Abstract
Human phenol sulfotransferases mediate the transfer of a sulfuryl moiety from the activated sulfate donor PAPS to hydroxy-containing substrates, altering substrate solubility and charge to affect phase II metabolism and cell signaling. Here, we present the development, computational modeling, in vitro enzymology, and biological application of STS-3, an activity-based fluorescent sensor for the SULT1A1 isoform.
Here, we report the development, computational modeling, in vitro enzymology, and biological application of an activity-based fluorescent sensor for the human phenol sulfotransferase SULT1A1.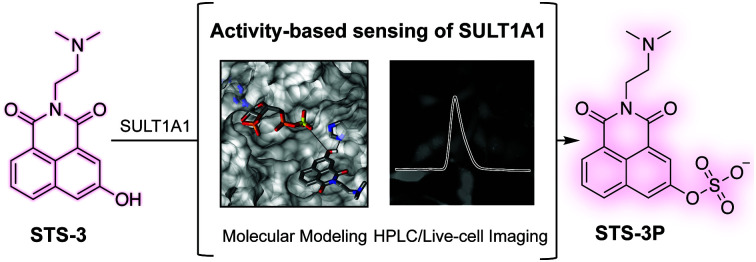
Human phenol sulfotransferases (SULT1 family) are essential phase II metabolic enzymes that mediate sulfuryl group transfer from the activated sulfate donor 3′-phosphoadenosine-5′-phosphosulfate (PAPS) primarily to hydroxy-containing small molecules.1–6 Sulfurylation enhances the water solubility to increase the clearance of xenobiotics, to recycle endogenous metabolites (e.g. estrogen, dopamine), and, in some cases, to (in)activate drugs.2–4,6,7 As such, phenol sulfotransferases are not only linked to cellular signaling in normal physiology but also in disease states ranging from cancer to neurodegeneration.1,3,4,6–12 Our understanding of the substrate scope and activities of these enzymes in biological contexts has been significantly advanced by parallel efforts using computational modeling, structural characterization, mechanistic enzymology, and assay methods.10,13–23
Along these lines, one of the most widely used approaches to monitor phenol sulfotransferase activity with purified protein or cell lysates relies on radiolabeling of substrates, PAP35S, or sulfate (35SO2−4), coupled to chromatographic detection of the sulfurylated products.24,25 Alternatively, coupled-enzyme assays with colorimetric and fluorescent substrates provide a safer, cost-effective, and continuous readout of enzyme activity.24–28 If radiolabeling is not preferred or if the sulfurylated product does not have an optical signature, mass spectrometry or nuclear magnetic resonance spectroscopy can be used.20,24,25,29,30 A limited number of these approaches have been translated to living cells but do not provide a direct readout with a spatially and temporally-resolved map of activity.31 We envision that activity-based fluorescent sensors can address this gap.32–39 This strategy affords the ability to chemically tune and transform an enzyme's substrate into a fluorescent imaging platform for live-cell applications. To our knowledge, activity-based sensing has not been widely exploited for phenol sulfotransferases in living cells. The SULT1 family consists of 9 isoforms, all with a high degree of sequence and structural similarity (≥55%).15 Of these isoforms, SULT1A1 is the most widely expressed in the human body and promiscuous, thus making it an ideal target for this proof-of-concept study.40,41
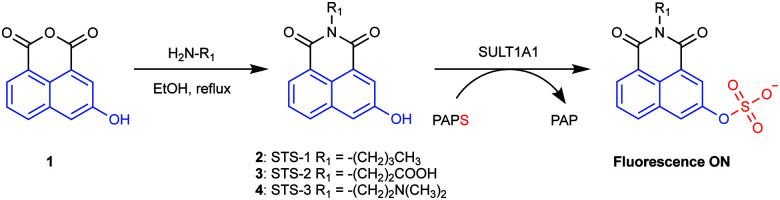
To generate an activity-based fluorescent sensor, we first selected the substrate 2-naphthol as it has been demonstrated to undergo sulfurylation by SULT1A1.15,18 Even though 2-naphthol and the resulting 2-naphthyl sulfate product are fluorescent in water, there are negligible differences in the emission spectra of these compounds at physiologically relevant pH.15,16 As such, our strategy to convert 2-naphthol into an activity-based fluorescent sensor relied on its structural similarity with 3-hydroxy-1,8-naphthalic anhydride (compound 1, Fig. 1). The latter can be readily functionalized with primary amines to generate naphthalimide fluorophores.42–47 Specifically, we selected three electronically distinct amines: butylamine (STS-1), 3-aminopropanoic acid (STS-2), and N,N-dimethylethylenediamine (STS-3) (Fig. 1). We reasoned that all three sensors would be weakly fluorescent because the non-bonding electrons on the oxygen atom of the phenol could quench the excited state of the naphthalimide fluorophore. However, the quenched state could be relieved upon sulfurylation to generate a turn-on or ratiometric fluorescence response, as previously reported for 3- and 4-substituted-1,8-naphthalimide-based sensors.42–44,48 We note that STS-1 has been evaluated as a fluorescent sensor for a plant glucosyltransferase in vitro,48 and STS-3 has been tested as an anti-cancer agent.45–47,49
Fig. 1. Synthesis of the sulfotransferase sensors STS-1, STS-2, and STS-3 and proposed sulfurylation of the embedded 2-naphthol (shown in blue) to generate a turn-on fluorescence response. The sulfuryl group is shown in red and is transferred from the PAPS (3′-phosphoadenosine-5′-phosphosulfate) cofactor to the sensor, generating the PAP (3′-phosphoadenosine-5′-phosphate) byproduct.
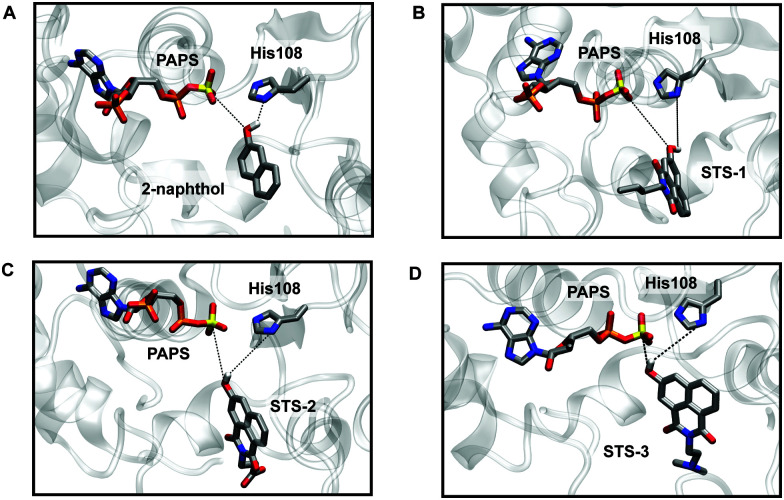
Preparatory docking calculations and constrained MD simulations were first carried out, followed by extensive equilibrium MD simulations to see if the sensors could bind SULT1A1 in a similar fashion to 2-naphthol.21,50 During the course of the MD simulations, two stabilizing interactions were monitored: one between the phenol group of the substrate and the sulfur atom of the PAPS cofactor (S–O distance) and one between the phenol group of the substrate and the δ-nitrogen atom (N–H distance) of the catalytic histidine residue (His108) in the active site (Fig. 2 and Fig. S9–S11, Movies smov1–smov8, ESI†). Like 2-naphthol, STS-1, STS-2, and STS-3 were maintained in catalytically productive orientations and had stabilizing interactions with PAPS and His108 (distances <6 Å).21,50 For each sensor, the functionalized naphthalimide backbone was exposed to the bulk water and did not interfere with the orientation of the phenol group in proximity to the site of catalysis. The importance of the phenol group for optimal substrate positioning and reactivity was further demonstrated with control substrates lacking the hydroxy functional groups, namely naphthalene and STS-3C (Fig. S9–S11, ESI†). The simulations revealed that water was able to interact with the PAPS sulfuryl group, the polar N and O atoms of the catalytic His108, and the control substrates in such a way that both naphthalene and STS-3C were separated from the binding pocket.
Fig. 2. MD simulations show that STS-1, STS-2, and STS-3 bind SULT1A1 in catalytically competent orientations like 2-naphthol. Snapshots from the MD simulations of SULT1A1 and (A) 2-naphthol, (B) STS-1, (C) STS-2 (anionic carboxylic acid), and (D) STS-3 (neutral dimethylamine). In each snapshot, the PAPS cofactor, catalytic His108 residue, and substrates are shown as sticks in gray with heteroatoms colored as follows: hydrogen (white), oxygen (red), nitrogen (blue), phosphorous (orange), and sulfur (yellow). Other hydrogen atoms are omitted for clarity.
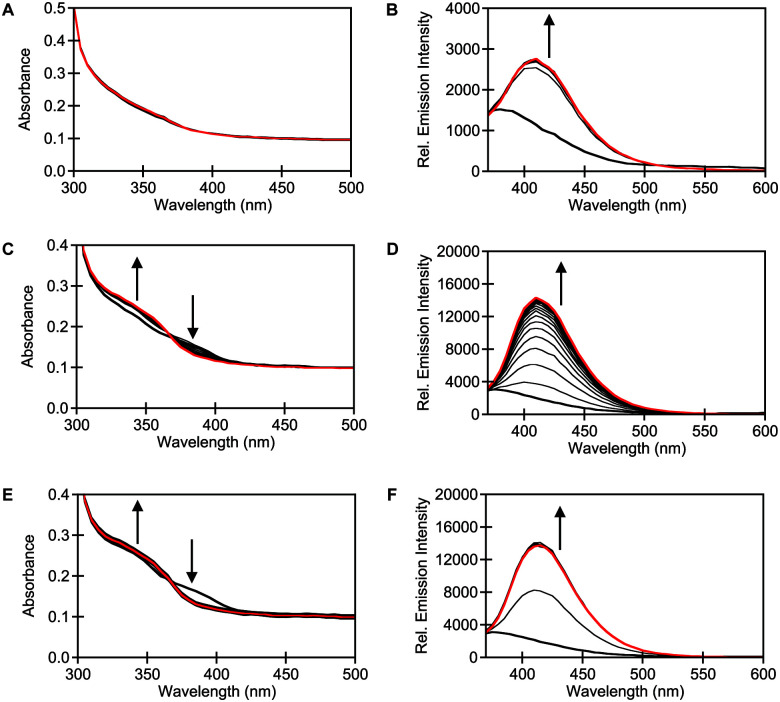
In parallel, the spectroscopic properties of STS-1, STS-2, and STS-3 were evaluated with purified SULT1A1. Spectral changes were observed when 10 μM of each sensor was incubated with 0.015 μg μL−1 (0.4 μM) of purified SULT1A1 and 60 μM of the PAPS cofactor at 37 °C in 50 mM Tris buffer at pH 7.4 (Fig. 3). At t = 0 min, the absorption spectrum for STS-1 was featureless, but STS-2 and STS-3 showed two broad maxima at ∼340 nm and ∼390 nm. Excitation at both absorption maxima resulted in no emission above the background of the buffer for all three sensors, which is consistent with a quenching mechanism. However, clear spectral changes were observed within t = 10 min of incubation with STS-2 and STS-3. Notably, the absorption maxima at ∼340 nm increased in intensity whereas the absorption maxima at ∼390 nm decreased in intensity, suggesting the formation of new products. Upon excitation at 340 nm, robust turn-on emission responses were observed at ∼415–420 nm within t = 1 h (average ± standard deviation): STS-1 (5.7 ± 0.9), STS-2 (6.3 ± 1.7), STS-3 (7.1 ± 0.2) (Fig. 3 and Table S1, ESI†). These spectral changes did not occur in the absence of the enzyme or PAPS cofactor (Fig. S14–S22, ESI†). Similarly, co-incubation with 10 μM of 2,6-dichloro-4-nitrophenol (DCNP), a substrate inhibitor for SULT1A1 (Ki = 2 μM)51 attenuated the turn-on fluorescence response of each sensor to varying degrees (Fig. S14–S22, ESI†).15,51–53 Encouraged by these results, we used liquid chromatography–mass spectrometry (LC–MS) to confirm that the turn-on fluorescence response for each sensor was attributed to the formation of the sulfurylated product (Fig. S23, ESI†).
Fig. 3. STS-1, STS-2, and STS-3 are turn-on fluorescent sensors for SULT1A1. Absorbance responses of 10 μM (A) STS-1, (C) STS-2, and (E) STS-3 to 0.015 μg μL−1 (0.4 μM) SULT1A1 at 0 (bold black) and 180 (red) min. Emission responses of 10 μM (B) STS-1, (D) STS-2, and (F) STS-3 to 0.015 μg μL−1 (0.4 μM) SULT1A1 at 0 (bold black) and 180 (red) min with 10 min intervals (black). Reactions were carried out in 50 mM Tris buffer at pH 7.5 with 60 μM PAPS at 37 °C. Excitation was provided at 340 nm, and the emission was collected from 370–600 nm. Arrow direction corresponds to increasing time. A representative data set from three technical replicates is shown (Fig. S14–S22, ESI†).
To better understand the differences in the observed emission responses, we determined the kinetic parameters for each sensor with SULT1A1 using a Michaelis–Menten model (Fig. S24, ESI†). The low solubility of STS-1 above 10 μM prevented accurate measurements, so it was not further evaluated. The average KM values of STS-2 and STS-3 were determined to be 10 μM and 4 μM, respectively. Based on the kcat values (average ± standard deviation), STS-3 (21 ± 5 s−1) reacted faster than STS-2 (8 ± 4 s−1) with SULT1A1, thus allowing for the quick buildup of fluorescence signal (Table S1, ESI†). These differences also translated to the overall catalytic efficiencies. Based on these parameters and the robust turn-on fluorescence response of STS-3, we next established the ability of STS-3 to detect SULT1A1 activity in living cells.
To validate STS-3, we selected the human SK-N-MC neuroepithelial cancer cell line, which has been previously reported to express SULT1A1.54,55 First, SK-N-MC cells were treated with either a DMSO vehicle control or 10 μM of STS-3 for 4 h at 37 °C in serum-free media, followed by lysis and analysis with fluorescence-based high-performance liquid chromatography (HPLC) (Fig. 4 and Fig. S25–S31, ESI†). Extracts from STS-3 treated cells revealed a single fluorescent product with emission at 420 nm, whereas extracts from DMSO treated cells did not. Co-injection with the authentic sulfurylated product standard, STS-3P, confirmed that STS-3 does indeed undergo sulfurylation in living cells. Encouraged by these results, we turned to live-cell fluorescence microscopy to visualize endogenous SULT1A1 activity (Fig. 4 and Fig. S32–S35, ESI†). Cells treated with STS-3 and excited at 340 nm showed measurable levels of fluorescence signal above the autofluorescence of cells treated with DMSO. Co-incubation with the substrate inhibitor DCNP attenuated the intracellular fluorescence signal by ∼51%. Moreover, this change did not arise from any cytotoxic effects of co-treatment with STS-3 and DCNP. It is not surprising that STS-3 lowered cell viability as related derivatives have been used as anti-cancer agents.46,47,49 Interestingly, DCNP is known to lower PAPS levels56 and inhibit the SULT1A1 isoform, but not the SULT1A3 isoform,15,51,57 which is also expressed in SK-N-MC cells.12,54,55 As such, this provides strong evidence that STS-3 is a reporter of SULT1A1 activity in this cell line. Indeed, the fluorescence imaging also highlights how the reported activity can vary from cell-to-cell. This could arise from differences in the uptake, distribution, or reactivity of DCNP or STS-3 with SULT1A1 or other sulfotransferases, and even cofactor availability, all of which will be the subject of future investigations.
Fig. 4. Fluorescence-based HPLC analysis of extracts and live-cell fluorescence imaging with STS-3 reveal endogenous phenol sulfotransferase activity in SK-N-MC cells. (A) HPLC chromatograms of STS-3P and extracts from live intact SK-N-MC cells that were first treated with a DMSO vehicle control or 10 μM STS-3 in DMEM for 4 h at 37 °C, 5% CO2, followed by lysis and HPLC analysis. Excitation was provided at 340 nm, and the emission was collected at 420 nm. Representative fluorescence microscopy (left) and differential interference contrast (DIC) (right) images of live SK-N-MC cells treated with (B) 10 μM STS-3 and (C) 10 μM STS-3 and 100 μM DCNP in DMEM for 4 h at 37 °C, 5% CO2. Scale bar = 10 μm. Excitation was provided at 340 nm, and the emission was collected with a filter centered at 460 nm with a 50 nm bandwidth. (D) Single cell analysis of the median fluorescence intensity for SK-N-MC cells treated with 10 μM STS-3 (n = 587 regions of interest or ROIs) or 10 μM STS-3 and 100 μM DCNP (n = 608 ROIs). For each biological replicate (n = 3), five fields were sampled, and the median fluorescence intensity of each ROI was normalized to the median fluorescence intensity of STS-3 for that biological replicate. The lower and upper quartile data is enclosed by the gray box with a black line corresponding to the median value for all of the ROIs. The lines extending below and above the gray box represent the minimum and maximum values for each dataset with the outliers shown as open circles. (E) Alamar Blue assay to determine the cell viability of SK-N-MC cells treated with 10 μM STS-3 in the absence and presence of 100 μM DCNP in DMEM for 4 h at 37 °C, 5% CO2. Cells treated with the DMSO vehicle control were considered 100% viable. For each condition, the average of two technical replicates for each of the three biological replicates with the standard deviation is shown.
In summary, we have presented the development, computational modeling, in vitro enzymology, and biological application of STS-3, a first-generation activity-based fluorescent sensor for SULT1A1. This proof-of-concept study sets the stage to further develop and apply activity-based fluorescent sensors to discover how phenol sulfotransferase activity can intersect competitive metabolic pathways to modify endogenous metabolites, xenobiotics, or drugs. Along these lines, efforts are currently underway to develop high-throughput screening methods with STS-3 and generate a palette of sensors with improved reaction kinetics and expanded isoform preferences for applications in a range of cell types.
Conflicts of interest
The authors declare no competing financial interests.
Supplementary Material
Acknowledgments
We thank Dr Alexander Lippert, Dr Gabriele Meloni, Dr John Sibert, and members of the Dodani Lab for helpful discussions. We also thank the Lippert lab at SMU and the Gassensmith lab at UT Dallas for providing access to instrumentation. S. D. acknowledges support from the UT Proteomics Network and National Institute of General Medical Sciences of the National Institutes of Health (R35GM13375). S. C. D. acknowledges support from The University of Texas at Dallas startup funds, the Welch Foundation (AT-1918-20170325), and the National Institute of General Medical Sciences of the National Institutes of Health (R35GM128923).
Electronic supplementary information (ESI) available: Methods, synthetic schemes and characterization, SDS-page, spectra for in vitro enzymology assays, LC–MS spectra, fluorescence-based HPLC spectra, fluorescence microscopy images, and cell viability. See DOI: 10.1039/d0cb00231c
Notes and references
- Günal S. Hardman R. Kopriva S. Mueller J. W. J. Biol. Chem. 2019;294:12293–12312. doi: 10.1074/jbc.REV119.007422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung A. W. Y. Backstrom I. Bally M. B. OncoTargets Ther. 2016;7:55811–55827. doi: 10.18632/oncotarget.10046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamage N. Barnett A. Hempel N. Duggleby R. G. Windmill K. F. Martin J. L. McManus M. E. Toxicol. Sci. 2006;90:5–22. doi: 10.1093/toxsci/kfj061. [DOI] [PubMed] [Google Scholar]
- Langford R. Hurrion E. Dawson P. A. J. Genet. Genomics. 2017;44:7–20. doi: 10.1016/j.jgg.2016.08.001. [DOI] [PubMed] [Google Scholar]
- Coughtrie M. W. H. Pharmacogenomics J. 2002;2:297–308. doi: 10.1038/sj.tpj.6500117. [DOI] [PubMed] [Google Scholar]
- Wang L.-Q. James M. Curr. Drug Metab. 2005;7:83–104. doi: 10.2174/138920006774832596. [DOI] [PubMed] [Google Scholar]
- Strott C. A. Endocr. Rev. 2002;23:703–732. doi: 10.1210/er.2001-0040. [DOI] [PubMed] [Google Scholar]
- Butcher N. J. Horne M. K. Mellick G. D. Fowler C. J. Masters C. L. Minchin R. F. Pharmacogenomics J. 2018;18:209–214. doi: 10.1038/tpj.2017.4. [DOI] [PubMed] [Google Scholar]
- Runge-Morris M. Kocarek T. A. Falany C. N. Drug Metab. Rev. 2013;45:15–33. doi: 10.3109/03602532.2012.748794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suiko M. Kurogi K. Hashiguchi T. Sakakibara Y. Liu M.-C. Biosci., Biotechnol., Biochem. 2017;81:63–72. doi: 10.1080/09168451.2016.1222266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda S. Yasuda T. Hui Y. Liu M. Y. Suiko M. Sakakibara Y. Liu M. C. Neurosci. Res. 2009;64:273–279. doi: 10.1016/j.neures.2009.03.011. [DOI] [PubMed] [Google Scholar]
- Yasuda S. Liu M.-Y. Suiko M. Sakakibara Y. Liu M.-C. J. Neurochem. 2007;103:2679–2689. doi: 10.1111/j.1471-4159.2007.04851.x. [DOI] [PubMed] [Google Scholar]
- Berger I. Guttman C. Amar D. Zarivach R. Aharoni A. PLoS One. 2011;6:e26794. doi: 10.1371/journal.pone.0026794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman E. Best M. D. Hanson S. R. Wong C.-H. Angew. Chem., Int. Ed. 2004;43:3526–3548. doi: 10.1002/anie.200300631. [DOI] [PubMed] [Google Scholar]
- Allali-Hassani A. Pan P. W. Dombrovski L. Najmanovich R. Tempel W. Dong A. Loppnau P. Martin F. Thonton J. Edwards A. M. Bochkarev A. Plotnikov A. N. Vedadi M. Arrowsmith C. H. PLoS Biol. 2007;5:e97. doi: 10.1371/journal.pbio.0050097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arand M. Robertson L. W. Oesch F. Anal. Biochem. 1987;163:546–551. doi: 10.1016/0003-2697(87)90261-2. [DOI] [PubMed] [Google Scholar]
- Wang T. Cook I. Leyh T. S. J. Biol. Chem. 2017;292:20305–20312. doi: 10.1074/jbc.M117.817387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook I. Wang T. Leyh T. S. Biochemistry. 2015;54:6114–6122. doi: 10.1021/acs.biochem.5b00406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakers C. Schumacher F. Meinl W. Glatt H. Kleuser B. Wolber G. J. Biol. Chem. 2016;291:58–71. doi: 10.1074/jbc.M115.685610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi L. Dratter J. Wang C. Tunge J. A. Desaire H. Anal. Bioanal. Chem. 2006;386:666–674. doi: 10.1007/s00216-006-0495-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin E. B. Stangl H. Pichu S. Mather T. N. King R. S. ACS Chem. Biol. 2011;6:176–184. doi: 10.1021/cb100266g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny V. Y. Carbonell P. Lagorce D. Villoutreix B. O. Moroy G. Miteva M. A. PLoS One. 2013;8:e73587. doi: 10.1371/journal.pone.0073587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litsa E. E. Das P. Kavraki L. E. Chem. Sci. 2020;11:12777–12788. doi: 10.1039/D0SC02639E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffel M. W., Comprehensive Toxicology, ed. C. A. B. T. McQueen, Elsevier, Oxford, 2nd edn, 2010, pp. 367–384 [Google Scholar]
- Paul P. Suwan J. Liu J. Dordick J. S. Linhardt R. J. Anal. Bioanal. Chem. 2012;403:1491–1500. doi: 10.1007/s00216-012-5944-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W. Duckworth B. P. Geraghty R. J. Anal. Biochem. 2014;461:1–6. doi: 10.1016/j.ab.2014.05.025. [DOI] [PubMed] [Google Scholar]
- Zhou W. Wang Y. Xie J. Geraghty R. J. Biochem. Biophys. Res. Commun. 2017;482:1207–1212. doi: 10.1016/j.bbrc.2016.12.013. [DOI] [PubMed] [Google Scholar]
- Wang T. Cook I. Leyh T. S. Drug Metab. Dispos. 2016;44:481–484. doi: 10.1124/dmd.115.068205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballet C. Correia M. S. P. Conway L. P. Locher T. L. Lehmann L. C. Garg N. Vujasinovic M. Deindl S. Löhr J. M. Globisch D. Chem. Sci. 2018;9:6233–6239. doi: 10.1039/C8SC01502C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook I. Wang T. Leyh T. S. Biochem. Pharmacol. 2019;159:25–31. doi: 10.1016/j.bcp.2018.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole G. B. Keum G. Liu J. Small G. W. Satyamurthy N. Kepe V. Barrio J. R. Proc. Natl. Acad. Sci. U. S. A. 2010;107:6222–6227. doi: 10.1073/pnas.0914904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D. Sedgwick A. C. Gunnlaugsson T. Akkaya E. U. Yoon J. James T. D. Chem. Soc. Rev. 2017;46:7105–7123. doi: 10.1039/C7CS00240H. [DOI] [PubMed] [Google Scholar]
- Chyan W. Raines R. T. ACS Chem. Biol. 2018;13:1810–1823. doi: 10.1021/acschembio.8b00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K. Rotaru A. M. Beharry A. A. ACS Chem. Biol. 2018;13:1785–1798. doi: 10.1021/acschembio.8b00014. [DOI] [PubMed] [Google Scholar]
- Feng L. Ning J. Tian X. Wang C. Zhang L. Ma X. James T. D. Coord. Chem. Rev. 2019;399:213026. doi: 10.1016/j.ccr.2019.213026. [DOI] [Google Scholar]
- Ning J. Wang W. Ge G. Chu P. Long F. Yang Y. Peng Y. Feng L. Ma X. James T. D. Angew. Chem., Int. Ed. 2019;58:9959–9963. doi: 10.1002/anie.201903683. [DOI] [PubMed] [Google Scholar]
- Zhang J. Chai X. He X.-P. Kim H.-J. Yoon J. Tian H. Chem. Soc. Rev. 2019;48:683–722. doi: 10.1039/C7CS00907K. [DOI] [PubMed] [Google Scholar]
- Yadav A. K. Reinhardt C. J. Arango A. S. Huff H. C. Dong L. Malkowski M. G. Das A. Tajkhorshid E. Chan J. Angew. Chem., Int. Ed. 2020;59:3307–3314. doi: 10.1002/anie.201914845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner S. H. Reinhardt C. J. Chan J. Angew. Chem., Int. Ed. 2021;60:5000–5009. doi: 10.1002/anie.202003687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riches Z. Stanley E. L. Bloomer J. C. Coughtrie M. W. H. Drug Metab. Dispos. 2009;37:2255–2261. doi: 10.1124/dmd.109.028399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salman E. D. Kadlubar S. A. Falany C. N. Drug Metab. Dispos. 2009;37:706–709. doi: 10.1124/dmd.108.025767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L. W. Wang P. Qian X. K. Feng L. Yu Y. Wang D. D. Jin Q. Hou J. Liu Z. H. Ge G. B. Yang L. Biosens. Bioelectron. 2017;90:283–289. doi: 10.1016/j.bios.2016.11.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv X. Ge G.-B. Feng L. Troberg J. Hu L.-H. Hou J. Cheng H.-L. Wang P. Liu Z.-M. Finel M. Cui J.-N. Yang L. Biosens. Bioelectron. 2015;72:261–267. doi: 10.1016/j.bios.2015.05.003. [DOI] [PubMed] [Google Scholar]
- Dong H. Q. Wei T. B. Ma X. Q. Yang Q. Y. Zhang Y. F. Sun Y. J. Shi B. B. Yao H. Zhang Y. M. Lin Q. J. Mater. Chem. C. 2020;8:13501–13529. doi: 10.1039/D0TC03681A. [DOI] [Google Scholar]
- Hardebeck L. K. E. Johnson C. A. Hudson G. A. Ren Y. Watt M. Kirkpatrick C. C. Znosko B. M. Lewis M. J. Phys. Org. Chem. 2013;26:879–884. doi: 10.1002/poc.3184. [DOI] [Google Scholar]
- Johnson C. A. Hudson G. A. Hardebeck L. K. E. Jolley E. A. Ren Y. Lewis M. Znosko B. M. Bioorg. Med. Chem. 2015;23:3586–3591. doi: 10.1016/j.bmc.2015.04.030. [DOI] [PubMed] [Google Scholar]
- Braña M. F. Ramos A. Curr. Med. Chem.: Anti-Cancer Agents. 2001;1:237–255. doi: 10.2174/1568011013354624. [DOI] [PubMed] [Google Scholar]
- Feng L. Tian X. Yu Z. Zhao X. Sun C. Cui J. Ning J. Wang C. Zhang B. Ma X. Sens. Actuators, B. 2019;282:112–121. doi: 10.1016/j.snb.2018.11.045. [DOI] [Google Scholar]
- Zee-Cheng R. K. Y. Cheng C. C. J. Med. Chem. 1985;28:1216–1222. doi: 10.1021/jm00147a016. [DOI] [PubMed] [Google Scholar]
- Cook I. Wang T. Falany C. N. Leyh T. S. J. Biol. Chem. 2013;288:34494–34501. doi: 10.1074/jbc.M113.510974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Clemens D. L. Cole J. R. Anderson R. J. J. Endocrinol. 2001;171:525–532. doi: 10.1677/joe.0.1710525. [DOI] [PubMed] [Google Scholar]
- Veronese M. E. Burgess W. Zhu X. McManus M. E. Biochem. J. 1994;302:497–502. doi: 10.1042/bj3020497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster H. Scholtens E. Mulder G. J. Aust. J. Exp. Biol. Med. Sci. 1979;57:340–344. [PubMed] [Google Scholar]
- Sidharthan N. P. Minchin R. F. Butcher N. J. J. Biol. Chem. 2013;288:34364–34374. doi: 10.1074/jbc.M113.493239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idris M. Mitchell D. J. Gordon R. Sidharthan N. P. Butcher N. J. Minchin R. F. Drug Metab. Dispos. 2020;48:337–344. doi: 10.1124/dmd.119.089714. [DOI] [PubMed] [Google Scholar]
- Gulat-Marnay C. Lafitte A. Vargas F. Schwartz J.-C. J. Neurochem. 1987;49:1443–1448. doi: 10.1111/j.1471-4159.1987.tb01012.x. [DOI] [PubMed] [Google Scholar]
- Mulder G. J. Scholtens E. Biochem. J. 1977;165:553–559. doi: 10.1042/bj1650553. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.