Abstract
Natural products are important because of their significant pharmaceutical properties such as antiviral, antimicrobial, and anticancer activity. Recent breakthroughs in DNA sequencing reveal that a great number of cryptic natural product biosynthetic gene clusters are encoded in microbial genomes, for example, those of Streptomyces species. However, it is still challenging to access compounds from these clusters because many source organisms are uncultivable or the genes are silent during laboratory cultivation. To address this challenge, we develop an efficient cell-free platform for the rapid, in vitro total biosynthesis of the nonribosomal peptide valinomycin as a model. We achieve this goal in two ways. First, we used a cell-free protein synthesis (CFPS) system to express the entire valinomycin biosynthetic gene cluster (>19 kb) in a single-pot reaction, giving rise to approximately 37 μg/L of valinomycin after optimization. Second, we coupled CFPS with cell-free metabolic engineering system by mixing two enzyme-enriched cell lysates to perform a two-stage biosynthesis. This strategy improved valinomycin production ~5,000-fold to nearly 30 mg/L. We expect that cell-free biosynthetic systems will provide a new avenue to express, discover, and characterize natural product gene clusters of interest in vitro.
Keywords: Valinomycin, Cell-free systems, In vitro biosynthesis, Nonribosomal peptide, Natural product, Synthetic biology
1. Introduction
Natural products originating from living organisms (e.g., microbes, plants, and animals) have complex chemical structures and exhibit diverse biological activities (Koehn and Carter, 2005). Historically, natural products have served as a rich source for pharmaceuticals like antibiotics to treat human diseases (Newman and Cragg, 2016). During the so-called ‘Golden Era’ of antibiotic discovery from the 1940s to 1960s, many notable antibiotics were discovered. However, declining rates of new antibiotic discovery and rising rates of re-discovery through conventional, low-throughput fermentation and chemical/biological screening approaches have limited the field (Katz and Baltz, 2016). As a result, the accelerating problem of antibiotic resistance is projected to soon threaten up to 10 million lives annually (O’Neill, 2016). This motivates the need for new approaches to discover new antibiotics and natural products for human health.
Several approaches are already emerging. For example, next-generation DNA sequencing and synthesis (Smanski et al., 2016), improved bioinformatic tools for genome mining and prediction (Blin et al., 2019), high-throughput mass spectrometry for metabolite analysis (Doroghazi et al., 2014), and CRISPR-based technology for activation of silent biosynthetic gene clusters (Zhang et al., 2017) are all contributing new dimensions to natural product discovery. Even with these new approaches, the rate at which new biosynthetic pathways are identified still greatly outpaces the capacity to characterize the small molecules for which production is encoded (Aigle et al., 2014). Current approaches are constrained by limitations in cultivating natural product producing organisms in the laboratory. Moreover, establishing heterologous expression systems is difficult. While several heterologous hosts like Escherichia coli and Saccharomyces cerevisiae have been used for production and discovery of natural products (Li and Neubauer, 2014; Luo et al., 2015; Nielsen, 2019; Zhang et al., 2011), engineering these cells remains a time-consuming and laborious process that often requires design-build-test-learn iterations to obtain the optimal cell performance (Nielsen and Keasling, 2016). In addition, yields of most compounds are still not satisfactory in heterologous hosts (Li and Neubauer, 2014). Therefore, a new generation of heterologous expression systems for efficient, high-yielding production of bioactive molecules is highly desirable.
Recently, cell-free systems have emerged as a complementary platform for biomanufacturing (Bundy et al., 2018; Dudley et al., 2015; Liu et al., 2019; Swartz, 2018), with potential for natural product biosynthesis. For example, in vitro reconstitution strategies based on purified enzymes (Li et al., 2018b) have elegantly demonstrated the biosynthesis of polyketides (PKs) (Cheng et al., 2007), nonribosomal peptides (NRPs) (Balibar et al., 2007), and PK/NRP hybrids (Greunke et al., 2017). Yet despite its success, this strategy often suffers from laborious purification process and enzyme instability. To circumvent these shortcomings, we recently demonstrated the use of cell-free protein synthesis (CFPS) systems derived from crude cell lysates to synthesize a NRP molecule (diketopiperazine, DKP) by in vitro coexpression of two large nonribosomal peptide synthetases (NRPS), namely, GrsA (126 kDa) and GrsB1 (121 kDa, the first module of GrsB) (Goering et al., 2017). While this work represented the first step to apply crude extract-based CFPS to synthesize natural products, DKP is only a shunt product and not the final pathway biosynthesis product gramicidin S. Thus, to date, total biosynthesis of a natural product has not been shown with crude extract-based CFPS systems. Here, we aim to address this gap, using the nonribosomal peptide valinomycin as our model system.
Valinomycin is a 36-membered cyclododecadepsipeptide (see Fig. 1A for the chemical structure), which possesses a broad spectrum of bioactivities such as antifungal (Park et al., 2008), antimicrobial (Tempelaars et al., 2011), insecticidal (Heisey et al., 1988), antiviral (Wu et al., 2004), and anticancer efficacy (Ryoo et al., 2006). Naturally, valinomycin is synthesized by several Streptomyces strains via the NRPS enzyme valinomycin synthetase (Cheng, 2006; Magarvey et al., 2006; Matter et al., 2009). Valinomycin synthetase consists of two distinct large NRPSs (Vlm1 370 kDa and Vlm2 284 kDa), and each of them has two modules for the assembly of their dedicated substrates pyruvate, α-ketoisovalerate, and l-valine (Fig. 1A) (Jaitzig et al., 2014). Our previous studies achieved the heterologous biosynthesis of valinomycin in vivo by reconstitution of the NRPS genes (vlm1 and vlm2) in E. coli (Jaitzig et al., 2014). The yield of valinomycin reached a milligram per liter level after a systematic bioprocess optimization (Li et al., 2014, 2015a). In the valinomycin biosynthetic gene cluster, a discrete type II thioesterase (TEII) was also identified (Cheng, 2006). TEII is a repairing enzyme that usually is associated with NRPS enzymes, playing a key role in regenerating the functionality of NRPS through hydrolysis of either misacylated thiol groups (Schwarzer et al., 2002) or incorrectly loaded substrates on the thiolation (T) domains (Fig. 1B) (Yeh et al., 2004). Notably, coexpression of the cognate protein TEII with Vlm1 and Vlm2 resulted in the highest production of valinomycin (13 mg/L) in E. coli (Li et al., 2015b), demonstrating the dedicated function of TEII in heterologous valinomycin biosynthesis.
Fig. 1. Reconstitution of valinomycin biosynthetic pathway in vitro.
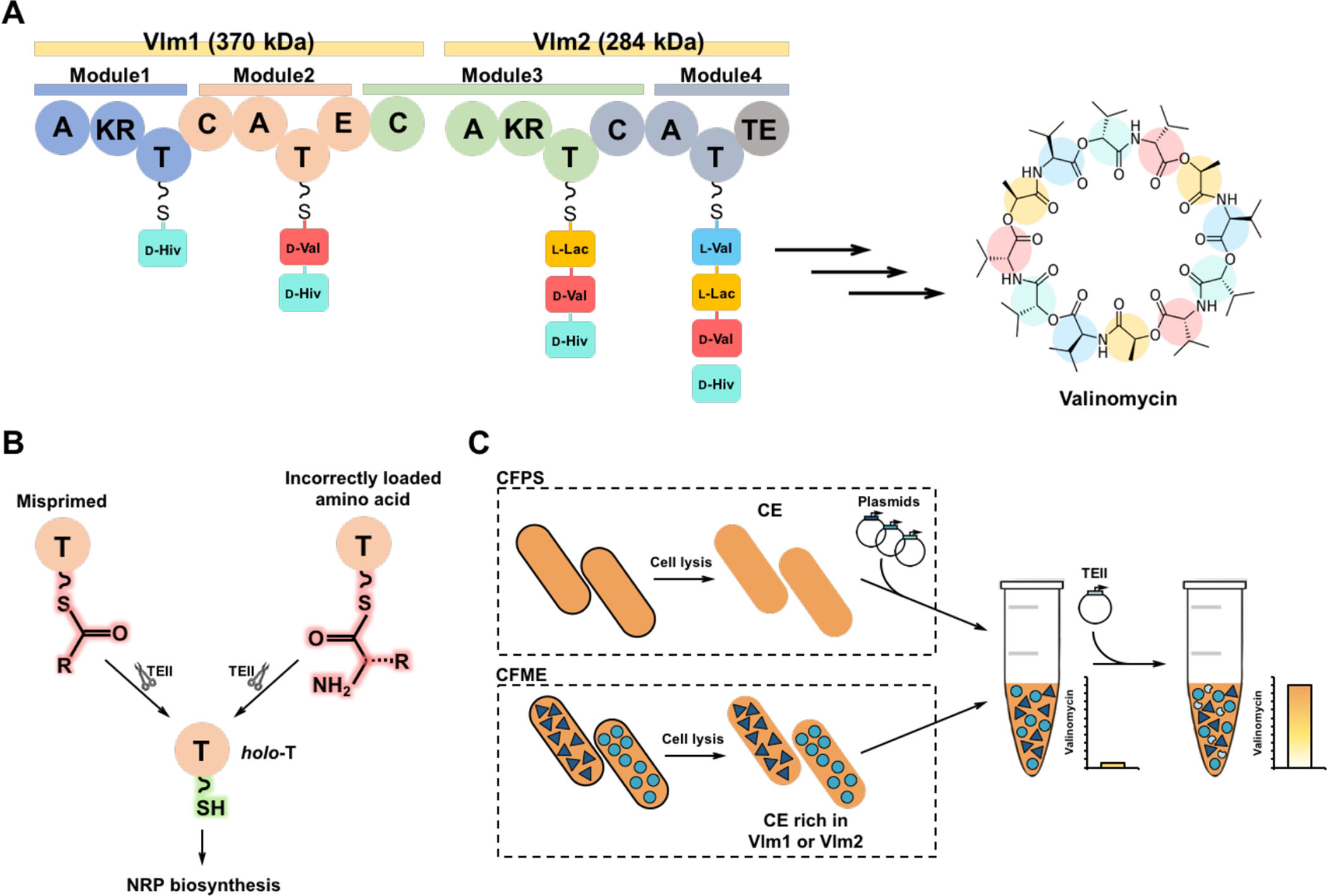
(A) Valinomycin synthetase contains two distinct NRPS enzymes Vlm1 (374 kDa) and Vlm2 (284 kDa) that are composed of adenylation (A), ketoreductase (KR), thiolation (T), condensation (C), epimerase (E), and thioesterase (TE) domains. A domains select and activate three substrates (pyruvate, α-ketoisovalerate (α-Kiv), and l-valine (l-Val)). T domains are responsible for the translocation of the bound aminoacyl or peptidyl intermediate between adjacent catalytic positions. C domains catalyze the formation of peptide bond and elongate the peptide chain. The KR domain in Module 1 reduces α-Kiv to d-hydroxyisovalerate (d-Hiv). The E domain in Module 2 converts l-Val to d-valine (d-Val). The KR domain in Module 3 reduces pyruvate to l-lactate (l-Lac). The four modules of valinomycin synthetase are iteratively reused to assemble three tetradepsipeptide monomers, which are eventually oligomerized and macrolactonized to form the 36-membered cyclododecadepsipeptide valinomycin. (B) Regeneration of the functionality of T domains in NRPS catalyzed by type II thioesterase (TEII). (C) Cell-free biosynthesis of valinomycin with different strategies: cell-free protein synthesis (CFPS), cell-free metabolic engineering (CFME), and a coupled CFPS-MS approach. CE, cell extract.
In this work, we demonstrate that simple cell-free systems enable the total biosynthesis of a complex NRP valinomycin (Fig. 1C). We show this in two ways. First, we use an E. coli-based CFPS system to coexpress two enzymes Vlm1 (370 kDa) and Vlm2 (284 kDa) for valinomycin synthesis. Second, cell lysates are enriched with Vlm1 or Vlm2 by cellular expression and then the full metabolic pathway is assembled in vitro to enable valinomycin biosynthesis by mixing two enzyme-enriched cell lysates. In both strategies, TEII substantially improves valinomycin production. Taken together, our work demonstrated three key advancements. First, to our best knowledge, this is the first report to use CFPS for coexpression of such large NRPS enzymes with catalytic activity. Second, in vitro total biosynthesis of valinomycin suggests the feasibility to produce other complex NRPs with cell-free systems. Third, fine tuning of cell-free reactions allows for enhanced production of target molecules. Looking forward, we envision that cell-free biosynthetic systems will provide a new avenue for the expression of complex natural product gene clusters (e.g., NRPs and PKs), enabling the rapid discovery and synthesis of novel natural products in vitro.
2. Materials and methods
2.1. Bacterial strains and plasmids
E. coli BL21 Star (DE3) was used for preparation of cell extracts to perform CFPS reactions. The strain E. coli BAP1, a generous gift from Prof. Yong Wang (Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, China), was used to overexpress Vlm1 and Vlm2 in vivo. E. coli BAP1 is a derivative of E. coli BL21 (DE3) with genomic integration of the sfp gene for posttranslational phosphopantetheinylation of NRPSs (Pfeifer et al., 2001). Plasmids pCTUT7-Vlm1 and pKS01-Vlm2 harboring genes vlm1 (~10 kb) and vlm2 (~8 kb) (Jaitzig et al., 2014), respectively, were kindly provided by Prof. Peter Neubauer (Technische Universität Berlin, Germany). Cell-free expression of Sfp and TEII were achieved with plasmids pET28a-Sfp (Goering et al., 2017) and pJL1-TEII (Li et al., 2017), respectively.
2.2. Preparation of cell extracts
All E. coli strains were grown in 2×YTPG medium, consisting of (per liter) 10 g yeast extract, 16 g tryptone, 5 g NaCl, 7 g K2HPO4, 3 g KH2PO4, and 18 g glucose. Cultivations of E. coli BL21 Star (DE3) were performed in 1 L of 2×YTPG in 2.5 L baffled Ultra Yield™ flasks (Thomson Instrument Company, USA). After inoculation (initial OD600 of 0.05), cultures were incubated in the shaker at 220 rpm and 34°C. When OD600 reached 0.6–0.8, cells were induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) to express T7 RNA polymerase. Then, cells were grown until an OD600 of 3.0 and harvested by centrifugation at 5,000 g and 4°C for 15 min. Afterwards, cell pellets were washed three times with cold S30 Buffer (10 mM Tris-acetate, 14 mM magnesium acetate, 60 mM potassium acetate, and 2 mM dithiothreitol (DTT)). After the final wash and centrifugation, the pelleted cells were resuspended in S30 Buffer (1 mL per gram of wet cell mass) and lysed by sonication (10 s on/off, 50% of amplitude, input energy ~600 Joules). The lysate was then centrifuged twice at 12,000 g and 4°C for 10 min. The resulting supernatant was flash frozen in liquid nitrogen and stored at −80° until use.
To prepare enzyme-enriched lysates, two strains of E. coli BAP1 containing the plasmid pCTUT7-Vlm1 and pKS01-Vlm2, respectively, were grown in 1 L of 2×YTPG and incubated at 30°C and 250 rpm. At OD600 of 0.6–0.8, the cultures were induced with 20 μM IPTG, followed by 5 h cultivation for Vlm1 and Vlm2 expression. Afterwards, cell collection, disruption, and extracts were prepared the same as described above. The total amount of protein in cell extracts was quantified by the Quick-Start Bradford Protein Assay Kit (Bio-Rad). Typically, the total protein concentration of cell extracts was 30–40 mg/mL. Overexpression of Vlm1 and Vlm2 was confirmed by SDS-PAGE protein gels.
2.3. CFPS reactions
CFPS reactions were performed at a total volume of 15 μL in 1.5 mL microcentrifuge tubes. Each reaction mixture contained the following components: 12 mM magnesium glutamate, 10 mM ammonium glutamate, 130 mM potassium glutamate, 1.2 mM ATP, 0.85 mM each of GTP, UTP, and CTP, 34 μg/mL folinic acid, 170 μg/mL of E. coli tRNA mixture, 2 mM each of 20 standard amino acids, 0.33 mM nicotinamide adenine dinucleotide (NAD), 0.27 mM coenzyme A (CoA), 1.5 mM spermidine, 1 mM putrescine, 4 mM sodium oxalate, 33 mM phosphoenolpyruvate (PEP), appropriate plasmids (see below), and 27% (v/v) of cell extract. In all 15 μL CFPS reactions, each plasmid was added individually or together as follows: 200 ng pCTUT7-Vlm1, 200 ng pKS01-Vlm2, 100 ng pET28a-Sfp, and 200 ng pJL1-TEII. In the case of coexpression of four proteins, pCTUT7-Vlm1, pKS01-Vlm2, and pET28a-Sfp were added together at the beginning for 6 h expression. Then, pJL1-TEII was added to the cell-free mixture for another 14 h reaction unless otherwise noted. Other CFPS reactions were incubated at 30°C for 20 h. All cell-free expressed proteins were analyzed by SDS-PAGE gels.
2.4. CFME reactions
Standard CFME reactions were carried out in 1.5 mL microcentrifuge tubes at 30°C for 12 h in 25 μL volumes. Reaction components consisted of 8 mM magnesium acetate, 10 mM ammonium acetate, 134 mM potassium acetate, 200 mM glucose, 10 mM dipotassium phosphate (pH 7.2), 1 mM NAD, 1 mM ATP, 1 mM CoA, and 35 mg/mL total protein. The total protein concentration was maintained by adjusting the mass ratio of two lysates (Vlm1 and Vlm2).
2.5. CFPS-ME reactions
CFPS synthesis of TEII was scaled up from 15 μL to a 25 μL reaction system. All reagent concentrations were the same as described in the section of “2.3 CFPS reactions”, except the plasmid (pJL1-TEII, 13.3 μg/mL) and cell extract (35 mg/mL, total protein of two mixed lysates). Unless otherwise noted, cell-free expression of TEII was performed at 30°C for 3 h, followed by spiking in glucose (200 mM) for valinomycin biosynthesis for another 12 h.
2.6. Valinomycin extraction and quantification
Valinomycin was extracted with three-fold volumes of ethyl acetate from cell-free reactions. After centrifugation at 16,000 g for 5 min, the organic fraction was transferred to a fresh 1.5 mL microcentrifuge tube, air dried, and resuspended in methanol for valinomycin analysis. Valinomycin quantification was performed with an Agilent 6470 Triple Quadrupole LC/MS System equipped with an Eclipse Plus C18 column (2.1×50 mm, 1.8 μm). For valinomycin detection, 2 μL of each sample was injected and separated at a flow rate of 0.4 mL/min with elution buffers A (water + 0.1% formic acid) and B (acetonitrile + 0.1% formic acid) through a linear gradient elution from 80 to 100% B over 2.5 min, a 100% B wash for 7.5 min, and a post time wash for 10 min with 80% B. Valinomycin concentrations were calculated according to a calibration curve prepared with commercial valinomycin (Sigma) as a standard. All measurements were performed in triplicate.
3. Results
3.1. Cell-free synthesis of active Vlm1 and Vlm2 for valinomycin biosynthesis
To establish an in vitro platform for valinomycin biosynthesis, we used an E. coli BL21 Star (DE3) based CFPS system (Kwon and Jewett, 2015) to synthesize the NRPS valinomycin synthetase. The successful expression of the large enzymes Vlm1 (370 kDa) and Vlm2 (284 kDa) was achieved in our cell-free system as shown clearly by SDS-PAGE with correct molecular weight bands (Fig. 2A). Most importantly, both enzymes can be coexpressed solubly in a single-pot cell-free reaction, suggesting the potential of valinomycin formation in vitro.
Fig. 2. In vitro biosynthesis of valinomycin using CFPS system.
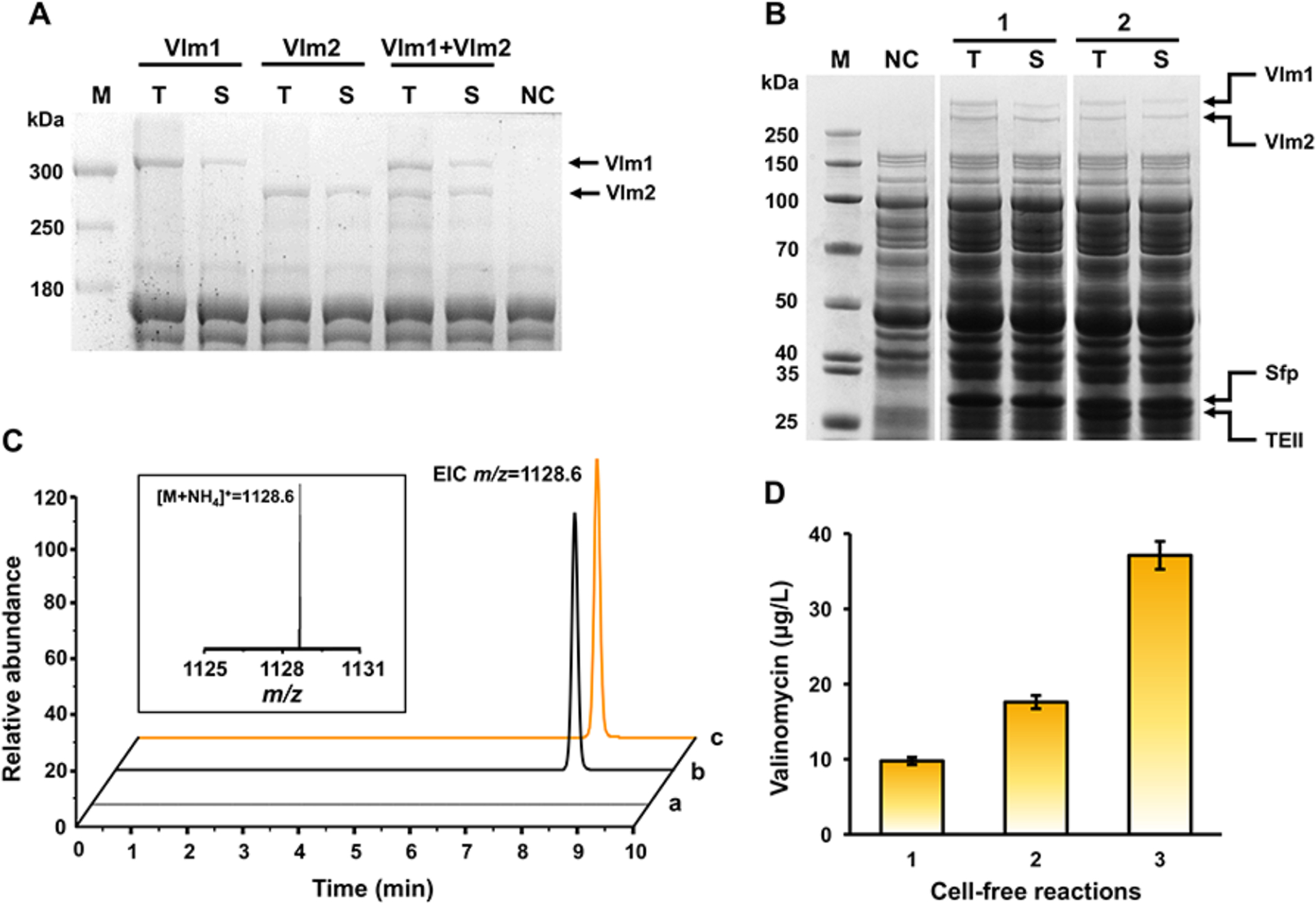
(A) SDS-PAGE (7.5% gel) analysis of Vlm1 and Vlm2 expressed individually or together. T, total protein. S, soluble protein. M, protein molecular weight marker. NC, negative control without plasmid in the reaction. (B) SDS-PAGE (4–12% gel) analysis of coexpressed proteins. 1, coexpression of Vlm1, Vlm2, and Sfp. 2, coexpression of Vlm1, Vlm2, Sfp, and TEII. (C) LC-MS analysis of valinomycin produced by the CFPS reaction. Insert, MS spectrum of valinomycin peak. EIC, extracted ion chromatogram. a, negative control without plasmid in the reaction. b, valinomycin standard. c, CFPS reaction sample. (D) Valinomycin yields from different CFPS reaction conditions. 1, valinomyicn synthesis by coexpression of Vlm1, Vlm2, and Sfp. Valinomyicn synthesis by coexpression of Vlm1, Vlm2, Sfp, and TEII for a total time of (2) 20 h reaction and (3) 24 h reaction, respectively. Values show means with error bars representing standard deviations (S.D.) of at least three independent experiments.
Once we had successfully expressed Vlm1 and Vlm2 in vitro, we next needed to convert the enzymes to their functional (holo), active form. To be functional the apo-T (thiolation) domains of NRPS enzymes have to be posttranslationally modified by transfer of a phosphopantetheine moiety from coenzyme A (CoA) to a conserved serine residue in the T domain (Fischbach and Walsh, 2006). This modification, called priming, of apo-T domains to functional, holo-T domains is carried out by phosphopantetheinyl transferases (PPTases) and consequently leads to active holo-NRPS (Lambalot et al., 1996). To enable priming and activate our Vlm1 and Vlm2 in vitro, we chose the promiscuous PPTase Sfp from Bacillus subtilis (Quadri et al., 1998), which has been frequently utilized for the phosphopantetheinylation of heterologous NRPS.
Previously, we added purified Sfp to cell-free reactions to modify NRPS (GrsA and GrsB1) (Goering et al., 2017). In this work, to avoid the laborious purification process, we directly coexpressed the sfp gene with Vlm1 and Vlm2 in a one-pot reaction. As shown in Fig. 2B, all three proteins were coexpressed and easily visible on an SDS-PAGE gel. Notably, cell-free expressed Sfp was able to activate Vlm1 and Vlm2 for the biosynthesis of valinomycin, which was successfully detected by LC-MS from the in vitro reaction mixture (Fig. 2C). While valinomycin was formed, the yield of 9.76±0.23 μg/L was relatively low (Fig. 2D). We next sought to enhance valinomycin biosynthesis yields.
3.2. Enhancing valinomycin biosynthesis by coexpression of type II thioesterase (TEII)
TEII is a discrete protein that often encoded within NRPS gene clusters, playing a repairing (editing) role by removal of nonreactive moieties and aberrant substrates that block the NRP assembly lines (Kotowska and Pawlik, 2014). In this way, TEII restores the activity of NRPS that allows for further rounds of product biosynthesis. Since coexpression of TEII with its related NRPS in host cells can be used to improve product yields (Li et al., 2015b), we wondered if a similar benefit would hold true in vitro. Thus, we set out to investigate the effect of TEII on valinomycin production in our cell-free system. To do this, we added the entire gene cluster of valinomycin (vlm1, vlm2, and TEII) as well as the sfp gene, each independently cloned on a single plasmid, to a one-pot CFPS reaction. We carried out the reaction in two steps. In step one, we expressed Vlm1, Vlm2, and Sfp together for an initial 6 h period, allowing for the expression and modification of Vlm1 and Vlm2. In step two, we added the TEII plasmid to the cell-free mixture. Following TEII plasmid addition, the reaction was allowed to continue for an addition 14 h. After the reaction, all four proteins were easily visible on an SDS protein gel (Fig. 2B). This, to the best of our knowledge, is the first example of expressing a whole natural product gene cluster (Vlm1, Vlm2, and TEII) and a heterologous modification enzyme (Sfp) together using an E. coli-based CFPS system. Furthermore, as we expected the valinomycin yield was significantly improved to 17.59±0.29 μg/L, which is nearly two times higher than that of the reaction without coexpression of TEII (Fig. 2D). The yield was further increased by two-fold up to 37.11±0.84 μg/L when the whole reaction time was extended from 20 to 24 h (Fig. 2D). Taken together, our data demonstrate the robustness (i.e., coexpression of four proteins including two large NRPSs, ~300 kDa) and flexibility (i.e., adding plasmids sequentially for timing tunability of gene expression) of the E. coli CFPS system for the rapid, total biosynthesis of complex natural products like NRPs in vitro. However, the yield of valinomycin was low. Specifically, yields were about three orders of magnitude lower than previous reports in E. coli cells.
3.3. In vitro biosynthesis of valinomycin using cell-free metabolic engineering (CFME)
While total biosynthesis was achieved in the cell-free biosynthesis approach above, we hypothesized that the low valinomycin yields resulted from resource limitations (e.g., energy) caused by the extended reaction duration and expression of four enzymes, especially the two large NRPSs (Vlm1 and Vlm2). In an attempt to bypass such a constraint, we next switched the expression of Vlm1 and Vlm2 from in vitro to in vivo. The key idea was to maximally express proteins in cells, and then mix lysates enriched with these enzymes to increase production of valinomycin (Fig. 1C). We previously used such an approach, which is called cell-free metabolic engineering (CFME), to efficiently synthesize small molecules (e.g., n-butanol, 2,3-butanediol) (Dudley et al., 2016, 2019; Karim and Jewett, 2016; Kay and Jewett, 2015). For example, glycolysis in crude E. coli lysates powered the production of 2,3-butanediol with high titers (>80 g/L) and productivities (>10 g/L/h).
To demonstrate CFME synthesis of valinomycin, we first introduced genes encoding Vlm1 and Vlm2 individually into the source strain E. coli BAP1, which has the sfp gene chromosomally integrated (Pfeifer et al., 2001). After heterologous expression of each enzyme in vivo, we lysed cell pellets to generate Vlm1 and Vlm2 enriched cell lysates, respectively. As expected, we observed that both enzymes were overexpressed in E. coli BAP1 as can be seen on the SDS-PAGE gel (Fig. 3A). In this case, Vlm1 and Vlm2 should be modified by Sfp that is encoded in the genome of BAP1 to be in their active holo form. We next initiated in vitro valinomycin biosynthesis by directly mixing two cell lysates (Vlm1 and Vlm2), acetate salts (Mg2+, NH4+, and K+), cofactors (CoA, NAD, and ATP), and other components. The CFME reaction was carried out for 12 h at 30°C and valinomycin was analyzed by LC-MS. However, the yield of 5.59±0.60 μg/L was unexpectedly low (Fig. 3B). We, therefore, next set out to optimize the CFME system for enhanced synthesis of valinomycin.
Fig. 3. In vitro biosynthesis of valinomycin using CFME system.
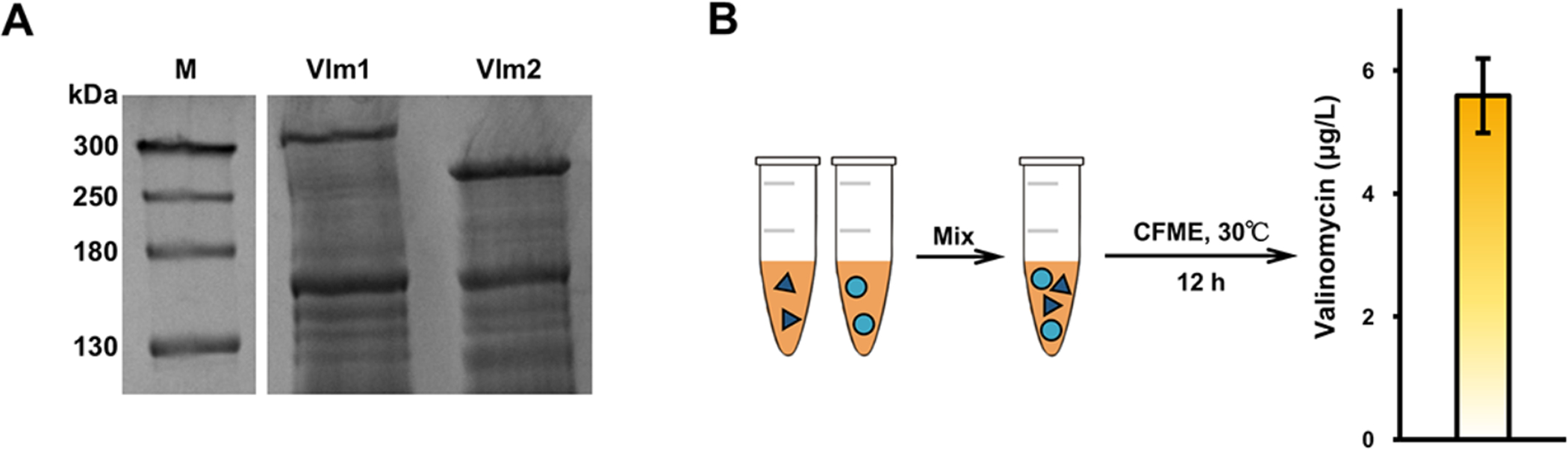
(A) SDS-PAGE (7.5% gel) analysis of Vlm1 (370 kDa) and Vlm2 (284 kDa) overexpressed individually in E. coli BAP1. M, protein molecular weight marker. (B) CFME reactions for valinomycin production by mixing two cell lysates. Error bar represents standard deviation (S.D.) of at least three independent experiments.
We began our optimization by investigating the effect of supplemental cofactors (CoA, NAD, and ATP) on valinomycin formation. Previous studies suggested that cofactors may play a positive role in the performance of cell-free metabolic pathways (Karim and Jewett, 2016; Kay and Jewett, 2015). When the concentration of CoA, NAD, and ATP were reduced from initial 1 mM to 0.1 mM in our CFME system, the yield of valinomycin was increased 1.5-fold up to 8.77±1.37 μg/L (Fig. 4A). Then, CoA, NAD, and ATP (each at 0.1 mM) were removed one-at-a-time, two-at-a-time, and altogether from CFME reactions. We found the reaction without any supplementary cofactors produced the highest valinomycin of 19.99±1.30 μg/L, which is nearly 4-fold higher than the initial yield (Fig. 4A). Our data are in agreement with previous reports (Dudley et al., 2016, 2019) that cofactors from the cell lysate are sufficient to drive product synthesis, in our case, valinomycin. Therefore, we omitted costly cofactors from our CFME system in the following investigations.
Fig. 4. Optimization of CFME reactions for enhanced valinomycin production.
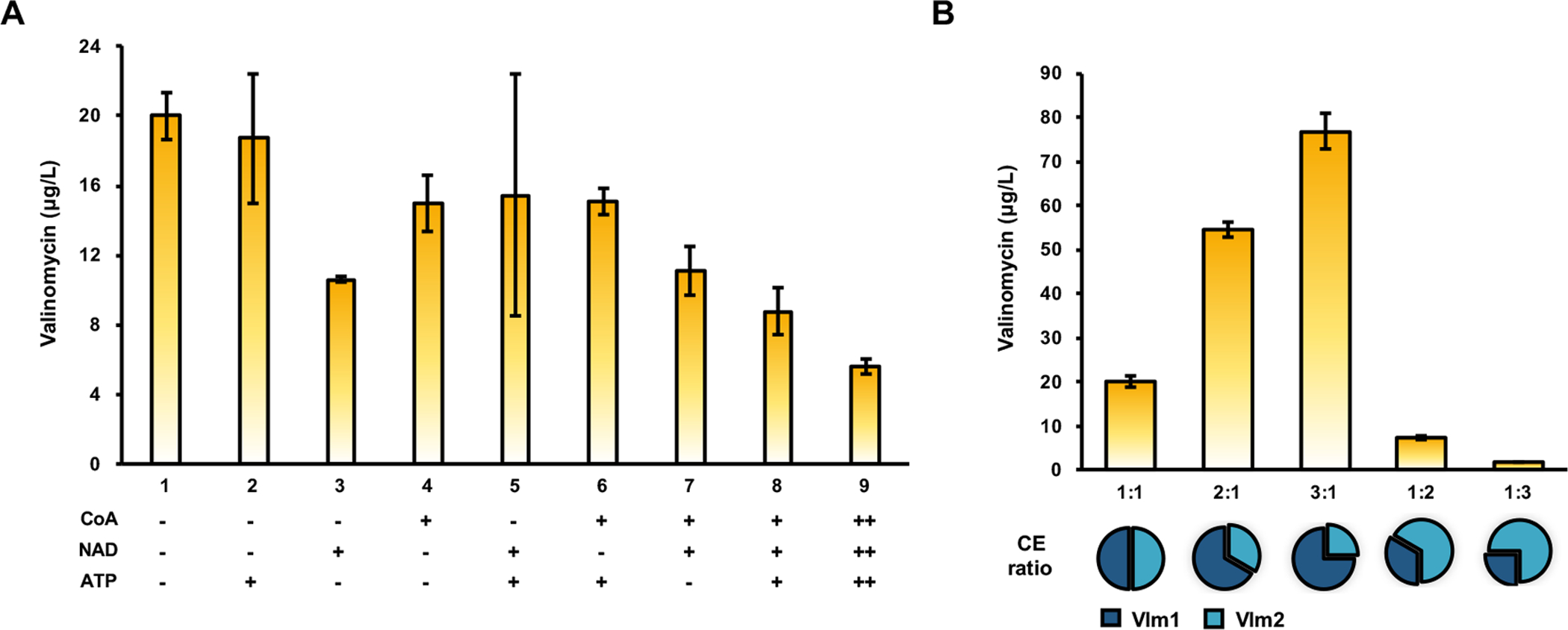
(A) Effect of supplementary cofactors (CoA, NAD, and ATP) on valinomycin formation. −, no cofactor added, +, cofactors were supplemented with 0.1 mM. ++, cofactors were supplemented with 1 mM. (B) Effect of the mass ratio of two cell extracts (CE) on valinomycin production. Values show means with error bars representing standard deviations (S.D.) of at least three independent experiments.
A key conceptual shift of our approach is that the design element is a lysate rather than a gene. Once in hand, selectively enriched lysates can be mixed in different ratios to optimize the pathway as this will impact the enzyme concentration. In each 25 μL CFME reaction, we added two cell lysates with a total protein mass of ~35 mg/mL. Since Vlm1 and Vlm2 were overexpressed in a similar level (Fig. 3A), we initially added two lysates at a mass ratio of 1:1, generating approximately 20 μg/L valinomycin (Fig. 4B). We next varied the mass ratio of the two lysates to find beneficial enzyme proportions. As shown in Fig. 4B, valinomycin formation in CFME reactions was notably impacted by the mass ratio of cell lysates. It is clear that more Vlm1 enriched cell lysates in the reaction produced more valinomycin with the highest yield of 76.9± 3.94 μg/L at a mass ratio of 3:1 (cell lysate-Vlm1:cell lysate-Vlm2). By contrast, valinomycin synthesis was almost abolished when the ratio was 1:3. The results demonstrate that the steps of valinomycin biosynthesis catalyzed by Vlm1 might be rate-limiting.
3.4. CFPS of TEII improves valinomycin biosynthesis in coupling with CFME
We next wondered if, as before, addition of TEII would enhance valinomycin yields. We therefore next used a coupled CFPS-CFME (CFPS-ME) system to perform a two-phase biosynthesis (Fig. 5A), whereby the repairing enzyme TEII was expressed by CFPS in the first reaction phase, which could be used to, as shown previously, regenerate the activity of Vlm1 and Vlm2 during the second CFME phase. We initially expressed TEII for 3 h and then added glucose (200 mM) to fuel the CFME process for another 12 h. With this strategy, the yield of valinomycin was dramatically improved to 29.32±1.37 mg/L (Fig. 5A), which is more than 5,200 times higher than the initial CFME yield of 5.59±0.60 μg/L. This result demonstrates that (i) a dedicated enzyme can be expressed by the upstream CFPS to efficiently drive the downstream CFME reactions; (ii) in vitro expressed TEII is active to restore the function of in vivo heterologously expressed, but misprimed NRPS enzymes (Vlm1 and Vlm2); and (iii) the CFPS-ME system is robust for the synthesis of complex natural product (valinomycin) from a simple, cheap precursor glucose. Further increase of the TEII expression time (>6 h) reduced the product yields (Fig. 5A), perhaps as a result of consuming more resources for TEII synthesis in the batch reaction that led to the decrease of valinomycin formation.
Fig. 5. Boosting valinomycin yields with a coupled CFPS-ME system.
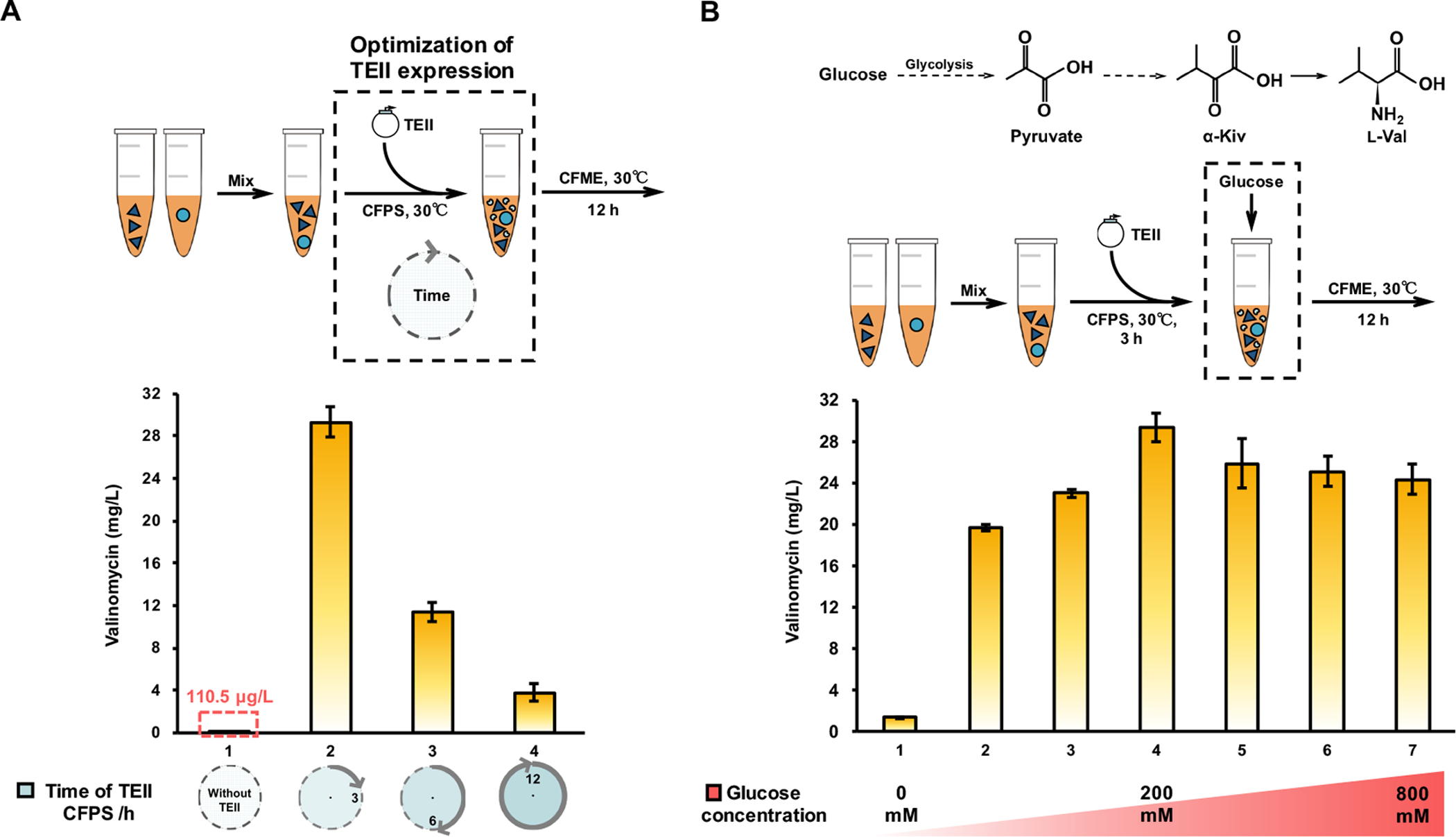
(A) Enhancement of valinomycin yields by CFPS of TEII in the upstream of CFME reactions. (B) Investigation of the glucose concentration on valinomycin production. Values show means with error bars representing standard deviations (S.D.) of at least three independent experiments.
To fuel the reaction, glucose is converted to pyruvate in glycolysis by enzymes in the extract, which is further converted in the branched chain amino acid l-valine biosynthetic pathway to α-ketoisovalerate (α-Kiv) and l-valine (Fig. 5B) (Jaitzig et al., 2014). To further enhance valinomycin synthesis yields, we next assessed the impact of glucose concentration on valinomycin synthesis, noting that glucose is the key reaction substrate. Specifically, we tested starting glucose concentrations from 0 – 800 mM. We observed that the product yield was substantially enhanced from 1.3±0.10 mg/L without feeding glucose to 19.6±0.33 mg/L with addition of 50 mM glucose. The yield reached the highest of approximately 30 mg/L at the glucose concentration of 200 mM. When glucose concentrations were increased to 400 – 800 mM, valinomycin yields were slightly reduced to a stable level of around 25 mg/L. Overall, our data demonstrate the ability to use glucose to fuel a highly active natural product biosynthetic pathway that is constructed by CFPS and CFME.
4. Discussion
Natural products play, and will continue to play, a significant role in the drug discovery engine (Katz and Baltz, 2016; Koehn and Carter, 2005; Newman and Cragg, 2016). DNA sequencing and genome mining have revealed that a far greater number of natural product biosynthetic gene clusters exist than we currently know products for (Aigle et al., 2014). To access and harness these natural products, rapid and robust approaches need to be developed for their synthesis. In this report, we show that cell-free biology is one such approach. This approach shifts the design-build-test unit from a genetic construct and its associated cell-line to a cell-free lysate. The utility of our approach is demonstrated by the ability to enable total synthesis of the nonribosomal macrolactone peptide valinomycin in a single CFPS system and a coupled CFPS-ME system.
Our results have several key features. First, the use of cell-free systems allows for fine tuning of reaction conditions, easy monitoring, and avoidance of mechanisms that have evolved to facilitate species survival. In recent years, cell-free systems have been utilized not only for the synthesis of various proteins (Liu et al., 2019), but also for the rapid and high-throughput prototyping of metabolic pathways (Karim and Jewett, 2016; O’Kane et al., 2019). Despite the wide applications of cell-free systems, total cell-free biosynthesis of both the enzymes and their natural product has not previously been reported. We show, somewhat surprisingly, that cell-free systems are capable large NRPS synthesis. Indeed, we succeeded in cell-free expression of the valinomycin biosynthetic gene cluster (>19 kb), which contains two large NRPSs (Vlm1, 370 kDa and Vlm2, 284 kDa) and one associate repairing enzyme TEII, as well as a heterologous modification enzyme (Sfp). Notably, four enzymes were actively coexpressed in a single-pot CFPS reaction, giving rise to valinomycin formation with a yield of nearly 40 μg/L (Fig. 2D).
Second, cell-free systems enable high titers of natural products. We were able to leverage the flexibility of the cell-free system to adjust the way enzymes were enriched in lysates (shifting from cell-free protein synthesis to heterologous overexpression) to facilitate increased natural product titers. Additionally, we could readily tune expression of the TEII enzyme to restore the activity of Vlm1 and Vlm2, as well as increase metabolic flux into pathway precursors (e.g., pyruvate, α-Kiv, and l-Val) by the addition of increased concentrations of glucose (200 mM). Of note, the optimized, coupled CFPS-ME system enabled synthesis of ~30 mg/L valinomycin, which compares favorably to the best previous reports. Several native Streptomyces organisms produce 4 to 32 mg/L (Matter et al., 2009) and a heterologous E. coli host was shown to make 13 mg/L (Li et al., 2015b). However, there may be situations where E. coli-based systems are not the best suited for expressing natural product biosynthetic gene clusters. To this point, new CFPS systems have recently been developed from Streptomyces species and others that have shown benefits for the soluble expression of high GC-content genes, which are often involved in natural product biosynthetic gene clusters (Li et al, 2017, 2018a).
Third, our cell-free approach is fast. It requires only hours to obtain a target compound, whereas days or weeks may otherwise be needed to cultivate strains for product production. With the price of DNA synthesis declining and assembly of large gene clusters increasing, we anticipate that the ability to directly input DNA to cell-free systems for accessing biosynthetic pathways from clusters assembled using metagenomics for uncultivable organisms could enable new approaches for discovering and studying natural products. In addition, CFPS platforms are shown to work with linear PCR templates (Schinn et al., 2016; Sun et al., 2014; Wang et al., 2018), highlighting the efficiency of cell-free systems for future high-throughput synthesis without laborious cloning work. As such, cell-free biosynthesis system offers a rapid and cost-effective way to synthesize molecules of interest.
In summary, our work demonstrates in vitro total biosynthesis of a complex NRP valinomyicn by cell-free expression of the entire gene cluster. Given the robustness and flexibility of cell-free systems, the reactions can be rationally optimized to increase the biosynthesis performance and thus give rise to high productivity. Looking forward, we believe that cell-free systems hold great potential to create easy-to-use platforms for studying and engineering natural product pathways.
Acknowledgments
We thank Prof. Peter Neubauer (Technische Universität Berlin, Germany) for kindly providing plasmids pCTUT7-Vlm1 and pKS01-Vlm2. We appreciate Prof. Yong Wang, from Shanghai Institutes for Biological Sciences (Chinese Academy of Sciences, Shanghai, China), for generously sharing the strain E. coli BAP1. We also acknowledge Min Yang, Analytical Instrumentation Center at School of Physical Science and Technology, for kind help with LC-MS analysis.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31971348 and 31800720), the Natural Science Foundation of Shanghai (19ZR1477200), and the Shanghai Pujiang Program (18PJ1408000). J.L. also thanks the starting grant of ShanghaiTech University. M.C.J. acknowledges support from the National Institutes of Health Grant 1U19AI142780-01, the David and Lucile Packard Foundation, and the Camille Dreyfus Teacher-Scholar Program.
Footnotes
Conflicts of interest
The authors declare that they have no competing interests.
References
- Aigle B, Lautru S, Spiteller D, Dickschat JS, Challis GL, Leblond P, Pernodet JL, Genome mining of Streptomyces ambofaciens. J. Ind. Microbiol. Biotechnol 41, 251–263 (2014). [DOI] [PubMed] [Google Scholar]
- Balibar CJ, Howard-Jones AR, Walsh CT, Terrequinone A biosynthesis through L-tryptophan oxidation, dimerization and bisprenylation. Nat. Chem. Biol 3, 584–592 (2007). [DOI] [PubMed] [Google Scholar]
- Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY, Medema MH, Weber T, antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 47, W81–W87 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundy BC, Hunt JP, Jewett MC, Swartz JR, Wood DW, Frey DD, Rao G, Cell-free biomanufacturing. Curr. Opin. Chem. Eng 22, 177–183 (2018). [Google Scholar]
- Cheng YQ, Deciphering the biosynthetic codes for the potent anti-SARS-CoV cyclodepsipeptide valinomycin in Streptomyces tsusimaensis ATCC 15141. ChemBioChem 7, 471–477 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q, Xiang L, Izumikawa M, Meluzzi D, Moore BS, Enzymatic total synthesis of enterocin polyketides. Nat. Chem. Biol 3, 557–558 (2007). [DOI] [PubMed] [Google Scholar]
- Doroghazi JR, Albright JC, Goering AW, Ju KS, Haines RR, Tchalukov KA, Labeda DP, Kelleher NL, Metcalf WW, A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nat. Chem. Biol 10, 963–968 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley QM, Anderson KC, Jewett MC, Cell-free mixing of Escherichia coli crude extracts to prototype and rationally engineer high-titer mevalonate synthesis. ACS Synth. Biol 5, 1578–1588 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley QM, Karim AS, Jewett MC, Cell-free metabolic engineering: biomanufacturing beyond the cell. Biotechnol. J 10, 69–82 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley QM, Nash CJ, Jewett MC, Cell-free biosynthesis of limonene using enzyme-enriched Escherichia coli lysates. Synth. Biol 4, ysz003 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach MA, Walsh CT, Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: logic, machinery, and mechanisms. Chem. Rev 106, 3468–3496 (2006). [DOI] [PubMed] [Google Scholar]
- Goering AW, Li J, McClure RA, Thomson RJ, Jewett MC, Kelleher NL, In vitro reconstruction of nonribosomal peptide biosynthesis directly from DNA using cell-free protein synthesis. ACS Synth. Biol 6, 39–44 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greunke C, Glöckle A, Antosch J, Gulder TA, Biocatalytic total synthesis of ikarugamycin. Angew. Chem. Int. Ed 56, 4351–4355 (2017). [DOI] [PubMed] [Google Scholar]
- Heisey RM, Huang J, Mishra SK, Keller JE, Miller JR, Putnam AR, D’Silva TDJ, Production of valinomycin, an insecticidal antibiotic, by Streptomyces griseus var. flexipertum var. nov. J. Agric. Food Chem 36, 1283–1286 (1988). [Google Scholar]
- Jaitzig J, Li J, Süssmuth RD, Neubauer P, Reconstituted biosynthesis of the nonribosomal macrolactone antibiotic valinomycin in Escherichia coli. ACS Synth. Biol 3, 432–438 (2014). [DOI] [PubMed] [Google Scholar]
- Karim AS, Jewett MC, A cell-free framework for rapid biosynthetic pathway prototyping and enzyme discovery. Metab. Eng 36, 116–126 (2016). [DOI] [PubMed] [Google Scholar]
- Katz L, Baltz RH, Natural product discovery: past, present, and future. J. Ind. Microbiol. Biotechnol 43, 155–176 (2016). [DOI] [PubMed] [Google Scholar]
- Kay JE, Jewett MC, Lysate of engineered Escherichia coli supports high-level conversion of glucose to 2,3-butanediol. Metab. Eng 32, 133–142 (2015). [DOI] [PubMed] [Google Scholar]
- Koehn FE, Carter GT, The evolving role of natural products in drug discovery. Nat. Rev. Drug Discov 4, 206–220 (2005). [DOI] [PubMed] [Google Scholar]
- Kotowska M, Pawlik K, Roles of type II thioesterases and their application for secondary metabolite yield improvement. Appl. Microbiol. Biotechnol 98, 7735–7746 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YC, Jewett MC, High-throughput preparation methods of crude extract for robust cell-free protein synthesis. Sci. Rep 5, 8663 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambalot RH, Gehring AM, Flugel RS, Zuber P, LaCelle M, Marahiel MA, Reid R, Khosla C, Walsh CT, A new enzyme superfamily—the phosphopantetheinyl transferases. Chem. Biol 3, 923–936 (1996). [DOI] [PubMed] [Google Scholar]
- Li J, Jaitzig J, Hillig F, Süssmuth R, Neubauer P, Enhanced production of the nonribosomal peptide antibiotic valinomycin in Escherichia coli through small-scale high cell density fed-batch cultivation. Appl. Microbiol. Biotechnol 98, 591–601 (2014). [DOI] [PubMed] [Google Scholar]
- Li J, Jaitzig J, Lu P, Süssmuth RD, Neubauer P, Scale-up bioprocess development for production of the antibiotic valinomycin in Escherichia coli based on consistent fed-batch cultivations. Microb. Cell Fact 14, 83 (2015a). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Jaitzig J, Theuer L, Legala OE, Süssmuth RD, Neubauer P, Type II thioesterase improves heterologous biosynthesis of valinomycin in Escherichia coli. J. Biotechnol 193, 16–22 (2015b). [DOI] [PubMed] [Google Scholar]
- Li J, Neubauer P, Escherichia coli as a cell factory for heterologous production of nonribosomal peptides and polyketides. New Biotechnol. 31, 579–585 (2014). [DOI] [PubMed] [Google Scholar]
- Li J, Wang H, Jewett MC, Expanding the palette of Streptomyces-based cell-free protein synthesis systems with enhanced yields. Biochem. Eng. J 130, 29–33 (2018a). [Google Scholar]
- Li J, Wang H, Kwon YC, Jewett MC, Establishing a high yielding Streptomyces-based cell-free protein synthesis system. Biotechnol. Bioeng 114, 1343–1353 (2017). [DOI] [PubMed] [Google Scholar]
- Li J, Zhang L, Liu W, Cell-free synthetic biology for in vitro biosynthesis of pharmaceutical natural products. Synth. Syst. Biotechnol 3, 83–89 (2018b). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WQ, Zhang L, Chen M, Li J, Cell-free protein synthesis: recent advances in bacterial extract sources and expanded applications. Biochem. Eng. J 141, 182–189 (2019). [Google Scholar]
- Luo Y, Li BZ, Liu D, Zhang L, Chen Y, Jia B, Zeng BX, Zhao H, Yuan YJ, Engineered biosynthesis of natural products in heterologous hosts. Chem. Soc. Rev 44, 5265–5290 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magarvey NA, Ehling-Schulz M, Walsh CT, Characterization of the cereulide NRPS α-hydroxy acid specifying modules: activation of α-keto acids and chiral reduction on the assembly line. J. Am. Chem. Soc 128, 10698–10699 (2006). [DOI] [PubMed] [Google Scholar]
- Matter AM, Hoot SB, Anderson PD, Neves SS, Cheng YQ, Valinomycin biosynthetic gene cluster in Streptomyces: conservation, ecology and evolution. PLoS One 4, e7194 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman DJ, Cragg GM, Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod 79, 629–661 (2016). [DOI] [PubMed] [Google Scholar]
- Nielsen J, Cell factory engineering for improved production of natural products. Nat. Prod. Rep 36, 1233–1236 (2019). [DOI] [PubMed] [Google Scholar]
- Nielsen J, Keasling JD, Engineering cellular metabolism. Cell 164, 1185–1197 (2016). [DOI] [PubMed] [Google Scholar]
- O’Kane PT, Dudley QM, McMillan AK, Jewett MC, Mrksich M, High-throughput mapping of CoA metabolites by SAMDI-MS to optimize the cell-free biosynthesis of HMG-CoA. Sci. Adv 5, eaaw9180 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill J, Tackling drug-resistant infections globally: Final report and recommendations (Wellcome Trust, UK, 2016). [Google Scholar]
- Park CN, Lee JM, Lee D, Kim BS, Antifungal activity of valinomycin, a peptide antibiotic produced by Streptomyces sp. strain M10 antagonistic to Botrytis cinerea. J. Microbiol. Biotechnol 18, 880–840 (2008). [PubMed] [Google Scholar]
- Pfeifer BA, Admiraal SJ, Gramajo H, Cane DE, Khosla C, Biosynthesis of complex polyketides in a metabolically engineered strain of E. coli. Science 291, 1790–1792 (2001). [DOI] [PubMed] [Google Scholar]
- Quadri LEN, Weinreb PH, Lei M, Nakano MM, Zuber P, Walsh CT, Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry 37, 1585–1595 (1998). [DOI] [PubMed] [Google Scholar]
- Ryoo IJ, Park HR, Choo SJ, Hwang JH, Park YM, Bae KH, Shin-Ya K, Yoo ID, Selective cytotoxic activity of valinomycin against HT-29 human colon carcinoma cells via down-regulation of GRP78. Biol. Pharm. Bull 29, 817–820 (2006). [DOI] [PubMed] [Google Scholar]
- Schinn SM, Broadbent A, Bradley WT, Bundy BC, Protein synthesis directly from PCR: progress and applications of cell-free protein synthesis with linear DNA. N. Biotechnol 33, 480–487 (2016). [DOI] [PubMed] [Google Scholar]
- Schwarzer D, Mootz HD, Linne U, Marahiel MA, Regeneration of misprimed nonribosomal peptide synthetases by type II thioesterases. Proc. Natl. Acad. Sci. U S A 99, 14083–14088 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smanski MJ, Zhou H, Claesen J, Shen B, Fischbach MA, Voigt CA, Synthetic biology to access and expand nature’s chemical diversity. Nat. Rev. Microbiol 14, 135–149 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun ZZ, Yeung E, Hayes CA, Noireaux V, Murray RM, Linear DNA for rapid prototyping of synthetic biological circuits in an Escherichia coli based TX-TL cell-free system. ACS Synth. Biol 3, 387–397 (2014). [DOI] [PubMed] [Google Scholar]
- Swartz JR, Expanding biological applications using cell-free metabolic engineering: an overview. Metab. Eng 50, 156–172 (2018). [DOI] [PubMed] [Google Scholar]
- Tempelaars MH, Rodrigues S, Abee T, Comparative analysis of antimicrobial activities of valinomycin and cereulide, the Bacillus cereus emetic toxin. Appl. Environ. Microbiol 77, 2755–2762 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Li J, Jewett MC, Development of a Pseudomonas putida cell-free protein synthesis platform for rapid screening of gene regulatory elements. Synth. Biol 3, ysy003 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CY, Jan JT, Ma SH, Kuo CJ, Juan HF, Cheng YS, Hsu HH, Huang HC, Wu D, Brik A, Liang FS, Liu RS, Fang JM, Chen ST, Liang PH, Wong CH, Small molecules targeting severe acute respiratory syndrome human coronavirus. Proc. Natl. Acad. Sci. U S A 101, 10012–10017 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Kohli RM, Bruner SD, Walsh CT, Type II thioesterase restores activity of a NRPS module stalled with an aminoacyl-S-enzyme that cannot be elongated. ChemBioChem 5, 1290–1293 (2004). [DOI] [PubMed] [Google Scholar]
- Zhang H, Boghigian BA, Armando J, Pfeifer BA, Methods and options for the heterologous production of complex natural products. Nat. Prod. Rep 28, 125–151 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang MM, Wong FT, Wang Y, Luo S, Lim YH, Heng E, Yeo WL, Cobb RE, Enghiad B, Ang EL, Zhao H, CRISPR-Cas9 strategy for activation of silent Streptomyces biosynthetic gene clusters. Nat. Chem. Biol 13, 607–609 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]


