Abstract
Whole-genome sequencing (WGS) is fundamental to Mycobacterium tuberculosis basic research and many clinical applications. Coverage across Illumina-sequenced M. tuberculosis genomes is known to vary with sequence context, but this bias is poorly characterized. Here, through a novel application of phylogenomics that distinguishes genuine coverage bias from deletions, we discern Illumina ‘blind spots’ in the M. tuberculosis reference genome for seven sequencing workflows. We find blind spots to be widespread, affecting 529 genes, and provide their exact coordinates, enabling salvage of unaffected regions. Fifty-seven pe/ppe genes (the primary families assumed to exhibit Illumina bias) lack blind spots entirely, while the remaining pe/ppe genes account for 55.1 % of blind spots. Surprisingly, we find coverage bias persists in homopolymers as short as 6 bp, shorter tracts than previously reported. While G+C-rich regions challenge all Illumina sequencing workflows, a modified Nextera library preparation that amplifies DNA with a high-fidelity polymerase markedly attenuates coverage bias in G+C-rich and homopolymeric sequences, expanding the ‘Illumina-sequenceable’ genome. Through these findings, and by defining workflow-specific exclusion criteria, we spotlight effective strategies for handling bias in M. tuberculosis Illumina WGS. This empirical analysis framework may be used to systematically evaluate coverage bias in other species using existing sequencing data.
Keywords: blind spots, coverage bias, homopolymers, Illumina, sequencing, tuberculosis
Data Summary
Code used to analyse the primary data and produce the figures and tables is available via GitLab (https://gitlab.com/LPCDRP/illumina-blindspots.pub/).
Data used in the analysis are included or referenced in the supplementary tables; Table S7 is available via Zenodo (https://zenodo.org/record/3701840#.Xma5TaaVtGo).
Impact Statement.
In 2018, Mycobacterium tuberculosis killed more human beings than any other single infectious agent. Whole-genome sequencing (WGS) is a major tool for molecular epidemiology and molecular diagnostics development and, thus, a critical component in curtailing the global tuberculosis burden. Illumina is the most common WGS platform for sequencing M. tuberculosis , but its biases and pitfalls are currently dealt with heuristically (such as excluding all pe/ppe genes), if at all. Here, we systematically identify coverage-biased regions in the primary M. tuberculosis reference genome. After filtering out true deletions from 1547 Illumina-sequenced M. tuberculosis genomes, we apply a probabilistic model to classify systematically under-covered positions as blind spots in the genome. We provide genome-wide, workflow-specific lists of blind spots for seven combinations of sequencing instrument+library preparation. These lists enable Illumina WGS studies to select sequencing workflows that maximize coverage for their genomic regions of interest, and set exclusion criteria from existing studies empirically, rather than heuristically. Workflow-stratified analysis of blind spot distribution across sequence features identified coverage-bias profiles dependent on workflow, and others that consistently pervade Illumina sequencing. One workflow markedly reduced coverage bias and reliably sequenced thousands of positions in pe/ppe genes, regions that have consistently evaded Illumina sequencing. All workflows had heightened coverage bias in homopolymers of shorter length than had been previously reported to impede Illumina sequencing. These findings and lists can inform design and analysis of future M. tuberculosis Illumina WGS studies.
Introduction
Mycobacterium tuberculosis is the leading cause of death from a single infectious agent, killing 1.5 million people globally in 2018 [1]. Drug-resistance in M. tuberculosis is a major challenge for tuberculosis (TB) control and effective treatment [1]. Today, whole-genome sequencing (WGS) is the most commonly used tool for establishing new markers for TB surveillance and identifying candidates for molecular diagnostics as M. tuberculosis evolves [2, 3]. Each sequencing technology has unique limitations, defined by both the sequencing instrument and library preparation methods (library prep), referred to together as ‘sequencing workflow’. While WGS presents many opportunities for understanding and controlling TB, workflow-specific shortcomings are poorly understood. In this paper, we empirically evaluate depth of coverage across seven common Illumina workflows to describe workflow-specific coverage bias, which affects genome assembly and variant calling [4].
Illumina sequencing by synthesis (SBS) is by far the most commonly used WGS technology [5]. Its library preparation includes a DNA amplification step, which significantly reduces the quantity of DNA required for sequencing. Illumina library preparation also allows many samples to be multiplexed in a single run, lowering sequencing costs substantially. These qualities make Illumina SBS desirable for many applications. However, some aspects of Illumina SBS limit its reliability for certain downstream analyses. Several genomic features cause biases that reduce coverage when sequenced on Illumina SBS technologies, particularly in segments of the genome where they are prevalent. The most well-characterized of these is G+C content [4, 6, 7]. GC bias originates primarily in library preparation, during amplification by PCR [8]. PCR is biased against amplifying GC- and AT-rich amplicons, which results in disproportionate read copy numbers [7, 8].
Repeat regions, homopolymers and palindromes also reportedly drive coverage bias in short-read sequencing [4, 7, 9–13]. The inability of short reads to unambiguously span repeat regions prevents them from being mapped confidently to the genome, thereby reducing coverage depth [7]. Homopolymers cause bias in some sequencing systems [4, 14], but Illumina states that homopolymers have virtually no effect on Illumina SBS [15]. Palindromic sequences can form hairpin and stem-loop structures during amplification, and have been shown to impede sequencing [9, 10, 16, 17]. Bias due to palindromes has been shown in sequencing by ligation (SBL) technologies and long-read SBS, but reportedly does not introduce bias in Illumina sequencing [10, 16–18]. Coverage bias due to these sequence attributes is influenced by two choices in the sequencing workflow: sequencing instrument and library preparation.
Many researchers are unaware that Illumina WGS data is affected by coverage bias, and take very low coverage to imply true deletions, ignoring coverage bias as a potential cause [19, 20]. Others are aware of this bias, and handle it by excluding large regions of the genome that meet field-standard criteria with limited knowledge of which locations are affected [7, 21, 22]. A common practice for handling Illumina sequencing bias in M. tuberculosis is to exclude all or part of the pe and ppe multigene families [22–25]. These genes make up 10 % of the coding capacity of the genome [26] and play roles in evading host immunity that are important, yet poorly understood [27]. Indiscriminate exclusion of these families needlessly obscures valuable information from sites unaffected by coverage bias. Bias is not limited to pe/ppe genes, yet they are the primary excluded segments of the genome. Apart from pe/ppe genes (whose exclusion is often attributed to repetitive segments rather than GC-richness), researchers rarely address GC bias, despite its well-characterized contribution to coverage bias. This fracture in how the M. tuberculosis WGS community handles Illumina bias highlights a need for specific, empirically determined exclusion criteria. In an initial step towards this, Tyler and colleagues [6] studied the fidelity of Illumina-sequenced M. tuberculosis genomes, and reported differential coverage bias between genomes prepared with Nextera and TruSeq library preps. They also reported that samples prepared with TruSeq resolved genomes into fewer contigs than Nextera and that certain regions, particularly GC-rich regions, could not be resolved with either library.
While these findings demonstrated differential coverage bias between library preps for M. tuberculosis , a systematic, single-base resolution analysis of positions in the M. tuberculosis genome that suffer from coverage bias is lacking. Here, we analyse coverage bias stratified across sequencing workflows to: (i) provide lists of blind spots in the M. tuberculosis reference genome with consistent coverage bias; (ii) characterize coverage bias for features of sequence composition known to be problematic.
We provide these deliverables for seven sequencing workflows separately, and in a pooled set. We find that blind spots distribute differently across the M. tuberculosis genome than accounted for by common practices in WGS analysis pipelines [7, 22–25] and overlap a variety of genes, including several implicated in drug resistance. Workflow-stratified analyses identify a modified Nextera library prep [28] as the least biased option and reveal distinct coverage biases across Illumina sequencing workflows, and the unexpected finding of coverage bias at homopolymers of a shorter length than previously thought. The provided blind spot list [29] enables more informed interpretation of short-read sequencing data and improved WGS design for genome-wide association (GWA) and phylogenomic studies.
Methods
Sequence processing
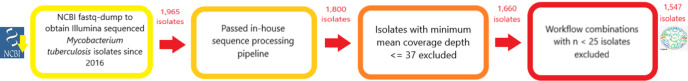
First, we searched the NCBI (National Center for Biotechnology Information) database for M. tuberculosis genomes uploaded between January 1st 2016 and March 24th 2019, and sequenced on an Illumina instrument, producing 5131 candidate genomes. Genomes without reported library prep, or prepared with a library prep other than a traditional or modified Illumina library, were excluded, leaving 1965. Candidate SRA IDs were pulled using SRA toolkit’s [30] --prefetch function and raw fastq files were obtained from NCBI using SRA-tools --fastq-dump. After each file was downloaded, reads were aligned to a reference genome using a parallelized in-house pipeline: first, the downloaded raw reads were trimmed to remove low-quality ends using TrimmomaticPE (-phred33 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15) [31]. After trimming, reads were aligned to the H37Rv reference genome (NC_000962.3) using bowtie2 (-x H37Rv −1–2 S) [32]. The SAMtools [33] package was then used to sort (--output-fmt BAM), index and produce an mpileup (-q 20 f) file. Only genomes from sequencing instrument and library prep combinations (sequencing workflows) with ≥25 total genomes were included. Aligned genomes with a low genome-wide mean depth of 37 or less were then excluded, leaving 1547 genomes (Fig. 1). A custom Python script was then implemented to find positions that met our low-coverage criteria for each genome. Finally, VarScan2 mpileup2cns (--min-avg-qual 20 --min-coverage 10 --variants --output-vcf 1 --strand-filter 0) [34] was used to identify variants for building phylogenies.
Fig. 1.
Isolate inclusion criteria. A total of 1965 Illumina sequenced isolates were downloaded from NCBI with fastq-dump. Of these isolates, 1800 passed through our in-house sequence processing pipeline. Isolates with ≤37 mean depth of coverage were excluded. Finally, excluding isolates sequenced with a workflow comprising fewer than 25 members left 1547 genomes to be used in downstream analyses.
The DNA extraction protocol and the software chosen for the sequence processing pipeline may affect the nature and number of the discovered low-coverage positions. While this pipeline could potentially be optimized to salvage blind spots by choosing a different alignment algorithm, adjusting parameters or by subsequently performing indel realignment, such optimization would be application specific. Our goal was to survey coverage bias in M. tuberculosis , so we have chosen common, field-standard software to represent typical M. tuberculosis sequencing applications. Irrespective of the sequencing processing pipeline selected, differences in extraction methods could influence coverage bias. Importantly, all 1547 samples included in this study were extracted using a N-cetyl-N,N,N-trimethylammonium bromide (CTAB) extraction protocol, thereby eliminating the confounding effects that different extraction methods across projects would have caused.
Identifying low-coverage positions
Sequencing depth or coverage refers to the number of reads that map to a position during alignment, and mean coverage is the mean of coverage across all positions in the genome. Instead of an absolute coverage threshold, we defined low coverage using a relative threshold specific to each genome. We express relative coverage of each position (Di) as the ratio of the detected coverage at that position (di) to the mean coverage (μCi) in the genome (Equation 1). We sought to determine which bases in each genome belonged to the set of low-coverage positions (KLC) for that genome. A given position k belonged to KLC when its relative coverage was ≤0.1 (Equation 2).
| (Equation 1) |
| (Equation 2) |
Phylogenetic filtering
When mapping reads to a reference, positions in the reference genome that are absent in the clinical genomes (true deletions) would be considered low coverage. To account for this, we implemented a phylogenomic filtering step to identify and exclude true deletions from consideration as low-coverage positions on a genome-specific basis. First, a maximum-likelihood phylogeny was created using RAxML [35] version 8.1.1 with a general time reversible model of evolution and 100 bootstrap replicates on a concatenation of 70 057 SNPs, gathered from each genome’s VCF file, which were generated using VarScan2 (from our reference-based assembly pipeline). Mycobacterium bovis and 'Mycobacterium canetti' were used as outgroups. The tree was visualized and manipulated using the iTOL [36, 37] web tool.
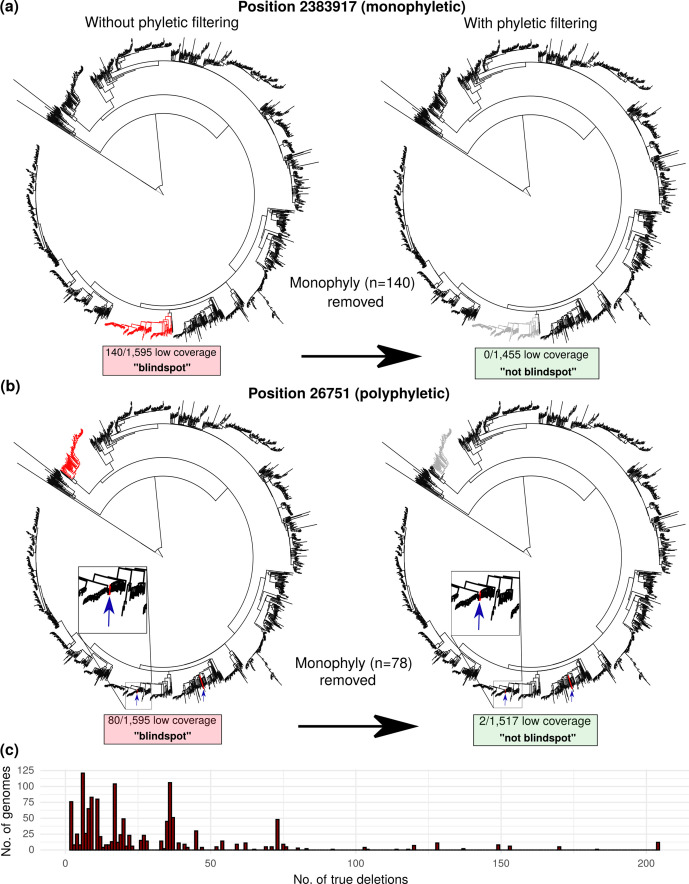
Next, we found which genomes shared each low-coverage position. For each position, the set of genomes with low coverage at the position was checked for monophyly on the phylogenetic tree, using the ete3 [38] Python package, run on the newick file generated by RAxML [39]. Each position with all low-coverage members belonging to a monophyletic group of a size larger than the minimum number required to qualify as a blind spot (n=5, calculated according to Equation 5) was considered a true deletion, rather than a potential blind spot. True deletions identified through these steps were removed from downstream analyses. Following filtering, the remaining low-coverage positions (Fig. 2) were screened for polyphyletic groups containing monophyletic subgroups at least as large as the threshold for counting as a blind spot (n=5; Equation 5). In such cases, the genomes comprising these monophyletic subgroups were excluded from G, the total number of genomes containing the position of interest, when determining blind spot classification (Equation 5). It is possible that an evolutionary event could result in a hard-to-sequence mutation, in which case these positions would be both monophyletic and genuine blind spots. The goal of this study was to minimize false positives and report a high-confidence set of blind spots. With this in mind, we filtered out all monophyletic positions, thereby possibly excluding some true blind spots to ensure high specificity.
Fig. 2.
Phylogenetic filtering of true deletions reduces blind spot false discovery rate. True deletions can confound naïve coverage-based analysis. Our phylogenetic filtering method is depicted, showing positions where low-coverage positions (red) were frequent enough to meet blind spot criteria when called naïvely. Example positions with (a) monophyletic and (b) polyphyletic distributions of genomes with low coverage (<10 % of mean genome coverage) are depicted prior (left) and following (right) phylogenetic filtering. (c) Distribution of genomes harbouring numbers of true deletions identified by phylogenetic filtering. The high number of genomes with identical numbers of true deletions suggests phylogenetic filtering captured clonal expansions harbouring deletions that would have otherwise inflated the blind spot count.
Classifying blind spots
We took a probabilistic approach to determine the threshold for how many genomes a low-coverage position had to occur in to be considered a blind spot. In a genome, each position, k, was considered to have low coverage if the coverage at position k was less than di (Equation 2). Each of the positions meeting this criterion were then included in set of positions with low coverage (KLC) for the genome. Positions that were true deletions in monophyletic groups and monophyletic subsets in polyphyletic groups were then excluded from KLC using the phylogenetic approach described above. Given the size of the set of positions included in KLC, we calculated the probability (E) that, by chance, a given position k belonged to KLC across genomes (Equation 3). Therefore, En is the probability that a given position k has low coverage in n genomes, by chance (Equation 4). This probability was calculated separately for each instrument/library prep workflow.
| (Equation 3) |
| (Equation 4) |
Next, we calculated the probability (PLC) that position k had low coverage in n of G total genomes in the workflow by random chance (Equation 5). The value of G was specific to each position because a base with low coverage could have been a true deletion in some genomes and a potential blind spot in others (Fig. 2b). In other words, for a given position, G excluded the number of genomes in which the position was truly deleted (determined using the monophyletic subsets in polyphyletic groups).
| (Equation 5) |
To determine the acceptable false-positive rate (Fp), we first set a threshold for the number of false positives we considered ‘acceptable’ to include. We sought to capture a minimal set of blind spots and be conservative in the number of false positives included. With this objective in mind, we accepted only 0.1 false positives in our set of classified blind spots. A false-positive rate of 6×10−7 yields 0.1 false blind spots; therefore, we set Fp to this value. Using this acceptable false-positive rate (6×10−7) and our known genome size (4 411 532 bp), we defined our blind spot detection threshold (F) as a function of the median (across genomes in each sequencing workflow) number of low-coverage positions (KLC) that were not deleted (Equation 6).
| (Equation 6) |
For each position that had low coverage in a genome, PLC was calculated and compared to the blind spot detection threshold F to determine whether the position had low coverage in more genomes than we would expect by chance. If the observed probability of a position appearing as low coverage in n genomes by chance out of G genomes in a workflow was lower than our detection threshold, we included that position in our final set of blind spots, Bs (Equation 7).
| (Equation 7) |
Annotating sequence attributes
Homopolymers
Homopolymers were defined as any sequence of consecutive (n >1) identical bases (guanine, cytosine, thymine or adenine). Homopolymers were retrieved from the H37Rv genome (NC_000962.3) with a custom shell script.
Repeats
To identify repeat regions, we used the Tandem Repeats Finder open source software (TRF) [40] with the following parameters: (2 7 7 80 10 50 500 -d –m). TRF uses Smith–Waterman alignment to detect repeats, and filters candidate repeats based on alignment score values the user inputs.
G+C content
Prior work has demonstrated that the primary driver of GC bias is the G+C content [(guanines+cytosines)/total bases] of fragments during PCR amplification [41]. For a given base, the probability that its flanking bases will co-occupy the same fragment diminishes as a function of the distance between them. However, relatively distant bases (up to the length of the fragment) will sometimes contribute to G+C content. To capture bases with extreme G+C content in fragments at multiple scales that might contribute to GC bias, we independently considered G+C content in variably sized windows, calculating G+C content around each base of the H37Rv genome for each window size (50 bp and between 100–1000 bp, in 100 bp intervals).
Palindromes
We identified palindromes in the H37Rv genome using the emboss [42] suite’s palindrome software. This component of the EMBOSS software package scans the genome for inverted matches, and filters based on user-defined match/mismatch and gap requirements. We included palindromes with stem length ≥7 bp [43] and allowed for mismatches and/or gaps based on emboss’s recommended settings to capture a wide range of candidate palindromes.
Defining thresholds to classify sequence attributes
We took an iterative, empirical approach to determine thresholds for the following attributes: homopolymer length, repeat period size (length of subunit being repeated), repeat length (total length of the repeated subunits) and ‘extreme’ G+C content. Within each iteration, sequencing attributes were binned on their criteria (e.g. length of homopolymer) and examined bin-wise for deviation from the ‘unexplained’ blind spot fraction (blind spots in positions qualifying for none of the attributes). The bases classified as one or more of the other attributes in the previous iteration were excluded from this set of unexplained positions. Thresholds for each criterion were set at the first (i.e. least extreme) bin/category where blind spots were significantly more prevalent than blind spots in unexplained sequences [two-sided Fisher’s exact test, 2.5th quantile of odds ratio (OR) >2] and increased monotonically thereafter. We iterated through this process until thresholds for all three attributes stabilized. All statistical tests for determining thresholds were implemented in R.
For the first iteration, homopolymers were binned by length and the fraction of blind spots in each bin was compared to the fraction of blind spots in all other positions in the genome. The minimum homopolymer length that was significantly enriched for blind spots was set as the threshold. The same process was performed on tandem repeat lengths, binned in intervals of two. Shorter repeats that did not meet the threshold for length were then separated on period size and investigated for enrichment of blind spots, though none were significantly enriched.
The threshold for extreme G+C content was determined empirically and calculated separately for each window size. G+C content was calculated for the number of bases in the window, half on each side flanking the base of interest. G+C content was then binned in 2% intervals for each window size. In each bin, the proportion of blind spots was calculated (no. of blind spots matching criteria/total positions in genome matching criteria) and compared to the proportion of blind spots in sites unexplained by homopolymers and repeats. While all bins were investigated for bias (high and low G+C content), only GC-rich bins had significantly disproportionate blind spot fractions. Therefore, we set thresholds only for high G+C content for each window size. Bases were classified as considered GC-rich if their G+C content exceeded thresholds for any of the windows.
Instrument and library preparation error profiles
We identified blind spots separately for each combination (n=7) of instrument and library prep (sequencing workflow). The instruments included were NextSeq 500, MiSeq, HiSeq 2000 and HiSeq2500. Library preps included were TruSeq, Shotgun Nextera, Nextera XT and modified Nextera. We defined two sets of blind spots to be used for different purposes throughout the analysis: (i) a ‘pooled’ set (n=1547 genomes) and (ii) a ‘comparison’ set (n=175 genomes, 25 per workflow). After using the blind spots classified separately within each workflow using all available genomes (Table S1, available with the online version of this article), these workflow-specific sets were pooled into a single pooled set (Table S2) to capture additional blind spots. Since the blind spot criteria is conservative, favouring false negatives over true positives (Equation 6), evaluating additional genomes is likely to increase true positives with a negligible increase in false positives. However, blind spot classification depends on the number of genomes considered, thereby adding the confounding effect of sample size when comparing the number of blind spots in each workflow. To enable fair comparison between workflows, we also determined the comparison set of blind spots by classifying blind spots using the same number of genomes within each workflow. The workflow with the smallest sample size (n=25) was the NextSeq 500 instrument paired with TruSeq library prep. Genome-wide mean coverage was then used to select 25 genomes from the other six workflows with the most similar mean coverage. Blind spots were classified using these curated sets of genomes. This comparison set was used for the stratified parts of our analysis when contrasting the factors that challenge each sequencing workflow.
Statistical tests – comparisons between sequencing instruments, library preparation methods and their combinations
When comparing blind spots between sequencing workflows (combinations of instrument and library prep), statistical tests were chosen according to the number of groups being compared, whether they distributed normally and whether they had similar variance. When more than two workflows were compared, ANOVA was used to compare all that distributed normally and had variances within fourfold of one another (the ‘rule-of-thumb’ for approximately equal variance of greatest variance must be no more than four times the smallest), and post-hoc Tukey tests were performed pairwise to estimate differences between means. Comparisons between non-normally distributed combinations were done pairwise using Wilcoxon signed rank sum test. Comparisons between the non-bootstrapped sequencing workflow were performed pairwise using one sample t-tests. Pairwise comparisons between normally distributing combinations of similar variance were evaluated with paired t-tests. All statistical tests were implemented in R [44].
Annotating blind spots
Overlap between pooled blind spots and annotated coding regions in the H37Rv reference genome (NC_000962.3), downloaded from MycoBrowser (https://mycobrowser.epfl.ch/), was used to determine genes affected by blind spots. Genes containing blind spots were grouped by gene family and the fraction of bases in each family that were blind spots was calculated, along with the fraction of blind spots that were bases in genes of each family. To find genes implicated in drug resistance, we joined the gene-based blind spot list with a curated list of resistance-implicated genes from recent publications (Table S3).
Results
The objective of this work was to precisely describe the coverage bias of common Illumina sequencing workflows for M. tuberculosis WGS and translate them into actionable knowledge for researchers designing or interpreting the results of M. tuberculosis WGS studies. We took a phylogeny-aware, probabilistic approach to classify blind spots, and stratified our analysis by sequencing instrument and library prep (sequencing workflow). We defined low coverage relative to each genome’s mean overall coverage (<10%), rather than an absolute threshold (e.g. <5 reads), mitigating the confounding effect of mean sequencing depth when applying an absolute threshold (Fig. 3b). By analysing coverage bias stratified across sequencing instrument and library preparation technique, we contrast how problematic sequence attributes challenge different sequencing workflows. We applied this approach to 1547 recently sequenced genomes (Table S1) across popular Illumina sequencing workflows to identify Illumina blind spots in the genome [29] that are systematically under-represented due to coverage bias.
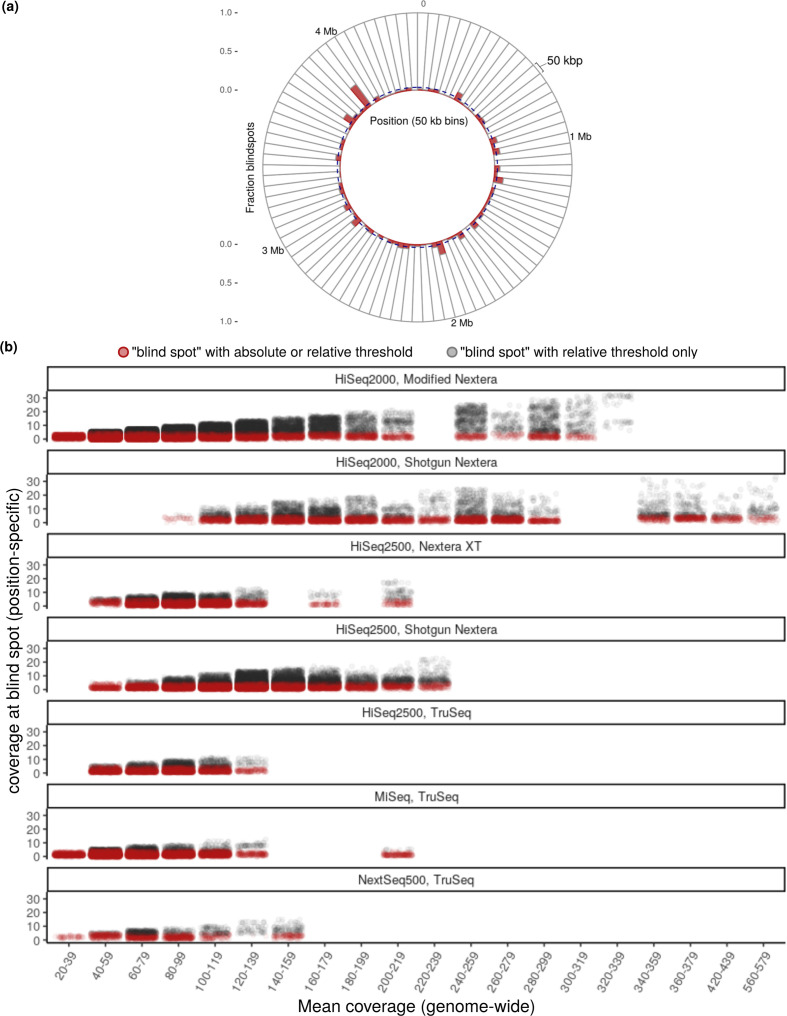
Fig. 3.
Illumina blind spots in M. tuberculosis WGS. (a) Distribution of blind spots from the pooled set (the union across all seven sequencing workflows) across the genome of M. tuberculosis virulent type strain H37Rv. The H37Rv genome is binned into 50 kb segments and the fraction of blind spots (red) is shown for each bin. The dashed blue line represents the baseline fraction of blind spots across the genome (3.6%), highlighting areas with disproportionate levels of blind spots. (b) Comparison of blind spot classification when only using our relative threshold (grey) versus when using either our relative threshold or the previously used absolute (red) threshold (coverage <5) [6] for classifying a position as having low coverage. Each position in the pooled set of blind spots is plotted according to its coverage (y-axis) and the mean coverage (x-axis) across the genome. Mean coverage is binned (bin width=20) and jittered within each bin, with each point rendered at 0.05 opacity to visualize density. Within any given sequencing workflow, the fraction of blind spots that are considered low coverage according to the relative threshold but not the absolute threshold increases in step with mean coverage. Therefore, when defining low coverage using an absolute threshold, genomes with higher mean coverage appear to have fewer blind spots, whether or not this is truly the case.
Phylogenetic filtering removes deletions masquerading as blind spots
If one assumes low coverage invariably indicates coverage bias during sequencing, true deletions would be included spuriously as blind spots. To remove such confounding phylogenetic events from our analysis, we filtered out positions where low coverage was localized to one portion of the phylogeny (monophyly) in more genomes than expected by chance. This filtered out positions both from monophyletic groups (Fig. 2a) of genomes and monophyletic subsets (≥5 genomes) within polyphyletic groups of genomes (Fig. 2b), removing true deletions to capture true blind spots. True-deletion frequency (mean=23) distributed irregularly among genomes (Fig. 2c), consistent with the clonal nature [45] of M. tuberculosis evolution. Following phylogenetic filtering, positions were classified as blind spots if they occurred in enough genomes within a sequencing workflow to meet our probability-based threshold (Equation 7).
Catalogue of blind spots in the M. tuberculosis genome
We defined two sets of blind spots: a pooled set (n=1547 genomes) and a comparison set (25 per each of the seven workflows, n=175 genomes). The pooled set is the union of blind spots across workflows, where blind spots are calculated separately for each workflow using all available genomes. This maximizes capture of true-positive blind spots, but is weighted unequally across sequencing workflows due to inequity in genomes per workflow [NextSeq 500, TruSeq (n=25); HiSeq 2000, Shotgun Nextera (n=61); HiSeq 2500, TruSeq (n=70); HiSeq 2500, Nextera XT (n=87); HiSeq 2500, Shotgun Nextera (n=159); MiSeq, TruSeq (n=284); HiSeq 2000, modified Nextera (n=861)] (Table S4). This set includes blind spots present when using any of the seven workflows, which is useful for researchers who are using data from multiple workflows or workflows not included in this study. This pooled set comprises 3.6 % (159 659 positions) of the H37Rv reference genome (Table S2), scattered across the genome (Fig. 3a) in 5888 regions. Only 1.1 % of blind spots appear as single positions, while the majority appear in consecutive positions, forming clusters (mean length 27 bp, range 1–1816 bp), consistent with the idea that coverage bias is driven by sequence context.
The comparison set of blind spots was created using the same number of genomes (n=25) from each sequencing workflow, accounting for the positive relationship between number of genomes in a group and number of blind spots classified in the group. In this comparison set (Table S5), 8379 (8 %) blind spots were shared among all seven workflows, while most sites only have coverage bias for a subset of workflows. Many blind spots were specific to only one workflow (uniquely present) or were present in all other workflows but one (uniquely absent) (Table 1). Notably, 10 519 of the blind spots in the comparison set are uniquely absent among genomes sequenced with the HiSeq2000/modified Nextera workflow, more than all blind spots uniquely absent in the other six workflows combined. The intersection of blind spots between the remaining six workflows would increase from 8379 to 18 898 if HiSeq2000/modified Nextera were not considered, a more than twofold increase. This workflow is able to resolve more than half of the blind spots present among all other workflows considered, suggesting it mitigates bias in regions that challenge other sequencing workflows.
Table 1.
Number of blind spots uniquely present or absent in each sequencing workflow
|
Instrument |
Library prep |
Uniquely present |
Uniquely absent |
|---|---|---|---|
|
NextSeq 500 |
TruSeq |
5359 |
3256 |
|
MiSeq |
TruSeq |
5726 |
1337 |
|
HiSeq 2500 |
TruSeq |
2590 |
451 |
|
HiSeq 2000 |
Modified Nextera |
3314 |
10 519 |
|
HiSeq 2000 |
Shotgun Nextera |
1869 |
158 |
|
HiSeq 2500 |
Nextera XT |
5387 |
2302 |
|
HiSeq 2500 |
Shotgun Nextera |
612 |
111 |
Tyler et al. reported 124 and 195 ‘ultra-low coverage’ (ULC) hotspots in genomes prepared with TruSeq and Nextera libraries [6], respectively. These ULC regions were defined by arbitrary absolute low coverage (<5×) and length (>10 bp) thresholds. While we used a different version of the reference genome for assembly and different methods for identifying coverage bias, our blind spots capture a majority of their ULC regions (83 % for TruSeq and 63 % for Nextera). Our approach captured more coverage-biased sites, but the overlap between blind spots and Tyler and colleagues’ ULC positions show congruent results despite methodological differences, attesting to the reproducibility of coverage-bias analyses.
Sequencing workflow affects prevalence of blind spots in Illumina WGS studies
To compare blind spots between workflows, we evaluated the number of blind spots in 100 bootstraps of 25 genomes (the minimum genomes in any given workflow; Table S6). Our methods for estimating the expected difference in the number of blind spots between sequencing workflows differed according to how the number of blind spots distributed among the bootstraps (Methods). However, when asking whether a given sequencing workflow produces significantly more/fewer blind spots than another workflow, we employed methods to capture what difference would be meaningful to researchers for WGS experimental design. When designing sequencing experiments, researchers often operate under significant financial or logistical constraints. When opting for a sequencing workflow that requires a more expensive library preparation or use of an instrument outside of their institution or trusted collaborators, they likely want to be confident that their sequencing experiments deliver fewer blind spots every time, or nearly so. Taking this into consideration when comparing sequencing workflows, we qualified the number of blind spots between workflows as significantly different only when the number of blind spots differ >99 % of the time, rather than considering an arbitrarily small difference between means as significantly different. We use the relationship described by Payton and colleagues [46] to estimate this from the overlap of the range in 2.5th–97.5th quantiles of two distributions under comparison.
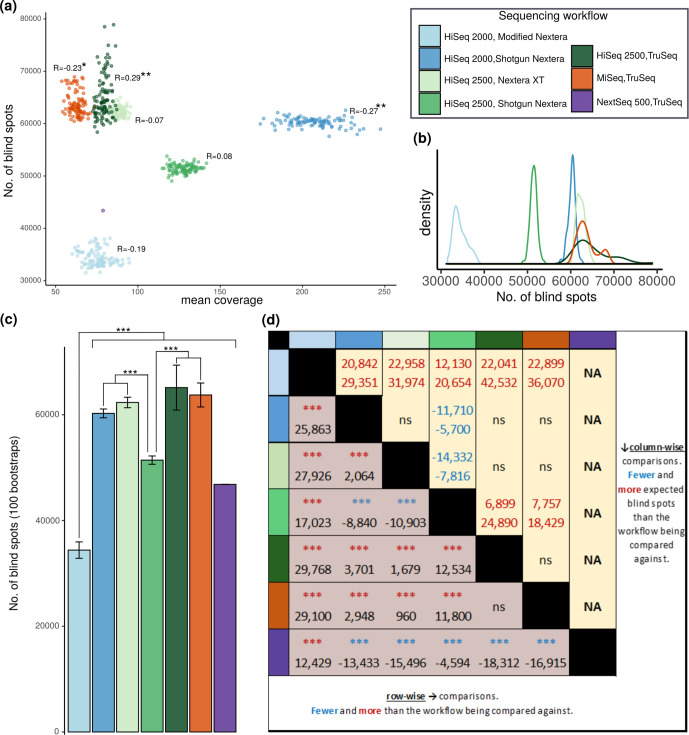
Before comparing sequencing workflows, we examined the relationship between mean coverage among genomes in bootstraps and blind spots. While 3/6 sequencing workflows correlated significantly (P <0.05, 2 negative, 1 positive) yet modestly with coverage (−0.27<R<0.29), the coverage–blind spot relationship does not appear to bias our conclusions regarding differences between workflows (Fig. 4a).
Fig. 4.
Blind spot prevalence across all instruments, library preps and their combinations. (a) Scatterplot and correlations between the number of blind spots (y-axis) and mean coverage (x-axis) among 25 genomes for each bootstrap (n=100 per instrument/library prep workflow). Correlation coefficients are displayed for each workflow. *P <0.05, **P <0.01. (b) Distribution of the number of blind spots across bootstraps for each workflow. (c) The number of blind spots (y-axis) across the seven sequencing workflows (x-axis) (n=175, 25 genomes for each instrument/library prep workflow). Error bars represent ±1 sd from the mean number of blind spots across bootstraps. ***Non-overlapping 95 % CI. (d) Pairwise comparison between sequencing workflows of estimated mean blind spots (row-wise, bottom left) and expected difference in number of blind spots (99 % interval, inferred by 95 % CI boundaries, as described elsewhere [46]; column-wise, top right). Statistical tests were chosen according to distribution and equality of variance. Normal distributions of equivalent variance were compared with Tukey multiple comparisons of means; comparisons involving one or more sequencing workflows with non-normal distributions were compared with Wilcoxon rank sum test; one sample t-tests were used to compare the blind spots in the single NextSeq 500/TruSeq set to the mean blind spots across bootstraps in other workflows. ***P <1×10−5. ns = not statistically significant, NA = notapplicable for non-bootstrapped sequencing workflow.
Coverage bias between instruments
We investigated the number of blind spots between libraries prepared with the same kit but sequenced on different instruments, focusing on evaluating differences in the expected number of blind spots when sequenced on each workflow (Fig. 4d). We first contrasted blind spots between samples prepared with TruSeq, allowing us to compare HiSeq 2500, MiSeq and NextSeq 500. Among these three instruments, NextSeq had the fewest mean blind spots (P <2.2×10−16, one-sample t-test), while HiSeq2500 had modestly fewer blind spots than MiSeq [mean difference=1397, confidence interval (CI) 522–2273, P=8.99×10−5, Tukey multiple comparisons of means], but an insignificant difference in the expected number of blind spots. While the relatively low representation (n=25) of NextSeq 500/TruSeq genomes in our study makes this conclusion tentative (with no bootstrapping, we cannot evaluate a CI), the number of blind spots in its single sampling of 25 genomes is fewer than in any of the 100 bootstraps for MiSeq or HiSeq 2500 workflows prepared with the TruSeq library prep (Fig. 4a). To evaluate the remaining instrument (HiSeq 2000), we compared it to HiSeq 2500 using workflows prepared with the same library prep kit (Shotgun Nextera). HiSeq 2500 had a lower mean number of blind spots than HiSeq 2000 (mean difference=8840, CI 7964–9716, Tukey multiple comparisons of means) and a significantly lower expected number of blind spots (difference between 95 % CI 6040–11 329) (Fig. 4c, d). By combining the results from these comparisons, we tentatively conclude that among the instruments we evaluated, sequencing with NextSeq leads to the fewest blind spots in the M. tuberculosis genome. However, examining more genomes sequenced on NextSeq, and evaluating its performance in combination with additional library prep kits, is needed to substantiate this conclusion. We cannot rule out non-additive combinatorial effects between instrument and library prep, which could condition instrument bias on library prep.
Coverage bias between library preparation methods
We investigated the number of blind spots between genomes sequenced with the same instrument but prepared with different kits. First, to compare Nextera XT, Shotgun Nextera and TruSeq library preps, we contrasted blind spots between workflows sequenced with HiSeq2500. Among these three kits, Shotgun Nextera had the lowest mean number of blind spots (estimated difference between means=10 903, CI 10 027–11 779, Tukey multiple comparisons of means) and a significantly lower expected number of blind spots (difference between 95 % CI 8228–13 987) (Fig. 4c). To evaluate the remaining library prep, modified Nextera, we compared blind spots between workflows sequenced with HiSeq2000. After combining these two comparisons, modified Nextera library prep appears to markedly reduce the number of positions impacted by coverage bias in M. tuberculosis . HiSeq2000/modified Nextera (the only workflow evaluated with modified Nextera) had the lowest number of expected blind spots among all workflows (difference between 95 % CI 12 740–20 114 compared to HiSeq 2500/Shotgun Nextera), and a lower mean number of blind spots than the workflow with the same instrument and Shotgun Nextera library prep (estimated difference between means=25 863, CI 24 987–26 738, Tukey multiple comparisons of means). This gap in blind spots between the HiSeq 2000/modified Nextera sequencing workflow and the next best performing sequencing workflow (Fig. 4c, d), HiSeq 2500/Shotgun Nextera, is particularly impressive considering that HiSeq 2500 outperforms HiSeq 2000 with the same library preparation, and that Shotgun Nextera was the best performing library prep on HiSeq 2500. Overall, this analysis demonstrates that choice of sequencing workflow can alter the range of blind spots in Illumina WGS studies by tens of thousands of positions.
Common sequence attributes across library preps and instruments challenge sequencing
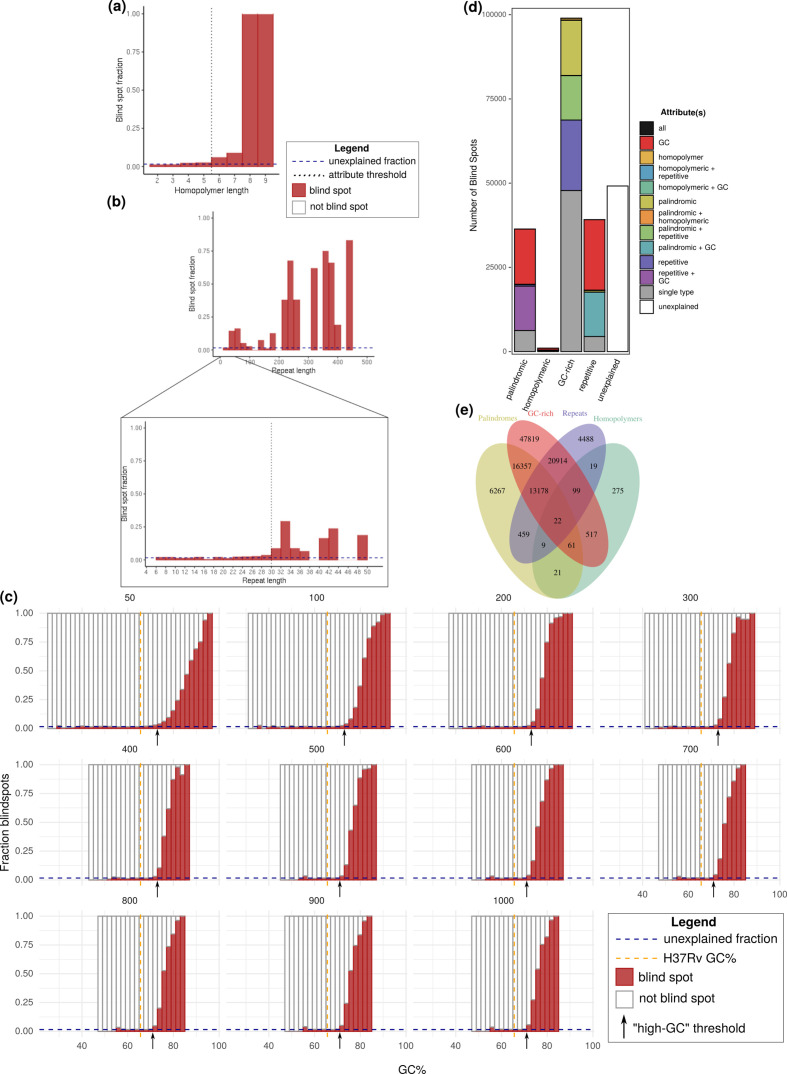
Next, we investigated how sequence attributes previously implicated in coverage bias associate with Illumina blind spots in M. tuberculosis . Extreme G+C content [4], tandem repeats [7], homopolymers [4, 11–13] and palindromes [9, 10] cause coverage bias for some SBS and SBL technologies, but only repeats and G+C content are mentioned by Illumina’s documentation and implicated in SBS chemistry biases [15]. After isolating the positions meeting the general criteria for each attribute, we took an iterative approach (Methods) to set thresholds for defining each sequence attribute and classified all positions in the H37Rv genome accordingly (Table S7; https://zenodo.org/record/3701840#.Xma5TaaVtGo) [29]. For all sequence attributes, blind spots became markedly more abundant as the extremity of each increased (Fig. 5a–c).
Fig. 5.
Relationship between blind spot prevalence and sequence attributes previously implicated in coverage bias. (a–c) The blind spot fraction (y-axis) among positions binned by attribute-specific parameters. For each attribute, only positions meeting criteria for no other attributes are considered. (a) Homopolymers (binned by length, bin size=1). (b) Tandem repeats binned at different lengths to show the general trend between blind spot fraction and repeat length (top main chart, range=0–500, bin size=20) and to show precise threshold value (pop-out expanded region, range=0–50, bin size=2). (c) G+C content (window sizes 50 bp and 100–1000 bp by increments of 100 bp). Positions were classified as ‘high GC’ if they exceeded the threshold for any window size. Palindromes were classified according to previously defined criteria [43]. (d) The total number of blind spots (y-axis) stratified by sequence attributes (x-axis). Blind spots are grouped and coloured according to the set of sequence attributes they meet criteria for. Bar segments are coloured as follows: groups of blind spots that only meet criteria for a single sequence attribute (grey), for at least two attributes (colours – see key for details), and those that meet criteria for none of the four sequence attributes (white). (e) Number of blind spots meeting criteria for each combination of sequence attributes.
Following classification, we asked how each sequence attribute changed the odds of a position being a blind spot. Over half (39 188/75 458, 51.9 %) of the bases within 1588 repetitive regions (≥30 bp) were blind spots, a staggering 37.8-fold (CI 37.2–38.4) greater frequency than non-repeat regions. G+C content was binned by G+C mol% within windows of length 50–1000 bp (at intervals of 100 bp other than the 50 bp window), and thresholds were calibrated separately for each window size. Because the G+C mol% within the amplified fragment during PCR can dictate coverage bias [4], positions were considered GC-rich if they exceeded the threshold in any of the window sizes (Table S8). Among 851 580 GC-rich positions, 98 967 (11.6 %) were blind spots, 7.58-fold the frequency of non-GC-rich positions (CI 7.50–7.66). In homopolymeric sequences (length ≥6 bp), blind spots were 5.5-fold (CI 5.28–5.93) more prevalent (1023/5961, 17.2 %) than in non-homopolymeric sequences. This could be viewed as surprising, considering Illumina maintains that homopolymers do not introduce coverage bias in their SBS technologies [15]. Nearly one in seven bases in the M. tuberculosis genome (651 837 bases) are within palindromic regions (length ≥7 bp), of which 36 374 (5.58 %) were blind spots, a 1.7-fold (CI 1.72–1.76) enrichment compared to non-palindromic sequences, considerably smaller than the other attributes. The modest blind spot enrichment in palindromic sequences also conflicts with previous literature, as they typically only challenge SBL technologies [10]. Of the remaining positions that met criteria for none of the four problematic attributes, only 1.6 % are blind spots (unexplained blind spots).
Next, we asked how being a blind spot changed the odds that a position would possess each sequence attribute, and evaluated overlap between sequencing attributes in the M. tuberculosis genome as a potential explanation for the blind spot enrichment in homopolymeric and palindromic sequences. High G+C content is considerably more prevalent among blind spots than the other sequencing attributes (62.0 % versus 24.5 % for repeats, the next highest; Fig. 5d). G+C content among blind spots (73.0 mol%) is significantly greater (P <2×10−16, Fisher’s exact test) than the G+C content of the H37Rv genome (65.6 mol%) (Fig. 5c), and alone ‘explains’ 30.0 % (47 819) of blind spots (Fig. 5e), dwarfing the number explained exclusively by repeats, homopolymers or palindromes. Unlike GC-rich blind spots, for other attributes, the overwhelming majority of blind spots are classified as multiple problematic sequence attributes, most often with high G+C content. Most (69.2 %) blind spots can be explained by at least one of these four attributes, yet 49 154 (30.8 %) remain unexplained.
While we have described the likelihood of a position being a blind spot given a sequence attribute, we have not done so when attributes are isolated, nor have we evaluated the effect of sequencing workflow on this relationship. To fill these gaps, we calculated the blind spot fraction for each problematic attribute, first by considering all positions meeting the sequence criteria (Fig. 6a) and then considering only positions that meet the criteria for one attribute (Fig. 6b). Other than for isolated palindromes, the blind spot fraction of all sequencing attributes exceeded the blind spot fraction for unexplained positions, both overall (Fig. 6a) and in isolation (Fig. 6b).
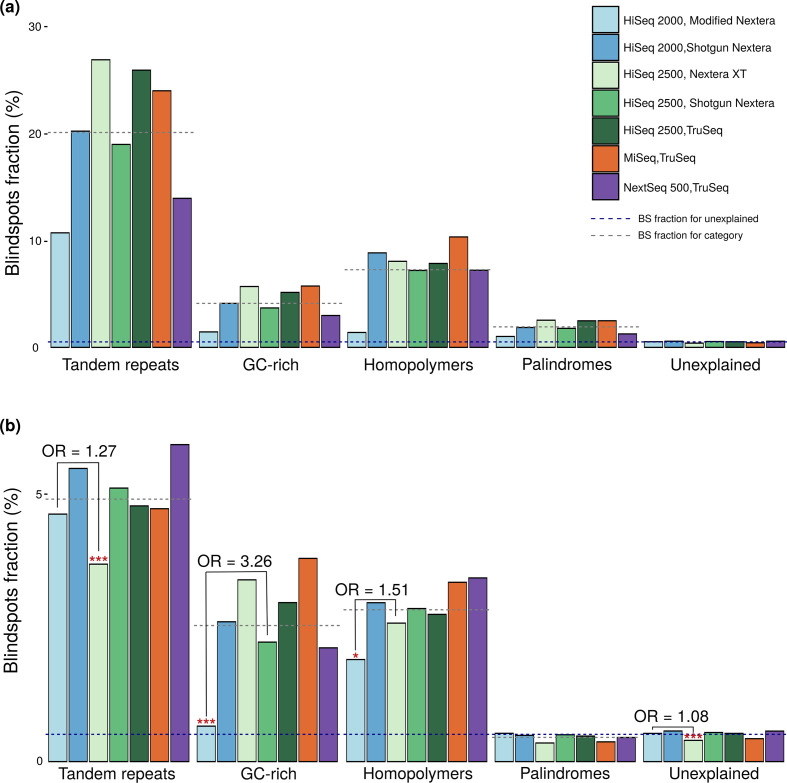
Fig. 6.
Blind spot prevalence in sequence attributes stratified by instruments and library preps. (a) The blind spot fraction (y-axis) in positions with sequence attributes (x-axis) across combinations of Illumina library preps and sequencing instruments (n=175, 25 genomes for each instrument/library prep workflow). Dashed lines denote the blind spot fraction for sequences not meeting criteria for any of the sequence attributes (blue line), and the mean blind spot fraction for each attribute across instrument/library prep workflows (grey lines). (b) Shows the same as (a), but only considering positions that meet criteria for a single sequence attribute. For each attribute, the workflow with the lowest blind spot fraction was compared to the workflow with the second lowest blind spot fraction, and indicated when their difference was significant (two-tailed Fisher’s exact test, *P <0.01, ***P <0.0001). ns = not statistically significant, NA = not applicapble for non-bootstrapped sequencing workflow.
Across all sequencing workflows, tandem repeats are the most problematic attribute, both in combination with other attributes (Fig. 6a), and in isolation (Fig. 6b). This result is consistent with the literature consensus that repeats introduce mapping ambiguity, creating assembly gaps and depleting coverage [6, 12, 47]. It is unsurprising that this problem persists irrespective of library preparation or sequencing instrument, as it is inherent to short-read assembly. While GC-rich regions explain the most blind spots, a given GC-rich position is less likely to be a blind spot than a given homopolymeric or tandem repeat position (Fig. 6). GC-richness accounts for such a large fraction of the blind spots because the entire H37Rv genome is GC-rich (65.6 %). The proportion of blind spots in homopolymeric regions was higher than the unexplained blind spot fraction, even in isolation (Fig. 6a, b). This departs from what has been previously described in the literature and refutes Illumina’s documentation that states homopolymers do not cause sequencing errors in Illumina SBS [15].
Isolated palindromic regions lacked coverage bias across all workflows (Fig. 6b), consistent with prior reports that palindromic sequences do not challenge SBS methods [10, 17]. Curiously, strictly palindromic positions had less coverage bias than unexplained positions (Fig. 6b), suggesting additional factors contribute bias and are absent or less prevalent in palindromic regions. This observation may be explained partially or wholly by homopolymers, GC-rich and repetitive regions with slight coverage bias beyond the resolution of our thresholding criteria (OR >2) and, therefore, considered unexplained.
Error profiles differ across sequencing protocols
To investigate the blind spot fraction within each attribute, we stratified our analysis across sequencing workflows, using the comparison set of blind spots (Fig. 6). By examining differences in coverage bias for the problematic attributes between workflows, we can gain an understanding of the source(s) of coverage bias. The HiSeq 2000/modified Nextera workflow exhibited the least coverage bias in GC-rich (P <2.2×10−16, OR 3.26, CI 3.14–3.37) and homopolymeric (P=0.0091, OR 1.51, CI 1.10–2.09) sequences, whereas HiSeq2500/Shotgun Nextera had the least bias (P <6.62×10−8, OR=1.27, CI 1.16–1.38) in repeat regions (two-sided Fisher’s exact test; Fig. 6b). HiSeq2000/modified Nextera also had marginally (OR 1.08, CI 1.05–1.11) yet significantly (P <1.06×10−9) less coverage bias in unexplained sequences (Fig. 6). This could be explained by marginal effects of sequences with GC-rich and shorter homopolymers that show signs of coverage bias (Fig. 5a–c), but did not qualify for these attributes by our definitions. One study that used other bacterial species with extreme overall G+C content to compare coverage bias for eight different library preparation kits found that Nextera XT introduces the most G+C content-associated sequencing bias. Their results are consistent with our finding that the HiSeq2500/Nextera XT exhibits more blind spots in GC-rich regions compared to other workflows with the same sequencing instrument (Fig. 6a, b).
Across Shotgun Nextera library preps, runs sequenced on HiSeq2000 had more severe coverage bias in all three problematic sequence attributes. This observation makes the comparatively low coverage bias in HiSeq2000/modified Nextera even more remarkable, though it is unclear whether the reductions in coverage bias by modified Nextera (compared to Shotgun Nextera on the same instrument) and HiSeq2500 (compared to HiSeq2000) are additive.
Common exclusion criteria are neither sensitive nor specific to Illumina coverage bias
While many researchers are unaware of coverage bias, some recognize the problem and address it by omitting large regions of the M. tuberculosis genome that are associated with known sequencing biases. These omitted regions are typically restricted to members of the pe, ppe and pe_pgrs gene families (pe/ppe genes), disregarded due to their hypervariable nature, repetitive elements and propensity for erroneous read mapping [22, 23, 48]. However, we identified blind spots beyond these regions. To identify which genes are affected by blind spots, we queried our pooled set of 159 659 blind spots against the H37Rv annotation (NC_000962.3). These blind spots are scattered throughout the genome and overlap 529 genes (Table S9). Cumulatively, the pe/ppe genes account for only 55.1 % of all blind spots (Table 2, bottom row), meaning almost half of blind spots fall in other regions that are not typically omitted from Illumina sequenced M. tuberculosis genomes. Meanwhile, over two-thirds (66.9 %) of positions in pe/ppe genes are not blind spots, meaning that many sites within pe/ppe genes are needlessly excluded. Beyond pe/ppe genes, other clinically important genes harbour blind spots, including effectors that subvert human immunity (esx genes) [49] and synthesize virulence lipids (pks genes) [50], among others (Table S9).
Table 2.
Blind spot prevalence in pe, ppe and pe_pgrs genes
pe_pgrs%=(pe_pgrs positions that are blind spots divided by total pe_pgrs positions). bs%=(pe_pgrs positions that are blind spots divided by total blind spot positions). Values for ppe and pe columns were calculated analogously (e.g. the blind spot fraction of positions within pe, ppe and pe_pgrs genes and the fraction of blind spots that fall within each gene set). Fractions were calculated using the comparison set of blind spots classified in each workflow (top rows) and the pooled set of total blind spots classified within any sequencing workflow (bottom row).
|
Platform |
Libraryprep |
pe_pgrs%(bs%) |
ppe%(bs%) |
pe%(bs%) |
|---|---|---|---|---|
|
NextSeq 500 |
TruSeq |
14.3 (38.5) |
2.54 (6.25) |
0.88 (0.46) |
|
MiSeq |
TruSeq |
32.6 (63.1) |
2.71 (4.78) |
2.00 (0.75) |
|
HiSeq 2500 |
TruSeq |
30.0 (59.4) |
2.54 (4.58) |
2.31 (0.89) |
|
HiSeq 2000 |
Modified Nextera |
7.42 (28.9) |
1.65 (5.87) |
0 |
|
HiSeq 2000 |
Shotgun Nextera |
22.6 (50.4) |
3.04 (6.20) |
2.30 (0.99) |
|
HiSeq 2500 |
Nextera XT |
35.2 (70.1) |
1.25 (2.27) |
0.93 (0.36) |
|
HiSeq 2500 |
Shotgun Nextera |
21.3 (51.6) |
2.44 (5.39) |
0 |
|
Pooled |
– |
61.3 (48.5) |
7.58 (5.47) |
7.45 (1.14) |
Next, we used the comparison set of blind spots (Table S5) to investigate which genes are affected by blind spots when using each sequencing workflow. A total of 92 genes contain blind spots regardless of sequencing workflow, while 98 genes contain blind spots only in a single workflow (Table S10). HiSeq2000/modified Nextera had the fewest genes affected by blind spots (152 genes) and NextSeq 500/TruSeq had the most (248 genes). We then compared how well the seven sequencing workflows capture the pe/ppe genes. When pe, ppe and pe_pgrs genes were considered together, the HiSeq 2000/modified Nextera workflow had the fewest blind spots in pe/ppe genes, capturing over 92 % of each (sub)family, while the HiSeq 2500/Nextera XT workflow had more than four times as many and the most among workflows (Table 2).
Blind spots affect genes implicated in drug resistance
To identify potential blind spots of clinical relevance, we screened the pooled set of blind spots against genes previously implicated in drug resistance (Table S3). Eight genes with blind spots have been previously implicated in resistance (Table 3), underscoring the importance of coverage bias in clinical WGS studies. Systematic coverage bias in these genes could obscure resistance signals in Illumina GWA studies, or potentially lead to false associations, if low coverage due to bias were taken to imply deletion. It should be noted that pks12 may sometimes be lost following prolonged subculturing [51], which would not be filtered out by our phyletic filtering if it arose convergently during culturing. Therefore, some positions in the pks12 gene (and possibly others) may not reflect true blind spots, but instead selection during culture between isolation and sequencing.
Table 3.
Resistance-implicated genes with blind spots
Genes implicated is resistance to anti-TB drugs that harbour blind spots. The absolute number of blind spots, proportion of bases in the gene affected (blind fraction) and the PubMed ID of the study that implicated the gene in resistance (PMID) are shown.
|
Gene |
No. of blind spots |
Blind fraction (%) |
Drug |
PMID |
|---|---|---|---|---|
|
pks12 |
3002 |
24.10 |
CIP, MDR |
15 328 105, 29 281 674 |
|
iniB |
170 |
11.80 |
INH, ETH |
27 665 704 |
|
Rv2820c |
45 |
4.90 |
ETH |
29 358 649 |
|
Rv0194 |
73 |
2.04 |
STR, TET |
18 458 127 |
|
mmpL4 |
47 |
1.62 |
RIF |
23 431 276 |
|
ppsA |
89 |
1.58 |
RIF |
23 002 228 |
|
ppsD |
2 |
0.04 |
RIF |
23 002 228 |
|
pks2 |
1 |
0.02 |
POA |
27 759 369 |
CIP, Ciprofloxacin; ETH, ethambutol; INH, isoniazid; MDR, multi-drug resistance; POA, pyrazinoic acid; RIF, rifampicin; STR, streptomycin; TET, tetracycline.
Discussion
Despite driving the TB pandemic and being among the most frequently sequenced prokaryotes, M. tuberculosis coverage bias is poorly understood, and dealt with in varied ways across research groups. Here, we have implemented a framework to identify high-confidence blind spots from publicly available genomes, stratified by sequencing workflow, demonstrating consistent platform-wide coverage bias in repetitive regions, and workflow-specific degrees of difficulty with GC-rich and homopolymeric sequences. We used a stringent false-positive rate to capture blind spots with high specificity. As a result, sensitivity is limited by the number of genomes examined. The number of blind spots we identified is a conservative estimate and could be expanded with additional genomes.
Comprising nearly 10 % of the genome’s coding capacity and unique to mycobacteria, pe/ppe genes were chief among the interests following publication of the H37Rv genome sequence in 1998 [26]. However, despite repeated implication of pe/ppe genes in TB pathogenesis, their recalcitrance to Illumina sequencing has led to their near uniform exclusion from many WGS studies. Our results refute the utility of this practice, demonstrating that while many pe/ppe genes contain blind spots (Table 2), numerous others – in some cases entire pe/ppe genes – do not suffer from severe coverage bias with Illumina sequencing. However, we find that tens of thousands of other positions typically included in Illumina WGS studies harbour positions with severe coverage bias and warrant exclusion (Tables S2 and S5). These findings provide more granular criteria for handling Illumina bias in the M. tuberculosis WGS.
GC bias is a known issue for Illumina sequencing, yet it is rarely addressed in M. tuberculosis WGS studies. Although repeats are the most problematic when present (Fig. 6), our results show that 10-fold more blind spots can be attributed exclusively to high G+C content than exclusively to repeats (Fig. 5e). While increasing the amount of native DNA can improve coverage across the genome and alleviate the effect of GC bias, it is a common misconception that increasing DNA through amplification can have a similar effect and improve coverage uniformly. Instead, amplification bias will further exacerbate the problem in GC-rich regions (Fig. 7a–f). Remarkably, however, the modified Nextera library preparation virtually eliminates blind spots in isolated GC-rich positions (Fig. 6b). The superior performance of this protocol (Fig. 6) was remarkable, particularly considering it was introduced more as a cost-saving measure than one to mitigate coverage bias [28]. The modified Nextera protocol substitutes the standard Nextera DNA polymerase with a high-fidelity DNA polymerase enzyme [28], which mitigates PCR amplification bias [52]. We recommend this modified Nextera library prep for short-read sequencing of M. tuberculosis , and encourage further development of methods that reduce amplification bias.
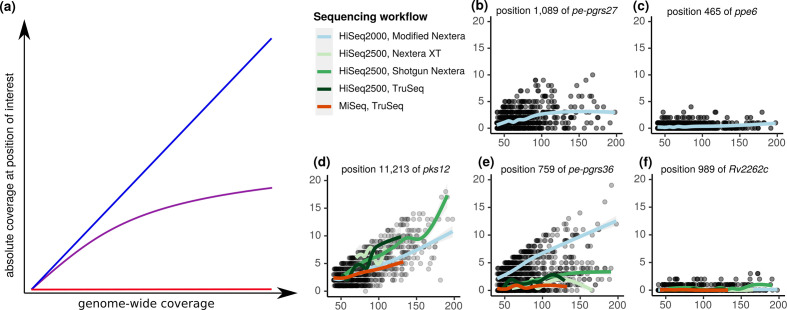
Fig. 7.
Relationship between mean coverage across the genome and coverage depth in blind spots. (a) Illustrations of theoretical linear (blue), sublinear (purple) and constant (red) functions of dependence between coverage depth in blind spots and genome-wide coverage. The type of function relating coverage depth in a blind spot and mean coverage is determined by (i) the nature and source of the coverage bias, and (ii) the method for increasing mean coverage. At positions with bias resulting from events with fixed likelihood, coverage depth would increase linearly (blue) with mean coverage, with slope equal to the frequency of the event biasing coverage. For instance, a position situated a number of base pairs away from a large repetitive element, such that 5 % of the time the position ends up on the same read as the repeat, would fail to map on those occasions. Therefore, the coverage at the position would be 5 % of the mean coverage, regardless of its magnitude (i.e. a linear relationship). Sublinear relationships (purple) can form when the source of bias compounds as mean depth increases. This could occur for positions in GC-rich regions during PCR, as the DNA fragments for which the polymerase has greater affinity will be preferentially amplified, increasing their relative abundance and, thus, the magnitude of bias for the next amplification cycle. The final relationship is a constant function (red), where the absolute coverage depth at the position of interest does not change as mean coverage changes. This can occur when large repeats cannot be mapped onto the reference genome unambiguously, as mappability does not depend on coverage. Which of the functions depicted in (a) predominates depends not only on the source of bias, but also on the method used for increasing coverage. For instance, GC-rich sequences will likely relate sublinearly to mean depth if PCR amplification is employed to increase mean coverage, whereas additional growth to generate more DNA would presumably increase linearly (assuming no additional attributes that would challenge mapping are present). (b–f) Observed relationships at representative blind spots in our study. While modified Nextera outperformed other workflows overall, there are still many positions where coverage follows a sublinear (b) or constant (c) function. Conversely, there are also positions that increase linearly with mean coverage across all workflows (d). Improved recoverability with modified Nextera is apparent at many high G+C positions (exemplified in e), ostensibly due to decreased amplification bias of the high-fidelity DNA polymerase used in the PCR step of the modified Nextera protocol. Lastly, there still remain unresolved sources of bias. The position in hypothetical gene Rv2262c (f) met criteria for none of the sequencing attributes we examined, yet demonstrates a constant function across all sequencing workflows.
Importantly, mean genome-wide coverage seems to affect blind spot coverage differentially (Fig. 7). For some blind spots, coverage increases linearly or sublinearly with mean genome-wide coverage (Fig. 7b), while others remain static (Fig. 7c, f). Depending on the attributes contributing to the bias, some may be salvageable with more amplification (particularly with the modified Nextera library preparation; Fig. 7e), while providing more native DNA is required to salvage others. Further experimentation and analysis are required to rigorously define recoverability of each type of blind spot.
Beyond species-specific insights, our findings clarify two contested notions about sequencing attributes contributing to coverage bias: we refute the assertion that homopolymeric sequences pose no issue for Illumina sequencing [15], and find no coverage bias in palindromic sequences. Our finding that homopolymers associate with coverage bias, even in isolation (Fig. 6b), is particularly noteworthy considering that M. tuberculosis homopolymers are invariably short (≤9 bp). Some researchers contend that homopolymers do not drive coverage bias in Illumina SBS [14, 15], and those who support homopolymer-driven coverage bias suggest it is exclusive to long homopolymers [4, 12, 13]. Here, we show that even positions within short homopolymers are manifold more often blind spots than non-homopolymeric positions (Figs 5a and 6b). This finding calls into question the prevailing notion that short homopolymers do not bias coverage [11, 15], suggesting instead that they systematically reduce coverage at lengths as short as 6 bp, and perhaps lower (Fig. 5a). Indel realignment could potentially mitigate some coverage bias in homopolymers, but was not implemented in the studies reporting homopolymer-driven bias exclusive to long tracts [4, 12, 13]. Moreover, indel realignment is uncommon in M. tuberculosis WGS studies [22]. Therefore, the concern of homopolymer-driven bias is still pertinent even if mitigated to some extent by indel realignment. A homopolymeric tract in glpK of M. tuberculosis has garnered recent attention for undergoing frequent, reversible, frameshifts [53, 54]. Variants with such rapid emergence and reversal could theoretically pass our phylogenetic filtering method, yet none of the positions comprising the phase-variable glpK homopolymeric tract were identified as blind spots (Table S7). Moreover, the persistence of this bias in the absence of other known attributes and its workflow-specific attenuation (Fig. 6b) strengthens our conviction that the bias is genuine and homopolymer-driven. Based on these observations, true variants that bypassed phylogenetic filtering are likely not the cause of low coverage at homopolymers. Yet we cannot rule out the possibility of flanking elements contributing to the bias [4]. In future work, systematic empirical testing of potential sources of homopolymer bias across multiple labs could pin down the contributing factors.
The observation that the modified Nextera library preparation attenuates homopolymer bias (Fig. 6) suggests the bias originates during PCR amplification, presumably by reducing polymerase slippage [55]. Unlike GC-rich and homopolymeric positions, modified Nextera did not improve coverage in repeats, consistent with ambiguous mapping driving coverage bias in repetitive elements, an inherent weakness of short-read technologies. Illumina SBS instruments beyond those examined here are available. However, they do not address the fundamental factors driving coverage bias, but rather improve sequencing affordability and throughput (e.g. NovaSeq and NextSeq 2000) or portability (MiniSeq and iSeq). Long-read technologies (LRTs) offer potential solutions for coverage bias, particularly for repeat regions [4]. While LRTs are still catching up to the affordability of Illumina short reads, they have recently made significant advances in single-read accuracy (Pacific Biosciences SMRT-sequencing) and portability (Oxford Nanopore). Applying this framework to identify blind spots across LRTs is an important next step toward a comprehensive knowledgebase of coverage bias across sequencing technologies.
By applying a phylogeny-aware coverage analysis framework to map Illumina blind spots onto the genome of virulent type strain H37Rv, we provide a comprehensive road map for setting workflow-specific exclusion criteria for M. tuberculosis WGS studies. The pooled set we report [29] encompasses blind spots present across any sequencing workflow, useful for interpreting Illumina WGS data where workflow is not specified and for studies analysing genomes sequenced with a variety of workflows, as is common in large-scale GWA studies [56, 57]. The comparison set of blind spots is useful for identifying which positions suffer from coverage bias on a workflow-specific basis. These lists both inform design of future Illumina WGS experiments and provide a lens through which existing data can be interpreted. While we applied this framework to M. tuberculosis, it can also be used to systematically evaluate coverage bias in other species without requiring expensive, time-consuming generation of new genomic data.
Supplementary Data
Funding information
This work was supported by the National Institutes of Health grant number R01AI105185 to F.V. The funding body had no role in the design of the study nor in collection, analysis and interpretation of data, nor in writing the paper. Funding for open access charge: National Institutes of Health.
Acknowledgements
We thank Afif Elghraoui for initially conceptualizing the project and for instructive discussions throughout the project. We also thank Derek Conkle-Gutierrez and Matthew Onorato for their careful review and thoughtful feedback that helped improve this manuscript.
Author contributions
Conceptualization: S. J. M., F. V., S. M. R.-B. and T. S. Data curation: C. M. and S. M. R.-B. Formal analysis and visualization: S. J. M. and C. R. Software: C. R., S. N. M., S. J. M., C. M. and S. M. R.-B. Writing – original draft: C. R., S. J. M., C. M. and S. N. M. Writing – review and editing: S. J. M., C. R., C. M. and F. V. Project administration and supervision: F. V., S. J. M. and S. M. R.-B. Funding acquisition: F. V.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: CI, confidence interval; GWA, genome-wide association; LRT, long-read technology; NCBI, National Center for Biotechnology Information; OR, odds ratio; SBL, sequencing by ligation; SBS, sequencing by synthesis; TB, tuberculosis; ULC, ultra-low coverage; WGS, whole-genome sequencing.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Ten supplementary tables are available with the online version of this article and via Zenodo (https://zenodo.org/record/3701840#.Xma5TaaVtGo).
References
- 1.WHO Global Tuberculosis Report. Geneva: World Health Organization; 2019. [Google Scholar]
- 2.CRyPTIC Consortium and the 100,000 Genomes Project. Allix-Béguec C, Arandjelovic I, Bi L, Beckert P, et al. Prediction of susceptibility to first-line tuberculosis drugs by DNA sequencing. N Engl J Med. 2018;379:1403–1415. doi: 10.1056/NEJMoa1800474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO The Use of Next-Generation Sequencing Technologies for the Detection of Mutations Associated with Drug Resistance in Mycobacterium tuberculosis Complex: Technical Guide. Geneva: World Health Organization; 2018. [Google Scholar]
- 4.Ross MG, Russ C, Costello M, Hollinger A, Lennon NJ, et al. Characterizing and measuring bias in sequence data. Genome Biol. 2013;14:R51. doi: 10.1186/gb-2013-14-5-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tyler AD, Christianson S, Knox NC, Mabon P, Wolfe J, et al. Comparison of sample preparation methods used for the next-generation sequencing of Mycobacterium tuberculosis . PLoS One. 2016;11:e0148676. doi: 10.1371/journal.pone.0148676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galagan JE. Genomic insights into tuberculosis. Nat Rev Genet. 2014;15:307–320. doi: 10.1038/nrg3664. [DOI] [PubMed] [Google Scholar]
- 8.Aird D, Ross MG, Chen W-S, Danielsson M, Fennell T, et al. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol. 2011;12:R18. doi: 10.1186/gb-2011-12-2-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Star B, Nederbragt AJ, Hansen MHS, Skage M, Gilfillan GD, et al. Palindromic sequence artifacts generated during next generation sequencing library preparation from historic and ancient DNA. PLoS One. 2014;9:e89676. doi: 10.1371/journal.pone.0089676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang Y-F, Chen S-C, Chiang Y-S, Chen T-H, Chiu K-P. Palindromic sequence impedes sequencing-by-ligation mechanism. BMC Syst Biol. 2012;6 (Suppl. 2):S10. doi: 10.1186/1752-0509-6-S2-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quail MA, Smith M, Coupland P, Otto TD, Harris SR, et al. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics. 2012;13:341. doi: 10.1186/1471-2164-13-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minoche AE, Dohm JC, Himmelbauer H. Evaluation of genomic high-throughput sequencing data generated on Illumina HiSeq and genome analyzer systems. Genome Biol. 2011;12:R112. doi: 10.1186/gb-2011-12-11-r112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laehnemann D, Borkhardt A, McHardy AC. Denoising DNA deep sequencing data-high-throughput sequencing errors and their correction. Brief Bioinform. 2016;17:154–179. doi: 10.1093/bib/bbv029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo C, Tsementzi D, Kyrpides N, Read T, Konstantinidis KT. Direct comparisons of Illumina vs. Roche 454 sequencing technologies on the same microbial community DNA sample. PLoS One. 2012;7:e30087. doi: 10.1371/journal.pone.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Illumina An Introduction to Next-Generation Sequencing Technology. San Diego, CA: Illumina; 2017. [Google Scholar]
- 16.Sabina J, Leamon JH. Bias in whole genome amplification: causes and considerations. Methods Mol Biol. 2015;1347:15–41. doi: 10.1007/978-1-4939-2990-0_2. [DOI] [PubMed] [Google Scholar]
- 17.Warris S, Schijlen E, van de Geest H, Vegesna R, Hesselink T, et al. Correcting palindromes in long reads after whole-genome amplification. BMC Genomics. 2018;19:798. doi: 10.1186/s12864-018-5164-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lasken RS, Stockwell TB. Mechanism of chimera formation during the multiple displacement amplification reaction. BMC Biotechnol. 2007;7:19. doi: 10.1186/1472-6750-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Advani J, Verma R, Chatterjee O, Pachouri PK, Upadhyay P, et al. Whole genome sequencing of Mycobacterium tuberculosis clinical isolates from India reveals genetic heterogeneity and region-specific variations that might affect drug susceptibility. Front Microbiol. 2019;10:00309. doi: 10.3389/fmicb.2019.00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zakham F, Laurent S, Esteves Carreira AL, Corbaz A, Bertelli C, et al. Whole-genome sequencing for rapid, reliable and routine investigation of Mycobacterium tuberculosis transmission in local communities. New Microbes New Infect. 2019;31:100582. doi: 10.1016/j.nmni.2019.100582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phelan JE, Coll F, Bergval I, Anthony RM, Warren R, et al. Recombination in pe/ppe genes contributes to genetic variation in Mycobacterium tuberculosis lineages. BMC Genomics. 2016;17:151. doi: 10.1186/s12864-016-2467-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meehan CJ, Goig GA, Kohl TA, Verboven L, Dippenaar A, et al. Whole genome sequencing of Mycobacterium tuberculosis: current standards and open issues. Nat Rev Microbiol. 2019;17:533–545. doi: 10.1038/s41579-019-0214-5. [DOI] [PubMed] [Google Scholar]
- 23.Mikheecheva NE, Melerzanov AV, Melerzanov AV, Danilenko VN. A nonsynonymous SNP catalog of Mycobacterium tuberculosis virulence genes and its use for detecting new potentially virulent sublineages. Genome Biol Evol. 2017;9:887–899. doi: 10.1093/gbe/evx053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Casali N, Broda A, Harris SR, Parkhill J, Brown T. Whole genome sequence analysis of a large isoniazid-resistant tuberculosis outbreak in London: a retrospective observational study. PLoS Med. 2016;13:e1002137. doi: 10.1371/journal.pmed.1002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farhat MR, Shapiro BJ, Sheppard SK, Colijn C, Murray M. A phylogeny-based sampling strategy and power calculator informs genome-wide associations study design for microbial pathogens. Genome Med. 2014;6:101. doi: 10.1186/s13073-014-0101-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 27.Fishbein S, van Wyk N, Warren RM, Sampson SL. Phylogeny to function: PE/PPE protein evolution and impact on Mycobacterium tuberculosis pathogenicity. Mol Microbiol. 2015;96:901–916. doi: 10.1111/mmi.12981. [DOI] [PubMed] [Google Scholar]
- 28.Baym M, Kryazhimskiy S, Lieberman TD, Chung H, Desai MM, et al. Inexpensive multiplexed library preparation for megabase-sized genomes. PLoS One. 2015;10:e0128036. doi: 10.1371/journal.pone.0128036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinhold C, Modlin S, Morrissey C, Valafar F. Table S7: blind spots and their attributes ( https://zenodo.org/record/3701840#.Xma5TaaVtGo) 2020.
- 30.NCBI SRA-Tools, NCBI. 2020
- 31.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 37.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huerta-Cepas J, Serra F, Bork P. ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol Biol Evol. 2016;33:1635–1638. doi: 10.1093/molbev/msw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tilak M-K, Botero-Castro F, Galtier N, Nabholz B. Illumina library preparation for sequencing the GC-rich fraction of heterogeneous genomic DNA. Genome Biol Evol. 2018;10:616–622. doi: 10.1093/gbe/evy022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 43.Varani G. Exceptionally stable nucleic acid hairpins. Annu Rev Biophys Biomol Struct. 1995;24:379–404. doi: 10.1146/annurev.bb.24.060195.002115. [DOI] [PubMed] [Google Scholar]
- 44.R Core Team Vienna: R Foundation for Statistical Computing; 2013. [Google Scholar]
- 45.Gagneux S. Ecology and evolution of Mycobacterium tuberculosis . Nat Rev Microbiol. 2018;16:202–213. doi: 10.1038/nrmicro.2018.8. [DOI] [PubMed] [Google Scholar]
- 46.Payton ME, Greenstone MH, Schenker N. Overlapping confidence intervals or standard error intervals: what do they mean in terms of statistical significance? J Insect Sci. 2003;3:34. doi: 10.1673/031.003.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsai IJ, Hunt M, Holroyd N, Huckvale T, Berriman M, et al. Summarizing specific profiles in Illumina sequencing from whole-genome amplified DNA. DNA Res. 2014;21:243–254. doi: 10.1093/dnares/dst054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ioerger TR, Koo S, No E-G, Chen X, Larsen MH, et al. Genome analysis of multi- and extensively-drug-resistant tuberculosis from KwaZulu-Natal, South Africa. PLoS One. 2009;4:e7778. doi: 10.1371/journal.pone.0007778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gröschel MI, Sayes F, Simeone R, Majlessi L, Brosch R. ESX secretion systems: mycobacterial evolution to counter host immunity. Nat Rev Microbiol. 2016;14:677–691. doi: 10.1038/nrmicro.2016.131. [DOI] [PubMed] [Google Scholar]
- 50.Quadri LEN. Biosynthesis of mycobacterial lipids by polyketide synthases and beyond. Crit Rev Biochem Mol Biol. 2014;49:179–211. doi: 10.3109/10409238.2014.896859. [DOI] [PubMed] [Google Scholar]
- 51.Domenech P, Reed MB. Rapid and spontaneous loss of phthiocerol dimycocerosate (PDIM) from Mycobacterium tuberculosis grown in vitro: implications for virulence studies. Microbiology. 2009;155:3532–3543. doi: 10.1099/mic.0.029199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van Dijk EL, Jaszczyszyn Y, Thermes C. Library preparation methods for next-generation sequencing: tone down the bias. Exp Cell Res. 2014;322:12–20. doi: 10.1016/j.yexcr.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 53.Vargas R, Farhat MR. Antibiotic treatment and selection for glpK mutations in patients with active tuberculosis disease. Proc Natl Acad Sci USA. 2020;117:3910–3912. doi: 10.1073/pnas.1920788117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Safi H, Gopal P, Lingaraju S, Ma S, Levine C, et al. Phase variation in Mycobacterium tuberculosis glpK produces transiently heritable drug tolerance. Proc Natl Acad Sci USA. 2019;116:19665–19674. doi: 10.1073/pnas.1907631116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gragg H, Harfe BD, Jinks-Robertson S. Base composition of mononucleotide runs affects DNA polymerase slippage and removal of frameshift intermediates by mismatch repair in Saccharomyces cerevisiae . Mol Cell Biol. 2002;22:8756–8762. doi: 10.1128/MCB.22.24.8756-8762.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Coll F, Phelan J, Hill-Cawthorne GA, Nair MB, Mallard K, et al. Genome-wide analysis of multi- and extensively drug-resistant Mycobacterium tuberculosis . Nat Genet. 2018;50:307–316. doi: 10.1038/s41588-017-0029-0. [DOI] [PubMed] [Google Scholar]
- 57.Farhat MR, Freschi L, Calderon R, Ioerger T, Snyder M, et al. GWAS for quantitative resistance phenotypes in Mycobacterium tuberculosis reveals resistance genes and regulatory regions. Nat Commun. 2019;10:2128. doi: 10.1038/s41467-019-10110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.