Abstract
Staphylococcus aureus chronic airway infection in patients with cystic fibrosis (CF) allows this pathogen to adapt over time in response to different selection pressures. We have previously shown that the main sequence types related to community-acquired methicillin-resistant S. aureus (MRSA) infections in Argentina – ST5 and ST30 – are also frequently isolated from the sputum of patients with CF, but in these patients they usually display multi-drug antimicrobial resistance. In this study, we sequenced the genomes of MRSA from four paediatric CF patients with the goal of identifying mutations among sequential isolates, especially those possibly related to antimicrobial resistance and virulence, which might contribute to the adaptation of the pathogen in the airways of patients with CF. Our results revealed genetic differences in sequential MRSA strains isolated from patients with CF in both their core and accessory genomes. Although the genetic adaptation of S. aureus was distinct in different hosts, we detected independent mutations in thyA, htrA, rpsJ and gyrA – which are known to have crucial roles in S. aureus virulence and antimicrobial resistance – in isolates recovered from multiple patients. Moreover, we identified allelic variants that were detected in all of the isolates recovered after a certain time point; these non-synonymous mutations were in genes associated with antimicrobial resistance, virulence, iron scavenging and oxidative stress resistance. In conclusion, our results provide evidence of genetic variability among sequential MRSA isolates that could be implicated in the adaptation of these strains during chronic CF airway infection.
Keywords: cystic fibrosis, microevolution , MRSA, ST5, ST30
Data Summary
Short reads for all sequenced isolates have been submitted to the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under project accession number PRJNA320874 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA320874).
Impact Statement.
Staphylococcus aureus is a commonly observed bacterium associated with cystic fibrosis (CF) lung infection. During chronic infection, S. aureus adapts in response to selective pressures and despite appropriate anti-staphylococcal therapy, it persists in the airways of these patients for long periods. Nevertheless, there is limited understanding of patterns of staphylococcal adaptation and evolution during CF pulmonary infections. We performed whole-genome sequencing of S. aureus sequential isolates recovered from four paediatric patients with CF, with the purpose of studying S. aureus intra- and inter-host variability and to identify mutations possibly related to antimicrobial resistance and virulence that might contribute to the adaptation of the pathogen in CF airways. The comparative genomics analyses among 20 CF isolates revealed genetic differences in sequential S. aureus isolates, assessing intra-host diversity. No non-synonymous mutation was shared among isolates recovered from the four patients. Moreover, in one patient, we detected mutations associated with antimicrobial resistance, virulence, iron scavenging and oxidative stress resistance in all of the isolates recovered after a certain time point that might provide evidence of convergent adaptation. Our results contribute towards a comprehensive understanding of genetic adaptation during chronic CF infection of airways.
Introduction
Cystic fibrosis (CF) is an inherited disease that primarily affects the lungs. The morbidity and mortality in patients with CF is associated with pulmonary failure caused by lung damage after recurrent pulmonary infections. Staphylococcus aureus is a commonly observed bacterium associated with CF lung infection and it is one of the first pathogens isolated from the airways of patients with CF [1]. Despite appropriate anti-staphylococcal therapy, S. aureus persists in the airways of these patients for months or even years [2]. During long-term infection, S. aureus is exposed to different selective pressures, such as numerous long-term repeated antibiotic treatments, dynamic adaptation of the host immune system, co-infection with other species and different oxygen levels that may select for adapted pathogens to this environment [3].
Genome plasticity allows S. aureus to undergo adaptations that contribute to its long-term persistence in the CF airways [4, 5]. Different adaptive strategies of S. aureus include acquisition of antimicrobial resistance (AMR), biofilm formation, deregulation of virulence genes and the appearance of persistent phenotypes [3, 5–8]. Whole-genome sequencing (WGS) has proven to be a very useful tool to track the microevolution of pathogens during their interactions with the host and to reconstruct the evolutionary events shaping populations [9–11].
We have previously shown that the main sequence types related to community-acquired methicillin-resistant S. aureus (MRSA) infections in Argentina – ST5 and ST30 – are also frequently isolated from the sputum of patients with CF, but in these patients they usually display multi-drug AMR [12]. In this study, we used WGS to investigate S. aureus intra- and inter-host variability in four paediatric CF patients and to identify mutations among sequential MRSA isolates present in CF chronic infection, especially those possibly related to AMR and virulence that might contribute to the adaptation of the pathogen in the airways of patients with CF.
Methods
Bacterial strains
The serial isolates analysed in this study were collected over two time periods (June 2012–May 2013 [12] and June 2014–November 2015) from respiratory samples obtained from four different paediatric patients with CF attending the Respiratory Center at the Hospital de Niños ‘Dr Ricardo Gutiérrez’. The Respiratory Center is a national reference centre in Buenos Aires for the treatment of patients with CF and has the largest current enrollment of patients with CF in Argentina. One colony was obtained from each sample unless differences in morphology, i.e. size and/or pigmentation, appeared in the same sample. Isolates included those recovered during severe pulmonary exacerbations as well as from clinically stable periods (routine visits). Briefly, S. aureus isolates were sub-cultured in brain heart infusion (BHI) agar and Columbia blood agar for small-colony variants (SCV), from primary cultures and incubated for 24–48 h at 37 °C. Isolates were frozen at −80 °C in BHI+20 % glycerol before further characterization.
Antimicrobial susceptibility testing
The minimal inhibitory concentrations (MICs) of oxacillin, cefoxitin, gentamicin, erythromycin, clindamycin, minocycline, tetracycline, trimethoprim/sulfamethoxazole, ciprofloxacin, rifampicin, vancomycin and linezolid were determined by the agar dilution procedure according to Clinical and Laboratory Standards Institute (CLSI) recommendations and interpreted based on the same guidelines [13]. For SCVs that were unable to grow on Mueller–Hinton agar either with or without 5 % sheep blood, Columbia blood agar was used instead. The MIC for tigecycline was determined by the E-test epsilometric strip method (bioMérieux, Montreal, QC, Canada) and MIC results were interpreted based on the 2017 European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoints [14].
Growth rate measurements
Growth studies for all isolates were performed in triplicate by inoculating BHI broth with an overnight BHI culture (dilution 1/1000) and incubated at 37 °C and 180 r.p.m. The optical density (OD) at 620 nm was measured every 30 min using a BioTek Synergy 2 plate reader (BioTek Instruments, Inc., Winooski, VT, USA). Growth curves were constructed plotting the OD variations over time. The growth rates were compared statistically through slope analysis by linear regression using GraphPad Prism 6.0. The significance level was set at P <0.05.
DNA extraction
Total bacterial DNA was extracted from overnight cultures using the MasterPure Complete DNA and RNA Purification kit (Epicenter, Madison, WI, USA) according to the manufacturer’s instructions. During the cell lysis step, 30 min incubation with lysostaphin (0.03 µg µl−1) was administered for regular colony-forming S. aureus , whereas up to 1 h of incubation with lysostaphin (0.09 µg µl−1) was necessary to obtain a clear lysate for SCV.
Genotyping
Molecular typing of the isolates was initially performed using pulsed-field gel electrophoresis (PFGE) [15], spa typing [16] and multilocus sequence typing (MLST) [17]. Characterization of the staphylococcal cassette chromosome mec (SCCmec) element and the detection of Panton–Valentine leukocidin (PVL) coding genes were carried out as described previously [16, 18].
Library preparation and whole-genome sequencing
Genome DNA sequencing of 20 S. aureus isolates was performed at the National Microbiology Laboratory (Winnipeg, Canada) using the Illumina MiSeq platform. Sequencing libraries were constructed using the Illumina TruSeq Nano DNA HT Sample Preparation kit. Paired-end (2×300 bp) sequencing was performed using the 600-cycle sequencing format kit (MiSeq reagent kit v.3), yielding an average of 1.6 million reads per genome and an average genome coverage of 170× (range: 61–505×).
Phylogenomic analyses
To analyse the phylogenomic relationships among the isolates, a tree based on single-nucleotide variants (SNVs) in the core genome was constructed using SNVPhyl pipeline [19]. In the first step of the pipeline, sequenced reads were aligned to the reference genome of S. aureus strain MRSA252 (ST36) belonging to clonal complex 30 (CC30) (GenBank accession #BX571856) [20]. In addition, to identify SNVs and to construct a distance matrix between isolates recovered from each patient, reads were mapped to the first isolate recovered from each patient. Only high-quality SNVs were retained to build the tree (minimum 10× coverage, minimum mean mapping quality score of 30, minimum of 75 % of mapped reads supporting the SNV). Insertions/deletions (indels) were identified using Snippy [21]. Annotation of SNVs was performed using SNPEff (v4.2) [22]. Manual inspection of SNVs and indels, and the analysis of the relative abundance of SNVs in those genes with more than one copy across the chromosome, were performed using Integrative Genomics Viewer (IGV) [23, 24].
Maximum-likelihood (ML) trees were constructed based on total SNVphyl-generated SNVs, using PHyML [25] with the generalized time-reversible (GTR)+γ model as default and tree support values were estimated using PhyML’s approximate likelihood ratio test. Variants occurring in regions of high SNV density (2 per 500 bp sliding window) were excluded from the phylogenetic tree construction.
Genome assembly and annotation
Sequenced reads were de novo assembled using SPAdes (v1.2) [26]. The assembled draft genomes yielded average total genome sizes of 2.8 and 2.7 Mb for ST30 and ST5 isolates, respectively. The average number of contigs that were > 1 kb was 30 (28–33) and 21 (19–28) for ST30 and ST5 genomes, respectively. The average N50 contig length was 222 086 bp (ST30) and 444 852 bp (ST5). Table S1 (available in the online version of this article) adds further information regarding the metrics of the assemblies. Gene annotation was performed using Prokka (v1.4.0) [27].
Pangenome analysis and detection of AMR determinants and mobile genetic elements
Pangenome was determined with Roary [28] and visualized with Phandango [29]. To seek AMR determinants, contigs were analysed using ARIBA [30] with customized databases plus three publicly available databases: the Comprehensive Antibiotic Resistance Database (CARD) [31], RESFinder [32] and ARG-ANNOT [33].
NCBI blastn [34] and ISseeker [35] were used to find possible insertion sites for different staphylococcal insertion sequences (ISs). Plasmids were detected from assemblies using the PlasmidFinder database [36] and bacteriophages were identified using PHASTER [37]. IslandViewer [38] and ARIBA were used for the detection of S. aureus pathogenicity islands (SAPIs) using a custom database including known SAPI-associated integrases.
All of the bioinformatics software used for this study was run using default parameters.
Results
Patients and isolates main features
To analyse the intra- and inter-patient diversity of S. aureus in CF, we selected sequential isolates from four paediatric patients with CF, who were followed at the Respiratory Center of the paediatric hospital ‘Hospital de Niños Dr Ricardo Gutierrez’. The median age for the patients at the first-time report of positive MRSA culture during the study period was 7.3 years (3.0–9.3 years). All patients, except patient 2, were chronically infected with MRSA (≥3 positive consecutive cultures in 6 months) and had established MRSA colonization since 2008. Patient 2’s isolate RGP2.1 was first-time MRSA positive. Patient demographics and their clinical courses are outlined in Table S2. A minimum of three and maximum of eight isolates were obtained from each patient. Two morphotypes each were isolated from the same sample for patients 1 (RGP1.1 and RGP1.2) and 3 (RGP3.3 and RGP3.4). Sequential isolates from each patient were undistinguishable/closely related [39] by PFGE and three out of four patients presented differences in their AMR pattern (Table 1). Isolates recovered from patients 1 and 2 belong to sequence type ST30; isolates from patients 3 and 4 belong to sequence type ST5. Collection periods varied from 52 days to 40 months. Additionally, when comparing the growth rates for all the sequential isolates, the values from the first and the last isolate were significantly different for 3/4 patients (Fig. S1, Table S3).
Table 1.
Genotyping and antimicrobial resistance profiles of sequential S. aureus isolates
|
Isolate ID |
Patient |
Genotyping |
SCV |
MIC (mg l−1) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
FOX |
OXA |
GEN |
CIP |
ERY |
CLI |
LZD |
MIN |
TET |
TGC |
RIF |
SXT |
VAN |
TEC |
||||
|
RGP1.1 |
1 |
ST30-IV-t019-PVL+ |
No |
8 |
8 |
>512 |
8 |
16 |
0.25 |
1 |
0.5 |
1 |
nd |
0.008 |
nd |
1 |
1 |
|
RGP1.2 |
1 |
ST30-IV-t019-PVL+ |
Yes |
8 |
8 |
>512 |
8 |
16 |
0.25 |
1 |
0.5 |
1 |
nd |
0.008 |
nd |
1 |
1 |
|
RGP1.3 |
1 |
ST30-IV-t019-PVL- |
Yes |
8 |
4 |
>512 |
8 |
16 |
0.25 |
1 |
0.5 |
1 |
nd |
0.008 |
nd |
1 |
1 |
|
RGP2.1 |
2 |
ST30-IV-t019-PVL+ |
No |
16 |
32 |
0.5 |
0.5 |
0.5 |
≤0.063 |
4 |
≤0.125 |
0.25 |
nd |
0.008 |
0.125 |
1 |
1 |
|
RGP2.2 |
2 |
ST30-IV-t019-PVL+ |
No |
16 |
32 |
0.5 |
0.5 |
0.5 |
≤0.063 |
4 |
≤0.125 |
0.25 |
nd |
0.004 |
0.125 |
1 |
1 |
|
RGP2.3 |
2 |
ST30-IV-t019-PVL+ |
No |
16 |
32 |
0.5 |
1 |
0.5 |
0.125 |
4 |
≤0.125 |
0.25 |
nd |
0.008 |
0.125 |
1 |
4 |
|
RGP3.1 |
3 |
ST5-IV-t311-PVL- |
No |
>32 |
>32 |
0.25 |
1 |
256 |
≤0.063 |
2 |
1 |
2 |
0.25 |
0.008 |
0.125 |
1 |
1 |
|
RGP3.2 |
3 |
ST5-IV-t311-PVL- |
No |
>32 |
>32 |
32 |
1 |
256 |
≤0.063 |
2 |
2 |
4 |
0.75 |
0.008 |
0.125 |
1 |
1 |
|
RGP3.3 |
3 |
ST5-IV-t311-PVL- |
No |
>32 |
32 |
16 |
1 |
32 |
≤0.063 |
4 |
1 |
2 |
0.5 |
0.008 |
0.125 |
2 |
2 |
|
RGP3.4 |
3 |
ST5-IV-t311-PVL- |
No |
>32 |
>32 |
16 |
2 |
256 |
0.125 |
16 |
2 |
4 |
1 |
0.008 |
0.125 |
2 |
1 |
|
RGP3.5 |
3 |
ST5-IV-t311-PVL- |
No |
>32 |
>32 |
32 |
2 |
256 |
0.125 |
16 |
2 |
4 |
1 |
0.008 |
≤0.063 |
1 |
1 |
|
RGP3.6 |
3 |
ST5-IV-t311-PVL- |
No |
16 |
16 |
16 |
1 |
256 |
≤0.063 |
8 |
2 |
4 |
1 |
0.008 |
0.125 |
1 |
0.5 |
|
RGP3.7 |
3 |
ST5-IV-t311-PVL- |
No |
>32 |
>32 |
16 |
4 |
256 |
0.125 |
16 |
2 |
4 |
1.5 |
0.008 |
≤0.063 |
1 |
1 |
|
RGP3.8 |
3 |
ST5-IV-t311-PVL- |
No |
>32 |
>32 |
32 |
2 |
128 |
0.125 |
8 |
2 |
4 |
2 |
0.004 |
0.125 |
1 |
1 |
|
RGP4.1 |
4 |
ST5-IV-t311-PVL+ |
No |
16 |
32 |
0.5 |
16 |
>512 |
MLSi ≤0.063 |
2 |
≤0.125 |
0.25 |
nd |
0.008 |
≤0.063 |
1 |
1 |
|
RGP4.2 |
4 |
ST5-IV-t311-PVL+ |
No |
16 |
32 |
0.5 |
16 |
>512 |
MLSi ≤0.063 |
2 |
≤0.125 |
0.25 |
nd |
0.008 |
≤0.063 |
1 |
1 |
|
RGP4.3 |
4 |
ST5-IV-t311-PVL+ |
No |
16 |
32 |
0.5 |
16 |
>512 |
MLSi ≤0.063 |
1 |
≤0.125 |
0.25 |
nd |
0.008 |
0.125 |
1 |
1 |
|
RGP4.4 |
4 |
ST5-IV-t311-PVL+ |
No |
16 |
>32 |
0.5 |
16 |
128 |
MLSi ≤0.063 |
2 |
≤0.125 |
0.25 |
nd |
0.008 |
≤0.063 |
1 |
0.5 |
|
RGP4.5 |
4 |
ST5-IV-t311-PVL+ |
Yes |
16 |
16 |
64 |
16 |
>512 |
MLSi 0.125 |
2 |
0.5 |
2 |
nd |
0.008 |
nd |
1 |
1 |
|
RGP4.6 |
4 |
ST5-IV-t311-PVL- |
Yes |
16 |
8 |
>512 |
16 |
32 |
MLSi 0.125 |
2 |
2 |
4 |
nd |
0.008 |
nd |
0.5 |
≤0.5 |
SCV, small colony variant; nd, not determined; FOX: cefoxitin; OXA: oxacillin; CIP, ciprofloxacin; CLI, clindamycin; ERY, erythromycin; GEN, gentamicin; LZD, linezolid; MIN, minocycline; RIF, rifampin; SXT, trimethoprim/sulfamethoxazole; TEC, teicoplanin; TGC, tigecycline; TOB, tobramycin; VAN, vancomycin. MLSi, macrolide, lincosamide and streptogramin inducible resistance phenotype.
Phylogenomic relatedness and genetic differences in CF isolates
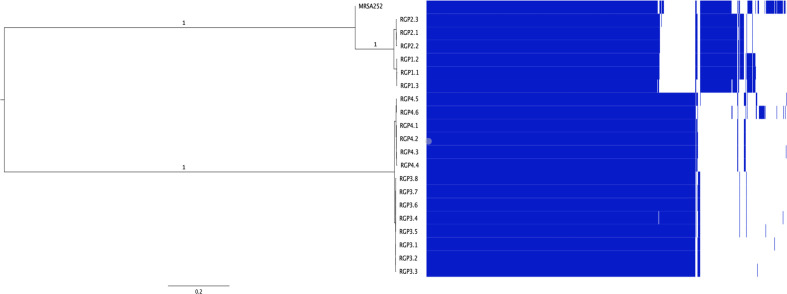
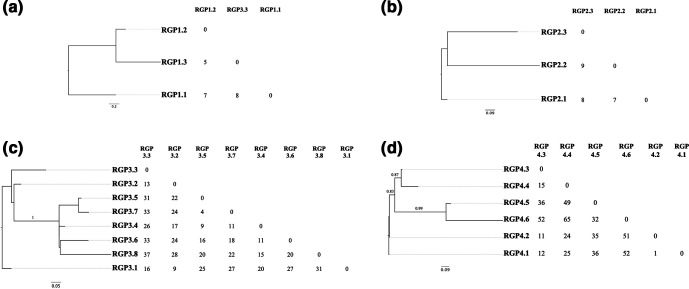
To study the within-host diversity of S. aureus populations arising in CF airways, the SNV-based core-genome phylogenetic relatedness of sequenced isolates was determined. Whole-genome phylogenetic analysis grouped isolates into two distinct clades consistent with PFGE and MLST data: ST5 (patients 1 and 2) and ST30 isolates (patients 3 and 4). Both clades were separated by more than 2000 SNVs in the core genome and additional differences in the accessory genome (Fig. 1). To maximize the size of the core genome analysed, we examined SNVs using the first isolate recovered from each patient as a reference. Despite the isolates belonging to the same ST, the tree topology reveals heterogeneity among sequential CF isolates and the number of SNVs between the isolates and the reference genome increased with time, except in patient 2 (Fig. 2).
Fig. 1.
Maximum-likelihood phylogenetic tree and the pangenome of isolates from patients with CF. Comparison between the tree and gene content (pangenome) among all the isolates. The pangenome is represented as a matrix with the presence (blue blocks)/absence (white blocks) of core and accessory genes. The tree was based on the alignment of all genomes against the MRSA252 (ST36), using 87.71 % of the reference as the core genome (defined as the percentage of the reference present in all the isolates). Branch support values were estimated using PhyML’s approximate likelihood ratio test (aLRT).
Fig. 2.
Maximum-likelihood trees based on core genome SNVs and pairwise SNV comparisons. The matrices exclude SNVs occurring in regions of high SNV density. (a) Isolates recovered from patient 1 using a 97.02 % core genome and RGP1.1 as the reference. (b) Isolates recovered from patient 2 using a 98.72 % core genome and RGP2.1 as the reference. (c) Isolates recovered from patient 3 using a 98.74 % core genome and RGP3.1 as the reference. (d) Isolates recovered from patient 4 using a 98.06 % core genome and RGP4.1 as the reference. Branch support values were estimated using PhyML’s approximate likelihood ratio test (aLRT).
Table 2 summarizes the genetic differences found between the first and the last isolate recovered from each patient considering all SNVs, including SNVs in high SNV-density regions. The differences in the genomes of the isolates belonging to ST5 and ST30 are mainly in the accessory genome (see the Mobile genetic elements section and Tables S1 and S4).
Table 2.
Total SNVs and indels observed between the first and last isolate recovered from each CF patient without SNVphyl’s density filtering
|
First/Last isolate |
Patient 1 RGP1.1/RGP1.3 |
Patient 2 RGP2.1/RGP2.3 |
Patient 3 RGP3.1/RGP3.8 |
Patient 4 RGP4.1/RGP4.6 |
|---|---|---|---|---|
|
Total SNVs |
8 |
4 |
32 |
75 |
|
Synonymous |
2 |
1 |
5 |
19 |
|
Non-synonymous |
3 |
1 |
22 |
40 |
|
Stop generated |
0 |
0 |
1 |
0 |
|
Stop lost |
1 |
0 |
0 |
0 |
|
Intergenic |
2 |
2 |
4 |
16 |
|
Total indels |
2 |
1 |
17 |
21 |
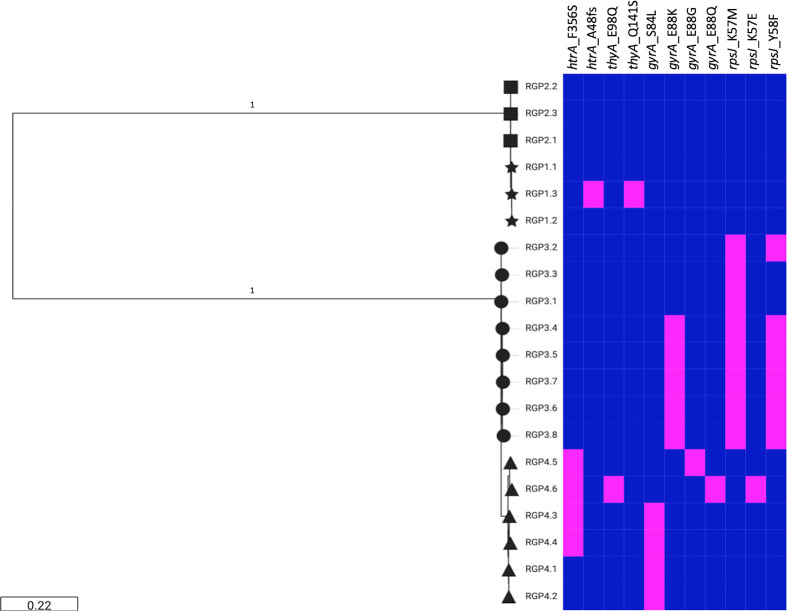
The total numbers of SNVs and indels present between the first and the last isolate recovered from each patient are presented in Table S5. Genetic variants were found in genes associated with different functions, including AMR, global regulation, virulence and metabolism, with AMR and virulence genes harbouring higher numbers of mutations. No non-synonymous mutations were shared between the isolates recovered from all four patients. Interestingly, some genes contained variations in isolates recovered from more than one patient, including htrA (encoding the HtrA surface protease), thyA (encoding thymidylate synthase), rpsJ (encoding 30S ribosomal S10 protein) and gyrA (encoding DNA gyrase subunit A) (Fig. 3).
Fig. 3.
Virulence and AMR genes that contained mutations in isolates recovered from more than one patient with CF. Midpoint-rooted SNV-based maximum-likelihood tree. The presence (magenta) or absence (blue) of each mutation is shown for patient 1 (stars), patient 2 (squares), patient 3 (circles) and patient 4 (triangles).
Genetic diversity in ST5 MRSA sequential isolates associated with AMR differences and virulence
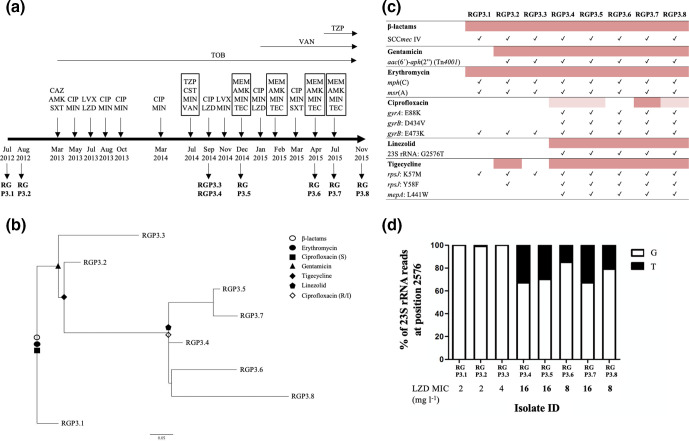
We found heterogeneity in AMR among isolates recovered from patients 3 and 4 (Table 1). Analysing S. aureus ’s population dynamics in patient 3, the two isolates recovered on September 2014 (RGP3.3 and RGP3.4) with different colony morphology also harboured different AMR profiles (Fig. 4). These isolates differed by 51 total SNVs and 4 indels. The sequential isolates recovered from this patient displayed a well-resolved phylogeny with non-synonymous mutations occurring on internal branches that could be associated with the acquisition of resistance to different antimicrobials (Fig. 4b). Remarkably, isolates recovered after September 2014 grouped with RGP3.4 and shared additional mutations not related to AMR that were present in all of the isolates recovered after that time point (Table 3). Fig. 4c, d show possible phenotypic and genotypic correlations for isolates recovered from patient 3.
Fig. 4.
Isolates recovered from patient 3: genetic changes related to AMR. (a) Antibiotic therapy received by the patient during 2012–2015 and MRSA isolation dates (modified from Haim et al. [62]). Antimicrobial therapy (oral, inhaled and intravenous) is represented above the timeline. Intravenous therapies appear framed and inhaled therapies are illustrated above horizontal arrows (if more than one inhaled antibiotic is prescribed, treatment consists of alternating the antibiotics every month). AMK, amikacin; CAZ, ceftazidime; CIP, ciprofloxacin; CST, colistin; LVX, levofloxacin; LZD, linezolid; MEM, meropenem; MIN, minocycline; SXT, trimethoprim/sulfamethoxazole; TEC, teicoplanin; TOB, tobramycin; TZP, piperacillin/tazobactam; VAN, vancomycin. (b) Maximum-likelihood tree based on SNV variations. Changes in AMR profiles are represented by different shapes. (c) AMR determinants related to resistance phenotype in patient 3 showing possible phenotypic and genotypic correlations. Resistant isolates are highlighted in dark pink and intermediate isolates in pale pink. Check marks represent the presence of the genetic feature. (d) Relative abundance of the G2576T mutation in 23S rRNA gene copies. White represents a G in that position (wild-type) and black a T. LZD MIC values in bold represent a resistant phenotype (2017 CLSI breakpoints for linezolid, S: ≤4 µg ml−1; R: ≥8 µg ml−1).
Table 3.
Mutations present in RGP3.4 and consecutive isolates recovered after September 2014 (patient 3)
|
Locus_tag |
Gene |
Product |
Mutation type |
Effect |
|---|---|---|---|---|
|
RGP3.1_00109 |
SA1292 |
Nucleoside triphosphate pyrophosphohydrolase, MazG |
Ins 1 bp |
Frameshift variant |
|
RGP3.1_00210 |
gyrB |
DNA topoisomerase subunit B |
SNV |
Missense variant: D432V |
|
RGP3.1_00449 |
SA0966 |
Hypothetical protein |
SNV |
Missense variant: I59N |
|
RGP3.1_00481 |
ptsA |
Phosphoenolpyruvate–protein phosphotransferase |
SNV |
Missense variant: V269A |
|
RGP3.1_00533 |
SA0886 |
Bacteriocin-associated integral membrane (putative immunity) protein |
SNV |
Missense variant: A189S |
|
RGP3.1_00573 |
opp-4A |
Oligopeptide ABC transporter substrate-binding protein |
SNV |
Stop gained |
|
RGP3.1_00697 |
gyrA |
DNA topoisomerase (ATP-hydrolyzing) subunit A, GyrA |
SNV |
Missense variant: E88K |
|
RGP3.1_2 : 148 268 |
|
Intergenic between acsA2 and mqo2 |
SNV |
|
|
RGP3.1_01053 |
hssR |
Winged helix family two-component transcriptional regulator |
SNV |
Missense variant: I109F |
|
RGP3.1_01160 |
rpsJ |
30S ribosomal protein S10 |
SNV |
Missense variant: Y58F |
|
RGP3.1_01198 |
|
Hypothetical protein |
Del 1 bp |
Frameshift variant |
|
RGP3.1_01453 |
sstA |
Iron (Fe3+) ABC superfamily ATP-binding cassette transporter, membrane protein |
SNV |
Missense variant: L18S |
|
RGP3.1_01473 |
secA |
Sec family type I general secretory pathway preprotein translocase subunit SecA |
SNV |
Synonymous variant |
|
RGP3.1_01576 |
mepA |
MATE family multi-antimicrobial extrusion protein |
SNV |
Missense variant: L441W |
|
RGP3.1_4 : 142 745 |
|
Intergenic between lytR and lrgA |
Del 1 bp |
|
|
RGP3.1_02037 |
argE |
Succinyl-diaminopimelate desuccinylase |
Del 8 bp |
Frameshift variant |
|
RGP3.1_6 : 132 865 |
|
Intergenic between groEL and trkG |
Ins 2 bp |
|
|
RGP3.1_02295 |
pdp |
Putative pyrimidine nucleoside phosphorylase |
Del 2 bp |
Frameshift variant |
|
RGP3.1_02339 |
hemY |
Protoporphyrinogen oxidase |
SNV |
Missense variant: M393I |
Similarly, resistance to aminoglycosides in the last two isolates recovered from patient 4 was associated with the presence of aac(6′)-aph(2′), which codes for a bifunctional aminoglycoside-modifying enzyme (acetyltransferase and phosphotransferase) and is harboured in Tn4001 transposon. Fig. S2 shows the AMR mechanisms found in all isolates recovered from patient 4.
Other mutations found that might contribute to the adaptation of S. aureus to the CF airways were related to proteins involved in iron utilization. In patient 3, all the isolates recovered after RGP3.1 had a non-synonymous mutation in a siderophore coded by sbnC (Table S6). RGP3.4 and the subsequent isolates presented non-synonymous mutations in sstA and hssR. Moreover, mutations in the haptoglobin-binding surface anchored protein IsdH/HarA were found in the last two isolates recovered from patient 4.
Mutations in genes related to oxidative stress resistance were found in isolates recovered from patients 3 and 4. In patient 3, an S10P mutation in AhpC, a peroxide-reducing protein, was found in RGP3.2 and subsequent isolates, whereas in the last two isolates of patient 4, a non-synonymous mutation was found in the perR gene, which codes for a peroxide-sensing protein. Moreover, the HtrA surface protease, which is involved in the virulence of many pathogens due to its roles in survival and stress resistance [40–42], was mutated in isolates recovered from patients 1 and 4.
Mobile genetic elements
A plasmid of approximately 20 kb carrying the β-lactamase gene blaZ and genes associated with cadmium resistance, cadD and cadX (plasmid replicons rep16, rep19 and rep21) [36], was identified in three isolates from patients 1 and 2. We also identified a ~20 kb plasmid in all isolates recovered from patient 3 (rep16, rep19 and rep20). This plasmid carried genes coding for erythromycin resistance msr(A) and mph(C), multidrug efflux pump qacA, and with the exception of the plasmid found in RGP3.1, aminoglycoside resistance aac(6´)-aph(2′), with their presence correlating with the AMR profile of isolates. Regarding the mobilization of this plasmid, no mating-pore genes (tra genes) were found when mapping sequenced reads to reference tra sequences. Moreover, all isolates from patient 4 carried a plasmid of approximately 2.6 kb carrying the MLS resistance gene ermC (rep20). Table S4 lists the different mobile genetic elements found in all the isolates.
Differences in IS content were detected between some of the sequential isolates (Table S7, Fig. S3). Widely distributed in S. aureus , IS256 was found in at least one isolate from each patient except for patient 2. Moreover, the composite transposon Tn4001 was found in all patient 3 isolates except for RGP3.1 (Fig. 4c). The last two isolates recovered from patient 4 harboured a Tn4001–IS257 hybrid structure [43] that was absent in other isolates recovered from this patient.
Identification of phage regions revealed differences in the phage content in isolates recovered from two patients. Specifically, in patient 1, a PVL-coding phage similar to phiPVL-CN125 (NC_012784) was present in RGP1.1 and RGP1.2, but was absent in RGP1.3. In patient 4, a phage similar to phiSa119 (NC_025460) was present in all except the last (RGP4.6) isolate recovered from this patient. RGP4.6 had an additional 41.6 kb phage, similar to phiJB (NC_028669), that was absent in the rest of the isolates recovered from this patient (Fig. S4).
Using ARIBA and a customized database containing different SAPI integrases, we detected intSaPI3-like that shared 96 % identity with intSaPI3 from the S. aureus COL reference sequence in all the isolates belonging to ST30, whereas no SAPI integrases were found in ST5 isolates.
Discussion
Despite the high prevalence of S. aureus in CF [44], there is limited understanding of S. aureus genomic epidemiology and intra-host evolution. In this study, comparative genomics of 20 MRSA isolates recovered from four paediatric patients revealed genetic variation among them, in both the core and the accessory genome, that could affect the way in which bacteria persist in the CF airways and how they respond to antimicrobial treatment.
Our study is focused on young children with CF [median age 7.3 years (3.0–9.3 years)]. This population has unique features, including more diverse airway bacterial communities associated with lower markers of airway inflammation (i.e. BALF total cell count, absolute neutrophil count and percentage of neutrophils), lower cumulative lifetime exposure to antibiotics and increased lung function compared with older children/adults with CF [45, 46]. Still, host-specific evolutionary pressures in paediatric patients and the genetic background of the infecting pathogen have been shown to affect initial adaptation of other bacterial species, such as Pseudomonas aeruginosa , within 3 years of colonization through distinct evolutionary modes [47].
Core SNV-based phylogenetic analysis grouped isolates from each patient in different clusters and genetic differences were observed among isolates recovered from the same subject, defining subject-specific genetic fingerprints. As described previously [48], pairwise SNV distances generally increased with time separating the collection date of isolates. No common mutation was acquired independently by the isolates recovered from all four patients, suggesting that, in this study, there is no specific mutation that could be broadly associated with all CF S. aureus isolates. These results are in line with the idea of patient-specific evolution as a reflection of spontaneous mutations followed by their selection by patient-specific selective pressures.
However, non-synonymous mutations were detected in the same gene in multiple patients, which might contribute to similar phenotypes. Those genes included S. aureus virulence-associated genes thyA and htrA, and tigecycline and quinolones resistance genes rpsJ and gyrA, respectively [42, 49]. Several intra-host studies have studied convergent evolution events among loci across multiple patients in S. aureus and other pathogens associated with CF [48, 50, 51]. Nonetheless, the mutations described herein were different from those reported by Azarian et al. Mutations in these genes, involved in virulence and AMR, should be further studied with regard to the persistence of S. aureus in CF airways.
High phenotypic and/or genotypic diversity, including antimicrobial susceptibility, among S. aureus sequential isolates recovered from patients with CF as well as among variants from the same sputum sample have been described previously [5, 6, 48, 52, 53]. Even though establishing relationships between mutations and the related phenotypic change in resistance was not the aim of this study, differences in AMR profiles among isolates have been associated with previously reported point mutations in the target genes of the antimicrobials and the acquisition of Tn4001 and Tn4001-like elements [43]. In agreement with the findings of Rouard et al. [53], the level of resistance to linezolid in isolates recovered from patient 3 showed a possible correlation with the percentage of reads harbouring the G2576T mutation in the 23S rRNA gene, and thus with the number of 23S rRNA gene copies carrying this mutation. In addition to the differences in AMR profiles, we also detected decreases in the growth rates of sequential isolates in three out of four patients, when compared to the first isolate (Fig. S1, Table S3). However, due to the numerous selective pressures acting in CF lungs that could influence bacterial fitness we are cautious about the interpretation of these results, which require deeper analysis.
Previous whole-genome sequencing studies on bacterial adaptation have provided strong evidence for the coexistence of different lineages in the CF airways: significant intra-host diversity between pairs of isolates collected on the same day [48] and mutations did not become fixed in the bacterial population, i.e. there were different variants for a particular gene throughout the population [54]. High diversity was also reported for S. aureus in other chronic infections [10]. In our study, differences in phenotypic characteristics (AMR patterns and morphology) and SNVs among isolates recovered on the same day suggest the possibility of the coexistence of different lineages. Furthermore, the finding that most polymorphisms among sequential S. aureus were not shared among all the isolates recovered from each patient are in line with the idea of multiple beneficial genotypes coexisting, making it difficult for one of them to reach fixation and thus enhancing the survival of the bacterial community [55, 56]. This strategy is of special importance in the airways of patients with CF, where S. aureus is subjected to different selective pressures resulting from the host immune system, numerous medical interventions, various co-infecting species and different oxygen pressure levels.
Nonetheless, particularly in isolates recovered from patient 3, we identified polymorphisms at internal branches of the phylogeny in genes associated with AMR, virulence, iron scavenging and oxidative stress resistance that were present in several isolate genomes from this patient (Table 3). Among them, we identified a frameshift mutation in a pyrophosphatase gene, known to contribute to S. aureus virulence in a silkworm model [57], and non-synonymous mutations in rpsJ, gyrA, gyrB and mepA as well as mutations in the 23S rRNA gene, all contributing to AMR. Even though the sampling method (one colony per sample) and the short period of time of the current study does not allow us to make any assumptions about the emergence of a persistent lineage, we believe that polymorphisms appearing at internal branches of the phylogeny might support the hypothesis of specific genetic variants that may be present at any given time in patient 3, as these SNVs seem to be stable in the population.
It has been difficult to pinpoint overarching drivers of adaptation in the complex niche of the lungs of patients with CF where multiple regulatory networks exist. Still, antimicrobial treatment has been considered one of the main selective pressures shaping lung microbial diversity in these patients and some of the phenotypic and genotypic patterns observed in this study may have been influenced by the treatments received by each patient (Table S2).
In addition to clonally acquired point mutations, horizontal gene transfer represents another key mechanism of S. aureus evolution [58] and in the CF context it was proven to be the major adaptation mechanism driven by inter-bacterial competition [59]. In agreement with previous findings, comparative genomic analysis showed differences in phage content (Fig. S4) [4, 5], in the number of copies and in the location sites of IS256 [60] and IS1811 (Fig. S3) among isolates recovered from two different patients. Cured S. aureus strains lacking the phage carrying the PVL and the sea enterotoxin genes showed reduced superantigenic responses and were significantly less cytotoxic to neutrophils than the wild-type [61]. Further studies should be performed to evaluate the impact of these ISs in-between genes such as walR, scn and chs.
Two limitations of our study are the small sample size and the analysis of only one colony from each clinical specimen at each different time point. Even though several studies that analyse intra-host evolution of MRSA in the setting of CF have used the one-colony-per-sample method [5, 53, 59], it underestimates the heterogeneity of S. aureus in the CF airways at each sampling moment, as we now know that host-adapted bacterial populations are not homogeneous. Studies that analyse multiple isolates from a single sample have shown great diversity among them and represent a more appropriate way to study the complexity of the adaptation of CF pathogens thoroughly [48]. Nonetheless, our results show genetic variations that might contribute to the adaptation of S. aureus in CF airways.
Finally, this study highlights the power of high-throughput sequencing in tracking genomic variants of bacterial pathogens during the infection in their human host. Our comparative genomic analyses shed light on the significant genomic differences found among S. aureus sequential isolates recovered from the airways of four CF patients, working towards a comprehensive understanding of genetic adaptation during pathogenesis. Further investigation will be required to understand the potential roles of some of the genetic variations identified in the present study and how they impact on antibiotic resistance, fitness and other aspects of pathogenesis.
Supplementary Data
Funding information
This work was supported in part by grants from University of Buenos Aires, Argentina (UBACYT 20020130100381BA), Agencia Nacional de Promoción Científica y Tecnológica (PICT 2012 2362) and Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) PIP 11220150100694CO to MM; the Public Health Agency of Canada; and University of Manitoba, Faculty of Science Interdisciplinary / New Directions Research Collaboration Initiation Grant and Discovery Grant No 684104 from the Natural Sciences and Engineering Research Council of Canada awarded to S.T.C. A travel award (ELAP scholarship) to MSH was granted by the Department of Foreign Affairs, Trade and Development (DFATD) of Canada.
Acknowledgements
The authors thank NML Genomics Core staff for their help and knowledge of NGS as well as the NML Bioinformatics Core for the development of the necessary bioinformatics tools for data analysis. We would also like to thank Anna Motnenko for technical support.
Author contributions
L. G. and S. L., collected the samples and clinical data. M. S. H. performed phenotypic and genotyping assays. M. S. H., M. M., G. V. D., M. G., G. G., S. C., R. Z. and A. R. critically designed the sequencing project and actively discussed results. M. G. and G. V. D. generated sequencing data. M. S. H. performed bioinformatics analyses and wrote the manuscript. R. Z. and A. B. contributed to bioinformatics analyses. S. D.G. and J. D. C. participated in the discussion of results and made an intellectual contribution. M. M. conceived the main idea and edited the manuscript. All authors reviewed, edited and approved the final version of the manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
The study was reviewed and approved by the Research Ethics Committee of the Hospital de Niños ‘Dr Ricardo Gutiérrez’ (CEI no. 16.10) and by the Committee of Ethics in Clinical Research (CEIC) of the Faculty of Pharmacy and Biochemistry, Universidad de Buenos Aires ((CD) no. 1463/16). Both ethics committees agreed that patient parental consent was not required as the samples were collected as part of routine procedures.
Footnotes
Abbreviations: AMR, antimicrobial resistance; BALF, bronchoalveolar lavage fluid; BHI, brain heart infusion; CC, clonal complex; CF, cystic fibrosis; indels, insertions / deletions; IS, insertion sequence; MIC, minimal inhibitory concentration; ML, maximum likelihood; MLST, multilocus sequence typing; MRSA, methicillin resistant Staphylococcus aureus; NCBI, National Center for Biotechnology Information; NGS, next-generation sequencing; OD, optical density; PFGE, pulsed-field gel electrophoresis; PVL, panton-valentine leucocidin; SAPI, S. aureus pathogenicity island; SCCmec, staphylococcal cassette chromosome mec; SCV, small-colony variants; SNV, single nucleotide variant; ST, sequence type; WGS, whole-genome sequencing.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Seven supplementary tables and four supplementary figures are available with the online version of this article.
References
- 1.O'Sullivan BP, Freedman SD. Cystic fibrosis. Lancet. 2009;373:1891–1904. doi: 10.1016/S0140-6736(09)60327-5. [DOI] [PubMed] [Google Scholar]
- 2.Kahl BC, Duebbers A, Lubritz G, Haeberle J, Koch HG, et al. Population dynamics of persistent Staphylococcus aureus isolated from the airways of cystic fibrosis patients during a 6-year prospective study. J Clin Microbiol. 2003;41:4424–4427. doi: 10.1128/jcm.41.9.4424-4427.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goerke C, Wolz C. Adaptation of Staphylococcus aureus to the cystic fibrosis lung. Int J Med Microbiol. 2010;300:520–525. doi: 10.1016/j.ijmm.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 4.Goerke C, Matias y Papenberg S, Dasbach S, Dietz K, Ziebach R, et al. Increased frequency of genomic alterations in Staphylococcus aureus during chronic infection is in part due to phage mobilization. J Infect Dis. 2004;189:724–734. doi: 10.1086/381502. [DOI] [PubMed] [Google Scholar]
- 5.McAdam PR, Holmes A, Templeton KE, Fitzgerald JR. Adaptive evolution of Staphylococcus aureus during chronic endobronchial infection of a cystic fibrosis patient. PLoS One. 2011;6:e24301. doi: 10.1371/journal.pone.0024301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirschhausen N, Block D, Bianconi I, Bragonzi A, Birtel J, et al. Extended Staphylococcus aureus persistence in cystic fibrosis is associated with bacterial adaptation. Int J Med Microbiol. 2013;303:685–692. doi: 10.1016/j.ijmm.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 7.Schwartbeck B, Birtel J, Treffon J, Langhanki L, Mellmann A, et al. Dynamic in vivo mutations within the ica operon during persistence of Staphylococcus aureus in the airways of cystic fibrosis patients. PLoS Pathog. 2016;12:e1006024. doi: 10.1371/journal.ppat.1006024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Treffon J, Block D, Moche M, Reiss S, Fuchs S, et al. Adaptation of Staphylococcus aureus to airway environments in patients with cystic fibrosis by upregulation of superoxide dismutase M and iron-scavenging proteins. J Infect Dis. 2018;217:1453–1461. doi: 10.1093/infdis/jiy012. [DOI] [PubMed] [Google Scholar]
- 9.Brockhurst MA, Colegrave N, Rozen DE. Next-Generation sequencing as a tool to study microbial evolution. Mol Ecol. 2011;20:972–980. doi: 10.1111/j.1365-294X.2010.04835.x. [DOI] [PubMed] [Google Scholar]
- 10.Harkins CP, Pettigrew KA, Oravcová K, Gardner J, Hearn RMR, et al. The microevolution and epidemiology of Staphylococcus aureus colonization during atopic eczema disease flare. J Invest Dermatol. 2018;138:336–343. doi: 10.1016/j.jid.2017.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Azarian T, Daum RS, Petty LA, Steinbeck JL, Yin Z, et al. Intrahost evolution of methicillin-resistant Staphylococcus aureus USA300 among individuals with reoccurring skin and soft-tissue infections. J Infect Dis. 2016;214:895–905. doi: 10.1093/infdis/jiw242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pena Amaya P, Haim MS, Fernández S, Di Gregorio S, Teper A, et al. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in cystic fibrosis patients from Argentina. Microb Drug Resist. 2018;24:613–620. doi: 10.1089/mdr.2017.0162. [DOI] [PubMed] [Google Scholar]
- 13.Clinical and Laboratory Standards Institute Performance Standards for Antimicrobial Susceptibility Testing. M100. 27th Edition. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.European Committee on Antimicrobial Susceptibility Testing (EUCAST Clinical breakpoints and dosing of antibiotics. 2017.
- 15.Chung M, de Lencastre H, Matthews P, Tomasz A, Adamsson I, et al. Molecular typing of methicillin-resistant Staphylococcus aureus by pulsed-field gel electrophoresis: comparison of results obtained in a multilaboratory effort using identical protocols and MRSA strains. Microb Drug Resist. 2000;6:189–198. doi: 10.1089/mdr.2000.6.189. [DOI] [PubMed] [Google Scholar]
- 16.Fernandez S, de Vedia L, Lopez Furst MJ, Gardella N, Di Gregorio S, et al. Methicillin-resistant Staphylococcus aureus ST30-SCCmec IVc clone as the major cause of community-acquired invasive infections in Argentina. Infect Genet Evol. 2013;14:401–405. doi: 10.1016/j.meegid.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 17.Enright MC, Knox K, Griffiths D, Crook DW, Spratt BG. Molecular typing of bacteria directly from cerebrospinal fluid. Eur J Clin Microbiol Infect Dis. 2000;19:627–630. doi: 10.1007/s100960000321. [DOI] [PubMed] [Google Scholar]
- 18.Lina G, Piémont Y, Godail-Gamot F, Bes M, Peter MO, et al. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. 1999;29:1128–1132. doi: 10.1086/313461. [DOI] [PubMed] [Google Scholar]
- 19.Aaron Petkau PM, Sieffert C, Knox N, Cabral J, et al. The IRIDA Consortium SNVPhyl: a single nucleotide variant phylogenomics pipeline for microbial genomic epidemiology. Microbial genomics. 2016 doi: 10.1099/mgen.0.000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holden MT, Feil EJ, Lindsay JA, Peacock SJ, Day NP, et al. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc Natl Acad Sci U S A. 2004;101:9786–9791. Reference sequences used in this project were downloaded from the NCBI genome database. doi: 10.1073/pnas.0402521101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seemann T. Snippy: fast bacterial variant calling from NGS reads. 2015.
- 22.Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thorvaldsdottir H, Robinson JT, Mesirov JP, Viewer IG. IGV): high-performance genomics data visualization and exploration. Briefings in bioinformatics. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 26.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 28.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hadfield J, Croucher NJ, Goater RJ, Abudahab K, Aanensen DM, et al. Phandango: an interactive viewer for bacterial population genomics. Bioinformatics. 2017 doi: 10.1093/bioinformatics/btx610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hunt M, Mather AE, Sánchez-Busó L, Page AJ, Parkhill J, et al. ARIBA: rapid antimicrobial resistance genotyping directly from sequencing reads. Microb Genom. 2017;3:e000131. doi: 10.1099/mgen.0.000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, et al. Card 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017;45:D566–D73. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta SK, Padmanabhan BR, Diene SM, Lopez-Rojas R, Kempf M, et al. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother. 2014;58:212–220. doi: 10.1128/AAC.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 35.Adams MD, Bishop B, Wright MS. Quantitative assessment of insertion sequence impact on bacterial genome architecture. Microb Genom. 2016;2:e000062. doi: 10.1099/mgen.0.000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arndt D, Grant JR, Marcu A, Sajed T, Pon A, et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44:W16–21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bertelli C, Laird MR, Williams KP, Lau BY, et al. Simon Fraser University Research Computing Group IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017;45:W30-W35. doi: 10.1093/nar/gkx343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/JCM.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cortés G, de Astorza B, Benedí VJ, Albertí S. Role of the htrA gene in Klebsiella pneumoniae virulence. Infect Immun. 2002;70:4772–4776. doi: 10.1128/iai.70.9.4772-4776.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jones CH, Bolken TC, Jones KF, Zeller GO, Hruby DE. Conserved DegP protease in gram-positive bacteria is essential for thermal and oxidative tolerance and full virulence in Streptococcus pyogenes . Infect Immun. 2001;69:5538–5545. doi: 10.1128/iai.69.9.5538-5545.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rigoulay C, Entenza JM, Halpern D, Widmer E, Moreillon P, et al. Comparative analysis of the roles of HtrA-like surface proteases in two virulent Staphylococcus aureus strains. Infect Immun. 2005;73:563–572. doi: 10.1128/IAI.73.1.563-572.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Byrne ME, Gillespie MT, Skurray RA. Molecular analysis of a gentamicin resistance transposonlike element on plasmids isolated from North American Staphylococcus aureus strains. Antimicrob Agents Chemother. 1990;34:2106–2113. doi: 10.1128/aac.34.11.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cystic Fibrosis Foundation Patient Registry Annual Data Report.Bethesda. MD: Cystic Fibrosis Foundation; 2017. [Google Scholar]
- 45.Cuthbertson L, Walker AW, Oliver AE, Rogers GB, Rivett DW, et al. Lung function and microbiota diversity in cystic fibrosis. Microbiome. 2020;8:45. doi: 10.1186/s40168-020-00810-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zemanick ET, Wagner BD, Robertson CE, Ahrens RC, Chmiel JF, et al. Airway microbiota across age and disease spectrum in cystic fibrosis. Eur Respir J. 2017;50 doi: 10.1183/13993003.00832-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bartell JA, Sommer LM, Haagensen JAJ, Loch A, Espinosa R, et al. Evolutionary highways to persistent bacterial infection. Nat Commun. 2019;10:629. doi: 10.1038/s41467-019-08504-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Azarian T, Ridgway JP, Yin Z, David MZ. Long-Term intrahost evolution of methicillin resistant Staphylococcus aureus among cystic fibrosis patients with respiratory carriage. Front Genet. 2019;10:546. doi: 10.3389/fgene.2019.00546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kriegeskorte A, Block D, Drescher M, Windmüller N, Mellmann A, et al. Inactivation of thyA in Staphylococcus aureus attenuates virulence and has a strong impact on metabolism and virulence gene expression. mBio. 2014;5:e01447–14. doi: 10.1128/mBio.01447-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marvig RL, Sommer LM, Molin S, Johansen HK. Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis. Nat Genet. 2015;47:57–64. doi: 10.1038/ng.3148. [DOI] [PubMed] [Google Scholar]
- 51.Lieberman TD, Michel JB, Aingaran M, Potter-Bynoe G, Roux D, et al. Parallel bacterial evolution within multiple patients identifies candidate pathogenicity genes. Nat Genet. 2011;43:1275–1280. doi: 10.1038/ng.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goerke C, Gressinger M, Endler K, Breitkopf C, Wardecki K, et al. High phenotypic diversity in infecting but not in colonizing Staphylococcus aureus populations. Environ Microbiol. 2007;9:3134–3142. doi: 10.1111/j.1462-2920.2007.01423.x. [DOI] [PubMed] [Google Scholar]
- 53.Rouard C, Garnier F, Leraut J, Lepainteur M, Rahajamananav L, et al. Emergence and within-host genetic evolution of methicillin-resistant Staphylococcus aureus resistant to linezolid in a cystic fibrosis patient. Antimicrob Agents Chemother. 2018;62 doi: 10.1128/AAC.00720-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lieberman TD, Flett KB, Yelin I, Martin TR, McAdam AJ, et al. Genetic variation of a bacterial pathogen within individuals with cystic fibrosis provides a record of selective pressures. Nat Genet. 2014;46:82–87. doi: 10.1038/ng.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yachi S, Loreau M. Biodiversity and ecosystem productivity in a fluctuating environment: the insurance hypothesis. Proc Natl Acad Sci U S A. 1999;96:1463–1468. doi: 10.1073/pnas.96.4.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boles BR, Thoendel M, Singh PK. Self-generated diversity produces "insurance effects" in biofilm communities. Proc Natl Acad Sci U S A. 2004;101:16630–16635. doi: 10.1073/pnas.0407460101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Imae K, Saito Y, Kizaki H, Ryuno H, Mao H, et al. Novel nucleoside diphosphatase contributes to Staphylococcus aureus virulence. J Biol Chem. 2016;291:18608–18619. doi: 10.1074/jbc.M116.721845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lindsay JA. Genomic variation and evolution of Staphylococcus aureus . Int J Med Microbiol. 2010;300:98–103. doi: 10.1016/j.ijmm.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 59.Langhanki L, Berger P, Treffon J, Catania F, Kahl BC, et al. In vivo competition and horizontal gene transfer among distinct Staphylococcus aureus lineages as major drivers for adaptational changes during long-term persistence in humans. BMC Microbiol. 2018;18:152. doi: 10.1186/s12866-018-1308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Di Gregorio S, Fernandez S, Perazzi B, Bello N, Famiglietti MM, et al. Increase in IS256 transposition in invasive vancomycin heteroresistant Staphylococcus aureus isolate belonging to ST100 and its derived VISA mutants. Infect Genet Evol. 2016;43:197–202. doi: 10.1016/j.meegid.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 61.Prabhakara S, Khedkar S, Shambat SM, Srinivasan R, Basu A, et al. Genome sequencing unveils a novel sea enterotoxin-carrying PVL phage in Staphylococcus aureus ST772 from India. PLoS One. 2013;8:e60013. doi: 10.1371/journal.pone.0060013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Haim MS, Di Gregorio S, Galanternik L, Lubovich S, Vázquez M, et al. First description of rpsJ and mepA mutations associated with tigecycline resistance in Staphylococcus aureus isolated from a cystic fibrosis patient during antibiotic therapy. Int J Antimicrob Agents. 2017;50:739–741. doi: 10.1016/j.ijantimicag.2017.10.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.