Abstract
MicroRNA (miR)-497 has been reported as a tumor suppressor in various cancer types. Nonetheless, the regulation of triple-negative breast cancer (TNBC) by miR-497 remains poorly understood. The present study aimed to investigate the potential function and mechanism of miR-497 in TNBC. A total of 36 TNBC and matched non-cancerous tissue samples were collected for analysis. Reverse transcription-quantitative PCR was performed to detect the miR-497 levels in TNBC tissue. The association between miR-497 expression, clinical characteristics and survival was then analyzed. To investigate the role of miR-497 in TNBC, MTT, colony formation, Transwell invasion, cell cycle and cell apoptosis assays were conducted following transfection of miR-497 mimics into the MDA-MB-231 and MDA-MB-468 cell lines. Luciferase reporter assays and western blot analysis were used to confirm the regulation of a putative target of miR-497. The results indicated that the expression of miR-497 was downregulated in the TNBC specimens. Further analysis demonstrated that the expression of miR-497 was downregulated in patients with advanced TNBC stages and that low miR-497 was associated with poor prognosis in patients with TNBC. Transfection of miR-497 mimics inhibited TNBC cell proliferation and increased cell apoptosis in MDA-MB-231 and MDA-MB-468 cells. Moreover, cell migration was inhibited following overexpression of miR-497, which also led to the arrest of the breast cancer cells in the G0/G1 phase of the cell cycle. Yes-associated protein 1 (YAP1), a critical molecule in the Hippo pathway, was identified as a target of miR-497. Notably, the protein and mRNA expression levels of YAP1 in MDA-MB-231 and MDA-MB-468 cells were downregulated following overexpression of miR-497. Overall, the findings of the present study indicated that miR-497 inhibited TNBC cell proliferation and migration and induced cell apoptosis by negatively regulating YAP1 expression. Thus, targeting miR-497 may represent a potential strategy for the treatment of TNBC.
Keywords: microRNA-497, breast cancer; YAP1, proliferation, metastasis
Introduction
Breast cancer is one of the most common malignant tumors in female patients (1). Based on molecular characterization of the estrogen receptor (ER), progesterone receptor (PR), Ki67 antigen and human epidermal growth factor receptor-2 (HER2), breast cancer is typically divided into different types, including luminal A, luminal B, HER-2-positive, basal-like and ‘normal-like’ breast tumors. Triple-negative breast cancer (TNBC) is defined as a subset of basal-like breast cancer, which is negative for ER, PR and HER2. Due to the lack of a specific hormonal biomarker, surgical resection and chemotherapy are currently the mainstays of systemic treatment for patients with TNBC (2,3). A previous study has demonstrated that TNBC exhibits higher rates of metastasis and recurrence compared with other breast cancer subtypes (4). Thus, the development of new therapeutic targets for the treatment of TNBC is essential.
MicroRNA (miRNA/miR) is a class of small non-coding RNA molecules of ~22 nucleotides typically acting as vital regulators of target mRNA transcripts by binding to 3′-untranslated regions (3′-UTRs) (5). miRNA molecules have been reported to be deregulated in various cancer types, including TNBC, suggesting that they may be involved in cancer development and progression (6). A number of studies have demonstrated that miR-497 is downregulated in various malignant tumors, such as thyroid cancer (7), lung cancer (8) and hepatocellular carcinoma (9). In addition, miR-497 levels have been shown to be significantly reduced in breast cancer tissue and cell lines compared with matched non-cancerous breast tissue samples and MCF-10A cells, respectively (10). Several downstream targets of miR-497 have only recently been identified, such as Bcl2, SRY-box transcription factor 5, proline, glutamate and leucine rich protein 1, and insulin-like growth factor 1 receptor (IGF1R) (11–13). Moreover, the function and mechanism of miR-497 in TNBC remain largely undetermined. Yes-associated protein 1 (YAP1) is a major downstream transducer of the Hippo pathway commonly identified as an oncogene (14). Previous studies have demonstrated that YAP1 is involved in various physiological process, including cell proliferation, cell apoptosis, stem cell differentiation and tumorigenesis (15,16). Specifically, when the Hippo pathway is inactive, YAP1 translocates to the nucleus, binds to other transcription factors, such as TEA domain transcription factor 4 (TEAD4), and drives the expression of target anti-apoptotic and proliferation genes (17). Furthermore, upon activation of the pathway, YAP1 is phosphorylated by the phosphorylated and activated form of large tumor suppressor (LATS) 1/2, leading to cytoplasmic retention of YAP1 by the 14-3-3 protein or degradation (17). Previous studies, including own research, have confirmed that dysregulation of YAP1 results in tumorigenesis, including in TNBC (18–21). Several miRNA molecules, such as miR-195-5p (22), miR-630 (23) and miR-1285-3p (24) were identified as YAP1 regulators. Nevertheless, further understanding of the post-transcriptional control of YAP1 in TNBC is necessary.
Therefore, the aim of the present study was to examine the function and mechanism of miR-497 in TNBC. The expression of miR-497 in was evaluated in tissue samples from patients with TNBC. The findings of the current study may provide a potential target for the treatment of TNBC.
Materials and methods
Clinical samples
In the present study, 36 pairs of TNBC and adjacent non-cancerous tissue samples were collected from patients in the Department of Breast and Thyroid Surgery of Shanghai No. 10 People's Hospital (Shanghai, China). All samples were snap-frozen in liquid nitrogen. None of the patients received any cancer treatment prior to surgery. TNBC diagnosis was based on a pathological report of ER, PR and cerbb-2 expression status, as well as a fluorescence in situ hybridization report of HER2 expression status. TNBC is defined as a tumor that is ER−PR−HER2−. When the cerbb-2 status in a pathological report is negative or 1+, HER2− is indicated. When the cerbb-2 status is 2+ or 3+, the HER2 status is detected by FISH. The study protocols were approved by the Institutional Ethics Committees of Shanghai No. 10 People's Hospital (approval no. SHSY-IEC-KY-4.0/17-83/01).
Samples were collected from 36 female patients with TNBC between December 2014 and December 2018 randomly selected from the Department of Thyroid and Breast Surgery of Shanghai No. 10 People's Hospital. The patients had a median age of 57 years (age range, 32–87 years). These patients did not receive any chemotherapy, radiotherapy or other comprehensive treatment before operation. The adjacent normal breast tissues were >5 cm away from the edge of the tumor. The blood vessels and adipose tissue around the tissues were removed. The tissues were chopped up with a scalpel and quickly put into a cryopreservation tube and into a portable liquid nitrogen tank. At the end of the day, the specimens were transferred to the laboratory liquid nitrogen tank for preservation. In order to avoid the degradation of tissue RNA by exogenous RNase, the samples were stored at −80°C immediately after tissue collection.
For survival analysis, the identification of the cut-off point to distinguish between high or low miR-497 expression was obtained by drawing a receiver operating characteristic curve. The sensitivity, specificity and Youden index were calculated (sensitivity, 0.909; specificity, 0.778; area under the curve, 0.854). The cut-off point (cut-off, 3.68) of miR-497 expression was located in the maximum of the Youden index, and this value was considered as the cut-off point to distinguish between high or low miR-497 expression. Kaplan-Meier analysis and log-rank test were used to evaluate the association between miR-497 expression and the overall survival of patients with TNBC.
Cell culture and transfection
The human TNBC cell lines MDA-MB-231, MDA-MB-468, MCF-7 and SKBR3, and 293T cells were purchased from The Cell Bank of Type Culture Collection of The Chinese Academy of Sciences. The cells were maintained in Dulbecco's Modified Eagle's Medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), penicillin (100 units/ml) and streptomycin (100 µg/ml). The human normal breast epithelial cell line, MCF-10A, was purchased from Shanghai Zhongqiao Xinzhou Biotechnology Co., Ltd. The MCF-10A cells were cultured in Mammary Epithelial Cell Medium (ScienCell Research Laboratories, Inc.). All cells were incubated at 37°C with 5% CO2. miR-497 mimics (sense, 5′-UGUUUGGUGUCACACGACGAC-3′ and antisense, 5′-GUCGUCGUGUGACAACCAAACA-3′) and non-specific negative control (NC; sense, 5′-UUCUCCGAACGUGUCACGUTT-3′, and antisense, 5′-ACGUGACACGUUCGGAGAATT-3′) oligos were purchased from Guangzhou RiboBio Co., Ltd. For transfection, actively growing MDA-MB-231 cells (25×105 cells/well) were plated into 6-well plates (BD Biosciences) and cultured with serum- and antibiotic-free DMEM. When cell density achieved 30–40% confluence, cells were transfected with 100 nM miR-497 mimics or NC using Lipofectamine® (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. All cells were incubated at 37°C with 5% CO2 for 5 h, and complete media was changed 5 h after transfection. All cells were incubated at 37°C with 5% CO2 for 48 h before subsequent experimentation.
RNA extraction and reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from cells or tissues using the TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). cDNA was synthesized using the PrimeScript RT kit for mRNA and Prime-Script miRNA cDNA Synthesis kit (both Takara Bio, Inc.) for miRNA, according to the manufacturer's instructions. qPCR was carried out using the SYBR® FAST qRT-PCR Master Mix kit (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol on a 7900HT fast RT-PCR instrument (Applied Biosystems; Thermo Fisher Scientific, Inc.). The primer sequences were designed and synthesized by Guangzhou RiboBio Co., Ltd. The primer sequences were as follows: miR-497 forward, 5′-GTGCAGGGTCCGAGGT-3′ and reverse, 5′-TAGCCTGCAGCACACTGTGGT-3′; U6 forward, 5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse, 5′-CGCTTCACGAATTTGCGTGTCAT-3′; YAP1 forward, 5′-AGAACAATGACGACCAATAGCTC-3′ and reverse, 5′-GTCGATGGCTAGTCGTAGCATCGAT-3′; and GAPDH forward, 5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse, 5′-TGCTAGCTGGCATGCCCGATCGATC-3′. Relative mRNA and miRNA levels were normalized to GAPDH and U6, respectively, and were obtained from the threshold cycle (Cq) values using the 2−ΔΔCq method (25). qPCR parameters for mRNA and miRNA quantification were as follows: 2 min at 95°C, followed by 40 cycles of 30 sec at 95°C and 45 sec at 60°C. Each sample was tested in triplicate.
Cell proliferation assays
For the MTT assays, transfected cells (2×103 cells/well) were first plated into 96-well plates. Cell proliferation was detected at 24, 48, 72 and 96 h detected using an MTT kit (Sigma-Aldrich; Merck KGaA) according to the manufacturer's instructions. The 490-nm optical density was measured using a microplate reader.
For the colony formation assays, transfected cells (500 cells/well) were transferred to six-well plates. The cell culture was terminated after 7–10 days or when the colonies were visible. The plates were washed twice with PBS, and the colonies were fixed in 95% ethanol for 10 min at room temperature, dried and stained with 0.1% crystal violet solution for 10 min at room temperature, and each plate was washed three times with water.
Transwell assays
After 48 h from transfection, cells of each group were harvested and made into single cell suspension at a density of 5×103/ml using serum-free medium. The Transwell assays were carried out using a 24-well insert with Matrigel-coated upper chambers (BD Biosciences). The transfected cells were plated into the upper chamber in serum-free medium. Medium containing 10% FBS was added to the lower chamber. The 24-well Matrigel-coated chambers were incubated at 37°C for 24 h. Subsequently, cells were fixed with 4% paraformaldehyde for 10 min at room temperature and stained with a 0.05% crystal violet solution for 10 min at room temperature. Representative images were obtained using a light microscope (magnification, ×200) and stained cells were counted in five randomly selected fields.
Cell cycle analysis
The transfected cells were harvested and fixed in 70% ethanol overnight at 4°C. After washing twice with cold PBS, 250 µl of a 0.05 g/l propidium iodide (PI; Beyotime Institute of Biotechnology) staining solution was added into each sample, followed by incubation for 30 min at room temperature. The cell cycle distribution data was acquired using a FACSCanto II flow cytometer (BD Biosciences) and analyzed using ModFit LT 3.2 (Verity Software House, Inc.).
Apoptosis assay
Cells transfected with miR-497 mimics or NC were incubated in six-well plates for 24 h. The cells were subsequently stained with fluorescein (FITC)-conjugated Annexin V and propidium iodide (BD Biosciences) for 30 min at room temperature. The Annexin V incubation reagent (100 µl) was prepared by combining 10 µl of the 10X binding buffer (BD Biosciences), 10 µl of propidium iodide (PI), 1 µl of Annexin V-FITC and 79 µl of deionized, distilled H2O. The cells were gently resuspended in the Annexin V incubation reagent at a concentration of 105−106 cells/100 µl. The rate of apoptosis was detected using a FACSCanto II flow cytometer (BD Biosciences) and analyzed using the CellQuest Pro software v1.0 (BD Biosciences).
Dual-luciferase reporter assay
293T cells were seeded in 12-well plates (BD Biosciences) and cultured until the cells reached 80–90% confluence. The 3′-UTR segments of the YAP1 mRNA sequence containing the predicted miR-497 binding sites were amplified by PCR using the PrimerStar kit (Takara Bio, Inc.) according to the manufacturer's protocol. The corresponding mutant constructs were created by mutating the seed regions of the miR-497-binding sites (5′-UGC UGC U-3′ to 5′-ACG AGC A-3′). Fragments were subcloned into the XhoI site in the 3′-UTR of Renilla luciferase of the psiCHECK-2 reporter vector (Shanghai Aibo Si Biological Technology Co., Ltd.). Cells were transiently co-transfected with 0.2 µg psiCHECK-2/YAP1 3′-UTR or psiCHECK-2/YAP1 3′-UTR mutant reporter plasmids together with 100 nmol/l miR-497 mimic or miR-NC using Lipofectamine 2000, according to the manufacturer's instructions. After 48 h, firefly and Renilla luciferase activities were measured using a Dual Luciferase Assay (Promega Corporation). Firefly luciferase activity was normalized to Renilla luciferase activity, and the ratio of firefly/Renilla luciferase activity was presented.
Western blot analysis
Cells were washed in ice-cold PBS and resuspended in RIPA lysis buffer (100 µl/well; Beyotime Institute of Biotechnology). Subsequently, the cells were collected and centrifuged at 12,000 × g for 30 min at 4°C (Eppendorf 5804R; Eppendorf). Supernatants were collected and the protein concentrations were quantified using a BCA protein assay kit (Beyotime Institute of Biotechnology). Protein samples were denatured with 5X SDS loading buffer (Beyotime Institute of Biotechnology) at 100°C for 10 min. Total protein (20 µg/lane)was separated by 8% SDS-PAGE (Beyotime Institute of Biotechnology) and transferred to a 0.45-µm nitrocellulose membrane (Beyotime Institute of Biotechnology). Following 60 min of blocking at room temperature with 5% skimmed milk, the membrane was incubated overnight at 4°C with primary antibodies against YAP1 (1:1,000; cat. no. BS1701; Bioworld Technology, Inc.) and β-actin (1:1,000; cat. no. AP0060; Bioworld Technology, Inc.). After washing with PBS-Tween (0.2%), blots were washed and incubated for 1 h at room temperature with the anti-mouse secondary fluorescence antibody (1:2,000; cat. no. 00002-1; Bioworld Technology, Inc.). Finally, immunoreactive protein bands were detected with an Odyssey Scanning system (LI-COR Biosciences).
Database analysis
Online databases were used to elucidate the target genes of miR-497 in breast cancer. Database analysis was performed using starBase3.0 (http://starbase.sysu.edu.cn) and TargetScan 7.2 (http://www.targetscan.org) according to the guidelines of the websites.
Statistical analysis
Data were presented as the mean ± SD from at least three independent experiments. All statistical analyses were carried out using GraphPad Prism version 6.0 (GraphPad Software, Inc.) and SPSS version 20.0 (IBM Corp.). All experiments were independently repeated three times. The differences between two paired groups (normal vs. tumor tissues) were assessed using paired Student's t-test. The differences between two unpaired groups were assessed using unpaired Student's t-test. Differences between multiple cell lines were assessed using one-way ANOVA followed by the Least Significant Difference test. Differences between tumor stages were assessed using Kruskal-Wallis test followed by Steel-Dwass' post-hoc test. Survival curves were analyzed using the Kaplan-Meier method and the log-rank test. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-497 expression is reduced in TNBC tissue samples, which is associated with poor prognosis in patients with TNBC
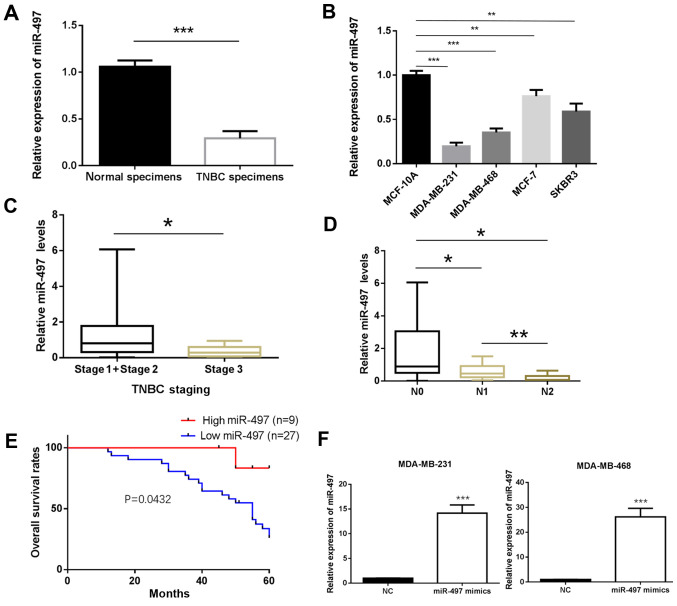
The expression of miR-497 in the TNBC and matched non-cancerous breast tissue samples was determined by RT-qPCR. The results showed that miR-497 was significantly downregulated in TNBC tissue compared with normal tissue samples (P<0.001; Fig. 1A). Furthermore, the expression of miR-497 was significantly decreased in the MDA-MB-231 and MDA-MB-468 cell lines compared with MCF-10A (P<0.01; Fig. 1B). The experiments carried out in the MCF-7 (Luminal A type) and SKBR3 (HER-2-overexpressing type) cells also exhibited a similar trend (P<0.001; Fig. 1B). To further investigate the clinicopathological and prognostic significance of miR-497, the clinical data of the 36 patients with TNBC was collected and analyzed together with the miR-497 levels. As demonstrated in Fig. 1C and D, the expression of miR-497 was significantly lower in the TNBC patients with an advanced TNM stage of the disease (P<0.05; Fig. 1C) and in those with lymph node metastasis compared with those without (P<0.05; Fig. 1D). The Kaplan-Meier method was used to evaluate the association between the expression of miR-497 and the survival rate of patients with TNBC. Patients with low expression of miR-497 had a shorter survival compared to those with high expression (P<0.05; Fig. 1E). The MDA-MB-231 and MDA-MB-468 cells were transfected with miR-497 mimics for subsequent experiments. RT-qPCR analysis confirmed that miR-497 expression was upregulated in miR-497 mimics-transfected MDA-MB-231 and MDA-MB-468 cells compared with in cells transfected with miR-NC (P<0.001; Fig. 1F).
Figure 1.
Low miR-497 expression in TNBC is associated with poor survival. (A) Expression of miR-497 in 36 samples from patients with TNBC tissues and matched non-cancerous breast tissue samples. (B) Expression of miR-497 in breast cancer cell lines and MCF-10A cells. The data are normalized to the MCF-10A group. (C and D) Expression of miR-497 in patients with TNBC with (C) advanced stage or (D) different N stages. (E) Association of high or low expression of miR-497 with overall survival in patients with TNBC patients. (F) miR-497 expression is upregulated in MDA-MB-231 and MDA-MB-468 cells following transfection with miR-497 mimics. *P<0.05, **P<0.01, ***P<0.001. TNBC, triple-negative breast cancer; miR, microRNA; N, lymph node metastasis; NC, negative control.
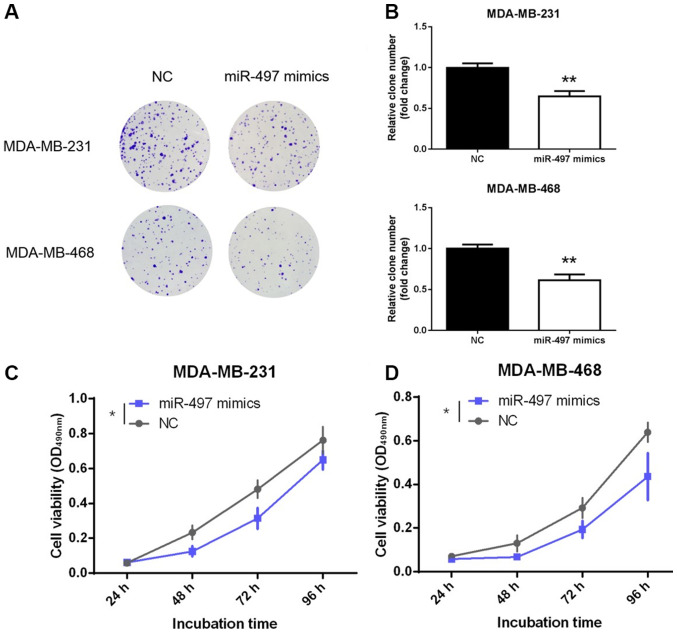
miR-497 inhibits proliferation of TNBC cells
Decreased expression of miR-497 in TNBC tissues may reflect tumor-suppressing role in TNBC. To further explore the function of miR-497, its effect on the proliferation of TNBC cells was evaluated. MDA-MB-231 and MDA-MB-468 cells were transfected with miR-497 mimics or NC. Notably, the representative results obtained from the colony formation assay showed fewer colonies in the miR-497 mimics group compared with the NC group (P<0.01; Fig. 2A and B). Moreover, the MTT assay also demonstrated that overexpression of miR-497 significantly inhibited the viability of the MDA-MB-231 and MDA-MB-468 cell lines compared with NC at the 96-h time point (P<0.05; Fig. 2C and D). These findings confirmed that miR-497 inhibited the proliferation and viability of the TNBC cells in vitro.
Figure 2.
miR-497 inhibits the proliferation of triple-negative breast cancer cells. (A) Effects of overexpression of miR-497 on colony formation in MDA-MB-231 and MDA-MB-468 cells. (B) Colony-forming efficiency in MDA-MB-231 and MDA-MB-468 cells. (C and D) Cell viability of MDA-MB-231 and MDA-MB-468 cells following overexpression of miR-497. *P<0.05, **P<0.01. miR, microRNA; NC, negative control; OD, optical density.
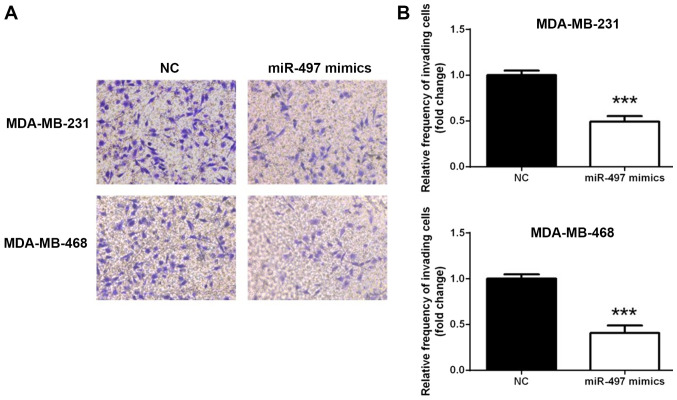
miR-497 inhibits the invasion of TNBC cells
To investigate whether miR-497 overexpression affected the TNBC cell invasion, a Transwell assay was conducted. As shown in Fig. 3, the MDA-MB-231 and MDA-MB-468 cell lines exhibited similar trends in terms of invasion (Fig. 3A). The number of cells penetrating the membrane significantly decreased 24 h following transfection with miR-497 mimics compared with the NC (P<0.001; Fig. 3B). This observation indicated that overexpression of miR-497 suppressed TNBC cell invasion in vitro.
Figure 3.
miR-497 inhibits the invasion of triple-negative breast cancer cells. (A) Representative photomicrographs of the Transwell assay using the MDA-MB-231 and MDA-MB-468 cells following overexpression of miR-497 (magnification, ×200). (B) Changes in the number of cells penetrating the membrane in the Transwell assay. ***P<0.001. miR, microRNA; NC, negative control.
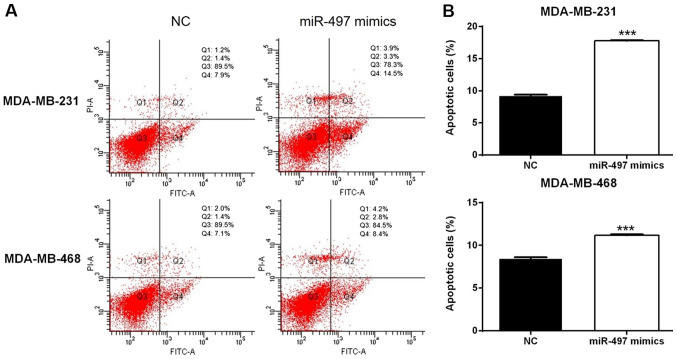
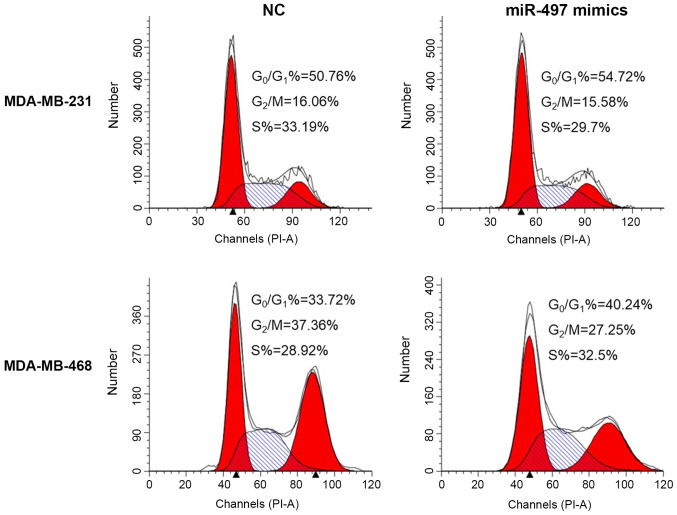
miR-497 disrupts the cell cycle and induces apoptosis in TNBC cells
Flow cytometry suggested that overexpression of miR-497 significantly increased the apoptosis rate in the MDA-MB-231 and MDA-MB-468 cells compared with the respective NC cells (Fig. 4). In addition, the cell cycle evaluation demonstrated that the percentage of cells in the G0/G1 phase increased in the miR-497 mimics group compared with the NC group in both MDA-MB-231 and MDA-MB-468 cell lines (Fig. 5). Thus, the upregulation of miR-497 could impact the cell cycle distribution and induce apoptosis in TNBC cells.
Figure 4.
Effects of miR-497 on cell apoptosis in the triple-negative breast cancer cells. miR-497 induced early (Q4) and late (Q2) apoptosis of MDA-MB-231 and MDA-MB-468 cells, compared with NC. (A) Effects of overexpression of miR-497 on cell apoptosis in MDA-MB-231 and MDA-MB-468 cells. (B) Percentage of apoptotic cells in MDA-MB-231 and MDA-MB-468 cells following overexpression of miR-497. ***P<0.001. miR, microRNA; NC, negative control; PI, propidium iodide; FITC, fluorescein isothiocyanate.
Figure 5.
Effects of miR-497 on cell cycle progression of triple-negative breast cancer cells. The relative proportions of cells in the G0/G1, S and G2/M phases of each group are shown. The percentage of cells in the G0/G1 phase was increased in the miR-497 mimics group compared with the NC group, both in the MDA-MB-231 and MDA-MB-468 cell lines. miR, microRNA; NC, negative control; PI, propidium iodide.
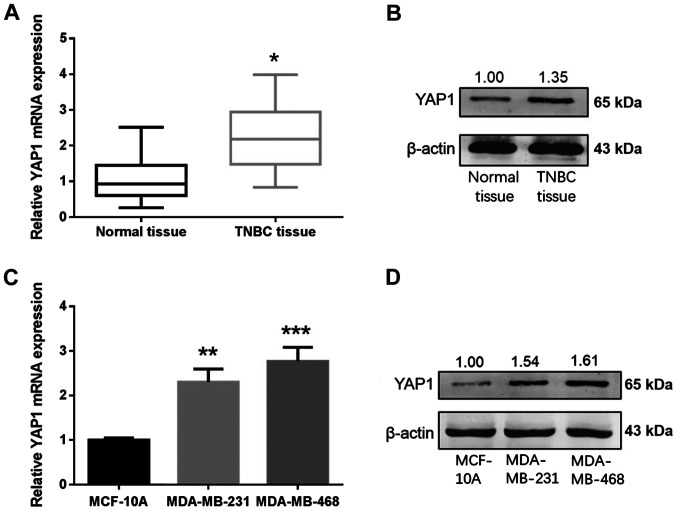
YAP1 is upregulated in TNBC and is a direct target for miR-497 in TNBC cells
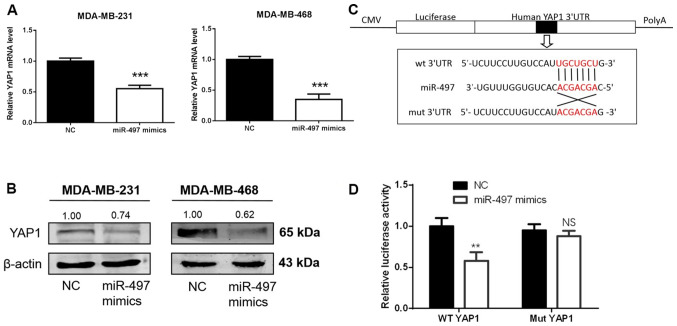
Online databases were used to elucidate the mechanism of miR-497 in breast cancer. Based on the analysis carried out in databases, miR-497 exhibited a high prediction score for interaction with YAP1. The expression levels of YAP1 in 36 paired samples from patients with TNBC and cell lines were measured using RT-qPCR and western blotting. YAP1 mRNA and protein levels were increased in the TNBC tissues in comparison with normal tissues (Fig. 6A and B). YAP1 mRNA and protein levels were higher in the TNBC cells (MDA-MB-231 and MDA-MB-468) compared with MCF-10A cells (Fig. 6C and D). To determine whether miR-497 regulates endogenous YAP1 at the mRNA or protein levels, miR-497 mimics or NC were transfected into the MDA-MB-231 and MDA-MB-468 cells, and the levels of YAP1 mRNA and protein were detected 48 h following transfection. YAP1 mRNA and protein levels in the MDA-MB-231 and MDA-MB-468 cells were markedly downregulated after transfection with miR-497 mimics (Fig. 7A and B). To establish whether miR-497 directly targeted YAP1, a luciferase reporter assay was carried out in 293T cells (Fig. 7C). In the wt YAP1 group, luciferase activity decreased following transfection with the miR-497 mimics compared with NC, while no evident differences were found in the mut YAP1 groups (Fig. 7D). Thus, miR-497 binds to the 3′-UTR of YAP1 mRNA, suggesting that YAP1 is a direct target of miR-497 in TNBC cells.
Figure 6.
YAP1 is upregulated in TNBC tissue samples and cell lines. (A) YAP1 mRNA in 36 samples from patients with TNBC tissues and matched non-cancerous breast tissue samples. (B) Expression of the YAP1 protein in TNBC tissue samples and matched non-cancerous breast tissue. (C and D) YAP1 (C) mRNA and (D) protein levels in the MCF-10A and TNBC cell lines. *P<0.05, **P<0.01, ***P<0.001. TNBC, triple-negative breast cancer; miR, microRNA; NC, negative control; YAP1, Yes-associated protein 1.
Figure 7.
YAP1 is a direct target of miR-497 in triple-negative breast cancer cells. (A and B) Expression of YAP1 mRNA and protein in MDA-MB-231 and MDA-MB-468 cells following miR-497 mimics transfection. miR-497 downregulates the expression of YAP1 at the mRNA and protein levels. (C) Binding site for miR-497 in the 3′-UTR of the YAP1 mRNA. (D) Relative luciferase activity was measured in 293T cells following co-transfection of the WT or Mut YAP1 3′-UTR luciferase constructs with either miR-497 mimics or NC. **P<0.01, ***P<0.001. miR, microRNA; NC, negative control; WT, wild-type; Mut, mutant; UTR, untranslated region; CMV, cytomegalovirus.
Discussion
Breast cancer is the most common malignant tumor in female patients. The incidence and mortality of breast cancer in China has markedly increased in recent years (26). It is noteworthy that the five-year disease-free survival rate of TNBC is the lowest among all molecular types of breast cancer (26). The health and quality of life of patients with TNBC is significantly challenged upon the occurrence of recurrence or metastasis. Thus, the determination of new targets to classify TNBC as well as the development of appropriate treatment are important. Recently, various miRNA molecules have been reported to be aberrantly expressed during the occurrence and development of TNBC. Some of these miRNA molecules are upregulated in TNBC tissue, such as miR-155, miR-21, whereas others are downregulated, including miR-10b, miR-125b and miR-145 (27). Sánchez-González et al (28) found that low miR-149 expression was associated with reduced macrophage infiltration and patient survival in lymph node-positive TNBC. Moreover, Lee et al (29) reported that miR-137 was markedly downregulated in TNBC tissue, and proposed this miRNA as a target for the treatment of TNBC.
The purpose of the present study was to explore the potential function and mechanism of miR-497 in TNBC. The results indicated that expression of miR-497 was downregulated in TNBC specimens and in the MDA-MB-231 and MDA-MB-468 cell lines. The clinical outcome analysis demonstrated that low expression of miR-497 was associated with advanced TNM stage, lymph node metastasis and reduced patient survival. Therefore, it was hypothesized that miR-497 may function as a tumor suppressor in TNBC. To confirm this, miR-497 mimics were transfected into the MDA-MB-231 and MDA-MB-468 cells to overexpress miR-497. miR-497 markedly suppressed the growth and invasion of the transfected TNBC cells in vitro. Furthermore, overexpression of miR-497 also induced apoptosis and arrested the cell cycle in the G0/G1 phase. Overall, the results of the current study demonstrated that miR-497 exhibited anti-cancer properties, which might attenuate the progression of TNBC. These results were partly analogous to those of previous studies concerning other types of cancer and molecular types of breast cancer (30–33).
Online databases were used to establish the molecular mechanisms of miR-497 in the progression of TNBC. YAP1 was identified as the potential targets of miR-497. A luciferase reporter assay to validate the regulation of the putative target YAP1 by miR-497. The results suggested that miR-497 specifically bound to the 3′-UTR of the YAP1 mRNA. Several targets of miR-497 have been previously identified in various cancer types. For instance, Shen et al (30) found that miR-497 significantly inhibited Bcl-w levels in breast cancer. Subsequently, Bcl-2 (10), cyclin E1 (31), VEGFR2 (32) and IGF-1R (33) were identified as targets of miR-497. Moreover, YAP1 was identified as a target of miR-497 in thyroid papillary carcinoma (7). It has also been reported that some long non-coding RNA molecules could bind to miR-497. For example, Li et al (34) suggested that DLX6-AS1 regulated the progression of neuroblastoma by targeting YAP1 via miR-497. Moreover, Duan et al (35) reported that LINC02476 promoted the malignant phenotype of hepatocellular carcinoma by sponging miR-497. However, our study is the first to show the relevance of miR-497 modulation in TNBC.
YAP1 is a major downstream transducer of the Hippo signaling pathway, which is known as a critical player in multiple human cancer types, including TNBC (36). The Hippo signaling pathway comprises numerous components and its upstream genes include mammalian Ste20-like kinases 1/2 (MST1/2) and LATS1/2. Moreover, the downstream genes include YAP1 and transcriptional coactivator with the PDZ-binding motif (37). Guo et al (38) determined that the overexpression of YAP1 was associated with poor prognosis of breast cancer patients and induces breast cancer cell growth by inhibiting PTEN. A recent study also reported that YAP-independent mechanotransduction drives breast cancer progression, suggesting a new mechanism underlying the role of YAP1 in breast cancer (39). In the present study, YAP1 mRNA and protein levels were upregulated in TNBC tissue and cell lines. Several miRNA molecules have been previously found to be critical upstream regulators of YAP1. For instance, Li et al (40) established that miR-141-3p regulated the proliferation and senescence of stem cells from apical papilla by targeting YAP1. Furthermore, Chen et al (41) demonstrated that miR-590-5p suppressed chemoresistance of hepatocellular carcinoma by targeting the expression of YAP1. Our previous study also suggested that miR-506 inhibited cell growth and disrupted the cell cycle by targeting YAP in breast cancer cells (19). In the current study, YAP1 mRNA and protein levels in the MDA-MB-231 and MDA-MB-468 cells were downregulated following overexpression of miR-497, indicating that miR-497 was an upstream regulator of YAP1.
This study had certain limitations. Firstly, only 36 cases of TNBC tissues could be obtained due to the limited number of TNBC patients in our hospital. The reliability based on these samples is relatively low and the results require validation in a larger number of samples. Secondly, in vivo experiments were not conducted due to the limitations of the laboratory conditions. Lastly, the downstream proteins of YAP1 should be explored to gain more comprehensive understanding of the role of miR-497 in TNBC.
In conclusion, the present study demonstrated that miR-497 was downregulated in TNBC tissue samples and cells. It was also confirmed that miR-497 inhibited cell proliferation and migration via direct regulation of YAP1 expression. This suggests that miR-497 may represent a potential therapeutic target for TNBC.
Acknowledgements
Not applicable.
Funding Statement
The present study was supported by grants from The National Natural Science Foundation of China (grant. no. 82172240), Shanghai Municipal Health Bureau of Shanghai (grant. no. 201640097) and Shanghai Science and Technology Commission Guidance Project (grant. no. 17411967200).
Funding
The present study was supported by grants from The National Natural Science Foundation of China (grant. no. 82172240), Shanghai Municipal Health Bureau of Shanghai (grant. no. 201640097) and Shanghai Science and Technology Commission Guidance Project (grant. no. 17411967200).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YL, KH and LF made substantial contributions to conception and design. JJ performed the experiments. YL drafted the manuscript. YL and KH confirmed the authenticity of the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study protocols were approved by the Institutional Ethics Committees of Shanghai No. 10 People's Hospital (approval no. SHSY-IEC-KY-4.0/17-83/01). Written informed consent was obtained from the participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Cardoso F, Harbeck N, Barrios CH, Bergh J, Cortés J, El Saghir N, Francis PA, Hudis CA, Ohno S, Partridge AH, et al. Research needs in breast cancer. Ann Oncol. 2017;28:208–217. doi: 10.1093/annonc/mdw571. [DOI] [PubMed] [Google Scholar]
- 2.Zhai Q, Li H, Sun L, Yuan Y, Wang X. Identification of differentially expressed genes between triple and non-triple-negative breast cancer using bioinformatics analysis. Breast Cancer. 2019;26:784–791. doi: 10.1007/s12282-019-00988-x. [DOI] [PubMed] [Google Scholar]
- 3.Mouh FZ, Mzibri ME, Slaoui M, Amrani M. Recent progress in triple negative breast cancer research. Asian Pac J Cancer Prev. 2016;17:1595–1608. doi: 10.7314/APJCP.2016.17.4.1595. [DOI] [PubMed] [Google Scholar]
- 4.Yuan Q, Zheng L, Liao Y, Wu G. Overexpression of CCNE1 confers a poorer prognosis in triple-negative breast cancer identified by bioinformatic analysis. World J Surg Oncol. 2021;19:86. doi: 10.1186/s12957-021-02200-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pratama MY, Pascut D, Massi MN, Tiribelli C. The role of microRNA in the resistance to treatment of hepatocellular carcinoma. Ann Transl Med. 2019;7:577. doi: 10.21037/atm.2019.09.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding L, Gu H, Xiong X, Ao H, Cao J, Lin W, Yu M, Lin J, Cui Q. MicroRNAs involved in carcinogenesis, prognosis, therapeutic resistance and applications in human triple-negative breast cancer. Cells. 2019;8:1492. doi: 10.3390/cells8121492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng H, Dong H, Feng J, Tian H, Zhang H, Xu L. miR-497 inhibited proliferation, migration and invasion of thyroid papillary carcinoma cells by negatively regulating YAP1 expression. Onco Targets Ther. 2018;11:4711–4721. doi: 10.2147/OTT.S164052. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Huang Q, Li H, Dai X, Zhao D, Guan B, Xia W. miR-497 inhibits the proliferation and migration of A549 non-small-cell lung cancer cells by targeting FGFR1. Mol Med Rep. 2019;20:3959–3967. doi: 10.3892/mmr.2019.10611. [DOI] [PubMed] [Google Scholar]
- 9.Xu GS, Li ZW, Huang ZP, Brunicardi FC, Jia F, Song C, Zou HJ, Sun RF. MiR-497-5p inhibits cell proliferation and metastasis in hepatocellular carcinoma by targeting insulin-like growth factor 1. Mol Genet Genomic Med. 2019;7:e00860. doi: 10.1002/mgg3.860. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Wei C, Luo Q, Sun X, Li D, Song H, Li X, Song J, Hua K, Fang L. MicroRNA-497 induces cell apoptosis by negatively regulating Bcl-2 protein expression at the posttranscriptional level in human breast cancer. Int J Clin Exp Pathol. 2015;8:7729–7739. [PMC free article] [PubMed] [Google Scholar]
- 11.Li G, Wang K, Wang J, Qin S, Sun X, Ren H. miR-497-5p inhibits tumor cell growth and invasion by targeting SOX5 in non-small-cell lung cancer. J Cell Biochem. 2019;120:10587–10595. doi: 10.1002/jcb.28345. [DOI] [PubMed] [Google Scholar]
- 12.Wang L, Li K, Lin X, Yao Z, Wang S, Xiong X, Ning Z, Wang J, Xu X, Jiang Y, et al. Metformin induces human esophageal carcinoma cell pyroptosis by targeting the miR-497/PELP1 axis. Cancer Lett. 2019;450:22–31. doi: 10.1016/j.canlet.2019.02.014. [DOI] [PubMed] [Google Scholar]
- 13.Ma W, Feng W, Tan J, Xu A, Hu Y, Ning L, Kang Y, Wang L, Zhao Z. miR-497 may enhance the sensitivity of non-small cell lung cancer cells to gefitinib through targeting the insulin-like growth factor-1 receptor. J Thorac Dis. 2018;10:5889–5897. doi: 10.21037/jtd.2018.10.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Overholtzer M, Zhang J, Smolen GA, Muir B, Li W, Sgroi DC, Deng CX, Brugge JS, Haber DA. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci U S A. 2006;103:12405–12410. doi: 10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson R, Halder G. The two faces of hippo: Targeting the hippo pathway for regenerative medicine and cancer treatment. Nat Rev Drug Discov. 2014;13:63–79. doi: 10.1038/nrd4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng Y, Pan D. The hippo signaling pathway in development and disease. Dev Cell. 2019;50:264–282. doi: 10.1016/j.devcel.2019.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng X, Fang L. VGLL4 is a transcriptional cofactor acting as a novel tumor suppressor via interacting with TEADs. Am J Cancer Res. 2018;8:932–943. [PMC free article] [PubMed] [Google Scholar]
- 19.Hua K, Yang W, Song H, Song J, Wei C, Li D, Fang L. Up-regulation of miR-506 inhibits cell growth and disrupt the cell cycle by targeting YAP in breast cancer cells. Int J Clin Exp Med. 2015;8:12018–12027. [PMC free article] [PubMed] [Google Scholar]
- 20.Cao L, Sun PL, Yao M, Jia M, Gao H. Expression of YES-associated protein (YAP) and its clinical significance in breast cancer tissues. Hum Pathol. 2017;68:166–174. doi: 10.1016/j.humpath.2017.08.032. [DOI] [PubMed] [Google Scholar]
- 21.Zhu C, Li L, Zhang Z, Bi M, Wang H, Su W, Hernandez K, Liu P, Chen J, Chen M, et al. A non-canonical Role of YAP/TEAD is required for activation of estrogen-regulated enhancers in breast cancer. Mol Cell. 2019;75:791–806.e8. doi: 10.1016/j.molcel.2019.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X, Zhou Y, Ning YE, Gu H, Tong Y, Wang N. MiR-195-5p inhibits malignant progression of cervical cancer by targeting YAP1. Onco Targets Ther. 2020;13:931–944. doi: 10.2147/OTT.S227826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Q, Ding J, Nan G, Lyu Y, Ni G. LncRNA NOC2L-4.1 functions as a tumor oncogene in cervical cancer progression by regulating the miR-630/YAP1 pathway. J Cell Biochem. 2019;120:16913–16920. doi: 10.1002/jcb.28949. [DOI] [PubMed] [Google Scholar]
- 24.Hu XH, Dai J, Shang HL, Zhao ZX, Hao YD. miR-1285-3p is a potential prognostic marker in human osteosarcoma and functions as a tumor suppressor by targeting YAP1. Cancer Biomark. 2019;25:1–10. doi: 10.3233/CBM-180013. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Thakur V, Kutty RV. Recent advances in nanotheranostics for triple negative breast cancer treatment. J Exp Clin Cancer Res. 2019;38:430. doi: 10.1186/s13046-019-1443-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piasecka D, Braun M, Kordek R, Sadej R, Romanska H. MicroRNAs in regulation of triple-negative breast cancer progression. J Cancer Res Clin Oncol. 2018;144:1401–1411. doi: 10.1007/s00432-018-2689-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sánchez-González I, Bobien A, Molnar C, Schmid S, Strotbek M, Boerries M, Busch H, Olayioye MA. miR-149 suppresses breast cancer metastasis by blocking paracrine interactions with macrophages. Cancer Res. 2020;80:1330–1341. doi: 10.1158/0008-5472.CAN-19-1934. [DOI] [PubMed] [Google Scholar]
- 29.Lee SJ, Jeong JH, Kang SH, Kang J, Kim EA, Lee J, Jung JH, Park HY, Chae YS. MicroRNA-137 inhibits cancer progression by targeting Del-1 in triple-negative breast cancer cells. Int J Mol Sci. 2019;20:6162. doi: 10.3390/ijms20246162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen L, Li J, Xu L, Ma J, Li H, Xiao X, Zhao J, Fang L. miR-497 induces apoptosis of breast cancer cells by targeting Bcl-w. Exp Ther Med. 2012;3:475–480. doi: 10.3892/etm.2011.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo QF, Li XY, Gao Y, Long Y, Chen L, Huang YX, Fang L. MiRNA-497 regulates cell growth and invasion by targeting cyclin E1 in breast cancer. Cancer Cell Int. 2013;13:95. doi: 10.1186/1475-2867-13-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tu Y, Liu L, Zhao D, Liu Y, Ma X, Fan Y, Wan L, Huang T, Cheng Z, Shen B. Overexpression of miRNA-497 inhibits tumor angiogenesis by targeting VEGFR2. Sci Rep. 2015;5:13827. doi: 10.1038/srep13827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo ST, Jiang CC, Wang GP, Li YP, Wang CY, Guo XY, Yang RH, Feng Y, Wang FH, Tseng HY, et al. MicroRNA-497 targets insulin-like growth factor 1 receptor and has a tumour suppressive role in human colorectal cancer. Oncogene. 2013;32:1910–1920. doi: 10.1038/onc.2012.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li C, Wang S, Yang C. Long non-coding RNA DLX6-AS1 regulates neuroblastoma progression by targeting YAP1 via miR-497-5p. Life Sci. 2020;11:117657. doi: 10.1016/j.lfs.2020.117657. [DOI] [PubMed] [Google Scholar]
- 35.Duan Y, Zhao M, Jiang M, Li Z, Ni C. LINC02476 promotes the malignant phenotype of hepatocellular carcinoma by sponging miR-497 and increasing HMGA2 expression. Onco Targets Ther. 2020;13:2701–2710. doi: 10.2147/OTT.S237069. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Andrade D, Mehta M, Griffith J, Panneerselvam J, Srivastava A, Kim TD, Janknecht R, Herman T, Ramesh R, Munshi A. YAP1 inhibition radiosensitizes triple negative breast cancer cells by targeting the DNA damage response and cell survival pathways. Oncotarget. 2017;8:98495–98508. doi: 10.18632/oncotarget.21913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng H, Zhang Z, Rodriguez-Barrueco R, Borczuk A, Liu H, Yu J, Silva JM, Cheng SK, Perez-Soler R, Halmos B. Functional genomics screen identifies YAP1 as a key determinant to enhance treatment sensitivity in lung cancer cells. Oncotarget. 2016;7:28976–28988. doi: 10.18632/oncotarget.6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo L, Chen Y, Luo J, Zheng J, Shao G. YAP1 overexpression is associated with poor prognosis of breast cancer patients and induces breast cancer cell growth by inhibiting PTEN. FEBS Open Bio. 2019;9:437–445. doi: 10.1002/2211-5463.12597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee JY, Chang JK, Dominguez AA, Lee HP, Nam S, Chang J, Varma S, Qi LS, West RB, Chaudhuri O. YAP-independent mechanotransduction drives breast cancer progression. Nat Commun. 2019;10:1848. doi: 10.1038/s41467-019-09755-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Z, Ge X, Lu J, Bian M, Li N, Wu X, Li Y, Yan M, Yu J. MiR-141-3p regulates proliferation and senescence of stem cells from apical papilla by targeting YAP. Exp Cell Res. 2019;383:111562. doi: 10.1016/j.yexcr.2019.111562. [DOI] [PubMed] [Google Scholar]
- 41.Chen M, Wu L, Tu J, Zhao Z, Fan X, Mao J, Weng Q, Wu X, Huang L, Xu M, Ji J. miR-590-5p suppresses hepatocellular carcinoma chemoresistance by targeting YAP1 expression. EBioMedicine. 2018;35:142–154. doi: 10.1016/j.ebiom.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.