins associate with genetic variations [21, 22], and we recently reported a robust signature of serum proteins that correlates with alleles of the APOE gene and changes in cognitive functions [23].
We therefore conducted a new GWAS of extreme human longevity that includes a large number of centenarians compared to previous studies [12], a dense panel of single nucleotide polymorphisms (SNPs) that includes common and rare variants and annotation of the most significant findings by their association with serum proteins. We also replicated both the genetic and SNP-protein associations in an independent study of centenarians and their offspring of Ashkenazi Jewish descent.
Materials and methods
Study populations and genetic data
New England Centenarian Study (NECS)
The NECS is a study of centenarians, their long-lived siblings, offspring, and additional unrelated controls selected because their parents died before reaching the median age survival of their birth-year cohort [24]. The study has enrolled participants since 1994 and it is still open for enrollment. Genome-wide genotype data of 2105 samples were previously generated using Illumina SNP arrays [24]. An additional 370 DNA samples including 284 centenarians were genotyped using Illumina Global Screening Array in 2019. The combined 2475 NECS subjects were imputed to the HRC panel (version r1.1 2016) of 64,940 haplotypes with 39,635,008 sites using the Michigan Imputation Server [25]. Eagle2 and European populations were selected for phasing and quality control respectively [26]. All subjects provided informed consent approved by the Boston University Medical Campus IRB.
Illumina controls
To increase statistical power, we included controls selected from the Illumina repository that comprises approximately 6000 samples of various races and ethnicities used as controls of a variety of GWASs (http://www.illumina.com/documents/icontroldb/document_purpose.pdf, March 2, 2020). We used this set of controls as a referent sample in the study of longevity since we expect that only a small portion of them would become centenarians, who have a prevalence of 1 per 5000 in the general US population. By pooling controls from different studies, we also do not expect sample-specific bias (e.g., controls not enriched for cardiovascular disease, or cancer). Genome-wide genotype data were generated with a variety of Illumina SNP arrays and data were carefully cleaned as described in [10]. We selected 3613 subjects to genetically match the European ethnicity of subjects from the NECS using genome-wide principal components. Genotype data were imputed to the HRC panel using the Michigan Imputation Server as in the NECS.
Longevity Genes Project (LGP)
This is a case-control study of individuals of Ashkenazi Jewish descent described in [27]. The study enrolled probands with exceptional longevity, defined as living independently at the age of 95, offspring of probands, and a third control group without a family history of exceptional longevity. Genotyping was completed using the HumanOmniExpress (Illumina, San Diego, CA, USA) array [13] and imputed to the HRC panel using the Michigan Imputation Server as in the NECS. For this study, 480 centenarians and ~800 controls of Ashkenazi Jewish descent with available genotype data were used. This study was approved by the Committee on Clinical Investigations at Albert Einstein College of Medicine.
LonGenity
LonGenity is an ongoing longitudinal study of health and longevity, initiated in 2008 [28]. The study enrolls older Ashkenazi Jewish adults who are free of significant cognitive impairment at enrollment. About half of the cohort has a parental history of exceptional longevity, defined as having one or more parents survive to 95 years of age or older while the other half does not. Participants undergo detailed evaluations at annual visits which include medical history, neurocognitive testing, collection of biological samples, and physical assessment. For this study, 733 individuals (342 males, 391 females, mean age 76 years at baseline) with available genotype and proteomic data were included. The LonGenity study was approved by the Institutional Review Board (IRB) at the Albert Einstein College of Medicine, and informed consent was obtained from all study participants.
GWAS analysis
GWAS in NECS
After removing duplicate, monomorphic, and ambiguous SNPs (AT, TA, GC, CG), we selected 9,039,731 SNPs of high imputation quality (Rsq > 0.7) for the genetic analysis. We designed a case-control study in the NECS and defined cases of extreme longevity those individuals who survived beyond an age reached by less than 1% of individuals in their sex and birth-year cohort (males: 96 years for 1900, 97 years for 1910, 98 years for 1920; females: 100 years) based on the US social security administration cohort tables [29]. Controls comprised referent subjects enrolled in NECS (n=306) and Illumina controls (n=2593). Participant characteristics are in Table 1. The association between SNPs and extreme longevity was tested using a mixed-effect logistic regression model using the case, control status as the outcome, and the SNP as the covariate. A random effect on the logit scale was included to account for cryptic relations. The random effect was assumed to be normally distributed with 0 mean and variance-covariance matrix proportional to the genetic relation matrix that was estimated using the R package PC-Relate [30]. The regression was adjusted by sex and 4 genome-wide principal components that were estimated using the R package PC-AIR. The analyses were conducted using the program GENESIS [31], and the significance of the associations was tested using the score test. Genome-wide significance was based on p-value < 5E−08. Significant results were annotated using ANNOVAR. See Table 2 for the top significant associations.
Table 1.
Characteristics of patients included in the GWAS
| s | Cases | Controls | Total | |
|---|---|---|---|---|
| NECS | Number of subjects | 1320 | 2899 | 4219 |
| Percent females | 73% | 59% | 63% | |
| Median age (range) | 104 (97–119) | --- | --- | |
| LGP | Number of subjects | 312 | 638 | 950 |
| Percent females | 63% | 51% | 55% | |
| Median age (range) | 102 (96–113) | --- | --- |
Table 2.
Top SNPs from the genome-wide significant loci in NECS
| SNP | Chr | Position | Ref/Alt | CAF NECS (cases/controls) | Score.Stat NECS | Score.pval NECS | CAF LGP (cases/controls) | Score.Stat LGP | Score.pval LGP | Genes |
|---|---|---|---|---|---|---|---|---|---|---|
| rs429358 | 19 | 45411941 | T/C | 0.06/0.14 | −11 | 3E−28 | 0.07/0.1 | −4 | 5E−04 | APOE |
| rs10973748 | 9 | 38322516 | G/T | 0.06/0.03 | 6 | 2E−09 | 0.04/0.03 | 1 | 0.3 | LOC107987065, SHB, ALDH1B1 |
| rs77546126 | 18 | 71518518 | T/G | 0.01/0.002 | 6 | 3E−09 | 0.007/0.006 | 0 | 0.9 | LOC10050581, FBXO15 |
| rs78043944 | 3 | 26368500 | G/A | 0.02/0.007 | 6 | 4E−09 | 0.0001/0.00007 | 0 | 0.7 | LINC00692, LRRC3B |
| rs77184423 | 15 | 29322932 | A/G | 0.01/0.004 | 6 | 1E−08 | 0.03/0.02 | 1 | 0.4 | APBA2 |
| rs114658003* | 6 | 100051300 | G/A | 0.007/0.0004 | 6 | 4E−08 | 0/0.000002 | −1 | 0.6 | CCNC, PRDM13 |
| rs649357 | 5 | 29426436 | A/C | 0.58/0.64 | −6 | 4E−08 | --- | --- | --- | LINC02064, LOC105374704 |
| rs78441534 | 18 | 47609371 | C/T | 0.005/0.0003 | 5 | 5E−08 | --- | --- | --- | MYO5B |
Chr, chromosome; Ref/Alt, reference and alternative alleles; Position, position of SNP on Grch27 reference panel; CAF (cases/controls) , coded allele frequencies in cases and controls (alternative allele (Alt) is the coded allele in all the analyses); Score.Stat, the score Z test statistic; Score.pval, the score p-value based on the score test statistic; Genes, closest gene/genes.
*The frequency of this SNP in dbSNP suggests that this variant is more frequent in Europeans.
Replication of GWAS results in LGP
Genetic analyses in the LGP were conducted as in the NECS using the same computational pipeline. We identified 56 SNPs that reached a 5E−06 suggestive level of significance in the NECS and replicated the association with nominal significance in the LGP. This list included rs7412 and rs429358 that define the APOE alleles (e2 : rs7412 = T, rs429358 = T; e3 : rs7412 = C, rs429358 = T; e4 : rs7412 = C, rs429358 = C). The set of APOE alleles and the remaining 54 SNPs were tested as pQTLs in the analyses described next.
Proteomic analysis
NECS proteomic data
Serum samples of 77 centenarians (mean age 106 years (SD 3.7), 82 centenarians’ offspring: mean age 71 years (SD 9.1), and 62 controls, mean age 71 years (SD 5)), were profiled using a SOMAscan array custom-made for Novartis that included 5034 aptamers linked to 4387 unique proteins [22] (Table 3). NECS participants were selected to be alive at least 1 year after the blood draw, and free of major aging-related diseases and treatment at least 1 year from the time of the blood draw. The samples were randomized into analytic batches of 84 samples or less, and the data of 4785 aptamers targeting 4116 unique human proteins passed a quality control assessment for median intra- and inter-assay variability similar to variability previously reported in the SOMAscan assays [32]. The data readout from the SOMAscan-based proteomics is relative fluorescence units (RFUs) and is proportional to the reported relative abundances of the targets of the SOMAmer reagents.
Table 3.
Subjects with SOMAscan protein data—NECS
| Centenarians | Offspring | Controls | |
|---|---|---|---|
| Numbers | 77 | 82 | 62 |
| Mean age at serum yrs (SD) | 105.7 (3.7) | 71.0 (9.1) | 70.6 (5.2) |
| Mean age at last contact | 107.5 | 80.2 | 78.7 |
| % alive (as of December 2017) | 31% | 84% | 84% |
| % Female | 66% | 66% | 55% |
Mean age at serum yrs (SD): mean age at serum draw in years with standard deviation; mean age at last contact: mean age at death for deceased subjects and age at the last follow-up for those who are still alive (based on the 2017 follow-up).
LonGenity proteomic data
Proteomics samples from 733 human plasma samples (342 males, 391 females, mean age 76, SD 6.8) collected at the initial baseline visit were used for this study. A SOMAscan platform with 5284 aptamers was used to measure 4953 unique proteins [33]. The platform used in this study shared 3642 aptamers with good quality data with the platform used in the NECS.
APOE protein signature
The association between the APOE genotypes (e2e2, e2e3, e2e4, e3e3, e3e4, e4e4) with protein abundance were estimated using an ANOVA type analysis, in which the aptamer RFU in log scale was the outcome, and the APOE genotypes were the covariate. All analyses were adjusted for age at blood draw and sex, in both the NECS and the LGP.
pQTL analyses in NECS
Each of 54 SNPs that passed a level of significance < 5E−06 in the GWAS conducted in the NECS was annotated by their associations with all 4785 aptamers, resulting in 258,390 SNP by aptamer pairwise associations. The expression data (RFU) of 4785 aptamers were log-transformed and, for each aptamer, values that differed from the mean by more than three standard deviations were removed, resulting in approximately 1% missing RFU values. SNP-to-aptamer associations were tested using a linear regression of log-transformed RFU versus SNP, and adjusted by gender and age at the time of blood draw. To distinguish aptamers associated with the APOE alleles from those associated with SNPs in the APOE locus, the analysis for SNPs in this region was adjusted by the APOE genotypes (with each of the six possible diploid genotypes coded as a binary variable).
pQTL analysis in LGP
We identified 3642 aptamers shared between the SOMAscan platforms used in the NECS and the LonGenity cohorts. PQTL analyses were conducted as described above using Python 3.6 and resulted in 196,668 SNP, aptamer pairwise associations.
Meta-analysis of pQTLs
The estimates of the regression coefficients and standard errors of the 196,668 common pQTL analyses in NECS and LGP were aggregated using a fixed-effect meta-analysis (package rmeta in R). We selected significant aptamer-SNP pairs using a false discovery rate (FDR) < 0.05 as level of significance to correct for multiple testing.
Enrichment analysis
Enrichment for various biological pathways was conducted using the program hyper [34], using the list of proteins shared in the two SOMAscan arrays as background. We choose two reference sets: the “Hallmarks” compendium, which comprises 50 sets of genes defining specific well-defined biological states or processes and displaying coherent expression and is available through MSigDB (http://software.broadinstitute.org/gsea/msigdb/index.jsp), and REACTOME v7.2.1.
All analyses were conducted using the statistical software R version 3.6.0 and Rstudio version 1.3.1073.
Results
Genome-wide association study
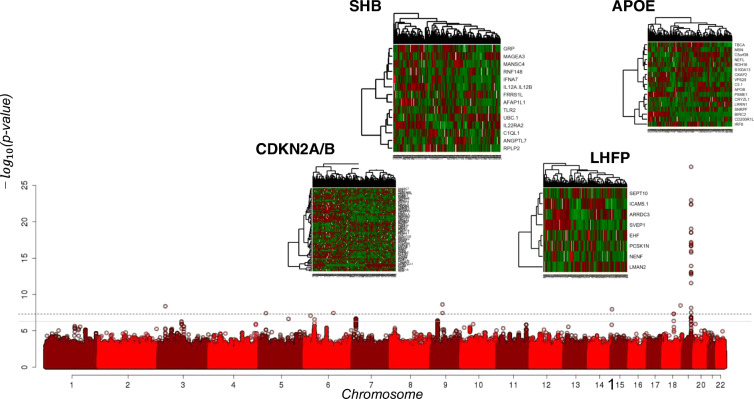
The GWAS in the NECS included 1320 individuals who survived past the age reached by less than 1% of their birth-year cohort (median age at death 104 years), and 2899 controls (Table 1). The genetic analyses identified 33 SNPs in 8 loci in chromosomes 3, 5, 6, 9, 15, 18, and 19 that passed the genome-wide significance threshold p=5 × 10−8 (Fig. 1). The full list of 33 SNPs is described in Supplement Table 1, while Table 2 lists the most significant SNP in each locus. Twenty-two of the SNPs tagged a locus in chromosome 19 that includes APOE, thus confirming a well-established association of SNPs in this locus with extreme human longevity [14]. Four SNPs were uncommon variants in the non-coding RNA LOC107987065 on chromosome 9, between SHB and ALDH1B1; three SNPs were rare variants in a region in chromosome 18 that includes MYO5B, LOC10050581, and FBXO15. Additional SNPs are listed in Table 2 and include one common variant and additional rare SNPs. Only the SNPs in APOE replicated in the LGP study, while the other associations did not reach statistical significance in the LGP (Table 2).
Fig. 1.
Manhattan plot of the genome-wide association with extreme human longevity in the New England Centenarian Study. The x-axis reports chromosomes and coordinates within chromosomes. The y-axis reports the −log10 (p-value). The 4 heatmaps report the log-transformed standardized protein expression data (rows) for the 224 individuals (columns) included in the serum protein SOMAscan experiment. The heatmap for the APOE locus includes only the top significant proteins
We noticed that among the 219 SNPs that reached a 5E−06 suggestive level of significance in the NECS, 56 replicated the association with nominal significance in the LGP, which is a 5-fold increase compared to the number of associations expected by chance. This set of 56 SNPs comprised 23 SNPs in the locus of APOE, including rs7412 and rs429358 that define the APOE alleles, two SNPs in the locus tagged by rs10973748 in chromosome 9, one SNP in the gene LHFPL6 in chromosome 13, and 30 common SNPs in the gene CDKN2B in chromosome 9. These 56 SNPs are summarized in Supplement Table 2. Interestingly, the SNP rs9576827 in LHFPL6 was in a region enriched for SNPs associated with aging-related traits such as hippocampal degeneration (rs9315702 [35]) and age at menarche (rs6563739 [36]). The most significant SNPs in the gene CDKN2B included rs1556515 that was associated with parental longevity in the UK Biobank with consistent effects (less common allele increased the odds for parental longevity) [37] and rs2184061 that also showed a consistent association with parental lifespan in a large meta-analysis of human longevity [13].
APOE signature
We investigated the association of aptamers with APOE genotypes in the LGP study and replicated 10 of the 16 aptamers included in the serum protein signature of APOE that we published in [23]. Five aptamers were not available on the SOMAscan array used in the LGP study (Table 4), and the association of CKAP2 failed to reach statistical significance (p=0.1243).
Table 4.
Replication of the APOE protein signature
| SomaID | UniProt | GeneID | NECS | LGP | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| e2e2 | e2e3 | e2e4 | e3e4 | p | e2e2 | e2e3 | e2e4 | e3e4 | e2e2 | p | |||
| 10046-55 | Q13490* | BIRC2 | 5.87 | 3.23 | 3.67 | 0.90 | 1.55E−61 | 1.94 | 2.03 | 0.95 | 0.84 | 5.09E−134 | |
| 11276-1 | Q86XR8* | CEP57 | 1.62 | 1.23 | 1.22 | 1.01 | 1.96E−28 | ||||||
| 7223-60 | Q99584 | S100A13 | 0.51 | 0.78 | 0.47 | 0.67 | 2.69E−23 | 0.18 | 0.53 | 0.16 | 0.46 | 0.16 | 2.26E−108 |
| 11293-14 | Q6UXK5 | LRRN1 | 0.89 | 0.96 | 1.17 | 1.4 | 1.17E−19 | 1.00 | 0.97 | 1.46 | 1.55 | 2.10 | 5.61E−105 |
| 14318-1 | Q9UBQ0 | VPS29 | 1.55 | 1.18 | 1.26 | 0.98 | 2.84E−19 | 1.80 | 1.58 | 0.90 | 0.90 | 3.52E−98 | |
| 5918-5 | Q06323* | PSME1 | 1.51 | 1.27 | 1.22 | 0.99 | 5.23E−19 | 1.27 | 1.07 | 0.99 | 1.01 | 0.95 | 0.174632 |
| 2418-55 | P02649* | APOE | 0.86 | 0.77 | 1.15 | 1.16 | 9.19E−11 | ||||||
| 12501-10 | O75347* | TBCA | 1.08 | 1.02 | 0.88 | 0.83 | 1.60E−10 | 0.81 | 0.83 | 0.43 | 0.56 | 0.31 | 1.09E−89 |
| 12500-88 | Q9UBT2* | UBA2 | 1.77 | 1.13 | 1.31 | 0.98 | 5.44E−10 | ||||||
| 6378-2 | Q86SI9 | C5orf38 | 0.73 | 0.84 | 0.88 | 1.29 | 7.68E−10 | 0.90 | 0.75 | 0.84 | 1.08 | 1.31 | 1.30E−29 |
| 13732-79 | Q16619 | CTF1 | 0.94 | 0.95 | 1.2 | 1.11 | 9.63E−09 | ||||||
| 11402-17 | Q8NEZ4* | KMT2C | 1.33 | 1.11 | 1.06 | 0.99 | 2.15E−08 | 0.73 | 1.06 | 0.75 | 0.94 | 1.14 | 0.035255 |
| 14643-27 | O60870* | KIN | 1.23 | 1.08 | 1.22 | 1.02 | 3.10E−07 | ||||||
| 2797-56 | P04114* | APOB | 0.5 | 0.86 | 0.97 | 1.07 | 3.36E−06 | 0.83 | 0.95 | 0.98 | 1.05 | 1.22 | 0.049591 |
| 9207-60 | O95825 | CRYZL1 | 0.89 | 0.88 | 0.7 | 1.11 | 7.07E−06 | 1.12 | 0.68 | 0.49 | 0.83 | 0.79 | 2.68E−23 |
| 5345-51 | Q8WWK9 | CKAP2 | 1.33 | 1.12 | 1.08 | 0.96 | 8.12E−06 | 0.94 | 1.05 | 0.98 | 0.99 | 1.08 | 0.124289 |
Columns e2e2, e2e3, e2e4, and e3e4 report fold change of protein level relative to e3e3 in the NECS and LGP. P is p-value from F test with 4 and 214 degrees of freedom, after adjusting for sex, age at blood draw, and length of sample storage. The aptamers 11276-1, 2418-55, 12500-88, 13732-79, and 14643-27 were not available in the SOMAscan array used in the LGP study
SNP × protein associations
We next looked for serum proteins that associated with the 54 SNPs associated with extreme longevity (21 in the APOE locus after the exclusion of SNPs rs7412 and rs429358 that define the APOE alleles; 30 in the CDKN2B locus; two in the locus tagged by rs10973748 in chromosome 9; and one in the gene LHFPL6 in chromosome 13). We analyzed the associations between these 54 SNPs in the NECS and LGP data separately. We then used a fixed-effect meta-analysis to aggregate the results from the two studies and selected aptamers that correlated with each cluster of SNPs using a 5% FDR. The analyses of the 21 SNPs in the locus of APOE were adjusted by the APOE genotypes and no significant pQTL was found in this list after correction for multiple comparisons (Supplement Table 3). The analysis of the other 33 SNPs identified 3 new serum protein signatures and the heatmaps of these signatures are in Fig. 1.
Eight aptamers targeting 8 unique proteins were associated with the SNP rs9576827 in LHFPL6 (Supplement Table 4), including SVEP1 and ARRDC3 that are overexpressed in centenarians, and LMAN2 that is low in centenarians compared to younger individuals [38]. The analysis of the two SNPs rs341466 and rs341467 in the locus tagged by rs10973748 in chromosome 9 generated 24 significant SNP by aptamer associations, for a total of 14 proteins with one or two pQTLs (Supplement Table 5). Using the list of 30 SNPs in the locus of the CDKN2B gene, we identified 330 pairs of significant SNP by aptamer associations, for a total of 69 unique proteins with one or more pQTLs (Supplement Table 6). The set of 69 proteins was enriched for genes in the allograft rejection pathway (p=0.0037, 19% FDR), estrogen response late (p=0.01, 29%FDR), and nominally enriched of genes involved in mitotic spindle check-point (p=0.0037), with digestion and absorption (p=0.0092), and digestion of carbohydrate (p=0.0092).
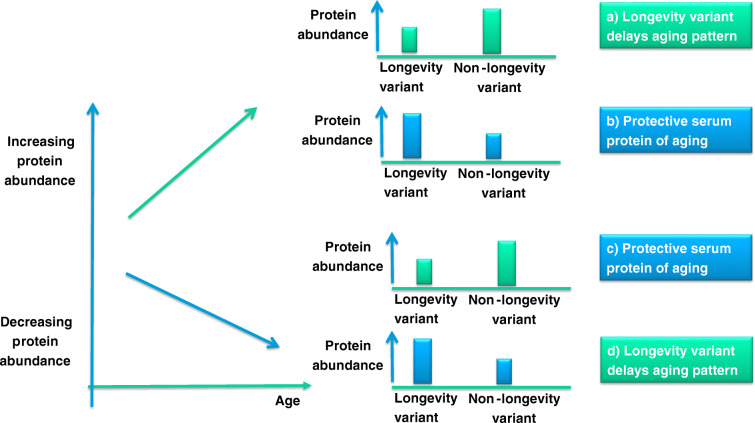
Longevity variants and aging biomarkers
In each of the four signatures, we selected the subset of proteins that change with age and/or with extreme longevity based on the analysis in [38]. We grouped these proteins into those that increase or decrease with older age, and those that are higher or lower in carriers of the longevity variants (see schematic in Fig. 2). By combining the age and longevity-allele patterns, we identified two groups of protein/genotype relations. The first group included the proteins that increase with older age and are lower in carriers of the longevity variant and proteins that decrease with older age and are higher in carriers of the longevity variant. The discordant change patterns in aging and longevity alleles in this group suggest that carriers of the longevity variants delay changes related to aging and maintain a more youthful profile of these serum proteins for a longer time. The second group included proteins that increase with older age and are higher in carriers of the longevity variants and proteins that decrease with older age and are lower in carriers of the longevity variants. The concordant change patterns in aging and longevity alleles in this group and the fact that the abundance of these proteins at older age matches that of the longevity variant carriers suggest that these proteins may be protective biomarkers of aging.
Fig. 2.
Summary of concordant and discordant protein effects. The figure summarizes 4 possible relations between longevity genetic variants and serum proteins of aging. Serum proteins of aging can increase (green) or decrease (blue) with older age, and carriers of the longevity variants can have lower or higher values of these proteins. When the age and longevity change patterns are disconcordant (a and d), we hypothesize that the longevity variants delay age-related accumulation of damage. When the age and longevity patterns are concordant (b and c), then we hypothesize that changes of those proteins with age may actually be protective
The APOE protein signature included 4 proteins that are very different in the centenarian serum compared to centenarians’ offspring and controls and are part of a proteomic signature of centenarians described in [23]. This set is summarized in Table 4 and detailed in Supplement Table 7, and included S100 calcium-binding protein 13 (S100A13), which increases with older age (Table 5) and is lower in carriers of the e2 allele compared to e3 carriers; leucine-rich repeat neuronal 1 (LRR1) that decreases with older age and is lower in carriers of the e2 allele compared to e3; tubulin-folding cofactor A (TBCA) that increases with older age and is also higher in carriers of the e2 variant; and apolipoprotein B (APOB) that is lower in carriers of the e2 allele compared to e3, and it increases with older age. The results suggest that the e2 allele of APOE promotes longevity by delaying aging-related changes of some serum proteins (e.g., lower S100A13, LRR1, and APOB) and possibly correlates with protective serum protein changes (e.g., higher TBCA).
Table 5.
Aging and longevity pQTLs
| Locus | SNP | SOMAmer ID | UniProt | Targeted protein | Longevity allele | Longevity variant trend | Aging trend | Replicated | Gene name |
|---|---|---|---|---|---|---|---|---|---|
| APOE | APOE alleles | 11293-14_3 | Q6UXK5 | LRRN1 | e2 | D | D | Yes | Leucine-rich repeat neuronal 1 |
| 7223-60_3 | Q99584 | S100A13 | e2 | D | U | Yes | S100 calcium-binding protein A13 | ||
| 2797-56_2 | P04114 | APOB | e2 | D | U | Yes | Apolipoprotein B | ||
| 12501-10_3 | O75347 | TBCA | e2 | U | U | Yes | Tubulin-folding cofactor | ||
| LHFPL6 | rs9576827 | 12352-70_3 | Q96B67 | ARRDC3 | G | U | U | Yes | Arrestin domain containing 3 (ARRDC3) |
| rs9576827 | 7638-30_3 | Q12907 | LMAN2 | G | D | D | Yes | Lectin, mannose binding 2 | |
| rs9576827 | 9219-70_3 | Q9UMX5 | NENF | G | U | U | No | Neudesin neurotrophic factor | |
| rs9576827 | 11109-56_3 | Q4LDE5 | SVEP1 | G | D | U | Yes | Sushi, von Willebrand factor type A, EGF, and pentraxin domain containing 1 (SVEP1) | |
| SHB-ALDH1B1 | rs341467 | 6647-55_3 | P0CG48 | UBC | A | U | D | Yes | Ubiquitin C (UBC) |
| rs341467 | 5087-5_3 | Q969J5 | IL22RA2 | A | D | D | Yes | Interleukin 22 receptor subunit alpha 2 | |
| rs341466 | 6404-20_3 | O75973 | C1QL1 | C | U | U | Complement C1q like 1 (C1QL1) | ||
| CDKN2B | rs10757275 | 4546-27_3 | Q9UHX3 | ADGRE2 | G | D | U | No | Adhesion G protein-coupled receptor E2 (ADGRE2) |
| rs6475609 | 12352-70_3 | Q96B67 | ARRDC3 | A | D | U | Yes | Arrestin domain containing 3 (ARRDC3) | |
| rs7857345 | 7100-31_3 | P06729 | CD2 | T | D | U | No | CD2 molecule (CD2) | |
| rs1537375 | 11696-7_3 | P29373 | CRABP2 | T | D | U | No | Cellular retinoic acid binding protein 2 (CRABP2) | |
| rs10757275 | 7968-15_3 | O95727 | CRTAM | G | D | U | No | Cytotoxic and regulatory T-cell molecule (CRTAM) | |
| rs7341786 | 4374-45_2 | Q99988 | GDF15 | A | D | U | Yes | Growth differentiation factor 15 (GDF15) | |
| rs8181050 | 2771-35_2 | P08833 | IGFBP1 | G | D | U | No | Insulin-like growth factor–binding protein 1 (IGFBP1) | |
| rs1412834 | 14054-17_3 | Q13261 | IL15RA | T | D | U | Yes | Interleukin 15 receptor subunit alpha (IL15RA) | |
| rs7857345 | 6227-1_3 | O43240 | KLK10 | T | D | U | No | Kallikrein-related peptidase 10 (KLK10) | |
| rs1537375 | 7551-33_4 | Q14392 | LRRC32 | T | D | U | No | Leucine-rich repeat containing 32 (LRRC32) | |
| rs7030641 | 3628-3_4 | P36507 | MAP2K2 | C | D | U | No | Mitogen-activated protein kinase kinase 2 (MAP2K2) | |
| rs7857345 | 8479-4_3 | P09238 | MMP10 | T | D | U | No | Matrix metallopeptidase 10 (MMP10) | |
| rs1412834 | 2944-66_2 | P41271 | NBL1 | T | D | U | No | Neuroblastoma 1, DAN family BMP antagonist (NBL1) | |
| rs1537374 | 9765-4_3 | Q9NXR1 | NDE1 | A | D | U | No | nudE neurodevelopment protein 1 (NDE1) | |
| rs10757275 | 5646-20_3 | Q93091 | RNASE6 | G | D | U | No | Ribonuclease A family member k6 (RNASE6) | |
| rs6475609 | 6990-44_3 | Q9HCN8 | SDF2L1 | A | D | U | Yes | Stromal cell derived factor 2 like 1 (SDF2L1) | |
| rs6475609 | 9805-51_3 | Q9NPR2 | SEMA4B | A | D | U | No | Semaphorin 4B (SEMA4B) | |
| rs1537375 | 9191-8_3 | Q03403 | TFF2 | T | D | U | Yes | Trefoil factor 2 (TFF2) | |
| rs10757275 | 9409-11_3 | Q15661 | TPSAB1 | G | D | U | No | Tryptase alpha/beta 1 (TPSAB1) | |
| rs10757275 | 3181-50_2 | P25774 | CTSS | G | D | D | No | CATHEPSIN S | |
| rs4451405 | 7156-2_3 | Q6P4F1 | FUT10 | C | D | U | No | FUCOSYLTRANSFERASE 10 | |
| rs7857345 | 5465-32_3 | O60243 | HS6ST1 | T | D | D | No | Heparan sulfate 6-O-sulfotransferase 1 | |
| rs1360589 | 11237-49_3 | Q15113 | PCOLCE | C | D | U | No | Procollagen C-endopeptidase enhancer | |
| rs10757275 | 9190-7_3 | P08962 | CD63 | G | D | D | Yes | cubilin (CUBN) | |
| rs10757275 | 9111-40_3 | Q96JJ6 | JPH4 | G | D | D | No | RNA guanylyltransferase and 5′-phosphatase (RNGTT) | |
| rs8181050 | 3310-62_1 | P31994 | FCGR2B | G | U | U | No | NGFI-A binding protein 1 (NAB1) | |
| rs10757275 | 7918-114_3 | P04745 | AMY1A | G | U | U | No | Amylase, alpha 1A (salivary) | |
| rs1537375 | 3600-2_3 | Q13231 | CHIT1 | T | U | U | No | Chitinase 1 | |
| rs7341791 | 7089-42_3 | Q9BT88 | SYT11 | A | U | U | No | Synaptotagmin 11 | |
| rs8181050 | 2333-72_1 | P01137 | TGFB1 | G | U | U | No | Transforming growth factor beta 1 |
Locus, genes with SNPs used in the pQTL analysis; SNP, used in the pQTL analysis; SOMAmer ID of the SomaLogic aptamer; UniProt ID; Targeted protein, protein targeted by the aptamer; Longevity allele, allele more prevalent in centenarians; Longevity variant trend: change of the protein in carriers of the longevity allele versus the non-longevity allele. U denotes a higher value is associated with the longevity variant, D denotes a lower value is associated with the longevity variant. Aging trend, U denotes older people have increased values compared to younger people while D denotes older people have lower values compared to younger controls. Replicated, yes if the aging trend was replicated in multiple studies
The protein signature associated with the SNP rs9576827 in LHFPL6 (Supplement Table 8) included Sushi, von Willebrand factor type A, EGF, and pentraxin domain containing 1 (SVEP1), arresting domain containing 3 (ARRDC3), and neudesin neutrophic factor (NENF) that are overexpressed in centenarians and increase with older age, and lectin, mannose binding 2 (LMAN2) that is lower in centenarians compared to younger individuals and decrease with age [38]. Serum level of SVEP1 was lower in carriers of the longevity variants, while serum levels of NENF, ARRDC3, and LMAN2 in carriers of the longevity variants matched the patterns of older individuals (Table 5).
The signature associated with the SNPs in the locus tagged by rs10973748 in chromosome 9 included ubiquitin C (UBC) and interleukin 22 receptor subunit alpha 2 (IL22RA2) that decrease with age, and C1QL1 that increases with age. Carriers of the longevity variants had lower levels of the cytokine IL22RA and C1QL1, and higher levels of UBC (Table 5).
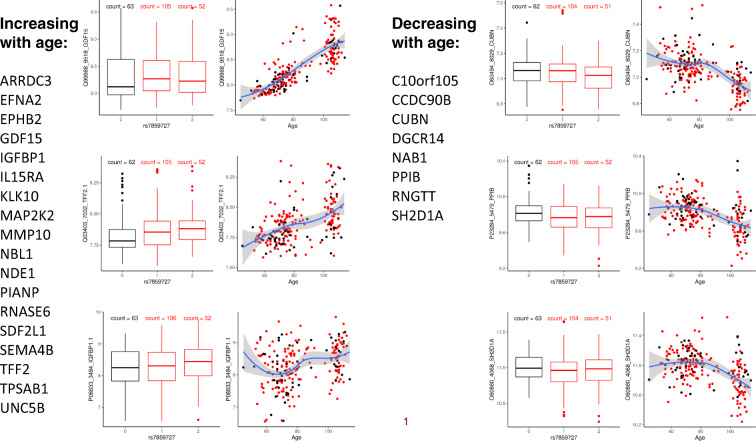
The protein signature of the locus in CDKN2B included 38 (40%) proteins that are strongly differentially expressed in centenarians and older age [38] and, for 29 of the 38, the change of protein expression in carriers of the longevity variants was in the opposite direction of the changes of expression in centenarians compared to younger individuals (Table 5). The levels of 21 proteins that were overexpressed at older age were lower in carriers of the longevity variants, and the levels of 8 proteins that were under-expressed at older age were higher in carriers of the longevity variants. Some examples are in Fig. 3 and include for example GDF15. This protein increases with older age, consistent with results from other studies [20], but it is on average lower in carriers of the longevity variants that we found in CDKN2B. The pattern suggests that individuals with the longevity variants tend to have lower values of this protein and their serum protein profile remains younger for a longer time.
Fig. 3.
Examples of pQTLs in CDKN2B. Example of 6 proteins that correlate with genotypes of the SNP rs7857345 in the CDKN2B that was associated with extreme human longevity in the genome-wide association studies in the New England Centenarian Study and LonGenity Gene Project. For each protein: the boxplots on the left show the distribution of the log-transformed protein data by genotype group (black = homozygote genotype for the longevity allele; red = genotypes on carriers of 1 or 2 non-longevity alleles); the scatter plot on the right shows the distribution of the log-transformed protein data (y-axis) versus the age of study participants (x-axis). The 3 plots on the left show 3 proteins that increase with age and are on average lower in carriers of the longevity-associated variant (black). The 3 plots on the right show 3 proteins that decrease with age and are on average higher in carriers of the longevity-associated variant
Discussion
We conducted a GWAS of extreme human longevity on a large number of centenarians from the NECS. The analysis discovered 33 genome-wide significant SNPs (p < 5E−08) in 8 loci in chromosomes 3, 5, 6, 9, 15, 18, and 19. Among the significant SNP associations was the well-established APOE locus [14] that replicated in the LGP. We then identified a set of 56 SNPs with significant and consistent associations with extreme human longevity in both studies (p< 5E−06 in NECS and p<0.05 in LGP) and annotated these SNPs with their association with serum proteins that were profiled using the SomaLogic technology. With this analysis, we replicated a protein signature of APOE alleles [23] and identified 3 new serum protein signatures of longevity-associated loci in chromosomes 9 and 13.
The genetic analysis identified a few uncommon and some rare variants that were associated with extreme human longevity in the NECS. All of these variants were more prevalent in centenarians—although still uncommon. The variant rs10973748 in the non-coding RNA LOC107987065, between genes SHB and ALDHB1, was in a region enriched for GWAS findings linked in particular to metabolism and cardiovascular disease in early GWASs, although those findings were not replicated [39]. The variants rs77546126 and rs78043944 were intragenic, in regions with little annotation. The variant rs77184423 was in the gene Amyloid beta precursor protein-binding family A member 2 (APB2) that is potentially very interesting for extreme human longevity. This gene encodes a neuronal adapter protein that stabilizes the amyloid precursor protein (APP) and inhibits the production of proteolytic APP fragments that are found in the brains of Alzheimer’s disease patients [40]. The SNP was also more prevalent in Ashkenazi Jewish centenarians than controls, but the association did not reach statistical significance. The rare SNP rs78441534 in myosin VB (MYO5B) was present in 2/10,000 chromosomes in gnomAD and absent in the Ashkenazi Jewish sample. This gene was linked to old age and cancer resistance in giant tortoises [41], but no evidence of a role in human longevity is known. These results point to possible novel genes involved with human extreme longevity and support the hypothesis that rare variants contribute to the susceptibility to extreme human longevity.
Most of the GWASs of human longevity to date have focused on common variants, and the yield of results has been limited [13]. In our study, we leveraged the mean-oldest sample published to date and improved techniques of imputation to analyze the association between extreme human longevity and rare variants imputed with high quality. The results are promising but also show the difficulty to replicate the finding of rare variants in independent samplers. In our replication study, the number of carriers of rare variants was too small to reach statistical significance. Ideally, large families enriched for longevity would be more powerful to discover and/or replicate the effects of rare variants. In addition, the data for SNP rs114658003 were imputed with good quality (score > 0.7) but the allele frequency of the imputed SNP is substantially different from that reported in dbSNPs for Europeans, thus lowering the confidence that this association is a true positive. Sequence data will be necessary to validate this association.
Despite the relatively small sample size, the yield of findings in the pQTL analysis was substantial. Other studies have shown that many SNPs are QTLs for serum proteins and the data can be used to suggest biological mechanisms that link genotype to phenotype [22]. Our analysis replicated 10 of the associations between APOE alleles and serum proteins that we reported in [23], with an almost perfect concordance of effects for the more common alleles. In addition, the current analysis extended the signature pattern to include carriers of the e4e4 genotype that was not represented in our original work, since no study participants had this genotype. Of particular note is the replication of the substantially lower expression of APOB in carriers of e2 versus carriers of e3e3, and the estimated 22% increase in carriers of e4e4. These results agree with findings found in other populations [42, 43]. Serum level of APOB is an important biomarker of cardiovascular disease, and although it correlates with other lipoproteins, it increases the risk assessment for cardiovascular disease [44]. Interestingly, serum level of APOB increases with older age but is lower in people with extreme longevity than in controls (Table 5, and [38]). These results agree with the beneficial effect of a lower level of APOB and a genetic regulation of this biomarker through the alleles of the APOE gene. Higher serum levels of the putative neuroprotectors BIRC2 and PSME1 in carriers of the e2 allele were also strongly replicated as well as the strong association between levels of LRRN1 and the e4 allele. In our original analysis, we detected overexpression of LRRN1 in e3e4 carriers, and the current analysis shows a substantial 2-fold increase in carriers of e4e4. Little annotation is available about this protein but several results, including data in the protein atlas suggesting that LRRN1 levels in serum decrease with age (Blood atlas - LRRN1 - The Human Protein Atlas), are significantly lower in healthy centenarians (Supplement Table 1 in [38]), and in carriers of the e2 allele. The results point to a protective mechanism possibly linked to APOE alleles that results in lower abundance of this protein in blood serum. Whether LRRN1 is simply a biomarker or an interesting target for therapeutics remains to be established. We also found BIRC2 to be associated with longevity in a prior study, associating protein changes with the APOE signature in centenarians [38]. As noted in that paper, baculoviral IAP repeat containing 2 (BIRC2) has been reported to be neuroprotective, and it can inhibit apoptosis [45, 46]. BIRC2 has also been characterized for its ability to regulate the noncanonical NF-kappaB pathway [47], though it also activates canonical NF-kappaB signaling [48]. The reproducibility of this marker, and its upregulation in e2 carriers, certainly invites some particular scrutiny as to using it as a biomarker for “healthy aging,” and potentially a mediator of preserved function. PSME1 is the proteasome activator subunit 1—its upregulation may simply be a readout of enhanced proteasome function in e2 carriers, consistent with the need to continue to clear misfolded proteins—perhaps, this is evidence of maintenance of elevated proteasome function, and thus preserved ability to degrade proteins that need to be cleared from the system.
The protein signatures associated with the rare variants in chromosomes 9 and 13 suggest potentially interesting biomarkers of longevity that are genetically regulated. The rare SNP rs9576827 in LHFPL6 correlated with higher levels of NENF that is also higher in centenarians and increases with age (Table 5). NENF may increase neuronal survival [49], and higher levels could be associated with the ability of centenarians to preserve good cognitive function as they age. The SNP is also a pQTL for Sushi, von Willebrand factor type A, EGF, and pentraxin domain containing 1 (SVEP1), which is a serum protein that increases with age [38], but it is lower in carriers of the rare longevity variant. The associated gene is highly expressed in fat, lung, platelets, and placenta, and variants of the gene are associated with cardiovascular disease risk, although the mechanism is still unclear. The association between rare variants in the SHB locus and IL22RA2 suggests a genetic basis for enhanced immune response in people predisposed to extreme longevity.
The SNPs in the CDKN2B gene were associated with the largest signature of 69 proteins (Supplement Table 6) that included 54% aging-related proteins (Supplement Table 7). An initial GWAS of centenarians provided suggestive evidence that SNPs in this gene are associated with extreme human longevity but the results did not reach a genome-wide level of significance [10]. A study of parental longevity in the UK Biobank with 500K DNA samples recently showed an association of rs2184061 with parental longevity with genome-wide significance [37]. In the current analysis, we did not detect a genome-wide significant level of association for any of the 30 SNPs selected for the pQTL analysis, but our sample size is substantially smaller. All these results however provide strong evidence that variants of CDKN2B have a role in human longevity although the biological mechanism is still unknown. Standard enrichment analysis based on the analysis of the proteins with significant pQTL did not produce strong results. It is possible that alternative procedures that include information about differential expression, like GSEA [50], may be more powerful but more work is needed to adapt them to QTL analysis in which a single protein may be significantly associated with multiple genetic variants.
The serum protein signature that we discovered for this locus shows that carriers of the longevity variants of this gene have a more youthful profile of known aging biomarkers including lower levels of GDF15 [51], insulin growth factor binding protein 1 (IGFBP1), and matrix metallopeptidase 10 (MMP10) (Table 5). GDF15 is a marker of cell senescence [51], and higher levels of GDF15 predict an increased risk for mortality [52]. High levels of GDF15 are also associated with cachexia [53]; it induces anorexia and thus synergizes with other cytokines that might induce muscle atrophy, cachexia, or sarcopenia. We showed in [38] that lower levels of GDF15 are associated with longer survival, which is also consistent with the idea that high levels of GDF15 are pathological. Higher levels of IGFBP1 also predict shorter survival [28, 54]. This finding adds to the debate as to whether IGF1 is positive or negative for longevity, since IGFBP1 was originally characterized as an inhibitor of IGF1 [55]; thus, the finding that higher levels of IGFBP1 are deleterious might be interpreted as indicating IGF1 is helpful for a longer lifespan; of course, IGF1-independent effects of IGFBP1 are also possible, and have been reported. Higher levels of MMP10 are associated with increased disease severity and mortality in patients with peripheral artery disease [56]. Interestingly, TGFB1 increases with older age but it is higher in carriers of the longevity variants of CDKN2B, thus suggesting that the aging-related increase may represent a protective or compensatory mechanism.
Overall, the results show that several longevity-associated variants are pQTLs for important aging biomarkers and suggest two possible mechanisms. One mechanism is to reduce the accumulation of damage that occurs with aging. This is shown by profiles of the biomarkers that remain more youthful in carriers of the longevity variants. Important examples are the lower level of APOB in carriers of the e2 allele of APOE, or the lower level of GDF15 in carriers of the longevity variants of CDKN2B. The second mechanism is to enhance potential compensatory processes that are activated with aging, as shown by carriers of the longevity variants that have patterns consistent with older age. Examples are the higher level of TGFB1 in carriers of the longevity variants of CDKN2B, or higher levels of NENF in carriers of the rare SNP rs9576827 in LHFPL6. Although most of our results need to be replicated and experimentally validated, our analysis shows the importance of linking genetics to molecular changes in order to distinguish between these two mechanisms. Particularly, it is easy to assume that age-related changes are “bad” and that healthy-aging interventions that target specific biomarkers should reverse the trends observed with aging. The patterns of these biomarkers in carriers of the longevity variants can help to identify these “protective” changes that should be enhanced rather than reversed.
Though a relatively small sample, these results suggest that rare genetic variants may play an important role in the ability of some individuals to reach extreme old ages but larger family-based studies and next-generation sequencing technology will be necessary to expand the yield of discoveries. In addition, our studies suggest that integration of genetics and proteomics will likely help to identify specific targets for healthy aging therapeutics. High throughput proteomics technology is still in its infancy, and the challenges ahead include the validation of the genotype-protein associations using an alternative technology to the aptamer-based approach provided by SomaLogic, and the assessment of the functional roles of these associations.
Supplementary Information
(DOCX 28 kb)
(XLSX 8.25 mb)
Funding
This work was supported by the National Institute on Aging (NIA cooperative agreements U19-AG023122 and UH2AG064704, and grant R01 AG061844), the Nathan Shock Center of Excellence for the basic Biology of Aging (P30AG038072) (N. B.), the Einstein-Paul Glenn Foundation for Medical Research Center for the Biology of Human Aging (N. B.), NIH/NIA 1 R01AG044829 (PIs-Veghese/Barzilai), NIH/NIA1 NIH-1 R01AG 046949-R01 AG057909-01 (PIs Barzilai & Zhang), U19 AG056278 (Vjig), K23AG051148 (SM), R01AG061155 (SM), the National Institute of General Medical Sciences (grant R24 GM134210), and the NIH Office of the Director (grant S10 OD021728).
Declarations
Conflict of interest
L.L.J. is an employee and stock-holder of Novartis. D.J.G is an employee and stock-holder of Regeneron Pharmaceuticals.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
6/18/2021
A Correction to this paper has been published: 10.1007/s11357-021-00400-7
Contributor Information
Lori L. Jennings, Email: lori.jennings@novartis.com
David J. Glass, Email: david.glass@regeneron.com
Paola Sebastiani, Email: psebastiani@tuftsmedicalcenter.org.
References
- 1.Hitt R, Young-Xu Y, Silver M, Perls T. Centenarians: the older you get, the healthier you have been. Lancet. 1999;354(9179):652. doi: 10.1016/S0140-6736(99)01987-X. [DOI] [PubMed] [Google Scholar]
- 2.Terry DF, Sebastiani P, Andersen SL, Perls TT. Disentangling the roles of disability and morbidity in survival to exceptional old age. Arch Intern Med. 2008;168(3):277–283. doi: 10.1001/archinternmed.2007.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersen SL, et al. Health span approximates life span among many supercentenarians: compression of morbidity at the approximate limit of life span. J Gerontol A Biol Sci Med Sci. 2012;67(4):395–405. doi: 10.1093/gerona/glr223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pignolo RJ. Exceptional human longevity. Mayo Clin Proc. 2019;94(1):110–124. doi: 10.1016/j.mayocp.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 5.Melzer D, Pilling LC, Ferrucci L. The genetics of human ageing. Nat Rev Genet. 2020;21(2):88–101. doi: 10.1038/s41576-019-0183-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perls T, Shea-Drinkwater M, Bowen-Flynn J, Ridge SB, Kang S, Joyce E, Daly M, Brewster SJ, Kunkel L, Puca AA. Exceptional familial clustering for extreme longevity in humans. J Am Geriatr Soc. 2000;48(11):1483–1485. [PubMed] [Google Scholar]
- 7.Perls TT, Wilmoth J, Levenson R, Drinkwater M, Cohen M, Bogan H, Joyce E, Brewster S, Kunkel L, Puca A. Life-long sustained mortality advantage of siblings of centenarians. Proc Natl Acad Sci U S A. 2002;99(12):8442–8447. doi: 10.1073/pnas.122587599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sebastiani P, Nussbaum L, Andersen SL, Black MJ, Perls TT. Increasing sibling relative risk of survival to older and older ages and the importance of precise definitions of “aging,” “life span,” and “longevity”. J Gerontol A Biol Sci Med Sci. 2016;71(3):340–346. doi: 10.1093/gerona/glv020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deelen J, Beekman M, Uh HW, Helmer Q, Kuningas M, Christiansen L, Kremer D, van der Breggen R, Suchiman HED, Lakenberg N, van den Akker EB, Passtoors WM, Tiemeier H, van Heemst D, de Craen AJ, Rivadeneira F, de Geus EJ, Perola M, van der Ouderaa FJ, Gunn DA, Boomsma DI, Uitterlinden AG, Christensen K, van Duijn CM, Heijmans BT, Houwing-Duistermaat JJ, Westendorp RGJ, Slagboom PE. Genome-wide association study identifies a single major locus contributing to survival into old age: the APOE locus revisited. Aging Cell. 2011;10(4):686–698. doi: 10.1111/j.1474-9726.2011.00705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sebastiani P, Solovieff N, DeWan AT, Walsh KM, Puca A, Hartley SW, Melista E, Andersen S, Dworkis DA, Wilk JB, Myers RH, Steinberg MH, Montano M, Baldwin CT, Hoh J, Perls TT. Genetic signatures of exceptional longevity in humans. PLoS One. 2012;7(1):e29848. doi: 10.1371/journal.pone.0029848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Broer L, Buchman AS, Deelen J, Evans DS, Faul JD, Lunetta KL, Sebastiani P, Smith JA, Smith AV, Tanaka T, Yu L, Arnold AM, Aspelund T, Benjamin EJ, de Jager PL, Eirkisdottir G, Evans DA, Garcia ME, Hofman A, Kaplan RC, Kardia SLR, Kiel DP, Oostra BA, Orwoll ES, Parimi N, Psaty BM, Rivadeneira F, Rotter JI, Seshadri S, Singleton A, Tiemeier H, Uitterlinden AG, Zhao W, Bandinelli S, Bennett DA, Ferrucci L, Gudnason V, Harris TB, Karasik D, Launer LJ, Perls TT, Slagboom PE, Tranah GJ, Weir DR, Newman AB, van Duijn CM, Murabito JM. GWAS of longevity in CHARGE consortium confirms APOE and FOXO3 candidacy. J Gerontol A Biol Sci Med Sci. 2015;70(1):110–118. doi: 10.1093/gerona/glu166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sebastiani P, Gurinovich A, Bae H, Andersen S, Malovini A, Atzmon G, Villa F, Kraja AT, Ben-Avraham D, Barzilai N, Puca A, Perls TT. Four genome-wide association studies identify new extreme longevity variants. J Gerontol A Biol Sci Med Sci. 2017;72(11):1453–1464. doi: 10.1093/gerona/glx027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deelen J, Evans DS, Arking DE, Tesi N, Nygaard M, Liu X, Wojczynski MK, Biggs ML, van der Spek A, Atzmon G, Ware EB, Sarnowski C, Smith AV, Seppälä I, Cordell HJ, Dose J, Amin N, Arnold AM, Ayers KL, Barzilai N, Becker EJ, Beekman M, Blanché H, Christensen K, Christiansen L, Collerton JC, Cubaynes S, Cummings SR, Davies K, Debrabant B, Deleuze JF, Duncan R, Faul JD, Franceschi C, Galan P, Gudnason V, Harris TB, Huisman M, Hurme MA, Jagger C, Jansen I, Jylhä M, Kähönen M, Karasik D, Kardia SLR, Kingston A, Kirkwood TBL, Launer LJ, Lehtimäki T, Lieb W, Lyytikäinen LP, Martin-Ruiz C, Min J, Nebel A, Newman AB, Nie C, Nohr EA, Orwoll ES, Perls TT, Province MA, Psaty BM, Raitakari OT, Reinders MJT, Robine JM, Rotter JI, Sebastiani P, Smith J, Sørensen TIA, Taylor KD, Uitterlinden AG, van der Flier W, van der Lee SJ, van Duijn CM, van Heemst D, Vaupel JW, Weir D, Ye K, Zeng Y, Zheng W, Holstege H, Kiel DP, Lunetta KL, Slagboom PE, Murabito JM. A meta-analysis of genome-wide association studies identifies multiple longevity genes. Nat Commun. 2019;10(1):3669. doi: 10.1038/s41467-019-11558-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sebastiani P, Gurinovich A, Nygaard M, Sasaki T, Sweigart B, Bae H, Andersen SL, Villa F, Atzmon G, Christensen K, Arai Y, Barzilai N, Puca A, Christiansen L, Hirose N, Perls TT. APOE alleles and extreme human longevity. J Gerontol A Biol Sci Med Sci. 2019;74(1):44–51. doi: 10.1093/gerona/gly174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bae H et al. Associations of FOXO3A polymorphisms with extreme human longevity in four longevity studies. J Gerontol A Biol Sci Med Sci. 2017; p. E-pub ahead of print Jul 18, 2017.
- 16.Pilling LC, Atkins JL, Bowman K, Jones SE, Tyrrell J, Beaumont RN, Ruth KS, Tuke MA, Yaghootkar H, Wood AR, Freathy RM, Murray A, Weedon MN, Xue L, Lunetta K, Murabito JM, Harries LW, Robine JM, Brayne C, Kuchel GA, Ferrucci L, Frayling TM, Melzer D. Human longevity is influenced by many genetic variants: evidence from 75,000 UK Biobank participants. Aging (Albany NY) 2016;8(3):547–560. doi: 10.18632/aging.100930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCorrison J, et al. Genetic support for longevity-enhancing drug targets: issues, preliminary data, and future directions. J Gerontol. 2019;74(Supplement_1):S61–S71. doi: 10.1093/gerona/glz206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.King EA, Davis JW, Degner JF. Are drug targets with genetic support twice as likely to be approved? Revised estimates of the impact of genetic support for drug mechanisms on the probability of drug approval. PLoS Genet. 2019;15(12):e1008489. doi: 10.1371/journal.pgen.1008489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davies DR, Gelinas AD, Zhang C, Rohloff JC, Carter JD, O'Connell D, Waugh SM, Wolk SK, Mayfield WS, Burgin AB, Edwards TE, Stewart LJ, Gold L, Janjic N, Jarvis TC. Unique motifs and hydrophobic interactions shape the binding of modified DNA ligands to protein targets. Proc Natl Acad Sci U S A. 2012;109(49):19971–19976. doi: 10.1073/pnas.1213933109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanaka T, Biancotto A, Moaddel R, Moore AZ, Gonzalez-Freire M, Aon MA, Candia J, Zhang P, Cheung F, Fantoni G, CHI consortium. Semba RD, Ferrucci L. Plasma proteomic signature of age in healthy humans. Aging Cell. 2018;17:e12799. doi: 10.1111/acel.12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suhre K, Arnold M, Bhagwat AM, Cotton RJ, Engelke R, Raffler J, Sarwath H, Thareja G, Wahl A, DeLisle RK, Gold L, Pezer M, Lauc G, el-Din Selim MA, Mook-Kanamori DO, al-Dous EK, Mohamoud YA, Malek J, Strauch K, Grallert H, Peters A, Kastenmüller G, Gieger C, Graumann J. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat Commun. 2017;8:14357. doi: 10.1038/ncomms14357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emilsson V, et al. Co-regulatory networks of human serum proteins link genetics to disease: Science; 2018. [DOI] [PMC free article] [PubMed]
- 23.Sebastiani P et al. A serum protein signature of APOE genotypes in centenarians. Aging Cell. 2019; p. e13023. [DOI] [PMC free article] [PubMed]
- 24.Sebastiani P, Perls TT. The genetics of extreme longevity: lessons from the New England Centenarian Study. Front Genet. 2012;3:277. doi: 10.3389/fgene.2012.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Das S, Forer L, Schönherr S, Sidore C, Locke AE, Kwong A, Vrieze SI, Chew EY, Levy S, McGue M, Schlessinger D, Stambolian D, Loh PR, Iacono WG, Swaroop A, Scott LJ, Cucca F, Kronenberg F, Boehnke M, Abecasis GR, Fuchsberger C. Next-generation genotype imputation service and methods. Nature Genetics. 2016;48(10):1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loh P-R, Danecek P, Palamara PF, Fuchsberger C, A Reshef Y, K Finucane H, Schoenherr S, Forer L, McCarthy S, Abecasis GR, Durbin R, L Price A. Reference-based phasing using the haplotype reference consortium panel. Nature Genetics. 2016;48(11):1443–1448. doi: 10.1038/ng.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suh Y, Atzmon G, Cho M-O, Hwang D, Bingrong L, Leahy DJ, Barzilai N, Cohen P. Functionally significant insulin-like growth factor 1 receptor mutations in centenarians. Proc Natl Acad Sci U S A. 2008;105:3438–3442. doi: 10.1073/pnas.0705467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang WB et al. Insulin-like growth factor-1 and IGF binding proteins predict all-cause mortality and morbidity in older adults. Cells. 2020; 9(6). [DOI] [PMC free article] [PubMed]
- 29.Bell F, Miller M. Life tables for the United States Social Security Area 1900-2100. Actuarial Study No. 116; 2005.
- 30.Conomos MP, Reiner AP, Weir BS, Thornton TA. Model-free estimation of recent genetic relatedness. Am J Hum Genet. 2016;98(1):127–148. doi: 10.1016/j.ajhg.2015.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gogarten SM, Sofer T, Chen H, Yu C, Brody JA, Thornton TA, Rice KM, Conomos MP. Genetic association testing using the GENESIS R/Bioconductor package. Bioinformatics. 2019;35(24):5346–5348. doi: 10.1093/bioinformatics/btz567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Candia J, Cheung F, Kotliarov Y, Fantoni G, Sellers B, Griesman T, Huang J, Stuccio S, Zingone A, Ryan BM, Tsang JS, Biancotto A. Assessment of variability in the SOMAscan assay. Scientific Reports. 2017;7(1):14248. doi: 10.1038/s41598-017-14755-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lehallier B, Gate D, Schaum N, Nanasi T, Lee SE, Yousef H, Moran Losada P, Berdnik D, Keller A, Verghese J, Sathyan S, Franceschi C, Milman S, Barzilai N, Wyss-Coray T. Undulating changes in human plasma proteome profiles across the lifespan. Nat Med. 2019;25(12):1843–1850. doi: 10.1038/s41591-019-0673-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Federico A, Monti S. hypeR: an R package for geneset enrichment workflows. Bioinformatics. 2020;36(4):1307–1308. doi: 10.1093/bioinformatics/btz700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melville SA, Buros J, Parrado AR, Vardarajan B, Logue MW, Shen L, Risacher SL, Kim S, Jun G, DeCarli C, Lunetta KL, Baldwin CT, Saykin AJ, Farrer LA, the Alzheimer's Disease Neuroimaging Initiative Multiple loci influencing hippocampal degeneration identified by genome scan. Annals of Neurology. 2012;72(1):65–75. doi: 10.1002/ana.23644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perry JRB, et al. Parent-of-origin-specific allelic associations among 106 genomic loci for age at menarche. Nature. 2014;514(7520):92–97. doi: 10.1038/nature13545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Timmers PRHJ, Mounier N, Lall K, Fischer K, Ning Z, Feng X, Bretherick AD, Clark DW, eQTLGen Consortium. Agbessi M, Ahsan H, Alves I, Andiappan A, Awadalla P, Battle A, Bonder MJ, Boomsma D, Christiansen M, Claringbould A, Deelen P, van Dongen J, Esko T, Favé M, Franke L, Frayling T, Gharib SA, Gibson G, Hemani G, Jansen R, Kalnapenkis A, Kasela S, Kettunen J, Kim Y, Kirsten H, Kovacs P, Krohn K, Kronberg-Guzman J, Kukushkina V, Kutalik Z, Kähönen M, Lee B, Lehtimäki T, Loeffler M, Marigorta U, Metspalu A, van Meurs J, Milani L, Müller-Nurasyid M, Nauck M, Nivard M, Penninx B, Perola M, Pervjakova N, Pierce B, Powell J, Prokisch H, Psaty BM, Raitakari O, Ring S, Ripatti S, Rotzschke O, Ruëger S, Saha A, Scholz M, Schramm K, Seppälä I, Stumvoll M, Sullivan P, Teumer A, Thiery J, Tong L, Tönjes A, Verlouw J, Visscher PM, Võsa U, Völker U, Yaghootkar H, Yang J, Zeng B, Zhang F, Agbessi M, Ahsan H, Alves I, Andiappan A, Awadalla P, Battle A, Bonder MJ, Boomsma D, Christiansen M, Claringbould A, Deelen P, van Dongen J, Esko T, Favé M, Franke L, Frayling T, Gharib SA, Gibson G, Hemani G, Jansen R, Kalnapenkis A, Kasela S, Kettunen J, Kim Y, Kirsten H, Kovacs P, Krohn K, Kronberg-Guzman J, Kukushkina V, Kutalik Z, Kähönen M, Lee B, Lehtimäki T, Loeffler M, Marigorta U, Metspalu A, van Meurs J, Milani L, Müller-Nurasyid M, Nauck M, Nivard M, Penninx B, Perola M, Pervjakova N, Pierce B, Powell J, Prokisch H, Psaty BM, Raitakari O, Ring S, Ripatti S, Rotzschke O, Ruëger S, Saha A, Scholz M, Schramm K, Seppälä I, Stumvoll M, Sullivan P, Teumer A, Thiery J, Tong L, Tönjes A, Verlouw J, Visscher PM, Võsa U, Völker U, Yaghootkar H, Yang J, Zeng B, Zhang F, Shen X, Esko T, Kutalik Z, Wilson JF, Joshi PK. Genomics of 1 million parent lifespans implicates novel pathways and common diseases and distinguishes survival chances. eLife. 2019;8:e39856. doi: 10.7554/eLife.39856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sebastiani P et al. Protein signatures of centenarians and their offspring suggest centenarians age slower than other humans. Aging Cell. 2021; p. e13290. [DOI] [PMC free article] [PubMed]
- 39.Hwang SJ, et al. A genome-wide association for kidney function and endocrine-related traits in the NHLBI’s Framingham Heart Study. BMC Med Genet. 2007;8(Suppl 1):S10. doi: 10.1186/1471-2350-8-S1-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hao Y, Chai KH, McLoughlin DM, Chan HYE, Lau KF. Promoter characterization and genomic organization of the human X11β gene APBA2. NeuroReport. 2012;23(3):146–151. doi: 10.1097/WNR.0b013e32834f1934. [DOI] [PubMed] [Google Scholar]
- 41.Quesada V, Freitas-Rodríguez S, Miller J, Pérez-Silva JG, Jiang ZF, Tapia W, Santiago-Fernández O, Campos-Iglesias D, Kuderna LFK, Quinzin M, Álvarez MG, Carrero D, Beheregaray LB, Gibbs JP, Chiari Y, Glaberman S, Ciofi C, Araujo-Voces M, Mayoral P, Arango JR, Tamargo-Gómez I, Roiz-Valle D, Pascual-Torner M, Evans BR, Edwards DL, Garrick RC, Russello MA, Poulakakis N, Gaughran SJ, Rueda DO, Bretones G, Marquès-Bonet T, White KP, Caccone A, López-Otín C. Giant tortoise genomes provide insights into longevity and age-related disease. Nat Ecol Evol. 2019;3(1):87–95. doi: 10.1038/s41559-018-0733-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ehnholm C, Lukka M, Kuusi T, Nikkilä E, Utermann G. Apolipoprotein E polymorphism in the Finnish population: gene frequencies and relation to lipoprotein concentrations. J Lipid Res. 1986;27(3):227–235. [PubMed] [Google Scholar]
- 43.Haddy N, Bacquer DD, Chemaly MM, Maurice M, Ehnholm C, Evans A, Sans S, Martins MC, Backer GD, Siest G, Visvikis S. The importance of plasma apolipoprotein E concentration in addition to its common polymorphism on inter-individual variation in lipid levels: results from Apo Europe. Eur J Hum Genet. 2002;10(12):841–850. doi: 10.1038/sj.ejhg.5200864. [DOI] [PubMed] [Google Scholar]
- 44.Cao J, Nomura SO, Steffen BT, Guan W, Remaley AT, Karger AB, Ouyang P, Michos ED, Tsai MY. Apolipoprotein B discordance with low-density lipoprotein cholesterol and non-high-density lipoprotein cholesterol in relation to coronary artery calcification in the Multi-Ethnic Study of Atherosclerosis (MESA) J Clin Lipidol. 2020;14(1):109–121 e5. doi: 10.1016/j.jacl.2019.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marivin A, Berthelet J, Plenchette S, Dubrez L. The inhibitor of apoptosis (IAPs) in adaptive response to cellular stress. Cells. 2012;1(4):711–737. doi: 10.3390/cells1040711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Varfolomeev E, Blankenship JW, Wayson SM, Fedorova AV, Kayagaki N, Garg P, Zobel K, Dynek JN, Elliott LO, Wallweber HJA, Flygare JA, Fairbrother WJ, Deshayes K, Dixit VM, Vucic D. IAP antagonists induce autoubiquitination of c-IAPs, NF-kappaB activation, and TNFalpha-dependent apoptosis. Cell. 2007;131(4):669–681. doi: 10.1016/j.cell.2007.10.030. [DOI] [PubMed] [Google Scholar]
- 47.Mak PY, Mak DH, Ruvolo V, Jacamo R, Kornblau SM, Kantarjian H, Andreeff M, Carter BZ. Apoptosis repressor with caspase recruitment domain modulates second mitochondrial-derived activator of caspases mimetic-induced cell death through BIRC2/MAP3K14 signalling in acute myeloid leukaemia. Br J Haematol. 2014;167(3):376–384. doi: 10.1111/bjh.13054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hinz M, Stilmann M, Arslan SÇ, Khanna KK, Dittmar G, Scheidereit C. A cytoplasmic ATM-TRAF6-cIAP1 module links nuclear DNA damage signaling to ubiquitin-mediated NF-kappaB activation. Mol Cell. 2010;40(1):63–74. doi: 10.1016/j.molcel.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 49.Moutaoufik MT, et al. Rewiring of the human mitochondrial interactome during neuronal reprogramming reveals regulators of the respirasome and neurogenesis. iScience. 2019;19:1114–1132. doi: 10.1016/j.isci.2019.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Basisty N, Kale A, Jeon OH, Kuehnemann C, Payne T, Rao C, Holtz A, Shah S, Sharma V, Ferrucci L, Campisi J, Schilling B. A proteomic atlas of senescence-associated secretomes for aging biomarker development. PLoS Biol. 2020;18(1):e3000599. doi: 10.1371/journal.pbio.3000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wiklund FE, Bennet AM, Magnusson PKE, Eriksson UK, Lindmark F, Wu L, Yaghoutyfam N, Marquis CP, Stattin P, Pedersen NL, Adami HO, Grönberg H, Breit SN, Brown DA. Macrophage inhibitory cytokine-1 (MIC-1/GDF15): a new marker of all-cause mortality. Aging Cell. 2010;9(6):1057–1064. doi: 10.1111/j.1474-9726.2010.00629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnen H, Lin S, Kuffner T, Brown DA, Tsai VWW, Bauskin AR, Wu L, Pankhurst G, Jiang L, Junankar S, Hunter M, Fairlie WD, Lee NJ, Enriquez RF, Baldock PA, Corey E, Apple FS, Murakami MAM, Lin EJ, Wang C, During MJ, Sainsbury A, Herzog H, Breit SN. Tumor-induced anorexia and weight loss are mediated by the TGF-beta superfamily cytokine MIC-1. Nat Med. 2007;13(11):1333–1340. doi: 10.1038/nm1677. [DOI] [PubMed] [Google Scholar]
- 54.Salminen M, Viljanen A, Eloranta S, Viikari P, Wuorela M, Vahlberg T, Isoaho R, Kivelä SL, Korhonen P, Irjala K, Löppönen M, Viikari L. Frailty and mortality: an 18-year follow-up study among Finnish community-dwelling older people. Aging Clin Exp Res. 2020;32(10):2013–2019. doi: 10.1007/s40520-019-01383-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Croze F, et al. Expression of insulin-like growth factor-I and insulin-like growth factor-binding protein-1 in the rat uterus during decidualization. Endocrinology. 1990;127(4):1995–2000. doi: 10.1210/endo-127-4-1995. [DOI] [PubMed] [Google Scholar]
- 56.Martinez-Aguilar E, Gomez-Rodriguez V, Orbe J, Rodriguez JA, Fernández-Alonso L, Roncal C, Páramo JA. Matrix metalloproteinase 10 is associated with disease severity and mortality in patients with peripheral arterial disease. Journal of Vascular Surgery. 2015;61(2):428–435. doi: 10.1016/j.jvs.2014.09.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 28 kb)
(XLSX 8.25 mb)