Abstract
Traditional Chinese medicine (TCM) has been utilized for the treatment of colon cancer. Qizhen decoction (QZD), a potential compound prescription of TCM, possesses multiple biological activities. It has been proven clinically effective in the treatment of colon cancer. However, the molecular mechanism of anticolon cancer activity is still not clear. This study aimed to identify the chemical composition of QZD. Furthermore, a collaborative analysis strategy of network pharmacology and cell biology was used to further explore the critical signaling pathway of QZD anticancer activity. First, ultraperformance liquid chromatography–quadrupole time-of-flight/mass spectrometry (UPLC–Q-TOF/MS) was performed to identify the chemical composition of QZD. Then, the chemical composition database of QZD was constructed based on a systematic literature search and review of chemical constituents. Moreover, the common and indirect targets of chemical components of QZD and colon cancer were searched by multiple databases. A protein–protein interaction (PPI) network was constructed using the String database (https://www.string-db.org/). All of the targets were analyzed by Gene Oncology (GO) bioanalysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, and the visual network topology diagram of “Prescription–TCM–Chemical composition–Direct target–Indirect target–Pathway” was constructed by Cytoscape software (v3.7.1). The top molecular pathway ranked by statistical significance was further verified by molecular biology methods. The results of UPLC–Q-TOF/MS showed that QZD had 111 kinds of chemical components, of which 103 were unique components and 8 were common components. Ten pivotal targets of QZD in the treatment of colon cancer were screened by the PPI network. Targets of QZD involve many biological processes, such as the signaling pathway, immune system, gene expression, and so on. QZD may interfere with biological pathways such as cell replication, oxygen-containing compounds, or organic matter by protein binding, regulation of signal receptors or enzyme binding, and affect cytoplasm and membrane-bound organelles. The main antitumor core pathways were the apoptosis metabolic pathway, the PI3K–Akt signal pathway, and so on. Expression of the PI3K–Akt signal pathway was significantly downregulated after the intervention of QZD, which was closely related to the inhibition of proliferation and migration of colon cancer cells by cell biology methods. The present work may facilitate a better understanding of the effective components, therapeutic targets, biological processes, and signaling pathways of QZD in the treatment of colon cancer and provide useful information about the utilization of QZD.
1. Introduction
Colon cancer is the third most common malignant tumor in the world and the fourth leading cause of cancer-related deaths. It is estimated that there will be 2.2 million new cases by 2030.1 Although there are many treatments such as surgery, radiotherapy, chemotherapy, and targeted therapy to improve the prognosis of patients with colon cancer, colon cancer is still one of the important causes of cancer-related deaths.2 At present, there are some shortcomings in the treatment, such as the small number of beneficiaries, lack of selectivity to tumor cells, insufficient drug concentration in tumor tissue, emergence of drug-resistant tumor cells, and systemic toxicity.3 The treatment of colon cancer is still a major challenge, and it has become the key to the treatment that how to effectively treat cancer while reducing toxicity. There is an urgent need to find new treatments or adjuvant drugs for the treatment of colon cancer.
With the characteristics of multicomponents and multitargets, traditional Chinese medicine (TCM) has the advantages of low toxicity and less adverse reactions, reversing multidrug resistance, reducing drug dosage, improving patient compliance, and a curative effect. TCM has been used as a supplement and alternative therapy for colon cancer.4 Because the compound prescription of TCM has the characteristics of complex chemical composition, diverse structure, and unclear material basis of efficacy, these restrict the promotion and application of TCM in clinics. It is an important scientific problem to clarify the effective composition and action mechanism of TCM compound prescription. Qizhen decoction (QZD) is an important formula of TCM for the treatment of colon cancer, which is composed of Radix Astragali (RA), Codonopsis pilosula (CP), Ligustrum lucidum (LL), Herba Hedyotis (HH), Gynostemma Pentaphylla (GP), Salviae Miltiorrhizae (SM), Radix Paeoniae Rubra (RPR), and Polygonum cuspidatum (PC) (Table 1). All of the above TCM have anticancer active components and are widely used in basic and clinical research of antitumor mechanisms.5−11 Zhenqi Fuzheng capsules comprise RA and LL that have been demonstrated to improve immunity, promote the recovery of normal functions after surgical operations, and are an important adjuvant therapy in cancer.12 Based on Zhenqi Fuzheng capsules, Qizhen decoction adds anticancer drugs such as CP, HH, GP, SM, RPR, and PC to increase the effect of detoxification and promoting blood circulation. However, the exact antitumor mechanism of QZD has not been reported. In this study, the ultraperformance liquid chromatography–quadrupole time-of-flight/mass spectrometry (UPLC–Q-TOF/MS) technique was used to isolate, identify, and analyze the main components of QZD. Also, Network pharmacology was used as the breakthrough point to explore the effective active components and targets of QZD in the treatment of colon cancer and reveal its molecular mechanism.
Table 1. Eight Chinese Traditional Medical Herbs of QZD.
| abbreviation | medicinal herbs | original plants | proportion |
|---|---|---|---|
| RA | Radix Astragali | Astragalus membranaceus (Fisch.) Bge. var. mongholicus (Bge.) Hsiao or Astragalus membranaceus (Fisch.) Bge | 3 |
| CP | Codonopsis pilosula | Codonopsis pilosula (Franch.) Nannf. or Codonopsis pilosulaNannf. var.modesta (Nannf.) L. T. Shen or Codonopsis tangshen Oliv. | 3 |
| LL | Ligustrum lucidum | Ligustrum lucidum Ait | 3 |
| HH | Herba Hedyotis | Hedyotis diffusa Willd | 4 |
| GP | Gynostemma Pentaphylla | G. pentaphyllum (Thunb.) Makino | 4 |
| SM | Salviae Miltiorrhizae | Salvia miltiorrhiza Bge | 2 |
| RPR | Radix Paeoniae Rubra | Paeonia lactiflora PalL or Paeonia veitchii Lynch | 2 |
| PC | Polygonum cuspidatum | Polygonum cuspidatum Sieb. et Zucc. | 2 |
The chemical composition of QZD is the core link to explain its efficacy and mechanism. Given the complex composition and diverse structure of QZD, the analysis, identification and content determination of its effective components have become difficult problems. In recent years, researchers have successfully solved such problems by applying the UPLC–Q-TOF/MS technique to the composition analysis of natural medicines.13 UPLC–Q-TOF/MS is a modern analytical technique, which has fast and efficient separation characteristics of UPLC and qualitative analysis characteristics of high resolution and high sensitivity of Q-TOF/MS. It can realize the rapid separation of complex samples and accurate qualitative analysis of multicomponents. The UPLC–Q-TOF/MS technique can quickly and efficiently analyze the main components of QZD. It provides a scientific basis for controlling the quality of QZD and elucidating its mechanism.
Due to the lack of chemical composition analysis of QZD and the prediction of its therapeutic targets for colon cancer, network pharmacology has become a powerful tool for studying complex diseases and revealing the complex relationship between proteins, diseases, and drugs.14 Based on the analysis of the main chemical components of QZD by UPLC–Q-TOF/MS, combined with network pharmacology, exploring the effective active components and targets of QZD in the treatment of colon cancer and revealing its molecular mechanism are of great significance to the interpretation of the scientific connotation and clinical transformation of QZD. Colon cancer is a multigene and multifactor disease.15,16 Single target therapy is difficult to achieve the desired effect. Network pharmacology emphasizes the multitarget and multipathway regulation of signal pathways, which can play an important role in the research and development of new drugs of TCM. It is helpful to reveal the mechanism of QZD with multicomponent and multitarget characteristics in the treatment of colon cancer. With the combination of UPLC–Q-TOF/MS and network pharmacology, the chemical components of QZD were associated with the disease target of colon cancer by Gene Oncology (GO) bioanalysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis.17 Key target proteins and signaling pathways of QZD intervention in colon cancer were predicted by reverse pharmacophore matching, and the pathways with the strongest correlation were screened and verified by molecular biology methods. It further confirmed the feasibility of QZD in the treatment of colon cancer and systematically explained the regulatory effect of QZD on colon cancer, which makes the research more targeted, saves scientific resources, and has certain guiding significance for the interpretation of scientific connotation and clinical transformation of QZD.
2. Results
2.1. Optimization of Chromatographic Conditions
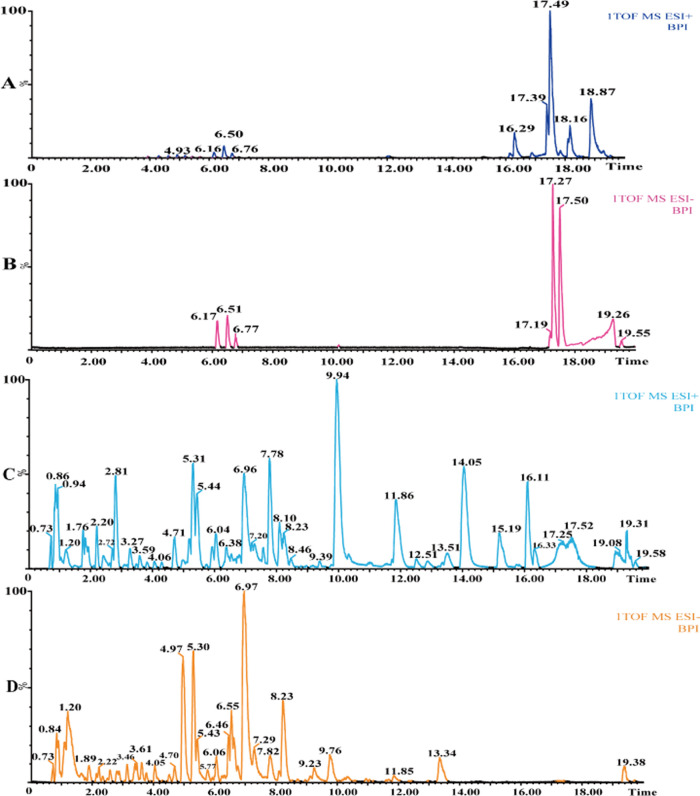
The chemical components in TCM prescriptions are critical to finding the active components and confirming the material basis of them. Traditional analytical methods usually use liquid chromatography–ultraviolet to detect complex compounds in TCM, but the fingerprint of the compounds was often low sensitivity and poor specificity so that the material structure information could not be confirmed. This study was based on the UPLC–Q-TOF/MS platform. To ensure the accuracy and reliability of the experimental results, the liquid conditions were optimized to a certain extent. This study was based on the composition information of eight main TCMs (Table 1) and the related literature. HSS C18 2.1 mm × 100 mm (Waters, America) and BEH C18 2.1 mm × 100 mm (Waters, America) columns were used to optimize the chromatographic conditions, and the injection volume and flow rate were adjusted at the same time. Finally, the BEH C18 column was used, and the optimal conditions were as follows: the column temperature was 40 °C, the flow rate was 0.3 mL·min–1, the injection volume was 5 μL, 0.1% formic acid was added to the mobile phase to obtain a better resolution and better peak shape. To comprehensively analyze the chemical components of QZD, the full scanning of positive and negative ion modes was carried out in this experiment. High-performance liquid chromatography (HPLC) combined with high-resolution mass spectrometry (HRMS) analysis showed that under positive and negative ionization modes, QZD contained abundant chromatographic peaks, with many kinds of chemical components and a good chromatographic profile, which was suitable for the follow-up study on the full characterization of chemical components (Figure 1). At the same time, according to the mass spectrometry behavior of the standard, two electrospray ionization (ESI) modes were selected for separating and conducting independent injection analysis.
Figure 1.
Base peak ion (BPI) chromatogram of chemical components of QZD based on UPLC–Q-TOF/MS. (A) BPI chromatogram of mixed standard in ESI+ mode, (B) BPI chromatogram of mixed standard in ESI– mode, (C) BPI chromatogram of QZD in ESI+ mode, and (D) BPI chromatogram of QZD in ESI– mode.
2.2. Identification and Confirmation of Chemical Components of QZD
As shown in Table 2, 111 chemical compositions (103 unique components and 8 common components) were identified in QZD. Among them, 24 were unique to RA, 11 to LL, 18 to SM, 16 to CP, 10 to PC, 6 to GP, 11 to RPR, and 7 to HH. Raffinose was the common component of RA and SM; tryptophan was the common component of RA and CP; caffeic acid was the common component of RA, LL, and SM; ferulic acid was the common component of RA, SM, CP, and HH; azelaic acid was the common component of RA and CP; oleanolic acid was the common component of LL and HH; ginsenoside Rb1 was the common component of SM and GP; and emodin was the common component of CP and PC.
Table 2. Chemical Composition Information of QZD Based on UPLC–Q-TOF/MSa.
| no | RT | ESI | components | formula | calcd | obsed | error (ppm) | MS/MS | source | category | references |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.83 | [M + Na]+ | hexose | C6H12O6 | 203.0532 | 203.0547 | 7.38 | 203.0547 [M + Na]+, 143.0261 [M + Na – C2H4O2]+ | RA | monosaccharide | (18) |
| 2 | 0.81 | [M – H]- | asparagine | C4H8N2O3 | 131.0457 | 131.0457 | 0 | 131.0457 [M – H]−; 114.0123 [M – H – NH3]−; 113.0207 [M – H – H2O]− | RA | amino acid | (18) |
| 3 | 0.85 | [M – H]− | choline | C5H13NO | 104.1075 | 104.1076 | 0.96 | 104.1076 [M – H]−, 60.0533 [M + H – C2H5O]+ | RA | choline | (18) |
| 4 | 0.97 | [M + Na]+ | raffinose | C18H32O16 | 527.1588 | 527.1575 | 2.46 | 527.1575 [M + Na]+, 161.0753 [M + H – 2glc]− | RA, CP | oligosaccharide | (18) |
| 5 | 1.14 | [M + H]+ | nicotinic acid | C6H5NO2 | 124.0399 | 124.0386 | 10.04 | 124.0386 [M + H]+, 106.0635 [M + H – H2O]+ | RA | pyridine carboxylic acid | (18) |
| 6 | 1.87 | [M + H]+ | leucine | C6H13NO2 | 132.1025 | 132.1033 | 6.05 | 132.1033 [M + H]+, 86.0575 [M + H – CH2O2]+ | RA | amino acid | (18) |
| 7 | 1.76 | [M + H]+ | adenine nucleoside | C10H13N5O4 | 268.1046 | 268.1045 | 0.37 | 268.1045 [M + H]+, 136.0638 [M + H-ribose]+, 119.0368 [M + H-ribose – NH3]+ | RA | nucleoside | (18) |
| 8 | 2.67 | [M + H]+ | phenylalanine | C9H11NO2 | 166.0868 | 166.0848 | 12.04 | 166.0848 [M + H]+, 149.0246 [M + H – NH3]+, 131.0436 [M + H – NH3 – H2O]+, 120.0823 [M + H – CH2O2]+, 103.0545 [M + H – NH3 – CH2O2]+ | RA | amino acid | (18) |
| 9 | 4.37 | [M – H]− | markhamioside F | C18H26O12 | 433.1346 | 433.1388 | 9.69 | 433.1388 [M – H]−, 301.0372 [M – H – C5H8O4]−, 139.0367 [M – H – C5H8O4 – glc]− | RA | glycoside | (18) |
| 10 | 3.25 | [M + H]+ | tryptophan | C11H12N2O2 | 205.0977 | 205.0963 | 6.82 | 205.0963 [M + H]+, 188.0700 [M + H – NH3]+, 170.0575 [M + H – NH3 – H2O]+, 159.0921 [M + H – CH2O2]+, 132.0802 [M + H – NH3 – CH2O2]+ | RA, CP | amino acid | (18−20) |
| 11 | 7.18 | [M + H]+ | hydroxybenzoic acid | C7H6O3 | 139.0395 | 139.0389 | 4.31 | 139.0389 [M + H]+, 121.0267 [M + H – H2O]+, 111.0443 [M + H – CO]+ | RA | organic acid | (18) |
| 12 | 7.20 | [M + H]+ | rhamnocitrin 3,4′-di-O-glucoside | C28H32O16 | 625.1769 | 625.1727 | 6.71 | 625.1727 [M + H]+, 301.0656 [M + H – 2glc]+ | RA | glycoside | (18) |
| 13 | 6.96 | [M + H]+ | caffeic acid | C9H8O4 | 181.0501 | 181.0524 | 12.7 | 181.0524 [M + H]+, 153.0596 [M + H – CO]+, 137.0588 [M + H – CO2]+ | RA, LL, SM | organic acid | (18) and (21−24) |
| 14 | 4.26 | [M + H]+ | emodin-di-O-glucoside | C29H38O16 | 595.1663 | 595.1713 | 8.40 | 595.1713 [M + H]+, 479.1435 [M – H – glc], 195.0919 [M – H – 2glc – RDA]− | RA | glycoside | (18) |
| 15 | 12.68 | [M + H]+ | syringaldehyde | C9H10O4 | 183.0657 | 183.0641 | 8.74 | 183.0641 [M + H]+, 155.0702 [M + H – CO]+, 140.9917 [M + H – CO – CH3]+, 125.0951 [M + H – CO – CH2O]+, 123.0428 [M + H – 2CH2O]+ | RA | phenol | (18) |
| 16 | 3.49 | [M + H]+ | ferulic acid | C10H10O4 | 195.0657 | 195.0675 | 9.23 | 195.0675 [M + H]+, 177.0573 [M + H – H2O]+, 149.0246 [M + H – CH2O2]+, 163.0428 [M + H – CH3OH]+, 117.0682 [M + H – CH2O2 – CH3OH]+ | RA, SM, CP, HH | organic acid | (18, 19, 22), and (25) |
| 17 | 4.51 | [M + H]+ | pratensein-7-O-glucoside | C22H22O11 | 463.1240 | 463.1282 | 9.06 | 463.1282 [M + H]+, 301.0700 [M + H – glc]+, 269.0430 [M + H – glc – CH3OH]+ | RA | glycoside | (18) |
| 18 | 5.39 | [M + H]+ | calycosin-7-O-glucoside | C10H10O4 | 447.1291 | 447.1266 | 5.59 | 447.1266 [M + H]+, 285.1270 [M + H – glc]+, 270.0525 [M + H – glc – CH3]+ | RA | glycoside | (18) and (26) |
| 19 | 5.54 | [M + H]+ | cosmosiin | C21H20O10 | 433.1135 | 433.1128 | 1.61 | 433.1128 [M + H]+, 271.0579 [M + H – glc]+ | RA | glycoside | (18) |
| 20 | 6.58 | [M – H]− | azelaic acid | C9H16O4 | 187.0970 | 187.0948 | 11.75 | 187.0948 [M – H]−, 143.1105 [M – H – CO2]−, 125.0977 [M – H – H2O – CO2]− | RA, CP | organic acid | (18) and (19) |
| 21 | 5.11 | [M + H]+ | 9,10-dimethoxypterocarp an-3-O-glucoside | C23H26O10 | 463.1604 | 463.1658 | 11.65 | 463.1658 [M + H]+, 301.0732 [M + H – glc]+ | RA | (18, 20, 26), and (27) | |
| 22 | 5.30 | [M + H]+ | isomucronulatol-7-O-glucoside | C23H28O10 | 465.1761 | 465.1788 | 5.81 | 465.1788 [M + H]+, 303.0537 [M + H – glc]+ | RA | (18) and (26) | |
| 23 | 6.38 | [M + H]+ | calycosin | C16H12O5 | 285.0763 | 285.0793 | 10.05 | 285.0793 [M + H]+, 270.0525 [M + H – CH3]+, 137.0588 [M + H – C7H3O3]+ | RA | flavonoids | (18, 20), and (26−28) |
| 24 | 8.91 | [M + H]+ | 6′-O-acetyl ononin | C24H24O10 | 473.1448 | 473.1415 | 6.97 | 473.1415 [M + H]+, 455.2506 [M + H – H2O]+, 269.0776 [M + H – glc – C2H4O2]+ | RA | (18, 20), and (26) | |
| 25 | 10.02 | [M + H]+ | formononetin | C16H12O4 | 269.0814 | 269.0847 | 12.26 | 269.0847 [M + H]+, 253.0517 [M + H – C15H7O4]+, 225.0556 [M + H – C14H7O3]+ | RA | flavonoids | (18) and (26−29) |
| 26 | 8.37 | [M + H]+ | astragaloside V | C47H78O19 | 947.5216 | 947.5292 | 8.02 | 947.5292 [M + H]+, 969.5027 [M + Na]+ | RA | triterpenoid saponin | (18) |
| 27 | 11.33 | [M + H]+ | soyasaponin I | C48H78O18 | 943.5266 | 943.5284 | 1.91 | 943.5284 [M + H]+, 923.5120, 733.4538, 615.4003, 457.3725, 247.0823, 205.7828 | RA | saponin | (18) |
| 28 | 13.05 | [M + H]+ | isoastragaloside I | C45H72O16 | 869.4899 | 869.4822 | 8.85 | 869.4822 [M + H]+, 807.4652, 765.4568, 179.0567 | RA | saponin | (18) and (30) |
| 29 | 8.56 | [M + H]+ | astragaloside IV (astragaloside A) | C41H68O14 | 784.4609 | 784.4692 | 10.58 | 784.4692 [M + H]+, 605.3468 [M + H – glc]+, 473.5548 [M + H – glc–Xyl]+ | RA | triterpenoid saponin | (18, 20, 26, 27), and (30) |
| 30 | 2.90 | [M – H]− | nuezhenidic acid | C17H24O14 | 451.1088 | 451.1080 | 1.77 | 451.1080 [M – H]−, 407.0549, 375.1001, 347.0675 | LL | iridoid | (31) |
| 31 | 3.41 | [M – H]− | 10-hydroxyl oleoside-dimethyl ester | C18H26O12 | 433.1346 | 433.1396 | 11.54 | 433.1396 [M – H]−, 401.0563, 209.0452, 177.0205, 165.0567, 119.0398 | LL | (31) | |
| 32 | 3.74 | [M – H]− | chlorogenic acid | C16H18O9 | 353.0873 | 353.0843 | 8.49 | 353.0843 [M – H]−, 191.0540 [M – H – C9H7O3]−, 161.0287 [M – H-quinineacyl]−, 173.0780 [M – H – C9H7O3 – H2O]−, 179.0434, 135.0371 | LL | phenolic acid | (31) |
| 33 | 4.36 | [M – H]− | oleoside 11-methyl ester | C17H24O11 | 403.1240 | 403.1240 | 0 | 403.1240 [M – H]−, 241.0643 [2M – H]−, 223.0502, 163.0365 | LL | (31) and (32) | |
| 34 | 4.61 | [M – H]− | oleuropein-aglycone | C16H26O10 | 377.1448 | 377.1423 | 6.62 | 377.1423 [M – H]−, 755.2076, 197.0797, 153.0949 | LL | (31) and (32) | |
| 35 | 0.84 | [M – H]− | 4-(2-acetoxyethyl)-1,2-hydroquinone | C10H12O4 | 195.0657 | 195.0634 | 11.79 | 195.0634 [M – H]−, 177.0263, 159.0113, 152.6951 | LL | (31) | |
| 36 | 5.61 | [M – H]− | oleuropein acid | C25H30O15 | 569.1506 | 569.1536 | 5.27 | 569.1536 [M – H]−, 447.0710, 417.1306, 255.0392 | LL | cyclienyl ether terpene glycosides | (33) |
| 37 | 6.08 | [M – H]− | specnuezhenide | C31H42O17 | 685.2344 | 685.2337 | 1.02 | 685.2337 [M – H]−, 523.1831, 453.1333, 365.1728, 299.1071 | LL | iridoid | (33−35) |
| 38 | 6.09 | [M – H]− | ligustroside | C25H32O12 | 523.1816 | 523.1831 | 2.86 | 523.1831 [M – H]−, 313.2710 | LL | iridoid | (31, 33), and (36−38) |
| 39 | 7.580 | [M – H]− | oleonuezhenide | C48H64O27 | 1071.3557 | 1071..3583 | 2.42 | 1071–3583 [M – H]−, 685.2255, 523.1731, 299.1071 | LL | (33) | |
| 40 | 17.26 | [M – H]− | oleanolic acid | C30H48O3 | 455.3525 | 455.3571 | 10.10 | 455.3571 [M – H]−, 437.6243, 411.5276 | LL,HH | pentacyclic triterpenes | (33) and (39) |
| 41 | 3.26 | [M – H]− | salidroside | C14H20O7 | 299.1131 | 299.1147 | 5.35 | [M + NH4]+ | LL | phenyl alkane | |
| 42 | 2.58 | [M – H]− | danshensu | C9H10O5 | 197.0450 | 197.0428 | 11.16 | 197.0428 [M – H]−, 135.0421 [M – H – H2O – CO2]−, 179.0434 [M – H – H2O]− | SM | phenolic acid | (22, 24), and (40) |
| 43 | 3.58 | [M – H]− | protocatechuic aldehyde | C7H6O3 | 137.0239 | 137.0245 | 4.37 | 137.0245 [M – H]− | SM | phenolic acid | (22, 24), and (40) |
| 44 | 6.53 | [M – H]− | rosmarinic acid | C9H8O4 | 359.0767 | 359.0783 | 4.45 | 359.0783 [M – H]−, 161.0232 [M – H – C9H10O5]−, 197.0428 [M – H – C9H6O3]− | SM | phenolic acid | (22−24), and (40) |
| 45 | 6.62 | [M – H]− | lithosperimic acid | C27H22O12 | 493.1135 | 493.1164 | 5.88 | 493.1164 [M – H]−, 295.0630 [M – H – CO2 – C9H10O5]− | SM | (40) | |
| 46 | 6.97 | [M – H]− | salvianolic acid B | C36H30O16 | 717.1456 | 717.1445 | 1.53 | 717.1445 [M – H]−, 519.0936 [M – H – C9H10O5]− | SM | phenolic acid | (22, 23), and (40) |
| 47 | 6.67 | [M – H]− | salvianolic acid A | C26H22O10 | 493.1135 | 493.1164 | 5.88 | 493.1164 [M – H]−, 295.0630 [M – H – C9H10O5]− | SM | phenolic acid | (40) |
| 48 | 8.00 | [M – H]− | salvianolic acid C | C26H20O10 | 491.0978 | 491.0907 | 14.45 | 491.0907 [M – H]−, 293.0401 [M – H – C9H10O5]− | SM | phenolic acid | (22) and (40) |
| 49 | 9.01 | [M – H]− | ginsenoside Rb1 | C54H92O23 | 1107.5951 | 1107.5990 | 3.52 | 1107.5990 [M – H]−, 599.5806 [M + 2HCOO]2–, 576.2953 [M – H + HCOO]2– | SM, GP | saponin | (40) |
| 50 | 13.45 | [M + H]+ | dihydrotanshinone I | C18H14O3 | 279.1021 | 279.1029 | 2.86 | 279.1029 [M + H]+, 301.0732 [M + Na]+ | SM | phenanthraquinone | (40) |
| 51 | 15.30 | [M + H]+ | cryptotanshinone | C18H12O3 | 277.0865 | 277.0861 | 1.44 | 277.0861 [M + H]+, 575.1473 [2M + Na]+, 299.0690 [M + Na]+ | SM | phenanthraquinone | (40) |
| 52 | 15.23 | [M + H]+ | tanshinone I | C19H20O3 | 297.1491 | 297.1467 | 8.07 | 297.1467 [M + H]+, 319.1161 [M + Na]+ | SM | phenanthraquinone | (40) |
| 53 | 16.32 | [M + H]+ | tanshinone IIA | C19H18O3 | 295.1334 | 295.1331 | 1.01 | 295.1331 [M + H]+, 611.1695 [2M + Na]+, 317.1148 [M + Na]+ | SM | phenanthraquinone | (22) and (40) |
| 54 | 10.09 | [M + HCOO]− | notoginsenoside R1 | C47H80O18 | 977.5321 | 977.5313 | 0.82 | 977.5313 [M + HCOO]−, 931.5224 [M – H]− | SM | saponin | (40) |
| 55 | 9.53 | [M + HCOO]− | sinsenoside Rg1 | C42H72O14 | 845.4899 | 845.4801 | 11.59 | 845.4801 [M + HCOO]− | SM | saponin | (40) |
| 56 | 10.58 | [M + HCOO]− | sinsenoside Re | C48H82O18 | 991.5478 | 991.5424 | 5.45 | 991.5424 [M + HCOO]−, 845.4880 [M – Rha + HCOO]− | SM | saponin | (40) |
| 57 | 12.35 | [M + HCOO]− | ginsenoside Rg3 | C42H72O13 | 829.4949 | 829.4977 | 3.38 | 829.4977 [M + HCOO]− | SM | saponin | (40) |
| 58 | 5.96 | [M – H]− | salvianolic acid I | C27H22O12 | 537.1033 | 537.1061 | 5.21 | 537.1061 [M – H]−, 339.0478, 321.0466, 313.0568, 295.0630 | SM | phenolic acid | (23) |
| 59 | 7.32 | [M – H]− | salvianolic acid E | C36H30O16 | 717.1456 | 717.1445 | 1.53 | 717.1445 [M – H]−, 537.1061, 519.0837, 493.1164, 339.0478, 321.0388, 295.0630, 185.0208 | SM | phenolic aci | (23) |
| 60 | 6.63 | [M – H]− | lithospermic acid | C27H22O12 | 537.1033 | 537.1061 | 5.21 | 537.1061 [M – H]−, 493.1164, 313.0645, 295.0630, 197.0428 | SM | phenolic acid | (23) |
| 61 | 1.77 | [M + H]+ | adenosine | C10H13N5O4 | 268.1046 | 268.1045 | 0.37 | 268.1045 [M + H]+, 136.0638 [C5H6N5]+, 119.0353 [M + H-ribose – NH3]+ | CP | nucleoside | (19) and (41) |
| 62 | 1.64 | [M – H]− | succinic acid | C4H6O4 | 117.0188 | 117.0177 | 9.40 | 117.0177 [M – H]− | CP | phenolic acid | (19) |
| 63 | 2.80 | [M + H]+ | codonopsine | C14H21NO4 | 268.1549 | 268.1545 | 1.49 | 268.1545 [M + H]+, 161.0586 [M + H – C4H11NO – H2O]+ | CP | alkaloid | (19, 20), and (42) |
| 64 | 7.78 | [M + H]+ | vanillic acid | C8H8O4 | 167.0344 | 167.0364 | 11.91 | 167.0364 [M + H]+, 152.0566 [M – H – CH3]−, 123.0428 [M – H – CO2]−, 108.0194 [M – H – CH3 – CO2]− | CP | organic acid | (19, 20), and (25) |
| 65 | 0.88 | [M – H]− | erigeside B sulfate | C12H22O9S | 341.0906 | 341.0944 | 11.14 | 341.0944 [M – H]− | CP | (19) | |
| 66 | 7.78 | [M + HCOO]− | lobetyol sulfate isomer I | C14H18O4S | 327.0914 | 327.0906 | 2.44 | 327.0906 [M + HCOO]− | CP | (19) | |
| 67 | 12.51 | [M – H]− | p-coumaric acid | C15H18O8 | 325.0923 | 325.0990 | 14.46 | 325.0990 [M – H]−, 163.0775 [M – H – C6H10O5]− | CP | (19) and (20) | |
| 68 | 9.06 | [M – H]− | (E)-2-hexenyl-α-larabinopyranosyl-(1/2)-β-d-glucopyranoside | C17H30O10 | 393.1761 | 393.1765 | 1.02 | 393.1765 [M – H]−, 179.0317 [C6H11O6]−, 149.0458 [C5H9O5]− | CP | (19) | |
| 69 | 5.56 | [M – H]− | hexyl-β-d-glucopyranosyl-(1/2)-β-d-glucopyranoside | C18H34O11 | 425.2023 | 425.2097 | 10.35 | 425.2097 [M – H]−, 263.1125 [M – H – C6H10O5]−, 245.1518 [M – H – C6H10O5 – H2O]−, 179.0555 [C6H11O6]− | CP | (19) and (20) | |
| 70 | 4.89 | [M – H]− | (6R,9S)-3-oxo-α-iono lβ-d-glucopyranoside sulfate | C19H30O10S | 449.1481 | 449.1428 | 11.80 | 449.1428 [M – H]− | CP | (19) | |
| 71 | 4.88 | [M – H]− | atractylenolide III sulfate | C15H20O6S | 327.0902 | 327.0952 | 12.23 | 327.0952 [M – H]− | CP | (19) | |
| 72 | 13.41 | [M – H]− | emodin | C15H10O5 | 269.0450 | 269.0422 | 10.41 | 269.0422 [M – H]−, 241.0508 [M – H – CO]−, 225.0580 [M – H – CO2]− | CP, PC | anthraquinone | (19) and (43) |
| 73 | 12.07 | [M – H]− | 6-methylgingediol | C18H30O4 | 309.2066 | 309.2034 | 10.35 | 309.2034 [M – H]−, 291.1657 [M – H – H2O]−, 210.0318 [M – H – C6H11O]− | CP | gingerol | (19) |
| 74 | 5.10 | [M + HCOO]− | lobetyol sulfate isomer II | C14H18O4S | 327.0902 | 327.0952 | 12.23 | 327.0952 [M + HCOO]− | CP | (19) | |
| 75 | 5.90 | [M + HCOO]− | codonopyrrolidium A | C19H28NO5 | 395.1944 | 395.1978 | 8.60 | 395.1978 [M + HCOO]−, 263.1505 [M + HCOO – C5H8O4]−, 131.0350 [C5H7O4]− | CP | alkaloid | (19) and (44) |
| 76 | 6.21 | [M + HCOO]− | lobetyolinin | C26H38O13 | 603.2289 | 603.2237 | 8.62 | 603.2237 [M + HCOO]−, 395.0762 [M – H – C6H10O5]−, 323.0775 [C12H19O10]−, 221.0643 [C8H13O7]−, 179.0434 [C6H11O6]− | CP | polyacetylene | (19) and (20) |
| 77 | 11.75 | [M + H]+ | ursolic acid | C30H48O3 | 457.3682 | 457.3677 | 1.09 | 457.3677 [M + H]+ | CP | triterpene | (19) and (20) |
| 78 | 10.15 | [M – H]− | chrysophanol | C15H10O4 | 253.0501 | 253.0525 | 9.48 | 253.0545 [M – H]−, 225.0514, 210.0761, 182.0552, 154.6341 | PC | quinone | (45) |
| 79 | 11.25 | [M – H]− | physcion | C16H12O5 | 283.0606 | 283.0635 | 10.24 | 283.0665 [M – H]−, 240.0275, 212.0630, 196.9999, 184.0414 | PC | anthraquinone | (45) |
| 80 | 7.92 | [M – H]− | rhein | C15H8O6 | 283.0606 | 283.0627 | 7.42 | 283.0627 [M – H]−, 239.0343, 211.0392, 183.0527 | PC | anthraquinone | (45) |
| 81 | 5.41 | [M – H]− | polydatin | C20H22O8 | 389.1236 | 389.1237 | 0.26 | 389.1237 [M – H]−, 227.0682 [M – H – glu]−, 185.0292 [M – H – glu – RDA]−, 157.0230 [M – H – glu – RDA – CO]−, 183.0376 [M – H – glu – CO2]−, 159.0682 [M – H – glu – RDA]− | PC | anthraquinone glycoside | (45) and (46) |
| 82 | 10.77 | [M – H]− | aloe-emodin | C15H10O5 | 269.0450 | 269.0422 | 10.41 | 269.0422 [M – H]−, 240.3443, 211.0191, 167.0329, 139.0447 | PC | anthraquinone | (45) |
| 83 | 5.47 | [M – H]− | resveratrol | C14H12O3 | 227.0708 | 227.0738 | 13.21 | 227.0748 [M – H]−, 183.0292 [M – H – CO2]−, 185.0623 [M – H – RDA]−, 157.0669 [M – H – RDA – CO]−, 159.0829 [M – RDA]− | PC | polyphenols | (45) |
| 84 | 3.87 | [M – H]− | resveratrol-4′-O-glucoside | C20H22O8 | 389.1236 | 389.1237 | 0.26 | 389.1237 [M – H]−, 227.0415 [M – H – glu]−, 185.0802 [M – H – glu – RDA]− | PC | (46) | |
| 85 | 6.23 | [M – H]− | trans-3,5,4′-trihydroxystilbene-4′-O-β-d-(2″O-galloyl)gulcopyranoside | C27H26O12 | 541.1346 | 541.1337 | 1.66 | 541.1337 [M – H]−, 313.0568 [M – H – C14H11O3]−, 125.0293 [M – H – C14H11O3 – C6H8O4 – CO2]− | PC | (47) and (48) | |
| 86 | 8.24 | [M – H]− | emodin-8-O-glucoside | C21H20O10 | 431.0978 | 431.0952 | 6.03 | 431.0952 [M – H]−, 269.0422 [M – H – glc]−, 241.0440 [M – H – glc – CO]− | PC | anthraquinone glycoside | (46) and (47) |
| 87 | 6.41 | [M – H]− | physcion-8-O-glucoside | C22H22O10 | 445.1135 | 445.1146 | 2.47 | 445.1146 [M – H]−, 283.0591 [M – H – glc]−, 268.0255 [M – H – glc – CH3]−, 240.0411 [M – H – glc – CH3 – CO]−, 212.0503 [M – H – glc – CH3 – 2CO]−, 184.0473 [M – H – glc – CH3 – 3CO]− | PC | anthraquinone glycoside | (46) and (47) |
| 88 | 11.08 | [M + HCOO]− | gypenoside LXIII | C53H90O22 | 1123.5900 | 1123.5935 | 3.12 | 1123.5935 [M + HCOO]−, 915.5429, 783.5080, 621.4413, 459.3957 | GP | saponin | (49) and (50) |
| 89 | 9.48 | [M + HCOO]− | gypenoside LXII | C53H90O23 | 1139.5849 | 1139.5847 | 0.18 | 1139.5847 [M + HCOO]−, 931.5030, 799.4846, 637.4455, 475.2202 | GP | saponin | (51) |
| 90 | 10.59 | [M + HCOO]− | gypenoside VIII | C48H82O18 | 991.5478 | 991.5424 | 5.45 | 991.5424 [M + HCOO]−, 783.5170, 621.1473, 459.3863 | GP | saponin | (51) |
| 91 | 11.03 | [M + HCOO]− | gypenoside LVII | C47H80O18 | 977.5321 | 977.5313 | 0.82 | 977.5313 [M + HCOO]−, 799.0768, 637.4455, 475.2444 | GP | saponin | (51) |
| 92 | 13.18 | [M + HCOO]− | gypenoside XXI | C41H70O13 | 815.4793 | 815.4760 | 4.04 | 815.4760 [M + HCOO]−, 637.3903, 475.3396, | GP | saponin | (52) and (53) |
| 93 | 11.74 | [M + HCOO]− | panaxadiol | C30H52O3 | 505.3893 | 505.3875 | 3.56 | 505.3875 [M + HCOO]− | GP | tetracyclic triterpenes | (49) |
| 94 | 7.53 | [M + HCOO]− | peoniflorin | C23H27O11 | 524.1530 | 524.1530 | 0 | 524.1530 [M + HCOO]−, 449.0316 [M – H – CH2O]−, 327.0658 [M – H – benzoic acid]− | RPR | monoterpene glycosides | (54) |
| 95 | 3.73 | [M – H]− | catechin | C15H14O6 | 289.0712 | 289.0745 | 11.42 | 289.0745 [M – H]−, 245.0903, 203.0370, 161.0287, 125.0146, 109.0339 | RPR | phenol | (54) |
| 96 | 2.13 | [M – H]− | Glucopyranos l-paeonisuffrone | C16H24O9 | 359.1342 | 359.1363 | 5.85 | 359.1363 [M – H]−, 179.0668, 197.0735 | RPR | (54) | |
| 97 | 3.18 | [M – H]− | mudanpioside F | C16H24O8 | 343.1393 | 343.1390 | 0.87 | 343.1390 [M – H]−, 181.0426, 159.0334, 151.0314 | RPR | glycoside | (54) |
| 98 | 5.00 | [M – H]− | desbenzoylpaeoniflorin | C16H24O8 | 479.1553 | 479.1577 | 5.01 | 479.1577 [M – H]−, 345.1086, 195.0729, 183.0580, 139.0470 | RPR | (54) | |
| 99 | 4.65 | [M – H]− | desbenzoylpaeoniflorin isomer I | C16H24O8 | 479.1553 | 479.1577 | 5.01 | 479.1577 [M – H]−, 345.1086, 195.0729, 183.0580 | RPR | (54) | |
| 100 | 3.66 | [M – H]− | oxypaeoniflorin | C23H28O12 | 495.1553 | 495.1561 | 1.62 | 495.1561 [M – H]−, 465.1166, 299.0694, 281.0632, 367.1208, 239.0537, 165.0567 | RPR | glycoside | (54) |
| 101 | 8.71 | [M – H]− | benzoylpaeoniflorin | C30H32O12 | 583.1816 | 583.1860 | 7.54 | 583.1860 [M – H]−, 553.1320, 431.1134, 291.1583, 329.0499, 314.0703, 298.9862 | RPR | glycoside | (54) |
| 102 | 7.71 | [M – H]− | 3,7- or 3,8-dimethyl ellagic acid | C16H10O8 | 329.0297 | 329.0262 | 10.64 | 329.0262 [M – H]−, 314.0394, 270.0321 | RPR | (54) | |
| 103 | 8.80 | [M – H]− | dihydroapigenin | C15H12O5 | 271.0606 | 271.0640 | 12.54 | 271.0640 [M – H]−, 177.0321, 165.0455, 151.0396, 119.0350, 107.6151 | RPR | (54) | |
| 104 | 5.90 | [M – H]− | galloylpaeoniflorin | C30H32O15 | 631.1663 | 631.1621 | 6.65 | 631.1621 [M – H], 613.1152, 509.1964, 491.0810, 271.0239 | RPR | glycoside | (54) |
| 105 | 7.59 | [M + H]+ | p-hydroxy coumaric acid | C10H10O4 | 165.0552 | 165.0560 | 4.84 | 165.0560 [M + H]+, 165.0560 [M + H – O2]+ | HH | phenylpropionic acid | |
| 106 | 4.93 | [M + H]+ | glucose-ethylglucoside | C8H16O6 | 209.1025 | 209.1052 | 12.91 | 209.1052 [M + H]+, 105.2247 [M + H – C4H8O3]+ | HH | ||
| 107 | 4.98 | [M + H]+ | methoxy cinnamic acid | C10H10O3 | 179.0708 | 179.0731 | 12.84 | 179.0731 [M + H]+, [M + H – C2H4]+ | HH | phenylpropionic acid | |
| 108 | 5.93 | [M + H]+ | quercetin-3-O-[2-O-(6-O-E-feruloyl)-β- d-glucopyranosyl]-β-d-glucopyranoside | C37H38O20 | 803.2035 | 803.2059 | 2.98 | 803.2059 [M + H]+, 175.0573 [M + H – C28H36O16]+ | HH | ||
| 109 | 6.39 | [M + H]+ | kaempferol-3-O-[2-O- (6-O-E-feruloyl)-β-d-glucopyranosyl]-β-d-galactopyranoside | C37H38O19 | 787.2086 | 787.2052 | 4.32 | 787.2052 [M + H]+, 591.2525 [M + H – C10H12O4]+, 575.1149 [M + H – C10H12O5]+, 307.0205 [M + H – C5H20O10]+ | HH | ||
| 110 | 10.15 | [M + H]+ | 2-hydroxy-1-methoxyanthraquinone | C15H10O4 | 255.0657 | 255.0629 | 10.98 | 255.0629 [M + H]+, 239.1759 [M + H – OH]+, 183.1154 [M + C3H4O2]+ | HH | ||
| 111 | 11.49 | [M + H]+ | 2-methyl-3-hydroxyanthraquinone | C15H10O3 | 239.0708 | 239.0747 | 16.31 | 239.0747 [M + H]+, 165.0728 [M + H – C6H2]+, 133.1008 [M + H – C7H6O]+ | HH |
Glc, glucosyl; RDA, −C2H2O; Xyl, β-d-xylose.
2.3. Construction of Chemical Components and Action Targets of QZD
According to the experiment results of Section 2.2, a total of 111 chemical components were identified by UPLC–Q-TOF/MS (Table 2), of which the conformations of 26 chemical components were not found in PubChem, so 85 chemical components were involved in this study. A total of 955 targets were obtained from the chemical components of RA, CP, LL, HH, GP, SM, RPR, and PC by reverse pharmacophore screening.
2.4. Screening of “Colon Cancer Targets”
Using “Colon cancer” as the keyword, 404 disease targets were found in CTD, 300 disease targets in Gene Cards, and 5 disease targets in TTD. Then, they were transformed into standard gene targets by the UniProtKB search function of the Uniprot database. A total of 608 disease targets were obtained after the above data were integrated and deduplicated.
2.5. Acquisition of “Common Targets”
A total of 955 chemical composition-related targets and 608 disease targets obtained from Sections 2.3 and 2.4 were uploaded to draw a Venn diagram for analysis, and 132 common targets were obtained (Figure 2).
Figure 2.
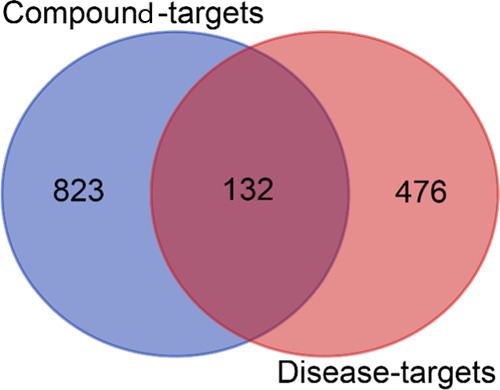
Venn diagram of chemical composition targets of QZD and colon cancer targets. Among them, the blue circle indicates that there are 955 chemical composition targets of QZD, the orange circle represents 608 Colon cancer targets, and the middle crossing part indicates 132 “common targets”.
2.6. Acquisition of “Indirect Targets”
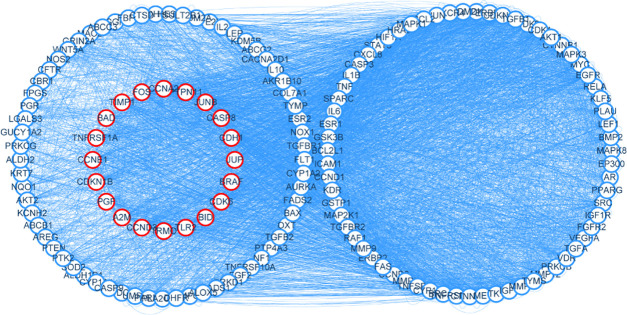
A total of 132 common targets obtained from Section 2.5 were uploaded to GeneMANIA. The source type was set as the human source, and 20 indirect targets were analyzed (Figure 3).
Figure 3.
Target map of QZD in the treatment of colon cancer. Among them, the nodes on the blue circle represent 132 common targets of QZD in the treatment of colon cancer, and the nodes on the red circle represent 20 indirect targets of QZD in the treatment of colon cancer, and the side represents the interaction between the targets.
2.7. Construction and Analysis of the Protein/Protein Interaction (PPI) Network
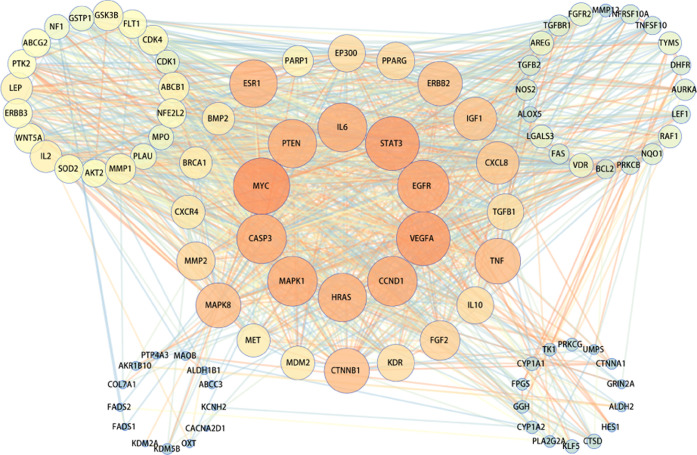
The targets obtained by Sections 2.5 and 2.6 were integrated and imported into the String database, the human source was defined, the protein interaction relationship was obtained, the TSV format file was saved, Cytoscape software for visualization was imported, and a total of 96 nodes were involved (Figure 4). Finally, 10 top targets of QZD in the treatment of colon cancer were obtained, which were MYC, VEGFA, STAT3, EGFR, CCND1, CASP3, MAPK1, IL6, HRAS, and PTEN (Table 3). It showed that these targets played an important role in the PPI network, suggesting that these targets played an important role in the anticolon cancer effect of QZD.
Figure 4.
Protein interaction network map of QZD in the treatment of colon cancer. In the figure, the node represents the target protein, the edge represents the interaction between the target protein, the size and color of the node represent the degree value, and the color of the edge represents the combined score value. The larger the node, the larger the degree value corresponding to the color from green to red, and the greater the combined score value corresponding to the color of the edge from green to red.
Table 3. Key Targets of QZD in the Treatment of Colon Cancera.
| no | name | degree | BC | CC | TC |
|---|---|---|---|---|---|
| 1 | MYC | 68 | 0.0528 | 0.7600 | 0.3427 |
| 2 | VEGFA | 64 | 0.0374 | 0.7364 | 0.3543 |
| 3 | STAT3 | 64 | 0.0389 | 0.7364 | 0.3528 |
| 4 | EGFR | 61 | 0.0285 | 0.7143 | 0.3648 |
| 5 | CCND1 | 58 | 0.0370 | 0.7037 | 0.3674 |
| 6 | CASP3 | 58 | 0.0391 | 0.7037 | 0.3654 |
| 7 | MAPK1 | 58 | 0.0462 | 0.7037 | 0.3602 |
| 8 | IL6 | 57 | 0.0415 | 0.6985 | 0.3605 |
| 9 | HRAS | 56 | 0.0273 | 0.6884 | 0.3762 |
| 10 | PTEN | 56 | 0.0221 | 0.6934 | 0.3794 |
“BC” means “betweenness centrality”, “CC” means “closeness centrality”, and “TC” means “topological coefficient”.
2.8. Molecular Docking Verification
The topological analysis of the PPI network was carried out by the “Network Analyzer” function of Cytoscape, and the attributes of “betweenness centrality”, “closeness centrality” and “topological coefficient” were retained. The top three targets of degree (MYC, VEGFA, STAT3) were selected as the key targets in the network and used as the receptors of molecular docking to verify the reliability of prediction. The results of molecular docking are shown in Table 4, of which 31 (12.2%) had docking score values greater than 7.0, 176 (69.0%) greater than 5.0, and 193 (75.7%) greater than 4.25, which proved that the targets obtained by virtual screening had a certain degree of credibility.
Table 4. Docking Results of Chemical Compositions and Key Targets.
| docking
score (pKd/pKi) |
||||
|---|---|---|---|---|
| no. | active ingredients | MYC (5I4Z) | VEGFA (1CZ8) | STAT3 (5AX3) |
| 1 | hexose | 5.077 | 5.320 | 5.386 |
| 2 | asparagine | 4.547 | 5.233 | 5.168 |
| 3 | choline | 3.373 | 3.362 | 4.239 |
| 4 | raffinose | 5.496 | 5.736 | 6.135 |
| 5 | nicotinic acid | 5.236 | 5.124 | 4.239 |
| 6 | leucine | 3.797 | 3.749 | 4.348 |
| 7 | adenine nucleoside | 5.311 | 5.468 | 5.445 |
| 8 | phenylalanine | 2.212 | 2.434 | 2.919 |
| 9 | markhamioside F | 5.192 | 5.448 | 6.212 |
| 10 | hydroxybenzoic acid | 2.053 | 2.525 | 2.526 |
| 11 | rhamnocitrin 3,4′-di-O-glucoside | 6.079 | 6.131 | 7.324 |
| 12 | caffeic acid | 3.661 | 4.048 | 4.248 |
| 13 | emodin-di-O-glucoside | 5.263 | 5.750 | 6.649 |
| 14 | syringaldehyde | 3.091 | 3.250 | 4.074 |
| 15 | ferulic acid | 3.128 | 3.289 | 3.762 |
| 16 | pratensein-7-O-glucoside | 5.269 | 6.086 | 6.417 |
| 17 | calycosin-7-O-glucoside | 6.096 | 5.774 | 6.222 |
| 18 | cosmosiin | 5.344 | 5.856 | 6.405 |
| 19 | azelaic acid | 4.496 | 4.740 | 4.720 |
| 20 | isomucronulatol-7-O-glucoside | 5.421 | 5.873 | 6.662 |
| 21 | calycosin | 3.002 | 2.333 | 4.297 |
| 22 | formononetin | 2.421 | 2.016 | 3.986 |
| 23 | astragaloside V | 7.540 | 4.998 | 7.954 |
| 24 | soyasaponin I | N/A | N/A | N/A |
| 25 | isoastragaloside I | 7.490 | 4.899 | 7.414 |
| 26 | astragaloside IV (astragaloside A) | N/A | N/A | N/A |
| 27 | nuezhenidic acid | 5.417 | 5.747 | 6.054 |
| 28 | chlorogenic acid | 5.329 | 5.577 | 6.116 |
| 29 | oleoside 11-methyl ester | 5.420 | 5.852 | 6.183 |
| 30 | oleuropein-aglycone | 5.405 | 6.233 | 6.611 |
| 31 | specnuezhenide | 6.161 | 6.121 | 7.925 |
| 32 | ligustroside | 5.299 | 6.106 | 7.087 |
| 33 | oleonuezhenide | 7.562 | 7.802 | 7.868 |
| 34 | oleanolic acid | 6.664 | 6.902 | 7.909 |
| 35 | salidrosid | 5.249 | 5.804 | 5.816 |
| 36 | danshensu | 3.898 | 4.283 | 4.720 |
| 37 | protocatechuic aldehyde | 5.378 | 5.432 | 5.102 |
| 38 | rosmarinic acid | 7.174 | 6.583 | 4.921 |
| 39 | lithosperimic acid | 5.346 | 5.941 | 7.321 |
| 40 | salvianolic acid B | 6.180 | 5.989 | 8.189 |
| 41 | salvianolic acid A | 5.512 | 5.760 | 5.034 |
| 42 | salvianolic acid C | 5.401 | 4.883 | 4.786 |
| 43 | ginsenoside Rb1 | 8.173 | 8.089 | 5.013 |
| 44 | dihydrotanshinone I | 5.885 | 6.524 | 6.553 |
| 45 | cryptotanshinone | 6.582 | 6.492 | 6.376 |
| 46 | tanshinone I | 6.701 | 6.335 | 6.460 |
| 47 | tanshinone IIA | 6.730 | 6.514 | 6.433 |
| 48 | notoginsenoside R1 | 7.494 | 8.077 | 7.957 |
| 49 | ginsenoside Rg1 | 6.872 | 7.357 | 8.062 |
| 50 | ginsenoside Re | N/A | N/A | N/A |
| 51 | ginsenoside Rg3 | 7.462 | 7.299 | 8.034 |
| 52 | salvianolic acid I | 5.312 | 5.197 | 4.909 |
| 53 | salvianolic acid E | 4.244 | 5.005 | 4.587 |
| 54 | lithospermic acid | 5.402 | 5.938 | 7.290 |
| 55 | adenosine | 5.690 | 5.704 | 5.770 |
| 56 | succinic acid | 3.980 | 3.968 | 4.041 |
| 57 | codonopsine | 5.514 | 5.856 | 6.351 |
| 58 | vanillic acid | 3.079 | 3.192 | 3.936 |
| 59 | p-coumaric acid | 2.915 | 3.186 | 3.603 |
| 60 | atractylenolide III sulfate | 5.817 | 5.530 | 6.389 |
| 61 | emodin | 6.640 | 5.945 | 6.594 |
| 62 | codonopyrrolidium A | 4.114 | 4.385 | 5.262 |
| 63 | lobetyolinin | 5.489 | 6.338 | 7.437 |
| 64 | ursolic acid | 6.069 | 6.976 | 8.059 |
| 65 | chrysophanol | 6.540 | 6.010 | 6.350 |
| 66 | physcion | 6.707 | 5.981 | 6.277 |
| 67 | rhein | 6.538 | 6.597 | 6.335 |
| 68 | polydatin | 5.314 | 5.741 | 6.626 |
| 69 | aloe-emodin | 6.466 | 5.989 | 6.480 |
| 70 | resveratrol | 2.759 | 3.362 | 3.720 |
| 71 | resveratrol-4′-O-glucoside | 5.450 | 5.798 | 6.820 |
| 72 | emodin-8-O-glucoside | 5.720 | 5.795 | 6.637 |
| 73 | physcion-8-O-glucoside | 5.227 | 5.925 | 6.603 |
| 74 | gypenoside LXIII | N/A | N/A | N/A |
| 75 | gypenoside VIII | N/A | N/A | N/A |
| 76 | gypenoside LVII | N/A | N/A | N/A |
| 77 | panaxadiol | 6.534 | 7.048 | 8.144 |
| 78 | peoniflorin | 5.235 | 5.767 | 6.703 |
| 79 | catechin | 6.505 | 6.437 | 6.595 |
| 80 | mudanpioside F | 5.380 | 5.846 | 6.497 |
| 81 | oxypaeoniflorin | 5.115 | 5.662 | 6.687 |
| 82 | galloylpaeoniflorin | 6.194 | 5.973 | 8.006 |
| 83 | p-hydroxy coumaric acid | 3.044 | 3.171 | 3.604 |
| 84 | methoxy cinnamic acid | 4.256 | 5.029 | 5.203 |
| 85 | stigmasterol | 5.848 | 6.804 | 8.149 |
2.9. GO Bioanalysis and KEGG Pathway Analysis
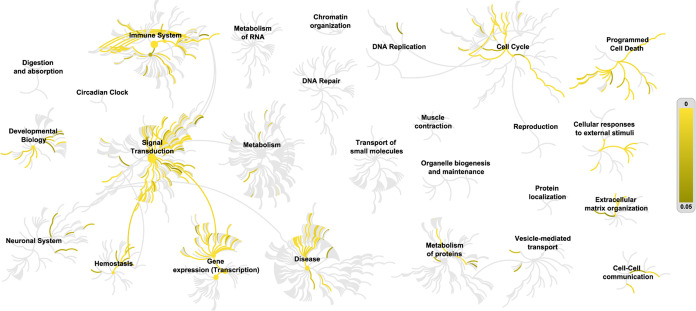
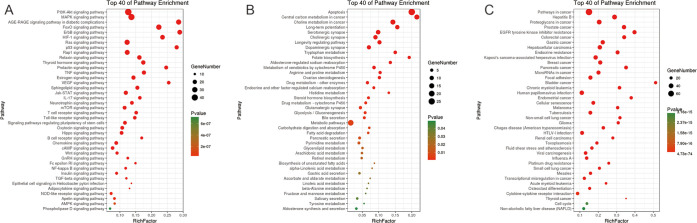
A variety of biological processes are involved in the mechanism of QZD in the treatment of colon cancer, reflecting the multiple therapeutic effects of QZD. Figure 5 shows that the target of QZD in the treatment of colon cancer involves biological processes such as signal transduction, immune system, gene expression, and so on. Figure 6 shows the results of pathway analysis of QZD in the treatment of colon cancer. As shown in Figure 6A, pathways such as the “PI3K–Akt signaling pathway”, “MAPK signaling pathway,” and “AGE-RAGE signaling pathway in diabetic complications” are highly associated with QZD in the treatment of colon cancer. As shown in Figure 6B, QZD is most likely to affect the higher-ranked pathways of “apoptosis”, “central carbon metabolism in cancer,” and “choline metabolism in cancer”. As shown in Figure 6C, the pathways of diseases such as “cancer”, “hepatitis B,” and “proteoglycans in cancer” may be disturbed by QZD.
Figure 5.
Tailless fireworks picture of QZD in the treatment of colon cancer. The yellow to brown lines indicate the important pathway of target enrichment, and the p-value increases gradually from yellow to brown.
Figure 6.
Important pathways of QZD in the treatment of colon cancer. (A) Signal pathway of the target, (B) metabolic pathway of the target, and (C) disease pathway of the target.
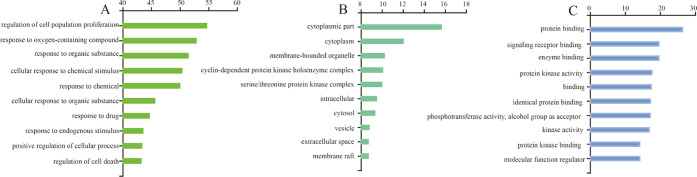
As shown in Figure 7A, the target of QZD may be involved in biological pathways such as “regulation of cell population proliferation”, “response to oxygen-containing compounds,” and “response to organic substances”. As shown in Figure 7B, the target of QZD plays an important role in the synthesis and decomposition of the “cytoplasmic part”, “cytoplasm,” and “membrane-bounded organelles”. As shown in Figure 7C, the processes of “protein binding”, “signaling receptor binding,” and “enzyme binding” play an important role in the treatment of colon cancer with QZD.
Figure 7.
Intracellular processes related to the target of QZD in the treatment of colon cancer. (A) Biological pathways involved in the target, (B) cellular components involved in the target, and (C) molecular functions involved in the target.
2.10. Construction and Analysis of “Prescription–TCM–Chemical Composition–Direct Target–Indirect Target–Pathway” of QZD in the Treatment of Colon Cancer
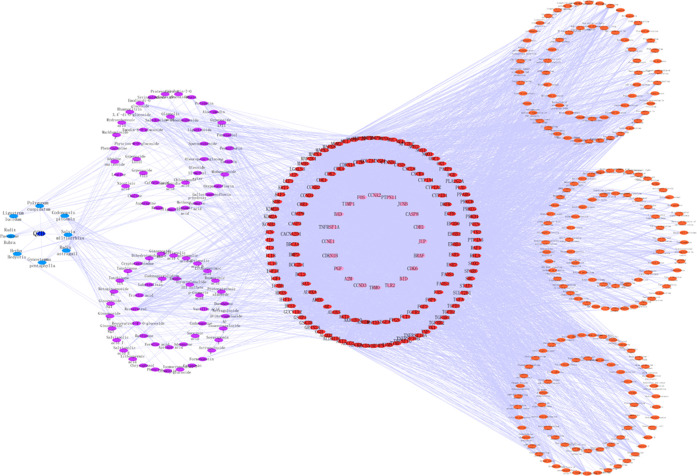
The prescription name, TCM, chemical composition, direct target, indirect target, and pathway information of QZD were introduced into Cytoscape software to construct the “Prescription–TCM–Chemical composition–Direct target–Indirect target–Pathway” network of QZD in the treatment of colon cancer (Figure 8). The results showed that 456 edges and 6165 points interact with each other, and the chemical composition of QZD could correspond to multiple targets. Of course, one target could also correspond to different chemical components, and different targets also correspond to multiple mechanism pathways. It could be seen that the mechanism of QZD against colon cancer had the characteristics of multicomponents, multitargets, and multimechanisms.
Figure 8.
Network diagram of “Prescription–TCM–Chemical composition–Direct target–Indirect target–Pathway” of QZD in the treatment of colon cancer. Dark blue nodes represent QZD, light blue nodes represent TCM, purple nodes represent chemical components, red nodes represent direct targets, pink nodes represent indirect targets, and orange nodes represent related pathways.
2.11. Effect of QZD on the PI3K–Akt Pathway and Proliferation and Migration of Colon Cancer LoVo Cells
2.11.1. Effect of QZD and Pathway Inhibitor LY294002 on PI3K–Akt Pathway
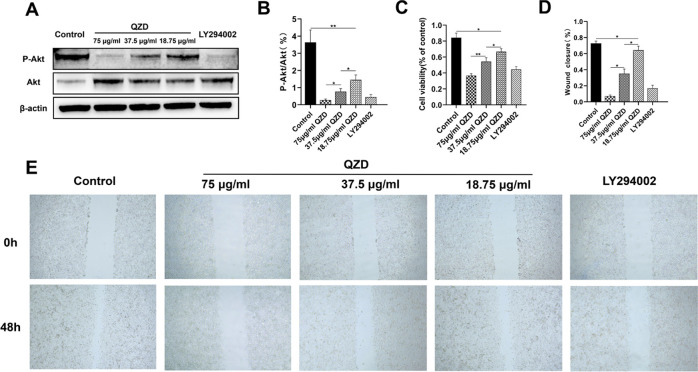
Compared with the control group, P-Akt/Akt of the QZD intervention group and LY294002 group decreased significantly, but there was no significant difference between the high-dose QZD group and LY294002 group (p > 0.05). Both QZD and inhibitor LY294002 could inhibit the PI3K–Akt pathway. QZD inhibited PI3K–Akt in a dose-dependent manner, and its inhibitory effect was similar to that of pathway inhibitor LY294002 (Figure 9A,B).
Figure 9.
(A) After LoVo cells were treated with LY294002 or different concentrations of QZD, the protein expression of P-Akt and Akt was detected by western blotting. (B) Quantitative histogram of P-Akt/Akt of LoVo cells treated with LY294002h or different concentrations of QZD. (C) MTT assay was used to detect the cell viability of LoVo cells treated with different concentrations of QZD or LY294002. (D) Cell wound scratch test was used to detect the quantitative histogram of the healing rate of LoVo cells treated with different concentrations of QZD or LY294002. (E) Cell wound scratch test was used to detect the representative diagram of cell migration ability of LoVo cells treated with different concentrations of QZD or LY294002 (40×).
2.11.2. Effects of QZD and LY294002 on LoVo Cell Proliferation
The results of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) detection showed that the cell proliferation activity of the QZD group and LY294002 group was lower than that of the control group (*p < 0.05, **p < 0.01). The cell proliferation activity of the QZD group was dose-dependent (Figure 9C). The effect of QZD and LY294002 on the migration of LoVo cells. The results of the scratch test showed that compared with the control group, the scratch healing rate of different concentrations of the QZD group and LY294002 group decreased (*p < 0.05), while the scratch healing rate of the high-dose QZD group was not significantly different than that of the LY294002 group (p > 0.05). QZD could significantly inhibit the migration of LoVo cells in vitro; the ability of high-dose QZD to inhibit the migration of LoVo cells in vitro was not significantly different than that of the LY294002 group (Figure 9D,E).
3. Discussion
Colon cancer has a high incidence and complicated pathogenesis.55 Finding a new comprehensive treatment method to improve clinical efficacy has always been a research hotspot in the treatment of colon cancer. Among the components of QZD, there are commonly used drugs for the treatment of tumors. As a compound prescription of TCM, QZD has the unique advantages of improving clinical symptoms, enhancing immunity, improving the sensitivity of chemotherapeutic drugs, and reducing recurrence and metastasis. Although complex TCM plays an important role in the treatment of colon cancer, its material basis and specific mechanism are still unknown.56 This study adopted a combination of the UPLC–Q-TOF/MS method and network pharmacology to deeply mine data and to conduct network analysis. Through the interaction between the chemical components of QZD and the molecular targets of colon cancer diseases, the mechanism of QZD against colon cancer was revealed.
3.1. Detection of Chemical Components of QZD
In this study, the chemical components of QZD were quickly separated, analyzed, and identified by the UPLC–Q-TOF/MS method. The structural formula was inferred according to the exact relative molecular weight and mass spectrometry fragments given by MassLynx v4.1 workstation, and a total of 111 chemical components in QZD were identified combined with the related literature and control substances, including 12 saponins (SM/GP), 11 phenolic acids (SM/CP), 10 glycosides (RA, RPR), 5 organic acids (RA, SM, CP), 4 anthraquinones (PC), 4 phenanthraquinones (SM), 4 amino acids (RA), 3 anthraquinone glycosides (PC), 3 iridoid terpenoids (LL), 2 alkaloids (CP), 2 triterpenoid saponins (RA), 2 flavonoids (RA), 2 phenylpropionic acid compounds (HH), and so on. The results of chemical composition attribution showed that the saponins mainly come from SM and GP. Phenolic acids mainly came from SM and CP. Anthraquinones and anthraquinone glycosides mainly came from PC. Glycosides mainly came from RA and RPR. Organic acids mainly came from RA, SM, and CP. Phenanthraquinones had the highest content in SM. Amino acids, flavonoids, and triterpenoid saponins were mainly derived from RA. Iridoid terpenoids were mainly from LL. Alkaloids and phenylpropionic acids were from CP and HH, respectively. This study provided an efficient and rapid analysis method for the qualitative analysis and quality control of chemical components in QZD and comprehensively expounded the chemical material basis of QZD, which laid the foundation for further pharmacology and mechanism research.
3.2. Effective Components of QZD to Improve the Therapeutic Effect of Radiotherapy, Chemotherapy, Targeted Drugs, and Other Drugs
The experimental results suggested that QZD has a variety of chemical components against colon cancer. Astragaloside could inhibit the proliferation of colon cancer cells and reverse cisplatin resistance.57 Notoginsenoside R1, resveratrol, and panaxadiol could inhibit intestinal tumor58−61 and combined with 5-fluorouracil could reduce the growth of colon cancer cells to a great extent and enhance their anticancer activity. The above chemical components might be effective adjuvants for clinical chemotherapy. Besides, both salidroside and salvianolic acid B could induce apoptosis of human colon cancer cells and increased the sensitivity of drug-resistant cells to chemotherapeutic drugs.62−64 Cryptotanshinone could induce autophagic death in the human colon cancer multidrug-resistant cell line.65,66 The combination of ursolic acid and oxaliplatin could significantly inhibit cell proliferation and the expression of drug-resistant genes. The above chemical components might be an adjuvant drug in the treatment of multidrug-resistant colon cancer. Besides, both polydatin and ginsenoside Rg3 could significantly improve the efficacy and sensitivity of fractionated radiotherapy.67,68 The effective components of QZD could not only improve the therapeutic effect of radiotherapy and chemotherapy but also improve the sensitivity of targeted drugs. For example, rhein could increase the sensitivity of human colon cancer cells to the targeted drug EGFR-TKIs.69 Tryptophan, the active ingredient of QZD, is an essential amino acid for T cell activation. The lack of local tryptophan can damage cellular immunity by reducing the proliferation of T cells.70 The metabolites of tryptophan have antiproliferation and antimigration activities on colon cancer cells.71 Tryptophan may be an adjuvant for immunotherapy
3.3. Effective Components of QZD Directly against Colon Cancer
The active components of QZD could resist angiogenesis, reduce cell viability, regulate the cell cycle, and inhibit the invasion and metastasis of colon cancer cells, including rosmarinic acid,72 caffeic acid,73 raffinose,74p-coumaric acid,75 emodin,76 ferulic acid,77 choline,78 calycosin,79 formononetin,80,81 tanshinone IIA,82 oleanolic acid,83 protocatechuic aldehyde.84 dihydrotanshinone I,85 aloe-emodin,86 and chlorogenic acid,87 which were the effective components that rely on reactive oxygen species (ROS) to produce cytotoxicity and have an anticolon cancer effect. In addition, other active ingredients exert their antitumor effects by inhibiting the epithelial mesenchymal transition (EMT) and the proliferation, migration, and invasion of colon cancer cells, such as ginsenoside Rb1,88 chrysophanol,89 physcion,90 peoniflorin.91
3.4. Gene Target and Signaling Pathway of QZD in the Treatment of Colon Cancer
The effective chemical components of TCM were the core links to explain the overall efficacy and the essence of the function of TCM. Because of the integrity of the effects of TCM and the complexity of its compositions and mechanisms, it had become complicated and difficult to study the material basis of the efficacy of TCM. A variety of active ingredients in QZD have been proved to promote apoptosis of colon cancer and improve the efficacy of chemotherapeutic drugs or radiotherapy in the treatment of colon cancer. It is worth noting that drugs produced from natural products often have the characteristics of multitargets, which is a characteristic of drugs for the treatment of complex diseases.
In addition to the active ingredients, we also successfully predicted the drug targets of QZD. Through the construction of a protein interaction network and Cytoscape visual analysis, we obtained the gene targets related to QZD in the treatment of colon cancer. Core gene targets such as MYC and VEGFA are highly sequenced. Cell proliferation, cell migration, and angiogenesis were considered to be very important factors in the development of colon cancer. Clinical studies suggested that the downregulation of MYC can change metabolism and inhibit the growth of colon cancer cells. The MYC target gene was a potential target for the treatment of colon cancer and could be used as an index to predict the prognosis of patients.92 VEGFA was a gene that promotes tumor growth, metastasis, and angiogenesis. Many cancers, including colon cancer, had angiogenic changes. At present, there were targeted drugs for VEGFA to inhibit angiogenesis of colon cancer cells.93,94 Because the targeting drugs of MYC and VEGFA still had some disadvantages, such as toxicity, poor pharmacokinetics, drug resistance, small beneficiary population, and so on.95−97 It was suggested that QZD can affect the key gene targets such as MYC, VEGFA. As a natural medicine, QZD might become an auxiliary drug for the above gene-targeting drugs, which could increase efficiency and reduce toxicity. QZD could be used as an auxiliary drug for sensitizing gene targeting, which provided a new research idea and choice.
In this study, GO and pathway analyses were used to explore the mechanism of QZD in the treatment of colon cancer, focusing on the biological process and signal pathway related to colon cancer. From the analysis of the reaction pathway, the high enrichment pathway of the chemical composition target of QZD was closely related to the highly ranked apoptosis metabolic pathway and PI3K–Akt signaling pathway. These pathways played an important role in the progress of QZD in the treatment of colon cancer.
Among the signal pathways screened in this study, PI3K–Akt ranks first. This experiment verified that QZD can affect this pathway and play a role in the treatment of colon cancer through molecular biology methods. Abnormal expression of the PI3K–Akt signal pathway was detected in many tumors. Activated Akt exists in colon cancer tumors, which has been proved to be an adverse factor affecting the prognosis of colon cancer patients. Akt phosphorylation represented the activation of this pathway, which promoted tumor cell proliferation, regulated cell cycle progression, and inhibited apoptosis. Therefore, inhibiting the activation of this pathway was beneficial to induce apoptosis, and other studies suggested that inhibition of this pathway could also enhance the sensitivity of chemotherapeutic drugs in the treatment of colon cancer.60,98,99 In this experiment, after the intervention of QZD on LoVo cells, it was found that the phosphorylation level of the Akt protein in the PI3K–Akt signal pathway decreased significantly. The cell scratch test and MTT test showed that QZD could inhibit the migration and proliferation of colon cancer LoVo cells. The results of this study suggested that QZD could inhibit the activation of the PI3K–Akt signal pathway. The PI3K–Akt signal pathway of QZD in the treatment of colon cancer was successfully verified by molecular biology experiments. It showed that the predicted targets and pathways based on this experiment could reveal the feasibility and accuracy of the mechanism of QZD in inhibiting colon cancer to a certain extent. The results of this experiment played a directive role in the in-depth study of QZD in the treatment of colon cancer, avoided unnecessary waste, and saved scientific research resources.
3.5. Prospects for Future Research
The network pharmacology method used in this study is based on the multilayer network of disease phenotypic gene drugs to predict drug targets in an overall way and promote effective drug discovery. This method was a breakthrough in the traditional Chinese herbal medicine research model “gene-target disease”, and embodied the new model of “multigene-multitarget-complex disease”.100 The results showed that QZD had the effect of multicomponents, multitargets, and multipathways in the treatment of colon cancer. It mainly exerted its antitumor effect from many aspects, such as inhibiting tumor cell differentiation and proliferation, promoting apoptosis and anti-neovascularization, reversing multidrug resistance, and so on. The main targets and pathways of QZD in the treatment of colon cancer were successfully predicted by this method for the first time, which provided a basis for further study of the antitumor effect of QZD. This experimental scheme had important reference value for the research of complex drugs and was worth popularizing and applying in future research.
The future of tumor therapy is combined therapy.101 A single drug cannot achieve the ideal antitumor effect. Combined drug therapy can affect multiple tumor targets, produce a synergistic effect, and reduce the occurrence of drug resistance.102 This study suggests that QZD may be used as an adjuvant for anticolon cancer therapies such as radiotherapy, chemotherapy, targeted drugs, and so on. In future research, to expand the application range of radiotherapy and chemotherapy drugs or targeted drugs, we can observe the anticolon cancer effect of QZD combined with radiotherapy and chemotherapy drugs or targeted drugs. Future experiments will accurately or optimize the best strategy of combined drug therapy, such as drug ratio, drug sequence, optimal application time, safety, toxicity management, and so on. This provides basic data for Qizhen decoction to become an adjuvant drug against multidrug resistance of colon cancer.
4. Conclusions
QZD has at least 111 chemical components, including a variety of effective components that had antiangiogenesis properties, inhibited the proliferation of Colon cancer, improved chemotherapy and radiotherapy, and reversed targeted drug resistance. The network pharmacology analysis showed that the corresponding 955 targets of 111 chemical components and 132 common targets were closely related to colon cancer. Further enrichment analysis showed that the signal pathways such as PI3K Akt, MAPK, AGE-RAGE, and other related signal pathways were overactivated. QZD can inhibit the proliferation and migration of colon cancer cells by inhibiting the PI3K Akt signaling pathway to achieve the biological anticolon cancer effect.
5. Materials and Methods
5.1. Reagents and Instruments
The herbs in Qizhen decoction are Radix Astragali (batch number: 20190408), Codonopsis Pilosula (batch number: 20190405), Ligustrum lucidum (batch number: 20190602), Herba Hedyotis (batch number: 20190511), Gynostemma Pentaphylla (batch number: 20190403), Salviae Miltiorrhizae (batch number: 20190513), Radix Paeoniae Rubra (batch number: 20190411), and Polygonum cuspidatum (batch number: 20190609) (Beijing Tong Ren Tang, China). The mixtures of the above herbs were prepared in the ratio of 15:15:15:20:20:10:10:10 g (Table 1). The mixture was thoroughly decocted in water for 45 min (1:7, w/v), decocted twice with water (1:5, w/v), 30 min for each time, and filtered through a gauze. It was rotated and evaporated (−45 °C, 30 min) to the liquid extract by an FDU-2100 freeze dryer (Tokyo Physicochemical EYELA Company, Japan) and then frozen in a refrigerator at −80 °C. The standard decoction was put to freeze-drying (−85 °C, 5–6 MPa) for 24 h, taken out, and ground to a get dry powder of QZD. Human colon cancer cell line LoVo (ATCC); P-Akt antibody, Akt antibody, LY294002 (Selleck Company); β-actin antibody, goat antirabbit IgG (Abcam, U.K.); MTT (Solarbio, China), dimethyl sulfoxide (DMSO) (Sigma), Roswell Park Memorial Institute (RPMI) 1640 medium, fetal bovine serum (FBS) (Gibco), bicinchoninic acid (BCA) protein quantitative analysis kit (Solarbi, China), a fluorescence inversion microscope (Olympus, Japan), an enzyme labeling instrument (Thermo), an electrophoresis instrument (Bio-Rad), a transmembrane instrument (Bio-Rad), a chemiluminescence gel imaging system (Aplegen), a Waters Acquity UPLC Class I ultrahigh-performance liquid chromatograph, a Waters SYNAPT G2-Si Q-TOF/MS in situ ion exposure high-resolution mass spectrometer, a Waters Acquity UPLC BEH C18 column (2.1 mm × 100 mm, 1.7 μm) were used.
5.2. Chromatographic Conditions
The chromatographic conditions are as follows: chromatographic column, Waters ACQUITY UPLC BEH C18 (2.1 mm × 100 mm, 1.7 μm); velocity of flow, 0.3 mL·min–1; column temperature, 40 °C; injection volume, 5 μL; mobile phase, phase A is 0.1% formic acid aqueous solution and phase B is 0.1% formic acid acetonitrile, using a gradient elution method; the specific elution conditions are shown in Table 5.
Table 5. Chromatographic Elution Conditions of QZD by UPLC–Q-TOF/MS.
| time (T) | A (0.1% formic acid aqueous solution) | B (0.1% acetonitrile formate) |
|---|---|---|
| 0 | 98 | 2 |
| 14 | 40 | 60 |
| 16.5 | 2 | 98 |
| 18 | 2 | 98 |
| 20 | 98 | 2 |
5.3. Mass Spectrometry Conditions
In the mode of electrospray positive and negative ionization, the mass spectrometry analysis was carried out respectively, using high-purity N2 as auxiliary spray ionization and desolvent gas. The dry air velocity was 10 mL·min–1. The temperature of N2 was 120 °C. The pressure of atomizing gas was 310 kPa. The flow rate of N2 was 900 L·h–1. The counterblow nitrogen of the taper hole was 50 L·h–1. The capillary ionization voltage was 500 V. The taper hole voltage was 40 V. The collision energy was 40–65 eV. The scanning range of quadrupole was 50–1500 Da.
5.4. UPLC–Q-TOF/MS Data Processing
Due to the complex chemical composition of QZD, the ESI+ and ESI– ionization modes were used to comprehensively scan its chemical composition to identify as many chemical compositions as possible. The BPI of the base peak chromatogram is shown in Figure 1. After the end of the UPLC–Q-TOF/MS analysis process, the original data were obtained by detecting, calibrating, and collecting the peak value by MassLynx software v4.1. The parameters are as follows: retention time (RT), 0–20; quality, 50–1500 Da; and noise elimination level, 6; then, these are converted to Excel format, including the retention time (RT), m/z value, peak area of each group, etc. The output data were processed to get the target compound. Finally, the target compounds were determined from the database, and the chemical components were identified by secondary fragments and standards.
5.5. Establishment of the Chemical Component Library of QZD
Based on the previous study, 111 chemical components were identified by UPLC–Q-TOF/MS, 26 of which had no conformation in PubChem. Therefore, 85 chemical components were included in the QZD chemical composition database in this study.
5.6. Construction of “Chemical Composition Target”
In this study, TCMSP (http://lsp.nwu.edu.cn/tcmsp.php), Pharmmapper (http://www.lilab-executive.cn/pharmmapper/), Swiss target prediction (http://www.swisstargetprediction.ch/), and BATMAN-TCM (http://bionet.ncpsb.org/batman-tcm/) were integrated to predict the related targets of the above 85 chemicals.
Based on the BATMAN-TCM database, the score off parameter was set to 20, and the p-value after the Benjamin–Hochberg multiple testing correction (adjusted p-value) parameter was set to 0.05 to search for related targets of chemical components. At the same time, the TCMSP database was used to screen out the target of chemical components.
Based on the PubChem (https://pubchem.ncbi.nlm.nih.gov/) database, the three-dimensional (3D) structures of the above 111 chemical components were found and the SDF format was downloaded. (Due to the unknown structure of some compounds, there are 85 kinds of chemical components involved in the research.) These structures were uploaded in Pharmapper, the human protein targets only formed in the source were selected, the “reserved target matching number” was set to 100, and the protein target was collected and organized. After that, the UniProt (https://www.uniprot.org/) database was used to search, and the target was transformed into the corresponding standard gene name, which was sorted out.
In the PubChem database, the Canonical SMILES form of the above 85 chemical components were found and sorted out. Then, the Swiss Target Prediction database was used, the Canonical SMILES of chemical composition were inputted, the property was set to Homo sapiens, and its related target was predicted and organized. Finally, the corresponding targets of 85 chemical components were integrated.
5.7. Acquisition of “Colon Cancer Targets”
With “Colon cancer” as the keyword, the related targets of colon cancer were found in the therapeutic target database, TTD (http://bidd.nus.edu.sg/group/cjttd/) and comparative genomics toxicology database, and CTD (http://ctdbase.org/) database. Among them, the disease target marked “approved” or “marker/mechanism” was further screened. In the gene cards (https://genecards.weizmann.ac.il/v3/) database, “Colon cancer” was used as the keyword to find the relevant target. Protein targets were transformed into standard gene names by the UniProt database. After that, the disease targets were integrated.
5.8. Acquisition of “Common Targets”
The chemical composition-related targets in Section 5.6 and the disease targets in Section 5.7 were uploaded to draw a Venn diagram (http://bioinformatics.psb.ugent.be/webtools/venn/) for analysis, and their common targets were obtained.
5.9. Acquisition of “Indirect Targets”
The common targets obtained in Section 5.8 above were uploaded to Gene MANIA (http://genemania.org/), the source type was set as the human source, and its indirect targets were analyzed.
5.10. Construction and Analysis of the PPI Network
The indirect targets and common targets were integrated using the string platform (https://string-db.org/) to build its PPI and then using the software of Cytoscape for topology analysis. The specific operations are as follows: upload all of the above-mentioned targets to the string platform, set the source as the human source, download its TSV format, import it into Cytoscape software for data analysis, use the function of “Network Analzyer” to carry out topology analysis on PPI network, and take the top three targets as the key targets in the network for molecular docking verification based on the degree. The color and brightness of nodes reflected the size of the degree in the network, and the brightness of the edge reflected the size of the combined score. The interaction network of QZD in the treatment of colon cancer target proteins was constructed.
5.11. Molecular Docking Verification
The PDB ID number of the key target selected in Section 5.10 was imported into the Systems Dock Web Site (http://systemsdock.unit.oist.jp/iddp/home/index) to conduct molecular docking verification with the above 85 chemical components. Based on the docking score, the binding capacity of the compounds to the key direct targets was evaluated. It was generally believed that when the docking score value is above 4.25; this meant that there was a certain binding activity between molecules and targets. If it was greater than 5.0, it meant that there was a good binding activity between molecules and targets. If it was greater than 7.0, it meant that it had an excellent binding activity.
5.12. GO Bioanalysis and KEGG Pathway Analysis
GO is the abbreviation of Gene Oncology. It annotates genes according to biological process, cellular component, and molecular function. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway could directly detect the pathways involved in the target and conduct enrichment analysis of the target with the help of the statistical analysis method. All of the targets integrated in Section 5.10 were uploaded to the string platform for analysis, and the obtained GO and KEGG data were downloaded. The KEGG data were classified into three aspects: signal pathway, metabolism, and disease. The core pathway was screened based on the Omicshare (http://www.omicshare.com/) database, and the results were presented in the form of an advanced bubble chart. The enrichment degree of the core pathway was analyzed according to the enrichment factor value, and the possible mechanism of QZD in the treatment of colon cancer was explored. The number of enriched targets was represented by the size of nodes, and the size of the p-value was represented by the color of nodes. The integrated target of Section 5.10 was uploaded to Reactome (https://realtome.org/), the forms of “Gene name list” and “Project to human” were set for analysis, and the biological process related to the treatment of colon cancer by QZD was obtained, which was presented in the form of a tailless fireworks map.
5.13. Network Construction and Analysis of “Prescription–TCM–Chemical Composition–Direct Target–Indirect Target–Pathway” in the Treatment of Colon Cancer with QZD
The prescription name, TCM, chemical composition, direct target, indirect target, and pathway of QZD were divided into “Prescription–TCM”, “TCM–Chemical composition”, “Chemical composition–Direct target”, “Direct target–Indirect target,” and “Target–Pathway” data, which were imported into Cytoscape software, and based on the merge function integration, the network of “Prescription–TCM–Chemical composition–Direct target–Indirect target–Pathway” was constructed.
5.14. Cell Culture
LoVo cells were cultured in RPMI 1640 containing 10% FBS, 100 IU/mL penicillin, and 100 IU/mL streptomycin. The cells were maintained in a 5% CO2 atmosphere at 37 °C.
5.15. Western Blotting
LoVo cells (2 × 106) were treated with LY294002 or different concentrations of QZD in a six-well culture plate. After 24 h, we harvested the cells on ice and cleaved them in radioimmunoprecipitation assay (RIPA) lysate with 1 mM phenylmethylsulfonyl fluoride (PMSF) and phosphatase inhibitor for 30 min. Then, the samples were centrifuged at 12 000 rpm for 20 min at 4 °C. After preparing the protein samples, we took the supernatant of each group for protein quantification. Each group of samples was mixed with the loading buffer and boiled for 10 min. After electrophoretic and membrane transfer, the poly(vinylidene difluoride) (PVDF) membrane containing the target protein was washed with TBST solution and incubated with 5% nonfat milk solution and primary antibodies: Akt, P-Akt, and β-actin (1:1000 dilution). The suspensions were gently shaken overnight at 4 °C. Then, we add horseradish peroxidase (HRP)-conjugated polymer-tagged secondary antibodies (1:10 000 dilution) and incubated them for 2 h on a shaker at room temperature. At last, the protein bands were captured using a Western blotting detection system. The gray value of the strip was calculated by ImageJ software.
5.16. MTT Cell Viability Assay
The antiproliferation effects of QZD against tumor cells were evaluated by the MTT cell viability assay. Briefly, the cells were cultured in 96-well plates (5 × 103 cells/well) for 12 h and then incubated with LY294002 or various concentrations of QZD (75, 37.5, and 18.75 μg/mL). The control group was added with the same volume of RPMI 1640 medium. After 24 h, 20 μL of MTT was added to each well and then cultured at 37 °C in a CO2 incubator for 4 h. The medium was abandoned and 150 μL of DMSO was added. The optical density was read at 490 nm using a microplate reader. Cells were counted, and cell viability was analyzed.
5.17. Cell Wound Scratch Assay
The wound-healing assay was performed to estimate the migration ability of different LoVo cell models. Cells were seeded and grown to 90% confluence in six-well culture plates. The wounds on the monolayer, which were created using 20 μL tips, were washed with phosphate-buffered saline (PBS) three times. A microscope was applied to image the scratch wound at 0 h and after 48 h under 40× magnification. The migration distance was measured by ImageJ software, and the healing rate was calculated. LoVo cell healing rate (%) = (T0h distance – T48h distance)/T0h distance × 100%.
5.18. Statistical Analysis
Statistical analysis was performed and presented with GraphPad Prism 8.0 software. Differences between two independent groups were evaluated by Student’s t-tests. Differences for multiple comparisons were evaluated using one-way analysis of variance (ANOVA). p < 0.05 was considered significant difference. Data were presented as mean ± standard deviation.
Acknowledgments
The study was supported by the National Natural Science Foundation of China (81973728) and the Natural Science Foundation of Tianjin (18JCZDJC36600). The authors are very grateful to Jingyan Meng for providing us with financial support. The authors also thank the editors and review experts of the ACS OMEGA for their guidance and advice in this manuscript. All of the listed authors have read and approved the submitted manuscript.
Author Contributions
X.K., C.L., and P.L. contributed equally to this work and were co-first authors. X.K.: conceptualization, writing-original draft, writing-review, and editing. C.L.: conceptualization. P.L.: conceptualization. Y.G.: writing-review and editing. C.Z.: writing-review and editing. Y.Y.: data curation. Y.P.: data curation. Z.B.: software, investigation. F.W.: formal analysis. J.M.: funding acquisition, project administration, validation. All authors have read and agreed to the published version of the manuscript.
The authors declare no competing financial interest.
References
- Buccafusca G.; Proserpio I.; Tralongo A. C.; Rametta Giuliano S.; Tralongo P. Early colorectal cancer: diagnosis, treatment and survivorship care. Crit. Rev. Oncol./Hematol. 2019, 136, 20–30. 10.1016/j.critrevonc.2019.01.023. [DOI] [PubMed] [Google Scholar]
- Arnold M.; Sierra M. S.; Laversanne M.; Soerjomataram I.; Jemal A.; Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017, 66, 683–691. 10.1136/gutjnl-2015-310912. [DOI] [PubMed] [Google Scholar]
- Geng F.; Wang Z.; Yin H.; Yu J.; Cao B. Molecular Targeted Drugs and Treatment of Colorectal Cancer: Recent Progress and Future Perspectives. Cancer Biother. Radiopharm. 2017, 32, 149–160. 10.1089/cbr.2017.2210. [DOI] [PubMed] [Google Scholar]
- Zhao H. D.; Xie H. J.; Li J.; Ren C. P.; Chen Y. X. Research Progress on Reversing Multidrug Resistance in Tumors by Using Chinese Medicine. Chin. J. Integr. Med. 2018, 24, 474–480. 10.1007/s11655-018-2910-1. [DOI] [PubMed] [Google Scholar]
- Deng S.; Wang A.; Chen X.; Du Q.; Wu Y.; Chen G.; Guo W.; Li Y. HBD Inhibits the Development of Colitis-Associated Cancer in Mice via the IL-6R/STAT3 Signaling Pathway. Int. J. Mol. Sci. 2019, 20, 1069 10.3390/ijms20051069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J.; Cheng B. C.; Fu X. Q.; Su T.; Li T.; Guo H.; Li S. M.; Wu J. F.; Yu H.; Huang W. H.; Cao H.; Yu Z. L. In vitro assays suggest Shenqi Fuzheng Injection has the potential to alter melanoma immune microenvironment. J. Ethnopharmacol. 2016, 194, 15–19. 10.1016/j.jep.2016.08.038. [DOI] [PubMed] [Google Scholar]
- Zuo H. J.; Liu S.; Yan C.; Li L. M.; Pei X. F. In Vitro and In Vivo Evaluation of Antitumor Activity of Ligustrum robustum, A Chinese Herbal Tea. Chin. J. Integr. Med. 2019, 25, 425–430. 10.1007/s11655-018-2983-5. [DOI] [PubMed] [Google Scholar]
- Kuo Y. T.; Liao H. H.; Chiang J. H.; Wu M. Y.; Chen B. C.; Chang C. M.; Yeh M. H.; Chang T. T.; Sun M. F.; Yeh C. C.; Yen H. R. Complementary Chinese Herbal Medicine Therapy Improves Survival of Patients With Pancreatic Cancer in Taiwan: A Nationwide Population-Based Cohort Study. Integr. Cancer Ther. 2018, 17, 411–422. 10.1177/1534735417722224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. W.; Yang L.; An L.; Li P.; Chen J. Discovery of cancer cell proliferation inhibitors from Salviae miltiorrhizae radix et rhizoma by a trace peak enrichment approach. J. Sep. Sci. 2019, 42, 534–546. 10.1002/jssc.201800895. [DOI] [PubMed] [Google Scholar]
- Lin M. Y.; Chiang S. Y.; Li Y. Z.; Chen M. F.; Chen Y. S.; Wu J. Y.; Liu Y. W. Anti-tumor effect of Radix Paeoniae Rubra extract on mice bladder tumors using intravesical therapy. Oncol. Lett. 2016, 12, 904–910. 10.3892/ol.2016.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law B. Y.; Mok S. W.; Wu A. G.; Lam C. W.; Yu M. X.; Wong V. K. New Potential Pharmacological Functions of Chinese Herbal Medicines via Regulation of Autophagy. Molecules 2016, 21, 359 10.3390/molecules21030359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. H.; Zhu R. J.; Hu F.; Guo L.; Yang Y. L.; Feng S. L. Tissue distribution of six major bio-active components after oral administration of Zhenqi Fuzheng capsules to rats using ultra-pressure liquid chromatography-tandem mass spectrometry. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2015, 986–987, 44–53. 10.1016/j.jchromb.2015.01.033. [DOI] [PubMed] [Google Scholar]
- Zhang Z.; Tang Y.; Yu B.; Ying J.; Wu B.; Wu J.; Zhao J.; Chen Z.; Xu J.; Tang C. Chemical composition database establishment and metabolite profiling analysis of Yangyin qingfei decoction. Biomed. Chromatogr. 2019, 33, e4581 10.1002/bmc.4581. [DOI] [PubMed] [Google Scholar]
- Lyu M.; Zhou Z.; Wang X.; Lv H.; Wang M.; Pan G.; Wang Y.; Fan G.; Gao X.; Feng Y.; Zhu Y. Network Pharmacology-Guided Development of a Novel Integrative Regimen to Prevent Acute Graft-vs.-Host Disease. Front. Pharmacol. 2018, 9, 1440 10.3389/fphar.2018.01440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song N.; Kim K.; Shin A.; Park J. W.; Chang H. J.; Shi J.; Cai Q.; Kim D. Y.; Zheng W.; Oh J. H. Colorectal cancer susceptibility loci and influence on survival. Genes, Chromosomes Cancer 2018, 57, 630–637. 10.1002/gcc.22674. [DOI] [PubMed] [Google Scholar]
- Bultman S. J. Interplay between diet, gut microbiota, epigenetic events, and colorectal cancer. Mol. Nutr. Food. Res. 2017, 61, 1500902 10.1002/mnfr.201500902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S.; Qian L.; Niu M.; Liu H.; Yang Y.; Wang Y.; Zhang L.; Zhou X.; Li H.; Wang R.; Li K.; Zhao Y. The Modulatory Properties of Li-Ru-Kang Treatment on Hyperplasia of Mammary Glands Using an Integrated Approach. Front. Pharmacol. 2018, 9, 651 10.3389/fphar.2018.00651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M.; Li P.; Zeng X.; Wu H.; Su W.; He J. Identification and pharmacokinetics of multiple potential bioactive constituents after oral administration of radix astragali on cyclophosphamide-induced immunosuppression in Balb/c mice. Int. J. Mol. Sci. 2015, 16, 5047–5071. 10.3390/ijms16035047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X. Q.; Leung A. K.; Chan C. L.; Su T.; Li W. D.; Li S. M.; Fong D. W.; Yu Z. L. UHPLC UHD Q-TOF MS/MS analysis of the impact of sulfur fumigation on the chemical profile of Codonopsis Radix (Dangshen). Analyst 2014, 139, 505–516. 10.1039/c3an01561k. [DOI] [PubMed] [Google Scholar]
- Liu M. H.; Tong X.; Wang J. X.; Zou W.; Cao H.; Su W. W. Rapid separation and identification of multiple constituents in traditional Chinese medicine formula Shenqi Fuzheng Injection by ultra-fast liquid chromatography combined with quadrupole-time-of-flight mass spectrometry. J. Pharm. Biomed. Anal. 2013, 74, 141–155. 10.1016/j.jpba.2012.10.024. [DOI] [PubMed] [Google Scholar]
- Li Y.; Zhou G.; Xing S.; Tu P.; Li X. Identification of Echinacoside Metabolites Produced by Human Intestinal Bacteria Using Ultraperformance Liquid Chromatography-Quadrupole Time-of-Flight Mass Spectrometry. J. Agric. Food. Chem. 2015, 63, 6764–6771. 10.1021/acs.jafc.5b02881. [DOI] [PubMed] [Google Scholar]
- Xie W.; Zhang H.; Zeng J.; Chen H.; Zhao Z.; Liang Z. Tissues-based chemical profiling and semi-quantitative analysis of bioactive components in the root of Salvia miltiorrhiza Bunge by using laser microdissection system combined with UPLC-q-TOF-MS. Chem. Cent. J. 2016, 10, 42 10.1186/s13065-016-0187-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S. Q.; Song H. P.; Li B.; Liu R. Z.; Yang H.; He L.; Li P. A fast and accurate method for the identification of peroxidase inhibitors from Radix Salvia miltiorrhizae by on-flow biochemical assay coupled with LC/Q-TOF-MS: comparison with ultrafiltration-based affinity selection. Anal. Bioanal. Chem. 2018, 410, 4311–4322. 10.1007/s00216-018-1081-z. [DOI] [PubMed] [Google Scholar]
- Miao J.; Sun W.; Huang J.; Liu X.; Li S.; Han X.; Tong L.; Sun G. Characterization of metabolites in rats after intravenous administration of salvianolic acid for injection by ultra-performance liquid chromatography coupled with quadrupole-time-of-flight mass spectrometry. Biomed. Chromatogr. 2016, 30, 1487–1497. 10.1002/bmc.3710. [DOI] [PubMed] [Google Scholar]
- Gruz J.; Novak O.; Strnad M. Rapid analysis of phenolic acids in beverages by UPLC-MS/MS. Food Chem. 2008, 111, 789–794. 10.1016/j.foodchem.2008.05.014. [DOI] [Google Scholar]
- Duan Y.; Pei K.; Cai H.; Tu S.; Zhang Z.; Cheng X.; Qiao F.; Fan K.; Qin K.; Liu X.; Cai B. Bioactivity evaluation-based ultra high-performance liquid chromatography coupled with electrospray ionization tandem quadrupole-time-of-flight mass spectrometry and novel distinction of multi-subchemome compatibility recognition strategy with Astragali Radix-Fructus Corni herb-pair as a case study. J. Pharm. Biomed. Anal. 2016, 129, 514–534. 10.1016/j.jpba.2016.07.043. [DOI] [PubMed] [Google Scholar]
- Qi L. W.; Li P.; Ren M. T.; Yu Q. T.; Wen X. D.; Wang Y. X. Application of high-performance liquid chromatography-electrospray ionization time-of-flight mass spectrometry for analysis and quality control of Radix Astragali and its preparations. J. Chromatogr. A 2009, 1216, 2087–2097. 10.1016/j.chroma.2008.02.095. [DOI] [PubMed] [Google Scholar]
- Dong J.; Zhu Y.; Gao X.; Chang Y.; Wang M.; Zhang P. Qualitative and quantitative analysis of the major constituents in Chinese medicinal preparation Dan-Lou tablet by ultra high performance liquid chromatography/diode-array detector/quadrupole time-of-flight tandem mass spectrometry. J. Pharm. Biomed. Anal. 2013, 80, 50–62. 10.1016/j.jpba.2013.02.011. [DOI] [PubMed] [Google Scholar]
- Wang X.; Liu X.; Xu X.; Zhu T.; Shi F.; Qin K.; Cai B. Screening and identification of multiple constituents and their metabolites of Fangji Huangqi Tang in rats by ultra-high performance liquid chromatography coupled with quadrupole time-of-flight tandem mass spectrometry basing on coupling data processing techniques. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2015, 985, 14–28. 10.1016/j.jchromb.2015.01.021. [DOI] [PubMed] [Google Scholar]
- Wang D.; Song Y.; Li S. L.; Bian Y. Y.; Guan J.; Li P. Simultaneous analysis of seven astragalosides in Radix Astragali and related preparations by liquid chromatography coupled with electrospray ionization time-of-flight mass spectrometry. J. Sep. Sci. 2006, 29, 2012–2022. 10.1002/jssc.200500486. [DOI] [PubMed] [Google Scholar]
- Pérez-Bonilla M.; Salido S.; van Beek T. A.; Altarejos J. Radical-scavenging compounds from olive tree (Olea europaea L.) wood. J. Agric. Food. Chem. 2014, 62, 144–151. 10.1021/jf403998t. [DOI] [PubMed] [Google Scholar]
- Chen Q.; Yang L.; Zhang G.; Wang F. Bioactivity-guided Isolation of antiosteoporotic compounds from Ligustrum lucidum. Phytother. Res. 2013, 27, 973–979. 10.1002/ptr.4820. [DOI] [PubMed] [Google Scholar]
- Guo N.; Yu Y.; Ablajan K.; Li L.; Fan B.; Peng J.; Yan H.; Ma F.; Nie Y. Seasonal variations in metabolite profiling of the fruits of Ligustrum lucidum Ait. Rapid Commun. Mass Spectrom. 2011, 25, 1701–1714. 10.1002/rcm.5036. [DOI] [PubMed] [Google Scholar]
- Shi L.; Ma Y.; Cai Z. Quantitative determination of salidroside and specnuezhenide in the fruits of Ligustrum lucidum Ait by high performance liquid chromatography. Biomed. Chromatogr. 1998, 12, 27–30. . [DOI] [PubMed] [Google Scholar]
- Di Donna L.; Mazzotti F.; Napoli A.; Salerno R.; Sajjad A.; Sindona G. Secondary metabolism of olive secoiridoids. New microcomponents detected in drupes by electrospray ionization and high-resolution tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2007, 21, 273–278. 10.1002/rcm.2830. [DOI] [PubMed] [Google Scholar]
- He Z. D.; Dong H.; Xu H. X.; Ye W. C.; Sun H. D.; But P. P. Secoiridoid constituents from the fruits of Ligustrum lucidum. Phytochemistry 2001, 56, 327–330. 10.1016/s0031-9422(00)00406-4. [DOI] [PubMed] [Google Scholar]
- Jensen S. R.; Franzyk H.; Wallander E. Chemotaxonomy of the Oleaceae: iridoids as taxonomic markers. Phytochemistry 2002, 60, 213–231. 10.1016/s0031-9422(02)00102-4. [DOI] [PubMed] [Google Scholar]
- Inoue K.; Nishioka T.; Tanahashi T.; Inouye H. Three secoiridoid glucosides from Ligustrum japonicum. Phytochemistry 1982, 21, 2305–2311. 10.1016/0031-9422(82)85196-0. [DOI] [Google Scholar]
- Liu H.; Shi Y.; Wang D.; Yang G.; Yu A.; Zhang H. MECC determination of oleanolic acid and ursolic acid isomers in Ligustrum lucidum Ait. J. Pharm. Biomed. Anal. 2003, 32, 479–485. 10.1016/s0731-7085(03)00235-8. [DOI] [PubMed] [Google Scholar]
- Xia L.; Liu H. L.; Li P.; Zhou J. L.; Qi L. W.; Yi L.; Chen J. Rapid and sensitive analysis of multiple bioactive constituents in Compound Danshen preparations using LC-ESI-TOF-MS. J. Sep. Sci. 2008, 31, 3156–3169. 10.1002/jssc.200800327. [DOI] [PubMed] [Google Scholar]
- Yang Z.; Chi S. Y.; Zhang C. H.; Wu A. Quantitative analysis of adenosine and cordycepin in Cordyceps sinensis and its substitutes with LC-MS-MS. Zhongguo Zhongyao Zazhi 2007, 32, 2018–2021. [PubMed] [Google Scholar]
- Ishida S.; Okasaka M.; Ramos F.; Kashiwada Y.; Takaishi Y.; Kodzhimatov O. K.; Ashurmetov O. New alkaloid from the aerial parts of Codonopsis clematidea. J. Nat. Med. 2008, 62, 236–238. 10.1007/s11418-007-0219-7. [DOI] [PubMed] [Google Scholar]
- Xu F. G.; Liu Y.; Song R.; Dong H. J.; Zhang Z. J. Correlation of material base between Da-Cheng-Qi decoction and its principal drug rhubarb by LC-MS/MS. J. China Pharm. Univ. 2008, 39, 136–141. [Google Scholar]
- Tsai T. H.; Lin L. C. Phenolic glycosides and pyrrolidine alkaloids from Codonopsis tangshen. Chem. Pharm. Bull. 2008, 56, 1546–1550. 10.1248/cpb.56.1546. [DOI] [PubMed] [Google Scholar]
- Yang Z.; Cai Q.; Chen N.; Zhou X.; Hong J. Selective separation and identification of metabolite groups of Polygonum cuspidatum extract in rat plasma using dispersion solid-phase extraction by magnetic molecularly imprinted polymers coupled with LC/Q-TOF-MS. RSC Adv. 2016, 6, 12193–12204. 10.1039/C5RA26695E. [DOI] [Google Scholar]
- Dong J.; Wang H.; Wan L.; Hashi Y.; Chen S. [Identification and determination of major constituents in Polygonum cuspidatum Sieb. et Zucc. by high performance liquid chromatography/electrospray ionization-ion trap-time-of-flight mass spectrometry]. Se Pu 2009, 27, 425–430. [PubMed] [Google Scholar]
- Jin W.; Wang Y. F.; Ge R. L.; Shi H. M.; Jia C. Q.; Tu P. F. Simultaneous analysis of multiple bioactive constituents in Rheum tanguticum Maxim. ex Balf. by high-performance liquid chromatography coupled to tandem mass spectrometry. Rapid. Commun. Mass. Spectrom. 2007, 21, 2351–2360. 10.1002/rcm.3086. [DOI] [PubMed] [Google Scholar]
- Jin W.; Ge R. L.; Wei Q. J.; Bao T. Y.; Shi H. M.; Tu P. F. Development of high-performance liquid chromatographic fingerprint for the quality control of Rheum tanguticum Maxim. ex Balf. J. Chromatogr. A 2006, 1132, 320–324. 10.1016/j.chroma.2006.08.022. [DOI] [PubMed] [Google Scholar]
- Takemoto T.; Arihara S.; Nakajima T.; Okuhira M. Studies on the constituents of Gynostemma pentaphyllum Makino. I. Structures of gypenoide I-XIV. Yakugaku Zasshi 1983, 103, 173–185. 10.1248/yakushi1947.103.2_173. [DOI] [Google Scholar]
- Yoshikawa K.; Takemoto T.; Arihara S. Studies on the constituents of Cucurbitaceae plants.X V. Yakugaku Zasshi 1986, 106, 758–763. 10.1248/yakushi1947.106.9_758. [DOI] [PubMed] [Google Scholar]
- Tsai Y. C.; Lin C. L.; Chen B. H. Preparative chromatography of flavonoids and saponins in Gynostemma pentaphyllum and their antiproliferation effect on hepatoma cell. Phytomedicine 2010, 18, 2–10. 10.1016/j.phymed.2010.09.004. [DOI] [PubMed] [Google Scholar]
- Yoshikawa K.; Mitake M.; Takemoto T.; Arihara S. Studies on the constituents of Cucurbitaceae plants. 17. On the saponin constituents of Gynostemma pentaphyllum Makino. 12. Yakugaku Zassh 1987, 107, 355–360. 10.1248/yakushi1947.107.5_355. [DOI] [Google Scholar]
- Liu X.; Ye W.; Mo Z.; Yu B.; Zhao S.; Wu H.; Che C.; Jiang R.; Mak T. C.; Hsiao W. L. Five new Ocotillone-type saponins from Gynostemma pentaphyllum. J. Nat. Prod. 2004, 67, 1147–1151. 10.1021/np034018+. [DOI] [PubMed] [Google Scholar]
- Liang J.; Xu F.; Zhang Y. Z.; Huang S.; Zang X. Y.; Zhao X.; Zhang L.; Shang M. Y.; Yang D. H.; Wang X.; Cai S. Q. The profiling and identification of the absorbed constituents and metabolites of Paeoniae Radix Rubra decoction in rat plasma and urine by the HPLC-DAD-ESI-IT-TOF-MS(n) technique: a novel strategy for the systematic screening and identification of absorbed constituents and metabolites from traditional Chinese medicines. J. Pharm. Biomed. Anal. 2013, 83, 108–121. 10.1016/j.jpba.2013.04.029. [DOI] [PubMed] [Google Scholar]
- Janney A.; Powrie F.; Mann E. H. Host-microbiota maladaptation in colorectal cancer. Nature 2020, 585, 509–517. 10.1038/s41586-020-2729-3. [DOI] [PubMed] [Google Scholar]
- Deng S.; Hu B.; An H. M. Traditional Chinese Medicinal Syndromes and Treatment in Colorectal Cancer. J. Cancer Ther. 2012, 03, 888–897. 10.4236/jct.2012.326114. [DOI] [Google Scholar]
- Xie T.; Li Y.; Li S. L.; Luo H. F. Astragaloside IV Enhances Cisplatin Chemosensitivity in Human Colorectal Cancer via Regulating NOTCH3. Oncol. Res. 2016, 24, 447–453. 10.3727/096504016x14685034103590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. Y.; Hsieh S. L.; Hsieh S.; Tsai C. C.; Hsieh L. C.; Kuo Y. H.; Wu C. C. Inhibition of human colorectal cancer metastasis by notoginsenoside R1, an important compound from Panax notoginseng. Oncol. Rep. 2017, 37, 399–407. 10.3892/or.2016.5222. [DOI] [PubMed] [Google Scholar]
- Wang C. Z.; Xie J. T.; Zhang B.; Ni M.; Fishbein A.; Aung H. H.; Mehendale S. R.; Du W.; He T. C.; Yuan C. S. Chemopreventive effects of Panax notoginseng and its major constituents on SW480 human colorectal cancer cells. Int. J. Oncol. 2007, 31, 1149–1156. 10.3892/ijo.31.5.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honari M.; Shafabakhsh R.; Reiter R. J.; Mirzaei H.; Asemi Z. Resveratrol is a promising agent for colorectal cancer prevention and treatment: focus on molecular mechanisms. Cancer Cell. Int. 2019, 19, 180 10.1186/s12935-019-0906-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. L.; Wang C. Z.; Mehendale S. R.; Sun S.; Wang Q.; Yuan C. S. Panaxadiol, a purified ginseng component, enhances the anti-cancer effects of 5-fluorouracil in human colorectal cancer cells. Cancer Chemother. Pharmacol. 2009, 64, 1097–1104. 10.1007/s00280-009-0966-0. [DOI] [PubMed] [Google Scholar]
- Fan X. J.; Wang Y.; Wang L.; Zhu M. Salidroside induces apoptosis and autophagy in human colorectal cancer cells through inhibition of PI3K/Akt/mTOR pathway. Oncol. Rep. 2016, 36, 3559–3567. 10.3892/or.2016.5138. [DOI] [PubMed] [Google Scholar]
- Li H.; Chen C. Inhibition of autophagy enhances synergistic effects of Salidroside and anti-tumor agents against colorectal cancer. BMC Complementary Altern. Med. 2017, 17, 538 10.1186/s12906-017-2046-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P.; Wang S.; Liang W.; Wang W.; Wang H.; Zhao M.; Liu X. Salvianolic acid B reverses multidrug resistance in HCT8/VCR human colorectal cancer cells by increasing ROS levels. Mol. Med. Rep. 2017, 15, 724–730. 10.3892/mmr.2016.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.; Chen C.; Duanmu J.; Wu Y.; Tao J.; Yang A.; Yin X.; Xiong B.; Gu J.; Li C.; Liu Z. Cryptotanshinone inhibits the growth and invasion of colon cancer by suppressing inflammation and tumor angiogenesis through modulating MMP/TIMP system, PI3K/Akt/mTOR signaling and HIF-1alpha nuclear translocation. Int. Immunopharmacol. 2018, 65, 429–437. 10.1016/j.intimp.2018.10.035. [DOI] [PubMed] [Google Scholar]
- Xu Z.; Jiang H.; Zhu Y.; Wang H.; Jiang J.; Chen L.; Xu W.; Hu T.; Cho C. H. Cryptotanshinone induces ROS-dependent autophagy in multidrug-resistant colon cancer cells. Chem. Biol. Interact. 2017, 273, 48–55. 10.1016/j.cbi.2017.06.003. [DOI] [PubMed] [Google Scholar]
- Chen Q.; Zeng Y. N.; Zhang K.; Zhao Y.; Wu Y. Y.; Li G.; Cheng H. Y.; Zhang M.; Lai F.; Wang J. B.; Cui F. M. Polydatin Increases Radiosensitivity by Inducing Apoptosis of Stem Cells in Colorectal Cancer. Int. J. Biol. Sci. 2019, 15, 430–440. 10.7150/ijbs.27050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T.; Duo L.; Duan P. Ginsenoside Rg3 Sensitizes Colorectal Cancer to Radiotherapy through Downregulation of Proliferative and Angiogenic Biomarkers. Evidence-Based Complementary Altern. Med. 2018, 2018, 1580427 10.1155/2018/1580427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y.; Bai Y.; Hu Y.; Guo Y.; Xu L.; Hu W.; Yang L.; Zhao C.; Li X.; Zhao H. Rhein sensitizes human colorectal cancer cells to EGFR inhibitors by inhibiting STAT3 pathway. OncoTargets Ther. 2019, 12, 5281–5291. 10.2147/OTT.S206833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramme F.; Crosignani S.; Frederix K.; Hoffmann D.; Pilotte L.; Stroobant V.; Preillon J.; Driessens G.; Van den Eynde B. J. Inhibition of Tryptophan-Dioxygenase Activity Increases the Antitumor Efficacy of Immune Checkpoint Inhibitors. Cancer Immunol. Res. 2020, 8, 32–45. 10.1158/2326-6066.CIR-19-0041. [DOI] [PubMed] [Google Scholar]
- Walczak K.; Langner E.; Szalast K.; Makuch-Kocka A.; Pożarowski P.; Plech T. A Tryptophan Metabolite, 8-Hydroxyquinaldic Acid, Exerts Antiproliferative and Anti-Migratory Effects on Colorectal Cancer Cells. Molecules 2020, 25, 1655 10.3390/molecules25071655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y.-H.; Kee J.-Y.; Hong S.-H. Rosmarinic Acid Activates AMPK to Inhibit Metastasis of Colorectal Cancer. Front. Pharmacol. 2018, 9, 68 10.3389/fphar.2018.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang E.-P. I.; Tsai S. Y.; Kuo Y. H.; Pai M. H.; Chiu H. L.; Rodriguez R. L.; Tang F. Y. Caffeic acid derivatives inhibit the growth of colon cancer: involvement of the PI3-K/Akt and AMPK signaling pathways. PLoS One 2014, 9, e99631 10.1371/journal.pone.0099631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agudelo C. D.; Luzardo-Ocampo I.; Campos-Vega R.; Loarca-Pina G.; Maldonado-Celis M. E. Bioaccessibility during In Vitro Digestion and Antiproliferative Effect of Bioactive Compounds from Andean Berry (Vaccinium meridionale Swartz) Juice. J. Agric. Food. Chem. 2018, 66, 7358–7366. 10.1021/acs.jafc.8b01604. [DOI] [PubMed] [Google Scholar]
- Sharma S. H.; Rajamanickam V.; Nagarajan S. Antiproliferative effect of p-Coumaric acid targets UPR activation by downregulating Grp78 in colon cancer. Chem. Biol. Interact. 2018, 291, 16–28. 10.1016/j.cbi.2018.06.001. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Huang L.; Shi H.; Chen H.; Tao J.; Shen R.; Wang T. Ursolic acid enhances the therapeutic effects of oxaliplatin in colorectal cancer by inhibition of drug resistance. Cancer Sci. 2018, 109, 94–102. 10.1111/cas.13425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa L. S.; Jordao N. A.; da Costa Pereira Soares N.; deMesquita J. F.; Monteiro M.; Teodoro A. J. Pharmacokinetic, Antiproliferative and Apoptotic Effects of Phenolic Acids in Human Colon Adenocarcinoma Cells Using In Vitro and In Silico Approaches. Molecules 2018, 23, 2569 10.3390/molecules23102569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M. S.; Fang Y. J.; Pan Z. Z.; Zhong X.; Zheng M. C.; Chen Y. M.; Zhang C. X. Choline and betaine intake and colorectal cancer risk in Chinese population: a case-control study. PLoS One 2015, 10, e0118661 10.1371/journal.pone.0118661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X.; Li X.; Ren Q.; Tian J.; Chen J. Calycosin induces apoptosis in colorectal cancer cells, through modulating the ERbeta/MiR-95 and IGF-1R, PI3K/Akt signaling pathways. Gene 2016, 591, 123–128. 10.1016/j.gene.2016.07.012. [DOI] [PubMed] [Google Scholar]
- Ong S. K. L.; Shanmugam M. K.; Fan L.; Fraser S. E.; Arfuso F.; Ahn K. S.; Sethi G.; Bishayee A. Focus on Formononetin: Anticancer Potential and Molecular Targets. Cancers 2019, 11, 611 10.3390/cancers11050611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auyeung K. K.; Law P. C.; Ko J. K. Novel anti-angiogenic effects of formononetin in human colon cancer cells and tumor xenograft. Oncol. Rep. 2012, 28, 2188–2194. 10.3892/or.2012.2056. [DOI] [PubMed] [Google Scholar]
- Xue J.; Jin X.; Wan X.; Yin X.; Fang M.; Liu T.; Zhao S. Effects and Mechanism of Tanshinone II A in Proliferation, Apoptosis, and Migration of Human Colon Cancer Cells. Med. Sci. Monit. 2019, 25, 4793–4800. 10.12659/MSM.914446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L.; Wei L.; Shen A.; Chu J.; Lin J.; Peng J. Oleanolic acid modulates multiple intracellular targets to inhibit colorectal cancer growth. Int. J. Oncol. 2015, 47, 2247–2254. 10.3892/ijo.2015.3198. [DOI] [PubMed] [Google Scholar]
- Zhong S.; Li Y. G.; Ji D. F.; Lin T. B.; Lv Z. Q. Protocatechualdehyde Induces S-Phase Arrest and Apoptosis by Stimulating the p27(KIP1)-Cyclin A/D1-CDK2 and Mitochondrial Apoptotic Pathways in HT-29 Cells. Molecules 2016, 21, 934 10.3390/molecules21070934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.; Hu T.; Shen J.; Zhang L.; Chan R. L.; Lu L.; Li M.; Cho C. H.; Wu W. K. Dihydrotanshinone I induced apoptosis and autophagy through caspase dependent pathway in colon cancer. Phytomedicine 2015, 22, 1079–1087. 10.1016/j.phymed.2015.08.009. [DOI] [PubMed] [Google Scholar]
- Cheng C.; Dong W. Aloe-Emodin Induces Endoplasmic Reticulum Stress-Dependent Apoptosis in Colorectal Cancer Cells. Med. Sci. Monit. 2018, 24, 6331–6339. 10.12659/MSM.908400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou N.; Liu N.; Han J.; Yan Y.; Li J. Chlorogenic acid induces reactive oxygen species generation and inhibits the viability of human colon cancer cells. Anticancer Drugs 2017, 28, 59–65. 10.1097/CAD.0000000000000430. [DOI] [PubMed] [Google Scholar]
- Deng S.; Wong C. K. C.; Lai H. C.; Wong A. S. T. Ginsenoside-Rb1 targets chemotherapy-resistant ovarian cancer stem cells via simultaneous inhibition of Wnt/β-catenin signaling and epithelial-to-mesenchymal transition. Oncotarget 2017, 8, 25897–25914. 10.18632/oncotarget.13071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng M.; Xue Y. J.; Xu L. R.; Wang Q. W.; Wei J.; Ke X. Q.; Wang J. C.; Chen X. D. Chrysophanol Suppresses Hypoxia-Induced Epithelial-Mesenchymal Transition in Colorectal Cancer Cells. Anat. Rec. 2019, 302, 1561–1570. 10.1002/ar.24081. [DOI] [PubMed] [Google Scholar]
- Han Y. T.; Chen X. H.; Gao H.; Ye J. L.; Wang C. B. Physcion inhibits the metastatic potential of human colorectal cancer SW620 cells in vitro by suppressing the transcription factor SOX2. Acta Pharmacol. Sin. 2016, 37, 264–275. 10.1038/aps.2015.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. W.; Li L. X.; Wu W. Z.; Pan T. J.; Yang Z. S.; Yang Y. K. Anti-Tumor Effects of Paeoniflorin on Epithelial-To-Mesenchymal Transition in Human Colorectal Cancer Cells. Med. Sci. Monit. 2018, 24, 6405–6413. 10.12659/MSM.912227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh K.; Yachida S.; Sugimoto M.; Oshima M.; Nakagawa T.; Akamoto S.; Tabata S.; Saitoh K.; Kato K.; Sato S.; Igarashi K.; Aizawa Y.; Kajino-Sakamoto R.; Kojima Y.; Fujishita T.; Enomoto A.; Hirayama A.; Ishikawa T.; Taketo M. M.; Kushida Y.; Haba R.; Okano K.; Tomita M.; Suzuki Y.; Fukuda S.; Aoki M.; Soga T. Global metabolic reprogramming of colorectal cancer occurs at adenoma stage and is induced by MYC. Proc. Natl. Acad. Sci. U.S.A. 2017, 114, E7697–E7706. 10.1073/pnas.1710366114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo J. S.; Park W. C.; Choi S. C.; Yun K. J.; Chae S. C. MicroRNA 452 Regulates Cell Proliferation, Cell Migration, and Angiogenesis in Colorectal Cancer by Suppressing VEGFA Expression. Cancers 2019, 11, 1613 10.3390/cancers11101613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apte R. S.; Chen D. S.; Ferrara N. VEGF in Signaling and Disease: Beyond Discovery and Development. Cell 2019, 176, 1248–1264. 10.1016/j.cell.2019.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beebe J. D.; Liu J. Y.; Zhang J. T. Two decades of research in discovery of anticancer drugs targeting STAT3, how close are we?. Pharmacol. Ther. 2018, 191, 74–91. 10.1016/j.pharmthera.2018.06.006. [DOI] [PubMed] [Google Scholar]
- Suh N.; Reddy B. S.; DeCastro A.; Paul S.; Lee H. J.; Smolarek A. K.; So J. Y.; Simi B.; Wang C. X.; Janakiram N. B.; Steele V.; Rao C. V. Combination of atorvastatin with sulindac or naproxen profoundly inhibits colonic adenocarcinomas by suppressing the p65/beta-catenin/cyclin D1 signaling pathway in rats. Cancer Prev. Res. 2011, 4, 1895–1902. 10.1158/1940-6207.CAPR-11-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mármol I.; Sánchez-de-Diego C.; Pradilla Dieste A.; Cerrada E.; Rodriguez Yoldi M. J. Colorectal Carcinoma: A General Overview and Future Perspectives in Colorectal Cancer. Int. J. Mol. Sci. 2017, 18, 197 10.3390/ijms18010197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng J.; Zhang X. T.; Liu X. L.; Fan L.; Li C.; Sun Y.; Liang X. H.; Wang J. B.; Mei Q. B.; Zhang F.; Zhang T. WSTF promotes proliferation and invasion of lung cancer cells by inducing EMT via PI3K/Akt and IL-6/STAT3 signaling pathways. Cell. Signalling 2016, 28, 1673–1682. 10.1016/j.cellsig.2016.07.008. [DOI] [PubMed] [Google Scholar]
- Jeyamohan S.; Moorthy R. K.; Kannan M. K.; Arockiam A. J. Parthenolide induces apoptosis and autophagy through the suppression of PI3K/Akt signaling pathway in cervical cancer. Biotechnol. Lett. 2016, 38, 1251–1260. 10.1007/s10529-016-2102-7. [DOI] [PubMed] [Google Scholar]
- Ma X.; Lv B.; Li P.; Jiang X.; Zhou Q.; Wang X.; Gao X. Identification of ″Multiple Components-Multiple Targets-Multiple Pathways″ Associated with Naoxintong Capsule in the Treatment of Heart Diseases Using UPLC/Q-TOF-MS and Network Pharmacology. Evidence-Based Complementary Altern. Med. 2016, 2016, 9468087 10.1155/2016/9468087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taieb J.; André T.; Auclin E. Refining adjuvant therapy for non-metastatic colon cancer, new standards and perspectives. Cancer Treat. Rev. 2019, 75, 1–11. 10.1016/j.ctrv.2019.02.002. [DOI] [PubMed] [Google Scholar]
- Housman G.; Byler S.; Heerboth S.; Lapinska K.; Longacre M.; Snyder N.; Sarkar S. Drug resistance in cancer: an overview. Cancers 2014, 6, 1769–1792. 10.3390/cancers6031769. [DOI] [PMC free article] [PubMed] [Google Scholar]