Summary
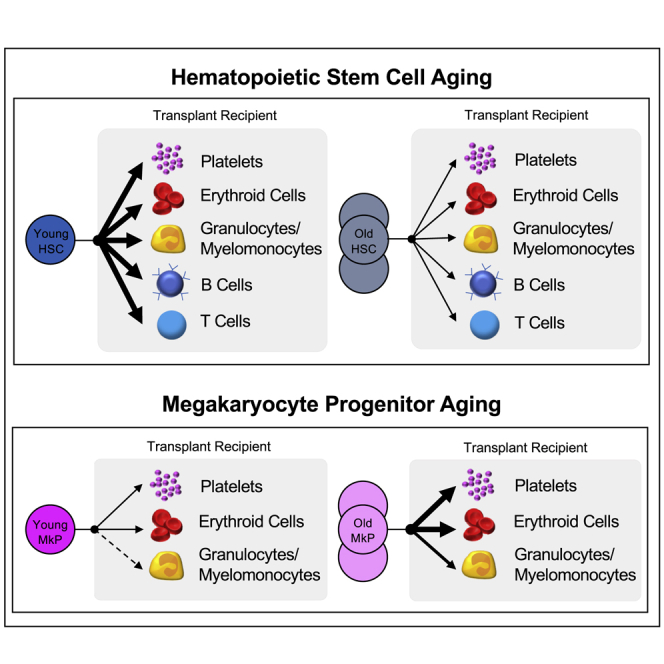
Age-related morbidity is associated with a decline in hematopoietic stem cell (HSC) function, but the mechanisms of HSC aging remain unclear. We performed heterochronic HSC transplants followed by quantitative analysis of cell reconstitution. Although young HSCs outperformed old HSCs in young recipients, young HSCs unexpectedly failed to outcompete the old HSCs of aged recipients. Interestingly, despite substantial enrichment of megakaryocyte progenitors (MkPs) in old mice in situ and reported platelet (Plt) priming with age, transplanted old HSCs were deficient in reconstitution of all lineages, including MkPs and Plts. We therefore performed functional analysis of young and old MkPs. Surprisingly, old MkPs displayed unmistakably greater regenerative capacity compared with young MkPs. Transcriptome analysis revealed putative molecular regulators of old MkP expansion. Collectively, these data demonstrated that aging affects HSCs and megakaryopoiesis in fundamentally different ways: whereas old HSCs functionally decline, MkPs gain expansion capacity upon aging.
Keywords: age-related hematopoiesis, hematopoietic stem cell, cell-intrinsic, cell-extrinsic, heterochronic HSC transplants, quantitative reconstitution, megakaryocyte progenitors, platelets, regenerative capacity, transcriptome
Graphical abstract
Highlights
-
•
Reconstitution deficit by old HSCs was observed by chimerism and absolute cell numbers
-
•
Young HSCs did not outcompete resident HSCs in aged recipient mice
-
•
Old MkPs display remarkable capacity to engraft, expand, and reconstitute platelets
-
•
Aging is associated with changes in MkP genome-wide expression signatures
Poscablo et al. explored age-related changes to hematopoietic stem cell and megakaryocyte progenitor (MkP) function. They found an unexpected gain of in vitro expansion and in vivo reconstitution potential of MkPs upon aging. These functional changes were accompanied by differences in transcriptome profiles between young and old MkPs, suggesting progenitor cell mechanisms contributing to hematopoietic aging.
Introduction
The elderly population is affected by numerous hematological abnormalities, including decreasing capacity to mount an immune response, increasing incidences of thrombotic cardiovascular disorders, and dramatically increased incidence of myelogenous diseases. Dysfunctions during chronological aging is paralleled by the declining functionality of hematopoietic stem cells (HSCs), leading to conclusions that the age-related pathologies of the blood manifests within the HSC compartment (Elias et al., 2017). HSCs self-renew and differentiate, giving rise to progenitor cells that are committed to either the myeloid or lymphoid lineage throughout the lifespan. However, there is convincing evidence that HSCs lose the full extent of these properties during aging. Evaluation of aged HSCs in transplantation studies has demonstrated reduced regenerative potential compared with young HSCs. Aged HSCs seem to be less effective at homing and engrafting in the host, suggesting that alterations to HSCs during aging persist (become intrinsic) on transplantation (Dykstra et al., 2011; Morrison et al., 1996; Rossi et al., 2005; Sudo et al., 2000). In those experiments, HSC multipotency has been traditionally defined by qualitative measurements of GM, B, and T cell donor-to-host chimerism; more quantitative measurements and direct tracking of erythroid cells and platelets (Plt) from hematopoietic stem and progenitor cells (HSPCs) were developed relatively recently to rectify limitations of transplantation experiments that focus on only white blood cell reconstitution (Boyer et al., 2019; Hamanaka et al., 2013; Schaefer et al., 2001; Yamamoto et al., 2013). In addition, analysis of HSCs at the single cell level has identified an increased molecular Plt priming and functional Plt bias in the old HSC compartment (Grover et al., 2016; Sanjuan-Pla et al., 2013; Yamamoto et al., 2018). These alterations raise the need to understand potential alterations in megakaryocyte differentiation in aged mice and humans.
The varying challenges to the blood system during aging may lead to alterations to multiple levels of differentiation, resulting in the dysregulation of homeostatic control maintained by committed progenitors. Previous studies have noted that the bone marrow (BM) frequencies of committed lymphoid and myeloid progenitors are altered with age (De Haan and Van Zant, 1999; Miller and Allman, 2003; Morrison et al., 1996; Rossi et al., 2005), but the functional properties and roles of hematopoietic progenitor cells during aging remains an enigma. Here, we investigated young and old HSCs and the contribution of both intrinsic changes and the environment on their differentiation. In our analysis, we included quantitative measurements of their erythroid and Plt production, as well as assessment of progenitor cells at steady-state and upon heterochronic HSC transplantation. These findings prompted us to further pursue age-associated changes in the Plt lineage, leading to unexpected revelations on the effect of aging on megakaryopoiesis.
Results
Both the frequencies and numbers of hematopoietic stem and progenitor cell populations are altered during aging
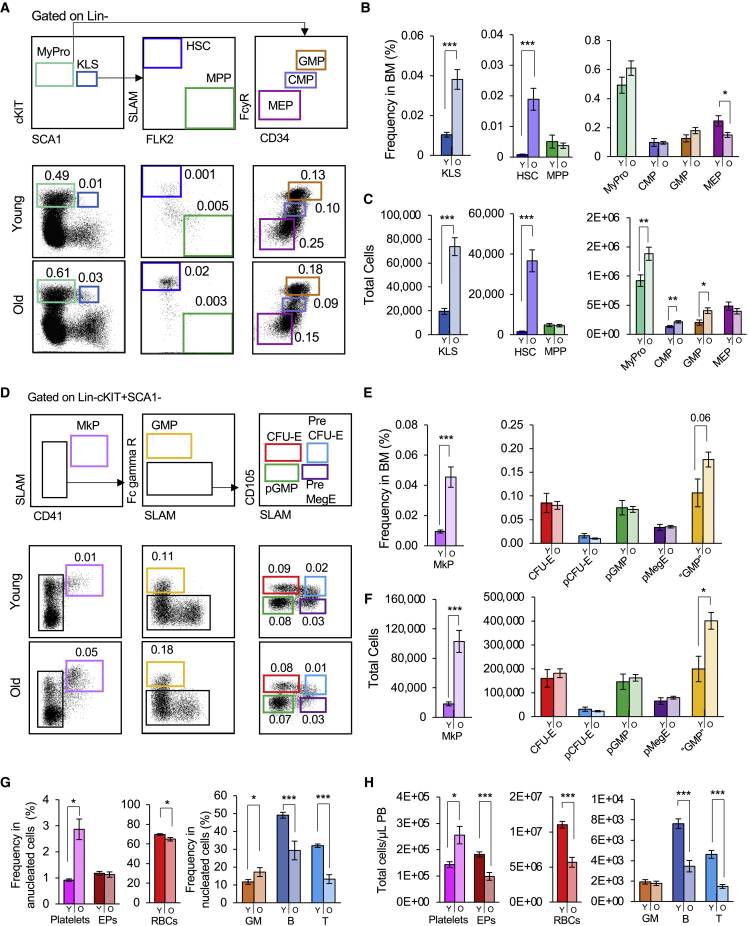
Aging has been shown to alter the frequencies of HSCs in the BM (De Haan and Van Zant, 1999; Morrison et al., 1996; Rossi et al., 2005; Sudo et al., 2000). To evaluate if similar patterns were observed in the total cell numbers and of more refined erythromyeloid populations (Pronk et al., 2007), we compared the absolute cell numbers of HSPC populations between young and old mice. Consistent with previous reports, the frequencies of the cKit + Lineage-Sca1+ (KLS) population increased dramatically with age (Figures 1A and 1B). This increase was also observed when presented as total cell numbers (Figure 1C). When we further fractionated the KLS compartment, we found that this KLS abundance in the old BM was due to a dramatic enrichment of old HSCs. By contrast, the frequency and number of MPPs remained consistent throughout life. The Myeloid Progenitor (MyPro) compartment increased in total cell numbers, but not frequency. Classically defined CMPs and GMPs (Akashi et al., 2000) were found at similar frequencies in young and old BM, while MEP frequencies were reduced with age (Figure 1B). Due to the overall increased cell numbers with age, however, absolute quantification revealed an increase in CMP and GMP numbers, but no change in MEPs, in old BM.
Figure 1.
Hematopoietic stem cells and megakaryocyte progenitor cells expand during aging
(A) Aging alters the composition of HSCs and classical myeloid progenitors. Gating and representative flow cytometry data of phenotypic HSCs, MPPs, and classical myeloid progenitor populations (phenotypic KLS, CMP, GMP, and MEP) in young and old BM.
(B and C) Increased frequencies and total cell numbers of HSCs of old mice compared wiwth young mice. MPP cell percentages and total cell numbers remained the same in young and old mice. Frequencies of CMPs and GMPs remained unchanged while MEPs decreased in frequencies (B), but not numbers (C). The total cell numbers of MyPros, CMPs, and GMPs were significantly increased (C).
(D) Aging alters the composition of erythromyeloid progenitors. Gating and representative flow cytometry data of MkPs and additional erythromyeloid progenitor cells (CFU-E, pCFU-E, pGMP, pMegE, “GMP”) in young and old BM.
(E and F) Frequency (E) and total cell (F) analysis revealed significantly higher levels of MkPs in the BM of old mice compared with young mice. There was also a slight increase in the frequency and total cell numbers of “GMPs”.
(G and H) Alterations in the frequencies (G) and total numbers (H) of circulating platelets, erythroid, and nucleated cells in old mice. Both the frequency and number of PB Plts platelets increased, whereas erythroid cells decreased, upon aging. The frequencies and total numbers of B and T cells decreased in old mice, leading to an increase in the frequency of myeloid cells.
Y, Young; O, Old; HSC, hematopoietic stem cells; MPP, multipotent progenitors; KLS, cKit + Lineage-Sca1+ cells; MyPro, myeloid progenitors; CMP, common myeloid progenitors; GMP, granulocyte/macrophage progenitor; MEP, megakaryocyte-erythroid progenitors; MkP, megakaryocyte progenitors; EPs, erythroid progenitor cells; RBCs, red blood cells, GM, granulocytes/myelomonocytes. BM analysis: Young n = 7, Old = 27 in 3 independent experiments. Blood analysis: Young n = 23, Old n = 17 in three independent experiments. Data are shown as mean ± SEM. p values were determined using unpaired two-tailed t test. ∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.0001.
We also investigated erythromyeloid populations more recently defined by alternative markers within the MyPro compartment (Figure 1D) (Pronk et al., 2007). Few age-related alterations in frequencies and numbers were observed: CFU-E, pCFU-E, pGMP, and pMegE frequencies and numbers were not significantly altered (Figures 1E and 1F). Alternatively defined “GMPs” trended toward an increase in frequency and were significantly increased in absolute numbers (Figures 1E and 1F), consistent with the highly overlapping classical GMPs (Figures 1B and 1C). Intriguingly, we observed a dramatic increase in both the frequency and total cell numbers of phenotypically defined MkPs in old mice (Figures 1E and 1F). The aging hematopoietic system has been reported to include alterations to circulating mature cells (Valletta et al., 2020; Young et al., 2020). Consistent with previous reports, we also observed age-related increase in Plt frequencies and numbers, whereas erythroid cells decreased (Figures 1G and 1H). Our data also recapitulated reported myeloid bias among nucleated cells, primarily driven by a decrease in B and T cells (Figures 1G and 1H). These data revealed that the composition of HSPC populations selectively changes during aging. Of note was the dramatic expansion of old HSCs and old MkPs, accompanied by a significant increase in circulating Plts, raising the need to explore megakaryopoiesis during the aging process.
Old HSCs exhibit impaired reconstitution potential compared with young HSCs
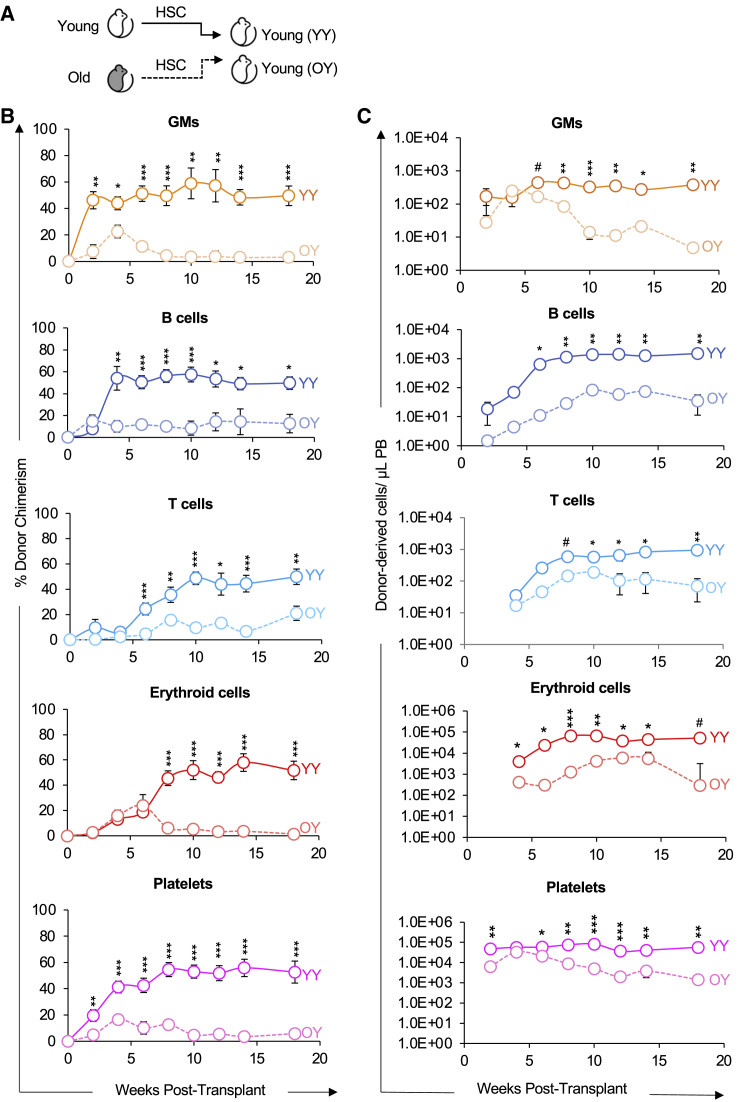
To directly interrogate the differences in reconstitution potential between young and old HSCs, we purified HSCs from fluorescent young or old mice and transplanted them into young recipients (Figure 2A). These experiments are analogous to those performed previously by others (Ergen et al., 2012; Rossi et al., 2005), except we expanded aforementioned investigations by also including direct detection of donor-derived Plts and circulating erythroid cells, in addition to the more commonly assayed granulocytes/myelomonocytes (GMs), and B and T cells.
Figure 2.
Old HSCs exhibited impaired reconstitution potential in young recipients
(A) Schematic of transplantation of HSCs from young or old mice into young recipient mice.
(B) Old HSCs contributed to donor-host-chimerism with lower efficiency compared with young HSCs. Analysis of donor-derived mature cells in PB of recipients presented as percent donor chimerism.
(C) Old HSCs produced fewer mature cells compared with young HSCs. Reconstitution data from (B) replotted as absolute numbers of donor-derived GMs, B cells, T cells, erythroid cells, and platelets per microliter of PB.
GM, granulocytes/myelomonocytes. YY, Young HSCs transplanted into young recipients; OY, Old HSCs transplanted into young recipients. Data are representative means ± SEM from nine recipient mice of young HSCs (four independent experiments) and 22 recipients of old HSCs (five independent experiments). p values were determined using unpaired two-tailed t test. #p < 0.06, ∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.0005.
First, also as commonly done, we determined the reconstitution kinetics of HSCs by measuring donor chimerism in the peripheral blood (PB) for the duration of 18 weeks posttransplantation (Figure 2B). Consistent with previous reports, analysis of lineage potential between young and old HSCs revealed that old HSCs have a diminished capacity to reconstitute GM, B, and T cells. Similarly, erythroid and Plt chimerism were also lower from old versus young HSCs (Figure 2B).
Second, we removed the host cell variable by quantifying the absolute numbers of donor-derived mature cells in the PB of recipient mice (Boyer et al., 2019; Cool et al., 2020; Rajendiran et al., 2020a, 2020b) to more quantitatively assess the differences in regenerative activity between young and old HSCs. Old HSCs generated fewer erythroid, GM, B, T, and Plt cells compared with young HSCs (Figure 2C); this observation is consistent with host cell production presented as donor-derived chimerism (Figure 2B). Our data demonstrated a pan-hematopoietic reconstitution deficit of HSCs during aging, which is consistent with previous reports in white blood cell reconstitution. Taken together, these results also support the notion that key changes intrinsically maintained by HSCs drive their feeble performance upon aging.
The old bone marrow environment impairs the reconstitution potential of HSCs
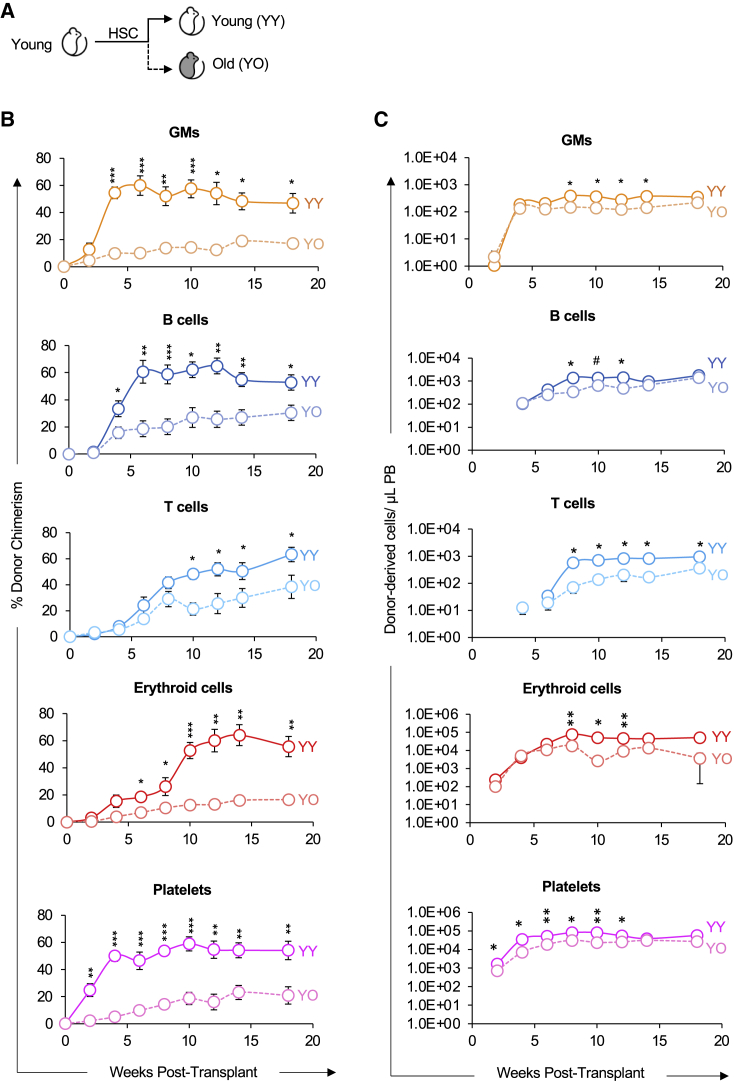
We next sought to determine how cues in the aging environment influence HSC reconstitution capacity. Because young HSCs outperformed old HSCs in young hosts (Figure 2), we reasoned that young HSCs would engraft with higher efficiency in old compared with young recipients. To test this directly, we purified HSCs from young mice and transplanted them into young or old recipient mice (Figure 3A). We then analyzed mature cell production of transplanted HSCs to quantitatively assess the role of host age on expansion capacity and lineage potential. Young HSCs were capable of long-term, multilineage reconstitution (LTMR) in both young and old hosts. However, in contrast to the young hosts, the old hosts allowed for significantly lower reconstitution chimerism of HSC-derived GM, B, T, erythroid, and Plt cells (Figure 3B). Overall, the differences were more significant at earlier timepoints (~4–14 weeks) than at later timepoints.
Figure 3.
Young HSCs exhibited lower reconstitution potential in old compared with young recipients
(A) Schematic of transplantation of HSCs from young mice into separate cohorts of young or old mice.
(B) Old recipients displayed lower HSCs donor-host-chimerism compared with young recipients. Analysis of donor-derived mature cells (GMs, B cells, T cells, erythroid cells, and platelets) in PB of recipients presented as percent donor chimerism.
(C) Donor HSCs produced fewer total mature cells in old recipients compared to in young recipients. Reconstitution data from (B) replotted as absolute number of donor-derived GMs, B cells, T cells, erythroid cells, and platelets per microliter of PB.
YY, Young HSCs transplanted into young recipients; YO, Young HSCs transplanted into old recipients. Data are representative means ± SEM from nine young recipient mice (four independent experiments) and 10 old recipient mice (three independent experiments). p values were determined using unpaired two-tailed t test. ∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.0005.
In the same cohorts, we also determined the absolute number of donor-derived mature cells in the PB after transplantation. The donor HSCs in the young host produced greater numbers of donor-derived cells per microliter of PB compared with the old host (Figure 3C). Although this pattern was overall consistent with the patterns observed by donor chimerism, the differences in total cell production was attenuated and, with the exception of T cells, was not sustained over time. This analysis revealed that the old host reduced donor chimerism by young HSCs. However, when we removed the host variable by measuring absolute mature cell production, our analysis revealed a less different reconstitution capacity between young HSCs placed in the young environment and the young HSCs in the old environment.
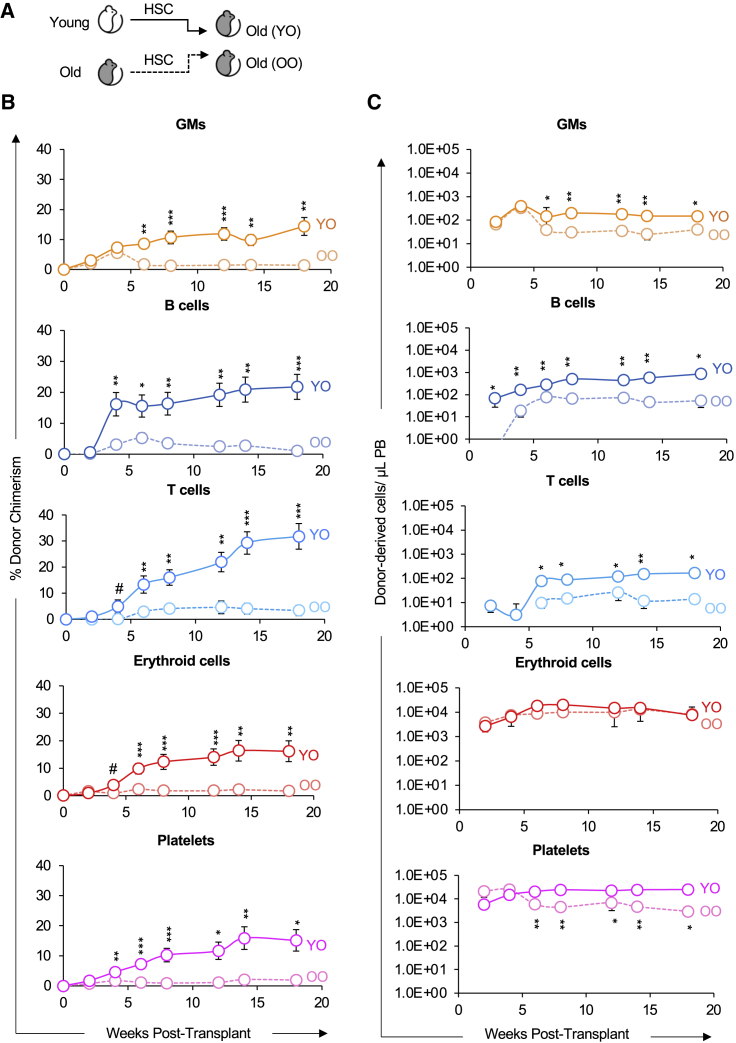
Because the isochronic Y-into-Y transplants led to higher chimerism than the two heterochronic (O-into-Y or Y-into-O) transplants (Figures 2 and 3), we hypothesized that an old environment would provide a more supportive environment for old HSCs. We therefore compared the reconstitution potential of young and old HSCs in an old host (Figure 4A) and assayed for donor chimerism (Figure 4B) and absolute number of donor-derived cells (Figure 4C). Surprisingly, transplantation of young HSCs led to higher reconstitution of old recipients compared with transplantation of old HSCs. We also examined the impact of reconstitution by old HSCs in a young or old environment; our analysis revealed that old HSCs exhibited poor reconstitution capacity in the old host compared with the young host (Figure S1). Therefore, while the aged BM environment impaired the reconstitution potential of transplanted young HSCs (Figure 3), HSCs from old mice performed even worse than young HSCs in an aged environment (Figure 4 and S1). In combination with the diminished reconstitution potential of old HSCs (Figure 2), these results indicate that aging also leads to reduction in HSC-intrinsic performance and in the ability of the BM environment to support transplanted HSCs (Figures 3, 4, and S1).
Figure 4.
Old HSCs exhibited impaired reconstitution potential compared to young HSCs in old recipients
(A) Schematic of transplantation of HSCs from young or old mice into separate cohorts of old mice.
(B) Old HSCs displayed lower donor-host-chimerism compared with young HSCs. Analysis of donor-derived mature cells (GMs, B cells, T cells, erythroid cells, and platelets) in PB of recipients presented as percent donor chimerism.
(C) Old HSCs produced fewer total mature cells in old recipients compared with in young HSCs. Reconstitution data from (B) replotted as absolute number of donor-derived GMs, B cells, T cells, erythroid cells, and platelets per microliter of PB.
YO, Young HSCs transplanted into old recipients; OO, Old HSCs transplanted into old recipients. Data are representative means ± SEM from 11 recipient mice of young HSCs (four independent experiments) and eight recipients of old HSCs (three independent experiments). p values were determined using unpaired two-tailed t test. ∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.0005.
Both old HSCs and the old niche impairs reconstitution of the stem and progenitor cell compartment
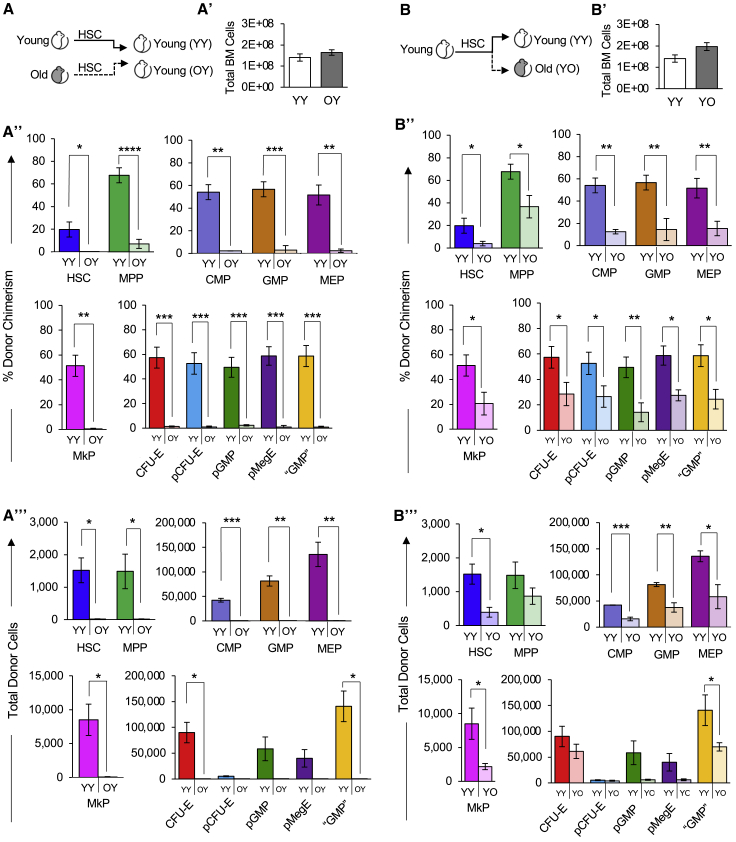
The reported Plt bias of old HSCs (Grover et al., 2016; Haas et al., 2015; Sanjuan-Pla et al., 2013; Yamamoto et al., 2013, 2018) had the potential to translate into maintained, or even increased, Plt reconstitution by old HSCs, possibly at the expense of the other lineages. However, Plt reconstitution by old HSCs in either young (Figure 2) or old (Figure 4) hosts, and by young HSCs in old hosts (Figure 3) was as impaired as reconstitution of erythroid cells and WBCs. To determine whether the observed reconstitution of the PB is reflected in the BM of the recipients, we also determined the capacity of young and old HSCs to regenerate stem and progenitor cells. In the cohorts analyzed for PB reconstitution in Figures 2 and 3, we determined donor chimerism in the BM stem and progenitor cell compartment >18 weeks posttransplantation (Figure 5A). All expected populations were detected, supporting the LTMR capability of HSCs. Quantitatively, young recipients contained similar total BM cell numbers (Figure 5A′) and, in comparison with young HSCs, old HSCs exhibited a much lower capacity to reconstitute classical HSPCs and erythromyeloid progenitor populations, with notably lower contribution to MkP reconstitution (Figure 5A″). Likewise, quantifying the total cell number of donor-derived HSPCs revealed a similar pattern (Figure 5A‴). When determining the role of the aging recipient environment on HSC reconstitution of BM populations (Figures 5B, S2B, and S2B‴), we also found that despite having similar total BM cell numbers (Figures 5B′ and S2B′), there were lower frequencies and total numbers of donor-derived classical myeloid progenitor cells and erythromyeloid progenitor populations in the old hosts compared with the young hosts (Figures 5B′ and B″). Similar observations were made for the old HSCs in old recipients (Figures 6A and 6A‴). Taken together, our data suggest that aging alters both HSC-intrinsic and recipient properties, and that these age-related changes affect the entire hematopoietic hierarchy and all lineages, including erythro- and megakaryopoiesis.
Figure 5.
Both old HSCs and the old niche impaired reconstitution of the stem and progenitor cell compartment
(A) Schematic of HSC transplantation from young or old mice into young mice.
(A′) The total number of BM cells was similar in young recipient mice >16 weeks after transplantation of either young or old HSCs.
(A″) Old HSCs contributed to significantly lower donor-to-host chimerism of BM stem and progenitor cells in young recipient mice compared with young HSCs. Percent donor chimerism of HSPCs and erythromyeloid progenitors in the recipient BM of young or old HSCs >16 weeks posttransplant.
(A‴) Old HSCs generated fewer total stem and progenitor cells than young HSCs in young recipient mice. Quantification of total donor-derived classical HSPCs and erythromyeloid progenitors in the BM of recipients >16 weeks posttransplant.
(B) Schematic of HSC transplantation from young mice into separate cohorts of young or old mice.
(B′) The total number of BM cells was similar in young and old recipient mice >18 weeks after transplantation of young HSCs.
(B″) Old recipients displayed lower donor-to-host HSPC chimerism than young recipient mice transplanted with young HSCs. BM analysis of young or old mice transplanted with young HSCs. HSPCs were analyzed >18 weeks posttransplant and presented as donor chimerism.
(B‴) Transplanted young HSCs produced fewer total HSPCs in the BM of old compared with young recipient mice. Quantification of total donor-derived HSPC numbers in the BM of recipients >18 weeks posttransplant.
YY, Young HSCs transplanted into young recipients; OY, Old HSCs transplanted into young recipients; YO, Young HSCs transplanted into old recipients. Data are representative means ± SEM. PB analysis of the same recipient mice are presented in Figures 2A′ and 2A‴ and Figure 3B′ and 3B‴. p values were determined using unpaired two-tailed t test. ∗p ≤ 0.05, ∗∗p < 0.005, ∗∗∗p < 0.0005, ∗∗∗∗p < 0.0001.
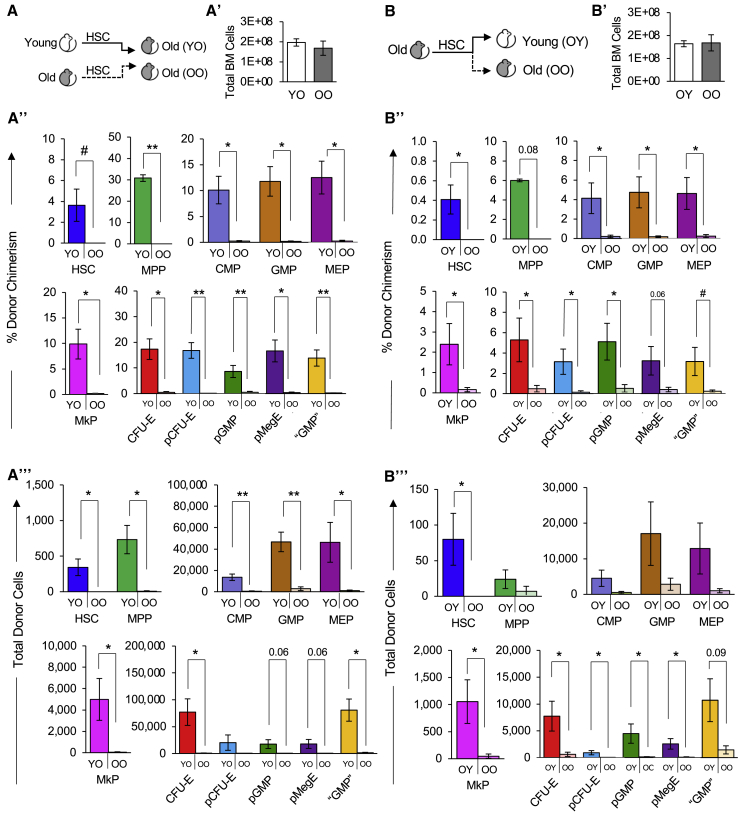
Figure 6.
Stem and progenitor cell reconstitution by old HSCs was impaired and exacerbated by the old niche
(A) Schematic of transplantation of HSCs from young or old mice into old mice.
(A′) Quantification of total BM cells in recipient mice in (A) >16 weeks posttransplant.
(A″) Old HSCs contributed to significantly lower donor-to-host chimerism of BM stem and progenitor cells compared with young HSCs in an old host. Percent donor chimerism of HSPCs and erythromyeloid progenitors in the old recipient BM of transplanted young or old HSCs >16 weeks posttransplant.
(A‴) Old HSCs generated fewer total stem and progenitor cells than young HSCs in old recipient mice. Quantification of total donor-derived classical HSPCs and erythromyeloid progenitors in the BM of recipients >16 weeks posttransplant.
(B) Schematic of transplantation of HSCs from old mice into separate cohorts of young or old mice.
(B′) Quantification of total BM cells in recipient mice in (B) >16 weeks posttransplant.
(B″) Old recipients displayed lower donor-to-host HSPC chimerism than young recipient mice transplanted with old HSCs. BM analysis of young or old mice transplanted with old HSCs. HSPCs were analyzed >18 weeks posttransplant and presented as donor chimerism.
(B‴) Transplanted old HSCs produced fewer total HSPCs in the BM of old compared with young recipient mice. Quantification of total donor-derived HSPC numbers in the BM of recipients >18 weeks posttransplant.
YO, Young HSCs transplanted into old recipients; OO, Old HSCs transplanted into old recipients; OY, Old HSCs transplanted into young recipients. Data are representative means ± SEM. PB analysis of the same recipient mice are presented in Figures 4A′ and A‴ and S1B′ and S1B‴. p values were determined using unpaired two-tailed t test. ∗p ≤ 0.05, ∗∗p < 0.005, ∗∗∗p < 0.0005, ∗∗∗∗p < 0.0001.
Old MkPs were functionally enhanced compared with young MkPs
The most profound and consistent change in old mice was alterations in megakaryopoiesis, denoted by a dramatic increase in MkPs and Plts (Figure 1). However, this age-dependent increase in MkPs was not accompanied by an age-dependent functional bias of HSCs toward the megakaryocyte lineage: transplanted old HSCs were as deficient in MkP (Figures 5A″, 5B″, 5A‴, 5B‴, 6A″, 6B″, 6A‴, and 6B‴) and Plt (Figures 2B, 2C, 4B, 4C, S1B, and S1C) reconstitution as they were in generating other lineages. Thus, we hypothesized that the dramatic increase in the frequency and total numbers of old MkPs and Plts (Figures 1D–1H) may be manifested by MkP-intrinsic changes, possibly in addition to increased contribution from the HSC compartment.
To begin to test this hypothesis, we determined functional differences between young and old MkPs in vitro. Interestingly, evaluation of MkP proliferative capacity by short-term culture assays revealed a significant, 2.5-fold increase in expansion of old MkPs compared with young MkPs (Figures 7A and 7B). These experiments showed that old MkPs displayed a surprising increased functional capacity compared with young MkPs in vitro.
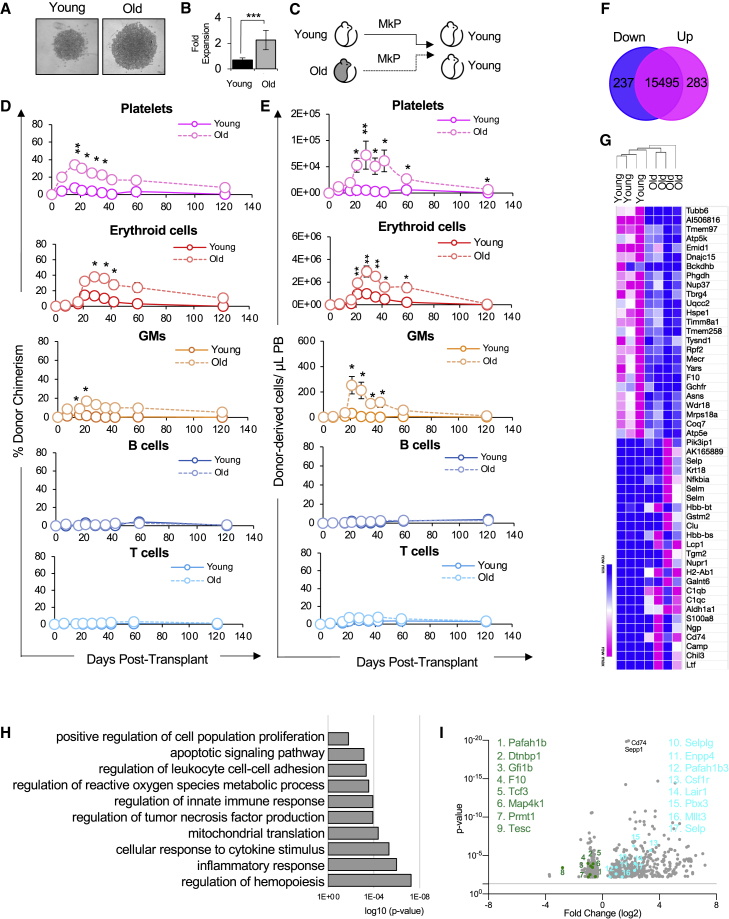
Figure 7.
Old MkPs had enhanced reconstitution potential and altered gene expression signatures compared with young MkPs
(A and B) Old MkPs have enhanced proliferative capacity compared to young MkPs in vitro. (A) Representative images of Y and O MkPs grown in vitro. To evaluate proliferative capacity, 2,000 MkPs were FACS sorted from young and old mice and grown in liquid culture. Three days after seeding, MkPs were microscopically evaluated for expansion efficiency. (B) MkP expansion in vitro. Flow cytometry was used to quantify the total fold expansion of cultured young and old MkPs from (A). n = 3 from three independent experiments.
(C) Schematic of transplantation of young or old MkP into young mice.
(D) Old MkPs demonstrated greater short-term contribution to platelet donor-to-host chimerism in the recipient mice compared to young MkPs. Old MkPs also contributed to short-term erythroid cells and GM donor-to-host chimerism, but not to B or T cells. Analysis of donor-derived mature cells (platelets, erythroid cells, GMs, B cells, T cells) in PB of recipients presented as percent donor chimerism.
(E) Old MkPs generated higher numbers of platelets compared with young MkPs in recipient mice. Old MkPs also produced more erythroid cells and GMs compared with young MkPs. Reconstitution data replotted as absolute numbers of donor-derived platelets, erythroid cells, GMs, B cells, and T cells per microliter of PB.
(F) RNA-seq analysis revealed transcriptional changes accompanying aging of the MkP compartment. A total of 237 genes were downregulated and 283 genes were upregulated in old MkPs. A Venn diagram showing the numbers of downregulated (blue) and upregulated (pink) differentially expressed genes (DEGs) in old versus young MkPs.
(G) Hierarchical clustering analysis confirmed the distinct gene expression characteristics of the young and old MkPs. Heatmap showing the scaled expression of the top 50 DEGs in young and old MkP clusters. Pink and blue indicate comparatively high and low expression, respectively.
(H) The expression patterns associated with aging MkPs revealed changes to important biological functions. Major GO biological process terms in which DEGs enriched for.
(I) Identifying specific DEGs highlights known and novel regulators of aging megakaryopoiesis. A volcano plot of DEGs between young and old MkPs. Genes described in text are highlighted.
Data in (A–E) are representative means ± SEM. Data from transplantation experiments are from five recipient mice of young MkPs and 11 recipient mice of old MkPs from at least three independent experiments. p values were determined by unpaired two-tailed Student's t test ∗p < 0.05, ∗∗p < 0.005, ∗∗∗p ≤ 0.001.
Old MkPs possessed greater platelet reconstitution potential compared with young MkPs
The notable increase in expansion capacity of old MkPs in vitro lead us to predict potential functional differences in vivo. To determine the reconstitution potential of old MkPs in vivo, we transplanted young or old MkPs into young recipient mice (Figure 7C). We evaluated MkP multilineage reconstitution in the host by measuring their contribution to mature cells (Plt, erythroid cells, GMs, B, T) in the PB for 16 weeks. Given that young MkPs have been shown to be strongly committed to the Plt lineage in in vitro assays (Nakorn et al., 2003; Pronk et al., 2007), we predicted that MkPs would primarily produce Plts. Indeed, young MkPs contributed to nominal donor-chimerism levels of GM, B, and T cells, with a more robust contribution to Plts, as well as, unexpectedly, a short burst of erythroid cells (Figure 7D). Surprisingly, relative to young MkPs, old MkPs exhibited a remarkable capacity to engraft and contribute to Plts in the recipient mice (Figure 7D). Although young MkPs repopulated a more modest 7.4% of Plts, the same number of old MkPs repopulated on average 34% of Plts in the recipient mice. Interestingly, old MkPs also contributed to significantly higher erythroid cell and GM chimerism in the recipient mice relative to young MkPs. It is also notable that unlike the long-term reconstitution capacity of HSCs (Figures 2, 3, 4, 5, and 6), the Plt, erythroid cell, and GM production from old MkPs was robust only in the first few weeks posttransplant and then declined over time.
These differences between young and old MkPs were reinforced by quantification of the absolute cell numbers from the same experiments (Figure 7E). The absolute quantification revealed that old MkPs engrafted and produced Plts, as well as erythroid cells and GMs, at a remarkably high efficiency compared with young MkPs. Taken together, our in vitro and in vivo results indicated that MkPs gain both expansion and reconstitution capacity during aging.
RNA-sequencing revealed different molecular patterns between young and old MkPs
We were surprised by the remarkable difference in expansion and reconstitution capacity between young and old MkPs. To test how aging affects the molecular profile of MkPs, we evaluated the molecular regulation of old MkPs by performing RNA-sequencing (RNA-seq) analysis of young and old MkPs. We found that young and old MkPs had very similar gene expression profiles as expected of two MkP populations, sharing over 15,000 transcripts. Importantly, we also identified 520 differentially expressed genes between young and old MkPs, 237 of which were downregulated and 283 were upregulated in old MkPs (Figure 7F). Hierarchical clustering of seven independent samples showed that independent biological replicates associated based on age, as expected (Figure 7G), with the top 50 differentially expressed genes shown as a heatmap (Figure 7G and Table S1).
To further evaluate the expression signatures of young and old MkPs, we performed Gene Ontology (GO) analysis. This analysis indicated that differentially expressed genes between young and old MkPs were highly enriched in categories related to proliferation (Figure 7H), inferring intrinsic regulation of the observed enhanced expansion capacity of old MkPs (Figures 7A–7E). Other GO terms associated with the differentially expressed genes included cell adhesion, inflammation, and mitochondrial regulation (Figure 7H). The latter is consistent with a recent publication of mitochondrial activity playing a role in age-related Plt function (Davizon-Castillo et al., 2019). Genes under each GO term are presented in Table S2. At the gene level, we observed both novel and well-known regulators of hematopoiesis and hematological processes (Figures 7G and 7I). For example, we identified regulators of lineage specification, including Csf1 and Tcf3 (also known as E2A) (Semerad et al., 2009; Stanley and Chitu, 2014). Also, previously described regulators of megakaryocyte differentiation were differentially expressed. These genes include Prmt1, Selp, and Tesc, a regulator of E26 transformation-specific (ETS) transcription factors (Banu et al., 2020; Jin et al., 2018; Levay and Slepak, 2007). We also observed differential expression of multiple genes associated with Plt regulation, production, and function: Coagulation Factor X, Dtnbp1, Gfi1b, Enpp4, Pafah1b3, Pafah1b1, Mmrn1, and Selplg. Mutations in Gfi1b and Dtnbp1 are associated with various Plt disorders and bleeding disorders (Ghiani and Dell’Angelica, 2011; Songdej and Rao, 2017). Interestingly, our analysis also revealed a small subset of upregulated genes previously implicated in acute myeloid leukemia (AML), including Pbx3, Lair1, and Mllt3 (Kang et al., 2015; Li et al., 2013; Marschalek, 2010), and is possibly linked to increased risk of AML in the elderly (Almeida and Ramos, 2016).
The results of the RNA-seq analysis corroborates the behaviors observed in our functional analyses of MkPs. Together, these data illuminate the fundamental difference between youthful and aging megakaryopoiesis. Importantly, our RNA-seq data revealed well-known genes implicated in the molecular mechanisms of megakaryopoiesis during aging, as well as a number of novel genes that can be pursued to understand, and possibly mitigate, the age-related changes that occur in megakaryopoiesis.
Discussion
Aging alters the reconstitution potential of HSCs
As previously reported by others, our characterization of the old BM populations revealed an expansion of HSCs, accompanied by alterations to descendant mature cells in the PB (Figure 1). These observations raised the need understand how HSC differentiation is modulated by the balance of both intrinsic changes to HSCs and extrinsic influence on HSCs during aging. Young HSCs have been suggested to have superior engraftment and reconstitution capacity compared with old HSCs (Dykstra et al., 2011; Morrison et al., 1996; Rossi et al., 2005; Sudo et al., 2000). Indeed, our experiments transplanting young or old HSCs indicated that old HSCs exhibited a diminished reconstitution capacity (Figures 2 and 5). To address whether the reduced reconstitution activity of HSCs during aging is influenced by the aged environment, we transplanted young HSCs into young or old hosts (Figures 3 and 5). Our data revealed that the old environment was less supportive of reconstitution by transplanted HSCs compared with the young environment. Therefore, transplanted young HSCs did not have an obvious competitive advantage over the resident old HSCs in the aged host. Notably, transplanted old HSCs had even lower capacity to reconstitute the old host compared with young HSCs (Figures 4 and 6), demonstrating that the poor performance of old HSCs in young recipients (Figures 2 and 5) was not due to the age-mismatch (heterochronic) between donor and host.
Because both young and old HSCs perform worse in old compared with young recipients, it is clear that age-related changes in the host impact donor HSC performance. This could be related to the reported increased proliferation of old HSCs (Kirschner et al., 2017), age-related changes to the distal and local microenvironment, or both. Our collective iso- and heterochronic transplantation data show that both HSC-intrinsic and extrinsic mechanisms during aging affect reconstitution efficiency (SanMiguel et al., 2020). Interestingly, we did not detect a selective retention of MkP or Plt reconstitution capacity of transplanted old HSCs, despite the increased frequencies and numbers of HSCs, MkPs, and Plts in unmanipulated aged mice (Figure 1). It remains possible that HSCs contribute to the greater production of MkPs in situ (Carrelha et al., 2018), with participation of the aging BM microenvironment (Ho et al., 2019). Together with the observed expansion of HSCs in situ (Figure 1), our transplantation results suggest that the impaired reconstitution performance of aged HSCs reflects reduced engraftment capacity rather than functional decline in situ. A better understanding of how young and old BM cells respond to niche-clearing methods could help to develop optimal systems for investigating reconstitution differences between young and old HSCs, and between young and old recipients.
Surprising reconstitution and differentiation potential of old MkPs
Our data provided evidence about the changes to the homeostatic control of blood cell production at the MkP level of hematopoietic differentiation during aging. Given the reconstitution deficit displayed by old HSCs (Figures 2, 4, 5A, and 5A‴), one might have expected that old MkPs would also display functional deficiencies. We were surprised to find from our in vitro experiments that old MkPs displayed greater proliferative potential (Figures 7A and 7B). Strikingly, we also observed from transplanting young and old MkPs that old MkPs harbored a remarkable capacity to engraft, expand, and reconstitute Plts (Figures 7C–7E). This age-associated elevation of MkP potential is reminiscent of the higher prevalence of thrombosis and other cardiovascular problems in the elderly. We hypothesize that age-related alterations made to MkP populations contribute to dysregulation and increased risk for age-related morbidities (Davizon-Castillo et al., 2019).
Our transplantation studies of old and young MkPs also revealed somewhat surprising evidence about the differentiation programs of immature progenitors during aging (Figures 7C–7E). Previous reports suggested that young MkPs are exclusively associated with megakaryocyte and Plt generation (Nakorn et al., 2003; Pronk et al., 2007). However, those conclusions were primarily based on transcription profiles and in vitro differentiation capacity, with no direct assessment of differentiation potential in vivo. We recently reported that classically (Akashi et al., 2000) and alternatively (Pronk et al., 2007) defined GMPs produce erythroid cells in vivo. This fits a model where erythropoiesis is a default HSC fate (Boyer et al., 2019). Under that model, it is not surprising that our experiments revealed that both young and old MkPs harbor in vivo erythroid capacity. Surprisingly, old MkPs also demonstrated greater capacity for the GM lineage compared with young MkPs. It is possible that our observations may be a result of poorly defined cellular phenotypes in old mice, raising the question of how aging might affect the immunophenotype of MkPs. Given that several markers are shared between MkPs and HSCs (Zhu et al., 2019), with a gain of CD41 expression as HSCs age (Gekas and Graf, 2013), the possibility that the MkP compartment in old BM contains significant numbers of HSCs was considered. However, old MkPs lacked B and T cell generation and the enhanced production of Plts, erythroid cells, and GMs did not persist long-term, unlike that of multipotent and self-renewing HSCs. Additionally, we did not observe any age-related differences in expression of megakaryopoiesis master regulators such as Fli1, Gabpa, Mpl, Pf4, and Runx1 (Bruns et al., 2014; De Sauvage et al., 1994; Doré and Crispino, 2011; Elagib et al., 2003; Kaushansky et al., 1994; Klimchenko et al., 2009; Zhu et al., 2018) (Figure 7). A high similarity between these genes suggest high purity of the MkP populations. Together, these findings indicate that the high expansion and oligopotency of old MkPs by transient production of Plts, erythroid cells, and GMs cannot be explained by contaminating HSCs, but rather due to enhanced potential in the aged MkP compartment.
Aging is associated with changes in MkP genome-wide expression signatures
Our transcriptome data provide important clues about the intrinsic molecular regulation of megakaryopoiesis during aging (Figure 7). RNA-seq analysis of young and old HSCs has provided a paradigm of molecular switches during ontogeny (Cabezas-Wallscheid et al., 2014; Grover et al., 2016; Kowalczyk et al., 2015; Sanjuan-Pla et al., 2013; Sun et al., 2014). Similarly, our data showed that young and old MkPs have different gene expression programs, reflecting divergence in the molecular control of Plt differentiation during aging. For example, differential expression of genes involved in proliferation reinforced the differences in proliferative capacity between young and old MkPs. The expansion of MkPs with age also suggest that MkPs themselves may serve as a reservoir for mutations promoting clonal progression to cardiovascular diseases and cancer (Pardali et al., 2020; Steensma and Ebert, 2020). In line with this model, we observed age-related increase in expression of genes involved in AML. Moreover, different transcriptional networks involving Plt function were observed between young and old MkPs, supporting a model in which MkPs propagate events toward age-related dysregulation of Plt biology. An important next step will be to identify key molecular regulators that cause or are a consequence of age-related changes to megakaryopoiesis.
In conclusion, our data revealed that profound perturbations in homeostatic control underlie megakaryopoiesis during the aging process. Given that HSCs gradually lose their regenerative potential during aging, it is reasonable to focus research on the HSC contribution to aging. There remains, however, a conceptual gap in the role of altered progenitor cells as potential perpetuators of aging. Our findings shed light on the mechanisms of aging within the Plt lineage that may dictate the etiology of Plt-related disorders, and other myeloid diseases that may be operating through committed progenitors rather than HSCs. Our study sets the stage for further exploration of mechanisms governing youthful and aging megakaryopoiesis and suggests that progenitor cell involvement should be considered when investigating normal and pathophysiological changes in the hematopoietic system during aging.
Experimental procedures
Mouse lines
All animals were housed and bred in the Association for Assessment and Accreditation of Laboratory Animal Care–accredited vivarium at UC Santa Cruz and maintained under approved Institutional Animal Care and Use Committee guidelines. Young adult mice were used between 8 and 16 weeks of age and old adult mice were 20+ months of age. Old recipient mice in transplantation experiments were 18+ months of age. All mice were randomized based on sex.
HSC and MkP isolation and analysis by flow cytometry
BM stem and progenitor cell populations and mature cell subsets were prepared and stained as previously described (Beaudin et al, 2014, 2016; Leung et al., 2019; Martin et al., 2021; Smith-Berdan et al, 2015, 2019; Ugarte et al., 2015). HSCs or MkPs from young or old mice were prospectively isolated using a fluorescence-activated cell sorting (FACS) Aria III (Becton Dickinson, San Jose, CA) as previously described (Boyer et al., 2019; Leung et al., 2019; Martin et al., 2021).
Transplantation reconstitution assays
Reconstitution assays were performed by transplanting double-sorted HSCs (200 per recipient) from young or old Ubc-GFP+ whole BM and transplanting into congenic C57BL/6 mice via retro-orbital intravenous transplant. We also transplanted double-sorted MkPs (22,000 per recipient) from C57Bl6 into Ubc-GFP+ hosts. Hosts were preconditioned with sub-lethal radiation (~750 rads) using a Faxitron CP160 X-ray instrument (Precision Instruments).
RNA-seq
The RNA-seq libraries were generated by sorting MkPs from young or old C57BL/6 mice using FACS Aria III (Becton Dickinson). Cells were stored in Trizol (Invitrogen) at −80°C until RNA isolation. RNA-seq libraries were generated using Nextera Library Prep, as we have previously done (Beaudin et al., 2016; Byrne et al., 2017). Libraries were validated using the Bioanalyzer (Agilent 2100), validated RNA-seq libraries were sequenced using Illumina HiSeq 4000 as Paired-end run reads at the QB3-Berkeley Genomics at University of California Berkeley, and DESeq analysis was done with the help of Dr. Sol Katzman at the UCSC Bioinformatics Core. Heatmap was generated using Morpheus, https://software.broadinstitute.org/morpheus. Go Term analysis was run using Panther.
Quantification and statistical analysis
Number of experiments, n, and what n represents can be found in the legend for each figure. Statistical significance was determined by two-tailed unpaired Student's t test. All data are shown as mean ± standard error of the mean (SEM) representing at least three independent experiments.
Data and code availability
The datasets generated in the current study are available in the Gene Expression Omnibus, accession number GEO: GSE166704.
Author contributions
D.M.P. and E.C.F conceived of the project, designed experiments, and co-wrote the manuscript. D.M.P., A.K.W., and S.S.-B. performed experiments, analyzed data, and edited the manuscript. The authors have no competing financial interests.
Acknowledgments
We thank Dr. Anne Brunet for reagents and the UCSC Flow Cytometry Core Facility for technical assistance. This work was funded by an NIH NIA Award (R01AG062879) to E.C.F.; by an NIH IMSD Training Grant (2R25GM058903-18), a Howard Hughes Medical Institute Gilliam Fellowship Award, and an American Heart Association Predoctoral Fellowship Award to D.M.P.; by an NIH NHLBI F31 Fellowship Award and a Tobacco-Related Disease Research Program Predoctoral Fellowship Award to A.K.W.; and by CIRM Shared Stem Cell Facilities (CL1-00506) and CIRM Major Facilities (FA1-00617-1) awards to UCSC.
Published: May 20, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.04.016.
Supplemental information
References
- Akashi K., Traver D., Miyamoto T., Weissman I.L. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- Almeida A.M., Ramos F. Acute myeloid leukemia in the older adults. Leuk. Res. Rep. 2016;6:1–7. doi: 10.1016/j.lrr.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banu A.N., Avraham S., Karsenty H. Regulates murine megakaryocytopoiesis P-selectin, and not E-selectin, negatively. J. Immunol. Ref. 2020;169:4579–4585. doi: 10.4049/jimmunol.169.8.4579. [DOI] [PubMed] [Google Scholar]
- Beaudin A.E., Boyer S.W., Forsberg E.C. Flk2/Flt3 promotes both myeloid and lymphoid development by expanding non–self-renewing multipotent hematopoietic progenitor cells. Exp. Hematol. 2014;42:218–229.e4. doi: 10.1016/j.exphem.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudin A.E., Boyer S.W., Perez-Cunningham J., Hernandez G.E., Derderian S.C., Jujjavarapu C., Aaserude E., MacKenzie T., Forsberg E.C. A transient developmental hematopoietic stem cell gives rise to innate-like B and T cells. Cell Stem Cell. 2016;19:768–783. doi: 10.1016/j.stem.2016.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer S.W., Rajendiran S., Beaudin A.E., Smith-Berdan S., Muthuswamy P.K., Perez-Cunningham J., Martin E.W., Cheung C., Tsang H., Landon M. Clonal and quantitative in vivo assessment of hematopoietic stem cell differentiation reveals strong erythroid potential of multipotent cells. Stem Cell Reports. 2019;12:801–815. doi: 10.1016/j.stemcr.2019.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns I., Lucas D., Pinho S., Ahmed J., Lambert M.P., Kunisaki Y., Scheiermann C., Schiff L., Poncz M., Bergman A. Megakaryocytes regulate hematopoietic stem cell quiescence through CXCL4 secretion. Nat. Med. 2014;20:1315–1320. doi: 10.1038/nm.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne A., Beaudin A.E., Olsen H.E., Jain M., Cole C., Palmer T., DuBois R.M., Forsberg E.C., Akeson M., Vollmers C. Nanopore long-read RNAseq reveals widespread transcriptional variation among the surface receptors of individual B cells. Nat. Commun. 2017;8:16027. doi: 10.1038/ncomms16027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabezas-Wallscheid N., Klimmeck D., Hansson J., Lipka D.B., Reyes A., Wang Q., Weichenhan D., Lier A., Von Paleske L., Renders S. Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA methylome analysis. Cell Stem Cell. 2014;15:507–522. doi: 10.1016/j.stem.2014.07.005. [DOI] [PubMed] [Google Scholar]
- Carrelha J., Meng Y., Kettyle L.M., Luis T.C., Norfo R., Alcolea V., Boukarabila H., Grasso F., Gambardella A., Grover A. Hierarchically related lineage-restricted fates of multipotent haematopoietic stem cells. Nature. 2018;554:106–111. doi: 10.1038/nature25455. [DOI] [PubMed] [Google Scholar]
- Cool T., Worthington A., Poscablo D., Hussaini A., Forsberg E.C. Interleukin 7 receptor is required for myeloid cell homeostasis and reconstitution by hematopoietic stem cells. Exp. Hematol. 2020;90:39–45.e3. doi: 10.1016/j.exphem.2020.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davizon-Castillo P., McMahon B., Aguila S., Bark D., Ashworth K., Allawzi A., Campbell R.A., Montenont E., Nemkov T., D’Alessandro A. TNF-a–driven inflammation and mitochondrial dysfunction define the platelet hyperreactivity of aging. Blood. 2019;134:727–740. doi: 10.1182/blood.2019000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doré L.C., Crispino J.D. Transcription factor networks in erythroid cell and megakaryocyte development. Blood. 2011;118:231–239. doi: 10.1182/blood-2011-04-285981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykstra B., Olthof S., Schreuder J., Ritsema M., Haan G. De. Clonal analysis reveals multiple functional defects of aged murine hematopoietic stem cells. J. Exp. Med. 2011;208:2691–2703. doi: 10.1084/jem.20111490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elagib K.E., Racke F.K., Mogass M., Khetawat R., Delehanty L.L., Goldfarb A.N. RUNX1 and GATA-1 coexpression and cooperation in megakaryocytic differentiation. Blood. 2003;101:4333–4341. doi: 10.1182/blood-2002-09-2708. [DOI] [PubMed] [Google Scholar]
- Elias H.K., Bryder D., Park C.Y. Molecular mechanisms underlying lineage bias in aging hematopoiesis. Semin. Hematol. 2017;54:4–11. doi: 10.1053/j.seminhematol.2016.11.002. [DOI] [PubMed] [Google Scholar]
- Ergen A.V., Boles N.C., Goodell M.A. Rantes/Ccl5 influences hematopoietic stem cell subtypes and causes myeloid skewing. Blood. 2012;119:2500–2509. doi: 10.1182/blood-2011-11-391730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gekas C., Graf T. CD41 expression marks myeloid-biased adult hematopoietic stem cells and increases with age. Blood. 2013;121:4463–4472. doi: 10.1182/blood-2012-09-457929. [DOI] [PubMed] [Google Scholar]
- Ghiani C.A., Dell’Angelica E.C. Dysbindin-containing complexes and their proposed functions in brain: from zero to (too) many in a decade. ASN Neuro. 2011;3:109–124. doi: 10.1042/AN20110010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover A., Sanjuan-Pla A., Thongjuea S., Carrelha J., Giustacchini A., Gambardella A., Macaulay I., Mancini E., Luis T.C., Mead A. Single-cell RNA sequencing reveals molecular and functional platelet bias of aged haematopoietic stem cells. Nat. Commun. 2016;7:1–12. doi: 10.1038/ncomms11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Haan G., Van Zant G. Dynamic changes in mouse hematopoietic stem cell numbers during aging. Blood. 1999;93:3294–3301. [PubMed] [Google Scholar]
- Haas S., Hansson J., Klimmeck D., Loeffler D., Velten L., Uckelmann H., Wurzer S., Prendergast Á.M., Schnell A., Hexel K. Inflammation-induced emergency megakaryopoiesis driven by hematopoietic stem cell-like megakaryocyte progenitors. Cell Stem Cell. 2015;17:422–434. doi: 10.1016/j.stem.2015.07.007. [DOI] [PubMed] [Google Scholar]
- Hamanaka S., Ooehara J., Morita Y., Ema H., Takahashi S., Miyawaki A., Otsu M., Yamaguchi T., Onodera M., Nakauchi H. Generation of transgenic mouse line expressing Kusabira Orange throughout body, including erythrocytes, by random segregation of provirus method. Biochem. Biophys. Res. Commun. 2013;435:586–591. doi: 10.1016/j.bbrc.2013.05.017. [DOI] [PubMed] [Google Scholar]
- Ho Y.H., del Toro R., Rivera-Torres J., Rak J., Korn C., García-García A., Macías D., González-Gómez C., del Monte A., Wittner M. Remodeling of bone marrow hematopoietic stem cell niches promotes myeloid cell expansion during premature or physiological aging. Cell Stem Cell. 2019;25:407–418.e6. doi: 10.1016/j.stem.2019.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S., Mi Y., Song J., Zhang P., Liu Y. PRMT1-RBM15 axis regulates megakaryocytic differentiation of human umbilical cord blood CD34+cells. Exp. Ther. Med. 2018;15:2563–2568. doi: 10.3892/etm.2018.5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang X., Lu Z., Cui C., Deng M., Fan Y., Dong B., Han X., Xie F., Tyner J.W., Coligan J.E. The ITIM-containing receptor LAIR1 is essential for acute myeloid leukaemia development. Nat. Cell Biol. 2015;17:665–677. doi: 10.1038/ncb3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushansky K., Lok S., Holly R.D., Broudy V.C., Lin N., Bailey M.C., Forstrom J.W., Buddle M.M., Oort P.J., Hagen F.S. Promotion of megakaryocyte progenitor expansion and differentiation by the c-Mpl ligand thrombopoietin. Nature. 1994;369:568–571. doi: 10.1038/369568a0. [DOI] [PubMed] [Google Scholar]
- Kirschner K., Chandra T., Kiselev V., Flores-Santa Cruz D., Macaulay I.C., Park H.J., Li J., Kent D.G., Kumar R., Pask D.C. Proliferation drives aging-related functional decline in a subpopulation of the hematopoietic stem cell compartment. Cell Rep. 2017;19:1503–1511. doi: 10.1016/j.celrep.2017.04.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimchenko O., Mori M., DiStefano A., Langlois T., Larbret F., Lecluse Y., Feraud O., Vainchenker W., Norol F., Debili N. A common bipotent progenitor generates the erythroid and megakaryocyte lineages in embryonic stem cell-derived primitive hematopoiesis. Blood. 2009;114:1506–1517. doi: 10.1182/blood-2008-09-178863. [DOI] [PubMed] [Google Scholar]
- Kowalczyk M.S., Tirosh I., Heckl D., Rao T.N., Dixit A., Haas B.J., Schneider R.K., Wagers A.J., Ebert B.L., Regev A. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 2015;25:1860–1872. doi: 10.1101/gr.192237.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung G.A., Cool T., Valencia C.H., Worthington A., Beaudin A.E., Camilla Forsberg E. The lymphoid-associated interleukin 7 receptor (IL7R) regulates tissue-resident macrophage development. Develpoment. 2019;146:dev176180. doi: 10.1242/dev.176180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levay K., Slepak V.Z. Tescalcin is an essential factor in megakaryocytic differentiation associated with Ets family gene expression. J. Clin. Invest. 2007;117:2672–2683. doi: 10.1172/JCI27465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Zhang Z., Li Y., Arnovitz S., Chen P., Huang H., Jiang X., Hong G.M., Kunjamma R.B., Ren H. PBX3 is an important cofactor of HOXA9 in leukemogenesis. Blood. 2013;121:1422–1431. doi: 10.1182/blood-2012-07-442004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschalek R. Mixed lineage leukemia: roles in human malignancies and potential therapy. FEBS J. 2010;277:1822–1831. doi: 10.1111/j.1742-4658.2010.07608.x. [DOI] [PubMed] [Google Scholar]
- Martin E.W., Krietsch J., Reggiardo R.E., Sousae R., Kim D.H., Forsberg E.C. Chromatin accessibility maps provide evidence of multilineage gene priming in hematopoietic stem cells. Epigenetics Chromatin. 2021;14:2. doi: 10.1186/s13072-020-00377-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.P., Allman D. The decline in B lymphopoiesis in aged mice reflects loss of very early B-lineage precursors. J. Immunol. 2003;171:2326–2330. doi: 10.4049/jimmunol.171.5.2326. [DOI] [PubMed] [Google Scholar]
- Morrison S.J., Wandycz A.M., Akashi K., Globerson A., Weissman I.L. The aging of hematopoietic stem cells. Nat. Med. 1996;2:1011–1016. doi: 10.1038/nm0996-1011. [DOI] [PubMed] [Google Scholar]
- Nakorn T.N., Miyamoto T., Weissman I.L. Characterization of mouse clonogenic megakaryocyte progenitors. Proc. Natl. Acad. Sci. U S A. 2003;100:205–210. doi: 10.1073/pnas.262655099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardali E., Dimmeler S., Zeiher A.M., Rieger M.A. Clonal hematopoiesis, aging, and cardiovascular diseases. Exp. Hematol. 2020;83:95–104. doi: 10.1016/j.exphem.2019.12.006. [DOI] [PubMed] [Google Scholar]
- Pronk C.J.H., Rossi D.J., Månsson R., Attema J.L., Norddahl G.L., Chan C.K.F., Sigvardsson M., Weissman I.L., Bryder D. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell. 2007;1:428–442. doi: 10.1016/j.stem.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Rajendiran S., Smith-Berdan S., Kunz L., Risolino M., Selleri L., Schroeder T., Forsberg E.C. Ubiquitous overexpression of CXCL12 confers radiation protection and enhances mobilization of hematopoietic stem and progenitor cells. Stem Cells. 2020;38:1159–1174. doi: 10.1002/stem.3205. [DOI] [PubMed] [Google Scholar]
- Rajendiran S., Boyer S.W., Camilla Forsberg E. A quantitative hematopoietic stem cell reconstitution protocol: accounting for recipient variability, tissue distribution and cell half-lives. Stem Cell Res. 2020;50:102145. doi: 10.1016/j.scr.2020.102145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi D.J., Bryder D., Zahn J.M., Ahlenius H., Sonu R., Wagers A.J., Weissman I.L. Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc. Natl. Acad. Sci. U S A. 2005;102:9194–9199. doi: 10.1073/pnas.0503280102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuan-Pla A., Macaulay I.C., Jensen C.T., Woll P.S., Luis T.C., Mead A., Moore S., Carella C., Matsuoka S., Jones T.B. Platelet-biased stem cells reside at the apex of the haematopoietic stem-cell hierarchy. Nature. 2013;502:232–236. doi: 10.1038/nature12495. [DOI] [PubMed] [Google Scholar]
- SanMiguel J.M., Young K., Trowbridge J.J. Hand in hand: intrinsic and extrinsic drivers of aging and clonal hematopoiesis. Exp. Hematol. 2020;91:1–9. doi: 10.1016/j.exphem.2020.09.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Sauvage F.J., Hass P.E., Spencer S.D., Malloy B.E., Gurney A.L., Spencer S.A., Darbonne W.C., Henzel W.J., Wong S.C., Kuang W.J. Stimulation of megakaryocytopoiesis and thrombopoiesis by the c-Mpl ligand. Nature. 1994;369:533–538. doi: 10.1038/369533a0. [DOI] [PubMed] [Google Scholar]
- Schaefer B.C., Schaefer M.L., Kappler J.W., Marrack P., Kedl R.M. Observation of antigen-dependent CD8+ T-cell/dendritic cell interactions in vivo. Cell. Immunol. 2001;214:110–122. doi: 10.1006/cimm.2001.1895. [DOI] [PubMed] [Google Scholar]
- Semerad C.L., Mercer E.M., Inlay M.A., Weissman I.L., Murre C. E2A proteins maintain the hematopoietic stem cell pool and promote the maturation of myelolymphoid and myeloerythroid progenitors. Proc. Natl. Acad. Sci. U S A. 2009;106:1930–1935. doi: 10.1073/pnas.0808866106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Berdan S., Nguyen A., Hong M.A., Forsberg E.C. ROBO4-mediated vascular integrity regulates the directionality of hematopoietic stem cell trafficking. Stem Cell Reports. 2015;4:255–268. doi: 10.1016/j.stemcr.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Berdan S., Bercasio A., Rajendiran S., Forsberg E.C. Viagra enables efficient, single-day hematopoietic stem cell mobilization. Stem Cell Reports. 2019;13:787–792. doi: 10.1016/j.stemcr.2019.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songdej N., Rao A.K. Inherited platelet dysfunction and hematopoietic transcription factor mutations. Platelets. 2017;28:20–26. doi: 10.1080/09537104.2016.1203400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley E.R., Chitu V. CSF-1 receptor signaling in myeloid cells. Cold Spring Harb. Perspect. Biol. 2014;6:a021857. doi: 10.1101/cshperspect.a021857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steensma D.P., Ebert B.L. Clonal hematopoiesis as a model for premalignant changes during aging. Exp. Hematol. 2020;83:48–56. doi: 10.1016/j.exphem.2019.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudo K., Ema H., Morita Y., Nakauchi H. Age-associated characteristics of murine hematopoietic stern cells. J. Exp. Med. 2000;192:1273–1280. doi: 10.1084/jem.192.9.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D., Luo M., Jeong M., Rodriguez B., Xia Z., Hannah R., Wang H., Le T., Faull K.F., Chen R. Epigenomic profiling of young and aged HSCs reveals concerted changes during aging that reinforce self-renewal. Cell Stem Cell. 2014;14:673–688. doi: 10.1016/j.stem.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugarte F., Sousae R., Cinquin B., Martin E.W., Krietsch J., Sanchez G., Inman M., Tsang H., Warr M., Passegué E. Progressive chromatin condensation and H3K9 methylation regulate the differentiation of embryonic and hematopoietic stem cells. Stem Cell Reports. 2015;5:728–740. doi: 10.1016/j.stemcr.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valletta S., Thomas A., Meng Y., Ren X., Drissen R., Sengül H., Di Genua C., Nerlov C. Micro-environmental sensing by bone marrow stroma identifies IL-6 and TGFβ1 as regulators of hematopoietic ageing. Nat. Commun. 2020;11:1–13. doi: 10.1038/s41467-020-17942-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto R., Morita Y., Ooehara J., Hamanaka S., Onodera M., Rudolph K.L., Ema H., Nakauchi H. Clonal analysis unveils self-renewing lineage-restricted progenitors generated directly from hematopoietic stem cells. Cell. 2013;154:1112–1126. doi: 10.1016/j.cell.2013.08.007. [DOI] [PubMed] [Google Scholar]
- Yamamoto R., Wilkinson A.C., Ooehara J., Lan X., Lai C.-Y., Nakauchi Y., Pritchard J.K., Nakauchi H. Large-scale clonal analysis resolves aging of the mouse hematopoietic stem cell compartment. Cell Stem Cell. 2018;22:600–607.e4. doi: 10.1016/j.stem.2018.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young K., Eudy E., Bell R., Loberg M., Stearns T., Velten L., Haas S., Trowbridge J. Hematopoietic stem and progenitor cell aging is initiated at middle age through decline in local insulin-like growth factor 1 (IGF1) BioRxiv. 2020 [Google Scholar]
- Zhu F., Feng M., Sinha R., Seita J., Mori Y., Weissman I.L. Screening for genes that regulate the differentiation of human megakaryocytic lineage cells. Proc. Natl. Acad. Sci. U S A. 2018;115:E9308–E9316. doi: 10.1073/pnas.1805434115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F., Feng M., Sinha R., Murphy M.P., Luo F., Kao K.S., Szade K., Seita J., Weissman I.L. The GABA receptor GABRR1 is expressed on and functional in hematopoietic stem cells and megakaryocyte progenitors. Proc. Natl. Acad. Sci. U S A. 2019;116:18416–18422. doi: 10.1073/pnas.1906251116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated in the current study are available in the Gene Expression Omnibus, accession number GEO: GSE166704.