Abstract
COVID-19 has emerged as global health threats. Chronic kidney disease (CKD) patients are immune-compromised and may have a high risk of infection by the SARS-CoV-2. We aimed to detect common transcriptomic signatures and pathways between COVID-19 and CKD by systems biology analysis. We analyzed transcriptomic data obtained from peripheral blood mononuclear cells (PBMC) infected with SARS-CoV-2 and PBMC of CKD patients. We identified 49 differentially expressed genes (DEGs) which were common between COVID-19 and CKD. The gene ontology and pathways analysis showed the DEGs were associated with “platelet degranulation”, “regulation of wound healing”, “platelet activation”, “focal adhesion”, “regulation of actin cytoskeleton” and “PI3K-Akt signalling pathway”. The protein-protein interaction (PPI) network encoded by the common DEGs showed ten hub proteins (EPHB2, PRKAR2B, CAV1, ARHGEF12, HSP90B1, ITGA2B, BCL2L1, E2F1, TUBB1, and C3). Besides, we identified significant transcription factors and microRNAs that may regulate the common DEGs. We investigated protein-drug interaction analysis and identified potential drugs namely, aspirin, estradiol, rapamycin, and nebivolol. The identified common gene signature and pathways between COVID-19 and CKD may be therapeutic targets in COVID-19 patients with CKD comorbidity.
Keywords: COVID-19, Chronic kidney disease, SARS-CoV-2, Systems biology, Transcriptional signature, Protein-protein interaction, Molecular pathways
1. Introduction
Novel severe acute respiratory syndrome-related coronavirus-2 (SARS-CoV-2) has appeared as global pandemic (Cui, 2019). SARS-CoV-2 is a highly infected single-stranded RNA coronavirus. The SARS-CoV-2 causes respiratory diseases dubbed as COVID-19. The symptoms of common cold, fever and pneumonia are observed in COVID-19 patients (Chen et al., 2020). In the mid-1960s coronavirus was first identified (Mahase, 2020). The severe acute respiratory syndrome-related coronavirus (SARS-CoV-1) affect almost 8096 peoples in 2002. The second coronavirus named Middle East respiratory syndrome (MERS) emerged in 2012 that infected almost 2494 peoples. The latest coronavirus known by severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) infected almost 166,492,108 peoples globally (as of May 22, 2021) (Worldometer, n.d.).
CKD is a disorder of kidneys caused by abnormal blood purification. Globally 10% of the population is affected by CKD (“World Kidney Day: Chronic Kidney Disease,” 2015). The CKD patients are more possibly to have other chronic diseases as well as hypertension, diabetes, cardiovascular complication, and anaemia (Robert Thomas et al., 2008). It has been previously (Guan et al., 2020) shown that about 173 out of 1099 confirmed COVID-19 patients had Chronic obstructive pulmonary disease (COPD) (3.5%), diabetes mellitus (16.2%), coronary heart diseases (5.8%), hypertension (23.7%), cerebrovascular disease (2.3%) and chronic renal disease (1.7%). Another report showed that among 5700 confirmed COVID-19 patients had comorbidities of hypertension (56.6%), CKD (5%), diabetes (33.8%) and COPD (5.4%) (Richardson et al., 2020). The CKD patients are immune-compromised and make the CKD patients more vulnerable to COVID-19. Although the main target of SARS-CoV-2 is to attack the respiratory systems, the other organs such as kidney might also be affected by the infection of the virus. The recent evidence of single-cell profiling showed that the kidneys are vulnerable to SARS-CoV-2 infection by the receptor ACE2 (Zou et al., 2020).
The mortality rate increases for COVID-19 patients with other comorbid diseases. There were several studies identified the comorbid influences on COVID-19 like, the cardiovascular and hypertensive (Kamyshnyi et al., 2020, Mahmud et al., 2021), Chronic obstructive pulmonary disease (COPD) comorbidities (Gerayeli et al., 2021, Lee et al., 2021) and so on. Despite critical important findings of the previous studies, the effect of COVID-19 on CKD has not been investigated yet. The transcriptomic analyses have been widely used in biomedical science to identify key genes and pathways. Several studies have been detected the transcriptomic alteration in lung epithelial cells and peripheral blood mononuclear cells (PBMC) in response to SARS-CoV-2 (Blanco-Melo et al., 2020, Islam et al., 2020) (Arunachalam et al., 2020, Auwul et al., 2021). To discover molecular mechanism of disease's biological condition, the Differentially Expressed (DE) genes should to identify for further system biological study (Crow et al., 2019). The DE genes are those whose expression levels are statistically different between groups. There were several statistical algorithms available in literature for finding the significant DE genes. Among them the DESeq2 algorithm is most popular in case of RNA-seq count data analysis (Love et al., 2015). Another popular method for microarray data is Limma-Linear Models for Microarray data (Gentleman et al., 2005, Ritchie et al., 2015). We used these two popular methods for finding DE genes in COVID-19 and CKD datasets respectively.
In this study, we used a systems biology approach to detect commonly dysregulated genes and associate pathways between COVID-19 and CKD. For this aim, we have used PBMC transcriptomic data of COVID-19 and CKD to identify common transcriptomic signatures followed by enrichment analysis to shed light into the potential common mechanisms that underlie the COVID-19 and CKD.
2. Materials and methods
2.1. Data pre-processing and differential expression analysis
We obtained the RNA-Sequencing (RNA-Seq) transcriptomic dataset of peripheral blood mononuclear cells (PBMC) infected upon SARS-CoV-2 with the accession number GSE152418 (Arunachalam et al., 2020) from the NCBI Gene Expression Omnibus (NCBI-GEO) (Barrett et al., 2013). This dataset consists of 34 samples (17 samples from SARS-CoV-2 infected PBMC and 17 samples from healthy controls). We collected the raw-count form of this dataset that was constructed under the platform GPL24676 via Illumina NovaSeq 6000 system. Then, we retrieved PBMC microarray gene expression dataset of CKD (with the accession number GSE15072). The dataset contained gene expression profiling data from patients with CKD of varying stages including 9 samples on stage II-III, 17 patients undergoing hemodialysis treatment (HD), and 8 healthy controls samples (Granata et al., 2009). For our study, we considered 9 CKD and 8 healthy control samples from this dataset and collect the normalized form of this dataset under the platform GPL570 [HG-U133_Plus_2] that was constructed via the Affymetrix Human Genome U133 Plus 2.0 Array system.
To identify significant differentially expressed genes (DEGs), we used the DESeq2 R package for COVID-19 RNA-Seq count data (Love et al., 2015). DESeq2 performed normalization process internally by calculating geometric mean of each gene over all samples. It used the negative binomial generalized linear models to fit for each gene and test the gene significance via Wald test. DESeq2 filtered low expressed and outlying genes automatically using cook’s distance. We analyzed the microarray CKD dataset via limma-Linear Models for Microarray data R package (Ritchie et al., 2015). It used statistical t-test for hypothesis testing the significance of each gene over samples. The significant DEGs were filtered based on adjusted p-value < 0.05 and |log2 FC|≥1. Venn analysis used to screen common DEGs between COVID-19 and CKD using the match function. These common DEGs were used for further downstream analysis.
2.2. Gene ontology and pathway enrichment analysis
The functional annotation and pathway enrichment analysis of the DEGs were executed through Enrichr web utility tools (Kuleshov et al., 2016). We considered Gene Ontology (GO) terms in three categories namely, ‘biological process (BP)’, ‘cellular component (CC)’, and ‘molecular functions (MF)’. For the pathway analysis, the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Reactome database were used as a data source. A statistical threshold criterion p-value < 0.05 were chosen for selecting significant GO and pathways.
2.3. Protein-protein interaction network analysis
We performed the protein–protein interactions (PPI) using the STRING database (Szklarczyk et al., 2017). The PPI network was processed and analyzed in Cytoscape (Smoot et al., 2011). We constructed the PPI network considering the proteins encrypted by the common DEGs between COVID-19 and CKD. The network comprised of seed proteins (i.e., common DEGs) and their neighboring interacting partners. We considered degree and Maximal Clique Centrality (MCC) parameters to detect the highly connected hub proteins from the PPI network via CytoHubba plugin in Cytoscape (Barabasi and Oltvai, 2004, Chin et al., 2014). We detected hub genes with MCC > 15. The higher the value of MCC of the nodes, the higher number of edges connected in those hub proteins.
2.4. Transcriptional and post-transcriptional network analysis
We performed TFs-Gene and Gene-miRNAs interaction via NetworkAnalyst webtools (Xia et al., 2015) to identify the significant TFs and miRNAs. We used the JASPAR (Khan et al., 2018) database to build the TFs-Gene network. The Tarbase (Sethupathy et al., 2006) and mirTarbase (Hsu et al., 2011) databases used to build Gene-miRNAs interactions network via NetworkAnalyst (Xia et al., 2015). The MCC >=15 cutoff criterion had been chosen for the selection of significant hub TFs and miRNAs by using CytoHubba plugin that were visualized in Cytoscape.
2.5. Drug prediction analysis
We used DSigDB (version 1.0) database to identify potential drugs which are significantly interacted with genes (Yoo et al., 2015). The DSigDB is a web-based freely accessible resource that contains the information of drugs and their target genes that have been used for the gene set enrichment analysis (GSEA). This database currently contains overall 22,527 gene sets including 17,389 drugs and 19,531 genes. We selected the enriched drugs for the DEGs by using Enrichr web server using DSigDB (Kuleshov et al., 2016) database based on the statistical criterion, adjusted p-value < 0.05.
3. Results
3.1. Transcriptomic signature
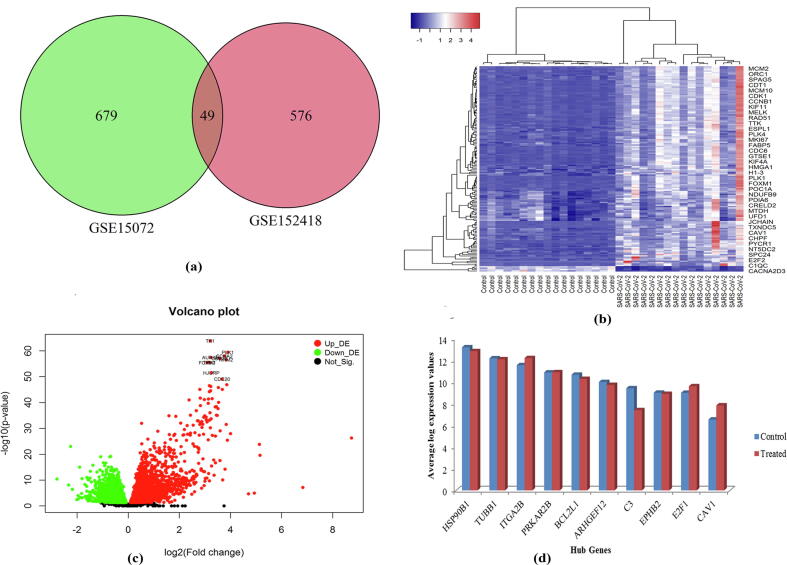
First of all, we analyzed the transcriptomic blood datasets of COVID-19 and identified 625 differentially expressed genes (DEGs) considering statistical significance criteria, adjusted p-value ≤0.05 and |log2 FC| ≥ 1. Then, we analyzed the gene expression profiling dataset of CKD matched with controls. Our analysis showed 728 DEGs in CKD compared to controls. We compared the significant DEGs between COVID-19 and CKD to identify common DEG. We found 49 common DEGs between COVID-19 and CKD (Fig. 1a). The heatmap analysis showed the expression patterns of DEGs between COVID-19 and healthy control samples that indicated the clustering of each DEGs in two groups (i.e., up-regulated and down-regulated) (Fig. 1b). The statistical measurements of the 49 common genes were summarized in Table S1.
Fig. 1.
DEGs and hub-gene expression profiles (a) Venn diagram shows 49 common DEGs between COVID-19 and chronic kidney disease datasets; (b) Heatmap of common DEGs (49 DEGs) between COVID-19 and chronic kidney disease datasets on COVID-19 dataset; (c) volcano plot of COVID-19 dataset, (d) Expression plot of the identified 10 hub-genes in COVID-19 dataset.
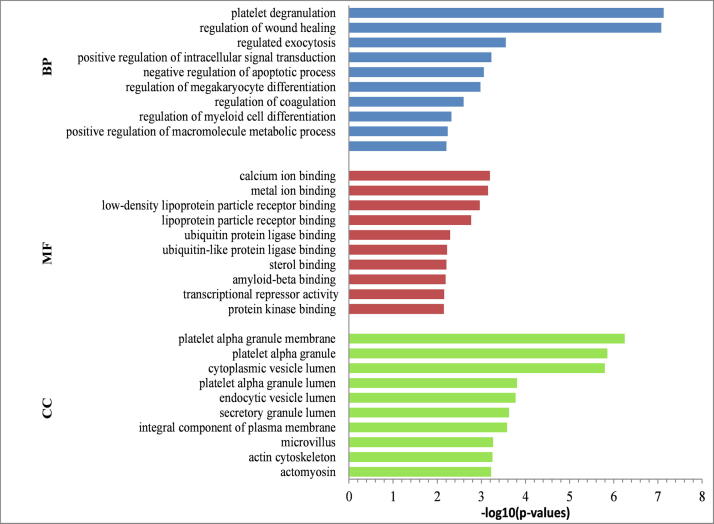
To obtain biological insights into common DEGs, we conducted a gene set enrichment analysis. Our analysis showed significant biological processes enriched by the DEGs were “platelet degranulation”, “regulation of wound healing” and “regulated exocytosis”. The significant molecular function (MF) terms enriched by the common DEGs included “calcium ion binding”, “metal ion binding” and “lipoprotein particle receptor binding”. The significant cellular components (CC) terms enriched in “platelet alpha granule membrane”, “cytoplasmic vesicle lumen”, “microvillus” and “actin cytoskeleton” for the common DEGs. The top 10 terms of gene ontology were summarized in Fig. 2.
Fig. 2.
Gene ontology enrichment analysis of 49 common DEGs identified in COVID-19 and CKD; The terms BP, CC and MF stand left side of this figure indicate the ‘biological function’, ‘cellular component’ and ‘molecular function’ enrichment categories, respectively.
3.2. Pathway enrichment analysis
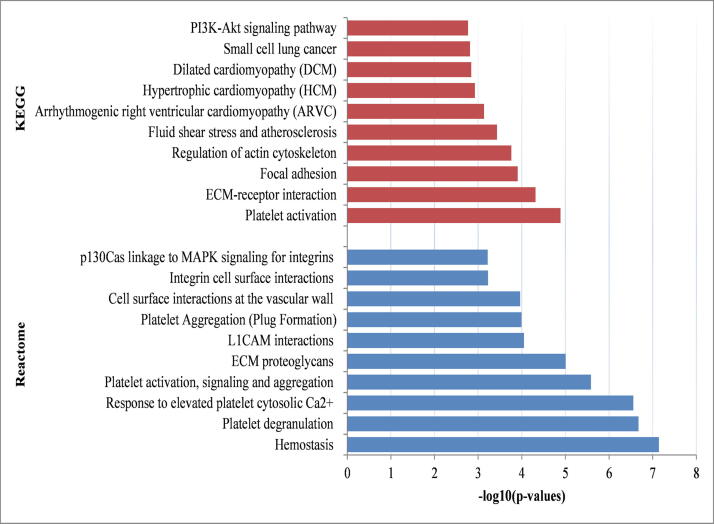
We conducted pathway analysis to identify dysregulated pathways enriched by the common DEGs identified in COVID-19 and CKD. Our analysis identified several pathways enriched by the common DEGs namely, “platelet activation”, “ECM-receptor interaction”, “regulation of actin cytoskeleton”, “PI3K-Akt signalling pathway”, “arrhythmogenic right ventricular cardiomyopathy”, “hypertrophic cardiomyopathy” “focal adhesion” and “small cell lung cancer”. The top significant pathways were summarized in Table 1 and visualized in Fig. 3.
Table 1.
Molecular pathways (top ten) enriched by the common differentially expressed genes.
| Source | Pathways | Adjusted p-value | # of Genes | Related Genes |
|---|---|---|---|---|
| KEGG | Platelet activation | 1.29E-05 | 5 | ARHGEF12;ITGB3;ITGA2B;GP1BA;MYLK |
| ECM-receptor interaction | 4.83E-05 | 4 | ITGB3;ITGA2B;ITGA7;GP1BA | |
| Focal adhesion | 1.24E-04 | 5 | ITGB3;CAV1;ITGA2B;ITGA7;MYLK | |
| Regulation of actin cytoskeleton | 1.74E-04 | 5 | ARHGEF12;ITGB3;ITGA2B;ITGA7;MYLK | |
| Fluid shear stress and atherosclerosis | 3.71E-04 | 4 | ITGB3;CAV1;ITGA2B;HSP90B1 | |
| Arrhythmogenic right ventricular cardiomyopathy (ARVC) | 7.32E-04 | 3 | ITGB3;ITGA2B;ITGA7 | |
| Hypertrophic cardiomyopathy (HCM) | 0.001185288 | 3 | ITGB3;ITGA2B;ITGA7 | |
| Dilated cardiomyopathy (DCM) | 0.001442953 | 3 | ITGB3;ITGA2B;ITGA7 | |
| Small cell lung cancer | 0.001536035 | 3 | ITGA2B;E2F1;BCL2L1 | |
| PI3K-Akt signalling pathway | 0.001700219 | 5 | ITGB3;ITGA2B;ITGA7;HSP90B1;BCL2L1 | |
| Reactome | Hemostasis | 7.21939E-08 | 10 | SELP;EHD3;SPARC;PRKAR2B;ITGB3;CAV1;KIF4A;ITGA2B;GP1BA;CLU;PF4 |
| Platelet degranulation | 2.11035E-07 | 5 | SELP;SPARC;ITGB3;ITGA2B;CLU;PF4 | |
| Response to elevated platelet cytosolic Ca2+ | 2.78284E-07 | 5 | SELP;SPARC;ITGB3;ITGA2B;CLU;PF4 | |
| Platelet activation, signalling and aggregation | 2.60487E-06 | 6 | SELP;SPARC;ITGB3;ITGA2B;GP1BA;CLU;PF4 | |
| ECM proteoglycans | 9.89058E-06 | 4 | SPARC;ITGB3;ITGA2B;ITGA7 | |
| L1CAM interactions | 8.94941E-05 | 4 | ITGB3;KIF4A;ITGA2B;EPHB2 | |
| Platelet Aggregation (Plug Formation) | 0.000101262 | 3 | ITGB3;ITGA2B;GP1BA | |
| Cell surface interactions at the vascular wall | 0.000109012 | 4 | SELP;ITGB3;CAV1;PF4 | |
| Integrin cell surface interactions | 0.000592926 | 3 | ITGB3;ITGA2B;ITGA7 | |
| p130Cas linkage to MAPK signalling for integrin | 0.000604968 | 2 | ITGB3;ITGA2B |
Fig. 3.
Barplot of KEGG and Reactome pathway enrichment analyses of 49 common DEGs identified in COVID-19 and chronic kidney disease datasets.
3.3. Proteomic signature
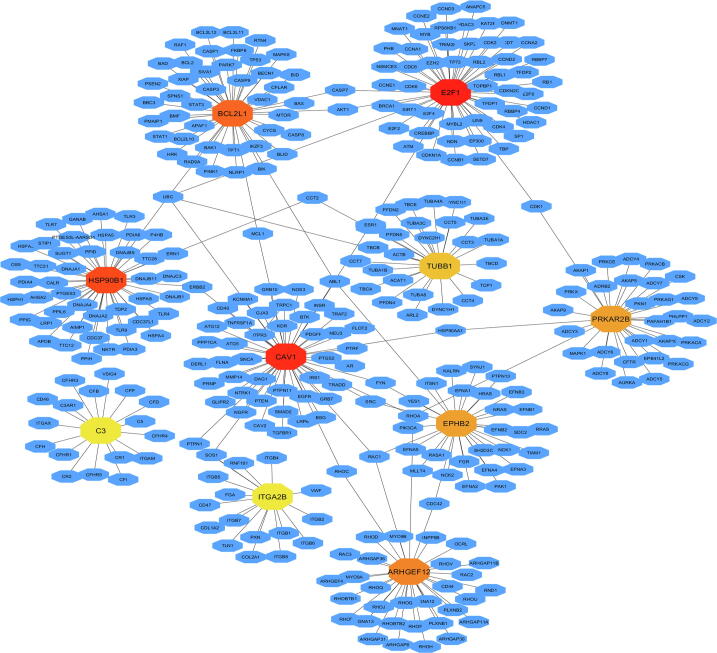
We investigated the protein–protein interaction (PPI) analysis of each proteins corresponding to the common DEGs identified between COVID-19 and CKD to identify critical molecules (Fig. 4). We employed Maximal Clique Centrality (MCC) based assessment of PPI network. We detected 10 hub proteins (EPHB2, PRKAR2B, CAV1, ARHGEF12, HSP90B1, ITGA2B, BCL2L1, E2F1, TUBB1 and C3) which may be considered as the proteomic signature. Fig. 1d showed the average log expression value of these hub genes. The functional associations of these hub genes were summarized in Table 2.
Fig. 4.
PPI sub-network analysis of DE genes using common differentially expressed genes identified in COVID-19 and chronic kidney disease datasets with ten hub genes.
Table 2.
Overview of the hub genes obtained from PPI network.
| Gene symbol | MCC value | Description | RelatedDiseasesand pathways | References |
|---|---|---|---|---|
| E2F1 | 56 | E2F Transcription Factor 1 | This gene plays a vital role in controlling the cell cycle and the tumor suppressed genes act and is also an aim of the converting proteins of small DNA tumor viruses and is also significantly associated with renal cell carcinoma. | (Ma et al., 2013, Neuman et al., 1996) |
| CAV1 | 54 | Caveolin 1 | CAV1 have a wide range of effect in all stage of viral infection like influenza A and an association with early-stage CKD cohort and in a cohort with more severe CKD. | (Chand et al., 2016, Xing et al., 2020) |
| HSP90B1 | 47 | Heat Shock Protein 90 Beta Family Member 1 | It have significant pathophysiological role in bipolar disorder and the high expression of HSP90B1 is related with bone metastasis in renal cell carcinoma. | (Kakiuchi et al., 2007) |
| BCL2L1 | 46 | BCL2 Like 1 | The gene is functionally associated with several cancer related process and its protein expression is associated with colorectal adenoma-to-carcinoma progression. The “absolute glaucoma” and “tongue carcinoma” diseases also associated with this gene. | (Boise et al., 1993, Sillars-Hardebol et al., 2012 |
| ARHGEF12 | 34 | Rho Guanine Nucleotide Exchange Factor 12 | Leukemia and Giant Axonal Neuropathy disease are associated clearly with ARHGEF12. | (Fagerberg et al., 2014) |
| EPHB2 | 31 | EPH Receptor B2 | EPHB2 gene has been shown to be up-regulated in glioblastoma and the mutation of this gene identified in colorectal, gastric, bleeding disorder and prostate cancer/brain cancer. The EPH family also associated with virus infection. | (Chan and Watt, 1991, Wang et al., 2020) |
| PRKAR2B | 31 | Protein Kinase CAMP-Dependent Type II Regulatory Subunit Beta | The PRKAR2B is an important protein kinase genes may associate with COPD, carney complex variant and with spinocerebellar ataxia. | (Mostafaei et al., 2018, Solberg et al., 1992) |
| TUBB1 | 27 | Tubulin Beta 1 Class VI | The gene is associated with virus infectious disease and is found as a hub gene in COVID-19 disease. It is also associated with macrothrombocytopenia, autosomal dominant, tubb1-related and autosomal dominant macrothrombocytopenia. | (Oh et al., 2020) |
| ITGA2B | 17 | Integrin Subunit Alpha 2b | This gene is significantly associated with renal cell carcinoma. It also related with glanzmann thrombasthenia and bleeding disorder, platelet-type, 16. | (Khoriaty et al., 2019, Lu et al., 2016) |
| C3 | 17 | Complement C3 | The genetic polymorphism in C3 is related to the progression in chronic kidney disease (CKD) patients and the C3 inhibitor also used to treat the first case of COVID-19 patient. | (Ibrahim et al., 2020, Mastaglio et al., 2020) |
The first column indicated the gene symbol of the ten hub genes, the second column indicated the Maximal Clique Centrality (MCC) values corresponding to each of the hub genes (the larger the value of MCC, the more significant the hub genes), the third column indicated the full name of each hub gene, the fourth column described the associated diseases and pathways of each hub genes and their corresponding reference given in column six.
3.4. Regulatory signature
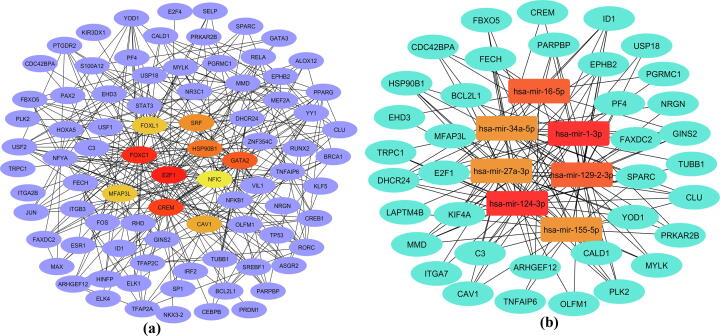
To detect transcriptional signatures and post-transcriptional regulatory signatures of the 49 common DEGs, the TFs-DEGs and miRNAs-DEGs interaction networks were reconstructed using the experimentally verified interactions. Considering the topological parameters, we detected significant transcription factors namely, FOXC1, CREM, GATA2, SRF and FOXL1 (Fig. 5a). We identified significant miRNAs namely, mir-124-3p, mir-1-3p, mir-27a-3, mir-129-2-3p, mir-155-5p and mir-34a-5p (Fig. 5b).
Fig. 5.
Regulatory signature identification for the common DEGs in COVID-19 and chronic kidney disease datasets: (a) Significant transcription factors were identified from TFs-Genes interaction analysis; (b) Significant miRNAs were identified from Genes-miRNAs interaction analysis.
3.5. Drug prediction analysis
To identify possible candidate drugs that may target the common DEGs, we performed drug target enrichment analysis with considering the DEGs as drug targets. The drug prediction analysis suggested several drugs that may target the DEGs (Table 3). Table 3 showed the top ten enriched drugs (tetradioxin, aspirin, estradiol, rapamycin, benzene, arachidonic acid, styrene, genistein, arsenous, nebivolol) by the DEGs that may be potential drugs for the treatment of SARS-CoV-2 infected CKD patients.
Table 3.
Candidate drugs (top ten) identified from gene-drug interaction enrichment analysis.
| Drugs | Adjusted p-values | Related Genes |
|---|---|---|
| Tetradioxin | 3.25E-04 | SPARC;TNFAIP6;PTGDR2;ITGB3;FAXDC2;RORC;CLU;USP18;NRGN;HSP90B1;C3; LAPTM4B;PGRMC1;PRKAR2B;TUBB1;E2F1;S100A12;FBXO5;MFAP3L;GINS2;CAV1; YOD1;DHCR24;VIL1;ID1;BCL2L1 |
| Aspirin | 0.001147 | SELP;SPARC;TNFAIP6;PTGDR2;ITGB3;ITGA2B;E2F1;GP1BA;PF4;BCL2L1 |
| Estradiol | 0.001461 | TNFAIP6;ITGB3;FAXDC2;RORC;CREM;CLU;USP18;MYLK;HSP90B1;C3;PRKAR2B; TUBB1;E2F1;FBXO5;MFAP3L;GINS2;MMD;PLK2;CAV1;DHCR24;CDC42BPA;VIL1; OLFM1;KIF4A;ID1;BCL2L1 |
| Rapamycin | 0.001491 | CALD1;FAXDC2;ID1;E2F1;FBXO5;MYLK;BCL2L1 |
| Benzene | 0.001575 | SPARC;TNFAIP6;PLK2;ITGA2B;CREM;GP1BA;ALOX12;CLU;USP18;PF4 |
| Arachidonic acid | 0.001916 | SELP;SPARC;ITGB3;ITGA2B;ALOX12;BCL2L1 |
| Styrene | 0.002021 | C3;SELP;BCL2L1;PF4 |
| Genistein | 0.0025 | GINS2;TNFAIP6;ITGB3;PLK2;CAV1;DHCR24;MYLK;C3;OLFM1;KIF4A;ID1; FBXO5;BCL2L1 |
| Arsenenous | 0.003526 | SPARC;TNFAIP6;ITGB3;PLK2;CAV1;CREM;ALOX12;CLU;MYLK;NRGN; PGRMC1;ID1;BCL2L1 |
| Nebivolol | 0.004022 | SELP;TNFAIP6;DHCR24 |
The first column indicated the names of the top ten drugs suggested in this study for the treatment of the COVID-19 patients with CKD comorbidity, the second column indicated the adjusted p-values (the p-value adjusted via Benjamini and Hochberg (FDR) of the corresponding drugs; the smaller the value of adjusted p-value, the more significancant the drug), the third column indicated the related genes of each drugs.
4. Discussion
The COVID-19 has become an immense threat to humankind. Higher death and critical COVID-19 conditions are observed in old patients with comorbid diseases including CKD. Investigations are being undertaken to identify the transcriptional signatures and pathways among COVID-19 and its comorbidities; however, the association between COVID-19 and CKD have not studied yet. In this study, we implemented a system biology approach to detect the common transcriptional signatures and pathways between COVID-19 and CKD that may clarify the increased critical COVID-19 condition in CKD patients. Our study suggests 49 DEGs which were common between COVID-19 and CKD suggesting common transcriptomic signature shared between two pathologies. To decipher the biological significance, we conducted the functional enrichment analysis of DEG(HK et al., 2020). Our analysis suggests the pathways enriched by the common DEGs involved in the platelet activation that is associated with CKD (Gremmel et al., 2013). The severe COVID-19 patients have significantly high association with platelet activation and platelet-dependent monocyte transcription factors expression (Hottz et al., 2020). The extracellular matrix (ECM) pathway was also identified as a critical pathway; ECM components include a combination of mechanical and functional macromolecules that have higher involvements in CKD and renal fibrosis (Genovese et al., 2014). The ECM play significant roles in lung fibrosis and it suggesting that the COVID-19 patients with lung fibrosis comorbidity may present greater complications (Sun et al., 2020). Our study suggests hemostasis is highly associated with COVID-19 and CKD as well(Lutz et al., 2014) (Ardy et al., 2020).
The hub genes have been identified from the PPI network to detect critical signaling molecules that may be therapeutic targets for the development of drugs to treat in COVID-19 patients with and CKD comorbidity. Among the hub genes, the Caveolin 1 (CAV1) is the key element of the caveolae plasma membranes that has an association with early-stage CKD cohort and severe CKD as well (Chand et al., 2016). In addition, the CAV1 has a wide range of effects in all stages of viral infection (Xing et al., 2020). The E2F Transcription Factor 1 (E2F1) gene plays a vital role in controlling the cell cycle and act as tumor suppressed genes. It aims also converting to proteins of small DNA tumor viruses (Neuman et al., 1996). The complement 3 (C3) gene activation has been implicated the aggravation of lung injury of SARS-CoV infection in preclinical models. The first COVID-19 patient was treated with the C3 inhibitor (Mastaglio et al., 2020). Moreover, the genetic polymorphism in C3 gene is also related to the progression of CKD (Ibrahim et al., 2020). The EPH family was also reported to be associated with viral infection (Chan and Watt, 1991, Wang et al., 2020). The PRKAR2B is an important protein kinase genes may associate with COPD (Mostafaei et al., 2018). The Tubulin Beta 1 Class VI (TUBB1) gene is associated with virus infectious disease and is being found as a hub gene in COVID-19 disease which was consistent with our findings (Oh et al., 2020). The Integrin Subunit Alpha 2b (ITGA2B) is significantly associated with clear renal cell carcinoma (Khoriaty et al., 2019, Lu et al., 2016).
A number of significant transcriptomics factors detected in this study that may regulate the common DEGs between COVID-19 and CKD datsets. Among the identified TFs, Forkhead Box C1 (FOXC1) and the Forkhead Box L1 (FOXL1) have been detected as differentially expressed in renal cell carcinoma (Yao et al., 2016). The expression of FOXC1 was reported as vital factor for increasing the hepatitis B virus X (Yang et al., 2017). One more transcription factor GATA2 was implicated in renal development (Yu et al., 2010). Besides, we detected several miRNAs as potential regulators of the DEGs. The mir-129-2-3p has been detected as a diagnostic and prognostic biomarker for renal cell carcinoma (Gao et al., 2016). It was detected as up-regulated signatures in human papillomavirus; the researcher suggested that the expression of mir-129-2-3p may be of considerable important signature for SRAS-CoV-2 (Arisan et al., 2020). Chronic renal inflammation has been developed with the altered mir-146a-5p expression (Osamu et al., 2012).
Next, the drugs that may target the common DEGs have been detected by using the DSigDB database. Among the detected significant drugs, the rapamycin has been proposed as a potential repurposable drug for the treatment of COVID-19 (Husain and Byrareddycde, 2020). Nebivolol is a drug for acute and chronic renal disorder and used to treat hypertension. The researchers also suggest using nebivolol to treat COVID-19 (Bocci et al., 2020). We propose further studies of biological and clinical investigation to evaluate the potential repurposing of these candidate drugs for potential treatment in COVID-19 patients with CKD comorbidity.
In this study, a popular system biology approach used to identify common signature genes between the COVID-19 and CKD. We chose both of the datasets originated from the same tissue (PBMC) that make more reliable on finding common signature between them. Instead of the advantages of this procedure, some limitations of this study would be noted as the samples of SARS-CoV-2 and CKD were not taken the same time, a small number of samples, and the dearth of clinical validation of the identified molecules. Thus cautions have to be taken while interpreting the findings of the study. The researcher may apply some existing machine learning algorithms to find the genetic effect of COVID-19 to Chronic Kidney Disease Patients for the future learning (Hasan et al., 2021, Hasan et al., 2020a, Hasan et al., 2020b, Hasan et al., 2020c).
5. Conclusions
In this study, we aimed to identify the potential pathogenic processes which were common between COVID-19 and CKD. We identified 49 common DEGs between COVID-19 and CKD by analyzing blood transcriptomic data. The neutrophil mediated immunity, neutrophil activation has been found significantly associated with the DEGs. We identified several significant pathways and hub proteins related to common DEGs. We identified several significant small molecules associated with the common DEGs that might be potential candidate drugs to treat COVID-19 patients with CKD comorbidity. The identified hub genes and pathways may be potential therapeutic targets. The findings of this study are based on bioinformatics analysis; further clinical investigations are suggested to validate the identified molecular signatures.
Data availability statement
All data utilized in this manuscript are available online from their respective NCBI GEO database with accession number GSE152418 (COVID-19) and GSE15072 (CKD).
CRediT authorship contribution statement
Md. Rabiul Auwul: Data curation, Formal analysis, Investigation, Methodology, Resources, Visualization, Writing - original draft, Writing - review & editing. Chongqi Zhang: Supervision, Writing - review & editing. Md Rezanur Rahman: Conceptualization, Data curation, Formal analysis, Resources, Software, Writing - original draft, Writing - review & editing. Md. Shahjaman: Data curation, Formal analysis, Writing - original draft, Writing - review & editing. Salem A. Alyami: Conceptualization, Funding acquisition, Writing - review & editing. Mohammad Ali Moni: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Software, Supervision, Validation, Writing - original draft, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We would like to thank the referees and the journal editorial team for providing valuable advice that improved the quality of the original manuscript.This work is supported by the National Nature Sciences Foundation of China (12071096).
Footnotes
Peer review under responsibility of King Saud University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.sjbs.2021.06.015.
Contributor Information
Md. Rabiul Auwul, Email: rauwul@e.gzhu.edu.cn.
Chongqi Zhang, Email: cqzhang@gzhu.edu.cn.
Salem A. Alyami, Email: saalyami@imamu.edu.sa.
Mohammad Ali Moni, Email: m.moni@unsw.edu.au.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- Ardy M., Lecompte T., Douxfils J. Management of the thrombotic risk associated with COVID-19: guidance for the hemostasis laboratory. Thromb. J. 2020;18 doi: 10.1186/s12959-020-00230-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arisan, E., Dart, A., Grant, G., Arisan, S., Cuhadaroglu, S., Lange, S., Uysal-Onganer, 2020. The Prediction of miRNAs in SARS-CoV-2 Genomes: hsa-miR Databases Identify 7 Key miRs Linked to Host Responses and Virus Pathogenicity-Related KEGG Pathways Significant for Comorbidities. Viruses 12, 614. https://doi.org/10.3390/v12060614 [DOI] [PMC free article] [PubMed]
- Arunachalam, P.S., Wimmers, F., Mok, C.K.P., Perera, R.A.P.M., Scott1, M., Hagan1, T., Sigal, N., Feng, Y., Bristow, L., T, O., Pulendran, B., 2020. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science (80-.). 369, 1210–1220. https://doi.org/10.1126/science.abc6261 [DOI] [PMC free article] [PubMed]
- Auwul M.R., Rahman R., Gov E., Shahjaman M., Moni M.A. Bioinformatics and machine learning approach identifies potential drug targets and pathways in COVID-19. Brief. Bioinform. 2021;bbab120:1–13. doi: 10.1093/bib/bbab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabasi A.-L., Oltvai Z.N. Network biology: understanding the cell’s functional organization. Nat. Rev. Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2013;41:991–995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo, D., Nilsson-Payant, B., Liu, W.-C., Moeller, R., Panis, M., Sachs, D., Albrecht, R., 2020. SARS-CoV-2 launches a unique transcriptional signature from in vitro, ex vivo, and in vivo systems. bioRxiv. https://doi.org/10.1101/2020.03.24.004655
- Bocci, G., B. Bradfute, S., Ye, C., J. Garcia, M., Parvathareddy, J., Reichard, W., Surendranathan, S., Bansal, S., G. Bologa, C., J. Perkins, D., B. Jonsson, C., A. Sklar, L., I. Oprea, T., 2020. Virtual and In Vitro Antiviral Screening Revive Therapeutic Drugs for COVID-19. ACS Pharmacol. Transl. Sci. https://doi.org/10.1021/acsptsci.0c00131 [DOI] [PMC free article] [PubMed]
- Boise, L., González-García, M., Postema, C., Ding, L., Lindsten, T., Turka, L., Mao, X., Nuñez, G., Thompson, C., 1993. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell 74, 597–608. https://doi.org/10.1016/0092-8674(93)90508-n [DOI] [PubMed]
- Chan J., Watt V. eek and erk, new members of the eph subclass of receptor protein-tyrosine kinases. Oncogene. 1991;6:1057–1061. [PubMed] [Google Scholar]
- Chand, S., Edwards, N., Chue, C., Jesky, M., Stringer, S., Simmonds, M., Duf, f C., Cockwell, P., Harper, L., Steeds, R., Townend, J., Ferro, C., Borrows, R., 2016. Caveolin-1 single-nucleotide polymorphism and arterial stiffness in non-dialysis chronic kidney disease. Nephrol. Dial. Transplant. 31, 1140–4. https://doi.org/10.1093/ndt/gfv350 [DOI] [PubMed]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y. Epidemiological and clinical characteristics of 99 cases of 2019 coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin C., Chen S., Wu H., Ho C., Ko M., Lin C. cytoHubba: identifying hub objects and sub- networks from complex interactome. BMC Syst. Biol. 2014;8:S11. doi: 10.1186/1752-0509-8-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow M., Lim N., Ballouz S., Pavlidis P., Gillis J. Predictability of human differential gene expression. Proc. Natl. Acad. Sci. U. S. A. 2019;116:6450–6491. doi: 10.1073/pnas.1802973116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerberg L., Hallström B., Oksvold P., Kampf C., Djureinovic D., Odeberg J., Habuka M., Tahmasebpoor S. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteomics. 2014;13:397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Feng B., Han S., Lu L., Chen Y., Chu X., Wang R., Chen L. MicroRNA-129 in Human Cancers: From Tumorigenesis to Clinical Treatment. Cell. Physiol. Biochem. 2016;39:2186–2202. doi: 10.1159/000447913. [DOI] [PubMed] [Google Scholar]
- Genovese F., Manresa A.A., Leeming D.J., Karsdal M.A., Boor P. The extracellular matrix in the kidney: a source of novel non-invasive biomarkers of kidney fibrosis? Fibrogenesis Tissue Repair. 2014;7:4. doi: 10.1186/1755-1536-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman, R., Carey, V., Huber, W., Irizarry, R., Dudoit, S., 2005. limma: Linear Models for Microarray Data, in: Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer, New York. https://doi.org/10.1007/0-387-29362-0_23
- Gerayeli F.V., Milne S., Cheung C., Li X., Yang C.W.T., Tam A. COPD and the risk of poor outcomes in COVID-19: A systematic review and meta-analysis. EClinicalMedicine. 2021;33 doi: 10.1016/j.eclinm.2021.100789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granata S., Zaza G., Simone S., Villani G., Latorre D., Pontrelli P., Carella M., Schena F., Grandaliano G., Pertosa G. Mitochondrial dysregulation and oxidative stress in patients with chronic kidney disease. BMC Genomics. 2009;10:388. doi: 10.1186/1471-2164-10-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gremmel T., Müller M., Steiner S., Seidinger D., Koppensteiner R., Kopp C.W., Panzer S. Chronic kidney disease is associated with increased platelet activation and poor response to antiplatelet therapy. Nephrol. Dial. Transplant. 2013;28:2116–2122. doi: 10.1093/ndt/gft103. [DOI] [PubMed] [Google Scholar]
- Guan, W., Ni, Z., Hu, Yu, Liang, W., Ou, C., He, J., Liu, L., Shan, H., Lei, C., Hui, D.S.C., Du, B., Li, L., Zeng, G., Yuen, K., Chen, R., Tang, C., Wang, T., Chen, P., Xiang, J., Li, S., Wang, Jin-lin, Liang, Z., Peng, Y., Wei, L., Liu, Y., Hu, Ya-hua, Peng, P., Wang, Jian-ming, Liu, J., Chen, Z., Li, G., Zheng, Z., Qiu, S., Luo, J., Ye, C., Zhu, S., 2020. Clinical Characteristics of Coronavirus Disease 2019 in China. Th e new Engl. J. o f Med. https://doi.org/10.1056/NEJMoa2002032
- H Sillars‐Hardebol, A., Carvalho, B., AM Beliën, J., de Wit, M., M Delis‐van Diemen, P., Tijssen, M., A van de Wiel, M., Pontén, F., JA Fijneman, R., A Meijer, G., 2012. BCL2L1 has a functional role in colorectal cancer and its protein expression is associated with chromosome 20q gain. J. Pathol. 226, 442–50. https://doi.org/10.1002/path.2983 [DOI] [PubMed]
- Hasan M., Alam M., Shoombuatong W., Deng H., Manavalan B., Kurata H. NeuroPred-FRL: an interpretable prediction model for identifying neuropeptide using feature representation learning. Brief. Bioinform. 2021;bbab167 doi: 10.1093/bib/bbab167. [DOI] [PubMed] [Google Scholar]
- Hasan M., Basith S., Khatun M., Lee G., Manavalan B., Kurata H. Meta-i6mA: an interspecies predictor for identifying DNA N6-methyladenine sites of plant genomes by exploiting informative features in an integrative machine-learning framework. Brief. Bioinform. 2020;bbaa202 doi: 10.1093/bib/bbaa202. [DOI] [PubMed] [Google Scholar]
- Hasan M., Khatun M., Kurata H. iLBE for Computational Identification of Linear B-cell Epitopes by Integrating Sequence and Evolutionary Features. Genomics Proteomics Bioinforma. 2020 doi: 10.1016/j.gpb.2019.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan M., Schaduangrat N., Basith S., Lee G., Shoombuatong W., Manavalan B. HLPpred-Fuse: improved and robust prediction of hemolytic peptide and its activity by fusing multiple feature representation. Bioinformatics. 2020;36:3350–3356. doi: 10.1093/bioinformatics/btaa160. [DOI] [PubMed] [Google Scholar]
- HK, R., MR, A., MB, I., MB, A., P, L., F, H., JM, Q., MA., M., 2020. Machine Learning and Bioinformatics Models to identify Pathways that Mediate Influences of Welding Fumes on Cancer progression. Sci. Rep. 17, 1–5. [DOI] [PMC free article] [PubMed]
- Hottz E., Azevedo-Quintanilha I., Palhinha L., Teixeira L., Barreto E., Pão C., Righy C., Franco S., Souza T., Kurtz P., Bozza F., Bozza P. Platelet activation and platelet-monocyte aggregate formation trigger tissue factor expression in patients with severe COVID-19. Blood. 2020;136:1330–1341. doi: 10.1182/blood.2020007252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu S.-D., Lin F.-M., Wu W.-Y., Liang C., Huang W.-C., Chan W.-L., Tsai W.-T., Chen G.-Z., Lee C.-J., Chiu C.-M., Chien C.-H., Wu M.-C., Huang C.-Y., Tsou A.-P., Huang H.-D. miRTarBase: a database curates experimentally validated microRNA–target interactions. Nucleic Acids Res. 2011;39:D163–D169. doi: 10.1093/nar/gkq1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain A., Byrareddycde S.N. Rapamycin as a potential repurpose drug candidate for the treatment of COVID-19. Chem. Biol. Interact. 2020;331 doi: 10.1016/j.cbi.2020.109282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim S., Chinnadurai R., Ali I., Payne D., Rice G., Newman W., Algohary E., Adam A., Kalra P. Genetic polymorphism in C3 is associated with progression in chronic kidney disease (CKD) patients with IgA nephropathy but not in other causes of CKD. PLoS One. 2020;15 doi: 10.1371/journal.pone.0228101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam T., Rahman R., Aydin B., Beklen H., Yalcin K. Integrative transcriptomics analysis of lung epithelial cells and identification of repurposable drug candidates for COVID-19. Eur. J. Pharmacol. 2020;887 doi: 10.1016/j.ejphar.2020.173594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakiuchi C., Ishiwata M., Nanko S. Association analysis of HSP90B1 with bipolar disorder. J. Hum. Genet. 2007;52:794–803. doi: 10.1007/s10038-007-0188-4. [DOI] [PubMed] [Google Scholar]
- Kamyshnyi A., Krynytska I., Matskevych V., Marushchak M., Lushchak O. Arterial Hypertension as a Risk Comorbidity Associated with COVID-19 Pathology. Int. J. Hypertens. 2020;2020:7. doi: 10.1155/2020/8019360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A., Fornes O., Stigliani A., Gheorghe A., Castro-Mondragon J.A., Van Der Lee R., Bessy A., Chèneby J., Kulkarni S.R., Tan G., Baranasic D., Arenillas D.J., Sandelin A., Vandepoele K., Lenhard B., Ballester B., Wasserman W.W., Parcy F., Mathelier A. JASPAR 2018: Update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2018;46:D260–D266. doi: 10.1093/nar/gkx1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoriaty R., Ozel A., Ramdas S., Ross C., Desch K., Shavit J., Everett L., Siemieniak D., Li J., Ginsburg D. Genome-wide linkage analysis and whole-exome sequencing identifies an ITGA2B mutation in a family with thrombocytopenia. Br. J. Haematol. 2019;186:574–579. doi: 10.1111/bjh.15961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., McDermott M.G., Monteiro C.D., Gundersen G.W., Ma’ayan A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S., Son, K., Han, C., et al., 2021. Impact of COPD on COVID-19 prognosis: A nationwide population-based study in South Korea. Sci. Rep. https://doi.org/10.1038/s41598-021-83226-9 [DOI] [PMC free article] [PubMed]
- Love M.I., Huber W., Anders S. oderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Geneome Biol. 2015;550 doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Wan F., Zhang H., Shi G., Ye D. ITGA2B and ITGA8 are predictive of prognosis in clear cell renal cell carcinoma patients. Tumor Biol. 2016;37:253–262. doi: 10.1007/s13277-015-3792-5. [DOI] [PubMed] [Google Scholar]
- Lutz J., Menke J., Sollinger D., Schinzel H., Thürmel K. Haemostasis in chronic kidney disease. Nephrol. Dial. Transplant. 2014;29:29–40. doi: 10.1093/ndt/gft209. [DOI] [PubMed] [Google Scholar]
- Ma X., Gao Y., Fan Y., Ni D., Zhang Y., Chen W., Zhang P., Song E., Huang Q., Ai Q., Li H., Wang B., Zheng T., Shi T., Zhang X. Overexpression of E2F1 promotes tumor malignancy and correlates with TNM stages in clear cell renal cell carcinoma. PLoS One. 2013;8 doi: 10.1371/journal.pone.0073436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahase, E., 2020. Covid-19: First coronavirus was described in The BMJ in 1965 1547, 2020. https://doi.org/10.1136/bmj.m1547 [DOI] [PubMed]
- Mahmud S., Al-Mustanjid M., Akter F., Rahman M.S., Ahmed K., Rahman M.H., Chen W., Moni M. Bioinformatics and system biology approach to identify the influences of SARS-CoV-2 infections to idiopathic pulmonary fibrosis and chronic obstructive pulmonary disease patients. Brief. Bioinform. 2021;bbab115 doi: 10.1093/bib/bbab115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastaglio S., Ruggeri A., Risitano A., Angelillo P., Yancopoulou D., Mastellos D., Huber-Lang M., Piemontese S., Assanelli A., Garlanda C., Lambris J., Ciceri F. The first case of COVID-19 treated with the complement C3 inhibitor AMY-101. Clin. Immunol. 2020;215 doi: 10.1016/j.clim.2020.108450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostafaei S., Kazemnejad A., Azimzadeh Jamalkandi S. Identification of Novel Genes in Human Airway Epithelial Cells associated with Chronic Obstructive Pulmonary Disease (COPD) using Machine-Based Learning Algorithms. Sci. Rep. 2018;8:15775. doi: 10.1038/s41598-018-33986-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman E., Sellers W., McNeil J., Lawrence J., Kaelin W.J. Structure and partial genomic sequence of the human E2F1 gene. Gene. 1996;173:163–169. doi: 10.1016/0378-1119(96)00184-9. [DOI] [PubMed] [Google Scholar]
- Oh, J., et al., 2020. dentification of biological correlates associated with respiratory failure in COVID-19. medRxiv. https://doi.org/10.1101/2020.09.29.20204289 [DOI] [PMC free article] [PubMed]
- Osamu I., Saori O., Nobuya S., Yuka N., Yoshiharu H., Kon Y. Altered expression of microRNA miR-146a correlates with the development of chronic renal inflammation. kidney Int. 2012;81:P280–P292. doi: 10.1038/ki.2011.345. [DOI] [PubMed] [Google Scholar]
- Richardson, S., Hirsch, J.S., Narasimhan, M., Crawford, J.M., Mcginn, T., Davidson, K.W., 2020. Presenting Characteristics, Comorbidities, and Outcomes Among 5700 Patients Hospitalized With COVID-19 in the New York City Area 323, 2052–2059. https://doi.org/10.1001/jama.2020.6775 [DOI] [PMC free article] [PubMed]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43 doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert Thomas, M.D., Abbas Kanso, M.D., John R. Sedor, M.D., 2008. Chronic Kidney Disease and Its Complications. Prim Care 35, 329–vii. [DOI] [PMC free article] [PubMed]
- Sethupathy P., Corda B., Hatzigeorgiou A.G. TarBase: A comprehensive database of experimentally supported animal microRNA targets. RNA. 2006;12:192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot M.E., Ono K., Ruscheinski J., Wang P.L., Ideker T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solberg R., Sistonen P., Träskelin A.L., Bérubé D., Simard J., Krajci P., Jahnsen T., de la Chapelle A. Mapping of the regulatory subunits RI beta and RII beta of cAMP-dependent protein kinase genes on human chromosome 7. Genomics. 1992;14:63–69. doi: 10.1016/s0888-7543(05)80284-8. [DOI] [PubMed] [Google Scholar]
- Sun P., Qie S., Liu Z., Ren J., Li K., Xi J. Clinical characteristics of hospitalized patients with SARS-CoV-2 infection: a single arm meta-analysis. J. Med. Virol. 2020;92:612–617. doi: 10.1002/jmv.25735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P., Jensen L.J., Mering C. Von. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45 doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Zheng X., Peng Q. Eph receptors: the bridge linking host and virus. Cell. Mol. Life Sci. 2020;77:2355–2365. doi: 10.1007/s00018-019-03409-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Kidney Day: Chronic Kidney Disease [WWW Document], 2015. URL http://www.worldkidneyday.org/faqs/chronic-kidney-disease/.
- Worldometer, n.d.
- Xia J., Gill E.E., Hancock R.E.W. NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat. Protoc. 2015;10:823–844. doi: 10.1038/nprot.2015.052. [DOI] [PubMed] [Google Scholar]
- Xing Y., Wen Z., Gao W., Lin Z., Zhong J., Jiu Y. Multifaceted Functions of Host Cell Caveolae/Caveolin-1 in Virus Infections. Viruses. 2020;12:487. doi: 10.3390/v12050487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Jiang S., Cheng Y., Li T., Hu W., Ma Z., Chen F., Yang Y. FOXC1 in cancer development and therapy: deciphering its emerging and divergent roles. Ther. Adv. Med. Oncol. 2017;9:797–816. doi: 10.1177/1758834017742576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T., Wang Q., Zhang W., Bian A., Zhang J. Identification of genes associated with renal cell carcinoma using gene expression profiling analysis. Oncol. Lett. 2016;12:73–78. doi: 10.3892/ol.2016.4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo M., Shin J., Kim J., Ryall K.A., Lee K., Lee S., Jeon M., Kang J., Tan A.C. DSigDB: drug signatures database for gene set analysis. Bioinformatics. 2015;31:3069–3071. doi: 10.1093/bioinformatics/btv313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Moriguchi T., Souma T., Takai J., Satoh H., Morito N., Engel J.D., Yamamoto M. GATA2 regulates body water homeostasis through maintaining aquaporin 2 expression in renal collecting ducts. Mol. Cell. Biol. 2010;34:1929–1941. doi: 10.1128/MCB.01659-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X., Chen K., Zou J., Han P., Hao J., Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front Med. 2020 doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data utilized in this manuscript are available online from their respective NCBI GEO database with accession number GSE152418 (COVID-19) and GSE15072 (CKD).