Abstract
Introduction
This study investigated the extent to which subjective and objective data from an online registry can be analyzed using machine learning methodologies to predict the current brain amyloid beta (Aβ) status of registry participants.
Methods
We developed and optimized machine learning models using data from up to 664 registry participants. Models were assessed on their ability to predict Aβ positivity using the results of positron emission tomography as ground truth.
Results
Study partner–assessed Everyday Cognition score was preferentially selected for inclusion in the models by a feature selection algorithm during optimization.
Discussion
Our results suggest that inclusion of study partner assessments would increase the ability of machine learning models to predict Aβ positivity.
1. INTRODUCTION
The global incidence of dementia caused by Alzheimer's disease (AD) is expected to increase to over 130 million people by 2050, and in the face of this growing health crisis, several world governments have set 2025 as a deadline for finding a way to treat or prevent AD. 1 Yet progress on finding a cure for AD has been slow. Difficulty in finding treatments for AD is most likely a combination of uncertainty over the cause of AD and the fact that individuals who are at risk for the disease cannot easily be identified prior to the onset of cognitive impairment. Considerable evidence indicates that abnormal levels of amyloid beta (Aβ) protein are one of the earliest biomarkers of AD, pre‐dating the appearance of clinically diagnosed dementia by up to 20 years. 2 High brain Aβ levels in older, cognitively normal patients are also associated with a more rapid progression to mild cognitive impairment (MCI) or AD‐associated dementia. 2 Consequently, monitoring Aβ levels in the brain is important for identifying patients at risk of developing AD before they become symptomatic. Because early detection of these at‐risk patients would allow disease‐modifying treatments to be commenced sooner rather than later, improved techniques for assessing amyloid burden could markedly improve clinical trial outcomes and would improve the ability to care for patients with AD dementia.
Amyloid burden can be assessed in vivo by positron emission tomography (PET) 3 , 4 or analysis of cerebrospinal fluid (CSF). 5 , 6 Nevertheless, the invasiveness and high cost of such procedures, coupled with the low availability of the needed equipment and technical expertise in primary care clinical settings, precludes their use in large‐scale patient screening. Thus, there is a need to develop noninvasive, easily accessible, and cost‐effective methodologies for identifying patients who are likely to have high Aβ burden. Several low‐cost markers that may be predictive of Aβ status in the brain have been suggested previously, including demographics, apolipoprotein E (APOE) genotype, cognition, 7 , 8 , 9 , 10 polygenic risk scores, 11 , 12 magnetic resonance imaging (MRI), 13 , 14 , 15 plasma Aβ, 16 , 17 , 18 and other blood biomarkers. 16 , 19 In addition, Ashford et al. 20 recently described a statistical model for predicting Aβ status that leveraged data collected in an online research and recruitment registry for cognitive aging, the Brain Health Registry (BHR). The data used by this model consisted solely of information collected by the BHR through online surveys from participants, demonstrating the feasibility of using low‐cost, self‐reported data to identify Aβ+ patients. However, because Ashford et al. focused exclusively on self‐reported, subjective assessments of cognition, the accuracy of models for predicting amyloid positivity based on information from the BHR may be improved if these models made use of the data from objective assessments of cognition that are also included in the registry 21 (as described in more detail below). Furthermore, although Ashford et al. used a linear model (logistic regression) to drive their statistical analysis, a number of other groups have selected and evaluated potential biomarkers for Aβ positivity by employing non‐linear machine learning techniques, including kernalized support vector machines and random forests. 7 , 19 , 22 , 23 , 24 , 25 In light of the complex, non‐linear nature of biological processes, non‐linear models may be better suited for uncovering relationships between putative disease biomarkers and clinical disease status. 26 However, to date, non‐linear models have not been used to analyze data from the BHR, and we were interested in whether non‐linear models would provide improved results relative to Ashford et al.
RESEARCH IN CONTEXT
Systematic review: The authors reviewed the literature using electronic databases (e.g., PubMed) and search engines (e.g., Google Scholar). Previous studies have described statistical or machine learning techniques for the prediction of brain amyloid beta (Aβ) status, including recent work using self‐reported data from the same online registry that is the basis for this study.
Interpretation: Although previous work revealed that online, self‐reported data may have utility in the prediction of Aβ status, this work suggests that inclusion of study partner assessments would increase the accuracy of such predictions.
Future directions: In order to further ascertain the ability of the machine learning models described in this article to make accurate predictions of Aβ status, future work may include evaluation of the models’ performance on other data sets. Additional research is also needed to determine whether higher predictive performance might be achieved through further optimization of these models, either as a result of improved feature selection processes or through inclusion of additional features.
HIGHLIGHTS
Data from an online registry can be used to predict brain amyloid beta (Aβ) positivity.
Partner assessments are key data for machine learning models of Aβ status.
Feature selection did not improve model performance, but certain features could be removed without lowering performance.
Imputation of missing scores not only did not improve performance but also did not lower performance.
We were particularly interested in exploring two types of objective data in the BHR that were not considered previously by Ashford et al., namely, results from the Cogstate Brief Battery (CBB), 27 which is a computerized cognitive assessment, and study partner–assessed Everyday Cognition metrics (SP‐ECog), 28 which are based on evaluations provided by people who are familiar with the participants rather than the participants themselves. Changes in both metrics are associated with cognitive decline in patients with AD, suggesting that they may be useful, either alone or combined, for predicting amyloid status. 29 , 30 Therefore, we hypothesized that analyses of CBB and SP‐ECog scores would increase the predictive ability of models targeting Aβ status. Because only a small subset of BHR patients had CBB and SP‐ECog data available, we were also interested in whether techniques such as imputation might help generate better models using these features.
The goals of this study were to (1) examine the extent to which data from the BHR, including objective measures of cognition such as CBB and SP‐ECog, can be analyzed using machine learning methodologies to provide a prediction of contemporaneous Aβ positivity; (2) develop predictive models from BHR data using random forests and kernelized support vector machines, two types of non‐linear machine learning classifiers; and (3) identify optimal groups of features present in the BHR data set that would maximize the predictive value of these models.
2. METHODS
2.1. BHR Dataset
Data were obtained from the BHR database. BHR is a public online registry for research recruitment, assessment, and longitudinal monitoring with a focus on cognitive aging. 30 , 31 Participants register online and, after agreeing to an online informed consent form, they complete several questionnaires and self‐administered cognitive assessments. At the time of the analyses described in this paper, 70,992 participants were enrolled in BHR. The data set used for this study included BHR participants with available Aβ status as of June 2019 (n = 930). BHR allows participants to be enrolled simultaneously in the BHR and certain other studies, with data linkage between the BHR and co‐enrolled studies. 30 Aβ data for BHR participants were obtained from two such co‐enrolled studies, the Imaging Dementia‐Evidence for Amyloid Scanning (IDEAS) study 32 , 33 and clinical studies at the San Francisco Veterans Affairs Medical Center (SFVAMC). Inclusion criteria for these studies have been described elsewhere. 20 Apart from having a known Aβ status, the key inclusion criterion for our analyses was that participants had completed certain BHR measures of interest, including age, gender, level of education, subjective memory concern, family history of AD, self‐reported ECog score, and a Geriatric Depression Scale Short Form score (GDS‐SF; all of which are further described below). A total of 664 participants met these criteria and were included in the modeling, although evaluations of certain BHR metrics (SP‐ECog and CBB scores) focused on subgroups of participants (n = 148‐449) whose records included one or both of such metrics. Of the 664 participants, 105 were listed in the BHR as having dementia, 459 as having mild cognitive impairment (MCI), and 92 as cognitively unimpaired (CU) (8 participants had no information on cognitive status). The impairment level for this subgroup was clinician‐rated for participants enrolled in the IDEAS study and self‐reported for participants enrolled in the SFVAMC studies. Clinician‐rated impairment level in IDEAS was determined prior to PET scans as described by Nosheny et al. 33 Impairment levels (whether self‐reported or clinician‐rated) were used only to monitor distribution of patients across subgroups and were not used to train any of the models. Figure 1 shows a flow diagram of the numbers of participants excluded from the entire BHR data set to create the various samples used in this study.
FIGURE 1.
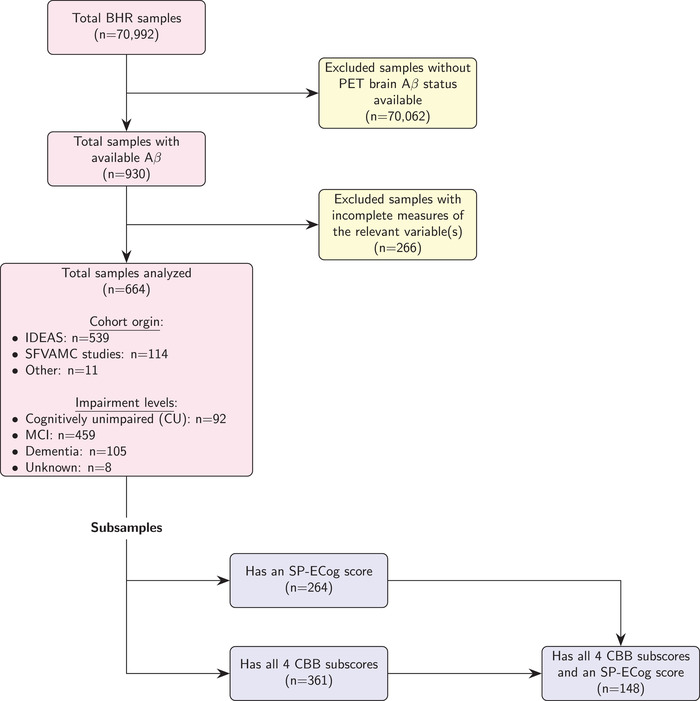
Generation of samples and subsamples
BHR participants complete a questionnaire during registration that asks for self‐reported sociodemographic information. For this analysis, we focused on the following sociodemographic variables: age, gender (male, female), and education. All of these variables are known to be associated with Aβ and are often included as predictors in Aβ prediction models that utilize data obtained in‐clinic. 32 , 34 , 35 The categorical variable education was converted to years of education (ranging from 6 to 20 years). Data on the distribution of these demographic markers (as well as some of the health‐related measurements described in Section 2.2 below) are included in Table 1.
TABLE 1.
Comparison of data subsets
| SP‐ECog+ / CBB+ | SP‐ECog+ / CBB‐ | SP‐ECog‐ / CBB+ | SP‐ECog‐ / CBB‐ | |
|---|---|---|---|---|
| Total subjects, n | 148 | 264 | 361 | 664 |
| Age, mean (SD) | 72.7 (5.3) | 73.4 (5.4) | 73.0 (5.4) | 73.5 (5.5) |
| Female, n (%) | 56 (37.8%) | 108 (40.9%) | 147 (40.7%) | 294 (44.3%) |
| Education, mean (SD) | 16.5 (2.7) | 16.2 (2.8) | 16.5 (2.6) | 16.2 (2.7) |
| FamHxAD, n (%) | 49 (33.1%) | 80 (30.3%) | 119 (33.0%) | 213 (32.1%) |
| Self_SMC, n (%) | 133 (89.9%) | 242 (91.7%) | 313 (86.7%) | 593 (89.3%) |
| Self‐ECog, mean (SD) | 1.78 (0.57) | 1.84 (0.64) | 1.70 (0.54) | 1.78 (0.61) |
| GDS_Score, mean (SD) | 2.14 (2.34) | 2.46 (2.68) | 2.17 (2.39) | 2.46 (2.68) |
| AD, n (%) | 11 (7.4%) | 52 (19.7%) | 30 (8.3%) | 105 (15.8%) |
| MCI, n (%) | 110 (74.3%) | 185 (70.1%) | 243 (67.3%) | 459 (69.1%) |
| Aβ+, n (%) | 84 (56.8%) | 168 (63.6%) | 177 (49.0%) | 365 (55.0%) |
Abbreviations: AD, diagnosed with Alzheimer's disease; Aβ+, amyloid beta positive.; CBB‐, subset does not includes Cogstate Brief Battery scores; CBB+, subset includes Cogstate Brief Battery scores; FamHxAD, family history of Alzheimer's disease; GDS_Score, Geriatric Depression Scale (short form) score; MCI, diagnosed with mild cognitive Impairment; SD, standard deviation; Self_SMC, self‐reported subjective memory concern; Self‐ECog, self‐assessed Everyday Cognition metric; SP‐ECog‐, subset does not include study partner–assessed Everyday Cognition scores; SP‐ECog+, subset includes study partner–assessed Everyday Cognition scores.
2.2. Subjective health‐related measures
BHR participants are invited to complete further online self‐report questionnaires including an assessment of detailed medical history and overall health. This analysis used subjective memory concern (“Are you concerned that you have a memory problem?”), family history of AD (“Have you, your sibling(s), or parent(s) ever been diagnosed with Alzheimer's disease?”), self‐reported and study partner‐reported Everyday Cognition Scale (or ECog) score (which are further described below), and Geriatric Depression Scale Short Form (or GDS‐SF) score, all of which have been shown previously to be correlated with Aβ levels. 28 , 34 , 36 , 37
2.3. Geriatric depression scale short form
The GDS‐SF is a 15‐item screening tool used to rate severity of depressive symptoms in older adults. 38 BHR participants complete an online version of the GDS‐SF that contains content that is identical to the paper version. Higher scores represent greater symptoms of depression.
2.4. Everyday cognition scale
The Everyday Cognition Scale (ECog) is a 39‐item assessment of the participant's self‐ or study partner–reported capability to perform everyday tasks in comparison to activity levels 10 years earlier. 28 BHR participants complete an online adaptation of the ECog. 30 This analysis used both the self‐reported (Self‐ECog) and study partner‐reported (SP‐ECog) scores.
2.5. Cogstate brief battery CBB scores
The CBB is a computerized cognitive assessment battery that has been validated under supervised and unsupervised conditions in various populations, including aging studies and studies on AD and related dementias. 39 , 40 , 41 Only participants whose CBB scores met preset integrity criteria were included. 42 The CBB consists of four cognitive tests 29 :
The detection test (DET) is a measure of psychomotor function and information‐processing speed that uses a simple reaction time paradigm with playing‐card stimuli. The subject is asked to press the “yes” key as soon as the card in the center of the screen turns faceup. The software measures the speed and accuracy of each response. The primary outcome variable for this test is reaction time in milliseconds for correct responses normalized using a logarithmic base 10 (Log 10) transformation.
The identification test (IDN) is a measure of visual attention and uses a choice reaction time paradigm with playing‐card stimuli. The subject is asked whether the card displayed in the center of the screen is red. The subject responds by pressing the “yes” key when the joker card is red and “no” when it is black. The primary outcome for this test is reaction time in milliseconds for correct responses normalized using a logarithmic base 10 (Log 10) transformation.
The one card learning test (OCL) is a measure of visual learning and memory that uses a pattern‐separation paradigm with playing‐card stimuli. In this task, the playing cards are identical to those found in a standard deck of 52 playing cards. The subject is asked whether the card displayed in the center of the screen was seen previously in this task. The subject responds by pressing the “yes” or “no” key. The primary outcome variable is the proportion of correct responses (accuracy) normalized using an arcsine transformation.
The one‐back test (ONB) is a measure of working memory and uses a well‐validated n‐back paradigm with playing‐card stimuli. In this task, the playing cards are identical to those found in a standard deck of 52 playing cards. The subject is asked whether the card displayed in the center of the screen is the same as the card presented immediately before. The subject responds by pressing the “yes” or “no” key. The primary outcome variable for this test is accuracy of correct response.
2.6. Amyloid Beta
Aβ PET scan results were provided directly by IDEAS (n = 539) or the SFVAMC studies (n = 114). Determination of participant Aβ status for IDEAS has been described previously. 43 In short, study imaging specialists interpreted Aβ PET images using approved reading methodologies for each tracer (fluorine 18 (18F)–labeled florbetapir, 18F‐labeled flutemetamol, and 18F‐labeled florbetaben). A “negative” interpretation meant that the retention of the Aβ tracer was in cerebral white matter only and a “positive” interpretation meant that Aβ tracer retention was also found in cortical gray matter. SFVAMC studies followed the same interpretation scheme, based on visual reads of PET scans performed using 18F‐labeled florbetapir. For the IDEAS group, PET scans preceded the BHR data collection, whereas in the SFVAMC studies, BHR online data collection was not restricted to time of PET scan.
2.7. Machine learning classifier construction and feature selection
BHR data were used to construct random forest (RF) and support vector machine (SVM) classifiers implemented in the Python library scikit‐learn (scikit‐learn.org). 44 All SVM classifiers used a radial basis function (rbf) kernel. In each case, the predictive target of the RF and SVM models was contemporaneous Aβ status of individual patients, and the predictive performance of the models was assessed by calculating the area under the receiver‐operating characteristic (ROC) curve (AUC). Optimal input features were selected using a greedy forward feature selection algorithm. Specifically, starting with a model having no features, each feature was added to the model in turn and the AUC score was determined. The model with the highest AUC score was retained, and this process was repeated until no features could be added without decreasing the AUC score, revealing the optimal feature set. The initial pool of features for this selection process consisted of a fixed set of seven sociodemographic and health‐related measurements (age, gender, years of education, family history of AD, subjective memory concern (SMC), Self‐ECog score, and GDS‐SF score), both with and without two additional types of features (SP‐ECog score and a set of four CBB scores). Thus, the starting pool of features used in this study ranged from 7 to 12. BHR participants who lacked data for a particular feature under investigation were excluded from that portion of the study, and therefore changes to the initial feature set altered the number of samples used to perform the feature selection and training. Sample numbers are reported in Tables 1 and 2 for each experimental condition. At the same time as feature selection, certain hyperparameters (number of trees for RF models; regularization parameter and gamma for SVM models) were optimized. Feature selection, hyperparameter optimization, and model evaluation were performed using nested cross‐validation. Specifically, feature selection and hyperparameter optimization were performed in the inner cross‐validation loop. The resulting models were validated in the outer, 10‐fold cross‐validation loop using held‐out data from each testing fold. This process was repeated three times, and the reported AUC scores are averages of the 30 AUC scores from the outer cross‐validation loop over 10 folds and three repetitions. Mean AUC scores for each experimental condition were compared using a Wilcoxon rank‐sum test, with P < .05 denoting statistical significance. In addition to AUC scores, sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were also calculated. Because the output of each model was the probability of a subject being Aβ+, an arbitrary cut‐off probability of 0.5 was used to separate positive and negative classes when calculating sensitivity, specificity, PPV, and NPV.
TABLE 2.
Results of feature selection
| Model | SP‐ECog | CBB | N | AUC, mean (SD) | Sens. | Spec. | PPV | NPV | AUC (n = 148), mean (SD) |
|---|---|---|---|---|---|---|---|---|---|
| RF | No | No | 664 | 0.5871 (0.0641) | 64.82% | 49.16% | 60.97% | 53.53% | 0.5574 (0.1836) |
| Yes | 361 | 0.5192 (0.0901) | 49.84% | 53.34% | 50.25% | 52.57% | 0.5019 (0.1554) | ||
| Yes | No | 264 | 0.6244 (0.1198) | 75.59% | 42.22% | 70.00% | 48.84% | 0.5970 (0.1294) | |
| Yes | 148 | 0.5790 (0.1467) | 65.37% | 44.76% | 61.11% | 49.00% | 0.5708 (0.1734) | ||
| SVM | No | No | 664 | 0.5864 (0.0659) | 75.37% | 37.44% | 59.49% | 56.11% | 0.4750 * (0.1699) |
| Yes | 361 | 0.4856 (0.0845) | 48.12% | 52.84% | 50.74% | 51.37% | 0.4499 (0.1615) | ||
| Yes | No | 264 | 0.6034 (0.1201) | 88.70% | 19.33% | 65.94% | 52.71% | 0.6606 (0.1215) | |
| Yes | 148 | 0.5138 (0.1326) | 79.77% | 22.22% | 57.36% | 48.37% | 0.4013* (0.1427) |
Final column (labeled as n = 148) reports scores generated using a data set consisting only of the 148 subjects with data for all 12 features. Values marked with * are significantly different (P < .05) from the corresponding means in the fifth column. Abbreviations: RF, random forest; SVM, support vector machine; SP‐ECog, study partner–assessed Everyday Cognition score; CBB, Cogstate Brief Battery score; AUC, area under the receiver‐operating characteristic curve, measured by 10‐fold cross‐validation; SD, standard deviation; Sens., sensitivity; Spec., specificity; PPV, positive predictive value; NPV, negative predictive value.
2.8. CBB and SP‐ECog imputation
For certain analyses, missing CBB scores and/or missing SP‐ECog scores were imputed using machine learning regressors. Missing scores were imputed using the IterativeImputer class from scikit‐learn, which applies a Bayesian ridge regressor to model each feature with missing values as a function of other features in a round‐robin fashion.
3. RESULTS
3.1. Feature selection
Table 2 and Figure 2 summarize the results of the feature selection process when applied to RF and SVM machine learning models trained to predict Aβ status, depending on whether SP‐ECog and/or CBB scores were made available to the feature selection algorithm. AUC scores ranged from 0.485 to 0.624. In the case of an RF model, including SP‐ECog scores in the pool of features available to the selection algorithm resulted in higher average AUC scores when compared to omitting SP‐ECog scores (0.624 vs 0.587, P = .015). Conversely, including CBB data in the pool of features available to the selection algorithm produced consistently lower scores (P < .006 in three of the four cases). Because sample sizes varied depending on the initial feature pool (as a result of missing values among subjects), we repeated the feature selection process using only those subjects who had data for all 12 features (n = 148). These results are presented in a separate column in Table 2. Note that the statistical difference seen in the SVM model trained using the seven core features (i.e., excluding SP‐ECog and CBB scores), which used the same subset of patients in either case, is likely due to random variation inherent in the cross‐validation process.
FIGURE 2.
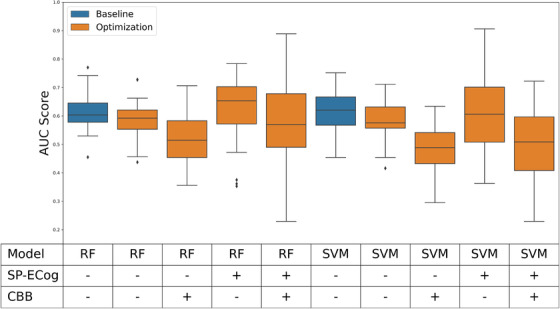
Performance of models after feature selection. Blue boxes represent baseline performance of models with hyperparameter optimization but no feature selection. Orange boxes represent performance of models with both hyperparameter optimization and feature selection. Abbreviations: RF, random forest; SVM, support vector machine; SP‐ECog, study partner–assessed Everyday Cognition score; CBB, Cogstate Brief Battery score; AUC, area under the receiver‐operating characteristic curve
Because models were trained and evaluated using a nested cross‐validation approach, feature selection was applied independently to each of the outer cross‐validation folds in order to prevent leakage of information from the testing folds. As a result, each run of the nested cross‐validation generated 30 locally optimal feature sets (10 folds x 3 repetitions). The frequency with which a given feature appears among these 30 sets provides an indication of feature importance. Figure 4 summarizes the occurrence of each feature in the 30 optimal feature sets generated under each starting condition. When starting with initial pools of features that included SP‐ECog, the most common or second most common feature in the optimal feature sets was always SP‐ECog. When SP‐ECog is not included in the initial feature sets, the most common features were usually Self_SMC and GDS_Score.
FIGURE 4.
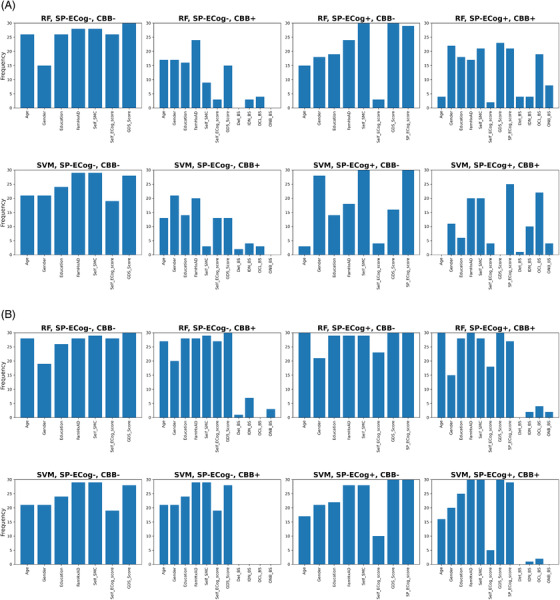
Frequency of feature selection. (A) Non‐imputed data. (B) Imputed data. Abbreviations: RF, random forest; SVM, support vector machine; SP‐ECog or SP_ECog_score, study partner–assessed Everyday Cognition score; CBB, Cogstate Brief Battery score; FamHxAD, family history of Alzheimer's disease; Self_SMC, self‐reported subjective memory concern; Self_ECog_score, self‐assessed Everyday Cognition metrics; GDS_Score, Geriatric Depression Scale (short form) score; Det_BS, Cogstate Detection test; IDN_BS, Cogstate Identification test; OCL_BS, Cogstate One Card Learning test; ONB_BS, Cogstate One Back test
3.2. CBB and SP‐ECog imputation
There was a significant amount of variability in whether any particular BHR participant reported SP‐ECog scores or results from each of the four CBB tests on any one assessment. Consequently, 400 of the samples had no SP‐ECog score and 215 of the samples were missing at least one CBB score. To include either or both of these metrics in the feature selection process, it was necessary to limit the data set to patients having these categories of data, which reduced the sample size considerably (see Figure 1 and Table 1). To explore an alternative method to address the missing data, imputation was used to generate predictions for missing CBB data and missing SP‐ECog data based on the available data for each patient. This allowed inclusion of CBB and/or SP‐ECog in the initial feature set without excluding as many participants from the analysis for missing data. However, there was a risk that imputation might negatively impact the models, since imputed data may not be as predictive as real data. The results from models trained using imputed data are summarized and compared to the original models in Figure 3. AUC scores ranged from 0.46 to 0.62. In three of the four models that used imputed CBB score, imputation resulted in a significant increase in AUC scores relative to excluding data from patients with missing data. In other cases, imputation had no significant effect on AUC scores.
FIGURE 3.
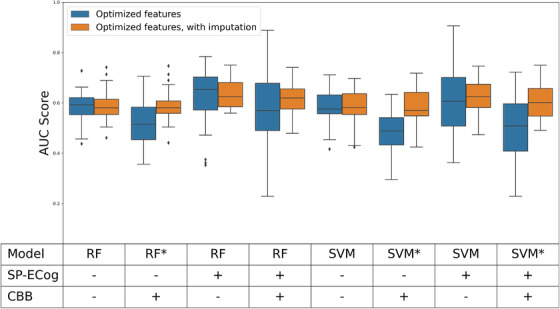
Effect of imputation of SP‐ECog and CBB data. Models having significantly different scores with and without imputation (P < .05) are denoted with an asterisk. No imputation was necessary for models without SP‐ECog and CBB scores, but these were built in parallel with rest as controls; minor differences between paired results for these models are the result of random variation inherent in cross‐validation process. Abbreviations: RF, random forest; SVM, support vector machine; SP‐ECog, study partner–assessed Everyday Cognition score; CBB, Cogstate Brief Battery score; AUC, area under the receiver‐operating characteristic curve
4. DISCUSSION
The main findings of this study are that (1) SP‐ECog is potentially a key feature when building machine learning models to predict Aβ status using data collected through the BHR; (2) including CBB scores generated by BHR participants had a statistically significant negative effect on the ability of these models to predict Aβ status; and (3) imputation of missing CBB scores and missing SP‐ECog data did not improve the predictive performance of models, but the models built using imputed data performed equivalently to those built using only actual data, suggesting that imputation may be a useful strategy for addressing missing data in the BHR data set.
Adding feature selection to the training pipeline did not have a statistically significant effect on the performance of the models (Figure 1). However, this is not entirely surprising. The benefits of feature selection stem from removing noisy data that can affect the ability of models to accurately learn relevant patterns. Because of the small number of features in this case, it is likely that noise is not a significant factor. On the other hand, it is notable that we were able to eliminate a significant proportion of the features (41% on average) and still retain the same levels of performance.
In addition, looking at what features were selected reveals some interesting patterns. When SP‐ECog was included in the starting feature pool prior to feature selection, it appeared more frequently in the final feature sets than any other feature. This was true regardless of the type of model (RF vs SVM), whether or not CBB scores were included in the initial features, and whether or not missing CBB or SP‐ECog scores were imputed. Moreover, in the case of an RF model trained on all of the available features other than the CBB scores, removing SP‐ECog caused a statistically significant decrease in the average AUC score (P = .015). A similar effect was seen in an SVM model (P = .038) when the various initial feature pools were analyzed using consistently sized data sets (n = 148; i.e., the population of subjects who have all 12 features). Taken together, these results suggest that the presence of SP‐ECog was a key contributor to the models’ ability to predict Aβ status. This result is consistent with previous work that found that SP‐ECog is statistically associated with cognitive decline in BHR participants. 30 , 33 Our results also suggest that SP‐ECog is more predictive than Self‐ECog. When models were trained on initial feature sets that included both SP‐ECog and Self‐ECog, Self‐ECog appeared in the final optimal feature sets less frequently than most of the other features. In contrast, when the same process was applied to initial feature sets that omitted SP‐ECog, the feature selection process was more likely to select Self‐ECog, as under these conditions Self‐ECog appeared in the final optimal features sets about as frequently as most of the other features. A likely explanation for these results lies in the fact that the greedy selection algorithm is designed to select non‐redundant features. Thus, Self‐ECog may be weakly predictive of Aβ status, but in the presence of SP‐ECog, the stronger correlation between SP‐ECog and Aβ positivity causes the selection algorithm to select SP‐ECog. Then, in later selection rounds, the algorithm continues to avoid Self‐ECog because the addition of Self‐ECog on top of the already present SP‐ECog does not provide enough added benefit.
On the other hand, inclusion of the CBB test scores in the initial feature set did not have a significant positive effect on the final AUC scores. In fact, including CBB scores in the initial feature set but omitting SP‐ECog consistently resulted in lower AUC scores than for any of the other initial feature sets we tested. This was somewhat surprising, as other studies have demonstrated the validity of the CBB as a screening tool for AD patients. 27 , 29 , 39 , 40 , 41 , 45 , 46 , 47 One possible explanation is that although CBB scores are accurate measures of cognitive decline, cognitive decline alone is insufficient to predict Aβ status. However, a recent analysis of data from another web‐based registry for Alzheimer's patients, the Alzheimer's Prevention Trial (APT) webstudy (aptwebstudy.org), concluded that one of the CBB tests (One Card Learning, or OCL), when administered remotely, was one of the top predictors of Aβ status. 24 Similarly, in another recent article, lower accuracy on the CBB OCL test was reported to be associated with an increased likelihood of being Aβ+ in a logistic regression model, but notably this association was masked when Self‐ECog and SP‐ECog scores were also included in the model. 33 Thus, contrary to our finding that adding CBB to the feature set results in lower AUC scores, these other articles suggest that scores from the CBB OCL are predictive of Aβ status.
Given that a number of patients in the original data set were missing SP‐ECog and CBB scores, we also examined whether imputation of these features might be a useful strategy for managing this missing data. Under three of the four starting conditions involving imputed CBB data, CBB imputation produced average AUC scores that were higher (P < .05) relative to our initial approach of eliminating samples with missing CBB data (Figure 3). However, in all of these cases, simply omitting the CBB scores from the initial feature set resulted in equivalent results. When we looked in detail at the features selected when imputed CBB data is present, we were surprised to see that CBB scores were almost never present in the final feature set. Thus, imputation of CBB data does not improve scores by increasing the performance of the CBB data itself. Rather, imputation appears to make it more likely that the feature selection process ignores the CBB data entirely, which for some reason causes the average AUC score to increase. We hypothesize that this effect may result from a limitation of the feature selection process. Feature selection is an NP‐hard problem, and no technique is guaranteed to identify the optimal feature set, other than an exhaustive search of all possible combinations of features. 48 Therefore, it is possible that, once it adds any of the CBB scores to the list of features, the feature selection algorithm becomes unable to reach the highest scoring combination of features.
Imputation of SP‐ECog data did not improve performance of the models, as the scores from models based on imputed SP‐ECog data were indistinguishable from models based on non‐imputed data. Here, the lack of effect of imputation was not because of a failure of the feature selection algorithm to pick SP‐ECog as one of the features. As shown in Figure 4, for both RF and SVM models trained using an initial feature pool that included imputed SP‐ECog data, SP‐ECog was nearly always present in the optimal group of features, which suggests that the imputed scores did provide some predictive value. However, comparing the feature distributions in Figure 4 for imputed SP‐ECog data with those for actual SP‐ECog data reveals that Self‐ECog appears more frequently in optimal feature sets when imputation is used. We speculate that imputation of SP‐ECog weakens the predictive power of the SP‐ECog scores, leading to both metrics being necessary for an optimal AUC score in many cases.
The range of AUC scores generated by the machine learning models used in this study in the absence of SP‐ECog and CBB metrics are lower than AUC scores previously reported by Ashford et al. for a statistical model that used logistic regression to analyze a similar set of features from the BHR. 20 In particular, Ashford et al. reported a cross‐validated AUC score of 0.66, and we obtained an average AUC score of 0.587 and 0.586 for RF and SVM classifiers, respectively, based on a similar set of BHR features. However, because we were interested in assessing multiple types of machine models (RF and SVM) and different hyperparameters (including optimal feature sets), we used nested cross‐validation to minimize the introduction of bias into this process. Because Ashford et al. did not need to use nested cross‐validation, their scores are not directly comparable to the ones reported here. In the absence of feature selection or hyperparameter optimization, where a single level of cross‐validation can be applied, our RF and SVM models achieved a score of 0.62 (Figure 1), which is similar to the score reported by Ashford et al. Our results are also in line with Langford et al., 24 which reported an AUC score of 0.598 when training an XGBoost classifier to predict Aβ status using data from remote, web‐based testing. Ansart et al. 25 also looked at possible low‐cost biomarkers for Aβ positivity, reporting AUC scores between 0.675 and 0.824 for random forests applied to a variety of data sets. However, their model included data on subjects’ APOE ε4 genotype, which is difficult to obtain reliably through an online registry. Because their models achieved AUC scores of 0.637‐0.751 using APOE ε4 genotype as the sole feature, it is likely that their models would have performed significantly worse if the APOE ε4 feature was omitted. Finally, Insel et al. 7 reported a random forest model for predicting Aβ positivity that had a positive predictive value (PPV) of 65%. Insel's models also include APOE ε4 genotype as a feature and therefore are not directly comparable to our models. Even so, their PPV figure is similar to the 50% to 70% PPV range reported here.
Some limitations that may operate to limit the generalizability of the current findings are worth noting. Elevated Aβ levels are considered necessary but not sufficient for an AD diagnosis, 49 and so by looking exclusively at Aβ status, our model may be ignoring other clinically relevant features. Moreover, the fact that our analysis was limited to patients in the BHR whose Aβ status had been determined by PET may have introduced a selection bias. Similarly, because our model only included patients who had completed BHR's online surveys, it might overestimate the ability of online metrics to predict Aβ positivity. Another potential source of error stems from the fact that most patients who participated in the IDEAS study were informed about their Aβ status. Because the BHR data rely heavily on self‐reported information and the results of self‐administered tests, many of the metrics used in our model could be affected by patients’ knowledge of their Aβ status. Finally, the lack of racial, ethnic, and educational diversity among the BHR participants may limit the generalizability of the study.
In conclusion, our results demonstrate the feasibility of predicting Aβ status in older adults through the use of an online registry involving subjective assessments from participants and study partners as well as computerized cognitive tests administered in an unsupervised manner. Such an online approach is inexpensive, non‐invasive, easily accessible, and scalable and therefore could be a useful method for pre‐screening older adults for clinical trials prior to assessing Aβ positivity using PET imaging. Our results also reveal that certain metrics, such as SP‐ECog, may be better suited than others to classifying patients as Aβ+. Additional research is needed to determine whether higher predictive performance might be achieved through further optimization of the machine learning models described in this article, either as result of improved feature selection processes or through inclusion of additional features.
CONFLICTS OF INTEREST
JA, MTA, CJ, JN, DT, and RLN have no interests to declare. RSM has received grant funding from the National Institute of Mental Health and has received research support from Johnson & Johnson. PM is an employee of Cogstate, Ltd. GDR is Study Chair for the IDEAS study and has received additional research support from National Institutes of Health (NIH), Alzheimer's Association, Tau Consortium, Avid Radiopharmaceuticals, and Eli Lilly. He is a consultant for Axon Neurosciences, General Electric (GE) Healthcare, Eisai, and Merck, and he is an associate editor for JAMA Neurology. MWW receives support for his work from the following funding sources: NIH, Department of Defense, Patient Centered Outcomes Research Institute (PCORI), California Department of Public Health, University of Michigan, Siemens, Biogen, Larry L. Hillblom Foundation, Alzheimer's Association, and the State of California. He also receives support from Johnson & Johnson, Kevin and Connie Shanahan, GE, Vrije Universiteit Medical Center Amsterdam, Australian Catholic University, The Stroke Foundation, and the Veterans Administration. He has served on Advisory Boards for Eli Lilly, Cerecin/Accera, Roche, Alzheon, Inc., Merck Sharp & Dohme Corp., Nestle/Nestec, PCORI, Dolby Family Ventures, National Institute on Aging (NIA), Brain Health Registry, and ADNI. He serves on the editorial boards for Alzheimer's & Dementia, Topics in Magnetic Resonance Imaging, and Magnetic Resonance Imaging. He has provided consulting and/or acted as a speaker/lecturer to Cerecin/Accera, Inc., Alzheimer's Drug Discovery Foundation (ADDF), Merck, BioClinica, Eli Lilly, Indiana University, Howard University, Nestle/Nestec, Roche, Genentech, NIH, Lynch Group GLC, Health & Wellness Partners, Bionest Partners, American Academy of Neurology (AAN), New York University, Japanese Government Alliance, National Center for Geriatrics and Gerontology (Japan), US Against Alzheimer's, Society for Nuclear Medicine and Molecular Imaging (SNMMI), The Buck Institute for Research on Aging, and FUJIFILM‐Toyama Chemical (Japan). He holds stock options with Alzheon, Inc., Alzeca, and Anven.
ACKNOWLEDGEMENTS
This work was funded by the National Institutes of Health, National Institute on Aging (K01AG055692). The authors gratefully acknowledge the following funding sources for the Brain Health Registry: National Institute on Aging, the Alzheimer's Association, California Department of Public Health, Patient Centered Research Outcomes Institute, Alzheimer's Drug Discovery Foundation, Genentech Health Equity Innovations Fund, Larry L. Hillblom Foundation, the Rosenberg Alzheimer's Project, the Ray and Dagmar Dolby Family Fund, Connie and Kevin Shanahan, General Electric, and The Drew Foundation. The authors would also like to acknowledge the contributions of the IDEAS study team, the SFVAMC study team, the entire Brain Health Registry staff, and all BHR participants. The authors are grateful for the support of all past and present BHR team members.
Albright J, Ashford MT, Jin C, et al. Machine learning approaches to predicting amyloid status using data from an online research and recruitment registry: The Brain Health Registry. Alzheimer's Dement. 2021;13:e12207. 10.1002/dad2.12207
REFERENCES
- 1. Cummings JL, Morstorf T, K. Alzheimer's disease drug‐development pipeline: few candidates, frequent failures. Alzheimers Res Ther. 2014;6:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baker JE, Lim YY, Pietrzak RH, et al. Cognitive impairment and decline in cognitively normal older adults with high amyloid‐β: a meta‐analysis. Alzheimers Dement Diagn Assess Dis Monit. 2017;6:108‐121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Klunk WE, Engler H, Nordberg A, et al. Imaging the pathology of Alzheimer's disease: amyloid‐imaging with positron emission tomography. Neuroimaging Clin N Am. 2003;13:781‐789. ix. [DOI] [PubMed] [Google Scholar]
- 4. Pontecorvo MJ, Mintun MA. PET amyloid imaging as a tool for early diagnosis and identifying patients at risk for progression to Alzheimer's disease. Alzheimers Res Ther. 2011;3:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Palmqvist S, Mattsson N, Hansson O. Cerebrospinal fluid analysis detects cerebral amyloid‐β accumulation earlier than positron emission tomography. Brain. 2016;139:1226‐1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tapiola T, Alafuzoff I, Herukka SK, et al. Cerebrospinal fluid β‐amyloid 42 and tau proteins as biomarkers of Alzheimer‐type pathologic changes in the brain. Arch Neurol. 2009;66:382‐389. [DOI] [PubMed] [Google Scholar]
- 7. Insel PS, Palmqvist S, Mackin RS, et al. Assessing risk for preclinical β‐amyloid pathology with APOE, cognitive, and demographic information. Alzheimers Dement Diagn Assess Dis Monit. 2016;4:76‐84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ko H, Ihm JJ, Kim HG. Cognitive profiling related to cerebral amyloid beta burden using machine learning approaches. Front Aging Neurosci. 2019;11:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Maserejian N, Bian S, Wang W, et al. Practical algorithms for amyloid β probability in subjective or mild cognitive impairment. Alzheimers Dement Diagn Assess Dis Monit. 2019;11:180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Voyle N, Patel H, Folarin A, et al. Genetic risk as a marker of amyloid‐β and Tau burden in cerebrospinal fluid. J Alzheimers Dis. 2017;55:1417‐1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Leonenko G, Shoai M, Bellou E, et al. Genetic risk for alzheimer disease is distinct from genetic risk for amyloid deposition. Ann Neurol. 2019;86:427‐435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tosun D, Chen YF, Yu P, et al. Amyloid status imputed from a multimodal classifier including structural MRI distinguishes progressors from nonprogressors in a mild Alzheimer's disease clinical trial cohort. Alzheimers Dement. 2016;12:977‐986. [DOI] [PubMed] [Google Scholar]
- 13. Casamitjana A, Petrone P, Tucholka A, et al. MRI‐based screening of preclinical Alzheimer's disease for prevention clinical trials. J Alzheimers Dis JAD. 2018;64:1099‐1112. [DOI] [PubMed] [Google Scholar]
- 14. Palmqvist S, Janelidze S, Stomrud E, et al. Performance of fully automated plasma assays as screening tests for Alzheimer disease‐related β‐amyloid status. JAMA Neurol. 2019;76:1060‐1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yasuno F, Kazui H, Morita N, et al. Use of T1‐weighted/T2‐weighted magnetic resonance ratio to elucidate changes due to amyloid β accumulation in cognitively normal subjects. NeuroImage Clin. 2017;13:209‐214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Park JC, Han SH, Lee H, et al. Prognostic plasma protein panel for Aβ deposition in the brain in Alzheimer's disease. Prog Neurobiol. 2019;183:101690. [DOI] [PubMed] [Google Scholar]
- 17. Schindler SE, Bollinger JG, Ovod V, et al. High‐precision plasma β‐amyloid 42/40 predicts current and future brain amyloidosis. Neurology. 2019;93:E1647‐E1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Verberk IMW, Slot RE, Verfaillie SCJ, et al. Plasma amyloid as prescreener for the earliest Alzheimer pathological changes. Ann Neurol. 2018;84:648‐658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ashton NJ, Nevado‐Holgado AJ, Barber IS, et al. A plasma protein classifier for predicting amyloid burden for preclinical Alzheimer's disease. Sci Adv. 2019;5:eaau7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ashford MT, Neuhaus J, Jin C, et al. Predicting amyloid status using self‐report information from an online research and recruitment registry: the Brain Health Registry. Alzheimers Dement Diagn Assess Dis Monit. 2020;12:e12102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nosheny RL, Camacho MR, Jin C, et al. Validation of online functional measures in cognitively impaired older adults. Alzheimers Dement. 2020;16:1426‐1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Burnham SC, Faux NG, Wilson W, et al. A blood‐based predictor for neocortical Aβ burden in Alzheimer's disease: results from the AIBL study. Mol Psychiatry. 2014;19:519‐526. [DOI] [PubMed] [Google Scholar]
- 23. Goudey B, Fung BJ, Schieber C, et al. A blood‐based signature of cerebrospinal fluid Aβ 1‐42 status. Sci Rep. 2019;9:1‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Langford O, Raman R, Sperling RA, et al. Predicting amyloid burden to accelerate recruitment of secondary prevention clinical trials. J Prev Alzheimers Dis. 2020;7:213‐218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ansart M, Epelbaum S, Gagliardi G, et al. Reduction of recruitment costs in preclinical AD trials: validation of automatic pre‐screening algorithm for brain amyloidosis. Stat Methods Med Res. 2020;29:151‐164. [DOI] [PubMed] [Google Scholar]
- 26. Jin T, Nguyen ND, Talos F, Wang D. ECMarker: interpretable machine learning model identifies gene expression biomarkers predicting clinical outcomes and reveals molecular mechanisms of human disease in early stages. Bioinformatics. 2020:btaa935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maruff P, Thomas E, Cysique L, et al. Validity of the CogState brief battery: relationship to standardized tests and sensitivity to cognitive impairment in mild traumatic brain injury, schizophrenia, and AIDS dementia complex. Arch Clin Neuropsychol. 2009;24:165‐178. [DOI] [PubMed] [Google Scholar]
- 28. Farias ST, Mungas D, Reed BR, et al. The Measurement of Everyday Cognition (ECog): scale development and psychometric properties. Neuropsychology. 2008;22:531‐544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mackin RS, Insel PS, Truran D, et al. Unsupervised online neuropsychological test performance for individuals with mild cognitive impairment and dementia: results from the Brain Health Registry. Alzheimers Dement Diagn Assess Dis Monit. 2018;10:573‐582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nosheny RL, Camacho MR, Insel PS, et al. Online study partner‐reported cognitive decline in the Brain Health Registry. Alzheimers Dement Transl Res Clin Interv. 2018;4:565‐574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Weiner MW, Nosheny R, Camacho M, et al. The Brain Health Registry: an internet‐based platform for recruitment, assessment, and longitudinal monitoring of participants for neuroscience studies. Alzheimers Dement. 2018;14:1063‐1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee JH, Byun MS, Yi D, et al. Prediction of cerebral amyloid with common information obtained from memory clinic practice. Front Aging Neurosci. 2018;10:309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nosheny RL, Camacho MR, Jin C, et al. Validation of online functional measures in cognitively impaired older adults. Alzheimers Dement. 2020:alz.12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Arenaza‐Urquijo EM, Bejanin A, Gonneaud J, et al. Association between educational attainment and amyloid deposition across the spectrum from normal cognition to dementia: neuroimaging evidence for protection and compensation. Neurobiol Aging. 2017;59:72‐79. [DOI] [PubMed] [Google Scholar]
- 35. Mielke MM, Wiste HJ, Weigand SD, et al. Indicators of amyloid burden in a population‐based study of cognitively normal elderly. Neurology. 2012;79:1570‐1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chung JK, Plitman E, Nakajima S, et al. Lifetime history of depression predicts increased amyloid‐accumulation in patients with mild cognitive impairment. J Alzheimers Dis. 2015;45:907‐919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nosheny RL, Camacho MR, Flenniken D, et al. F4‐01‐02: increasing the impact of the ideas study using brain health registry online data collection. Alzheimers Dement. 2019;15:P1216‐P1217. [Google Scholar]
- 38. Yesavage JA, Brink TL, Rose TL, et al. Development and validation of a geriatric depression screening scale: a preliminary report. J Psychiatr Res. 1982;17:37‐49. [DOI] [PubMed] [Google Scholar]
- 39. Cromer JA, Harel BT, Yu K, et al. Comparison of cognitive performance on the Cogstate Brief Battery when taken in‐clinic, in‐group, and unsupervised. Clin Neuropsychol. 2015;29:542‐558. [DOI] [PubMed] [Google Scholar]
- 40. Maruff P, Lim YY, Darby D, et al. Clinical utility of the cogstate brief battery in identifying cognitive impairment in mild cognitive impairment and Alzheimer's disease. BMC Psychol. 2013;1:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mielke MM, Machulda MM, Hagen CE, et al. Performance of the CogState computerized battery in the Mayo Clinic Study on Aging. Alzheimers Dement. 2015;11:1367‐1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Eckner JT, Kutcher JS, Richardson JK. Pilot evaluation of a novel clinical test of reaction time in National Collegiate Athletic Association Division i football players. J Athl Train. 2010;45:327‐332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rabinovici GD, Gatsonis C, Apgar C, et al. Association of amyloid positron emission tomography with subsequent change in clinical management among Medicare beneficiaries with mild cognitive impairment or dementia. JAMA ‐ J Am Med Assoc. 2019;321:1286‐1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit‐learn: machine Learning in Python. J Mach Learn Res. 2011;12:2825‐2830. [Google Scholar]
- 45. Kuiper JS, Voshaar RCO, Verhoeven FEA, Zuidema SU, Smidt N. Comparison of cognitive functioning as measured by the Ruff Figural Fluency Test and the CogState computerized battery within the LifeLines Cohort Study. BMC Psychol. 2017;5:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Maruff P, Schembri A, Petersen RC, Aisen PS, Weiner M, Albala B. [O2‐16‐02]: acceptability, usability, validity and sensitivity of cognition assessed in unsupervised settings. Alzheimers Dement. 2017;13:P596‐P597. [Google Scholar]
- 47. Sumner JA, Hagan K, Grodstein F, Roberts AL, Harel B, Koenen KC. Posttraumatic stress disorder symptoms and cognitive function in a large cohort of middle‐aged women. Depress Anxiety. 2017;34:356‐366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Guyon I, Elisseeff A. An introduction to variable and feature selection. J Mach Learn Res. 2003;3:1157‐1182. [Google Scholar]
- 49. Jack CR, Bennett DA, Blennow K, et al. NIA‐AA Research Framework: toward a biological definition of Alzheimer's disease. Alzheimers Dement. 2018;14:535–562. [DOI] [PMC free article] [PubMed] [Google Scholar]


