In this paper, we evaluate whether elimination or repopulation of microglia affects Aβ-induced tau seeding and spreading. Elimination of microglia increases NP-tau seeding and spreading. Interestingly, repopulated microglia cluster around amyloid plaques, exhibit a homeostatic gene expression signature, and also result in elevated NP-tau seeding and spreading.
Abstract
In Alzheimer’s disease (AD) models, AD risk variants in the microglial-expressed TREM2 gene decrease Aβ plaque–associated microgliosis and increase neuritic dystrophy as well as plaque-associated seeding and spreading of tau aggregates. Whether this Aβ-enhanced tau seeding/spreading is due to loss of microglial function or a toxic gain of function in TREM2-deficient microglia is unclear. Depletion of microglia in mice with established brain amyloid has no effect on amyloid but results in less spine and neuronal loss. Microglial repopulation in aged mice improved cognitive and neuronal deficits. In the context of AD pathology, we asked whether microglial removal and repopulation decreased Aβ-driven tau seeding and spreading. We show that both TREM2KO and microglial ablation dramatically enhance tau seeding and spreading around plaques. Interestingly, although repopulated microglia clustered around plaques, they had a reduction in disease-associated microglia (DAM) gene expression and elevated tau seeding/spreading. Together, these data suggest that TREM2-dependent activation of the DAM phenotype is essential in delaying Aβ-induced pathological tau propagation.
Introduction
Alzheimer’s disease (AD) is characterized by extracellular plaques composed of aggregated forms of the amyloid-β (Aβ) peptide and intraneuronal neurofibrillary tangles (NFTs), neuropil threads, and dystrophic neurites that contain aggregated forms of the tau protein (Holtzman et al., 2011; Castellani et al., 2010; Serrano-Pozo et al., 2011). Progression of tau pathology into the limbic cortex and neocortex appears to be driven by the presence of Aβ pathology (Hurtado et al., 2010; Bennett et al., 2017; Götz et al., 2001) and correlates with cognitive impairment in individuals with AD (Nelson et al., 2012). The presence of Aβ plaques facilitates local tau seeding in dystrophic neurites that leads to the spreading and formation of phosphorylated forms of tau in neuritic plaque (NP) tau aggregates and NFTs in mice (He et al., 2018). Beyond tau and Aβ pathologies, neuroinflammation is evident in the AD brain, including alterations in the morphology, activation state, and distribution of microglia characterized as disease-associated microglia (DAM; Keren-Shaul et al., 2017) or neurodegenerative disease microglia (Krasemann et al., 2017), which cluster around amyloid plaques. In DAM, there is a down-regulation in the expression of homeostatic microglial genes such as P2ry12, Tmem119, and Cx3cr1, while there is an increase in expression of several AD-associated activation markers, such as ApoE and TREM2 (Keren-Shaul et al., 2017; Krasemann et al., 2017). In amyloid mouse models, the clustering of DAM around plaques is dependent upon TREM2 activity, and partial loss-of-function variants in the TREM2 gene increase the risk of developing AD two- to fourfold (Guerreiro et al., 2013; Jonsson et al., 2013). Although the functional consequences of plaque-associated microgliosis are still unclear, loss of TREM2 or expression of AD-associated TREM2 variants increased neuritic dystrophy as well as the seeding and spreading of phosphorylated tau around NPs (NP-tau) in the APPPS1–21 amyloid depositing mouse model; in TauPS2APP mice, where loss of TREM2 also increased Aβ-driven tau pathology and degeneration; and in AD (Leyns et al., 2019; Prokop et al., 2019; Yuan et al., 2016; Wang et al., 2016; Lee et al., 2021). Whether the increased NP-tau in TREM2 deficiency results from decreased microglial clearance of pathological tau or increased neuritic dystrophy is unclear. However, the inverse correlation between microgliosis and NP-tau suggests that manipulating microglial function to decrease plaque-associated tauopathy is a potential therapeutic strategy to slow the early progression of AD pathology.
To decipher the precise role of microglia on AD pathogenesis, several studies have used colony stimulating factor 1 (CSF1) receptor inhibitors to deplete microglia in mouse models with AD pathology. Administration of CSF1 antagonists robustly decreases the number of microglia, which rapidly repopulate the brain following cessation of antagonist treatment. In tau mouse models without amyloid pathology, microglial depletion reduced tau seeding (Asai et al., 2015), tau pathology, and tau-mediated neurodegeneration (Shi et al., 2019; Mancuso et al., 2019). Interestingly, in mouse models with brain amyloidosis, microglial ablation in plaque-containing mice did not significantly affect Aβ deposition, whereas depleting microglia before amyloidosis impaired parenchymal plaque development but increased neuritic dystrophy around the remaining plaques (Spangenberg et al., 2019, 2016). Another potential method to modulate microglial function is to pharmacologically induce microglial turnover by administration and withdrawal of CSF1 antagonists to induce microglial repopulation. In aging studies, microglial repopulation was found to rescue age-associated reductions in synaptic densities and cognitive function (Elmore et al., 2018). It has been shown that microglial repopulation could increase synaptic number and dampen inflammation in a neuronal lesion model and facilitate neuroprotective effects during the acute phase of traumatic brain injury (Willis et al., 2020; Rice et al., 2017). Fewer studies have examined the function of repopulated microglia in mouse models with AD pathology. One report found that 4 wk following cessation of a CSF1 antagonist in 5XFAD mice, microglial repopulation varied depending on brain region, with decreased microgliosis within the cortex (Casali et al., 2020). Herein, we tested whether elimination or repopulation of microglia affected Aβ-induced NP-tau seeding and spreading. Surprisingly, we found that although repopulated microglia were able to cluster around amyloid plaques like WT microglia, they exhibited a strong homeostatic gene expression signature and had markedly elevated NP-tau seeding and spreading compared with unperturbed microglia.
Results and discussion
Resident microglial functions are essential to slowing down NP-tau seeding and spreading
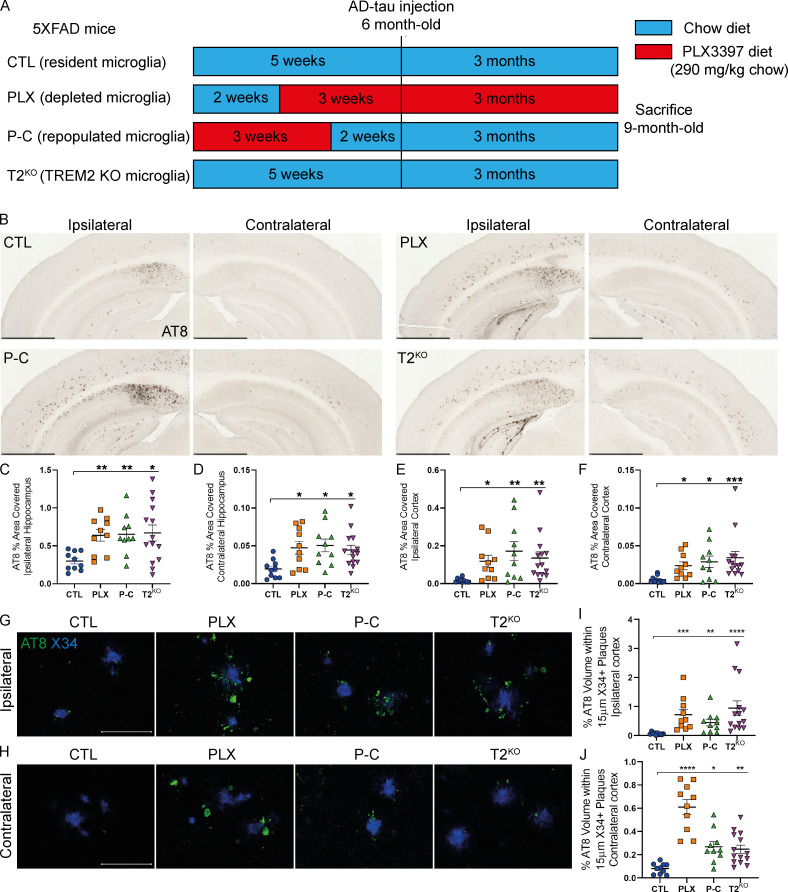
To investigate the role of microglia and microglial repopulation on NP-tau pathology, we used a previously described model of tau seeding and spreading by injecting brains with sarkosyl-insoluble tau aggregates isolated from human AD brain tissue (AD-tau; Leyns et al., 2019; He et al., 2018). 5XFAD mice were pretreated with PLX3397 (PLX group), a selective CSF1R inhibitor that has been shown to readily cross the blood–brain barrier and eliminate microglia via oral delivery in mouse chow (Elmore et al., 2014), or AIN-76A control chow diet (CTL group) for 3 wk before AD-tau injection (Fig. 1 A). Another group of mice were pretreated with PLX for 3 wk and subsequently fed with control diet for 2 wk before AD-tau injection to repopulate microglia in the brain (P-C group). This experimental design was first tested in WT mice (Fig. S1) to confirm that 3 wk of PLX removed most of the microglia (Fig. S1 B), followed by reversion to control diet for 2 wk, which allowed microglia to repopulate similarly to control mice (Fig. S1 C). 5XFAD mice were unilaterally injected at 6 mo of age in the dentate gyrus and the overlying cortex with AD-tau and analyzed 3 mo later. We observed widespread seeded NP-tau in the ipsilateral hippocampus (HC) and cortex of 5XFAD mice treated with control diet, as well as spread in the contralateral HC and cortex. This NP-tau seeding and spreading was strongly increased in mice with depleted microglia (Fig. 1, B–F). Surprisingly, NP-tau pathology was also significantly elevated in mice with repopulated microglia. We next wanted to compare the effect of microglial ablation and especially microglial repopulation to TREM2 deficiency in this assay to better understand the phenotypes observed in relation to the type of microglial dysfunction in these different states. When assessing Aβ-induced NP-tau seeding and spreading, we confirmed our previous findings from a different amyloid mouse model of elevated NP-tau seeding and spreading in TREM2-deficient mice that was very similar in extent to that seen in the PLX and P-C groups (Fig. 1, B–F; Leyns et al., 2019). To confirm that cortical NP-tau pathology was increased independently of the number of Aβ plaques, we performed confocal analysis and quantified the amount of NP-tau surrounding individual X34+ Aβ plaques (Fig. 1, G–J). We confirmed that NP-tau pathology that represents both seeding and spreading was significantly increased with loss of microglia or TREM2 function, as well as when microglia were repopulated. Our findings are in contrast with a previous study showing dramatically suppressed tau propagation after depleting microglia (Asai et al., 2015). Interestingly, the authors used a nonamyloid plaque depositing mouse model, suggesting that amyloid pathology modifies the role of microglia in tau seeding and spreading. Altogether, these data support that resident, TREM2-expressing microglia are essential to slowing down the burden and spreading of seeded periplaque NP-tau.
Figure 1.
Microglia depletion, TREM2 KO, or microglia repopulation significantly increased NP-tau seeding and spreading in 5XFAD mice. (A) Schematic of the experimental design. 6-mo-old 5XFAD mice were injected with AD-tau in the HC and overlaying cortex and sacrificed 3 mo later to evaluate tau seeding and spreading. Before AD-tau injection, 5XFAD mice were pretreated for 3 wk with PLX3397 to deplete microglia (PLX; n = 10) or pretreated for 3 wk with PLX3397 and then switched back to a control diet to allow microglia repopulation in the brain (P-C; n = 10). Both controls, 5XFAD (CTL; n = 10) and 5XFAD-T2KO mice (T2KO; n = 14), were fed a chow diet. (B) Representative images of ipsi- and contralateral hemisphere stained with AT8+ NP-tau pathology in AD-tau–injected 5XFAD mice. Scale bars, 1 mm. (C–F) Quantification of AT8+ staining in the ipsi- and contralateral hippocampi (C and D, respectively) and cortices (E and F, respectively) of AD-tau–injected 5XFAD mice. (G and H) Confocal images of cortical AT8+ NP-tau (green) around X34+ plaques (blue) in the ipsi- (G) and contralateral (H) cortices. Scale bars, 50 µm. (I and J) Quantification of percentage AT8+ volume within 15 µm of plaques in the ipsi- (I) and contralateral (J) cortices. Data are presented as mean ± SEM. Significance was determined using Welch’s and Brown–Forsythe ANOVA test (C and J) or Kruskal–Wallis test followed by a Dunn's post hoc test (D–F and I). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Figure S1.
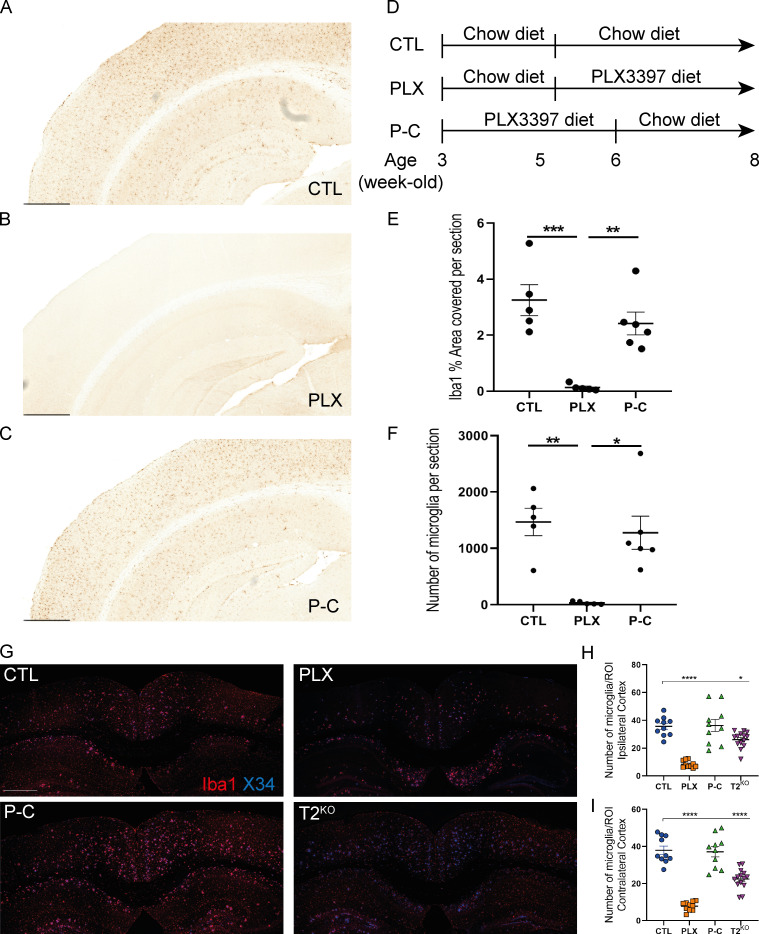
CSF1R inhibition eliminates microglia from the mouse brain.(D) Schematic of the experimental design. 5-wk-old WT mice (n = 5–6 per group) were treated with chow diet or PLX3397 (290 mg/kg chow; PLX) for 3 wk, and 3-wk-old WT mice were treated with PLX3397 (290 mg/kg chow) for 3 wk and then chow diet for 2 wk (P-C). (A–C) Representative sections from the hippocampal and cortical field for Iba1 immunostaining for the three groups. (E and F) Quantification of Iba1% coverage (E) and number of Iba1+ cells in full brain sections (F) shows that 3 wk of PLX3397 treatment showed robust decrease in microglial numbers and Iba1 coverage in PLX groups, with little to no microglia present after 3 wk of treatment. After 2 wk back to control chow diet, the microglia population was restored, showing that repopulating microglia are fully dependent upon CSF1R signaling. Scale bars, 500 µm. (G) Representative images of Iba1+ microglia (red) surrounding X34+ plaques (blue) in cortices of 5XFAD mice. (H and I) Quantification of the number of microglia per image in ipsi-(H, left) and contralateral (I, right) cortices. Data are presented as mean ± SEM. Significance was determined using a one-way ANOVA followed by a Tukey's post hoc test for Iba1 (E) coverage and with a Kruskal–Wallis test followed by a Dunn's post hoc test for the number of microglia (F). Welch’s and Brown–Forsythe ANOVA test was used for H and I. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Increased plaque-associated neuritic dystrophy associated with NP-tau pathology and depleted, repopulated, or T2KO microglia
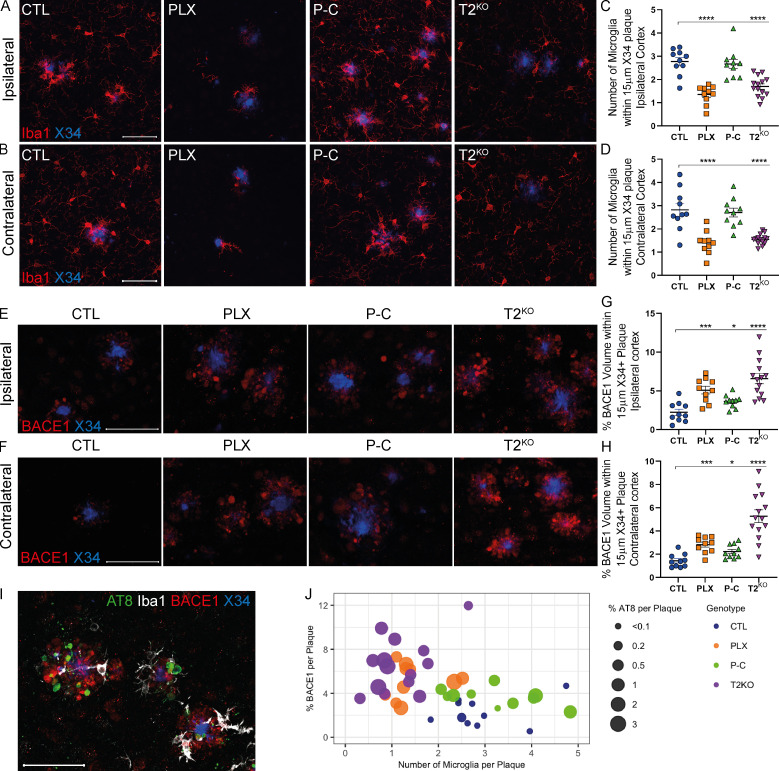
We next investigated why NP-tau pathology increased when microglia function or number was affected. First, we evaluated the number of microglia in the cortex of 9-mo-old 5XFAD mice. As previously described (Spangenberg et al., 2016), PLX treatment removed most of the microglia in both ipsilateral and contralateral cortex, although some microglia (∼20%) were resistant to PLX treatment (Fig. S1, G–I). When we evaluated the number of microglia clustering around Aβ plaques (Fig. 2, A–D), we also observed fewer microglia clustering around amyloid plaques in both PLX-treated and TREM2-deficient mice. Interestingly, 80% of the PLX-resistant microglia that remained were clustered around amyloid plaques. This suggests that a population of activated microglia in 5XFAD mice survive independently of CSF1R signaling (Spangenberg et al., 2016, 2019; Zhan et al., 2019 Preprint). In contrast, P-C mice exhibited robust microglial clustering around plaques similar to that observed in control (CTL) mice. We observed reduced plaque-associated microglia in T2KO mice as previously reported (Jay et al., 2015; Ulrich et al., 2014; Wang et al., 2015). Microglial depletion and TREM2 deficiency are associated with increased amyloid-dependent neuritic dystrophy, which in turn correlated with increased NP-tau seeding and spreading (Leyns et al., 2019). Whether microglial repopulation alters neuritic dystrophy is unclear. Therefore, we immunostained for BACE1, which accumulates in dystrophic neurites (Sadleir et al., 2016). We observed a strong increase in the amount of BACE1+ processes in PLX, P-C, and T2KO groups compared with CTL in both ipsi- and contralateral cortex, which also correlated with AT8 staining (Fig. 2, E–H). We further observed a strong correlations among AT8+ NP-tau pathology, BACE1+ neuritic dystrophy, and the number of Iba1+ microglia-clustering Aβ plaques, except in P-C mice, suggesting that microglia may be essential in mitigating plaque-induced neuronal toxicity, which in turn may promote NP-tau seeding (Fig. 2, I and J). Importantly, although the repopulated microglia clustered around plaques at a similar density to control-treated microglia, the amount of NP-tau pathology and neuritic dystrophy was markedly elevated. The neuritic dystrophy result is similar to a previous report that found elevated cortical neuritic dystrophy following 4 wk of microglial repopulation rather than the 14 wk of repopulation in our study (Casali et al., 2020). Overall, this suggests that the repopulated microglia are deficient in mitigating against plaque-associated toxicity and, importantly, Aβ-mediated tau seeding and spreading.
Figure 2.
Altered microglia, either depleted, repopulated, or TREM2 KO, is associated with increased neuritic dystrophy around plaques, which correlates with NP-tau pathology. (A and B) Confocal analysis of Iba1+ microglia (red) surrounding X34+ plaques (blue) in ipsi- (A) and contralateral (B) cortices of 5XFAD mice. (C and D) Quantification of the number of microglia surrounding plaques in ipsi- (C) and contralateral (D) cortices. (E and F) Confocal images of BACE1 (red) around X34+ plaques (blue) in ipsi- (E) and contralateral (F) cortices of 5XFAD mice. (G and H) Quantification of percentage BACE1+ volume within 15 µm of plaques in ipsi- (G) and contralateral (H) cortices. (I) Representative image of costain for X34 (blue), AT8 (green), BACE1 (red), and Iba1 (white). (J) Relationship between costained Iba1+ microglia (x axis), BACE1 dystrophic neurites (y axis), and seeded AT8+ NP-tau surrounding X34+ plaques in the ipsilateral cortex. Each dot represents an individual mouse, dot color indicates the different groups, and dot size corresponds to percentage of AT8+ staining around plaques (Fig. 1). Scale bars, 50 µm. Data are presented as mean ± SEM. Significance was determined using ordinary one-way ANOVA followed by a Tukey's post hoc test (C and D) or Welch’s and Brown–Forsythe ANOVA test (G and H). *, P < 0.05; ***, P < 0.001; ****, P < 0.0001.
Repopulated or T2KO microglia are associated with more amyloid plaques but reduced plaque-associated ApoE
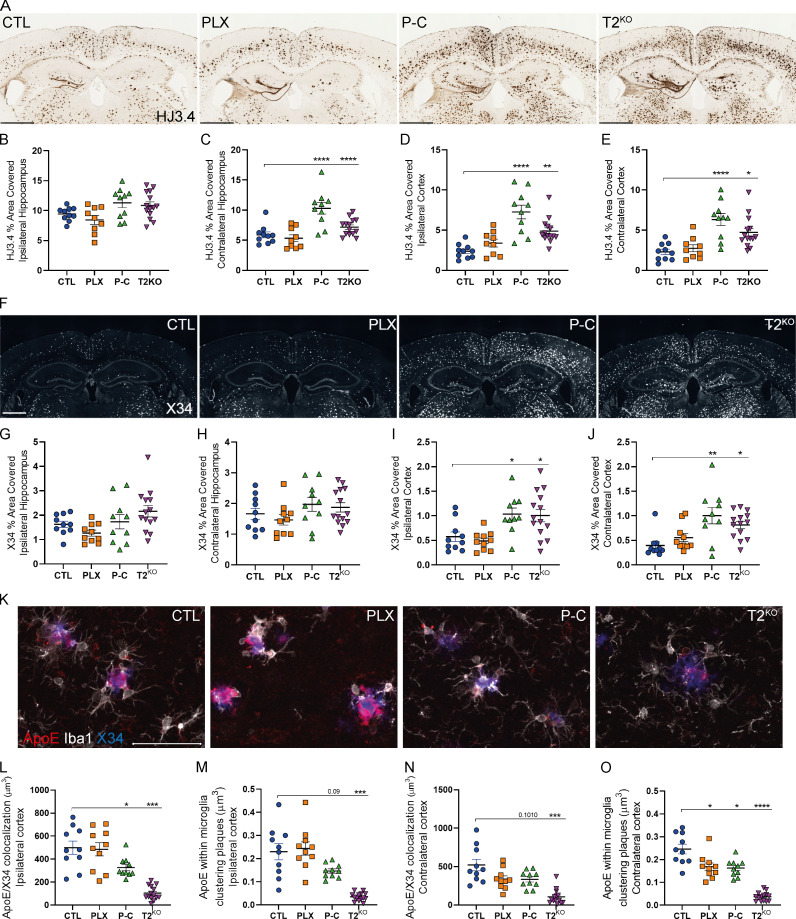
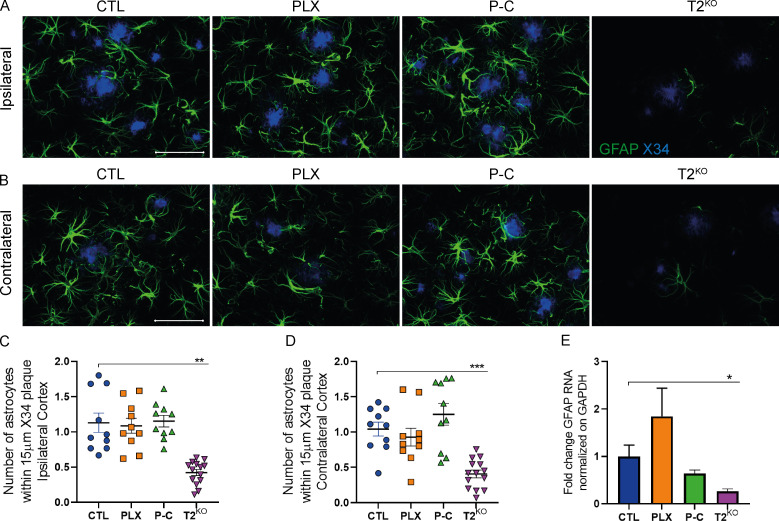
We next evaluated the effect of microglial depletion and repopulation on Aβ plaques using an anti-Aβ antibody (HJ3.4) and a stain for fibrillar plaques (X34; Fig. 3, A–J). Importantly, microglial depletion and TREM2 KO mouse models have reported conflicting results on the effect of microglia on amyloid plaque burden (Gratuze et al., 2018; Spangenberg et al., 2016, 2019; Casali et al., 2020). We found that Aβ plaques were comparable in CTL and PLX groups, but we observed increased Aβ pathology in 5XFAD mice with repopulated or T2KO microglia compared with CTL possibly due to impaired plaque compaction in the absence of TREM2 (Wang et al., 2016). Parhizkar et al. (2019) recently demonstrated that loss of TREM2 increased the seeding of amyloid pathology as well as reduced plaque-associated ApoE in TREM2-deficient mice or in a genetic microglial ablation model. We confirmed that T2KO mice exhibited strongly reduced ApoE colocalization with plaques and ApoE present within microglia (Fig. 3, K–O), suggesting a microglial origin of plaque-associated ApoE. Interestingly, the amount of ApoE in plaques and plaque-associated microglia was not significantly different between PLX-treated and CTL mice, suggesting that the plaque-associated microglia that persisted in PLX mice remained highly activated (Fig. 3, K–O). In contrast, we noted that ApoE colocalization with both plaques and microglia was decreased in P-C mice, further suggesting that repopulated microglia did not acquire a typical DAM phenotype, as previously described (Keren-Shaul et al., 2017; Krasemann et al., 2017), despite clustering around plaques. Given that astrocytes are the predominant cell type that expresses ApoE in the brain, we also assessed the amount of GFAP+ astrocytes around plaques. Microglia depletion or repopulation did not affect the number of GFAP+ astrocytes; however, TREM2 deletion strongly suppressed the clustering of astrocytes around plaques as well as GFAP mRNA level (Fig. S2).
Figure 3.
Repopulated microglia or TREM2 KO increase amyloid plaques and reduce plaque-associated ApoE in 5XFAD mice.(A) Representative images of HJ3.4+ plaques in 5XFAD mice. Scale bars, 1 mm. (B–E) Quantification of HJ3.4 staining in the ipsi- and contralateral hippocampi (B, left and C, right, respectively) and cortices (D and E, respectively). (F) Representative images of X34+ fibrillar plaques. Scale bars, 1 mm. (G–J) Quantification of X34 staining in the ipsi- and contralateral hippocampi (G and H, respectively) and cortices (I, left and J, right, respectively) of 5XFAD mice. (K) Confocal analysis of Iba1+ microglia (white), ApoE (red), and X34+ plaques (blue) in ipsilateral cortex of 5XFAD mice. (L–O) Quantification of colocalized ApoE and X34 volumes in ipsi- (L) and contralateral (N) cortices and quantification of ApoE volume within microglia clustering X34+ plaques in ipsi- (M) and contralateral (O) cortices. Scale bars, 50 µm. Data are presented as mean ± SEM. Significance was determined using ordinary one-way ANOVA followed by a Tukey's post hoc test (B, C, E, H, and I), Welch’s and Brown–Forsythe ANOVA test (G and L–O), or Kruskal–Wallis test followed by a Dunn's post hoc test (D and J). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Figure S2.
TREM2 deletion, but not microglia depletion or repopulation, decreases astrocyte clustering amyloid plaques and GFAP RNA expression in 5XFAD mice. (A and B) Confocal analysis of GFAP-labeled astrocytes (green) surrounding X34+ plaques (blue) in ipsilateral (A) and contralateral (B) cortices of 9-mo-old 5XFAD mice. Scale bars, 50 µm. (C and D) Quantification of the number of astrocyte cells surrounding plaques in ipsilateral (C) and contralateral (D) cortices. (E) Expression of cortical GFAP mRNA in 6-mo-old 5XFAD mice. Data are presented as mean ± SEM. Significance was determined using Welch’s and Brown–Forsythe ANOVA test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Repopulated microglia do not reestablish the DAM signature
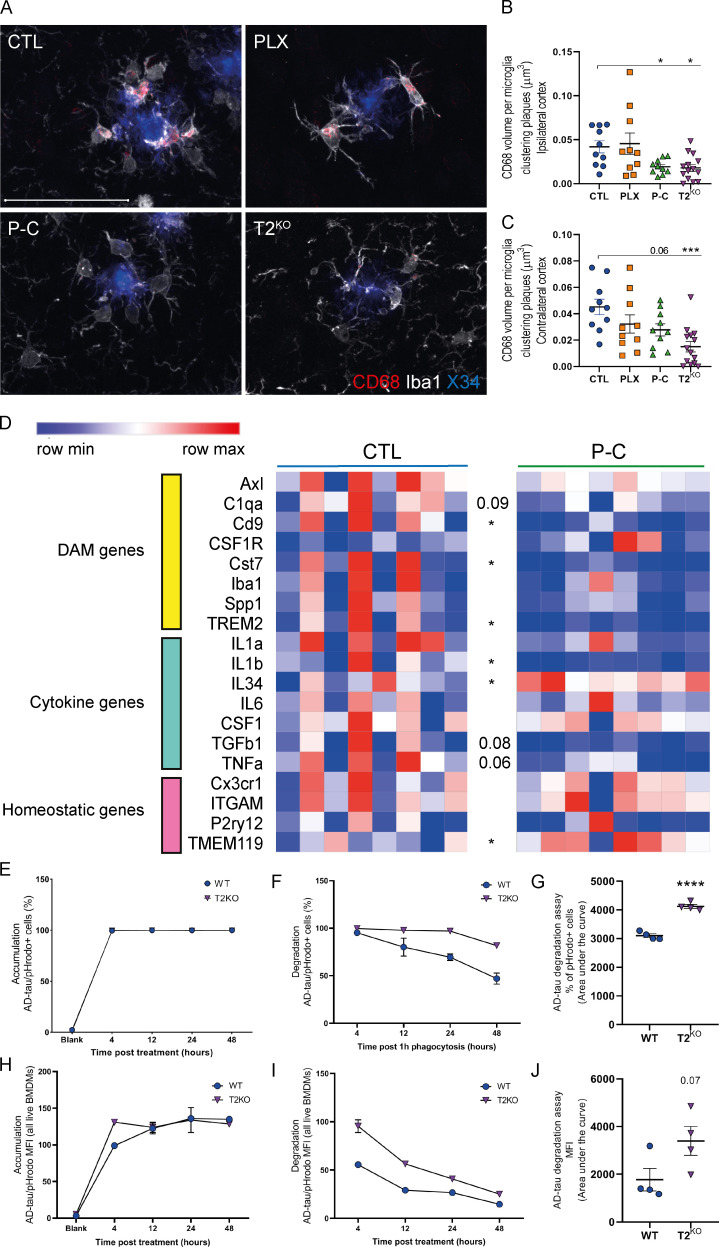
To evaluate microglial activation in our different groups, we first costained brain sections with Iba1 (microglia) and CD68 phagolysosomes in microglia around X34+ plaques (Fig. 4, A–C). We observed similar CD68+ vesicle volume per microglia around plaques in CTL and PLX-resistant microglia. In contrast, we observed a strong decrease in CD68+ vesicles within repopulated and T2KO microglia, suggesting decreased late-endosomal/lysosomal function in microglia and overall DAM activation state in these groups in both the ipsi- and contralateral cortices. Decreased late-endosomal/lysosomal activity in microglia could increase NP-tau seeding and spreading by failing to efficiently clear toxic tau species from the brain. Given that P-C mice exhibited elevated NP-tau pathology and decreased activation in plaque-associated microglia, we decided to compare the expression level of homeostatic and DAM genes in our experimental groups immediately before NP-tau injections using a second cohort of animals that were 6 mo of age. We assessed gene expression profile in the cortex (Fig. 4 D) and observed a decrease in DAM and proinflammatory cytokine gene expression (e.g., Cst7 and IL1β) in mice with repopulated microglia compared with CTL mice. Consistent with the reduced CD68 staining, repopulated microglia display raised homeostatic gene expression (e.g., TMEM119). Interestingly, we observed an increase of IL34 gene expression in mice with repopulated microglia, which might explain how microglia derived from the proliferation of residual PLX-resistant microglia were able to repopulate the brain. Altogether, we demonstrate that repopulated microglia have not built up a typical DAM signature yet despite the presence of Aβ pathology. Importantly, repopulated microglia also displayed lower TREM2 gene expression, which could explain similar results observed between mice with repopulated microglia and TREM2-deficient mice.
Figure 4.
Repopulated microglia exhibit homeostatic phenotype and T2KO BMDMs degrade AD-tau slower.(A) Confocal analysis of Iba1-labeled microglia (white) and CD68+ phagolysosomes (red) and X34+ plaques (blue) in ipsilateral cortex of 5XFAD mice. (B and C) Quantification of CD68+ phagolysosomes volume within microglia clustering X34+ plaques in ipsilateral (B) and contralateral (C) cortices of 5XFAD mice. Scale bars, 50 µm. (D) Heatmap analysis of bulk RNA in cortices of 6-mo-old CTL and P-C 5XFAD mice generated by hierarchical gene clustering based on groups (CTL, n = 8; and P-C, n = 8). (E–J) AD-tau phagocytosis assay using BMDMs cells and AD-tau–conjugated to pHrodo. (E, F, H, and I) Time-dependent representative AD-tau uptake (E and H) and degradation (F and I) curves of percentage of AD-tau pHrodo+ BMDMs (E and F) and mean fluorescence intensity (MFI; H and I). (G and J) Statistical differences of the area under the curve from four independent experiments in technical duplicate for the degradation of AD-tau obtained from the percentage of AD-tau pHrodo+ (G) and mean fluorescence intensity analysis (J). Data are presented as mean ± SEM. Significance was determined using Welch’s and Brown–Forsythe ANOVA test (B and C). For D, G and J, statistical analyses were performed using an unpaired t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
TREM2 regulates AD-tau clearance
Since we observed increased NP-tau seeding in T2KO mice and mice with repopulated microglia displayed decreased TREM2 expression, we tested the effect of TREM2 on the uptake and degradation of human tau using an in vitro phagocytosis assay by conjugating AD-tau with pHrodo (Fig. 4, E–J). We first treated bone marrow–derived macrophages (BMDMs) from WT or T2KO mice with AD-tau incubated for 4, 12, 24, and 48 h. We observed similar AD-tau uptake on both mean fluorescence intensity and percentage of positive cells between WT and T2KO BMDMs (Fig. 4, E and H). However, when we treated BMDMs for 1 h with AD-tau and then evaluated tau degradation over 48 h, we found that the pHrodo signal decayed more rapidly in WT BMDMs than in T2KO BMDMs (Fig. 4, F, G, I, and J), suggesting a defect in tau degradation in myeloid cells without TREM2. These results suggest that increased AD-tau seeding in mice with repopulated and T2KO microglia are likely a result of a defect in AD-tau degradation by microglia.
Conclusion
Thus far, the majority of studies have evaluated the role of microglia in AD in the context of Aβ or tau pathologies separately. Here, we examined the role of microglia in mitigating against amyloid-driven tau seeding and spreading. How Aβ augments tau pathology to drive AD pathogenesis is a critical step in disease progression, as it drives neurodegeneration, and needs to be better understood. We assessed whether pharmacologically induced microglial repopulation protected against NP-tau seeding and spreading. We found that microglial depletion or TREM2 deficiency increased susceptibility of NP-tau pathology in dystrophic neurons surrounding Aβ plaques as well as the spreading of tau pathology. Surprisingly, microglial repopulation also exhibited increased NP-tau pathology, most likely due in part to a failure of repopulated microglia to switch from a homeostatic to a DAM phenotype. Our data support a key role for TREM2-dependent microglial activation at the intersection of Aβ and tau pathologies. The ability of microglia to switch to a DAM phenotype appears essential to limit Aβ plaque–mediated tau pathogenesis in AD. Our data suggest that during the phase of AD when Aβ is driving tau seeding and spreading, TREM2 activation as well as other ways to stimulate the microglial DAM phenotype should be explored therapeutically.
Materials and methods
Mice
5XFAD mice (Oakley et al., 2006) overexpressing both mutant human Aβ precursor protein with the Swedish (K670N and M671L), Florida (I716V), and London (V717I) familial AD mutations and human PS1 harboring two familial AD mutations, M146L and L286V, both under the control of the Thy1 promoter, were purchased from The Jackson Laboratory (MMRRC; stock no. 34848-JAX). TREM2−/− mice were previously described (Turnbull et al., 2006). 5XFAD were crossed to TREM2−/− mice to generate TREM2+/− 5XFAD and TREM2−/− 5XFAD mice. Only males were used in this study, because male mice showed a higher level of microglial reduction than females when treated with the same PLX diet (Shi et al., 2019), and 5XFAD male and female mice exhibit differences in amyloid pathology (Bundy et al., 2019). All mice were on the C57BL/6 background and housed under a normal 12-h light/dark cycle. All animal studies were approved by the Animals Studies Committee at Washington University School of Medicine in St. Louis.
Diet
PLX3397 was purchased from SelleckChem. PLX3397 was formulated in the AIN-76A (Research Diets) chow at a concentration of 290 mg/kg chow as previously done (Elmore et al., 2014; Spangenberg et al., 2016). 5XFAD mice were treated with PLX3397 for 3 wk to eliminate microglia or control chow, or they were treated with PLX3397 for 3 wk and then control chow for 2 wk to repopulate microglia before AD-tau injection at 6 mo of age, creating four groups: control 5XFAD (CTL; n = 10), PLX3397-treated 5XFAD mice (PLX; n = 10), 5XFAD with repopulated microglia (P-C; n = 10), and 5XFAD-TREM2 KO (T2KO; n = 14). After AD-tau injection, mice were kept on their respecting diet (chow diet for CTL, P-C, and T2KO and PLX3397 for PLX mice) for 3 mo until harvesting brains. A second cohort of 5XFAD mice was treated similarly and at the same age compared with the first cohort, but these mice were instead sacrificed at 6 mo of age without AD-tau injection in order to assess the microglia profile before AD-tau injection for the previous cohort (CTL, n = 8; and P-C, n = 8).
Preparation of AD-tau aggregates from human AD brain tissue and stereotactic intracerebral injections
AD-tau was isolated as previously described (Guo et al., 2016) from a human AD brain. Using a bicinchoninic acid assay (Thermo Fisher Scientific; catalog no. 23225), the total protein concentration of the AD-tau preparation was 21.1 µg/µl. Tau-specific sandwich ELISA was used to determine the tau concentration in the preparation as previously described (Guo et al., 2016), which was found to be 5.4 µg/µl. AD-tau preparation was diluted at 0.4 µg/µl prior to the injections and sonicated in water bath sonicator (QSonica; Q700) for 30 s at 60% amplitude at 4°C. 6-mo-old 5XFAD mice were anesthetized with isoflurane, immobilized in a stereotactic frame (David Kopf Instruments), and unilaterally injected with a total of 2 µg AD-tau (1 µg at each injection site) in the dentate gyrus (bregma: −2.5 mm; lateral: −2.0 mm; depth: −2.2 mm) and overlying cortex (bregma: −2.5 mm; lateral: −2.0 mm; depth: −1.0 mm) using a Hamilton syringe (Hamilton; syringe: 80265–1702RNR; needle: 7803–07). Mice were allowed to recover on a 37°C heating pad and monitored for the first 48 h after surgery.
Preparation of brain samples
Mice were sacrificed by intraperitoneal injection of pentobarbital (200 mg/kg). Blood samples were collected in EDTA-treated tubes before cardiac perfusion with 3 U/ml heparin in cold Dulbecco’s PBS. Blood samples were spun down (10 min, 2,000 ×g, 4°C), and blood plasma was collected. For the 9-mo-old AD-tau–injected 5XFAD mice, whole brains were carefully extracted and immerse-fixed in 4% paraformaldehyde for 24 h before being transferring to 30% sucrose and stored at 4°C until sectioned. Brains were cut coronally into 30-µm sections on a freezing sliding microtome (Microm; HM 400) and stored in cryoprotectant solution (0.2 M PBS, 15% sucrose, and 33% ethylene glycol) at −20°C until use. A notch on the left hemisphere at the piriform cortex ensured proper identification of the ipsilateral injected side.
For cytokine and gene expression experiments, brains were carefully extracted and cut into two hemispheres. The left hemisphere was collected for immunostaining and immerse-fixed in 4% paraformaldehyde overnight before being transferring to 30% sucrose and stored at 4°C until sectioned. Brains were cut coronally into 50-µm sections on a freezing sliding microtome (Microm; HM 400) and stored in cryoprotectant solution at −20°C until use. The right hemisphere was dissected to isolate the HC and cortex for biochemical analysis, and the tissue was kept at −80°C until analyzed.
Immunohistochemistry (IHC) and immunofluorescence (IF)
For immunohistochemical staining of microglia (Wako; Iba1, rabbit polyclonal, 1:5,000, catalog no. 019–19741), NP-tau (Thermo Fisher Scientific; AT8, mouse monoclonal, 1:500; catalog no. MN1020B) and Aβ (HJ3.4 biotinylated, anti-Aβ1–13, mouse monoclonal, 2 µg/ml generated in-house), sections were washed three times in TBS for 5 min and blocked in 0.3% hydrogen peroxide for 10 min. After washing, sections were blocked in 3% milk in TBS with 0.25% Triton X-100 (TBSX) for 30 min. Primary antibody was diluted in 3% milk/TBSX, and the sections were incubated in the primary antibody overnight at 4°C. The next day, sections were washed three times. For AT8 and HJ3.4 staining, after washing, sections were incubated in ABC Elite solution (VectaStain; PK-6100) for 1 h, prepared following the manufacturer’s instructions, followed by another washing step. For Iba1 staining, after washing, sections were incubated with HRP-conjugated secondary antibodies diluted in 3% milk/TBSX for 1 h at room temperature (Jackson Laboratory; 1:500, 111–035-003). Sections were developed in DAB solution (Vector Laboratories; catalog no. SK4103 for Iba1; Sigma; catalog no. D5905 for AT8 and HJ3.4), washed, and mounted on slides. After drying overnight, the slides were dehydrated in increasing ethanol concentrations followed by xylene and coverslipped with Cytoseal 60 (Thermo Fisher Scientific; catalog no. 8310).
For IF staining, costains were performed for (1) X34, BACE1, AT8, and Iba1; (2) X34, GFAP, Iba1, and ApoE; or (3) X34, Iba1, and CD68. Fibrillar Aβ was stained by X34 dye (Sigma; SML-1954) and antibodies to AT8, Iba1 (Wako; catalog no. 011–27991, 1:2,000), BACE1 (Abcam; catalog no. ab108394, 1:500), GFAP (eBioscience; catalog no. 53–9892-82, 1:500), ApoE (HJ6.3 biotinylated, 1:300 generated in-house), and CD68 (AbD SeroTec; catalog no. MCA1957, 1:500) were used to evaluate periplaque pathologies.
Free-floating sections were washed three times and then permeabilized in 0.25% Triton X-100 PBS (PBS-X) for 30 min. Tissue sections were then incubated in X34 for 20 min, washed in X34 buffer (40% EtOH in PBS), and washed in PBS twice. Sections were incubated in blocking solution for 30 min (3% BSA, 3% normal donkey serum, and 0.1% PBS-X) before incubating in primary antibodies in blocking solution overnight at 4°C. The next day, sections were washed three times, placed in secondary antibodies for 4 h (Thermo Fisher Scientific; 1:500) at room temperature, and then washed again three times for 20 min. Lipofuscin was quenched with 0.1% Sudan black and washed once in 0.02% PBS–Tween 20 and again in PBS. Sections were mounted, sealed in ProLong Gold anti-fade (Thermo Fisher Scientific; catalog no. P36930), and stored in the dark at 4°C until imaging.
Image acquisition and analysis
Images were obtained from an average of sections per mouse for IHC and IF. For IHC stains, slides were scanned on the NanoZoomer 2.0-HT system (Hamamatsu Photonics). Images were further processed using NDP viewing software (Hamamatsu Photonics) and Fiji software version 1.51 (National Institutes of Health). For IF images, four to six z-stacks (20 µm) per section were acquired on an LSM 880 II Airyscan FAST confocal microscope (Zeiss) with a 20× objective and 1,024 × 1,024 resolution. Quantification of confocal images for AT8, BACE1, and ApoE around X34+ plaques was performed on a semiautomated platform using MATLAB and Imaris 9.5 software (Bitplane) to create surfaces of each stain based on a threshold applied to all images, dilate X34 surfaces 15 µm, and colocalize various immunostained surfaces and dilated X34 surfaces. For quantification of the number of plaque-associated microglia and astrocytes, a threshold was applied across all images to assign spots to each cell body. X34 surfaces were dilated 15 µm, and spots were counted within the X34+ extended surface. All staining experiments were imaged and quantified by a blinded investigator.
Quantification of confocal images for CD68 in Iba1+ microglia around X34+ plaques was performed by creating surface of CD68/Iba1 costaining and colocalize it with dilated X34 surfaces. R studio was used to generate plots comparing different AT8, BACE1, and microglia cells surrounding X34+ plaques.
BMDM culture and in vitro phagocytosis assay
BMDMs were differentiated from bone marrow cells isolated from WT and TREM2KO mice at same ages in BMDM differentiation media (RPMI 1640 medium, 10% FBS, 10% L929 supernatant, 1× Pen-Strep, 1× Glutamax, 1× sodium pyruvate, and 1× nonessential amino acids) for 4–5 d depending on confluence. When fully differentiated BMDMs were 70% confluent, non–tissue culture–treated 96-well plates were seeded with 30,000 cells in BMDM media for a phagocytosis/degradation assay the next day.
All AD-tau phagocytosis or degradation assays were conducted with prepared cell culture as described above. Briefly, AD-tau conjugated with pH-sensitive pHrodo Red dye was added into each well at final concentration of 50 nM at 48, 24, 12, and 4 h before harvesting cells. For degradation assay, all cells were treated with AD-tau for 1 h and switched to normal media for 4, 12, 24, and 48 h before being harvested. After washing with PBS twice, BMDMs were resuspended with MACS buffer (PBS, 1 mM EDTA, and 0.5% BSA), and flow cytometry was performed on a BD FACS Calibur System at the flow cytometry core at Washington University in St. Louis. pHrodo signal was determined by FL2 Yellow after pregating with FL3 Red Propidium Iodide staining and further analyzed by FlowJo.
Gene expression
We extracted total RNA from mouse cortex with the RNeasy Mini Kit (QIAGEN; catalog no. 74104) and prepared cDNA with the High-Capacity RNA-to-cDNA kit (Applied Biosystems; catalog no. 4388950) following the manufacturers’ instructions. Gene expression analysis was performed using microarray in collaboration with the Genome Technology Access Core at Washington University. Using TaqMan probes, the relative gene expression was quantitatively measured using Fluidigm Biomark HD with integrated fluidic circuits.
Statistics
Unless otherwise stated, all data are presented as mean ± SEM. GraphPad Prism 8.0 was used to perform statistical analyses. Gaussian distribution was evaluated using the D’Agostino–Pearson normality test. Statistical analysis was performed using ordinary one-way ANOVA followed by a Tukey’s post hoc test under normal distribution. In case of unequal variances, Welch’s and Brown–Forsythe test were used. If samples deviate from normal distribution, statistical analysis was performed using Kruskal–Wallis test followed by a Dunn’s post hoc test. For comparisons of two groups, statistical analyses were performed using unpaired t tests. Statistical significance was set as P < 0.05 (*, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001 versus CTL 5XFAD).
Online supplemental material
Fig. S1 shows microglial depletion and repopulation efficiency with the AIN-76A chow at a 290-mg/kg dose in WT and 5XFAD mice. Fig. S2 shows a decrease of GFAP+ astrocytes in 5XFAD-T2KO mice as well as lowered GFAP gene expression.
Acknowledgments
Scanning of IHC was performed on the NanoZoomer digital pathology system courtesy of the Hope Center Alafi Neuroimaging Laboratory. We thank the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine for help with genomic analysis.
This study was supported by the BrightFocus Foundation (grant A2020257F to M. Gratuze), the National Institutes of Health (NIH; grant AG047644), the JPB Foundation, the Charles and Helen Schwab Foundation (D.M. Holtzman), and the Edward N. and Della L. Thome Memorial Foundation, Bank of America, N.A., Trustee (D.M. Holtzman). Confocal data were generated on a Zeiss LSM 880 Airyscan Confocal Microscope, which was purchased with support from the Office of Research Infrastructure Programs, a part of the NIH Office of the Director under grant OD021629, and in part with support from the Washington University Center for Cellular Imaging, which is supported by Washington University School of Medicine, The Children’s Discovery Institute of Washington University, St. Louis Children’s Hospital (CDI-CORE-2015-505 and CDI-CORE-2019-813), and the Foundation for Barnes-Jewish Hospital (3770 and 4642). The Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine is partially supported by the National Cancer Institute (cancer center support grant P30 CA91842 to the Siteman Cancer Center) and the National Center for Research Resources, a component of the NIH, and NIH Roadmap for Medical Research (Institute of Clinical and Translational Sciences/Clinical and Translational Sciences Award grant UL1 TR000448). This publication is solely the responsibility of the authors and does not necessarily represent the official view of the National Center for Research Resources or NIH.
Author contributions: M. Gratuze, M. Colonna, J.D. Ulrich, and D.M. Holtzman designed the study. M. Gratuze, Y. Chen, S. Parhizkar, N. Jain, and M.R. Strickland performed the experiments and analyzed the data. M. Gratuze, J.D. Ulrich, and D.M. Holtzman wrote the manuscript. All authors discussed the results and commented on the manuscript.
References
- Asai, H., Ikezu S., Tsunoda S., Medalla M., Luebke J., Haydar T., Wolozin B., Butovsky O., Kügler S., and Ikezu T.. 2015. Depletion of microglia and inhibition of exosome synthesis halt tau propagation. Nat. Neurosci. 18:1584–1593. 10.1038/nn.4132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, R.E., DeVos S.L., Dujardin S., Corjuc B., Gor R., Gonzalez J., Roe A.D., Frosch M.P., Pitstick R., Carlson G.A., and Hyman B.T.. 2017. Enhanced Tau Aggregation in the Presence of Amyloid β. Am. J. Pathol. 187:1601–1612. 10.1016/j.ajpath.2017.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casali, B.T., MacPherson K.P., Reed-Geaghan E.G., and Landreth G.E.. 2020. Microglia depletion rapidly and reversibly alters amyloid pathology by modification of plaque compaction and morphologies. Neurobiol. Dis. 142:104956. 10.1016/j.nbd.2020.104956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellani, R.J., Rolston R.K., and Smith M.A.. 2010. Alzheimer disease. Dis. Mon. 56:484–546. 10.1016/j.disamonth.2010.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore, M.R.P., Najafi A.R., Koike M.A., Dagher N.N., Spangenberg E.E., Rice R.A., Kitazawa M., Matusow B., Nguyen H., West B.L., and Green K.N.. 2014. Colony-stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron. 82:380–397. 10.1016/j.neuron.2014.02.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore, M.R.P., Hohsfield L.A., Kramár E.A., Soreq L., Lee R.J., Pham S.T., Najafi A.R., Spangenberg E.E., Wood M.A., West B.L., and Green K.N.. 2018. Replacement of microglia in the aged brain reverses cognitive, synaptic, and neuronal deficits in mice. Aging Cell. 17:e12832. 10.1111/acel.12832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz, J., Chen F., van Dorpe J., and Nitsch R.M.. 2001. Formation of neurofibrillary tangles in P301l tau transgenic mice induced by Abeta 42 fibrils. Science. 293:1491–1495. 10.1126/science.1062097 [DOI] [PubMed] [Google Scholar]
- Gratuze, M., Leyns C.E.G., and Holtzman D.M.. 2018. New insights into the role of TREM2 in Alzheimer’s disease. Mol. Neurodegener. 13:66. 10.1186/s13024-018-0298-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro, R., Wojtas A., Bras J., Carrasquillo M., Rogaeva E., Majounie E., Cruchaga C., Sassi C., Kauwe J.S.K., Younkin S., et al. Alzheimer Genetic Analysis Group . 2013. TREM2 variants in Alzheimer’s disease. N. Engl. J. Med. 368:117–127. 10.1056/NEJMoa1211851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, J.L., Narasimhan S., Changolkar L., He Z., Stieber A., Zhang B., Gathagan R.J., Iba M., McBride J.D., Trojanowski J.Q., et al. 2016. Unique pathological tau conformers from Alzheimer’s brains transmit tau pathology in nontransgenic mice. J. Exp. Med. 213:2635–2654. 10.1084/jem.20160833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Z., Guo J.L., McBride J.D., Narasimhan S., Kim H., Changolkar L., Zhang B., Gathagan R.J., Yue C., Dengler C., et al. 2018. Amyloid-β plaques enhance Alzheimer’s brain tau-seeded pathologies by facilitating neuritic plaque tau aggregation. Nat. Med. 24:29–38. 10.1038/nm.4443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman, D.M., Morris J.C., and Goate A.M.. 2011. Alzheimer’s disease: the challenge of the second century. Sci. Transl. Med. 3:77sr1. 10.1126/scitranslmed.3002369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado, D.E., Molina-Porcel L., Iba M., Aboagye A.K., Paul S.M., Trojanowski J.Q., and Lee V.M.Y.. 2010. Aβ accelerates the spatiotemporal progression of tau pathology and augments tau amyloidosis in an Alzheimer mouse model. Am. J. Pathol. 177:1977–1988. 10.2353/ajpath.2010.100346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay, T.R., Miller C.M., Cheng P.J., Graham L.C., Bemiller S., Broihier M.L., Xu G., Margevicius D., Karlo J.C., Sousa G.L., et al. 2015. TREM2 deficiency eliminates TREM2+ inflammatory macrophages and ameliorates pathology in Alzheimer’s disease mouse models. J. Exp. Med. 212:287–295. 10.1084/jem.20142322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson, T., Stefansson H., Steinberg S., Jonsdottir I., Jonsson P.V., Snaedal J., Bjornsson S., Huttenlocher J., Levey A.I., Lah J.J., et al. 2013. Variant of TREM2 associated with the risk of Alzheimer’s disease. N. Engl. J. Med. 368:107–116. 10.1056/NEJMoa1211103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren-Shaul, H., Spinrad A., Weiner A., Matcovitch-Natan O., Dvir-Szternfeld R., Ulland T.K., David E., Baruch K., Lara-Astaiso D., Toth B., et al. 2017. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell. 169:1276–1290.e17. 10.1016/j.cell.2017.05.018 [DOI] [PubMed] [Google Scholar]
- Krasemann, S., Madore C., Cialic R., Baufeld C., Calcagno N., El Fatimy R., Beckers L., O’Loughlin E., Xu Y., Fanek Z., et al. 2017. The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases. Immunity. 47:566–581.e9. 10.1016/j.immuni.2017.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S.-H., Meilandt W.J., Xie L., Gandham V.D., Ngu H., Barck K.H., Rezzonico M.G., Imperio J., Lalehzadeh G., Huntley M.A., et al. 2021. Trem2 restrains the enhancement of tau accumulation and neurodegeneration by β-amyloid pathology. Neuron. 109:1283–1301.e6. 10.1016/j.neuron.2021.02.010 [DOI] [PubMed] [Google Scholar]
- Leyns, C.E.G., Gratuze M., Narasimhan S., Jain N., Koscal L.J., Jiang H., Manis M., Colonna M., Lee V.M.Y., Ulrich J.D., and Holtzman D.M.. 2019. TREM2 function impedes tau seeding in neuritic plaques. Nat. Neurosci. 22:1217–1222. 10.1038/s41593-019-0433-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso, R., Fryatt G., Cleal M., Obst J., Pipi E., Monzón-Sandoval J., Ribe E., Winchester L., Webber C., Nevado A., et al. NIMA Consortium . 2019. CSF1R inhibitor JNJ-40346527 attenuates microglial proliferation and neurodegeneration in P301S mice. Brain. 142:3243–3264. 10.1093/brain/awz241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, P.T., Alafuzoff I., Bigio E.H., Bouras C., Braak H., Cairns N.J., Castellani R.J., Crain B.J., Davies P., Del Tredici K., et al. 2012. Correlation of Alzheimer disease neuropathologic changes with cognitive status: a review of the literature. J. Neuropathol. Exp. Neurol. 71:362–381. 10.1097/NEN.0b013e31825018f7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley, H., Cole S.L., Logan S., Maus E., Shao P., Craft J., Guillozet-Bongaarts A., Ohno M., Disterhoft J., Van Eldik L., et al. 2006. Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: potential factors in amyloid plaque formation. J. Neurosci. 26:10129–10140. 10.1523/JNEUROSCI.1202-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parhizkar, S., Arzberger T., Brendel M., Kleinberger G., Deussing M., Focke C., Nuscher B., Xiong M., Ghasemigharagoz A., Katzmarski N., et al. 2019. Loss of TREM2 function increases amyloid seeding but reduces plaque-associated ApoE. Nat. Neurosci. 22:191–204. 10.1038/s41593-018-0296-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokop, S., Miller K.R., Labra S.R., Pitkin R.M., Hoxha K., Narasimhan S., Changolkar L., Rosenbloom A., Lee V.M.Y., and Trojanowski J.Q.. 2019. Impact of TREM2 risk variants on brain region-specific immune activation and plaque microenvironment in Alzheimer’s disease patient brain samples. Acta Neuropathol. 138:613–630. 10.1007/s00401-019-02048-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice, R.A., Pham J., Lee R.J., Najafi A.R., West B.L., and Green K.N.. 2017. Microglial repopulation resolves inflammation and promotes brain recovery after injury. Glia. 65:931–944. 10.1002/glia.23135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadleir, K.R., Kandalepas P.C., Buggia-Prévot V., Nicholson D.A., Thinakaran G., and Vassar R.. 2016. Presynaptic dystrophic neurites surrounding amyloid plaques are sites of microtubule disruption, BACE1 elevation, and increased Aβ generation in Alzheimer’s disease. Acta Neuropathol. 132:235–256. 10.1007/s00401-016-1558-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Pozo, A., Frosch M.P., Masliah E., and Hyman B.T.. 2011. Neuropathological alterations in Alzheimer disease. Cold Spring Harb. Perspect. Med. 1:a006189. 10.1101/cshperspect.a006189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, Y., Manis M., Long J., Wang K., Sullivan P.M., Remolina Serrano J., Hoyle R., and Holtzman D.M.. 2019. Microglia drive APOE-dependent neurodegeneration in a tauopathy mouse model. J. Exp. Med. 216:2546–2561. 10.1084/jem.20190980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangenberg, E.E., Lee R.J., Najafi A.R., Rice R.A., Elmore M.R.P., Blurton-Jones M., West B.L., and Green K.N.. 2016. Eliminating microglia in Alzheimer’s mice prevents neuronal loss without modulating amyloid-β pathology. Brain. 139:1265–1281. 10.1093/brain/aww016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangenberg, E., Severson P.L., Hohsfield L.A., Crapser J., Zhang J., Burton E.A., Zhang Y., Spevak W., Lin J., Phan N.Y., et al. 2019. Sustained microglial depletion with CSF1R inhibitor impairs parenchymal plaque development in an Alzheimer’s disease model. Nat. Commun. 10:3758. 10.1038/s41467-019-11674-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull, I.R., Gilfillan S., Cella M., Aoshi T., Miller M., Piccio L., Hernandez M., and Colonna M.. 2006. Cutting edge: TREM-2 attenuates macrophage activation. J. Immunol. 177:3520–3524. 10.4049/jimmunol.177.6.3520 [DOI] [PubMed] [Google Scholar]
- Ulrich, J.D., Finn M.B., Wang Y., Shen A., Mahan T.E., Jiang H., Stewart F.R., Piccio L., Colonna M., and Holtzman D.M.. 2014. Altered microglial response to Aβ plaques in APPPS1-21 mice heterozygous for TREM2. Mol. Neurodegener. 9:20. 10.1186/1750-1326-9-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Cella M., Mallinson K., Ulrich J.D., Young K.L., Robinette M.L., Gilfillan S., Krishnan G.M., Sudhakar S., Zinselmeyer B.H., et al. 2015. TREM2 lipid sensing sustains the microglial response in an Alzheimer’s disease model. Cell. 160:1061–1071. 10.1016/j.cell.2015.01.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Ulland T.K., Ulrich J.D., Song W., Tzaferis J.A., Hole J.T., Yuan P., Mahan T.E., Shi Y., Gilfillan S., et al. 2016. TREM2-mediated early microglial response limits diffusion and toxicity of amyloid plaques. J. Exp. Med. 213:667–675. 10.1084/jem.20151948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis, E.F., MacDonald K.P.A., Nguyen Q.H., Garrido A.L., Gillespie E.R., Harley S.B.R., Bartlett P.F., Schroder W.A., Yates A.G., Anthony D.C., et al. 2020. Repopulating Microglia Promote Brain Repair in an IL-6-Dependent Manner. Cell. 180:833–846.e16. 10.1016/j.cell.2020.02.013 [DOI] [PubMed] [Google Scholar]
- Yuan, P., Condello C., Keene C.D., Wang Y., Bird T.D., Paul S.M., Luo W., Colonna M., Baddeley D., and Grutzendler J.. 2016. TREM2 Haplodeficiency in Mice and Humans Impairs the Microglia Barrier Function Leading to Decreased Amyloid Compaction and Severe Axonal Dystrophy. Neuron. 90:724–739. 10.1016/j.neuron.2016.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan, L., Sohn P.D., Zhou Y., Li Y., and Gan L.. 2019. A Mac2-positive progenitor-like microglial population survives independent of CSF1R signaling in adult mouse brain. bioRxiv. 10.1101/722090 (Preprint posted August 1, 2019) [DOI]