Abstract
Background
Radiologists have difficulty distinguishing benign from malignant bone lesions because these lesions may have similar imaging appearances. The purpose of this study was to develop a deep learning algorithm that can differentiate benign and malignant bone lesions using routine magnetic resonance imaging (MRI) and patient demographics.
Methods
1,060 histologically confirmed bone lesions with T1- and T2-weighted pre-operative MRI were retrospectively identified and included, with lesions from 4 institutions used for model development and internal validation, and data from a fifth institution used for external validation. Image-based models were generated using the EfficientNet-B0 architecture and a logistic regression model was trained using patient age, sex, and lesion location. A voting ensemble was created as the final model. The performance of the model was compared to classification performance by radiology experts.
Findings
The cohort had a mean age of 30±23 years and was 58.3% male, with 582 benign lesions and 478 malignant. Compared to a contrived expert committee result, the ensemble deep learning model achieved (ensemble vs. experts): similar accuracy (0·76 vs. 0·73, p=0·7), sensitivity (0·79 vs. 0·81, p=1·0) and specificity (0·75 vs. 0·66, p=0·48), with a ROC AUC of 0·82. On external testing, the model achieved ROC AUC of 0·79.
Interpretation
Deep learning can be used to distinguish benign and malignant bone lesions on par with experts. These findings could aid in the development of computer-aided diagnostic tools to reduce unnecessary referrals to specialized centers from community clinics and limit unnecessary biopsies.
Funding
This work was funded by a Radiological Society of North America Research Medical Student Grant (#RMS2013) and supported by the Amazon Web Services Diagnostic Development Initiative.
Keywords: Deep learning, MRI, Bone tumor, Convolutional neural network, Bone lesion
Research in context.
Evidence before this study
Literature searches were conducted using the PubMed search engine using the following search terms: (“machine learning” OR “deep learning” OR “convolutional neural network”) AND (“bone tumor” OR “bone lesion”) AND (“diagnosis” OR “benign malignant”) AND (“imaging” OR “MRI”). Our search identified one previous study that utilized neural networks to distinguish benign and malignant bone tumors on radiographs, but this study involved manually rather than automatically encoded imaging features, did not study advanced imaging modalities, and did not include external validation. Another study utilized a Bayesian model for histological diagnosis of bone tumors, but this study was also based on radiographic imaging and did not include external validation. Both of these studies also suffered from small sample sizes. We did not find any studies involving deep learning techniques for characterization of bone lesions on MRI.
Added value of this study
In this study, we demonstrate that convolutional neural networks trained with MRI studies in combination with a logistic regression based upon clinical data are able to discern benign and malignant bone lesions with performance equivalent to that of expert musculoskeletal radiologists. Our study utilizes a multi-institutional dataset and includes external validation to ensure the generalizability of our findings.
Implications of all the available evidence
By providing a validated assessment of bone lesions on MRI, our approach has the potential to aid in diagnostic evaluation of bone lesions, particularly non-expert primary evaluation outside of specialist centers. Moreover, morbidity related to unnecessary biopsy of benign lesions can be reduced by enabling radiologists to rule out malignancy with greater confidence.
Alt-text: Unlabelled box
1. Introduction
Cancer of the bones and joints was the 3rd leading cause of cancer-related deaths in people under the age of 20 in the United States in 2016, with approximately 3,500 new bone cancer diagnoses in 2019 [1]. Outside of bone metastases (secondary bone tumors) and plasma cell myeloma, the most common bone malignancies are osteosarcoma, chondrosarcoma, and Ewing sarcoma [2], [3], [4]. While the incidence of benign bone tumors is more difficult to determine because they are rarely fully evaluated or biopsied, osteochondroma, enchondroma and osteoid osteoma are among the most common benign tumors [3].
Radiographs are the recommended first line imaging modality for the characterization of bone lesions, as it typically provides clear assessment of lesion location, internal matrix, margins, and associated periosteal reaction [3]. These lesion characteristics in combination with patient age are often sufficient to provide differential diagnoses of bone lesions [3], [5]. However, radiographs have limitations. Superimpositions, poorly-visualized partial cortex destruction, and difficulties analyzing flat and short bones and soft tissues can make radiographic diagnosis more challenging [6]. In addition, the clinical symptoms and radiographic appearance of infections (osteomyelitis) often mimic those of bone tumors [5,7]. Image diagnosis of primary bone lesions can be further complicated by the presence of pathologic fractures, which can increase the amount of fluid, hemorrhage, or edema in and around a lesion; this is particularly relevant for benign lesions such as non-ossifying fibroma, aneurysmal bone cyst, and fibrous dysplasia [2]. Chondroblastoma, osteoid osteoma, and Langerhans cell histiocytosis are among the benign bone lesions that can present with extra-lesional edema-like signal even in the absence of pathological fracture [8].
In cases with such complicating factors or any case in which a lesion is indeterminate or potentially aggressive, advanced imaging with MRI is warranted. MRI is highly sensitive for the detection of bone abnormalities due to its ability to characterize bone marrow involvement, soft tissue invasion, and fluid content of lesions [3,9]. The excellent tissue contrast provided by MR imaging can occasionally yield sufficient information to allow a specific histologic diagnosis to be made (e.g., intraosseous lipoma, enchondroma, hemangioma, or aneurysmal bone cyst) [3], [9], [10]. However, even when combining plain radiograph with MRI, radiologists were 100% sensitive but only 55% specific and 73% accurate in classifying bone malignancy in a small dataset [11]. Upon considering the limitations of advanced imaging and the rarity with which bone tumors are encountered clinically, a clear need emerges for technologies to aid in the diagnosis of bone tumors.
Artificial intelligence tools have been used to augment the ability of radiologists to assess the malignancy of tumors, including from MR images. Previous studies have showcased the ability of convolutional neural network (CNN) models to classify breast, prostate, kidney and brain lesions on MRI with high sensitivity and specificity [12], [13], [14], [15], [16]. While the body of literature is limited, some studies have employed such techniques for the classification of bone lesions. As early as 1994, rudimentary two-layer, feed-forward neural network models were used to distinguish benign from malignant bone tumors with 85% accuracy, 76% specificity and 89% sensitivity. However, this outcome was based upon a dataset of only 709 lesions, manually encoded radiographic characteristics, and, critically, training rather than validation performance [17]. In a more recent work, Do et al. used a Naïve Bayesian model trained upon 18 demographic and radiographic features to determine a top-3 histological differential diagnosis of 710 bone tumors, capturing the true diagnosis with 60% accuracy [18]. Our research group has recently demonstrated that convolutional neural networks can be used to achieve 3-class discrimination of bone tumors on radiographs according to histopathologic categories with 73% accuracy, comparable to expert radiologists [19].
With current state-of-the-art machine learning methods, it may be possible to achieve better performance in automated bone tumor characterization through analysis of advanced imaging. In this study, we utilized deep learning to develop a malignancy prediction algorithm for bone lesions on routine MRI.
2. Methods
2.1. Study participants
1368 lesions with pre-operative MRI demonstrating single or multiple lesions with apparent bone involvement and histologically confirmed diagnosis following biopsy or surgery were retrospectively identified from the 5 hospitals from 2006 to 2020 by consecutive sampling. Lesions were identified according to the World Health Organization (WHO) system for the classification of bone and soft tissue tumors. The World Health Organization classifies bone tumors into histological categories based upon the potential of the tumor to cause local tissue destruction and metastasize to distant sites. In order of disease severity, tumors can be classified as: 1) Benign, 2) Intermediate, locally aggressive (possibility of destructive local recurrence). 3) Intermediate, rarely metastasizing (as above, with the additional possibility of metastasis) or 4) Malignant [4,20,21]. Tumors classified as intermediate according to the WHO classification criteria were grouped as benign, as each of the diagnoses present in this group (osteoblastoma, desmoplastic fibroma, giant cell tumor, epithelioid hemangioma, myofibromatosis, Langerhans cell histiocytosis, and myoepithelioma) are in practice generally considered benign (henceforth collectively refer to as benign).
Age at time of imaging, gender, and the skeletal location of the lesion of interest were extracted from patients’ electronic medical record. Exclusion criteria were incomplete imaging protocols lacking a T1- or T2-weighted sequence (T1W or T2W, respectively), inconclusive involvement of osseous structures, and insufficient image quality for analysis. Inclusion and exclusion criteria are described in Supplementary Figure 1.
2.2. Image Segmentation and Preprocessing
The MR images were manually segmented by a radiologist with 3 years of experience reading musculoskeletal MRI using 3D Slicer (version 4.10). N4 bias correction and intensity normalization were performed upon each image using SimpleITK [22]. The intensity of each image was normalized relative to a reference image, with all images acquired using a given sequence (i.e. T1W or T2W) normalized with a single, high-resolution reference of the same sequence. Each image was cropped to a rectangular volume of interest delineated by the widest and tallest non-zero valued pixels in the segmentation. The largest sagittal, axial and coronal slices of each processed image volume were selected as inputs for the classification model; this 2.5D approach has been shown to have robust performance relative to 3D image classification approaches but with significantly reduced computational cost [23,24].
2.3. Model design
2.3.1. Imaging data models
Models for image classification were developed by adapting the EfficientNet deep learning architecture. EfficientNet is a state-of-the-art image classification network architecture that is an improvement upon previously developed convolutional neural network designs as it improves accuracy while significantly decreasing the number of network parameters and thereby substantially improving computational efficiency. EfficientNet models initialized with weights pre-trained on the ImageNet database were used for feature extraction from imaging data. The EfficientNet classifying layer was replaced with a series of fully connected layers of size 256, 128, 64, 32 and 16 nodes with interposing dropout layers and batch normalization layers. A final classification layer with a single node and sigmoid activation was used to perform the binary classification task.
2.3.2. Clinical data model
A logistic regression model using clinical variables was separately developed for the classification task. Inputs were patient age, sex, and lesion location. 21 locations (clavicle, cranium, proximal femur, distal femur, foot, proximal radius, distal radius, proximal ulna, distal ulna, hand, hip, proximal humerus, distal humerus, proximal tibia, distal tibia, proximal fibula, distal fibula, mandible, rib/chest wall, scapula, or spine) were one-hot encoded such that the model received 23 distinct quantified input variables.
2.3.3. Ensemble model
The imaging and clinical feature models were then combined using a stacked ensemble approach in which a voting ensemble received malignancy probabilities from the imaging and clinical feature models as inputs and created outputs based upon a summation of the predicted probabilities. Each ensemble classification model consisted of the outputs of an EfficientNet trained upon T1W imaging studies, an EfficientNet trained upon T2W imaging studies, and a logistic regression model based upon clinical features.
2.4. Model training and evaluation
2.4.1. Imaging data models
Binary classification models were trained to distinguish benign from malignant bone lesions on T1W and T2W images. 4-fold cross validation was used to evaluate the model building pipeline and select hyperparameters for the final trained models. The data from CHOP, HUP, RIH, and SXH were used for cross validation, as well as training, validation, and internal testing for the final models by a 7:2:1 split. Data from XH was reserved for external testing to assess generalizability of the created algorithms to data from separate institutions. Using the EfficientNet-B0 architecture, models were trained with a batch size of 64 for 200 epochs during cross-validation and 200 epochs with early stopping after 100 epochs of no loss improvements on the validation set during final model training. Models were implemented in Python (version 3.8) and trained on a machine with a NVIDIA Tesla V100 GPU.
During training, segmented images were scaled up or down to 200 × 200 pixels using bilinear interpolation. The training set was augmented with horizontal flip, vertical flip, shear, and zoom transformations to add variability. A predetermined probability of 0·5 was assigned to the final sigmoid activation neuron as a threshold for classification of malignancy. Loss on the validation set was monitored over each epoch and the model with the minimum validation loss was selected to represent a given training trial. The hyperparameters that produced the best average test performance in cross-validation were selected for training the final image classification models.
2.4.2. Clinical data model
The logistic regression model for clinical feature-based classification was trained with L2 regularization and a stochastic averaged gradient descent optimizer. Feature ranking with recursive feature elimination and 4-fold cross-validated selection of the best feature set was implemented for the creation of the clinical data model. Training involved all features initially then the least predictive feature was removed with each iteration until the desired feature set size was achieved. Feature set sizes from 1 to 23 (all features) were trialed. The feature set with maximum cross validation test performance was selected as the final feature set.
2.4.3. Ensemble model
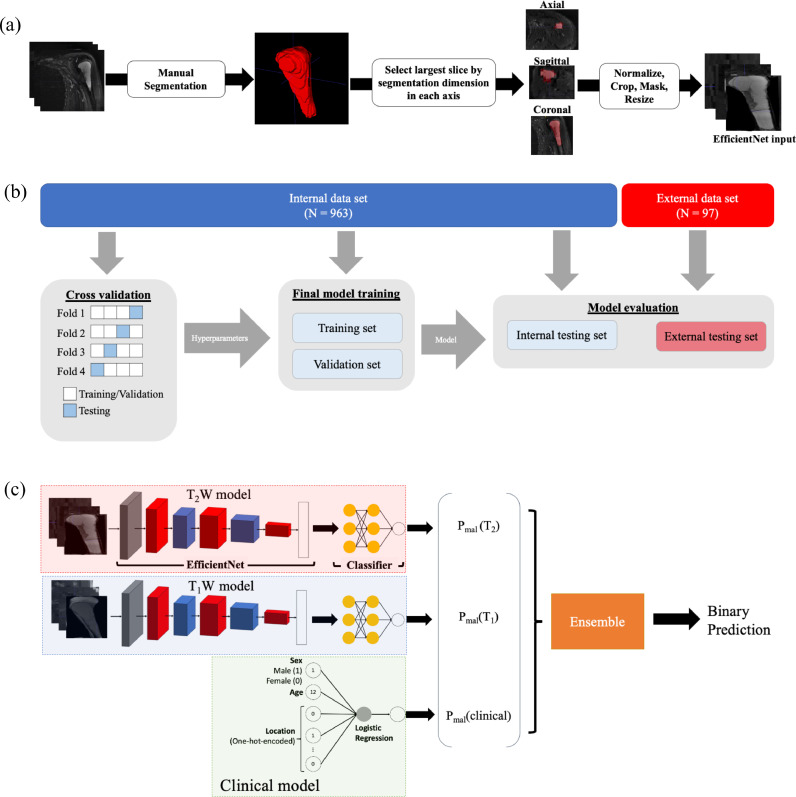
To incorporate sensitivity bias in the voting ensemble, malignancy thresholds for each of the constituent models were empirically determined via a grid search algorithm that maximized Youden's index while achieving at least 90% sensitivity on the validation set [25]. Figure 1 illustrates the data processing pipeline and model architecture.
Figure 1.
Schematic of the bone tumor classification deep learning pipeline. Top: Image segmentation. Raw image volumes were manually segmented to a region of interest focused upon the tumor. The largest axial, transverse, and coronal slices of the segmented volume were used as inputs for the imaging models (“2.5D” image representation). Middle: Training and evaluation scheme. Hyperparameters were selected based upon 4-fold cross validation scheme. Final models were trained using the training and validation data sets then evaluated using the internal and external testing sets, where the external testing set was from an independent institution. Bottom: Model architecture. An EfficientNet-B0 took T1-weighted images as an input and output a malignancy probability; another EfficientNet-B0 took T2-weight images as inputs. A logistic regression model accepted age, binary-encoded sex, and one-hot encoded lesion location as inputs and output a malignancy probability. A voting ensemble model used classifications from the T1W, T2W, and clinical features models as inputs and output a final classification by a soft, probability-based majority rule vote.
2.5. Radiologist Interpretation
Three expert radiologists (Y.H., R.S., Y.L.) with 3, 8, and 7 years of experience reading musculoskeletal MRI respectively, blind to histopathologic data, evaluated unsegmented MRI images of the bone lesions in the internal test set for malignancy. T1-weighted and T2-weighted images were made available to the evaluators for each lesion in the internal test set; T1 contrast-enhanced (T1C) images were also provided to the evaluators when available but were not used in the model training and evaluation due to limited availability among samples in the dataset. The evaluators were also given demographic information (age and sex) for each patient. The model's results were compared to these expert evaluations and a contrived “expert committee” (expert decision by majority rule) to assess model performance. Supplementary Table 1 shows information regarding the previous experience of the radiologists who evaluated the lesions in the internal test set in reading musculoskeletal MRI.
2.6. Statistics
The demographic and clinicopathologic features of the benign and malignant groups were compared using a chi-squared test for categorical variables (lesion location, sex) and a T-test or single-factor ANOVA for continuous variables (age). Post hoc chi-squared tests pairing lesions of each location against a subset comprised of lesions from all other locations were performed following indication of an overall statistically significant difference in location between the groups. These “one-vs-rest” comparisons were performed with Bonferroni-corrected p-values for significance. The same analyses were performed to compare the combined training and validation set to the internal testing and external testing datasets.
Accuracy, sensitivity, specificity, and area under the Receiver-Operator Characteristic curve (ROC AUC) were calculated for the classification task with 95% confidence intervals determined by the Wilson method [26]. ROC AUC on the validation set was used to empirically select final training hyperparameters for the imaging data models during cross validation. Fleiss’ κ was used to evaluate interrater reliability. Model binary classification performance was compared to expert committee performance using the McNemar test. Statistical significance was defined as P < 0·05. Statistical analyses were performed using Python (version 3.8) statistical libraries.
2.7. Code Availability
The image classification models were deployed with an implementation of the EfficientNet architecture using the Python Keras library (https://github.com/qubvel/efficientnet). The clinical feature-based logistic regression model was implemented using the Python scikit-learn library (version 0·24.1). All code for image preprocessing and malignancy prediction is publicly available at https://github.com/sopeeweje/Bone-MRI.
2.8. Ethics Statement
Our study received a waiver of informed consent and exempt status from the institutional review boards of the Hospital of the University of Pennsylvania (HUP) (protocol number 831582) and the Children's Hospital of Pennsylvania (CHOP) (protocol number 20-017327) in Philadelphia, PA, and Rhode Island Hospital (RIH) (protocol number 1747284) in Providence, Rhode Island. The study was also approved by the institutional review boards of the Xiangya Hospital (XH) and Second Xiangya Hospital (SXH) of Central South University in Hunan, China.
2.9. Role of the funding source
The study sponsors did not have any role in the study design; the collection, analysis and interpretation of data; in writing of the report; or in the decision to submit the paper for publication.
3. Results
3.1. Study Participants
The final cohort consisted of 1060 lesions – 185 from HUP, 464 from CHOP, 208 from SXH, 111 from RIH, and 97 from XH. Table 1 summarizes the clinical characteristics of the study participants and Supplementary Table 2 shows the detailed histopathological diagnoses. The sample had a mean age of 30±23 years and comprised 619 males and 441 females. 582 lesions (27±20 years, 342 males) were benign and 478 lesions (34±25 years, 277 males) were malignant. Comparing the benign and malignant lesion groups, there was a balanced gender distribution (p = 0·79) and significant differences in age (p < 0·001) and lesion location (p < 0·001). Upon “one-vs-rest” comparison, there was a statistically significant difference in malignancy distribution for lesions located in the cranium, hand, foot, hip, and spine. 678, 192, 93, and 97 lesions were allocated to the training, validation, internal testing, and external testing sets respectively. Supplementary Table 3 summarizes the characteristics of the training, validation, and internal test and external test sets.
Table 1.
Characteristics of patients included in the study. “One-vs-rest” tests for statistical significance in location distribution (e.g. Foot vs. Rest) were performed with Bonferroni-corrected p-values used for significance. ***Statistically significant
| Benign (N=582) | Malignant (N=478) | p-value | |
|---|---|---|---|
| Age (years ± SD) | 27 ± 20 | 34 ± 25 | <0·001*** |
| Sex (%) | 0·79 | ||
| Male | 342 (59%) | 277 (58%) | |
| Female | 240 (41%) | 201 (42%) | |
| Location (%) | <0·001*** | ||
| Clavicle | 3 (0.7%) | 5 (1.7%) | 0.52 |
| Cranium | 12 (2.6%) | 55 (18.3%) | <0·001*** |
| Proximal femur | 74 (16.1%) | 35 (11.6%) | 0.0055 |
| Distal femur | 80 (17.4%) | 57 (18.9%) | 0.43 |
| Foot | 89 (19.3%) | 5 (1.7%) | <0·001*** |
| Proximal radius | 0 (0%) | 1 (0.3%) | 0.92 |
| Distal radius | 13 (2.8%) | 0 (0%) | 0.0026 |
| Proximal ulna | 4 (0.9%) | 2 (0.7%) | 0.87 |
| Distal ulna | 1 (0.2%) | 0 (0%) | 0.92 |
| Hand | 29 (6.3%) | 1 (0.3%) | <0·001*** |
| Hip | 41 (8.9%) | 62 (20.6%) | 0.0017 |
| Proximal humerus | 34 (7.4%) | 33 (11%) | 0.56 |
| Distal humerus | 16 (3.5%) | 8 (2.7%) | 0.34 |
| Proximal tibia | 65 (14.1%) | 37 (12.3%) | 0.075 |
| Distal tibia | 11 (2.4%) | 4 (1.3%) | 0.24 |
| Proximal fibula | 6 (1.3%) | 10 (3.3%) | 0.24 |
| Distal fibula | 4 (0.9%) | 1 (0.3%) | 0.5 |
| Mandible | 3 (0.7%) | 7 (2.3%) | 0.2 |
| Rib/Chest wall | 6 (1.3%) | 19 (6.3%) | 0.0033 |
| Scapula | 10 (2.2%) | 7 (2.3%) | 0.93 |
| Spine | 81 (17.6%) | 129 (42.9%) | <0·001*** |
3.2. Model training and evaluation
Performance of the imaging data training algorithms in cross-validation is shown in Supplementary Figure 2. Results of the grid search for thresholds in the voting ensemble are shown in Supplementary Figure 3. Performance of the final T1W, T2W, clinical features and ensemble models on the internal test set in comparison to expert evaluations and the external test set is described in Table 2 and performance of the models on the training and validation sets is summarized in Supplementary Table 4.
Table 2.
Performance of T1W, T2W, clinical features and ensemble models on the internal test set (n = 93) compared with expert evaluation, as well as the external test set (n = 97). p-value as calculated by the McNemar test for each expert is for accuracy relative to the performance of the ensemble model. Abbreviations - ROC AUC, area under ROC curve; PPV, positive predictive value; NPV, negative predictive value; 95% CI, 95% confidence interval.
| Internal Test Set | ||||||||
|---|---|---|---|---|---|---|---|---|
| Modality | F1 Score | ROC AUC | Accuracy (95% CI) | Sensitivity (95% CI) | Specificity (95% CI) | PPV | NPV | p-value |
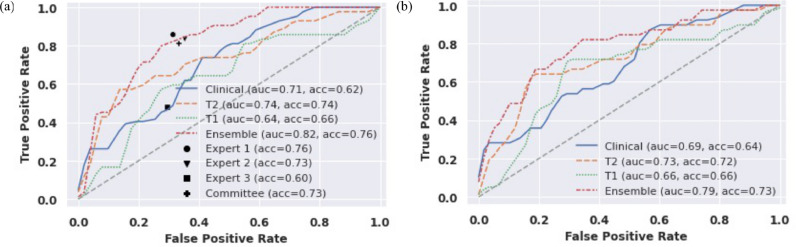
| Clinical | 0·58 | 0·71 | 0·62 (0·52-0·72) | 0·57 (0·42-0·71) | 0·67 (0·53-0·78) | 0·59 | 0·65 | - |
| T1W | 0·59 | 0·64 | 0·66 (0·55-0·74) | 0·55 (0·40-0·69) | 0·75 (0·61-0·85) | 0·64 | 0·67 | - |
| T2W | 0·67 | 0·74 | 0·74 (0·64-0·82) | 0·57 (0·42-0·71) | 0·88 (0·76-0·95) | 0·80 | 0·71 | - |
| Ensemble | 0·75 | 0·82 | 0·76 (0·67-0·84) | 0·79 (0·64-0·89) | 0·66 (0·53-0·78) | 0·72 | 0·81 | - |
| Expert 1 | 0·77 | - | 0·76 (0·66-0·84) | 0·86 (0·72-0·94) | 0·68 (0·54-0·79) | 0·69 | 0·85 | 1.0 |
| Expert 2 | 0·74 | - | 0·73 (0·63-0·81) | 0·83 (0·69-0·92) | 0·64 (0·50-0·76) | 0·66 | 0·82 | 0.66 |
| Expert 3 | 0·52 | - | 0·60 (0·50-0·69) | 0·48 (0·33-0·62) | 0·70 (0·56-0·81) | 0·57 | 0·61 | 0·02 |
| Expert Committee | 0·73 | - | 0·73 (0·63-0·81) | 0·81 (0·67-0·90) | 0·66 (0·52-0·78) | 0·67 | 0·81 | 0.7 |
| External Testing Set | ||||||||
| Modality | F1 Score | ROC AUC | Accuracy (95% CI) | Sensitivity (95% CI) | Specificity (95% CI) | PPV | NPV | |
| Clinical | 0·52 | 0·69 | 0·64 (0·54-0·73) | 0·49 (0·34-0·64) | 0·74 (0·62-0·84) | 0·56 | 0·68 | |
| T1W | 0·51 | 0·66 | 0·66 (0·56-0·75) | 0·44 (0·29-0·59) | 0·81 (0·69-0·89) | 0·61 | 0·68 | |
| T2W | 0·65 | 0·73 | 0·72 (0·62-0·80) | 0·64 (0·48-0·77) | 0·78 (0·65-0·87) | 0·66 | 0·76 | |
| Ensemble | 0·70 | 0·79 | 0·73 (0·64-0·81) | 0·77 (0·61-0·88) | 0·71 (0·58-0·81) | 0·63 | 0·82 | |
On internal testing, the clinical variable logistic regression achieved an accuracy of 0·62 (95% CI: 0·52-0·72), F1 score of 0·58, sensitivity of 0·57 (95% CI: 0·42-0·71), and specificity of 0·67 (95% CI: 0·53-0·78). On external testing, the logistic regression model based on clinical variables achieved an accuracy of 0·64 (95% CI: 0·54-0·73), F1 score of 0·52, sensitivity of 0·49 (95% CI: 0·34-0·64), and specificity of 0·74 (95% CI: 0·62-0·84). The generated regression equation was:0.566Age + 0.955 Cranium + 0.705Hip + 0.588Spine + 0.438RibChest + 0.438DistalFemur + 0.398ProximalTibia
Location in the hand (-0·505) and foot (-0·815) were the most heavily weighted determinants towards benign classification and location in the hip (+0·705) and cranium (+0·955) were the most heavily weighted determinants towards malignant classification.
The final T1W model was trained with stochastic gradient descent optimization with Nesterov momentum, learning rate of 0·001, and dropout rate of 0·2 in the classifier. On the internal test set, the T1W model achieved a test accuracy of 0·66 (95% CI: 0·55-0·74), F1 score of 0·59, sensitivity of 0·55 (95% CI: 0·40-0·69), and specificity of 0·75 (95% CI: 0·61-0·85). On the external test set, the T1W trained model achieved a test accuracy of 0·66 (95% CI: 0·56-0·75), F1 score of 0·51, sensitivity of 0·44 (95% CI: 0·29-0·59), and specificity of 0·81 (95% CI: 0·69-0·89).
The final T2W model was trained with Adam optimization and dropout rate of 0·4 in the classifier. On the internal test set, the T2W model achieved a test accuracy of 0·72 (95% CI: 0·62-0·80), F1 score of 0·75, sensitivity of 0·64 (95% CI: 0·48-0·77), and specificity of 0·78 (95% CI: 0·65-0·87). On the external test set, the T2W model achieved a test accuracy of 0·74 (95% CI: 0·62-0·80), F1 score of 0·65, sensitivity of 0·64 (95% CI: 0·48-0·77), and specificity of 0·78 (95% CI: 0·65-0·87).
On the internal test set, the ensemble model achieved a test accuracy 0·76 (95% CI: 0·67-0·84), F1 score of 0·75, and sensitivity of 0·79 (95% CI: 0·64-0·89), and specificity of 0·75 (95% CI: 0·61-0·85). On the external test set, the ensemble model achieved a test accuracy of 0·73 (95% CI: 0·64-0·81), F1 score of 0·70, sensitivity of 0·77 (95% CI: 0·61-0·88), and specificity of 0·71 (95% CI: 0·58-0·81). Adding an optimized T1C-trained model to the ensemble model neither supplemented nor decremented internal test set performance (Supplementary Table 5).
3.3. Radiologist Interpretation
In evaluating the internal test set, expert 1 achieved a test accuracy of 0·76 (95% CI: 0·66-0·84), F1 score of 0·77, and sensitivity of 0·86 (95% CI: 0·72-0·94), and specificity of 0·68 (95% CI: 0·54-0·79). Expert 2 achieved a test accuracy of 0·73 (95% CI: 0·63-0·81), F1 score of 0·74, and sensitivity of 0·83 (95% CI: 0·69-0·92), and specificity of 0·64 (95% CI: 0·50-0·76). Expert 3 achieved a test accuracy of 0·60 (95% CI: 0·50-0·69), F1 score of 0·52, and sensitivity of 0·48 (95% CI: 0·33-0·62), and specificity of 0·70 (95% CI: 0·56-0·81). Interrater reliability as calculated with Fleiss’ κ was 0·02.
The expert committee assembled by majority-rule achieved a test accuracy of 0·73 (95% CI: 0·63-0·81), F1 score of 0·73, and sensitivity of 0·81 (95% CI: 0·67-0·90), and specificity of 0·67 (95% CI: 0·53-0·78). Compared to the expert committee, the ensemble deep learning model achieved similar accuracy (0·76 vs. 0·73, p=0·7 [McNemar test]), sensitivity (0·79 vs. 0·81, p=1·0 [McNemar test]) and specificity (0·75 vs. 0·66, p=0·48 [McNemar test]). Figure 2 shows the ROC curve for each model overlaid with expert performance on the internal test set and the ROC curve for the models’ performance on the external test set.
Figure 2.
Receiver-Operator Characteristic (ROC) curves for all models on internal test data set (n = 93) compared to expert performance and on the external test data set (n = 97).
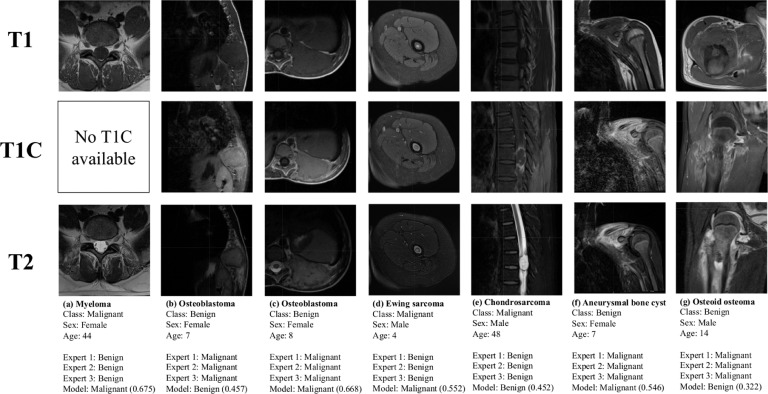
There were 7 tumors out of the 92 cases in the test set that were classified incorrectly by all 3 evaluators. These cases are depicted in Figure 3. 4 out of these 7 cases were benign entities incorrectly assessed as malignant by all 3 raters and the remaining 3 were malignant entities incorrectly assessed as benign. The model was correctly able to assess malignancy in 4 out of these 7 cases. Table 3 shows the performance of the expert evaluators and the model in classifying the benign and malignant lesion types that were most frequent in the test set. No statistically significant differences in performance by lesion were observed.
Figure 3.
Cases in the test set that were misclassified by all experts. Model classifications are displayed with the probability of malignancy determined by the model.
Table 3.
Performance of the experts and the ensemble model in classifying high frequency benign and malignant lesions in the internal test set.
| Malignant Tumors | N | Expert 1 accuracy | Expert 2 accuracy | Expert 3 accuracy | Expert committee | Model accuracy |
|---|---|---|---|---|---|---|
| Osteosarcoma | 11 | 90•9% | 100•0% | 81•8% | 100% | 90.9% |
| Ewing sarcoma | 12 | 83•3% | 91•7% | 41•7% | 83•3% | 91•7% |
| Multiple Myeloma | 8 | 87•5% | 62•5% | 62•5% | 75•0% | 75.0% |
| Chondrosarcoma | 5 | 60•0% | 80•0% | 20•0% | 60•0% | 60•0% |
| Benign Lesions | N | Expert 1 accuracy | Expert 2 accuracy | Expert 3 accuracy | Expert committee | Model accuracy |
| Giant cell tumor of the bone | 9 | 44•4% | 44•4% | 77•8% | 44•4% | 77•8% |
| Chondroblastoma | 7 | 100•0% | 85•7% | 42•9% | 85•7% | 85•7% |
| Enchondroma | 6 | 83•3% | 83•3% | 100•0% | 83•3% | 100% |
| Aneurysmal Bone Cyst | 6 | 50•0% | 33•3% | 66•7% | 50•0% | 50•0% |
| Osteomyelitis | 5 | 100•0% | 100•0% | 80•0% | 100•0% | 100% |
4. Discussion
MRI is the go-to advanced imaging modality for the evaluation of potentially suspicious bone lesions prior to biopsy or intervention. The diagnosis of bone lesions on imaging is complicated by the rarity with which they are encountered in clinical practice and the non-specific presentations of various benign and malignant entities. In this study, we utilized a deep learning method combining routine MRI images and clinical characteristics to develop a model to classify the malignancy of bone lesions. The model was a voting ensemble comprised of EfficientNets trained upon T1-weighted and T2-weighted images and a logistic regression trained upon patient age, sex, and tumor location. Generalizability was effectively demonstrated by showing a lack of significant decrement in performance on validation with an external data set. As shown in Table 3, the model was able to classify benign entities such as giant cell tumor and malignant entities such as Ewing sarcoma and multiple myeloma with higher accuracy than the experts. The analysis was not sufficiently powered to observe statistically significance differences in classification performance by lesion but expanding the test set may have allowed for such differences to be determined.
There is significant value in a model that can recapitulate the assessment of bone lesions on MRI by expert musculoskeletal radiologists. In one review of patients with equivocal findings on initial MRI that were subsequently referred to an orthopedic oncology clinic, radiologists at the clinic found that one-third of 390 referred patients had images that were clearly characteristic of non-neoplastic entities or benign tumors that did not in fact require follow-up with an orthopedic oncologist [27]. These unnecessary referrals complicate the task of identifying malignancy for specialist radiologists. In this context, a sensitive validated model for the characterization of suspicious bone lesions could perhaps reduce the rate of unnecessary referrals to higher levels of care and reduce patient anxiety regarding a potential cancer diagnosis. Both the experts and the model were highly sensitive, the former due to an inherent bias towards avoiding false negative diagnoses and the latter due to an encoded bias designed to mimic the expert approach. However, specific assessment of bone lesions would have also proven valuable. Unnecessary biopsy of benign lesions falsely considered malignant can create undue patient stress and leave patients at risk of post-operative complications, especially when managed outside of specialist multidisciplinary centers [28]. In addition, biopsy can be non-diagnostic in up to 30% of cases, subjecting patients to repeat biopsy procedures and a higher risk of complication [28,29]. A computer-aided diagnostic tool that can identify benign lesions with high specificity would be valuable in reducing the rate of unnecessary biopsies, by aiding radiologists in ruling in malignancy with greater certainty. By adjusting our thresholding approaches, we could easily create additional models that are biased towards high specificity performance to be used for this purpose.
Explainability is a significant barrier to the utilization of machine learning methods to support clinical practice. Our clinical features model represents a step towards a more explicit understanding of artificial decision-making for clinical diagnostics. The clinical model was correctly able to predict hand and foot locations as negative predictors of malignancy and cranial and spinal locations as positive predictors of malignancy. The majority of tumors affecting the hand, cranium and spine are enchondromas; chordomas and chondrosarcomas; and bone metastases and multiple myeloma, respectively [30], [31], [32], [33]. While the majority of bone tumors of the foot are benign, our clinical features model likely associated foot location with benign nature because the majority of foot-located lesions in our cohort were osteomyelitis, a bone tumor-mimicker that commonly affects the lower extremity. The clinical model also predicted increased probability of malignancy with increased age which is consistent with the epidemiology of benign bone tumors, most of which occur in the first two decades of life. Sex had no predictive value for the model, which is consistent with the observation that most bone lesions show no particular gender predilection [34].
There were select tumors that were misclassified by all expert evaluators but correctly assessed by the model (Figure 3). Ewing sarcoma (Figure 3d) has a heterogenous appearance on MRI and can be difficult to clinically diagnose in its earliest stages prior to the significant cortical destruction that occurs following spread beyond the bone marrow [35,36]. Osteoid osteoma (Figure 3g) frequently demonstrates an abnormally high peritumoral signal intensity on MRI due to hyperemia and consequent bone marrow edema, resulting in frequent misinterpretation [11,37]. There were also cases that were misclassified by all expert evaluators as well as the model. Osteoblastoma (Figure 3c) is an uncommon benign bone tumor with rib involvement in less than 5% of cases. The expansile growth pattern with well-defined margins exhibited by both of the present tumors is consistent with previously documented observations of rib osteoblastomas; however, these are somewhat aggressive features that are frequently considered to be on the borderline of osteoblastoma and low-grade osteosarcoma [38,39]. Aneurysmal bone cyst (Figure 3f) shares several clinical and imaging features with telangiectatic osteosarcoma, such as young age at presentation, large size, and heterogeneous to high T2 signal intensity corresponding to fluid levels [40]. The “black-box” nature of deep neural networks makes it challenging to explain why our model was able to achieve the correct classification in some of these cases but was similarly misled in others.
It is noteworthy that our model was able to achieve performance on par with the experts without the use of data from T1-weighted contrast-enhanced studies, which were available to the experts in 81 out of 93 lesions in the test set. This may have introduced a bias towards the experts. There is also a question of the utility of contrast-enhanced MRI imaging in bone tumor diagnosis. Review of the literature showed that in a one study, MR scans with gadolinium did not contribute to differential diagnosis or management in 89% of a cohort of 242 patients with musculoskeletal tumors and tumor mimickers [41]. Contrast imaging did however aid in guiding biopsy of bulky lesions and evaluating tumor beds for possible recurrence [41]. Another author which reviewed the use of gadolinium in MR imaging of solitary bone tumors found that the role of contrast imaging is limited outside of directing biopsy and planning tumor resection [42]. By maintaining diagnostic performance without the need for contrast imaging, our model is of utility in contexts where contrast imaging is not readily available to the radiologist (e.g., incidentally discovered lesions). Moreover, for intentional evaluation of suspicious bone lesions, protocols that are sufficiently informative without the use of contrast enhancement would be of significant benefit to the pediatric radiology community. Given the pain-related anxiety that can be provoked by IV placement and the unknown risks of gadolinium deposition in children, elimination of contrast imaging for bone lesion assessment could be valuable [43,44]. The present study represents a first step towards a validated computational method for this purpose.
The rarity of bone tumors presented a challenge in compiling a dataset that could effectively power the training of a deep neural network for this task. While the size of our dataset is larger than many others that have been used for tumor classification tasks, it is still orders of magnitude smaller than datasets that have been used for other medical image characterization tasks. A larger dataset could also allow for granular classification beyond binary, such as differentiating between types of bone sarcomas or other histopathological diagnoses.
The study was limited by the need to perform manual lesion segmentation prior to analysis using our deep learning method. Manual segmentation precludes the creation of a fully automated lesion characterization pipeline. Given the variability in bone lesion location and the non-uniform shape of bones based on anatomical location, automated bone lesion segmentation would ostensibly be a much more challenging task than, for example, automated segmentation of the breast or brain tumors on MRI, both of which have been previously demonstrated [45,46]. While automated segmentation of specific osseous structures has been demonstrated, such as the proximal and distal femur and the proximal tibia, fully automated segmentation of bone lesions has not been reported [47,48]. One study employed a semiautomatic segmentation technique for bone sarcomas on MRI that involved manual segmentation of slices at the extremes and the middle of the volume, followed by an interpolation to create the final segmented volume [49]. Achieving automated bone lesion segmentation will likely be critical to wide-spread adoption of deep learning techniques for lesion classification in clinical practice.
In summary, we have developed a deep learning model that can evaluate the malignancy of bone lesions with similar accuracy and improved specificity in comparison to expert evaluators. Future studies will seek to combine radiograph and MRI findings in the development of classification models using deep learning and accompanying radiologist interpretation, as this is the clinical standard of care for suspicious bone lesions. In addition, a future study with a larger cohort may allow for classification of bone lesions by specific diagnosis. Finally, the development of a fully automated bone lesion classification tool would be facilitated by establishing an automated segmentation technique and utilizing a tool to automatically query for and extract relevant imaging studies from hospitals’ picture archive and communication systems (PACS), such as the DICOM Image Analysis and Archive (DIANA) system previously developed by our group [50]. At present, this work demonstrates the promise of deep learning to aid radiologists in characterizing the malignancy of bone lesions with improved certainty.
5. Contributors
HXB, PZ, and RS conceptualized the study. JW, BB, WL, SL and XP contributed to the curation of the data. RS and HXB provided supervision. RS, YH, and YL performed the expert evaluations. DD and JW contributed to project administration. FRE, RS, HXB, and LS developed the study methodology. HXB provided computational resources and contributed to funding acquisition. FRE performed the statistical analyses, software development, data visualization, and wrote the original draft. HXB and FRE have verified the underlying data. All authors contributed to reviewing and editing the final manuscript.
6. Data sharing statement
MRI imaging volumes and clinical data will not be made publicly available to ensure patient confidentiality, but they are available upon reasonable request to the corresponding author. Source code used to perform this study is available at https://github.com/sopeeweje/Bone-MRI.
Declaration of Competing Interest
The authors have nothing to disclose.
Acknowledgements
This work was funded by a Radiological Society of North America Research Medical Student Grant (#RMS2013) and supported by the Amazon Web Services Diagnostic Development Initiative. The authors would like to acknowledge Andrew Delworth for his contributions to this work.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2021.103402.
Contributor Information
Xianjing Peng, Email: pengxianjing@sina.cn.
Harrison X. Bai, Email: Harrison_Bai@Brown.edu.
Lisa States, Email: states@email.chop.edu.
Appendix. Supplementary materials
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Picci P. Epidemiology of Bone Lesions. In: Picci P, Manfrini M, Donati DM, Gambarotti M, Righi A, Vanel D, editors. Diagnosis of Musculoskeletal Tumors and Tumor-like Conditions: Clinical, Radiological and Histological Correlations - The Rizzoli Case Archive [Internet] Springer International Publishing; Cham: 2020. pp. 3–9. In: editors. [cited 2020 Jul 18] [Google Scholar]
- 3.WHO Classification of Tumours of Soft Tissue and Bone. 5th ed. Vol. 3. International Agency for Research on Cancer (I A R C) (UN);
- 4.Wyers MR. Evaluation of pediatric bone lesions. Pediatr Radiol. 2010;40(4):468–473. doi: 10.1007/s00247-010-1547-4. Apr. [DOI] [PubMed] [Google Scholar]
- 5.Vanel D. General Principles of Imaging. In: Picci P, Manfrini M, Donati DM, Gambarotti M, Righi A, Vanel D, editors. Diagnosis of Musculoskeletal Tumors and Tumor-like Conditions: Clinical, Radiological and Histological Correlations - The Rizzoli Case Archive [Internet] Springer International Publishing; Cham: 2020. pp. 27–30. et al., editors. [cited 2020 Jul 18] [Google Scholar]
- 6.Huang P-Y, Wu P-K, Chen C-F, Lee F-T, Wu H-T, Liu C-L. Osteomyelitis of the femur mimicking bone tumors: a review of 10 cases. World J Surg Oncol. 2013;11(1):283. doi: 10.1186/1477-7819-11-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holden DM, Ilaslan H, Sundaram M. An Imaging Approach to Bone Tumors. In: Santini-Araujo E, Kalil RK, Bertoni F, Park Y-K, editors. Tumors and Tumor-Like Lesions of Bone [Internet] Springer International Publishing; Cham: 2020. pp. 13–59. In: editors. [cited 2020 Jul 18] [Google Scholar]
- 8.Wu JS, Hochman MG. Imaging Modalities. In: Wu JS, Hochman MG, Bone editors., editors. Tumors: A Practical Guide to Imaging [Internet] Springer; New York, NY: 2012. pp. 51–86. [Google Scholar]
- 9.Hwang S, Panicek DM. Imaging Techniques: Magnetic Resonance Imaging. In: Davies AM, Sundaram M, James SLJ, editors. Imaging of Bone Tumors and Tumor-Like Lesions: Techniques and Applications [Internet] Springer; Berlin, Heidelberg: 2009. pp. 31–52. [Google Scholar]
- 10.Nascimento D, Suchard G, Hatem M, de Abreu A. The role of magnetic resonance imaging in the evaluation of bone tumours and tumour-like lesions. Insights Imaging. 2014;5(4):419–440. doi: 10.1007/s13244-014-0339-z. Jul 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma LD, Frassica FJ, Scott WW, Fishman EK, Zerbouni EA. Differentiation of benign and malignant musculoskeletal tumors: potential pitfalls with MR imaging. RadioGraphics. 1995;15(2):349–366. doi: 10.1148/radiographics.15.2.7761640. Mar 1. [DOI] [PubMed] [Google Scholar]
- 12.Song Y, Zhang Y-D, Yan X, Liu H, Zhou M, Hu B. Computer-aided diagnosis of prostate cancer using a deep convolutional neural network from multiparametric MRI. J Magn Reson Imaging JMRI. 2018;48(6):1570–1577. doi: 10.1002/jmri.26047. [DOI] [PubMed] [Google Scholar]
- 13.Schelb P, Kohl S, Radtke JP, Wiesenfarth M, Kickingereder P, Bickelhaupt S. Classification of Cancer at Prostate MRI: Deep Learning versus Clinical PI-RADS Assessment. Radiology. 2019;293(3):607–617. doi: 10.1148/radiol.2019190938. Oct 8. [DOI] [PubMed] [Google Scholar]
- 14.Zhou J, Luo L-Y, Dou Q, Chen H, Chen C, Li G-J. Weakly supervised 3D deep learning for breast cancer classification and localization of the lesions in MR images. J Magn Reson Imaging. 2019;50(4):1144–1151. doi: 10.1002/jmri.26721. Oct 1. [DOI] [PubMed] [Google Scholar]
- 15.Xi IL, Zhao Y, Wang R, Chang M, Purkayastha S, Chang K. Deep learning to distinguish benign from malignant renal lesions based on routine MR imaging. Clin Cancer Res [Internet] 2020 doi: 10.1158/1078-0432.CCR-19-0374. Jan 1. [DOI] [PubMed] [Google Scholar]
- 16.Chang K, Bai HX, Zhou H, Su C, Bi WL, Agbodza E. Residual Convolutional Neural Network for Determination of IDH Status in Low- and High-grade Gliomas from MR Imaging. Clin Cancer Res Off J Am Assoc Cancer Res. 2018;24(5):1073–1081. doi: 10.1158/1078-0432.CCR-17-2236. Mar 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reinus WR, Wilson AJ, Kalman B, Kwasny S. Diagnosis of Focal Bone Lesions Using Neural Networks. Invest Radiol. 1994;29(6):606–611. doi: 10.1097/00004424-199406000-00002. Jun. [DOI] [PubMed] [Google Scholar]
- 18.Do BH, Langlotz C, Beaulieu CF. Bone Tumor Diagnosis Using a Naïve Bayesian Model of Demographic and Radiographic Features. J Digit Imaging. 2017;30(5):640–647. doi: 10.1007/s10278-017-0001-7. Oct. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He Y, Pan I, Bao B, Halsey K, Chang M, Liu H. Deep learning-based classification of primary bone tumors on radiographs: A preliminary study. EBioMedicine. 2020;62 doi: 10.1016/j.ebiom.2020.103121. Dec 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fletcher C, Bridge J, Hogendoorn P, Mertens F. WHO classification of tumours of soft tissue and bone. 4th ed. Lyon: IARC Press; 468 p.
- 21.Picci P, Gambarotti M, Righi A. Classification of Primary Bone Lesions. In: Picci P, Manfrini M, Donati DM, Gambarotti M, Righi A, Vanel D, editors. Diagnosis of Musculoskeletal Tumors and Tumor-like Conditions: Clinical, Radiological and Histological Correlations - The Rizzoli Case Archive [Internet] Springer International Publishing; Cham: 2020. pp. 11–12. In: et al., editors. [cited 2020 Jul 18] [Google Scholar]
- 22.Lowekamp BC, Chen DT, Ibanez L, Blezek D. The Design of SimpleITK. Front Neuroinformatics [Internet] 2013;7 doi: 10.3389/fninf.2013.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ziabari A, Ye DH, Srivastava S, Sauer K, Thibault J-B, Bouman C. 2.5D Deep Learning for CT Image Reconstruction using a Multi-GPU implementation. 2018.
- 24.Roth HR, Lu L, Seff A, Cherry KM, Hoffman J, Wang S. A New 2.5D Representation for Lymph Node Detection using Random Sets of Deep Convolutional Neural Network Observations. Med Image Comput Comput-Assist Interv MICCAI Int Conf Med Image Comput Comput-Assist Interv. 2014;17:520–527. doi: 10.1007/978-3-319-10404-1_65. (0 1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruopp MD, Perkins NJ, Whitcomb BW, Schisterman EF. Youden Index and Optimal Cut-Point Estimated from Observations Affected by a Lower Limit of Detection. Biom J Biom Z. 2008;50(3):419–430. doi: 10.1002/bimj.200710415. Jun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Erdoğan S, Gülhan OT. Alternative Confidence Interval Methods Used in the Diagnostic Accuracy Studies. Comput Math Methods Med [Internet] 2016 doi: 10.1155/2016/7141050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stacy GS, Dixon LB. Pitfalls in MR Image Interpretation Prompting Referrals to an Orthopedic Oncology Clinic. RadioGraphics. 2007;27(3):805–826. doi: 10.1148/rg.273065031. May 1. [DOI] [PubMed] [Google Scholar]
- 28.Trieu J, Sinnathamby M, Bella CD, Pianta M, Perera W, Slavin JL. Biopsy and the diagnostic evaluation of musculoskeletal tumours: critical but often missed in the 21st century. ANZ J Surg. 2016;86(3):133–138. doi: 10.1111/ans.13251. [DOI] [PubMed] [Google Scholar]
- 29.Traina F, Errani C, Toscano A, Pungetti C, Fabbri D, Mazzotti A. Current Concepts in the Biopsy of Musculoskeletal Tumors: AAOS Exhibit Selection. J Bone Jt Surg [Internet] 2015;97(2) doi: 10.2106/JBJS.N.00661. Jan 21. [DOI] [PubMed] [Google Scholar]
- 30.Kotnis NA, Davies AM, James SLJ. Hand and Wrist. In: Davies AM, Sundaram M, James SLJ, editors. Imaging of Bone Tumors and Tumor-Like Lesions: Techniques and Applications [Internet] Springer; Berlin, Heidelberg: 2009. pp. 621–636. [Google Scholar]
- 31.Kakkar A, Nambirajan A, Suri V, Sarkar C, Kale SS, Singh M. Primary Bone Tumors of the Skull: Spectrum of 125 Cases, with Review of Literature. J Neurol Surg Part B Skull Base. 2016;77(4):319–325. doi: 10.1055/s-0035-1570347. Aug. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ciftdemir M, Kaya M, Selcuk E, Yalniz E. Tumors of the spine. World J Orthop. 2016;7(2):109–116. doi: 10.5312/wjo.v7.i2.109. Feb 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tosi P. Diagnosis and Treatment of Bone Disease in Multiple Myeloma: Spotlight on Spinal Involvement [Internet] Scientifica. Hindawi; 2013. 2013 doi: 10.1155/2013/104546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Franchi A. Epidemiology and classification of bone tumors. Clin Cases Miner Bone Metab. 2012;9(2):92–95. [PMC free article] [PubMed] [Google Scholar]
- 35.Sun X, Lou Y, Wang X. The Diagnosis of Iliac Bone Destruction in Children: 22 Cases from Two Centres. BioMed Res Int [Internet]. 2016. 2016 doi: 10.1155/2016/2131859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tow B, Tan M. Delayed Diagnosis of Ewing's Sarcoma of the Right Humerus Initially Treated as Chronic Osteomyelitis: A Case Report. J Orthop Surg. 2005;13(1):88–92. doi: 10.1177/230949900501300117. Apr 1. [DOI] [PubMed] [Google Scholar]
- 37.Greenspan A, Jundt G, Remagen W. Lippincott Williams & Wilkins; 2007. Differential Diagnosis in Orthopaedic Oncology; p. 552. [Google Scholar]
- 38.Ye J, Liu L, Wu J, Wang S. Osteoblastoma of the rib with CT and MR imaging: a case report and literature review. World J Surg Oncol. 2012;10:49. doi: 10.1186/1477-7819-10-49. Mar 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Limaiem F, Byerly DW, Singh R. StatPearls [Internet] StatPearls Publishing; Treasure Island (FL): 2020. Cancer, Osteoblastoma. In: [Google Scholar]
- 40.Zishan US, Pressney I, Khoo M, Saifuddin A. The differentiation between aneurysmal bone cyst and telangiectatic osteosarcoma: a clinical, radiographic and MRI study. Skeletal Radiol. 2020;49(9):1375–1386. doi: 10.1007/s00256-020-03432-w. Sep 1. [DOI] [PubMed] [Google Scholar]
- 41.May DA, Good RB, Smith DK, Parsons TW. MR imaging of musculoskeletal tumors and tumor mimickers with intravenous gadolinium: experience with 242 patients. Skeletal Radiol. 1997;26(1):2–15. doi: 10.1007/s002560050183. Jan. [DOI] [PubMed] [Google Scholar]
- 42.Vol. 18. Semin Ultrasound; CT MRI: 1997. pp. 307–311. (The use of gadolinium in the MR imaging of bone tumors). Jan 1. [DOI] [PubMed] [Google Scholar]
- 43.Bhargava R, Hahn G, Hirsch W, Kim M-J, Mentzel H-J, Olsen ØE. Contrast-Enhanced Magnetic Resonance Imaging in Pediatric Patients: Review and Recommendations for Current Practice. Magn Reson Insights. 2013;6:95–111. doi: 10.4137/MRI.S12561. Oct 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Otero HJ, Bhatia A. How to address parents’ concerns about MRI contrast agent safety. AAP News [Internet] 2021 Jan 15. [Google Scholar]
- 45.Zeineldin RA, Karar ME, Coburger J, Wirtz CR, Burgert O. DeepSeg: deep neural network framework for automatic brain tumor segmentation using magnetic resonance FLAIR images. Int J Comput Assist Radiol Surg. 2020;15(6):909–920. doi: 10.1007/s11548-020-02186-z. Jun 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Benjelloun M, Adoui ME, Larhmam MA, Mahmoudi SA. 2018 4th International Conference on Cloud Computing Technologies and Applications (Cloudtech) 2018. Automated Breast Tumor Segmentation in DCE-MRI Using Deep Learning; pp. 1–6. In: [Google Scholar]
- 47.Deniz CM, Xiang S, Hallyburton RS, Welbeck A, Babb JS, Honig S. Segmentation of the Proximal Femur from MR Images using Deep Convolutional Neural Networks. Sci Rep. 2018;8(1):1–14. doi: 10.1038/s41598-018-34817-6. Nov 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gandhamal AP, Talbar SN, Gajre SS, Hani AM, Kumar D. Automatic and Unsupervised Femur and Tibia Segmentation Using Magnetic Resonance Images. Osteoarthritis Cartilage. 2017;25:S258. Apr 1. [Google Scholar]
- 49.Dionísio FCF, Oliveira LS, Hernandes MA, Engel EE, Rangayyan RM, Azevedo-Marques PM. Manual and semiautomatic segmentation of bone sarcomas on MRI have high similarity. Braz J Med Biol Res [Internet] 2020;53(2) doi: 10.1590/1414-431X20198962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yi TY, Bai H, Merck D. DICOM Image Analysis and Archive: Extensions to Clinical AI Applications. In: Society for Imaging Informatics in Medicine.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.