Abstract
Recent efforts have shown that structural variations (SVs) can disrupt the 3D genome organization and induce enhancer-hijacking, yet no computational tools exist to identify such events from chromatin interaction data. Here, we develop NeoLoopFinder, a computational framework to identify the chromatin interactions induced by SVs, including inter-chromosomal translocations, large deletions, and inversions. Our framework can automatically resolve complex SVs, reconstruct local Hi-C maps surrounding the breakpoints, normalize copy number variation and allele effects, and predict chromatin loops induced by SVs. We applied NeoLoopFinder in Hi-C data from 50 cancer cell lines and primary tumors and identified tens of recurrent genes associated with enhancer-hijacking. To experimentally validate NeoLoopFinder, we deleted the hijacked enhancers in prostate adenocarcinoma cells by CRISPR/Cas9, which significantly reduced the target oncogene expression. In summary, NeoLoopFinder enables to identify critical oncogenic regulatory elements that can potentially reveal therapeutic targets.
Introduction
Structural variations (SVs), such as deletions, inversions and translocations, frequently occur in cancer genomes, and have been shown to play a vital role in tumorigenesis1. Most of the previous studies have focused on how SVs affect protein-coding genes, including formation of the oncogenic fusion genes. However, recent efforts by the ENCODE2 Consortium and the Roadmap Epigenomics Project3 have shown that there are millions of potential distal regulatory elements such as enhancers in the human genome. SVs have been shown to juxtapose enhancers to key cancer genes and contribute to their aberrant elevated expression, which is termed “enhancer hijacking” 4,5. For example, inversion inv(3)(q21q26.2) has been recurrently observed in acute myeloid leukemia (AML), in which the GATA2 enhancer is repositioned near the oncogene EVI1 and ectopically activates EVI1 expression6. In adenoid cystic carcinoma (ACC), translocations reposition a super-enhancer in proximity to the MYB gene, activating its elevated expression7. Further, structural variations can induce “neo-TADs” in developmental diseases8 and a subgroup of medulloblastoma9.
Despite its importance, “enhancer hijacking” has only been mainly reported in case studies6,7,10-16. CESAM4 and PANGEA17 can predict enhancer-hijacking events genome-wide, by building a regression model of gene expression with SV profiles from hundreds or thousands of genomes. Such methods, while valuable for large cohorts of samples, cannot be directly applied when we only have one or a small number of samples.
Although chromatin interaction data have been used to validate enhancer-promoter linkages, to the best of our knowledge, there is no computational method to identify enhancer hijacking events directly from genome-wide chromatin interaction experiments such as Hi-C18. Recently, we and other groups showed that Hi-C can be used to systematically detect SVs genome-wide in the cancer genome19-21. Here, we present NeoLoopFinder, a computational framework aiming at the identification of chromatin interactions from Hi-C map in cancer and other diseases with genome rearrangements. NeoLoopFinder can correct CNV biases, assemble complex SVs whenever present, normalize allelic effects within local assemblies, and predict SV-induced neo-loops. To facilitate the visualization of neo-loops and integration with other omics data, NeoLoopFinder also provides a module for genome-browser-like region plotting. We applied this pipeline to 50 Hi-C datasets from cancer cell lines and patient samples spanning 17 cancer types (Supplementary Table 1). We identified distinct sets of neo-loop-involved genes in different cancer types, which are enriched in cancer type-specific pathways and closely related to patient survival. By integrating H3K27ac ChIP-Seq, chromatin accessibility (DNase-Seq), and RNA-Seq data from the ENCODE Consortium, we further annotated enhancer hijacking events within the neo-loops, and validated them by CRISPR knock-out experiments. Finally, we show that NeoLoopFinder is also applicable to developmental diseases with genomic rearrangements.
Results
Overall design of the NeoLoopFinder framework
Fig. 1 illustrates the overall framework design. The inputs of NeoLoopFinder are a Hi-C contact matrix and a list of SV breakpoints, which can be identified from different platforms such as Hi-C, whole-genome sequencing (WGS), and optical mapping20. NeoLoopFinder outputs the following: 1) genome-wide CNV profile; 2) genome-wide CNV segments; 3) a chain of linked SV events (local assembly); 4) corrected Hi-C matrix for the newly assembled regions; 5) chromatin loops in the rearranged regions. 6) Enhancer-hijacking events when H3K27ac data are available.
Figure 1 ∣. The overall design of the NeoLoopFinder framework.
a, Workflow of NeoLoopFinder. b, Reconstruction of an example complex SV in K562 cells using Hi-C map.
As copy number variations (CNVs) can distort Hi-C signals in cancer cells (STEP 1 in Fig. 1a), we proposed and implemented a modified matrix balancing algorithm to remove such effects along with other systematic biases including mappability, GC content, and restriction fragment sizes (Methods). The algorithm begins with the inference of genome-wide CNV profiles and CNV segments from Hi-C maps, and then applies matrix balancing to regions with different copy numbers separately. HiNT19 and HiCnv21 have been used to compute CNV profiles from Hi-C, and it has been reported that HiNT is more accurate than HiCnv. In this work, we developed a CNV segmentation module based on Hidden Markov Model (HMM) in NeoLoopFinder, which has a similar performance with HiNT in identifying CNV bins, but achieved a better performance when merging multiple bins into a larger CNV block (Extended Data Fig. 1a-g). By simulating CNV effects in a normal cell line (GM12878), we found our modified matrix balancing algorithm outperformed the standard matrix balancing method (ICE)22,23 when CNVs are present, as we observed lower variances between regions with different copy numbers and higher SCC (Stratum-adjusted Correlation Coefficient) scores24 compared with the gold-standard matrix (i.e., the ICE normalized GM12878 Hi-C) (Extended Data Fig. 2). Furthermore, we benchmarked our algorithm against three existing CNV-aware Hi-C normalization tools calCB25, OneD26 and CAIC27. While all methods can eliminate CNV bias and output an unbiased Hi-C map for regions with different copy numbers (Extended Data Fig. 1h-i), NeoLoopFinder is the fastest and consumes much less memory than other methods (Supplementary Table 2).
The next step is to reconstruct the Hi-C matrix for the rearranged genomic regions (STEP 2 in Fig. 1a). Submatrices from different parts of the reference map are flipped or rotated according to the types and orientations of the breakpoints (Extended Data Fig. 3a). To resolve the complex SVs formed by a chain of SVs, we developed a graph-based algorithm to assemble the correct order of each individual SVs (Fig. 1b and Extended Data Fig. 3b-f). The algorithm first determines the other endpoint of the DNA fragments affected by SVs, by using a principal component analysis (PCA) based procedure to scan for a change in the contact pattern near the SV breakpoints. We then build a connection network to connect the rearranged fragments and resolve the complex SVs, which are defined as local assemblies made up of multiple (≥3) fragments (Methods). Finally, the continuity of an output assembly is guaranteed by comparing local contact-distance decay with the global contact-distance decay pattern. We validated the algorithm by comparing the results with local complex SV structures manually reconstructed in our previous work20 . While the manual reconstruction required WGS, optical mapping, and Hi-C20, our algorithm automatically assembled all these complex SVs solely based on Hi-C data (Extended Data Fig. 4).
Another source of bias in the originally reconstructed Hi-C map is the allelic effect. Due to the heterozygosity of SVs and potential heterogeneity of patient samples, Hi-C signal intensity across the breakpoints tends to be lower than signals in the genomes not affected by SVs. Therefore, we designed a linear-regression model to scale signals from different regions into a similar range (STEP 3 in Fig. 1a and Extended Data Fig. 5a-c).
After the allele-normalized matrix has been attained, the next step is to identify loops. To do so, we integrated our recently developed machine-learning based framework Peakachu (STEP 4 in Fig. 1a)28. We included pre-trained models from the deeply sequenced GM12878 Hi-C library22, and to best handle the variable sequencing depths and data quality in different samples, we trained the models at various resolutions and window sizes, covering both in situ and dilution Hi-C protocols (Methods). The products from this framework include both loops in the regions not affected by SVs and chromatin loops induced by SVs, which we refer to as neo-loops throughout this work. These neo-loops will serve as the basis for the detection of enhancer hijacking events. We also tested chromatin loop predictors without removing CNV effect or reconstructing local Hi-C maps surrounding SVs (Extended Data Fig. 6). We observed that such practice resulted in extensive number of false positives, mainly due to the altered expected values induced by CNVs and SVs. This result suggests the necessity of considering CNVs and SVs in analyzing chromatin interaction data in rearranged genomes.
Finally, NeoLoopFinder also has a visualization module to facilitate the integration with other omics data, which provides functions such as plotting triangular contact heatmaps, genes, epigenomic tracks, and loops, using SV assembly coordinates with correct orientations. All genomic screenshots presented in this work were produced by this module.
Detection of neo-loops in 50 cancer samples
We applied the NeoLoopFinder pipeline to 50 Hi-C datasets spanning 17 cancer types (Supplementary Table 1). Among them, SVs in 8 cell lines have been identified in our previous work20. For other samples, we identified SVs with Hi-C breakfinder20. In this study, we only considered large SVs that can induce a valid local assembly where Hi-C contacts are continuous and attenuated along with the increasing genomic distance across all regions on the assembly (Methods, Extended Data Fig. 3). In total, we collected 1,510 such SVs across all samples (Supplementary Table 3). Using the graph-based algorithm described above, we detected complex SVs in 18 samples (Extended Data Fig. 7). The rearranged fragments in a complex SV are generally smaller than simple inversions but larger than simple deletions/duplications (Extended Data Fig. 7b).
Then we searched chromatin loops on local assemblies of either single SVs or complex SVs. The number of neo-loops detected in each sample ranges from 0 to 562, with a median value of 37 (Fig. 2a and Supplementary Table 4). The number of neo-loops is approximately proportional to the number of SVs in each sample (Fig. 2b). To estimate the false discovery rate (FDR), we randomly shuffled the SV breakpoints 50 times for each dataset, controlling for the distribution of SV types, the ratio of inter- vs. intra-chromosomal SVs, and the sizes of the SVs. The estimated FDRs are less than 1% for 97.9% (46 out of 47 samples that have neo-loops detected) of the samples (Fig. 2c), and 59.6% (28 / 47) of them have an FDR of 0. Importantly, the estimated FDRs and the number of neo-loops in corresponding samples are not correlated with each other (Fig. 2d). Therefore, we concluded that the neo-loop interactions detected in this study are not likely to be caused by data noise.
Figure 2 ∣. Detection of neo-loops in 50 cancer cell lines or patient samples.
a, Number of neo-loops detected in each sample. Samples with zero predicted neo-loops are not shown. b, Number of predicted neo-loops is proportional to the number of large SVs in the sample. Large SVs here refer to large deletions and inversions (>1Mb) and inter-chromosomal translocations that have great impacts on Hi-C signals. The details for SV filtering are described in Methods. c, False discovery rates (FDR) of predicted neo-loops in each sample. Each dot represents the FDR in one sample. d, Correlation between the estimated FDR and the number of detected neo-loops. e, Aggregate Peak Analysis (APA) for 4,672 predicted neo-loops in all the samples analyzed in this study at 10Kb resolution. f, Distribution of the distances between two loop anchors of the neo-loops. Data were combined from all samples. g, Predicted neo-loops are enriched for CTCF binding sites. Further, we observed that the loop anchors are also enriched for convergent CTCF motifs (pie chart). Data were plotted using the CTCF binding profiles in 11 cell lines (22Rv1, A549, C4-2B, HL-60, HepG2, K562, LNCaP, MCF7, PANC-1, PC3, and SK-N-SH) from the ENCODE consortium. h, Comparison of predicted neo-loops to 4C-Seq anchored at the MYC gene promoter in SK-N-MC cells. There are five neo-loops (purple arcs) in this region and they are formed due to a 1.5Mb deletion (chr8: 127.88M, +; chr8: 129.37M, −). Four of them match with 4C-Seq (green track, using MYC gene promoter as the bait) peaks, and all of them match with peaks on the virtual 4C track (orange track) extracted from SK-N-MC Hi-C for the same bait region. As a control, none of these peaks exist in the virtual 4C track from normal brain cells (gray track, astrocyte of the cerebellum).
Next, we performed Aggregate Peak Analysis (APA) by piling up 4,672 neo-loops predicted in all cancer samples and the results showed that there is a strong enrichment (Fig. 2e). For 89.5% of the neo-loops, their anchors are within 800Kb on the local assemblies (Fig. 2f). Furthermore, we found that neo-loops are also enriched with CTCF ChIP-Seq peaks, and for those with CTCF binding at both anchors, ~74% have convergent CTCF binding motifs, suggesting they are formed via the loop extrusion model (Fig. 2g)22,29,30. By utilizing a pileup analysis of breakpoint-crossing Hi-C signals, our previous work has shown that neo-TADs are frequently formed as the result of large-scale genomic rearrangements in cancer cells20. With the assembly reconstruction and normalization pipeline in this work, we can now detect neo-TADs systematically with a DI-based method (or any other traditional TAD identification tool, such as insulation score) (Extended Data Fig. 5)31,32. The number of detected neo-TADs in each sample ranges from 0 to 137, with a median value of 15.5. We found that most neo-loops (~57.6%) are located within a neo-TAD, supporting a critical role of neo-TADs in rewiring aberrant interactions.
Furthermore, we found the detected neo-loops frequently involved known cancer driver genes, such as MYC in brain tumors and breast cancers and ETV1 in prostate adenocarcinoma (Fig. 3a). For example, across breakpoints of a deletion (chr8: 127.88M, +; chr8: 129.37M, −) in SK-N-MC (neuroepithelioma), we detected nine neo-loops (Extended Data Fig. 5b), five of which were between the MYC gene promoter and cis-elements located more than 1.5Mb away on the reference genome. We further performed 4C-seq experiment, using the MYC gene promoter as the bait. We observed that four predicted neo-loops overlapped with peaks in the 4C track (Fig. 2h), while no visible peaks were noticed in the virtual 4C from normal brain cells (astrocyte of the cerebellum).
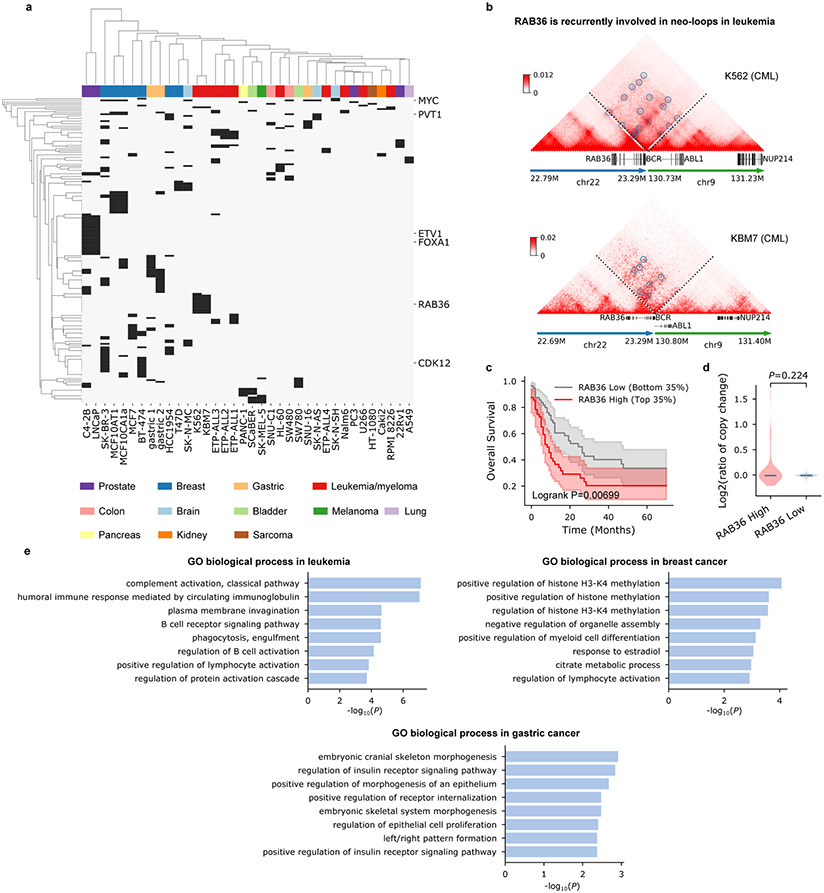
Figure 3 ∣. Cancer-type specificity of the neo-loop-involved genes.
a, Unsupervised clustering of the neo-loop-involved genes and cancer samples based on the occurrence of genes in each sample. Different cancer types are coded by different colors. Genes that are disrupted by an SV breakpoint (breakpoint located in the gene body) or only appear in one sample are removed from the list. b, RAB36 is recurrently associated with neo-loops in leukemia (Fig. 3a). c, Kaplan-Meier survival analysis of TCGA acute myeloid leukemia (AML) patients shows that the differential expression of RAB36 is associated with different survival rates. The means and the 95% confidential intervals are shown for each group of patients. The p-value was calculated from the two-sided log-rank test. d, Log2 converted copy number ratios of the RAB36 gene with high (top 35%) or low (bottom 35%) expression in corresponding patients. Changes in RAB36 expression are not due to gene amplification (two-sided Mann-Whitney U test). For the violin plots, the horizontal bar represents the median. e, Biological process enrichment of the neo-loop involved genes in different cancer types. The p-values were obtained from the Fisher’s exact test using EnrichR.
Discovery of novel cancer type-specific genes by neo-loops
In total, we identified 3,459 genes within neo-loops across 17 different cancer types. For ~89.6% (3,099 / 3,459) of them, their gene bodies are not disrupted by SV breakpoints. Besides MYC and ETV1, we also recognized many other known cancer-related genes, such as PVT1 (in SK-BR-3, MCF10AT, MCF10CA1, HCC1954, SK-N-AS, and SW480), a non-coding gene that regulates proliferation in a wide range of cancers33; CDK12 (in SK-BR-3 and BT-474), a therapeutic target in triple-negative breast cancer34; FOXA1 (in C4-2B and LNCaP), a pioneer transcription factor that is frequently mutated in hormone-receptor-driven tumors, and has been linked to enhancer hijacking in metastatic prostate cancers35. Overall, ~8.0% (247 / 3,099) of these genes have been reported as cancer-related genes and 3.8% (117 / 3,099) were identified as enhancer-hijacking genes by CESAM or PANGEA from different patient cohorts4,17 (Supplementary Table 5). Importantly, we found that samples of the same cancer type tend to be clustered together based on the occurrence of these neo-loop-involved genes in each sample (Fig. 3a). There are totally 99 recurrent neo-loop-involved genes from different cancer types (same gene appears in ≥ 2 samples of the same cancer type) (Supplementary Table 6). 19.2% of them (19 / 99) are located in the recurrent neo-loops.
In addition to known cancer-related genes, our framework also reported tens of novel genes that are non-fusion, unamplified, but might be involved in tumor proliferation or progression. For example, we found that the RAB36 gene is located inside predicted neo-loops in two chronic myeloid leukemia cell lines (K562 and KBM7) (Fig. 3b). Notably, the overexpression of RAB36 is significantly associated with poorer overall survival across all available leukemia data from TCGA (P = 0.00699, Log-rank test, Fig. 3c), and its overexpression cannot be explained by copy number variations in corresponding patients (Fig. 3d). These results indicate a prognostic role of RAB36 in leukemia and highlight enhancer hijacking as a potential regulatory mechanism. Similarly, we observed UPK2 in T-ALL and EYA1 in gastric cancer are involved in neo-loops (Extended Data Fig. 8).
Furthermore, we found that neo-loop-involved genes are enriched in both tumorigenesis-associated and tissue-specific processes (Fig. 3e)36. For example, the most significant processes involved in leukemia are related to the immune system such as the classical complement pathway, the B cell receptor signaling pathway, and the regulation of lymphocyte activation. In breast cancer, genes within neo-loops are mostly enriched in the regulation of histone methylation, including H3K4 methylation, whose elevated level has been associated with a poor clinical outcome in various cancers37. In gastric cancers, we found neo-loops involved genes in both the regulation of epithelial cell proliferation and the regulation of insulin receptor signaling pathway.
Detection of enhancer hijacking events induced by neo-loops
To identify genes with hijacked enhancers, we integrated H3K27ac ChIP-Seq, chromatin accessibility (DNase-Seq), and RNA-Seq data from the ENCODE Consortium (Supplementary Table 7). We defined potential enhancers as H3K27ac or DNase-Seq peaks, and defined enhancer hijacking as the following: a gene promoter located in one anchor of a predicted neo-loop, and there is an enhancer located in the other anchor of the neo-loop. We predicted the enhancer-hijacking events in 11 cell lines, including A549, K562, LNCaP, MCF7, T47D, HepG2, SK-MEL-5, NCI-H460, PANC-1, HT-1080 and C4-2B (full list of the events is provided in Supplementary Table 8). We observed that the genes involved in enhancer-hijacking events showed higher expression compared with other neo-loop-involved genes (Fig. 4a). These genes were also upregulated compared with their expression in control cell lines from the same tissue, further suggesting the role of enhancer-hijacking in gene dysregulation in cancer (Fig. 4b and Extended Data Fig. 9).
Figure 4 ∣. Analysis of the expression of the genes in neo-loops.
In each box plot, the center line indicates the median, the box limits represent the upper and lower quartiles, and the box whiskers indicate the 1.5x interquartile range. a, We categorized genes in the SV regions into three groups: G1) genes located in one anchor of neo-loops, and the other anchor contains a predicted enhancer (H3K27ac peak); G2) all genes in neo-loops; G3) genes not in any loops. Numbers in the bracket below indicate the number of genes in each category. Transcripts Per Kilobase Million, TPM. The p-values were computed using the two-sided Mann-Whitney U test. b, Quantile normalized gene expression signals for genes with hijacked enhancers (the first group in a) are compared between cancer cells and corresponding normal cells. We used the following cell lines for this analysis: C4-2B vs. RWPE-1 (prostate), MCF7 vs. HMEC (Mammary Epithelial), K562 vs. GM12878 (lymphoblastoid). The p-values were computed using the two-sided Wilcoxon signed-rank test.
Deletion of hijacked enhancers reduced oncogene expression
To further interrogate the role of hijacked enhancers in cancer, we performed genome editing experiment utilizing CRSPR/Cas9 system. ETV1 is a known driver gene in prostate cancer, whose over-expression by amplification or gene-fusion has been associated with aggressive disease and poorer outcomes38,39.
In this study, we observed that ETV1 is dramatically up-regulated in LNCaP (a prostate adenocarcinoma cell line), compared with normal prostate epithelial cells RWPE-1 (> 16-fold) (Figs. 5a-b). However, we did not observe gene fusion or amplification of ETV1 in LNCaP. Rather, we found that ETV1 is located inside neo-loops induced by a translocation between chromosome 7 and chromosome 14 (chr7: 14.15M, +; chr14: 37.51M, +), interacting with multiple potential enhancers on chromosome 14 (Fig. 5a, DNase-Seq, purple track). Moreover, we observed that these neo-loops coincide with convergent CTCF binding motifs (Fig. 5d). As a comparison, there is no interplay between ETV1 and the same regions in RWPE-1 cells (Fig. 5b), suggesting that ETV1 overexpression in LNCaP might be driven by enhancer hijacking.
Figure 5 ∣. Deletion of hijacked enhancers reduced oncogene expression in prostate adenocarcinoma.
a, A predicted enhancer-hijacking event for ETV1 in LNCaP cells. The Hi-C map (10kb), copy number segments, RNA-Seq and DNase-Seq tracks are reconstructed for the translocation event “chr7: 14.15M, +; chr14: 37.51M, +” in LNCaP cells. The yellow vertical bar highlights the hijacked enhancer region. Blue circles indicate the detected neo-loops. b, Hi-C, RNA-Seq, DNase-seq, and CNV segments for the same region in RWPE-1 cells (a normal prostate epithelial cell line). c, We deleted two candidate enhancers, M1-del (chr14: 37,278,706-37,279,509) and M2-del (chr14: 37,246,515-37,246,652), by CRISPR/Cas9 separately. The ETV1 expression in control (transfected with empty vector) and enhancer-deleted LNCaP cells were measured in three technical replicates for each clone using RT-qPCR. Each bar indicates the mean of the ETV1 expression level relative to the control cells, and the error bars represent the standard deviation for the technical replicates of each clone. d, CRISPR deletion of the hijacked enhancer M1 impaired the neo-loop formation. (Left panel) The reconstructed Hi-C map in the wild-type cells. The DNase-Seq profiles and CTCF binding motifs overlapped with DNase-Seq peaks for the same region are shown below by purple tracks and gray triangles, respectively. The neo-loop between the ETV1 promoter and the hijacked enhancer M1 involves pairs of CTCF binding sites with convergent motif orientations. (Right panel) We performed in situ Hi-C in the LNCaP cells (the M1-E10 clone in 5c) with hijacked enhancer M1 deleted. The deletion strongly reduced the contact frequency between the ETV1 promoter and M1.
To validate the prediction, we used the CRISPR/Cas9 system to delete the hijacked enhancers in these loci (Fig. 5a, yellow band; Fig. 5c). Within the neo-loop anchor, there are three DNase-Seq peaks, and one of them overlaps with a MIPOL1 exon. To rule out the potential effect of affecting the MIPOL1 gene, we only deleted the two intronic DNase-Seq peaks (M1-del and M2-del). We then performed RT-qPCR to compare the ETV1 expressions between the enhancer-deleted LNCaP cells and the control LNCaP cells (transfected with empty vector). The experiments were performed across 6 and 4 independent clones for M1-del and M2-del, respectively. We observed a dramatic reduction of ETV1 expression in the engineered LNCaP cells, with an average of 66.3% reduction of expression in M1-del and an average 80.5% reduction in M2-del clones, suggesting a previously unknown mechanism for ETV1 activation in prostate cancers by enhancer hijacking. To investigate the structural role of these hijacked enhancers, we performed in situ Hi-C for both control cells and the M1-deleted cells (M1-E10, Fig. 5c). Strikingly, we observed that the Hi-C signals in the neo-loop were greatly reduced (Fig. 5d), while the local interactions surrounding MIPOL1 were strengthened. This suggests that the deletion of the hijacked enhancers might have changed the overall structure in this region.
Detection of enhancer hijacking in developmental diseases
To investigate whether NeoLoopFinder can be used to predict potential enhancer hijacking in other diseases with rearranged genomes, we collected data for various structural variations causing limb malformations that have been functionally dissected in previous studies: (1) an inversion involving an enhancer (named “Pen”) that controls the Pitx1 gene expression40; (2) a duplication covering the Sox9 regulatory domain and the Kcnj2 gene8; (3) an inversion of the Sox9 regulatory domain including the TAD boundary41. As shown in Extended Data Fig. 10, in each case, NeoLoopFinder was able to automatically reconstruct the Capture Hi-C map for the local genome that is highly similar to the matrices presented in the original reports. Furthermore, NeoLoopFinder also identified potential hijacked enhancer-promoter interactions for the indicated disease-causal genes (Pitx1, Kcnj2, and Kcnj2). Note that these data are Capture Hi-C data, therefore, the proposed algorithm should work on otherchromatin interaction data with rearranged genome for different types of SVs, such as inversion and duplications.
Discussion
Recent years have seen a growth spurt of Hi-C data in cancer. However, most of the current studies have focused on regions not affected by SVs, mainly due to the lack of specialized Hi-C data analysis tools that can handle such events. Here, we developed the first computational framework dedicated to analysis of chromatin interaction data in rearranged genomes. We showed that NeoLoopFinder can remove CNV bias, resolve complex SVs, reconstruct local contact maps across the SV breakpoints, and predict neo-TADs and neo-loops. NeoLoopFinder also has a built-in module to generate browser-like visualization for local SV assemblies. Although the main body of this work is focused on enhancer-hijacking events, the predicted neo-loops can be used to define other functional chromatin interactions, such as repressor hijacking. Further, through extensive analysis of 50 Hi-C data in cancer, we identified recurrent genes and biological pathways for different cancer types.
We note that the accuracy of local assemblies and reconstructed Hi-C maps relies on the quality of the input SVs. Further, when heterozygous SVs are provided, NeoLoopFinder can reconstruct different Hi-C maps for different alleles. One such example is shown in Extended Data Fig. 4, where a fragment on chr9 is translocated to chr13 (Extended Data Fig. 4b) on one allele and in the other allele, the same fragment is translocated to chr22 (Extended Data Fig. 4e). As for predicting chromatin neo-loops, an advantage of NeoLoopFinder relies on its machine-learning based method, rather than testing for significant enrichment of Hi-C signals compared to a local or global background28. As many SV breakpoints in cancer are located within the repetitive regions, the abnormal signals can defeat the statistical assumptions of traditional enrichment-based methods and yield incorrect results22,42. Another advantage of machine-learning based methods is their robustness to sequencing depth, which enabled us to detect neo-loops in samples with less than 100 million read pairs.
Furthermore, the embedded visualization module is the first stand-alone automatic tool to display rearranged-chromatin structures along with genes and epigenomic tracks for any SVs or complex SVs. With the continuous growth of Hi-C data in cancer, we hope to build a 3D cancer browser upon our 3D genome browser43 to provide a comprehensive toolkit for interactive visualization and data integration of cancer 3D genomics data.
Methods
Data sources and data processing
All genomic datasets used in this work were mapped to the human genome assembly GRCh38 (hg38). Hi-C data were either downloaded from the 4DN data portal (https://data.4dnucleome.org/) or processed by the runHiC python package (https://pypi.org/project/runHiC/) (Supplementary Table 1). The CNV profile in each sample was directly estimated from Hi-C maps using a generalized additive model, and then an HMM-based segmentation was applied to determine the boundaries of CNV segments. In Extended Data Figures 1a-g, we calculated CNV profiles and segments from WGS reads by Control-FREEC44 in 8 cell lines for the benchmark. The ploidy parameter was set as follows in running Control-FREEC: 2 for pseudodiploid cells SK-N-MC and hypotriploid cells NCI-H460; 3 for triploid cells K562, hypotriploid cells A549 and T47D, and hypertriploid cells PANC-1; 4 for hypotetraploid cells LNCaP; 5 for hypopentaploid or hypohexaploid cells Caki2. As another input to the NeoLoopFinder framework, the breakpoints of large SVs were mostly identified from Hi-C maps using Hi-C break finder (https://github.com/dixonlab/hic_breakfinder), except for the aforementioned 8 cell lines A549, Caki2, K562, LNCaP, NCI-H460, PANC-1, SK-N-MC and T47D, where we used our published SV lists compiled from multiple experimental platforms20. The RNA-Seq data were processed using the ENCODE long-read RNA-Seq pipeline (https://github.com/ENCODE-DCC/long-rna-seq-pipeline). Data used for survival analysis in leukemia and gastric cancer were downloaded from the cBioPortal for Cancer Genomics (https://www.cbioportal.org)45. The list of cancer-related genes was obtained from the Bushman Lab (http://www.bushmanlab.org/assets/doc/allOnco_May2018.tsv).
4C-Seq Library preparation and sequencing
4C experiments were performed in SK-N-MC cells (obtained from ATCC, https://www.atcc.org/) to quantify genome-wide DNA-DNA interactions that involve promoter of the MYC gene in this cell line. Ten million SK-N-MC cells in cell culture were harvested, resuspended in 8.75ml 1X complete medium, and immediately cross-linked in 2% formaldehyde for 10 minutes. Nuclei were isolated from fixed cells and digested by restriction enzyme DpnII (NEB, #R0543) overnight. Digested DNA was ligated using T4 DNA ligase (NEB, #M2020) and de-crosslinked by incubating with proteinase K overnight (Qiagen, #19133). DNA was then purified and fragments larger than 30kDa were selected and digested by the second restriction enzyme Csp6i (Thermo, #ER0211) overnight. DNA was re-ligated using DNA ligase (1U/ul, Invitrogen, #15224017) at low concentration. DNA was further purified and DNA fragments that contain MYC promoter were amplified by PCR reaction with 30 cycles: A transcription start site of MYC gene flanked by DpnII and Csp6i cutting site at each end was chosen as an anchor (chr8: 127,735,189-127,735,615), and a pair of primers that contain Truseq indices were used to amplify regions located to the upstream and downstream of the anchor. The PCR products were further purified by size selection to remove small DNA fragments, and sent for sequencing on Illumina HiSeq 2500 with a depth of 2 million reads.
The sequences of primers are as following:
MYC-upstream: CAAGCAGAAGACGGCATACGAGAGATGTAGCCGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTGTTGCCTGCTCTCTGCCAGT
MYC-downstream: AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTTCCACTCTCCCTGGGACTCT
The 4C sequencing reads were first trimmed by 23 bp from the 5' end to remove the bait sequence. Trimmed reads were then mapped to human reference GRCh38 with "BWA MEM". Per-base read coverage was then calculated by "bedtools genomecov".
CRISPR/Cas9-mediated enhancer deletion
sgRNAs were designed by combining results from CRISPOR (http://crispor.tefor.net) and GuideScan (http://www.guidescan.com). Two pairs of sgRNA were chosen for each site. sgRNA sequences and PCR primers are listed in Supplementary Table 9. The gRNA oligos were inserted into pX458 vectors (Addgene) using the BbsI restriction enzyme (NEB). For target deletion, the LNCaP cell line (obtained from ATCC, https://www.atcc.org/) was transfected with each pair of gRNA vector aiming to upstream and downstream of the target region (named M1 and M2) and flow-sorted for GFP positive cells after 2 days. Individual cells were seeded into 96-well plates and expanded for 3-4 weeks. Single-cell clones were selected and amplified. The deletion was verified by genomic DNA PCR with the primers outside the target deletion region and Sanger sequencing.
RNA extraction and real-time PCR
RNA was extracted using RNeasy Mini kit (Qiagen) according to the manufacturer’s instructions. For cDNA preparation and DNA elimination, the SuperScript® III First-Strand Synthesis System (Invitrogen) was used according to the manufacturer’s instructions. qRT-PCR was performed on a Bio-Rad CFX Connect Real-time PCR Detection system using KAPA SYBR FAST qPCR Master Mix. PCR conditions were as follows: 1 cycle of 95°C for 3min and 40 cycles of 95°C for 15sec, 60°C for 15sec and 72°C for 35sec. The housekeeping gene GAPDH was used as a normalization control. The primer sequences are listed in Supplementary Table 9.
Hi-C library preparation and sequencing
Approximately 2M of LNCaP wild type and LNCaP M1-deleted (M1-E10) cells were pelleted and crosslinked with 2% formaldehyde in PBS at RT for 10 mins. ARIMA’s Hi-C kit (ARIMA Genomics) was applied for capturing proximally-ligated DNA. The biotinylated DNA was sheared to a size of 300–500 bp and libraries were constructed following Kapa Hyper Prep Kit (KAPA) protocols. The library products were quantified with Tapestation High Sensitivity D1000 Assay (Agilent Technologies, CA, USA) and then sequenced using Illumina’s NovaSeq platform (Illumina).
Copy number inference from Hi-C map
We have embedded a copy number inference module within NeoLoopFinder to detect copy number variations (CNVs) directly from Hi-C map at a given resolution. The whole process takes two steps:
In the first step, we implemented the same generalized additive model (GAM) proposed by HiNT-CNV19 to model the non-linear relationship between the one-dimensional coverage profile and different biases commonly observed in a Hi-C experiment, including GC content, mappability, and the density of restriction sites; the residuals from the GAM model are used as an initial estimate of the copy number ratio for each bin. We confirmed that our results for this step are nearly identical to the output from HiNT-CNV and well correlated with the profile calculated from whole-genome sequencing (Extended Data Fig. 1a-b).
In the second step, we proposed a Hidden Markov Model (HMM) based segmentation algorithm to determine the boundaries of CNV segments from the initial CNV profile. The motivation is that the original segmentation algorithm (BIC-Seq) used by HiNT-CNV tends to report large blocks of amplified/depleted regions (Extended Data Fig. 1f-g), which caused inaccurate CNV normalization of Hi-C matrix in our test. In this HMM model, the initial CNV profile outputted from the first step is taken as the observed sequence, while the real copy number of a fragment is treated as the hidden state. For each chromosome, we first determine the best number of possible states N (the maximum number was set to 15) with the Gaussian Mixture Model and the BIC criterion. Then an HMM model is built using the pomegranate (https://github.com/jmschrei/pomegranate) Python package, with each state approximated by a 2-component Gaussian mixture. Before training, we detect and remove the outliers within the raw CNV profile with the Hampel filter and split it by 0s. The Baum-Welch algorithm is used to estimate the parameters of transition and emission and the Viterbi algorithm is used to predict the state (copy number) of each bin. Finally, the consecutive bins assigned with the same state are merged together to form a CNV segment. By default, the copy number of a segment is defined as the average copy number ratios. If ploidy is known, the absolute copy number will be further calculated by multiplying the copy number ratio by the ploidy and rounding to the nearest integer.
Separate matrix balancing & CNV effect correction
In a traditional matrix balancing or ICE algorithm, the normalized Hi-C map is modeled identically for the whole genome as CiMijCj, where Ci is the bias vector corresponding to each bin, which can be solved via an iterative correction approach. This normalization algorithm can remove most biases in Hi-C, including mappability, GC content and restriction fragment sizes23. Its efficiency and easy-to-implementation features have made it a standard and necessary step in most Hi-C processing software. However, we found that applying ICE or matrix balancing naively to cancer Hi-C can yield biased values due to amplification/depletion of the genome. Therefore, we proposed a modified ICE procedure by applying ICE to regions with different copy numbers separately:
STEP 1. Initialize an intermediate matrix and the bias vector Ci filled with ones.
STEP 2. Calculate the marginal sums of
STEP 3. Extract all bins with copy number k, and calculate the bias vector as below:
where scalek represents the average of non-zero marginal sums of bins with copy number equaling to k.
STEP 4. Iterate all possible copy numbers and repeat STEP 3.
STEP 5. Update using following equation:
STEP 6. Repeat STEP 1-5 until the average of variance of marginal sums for bins with identical copy numbers is less than 1e-5.
STEP 7. Scale Ci by , where CNV(i) = k
STEP 8. The normalized Hi-C map can be calculated by multiplying the final bias vector .
Our implementation is built upon the cool format, and the bias vector returned by the modified ICE above is stored in the “sweight” column in the cool file.
Simulation of CNV effects on a normal Hi-C map
In order to benchmark the performance of our CNV normalization algorithm and the traditional ICE procedure, we proposed a mathematical framework to simulate the effect of abnormal karyotypes on a diploid Hi-C dataset. As shown in the Extended Data Fig. 2a, the inputs to the framework are the Hi-C matrix of a normal/diploid cell (GM12878), the Hi-C matrix of a cancer cell (K562), and the CNV profile of the same cancer cell. A scaling factor is estimated for both intra-chromosomal and inter-chromosomal contacts of different copy number regions in K562.
For inter-chromosomal contacts , we first calculate the average contact frequencies separately for different copy number pairs (a, b):
where n is the number of contacts with one locus having a genomic copies and the other loci having b copies. Then for each copy number pair (a, b), we calculate the linear factor by applying
For intra-chromosomal contacts , we first calculate the contact-distance decay curves for each copy number pair:
After validating the linear relationship between curves from different copy number regions, we propose a linear regression model to estimate the scaling factor as follows:
To simulate the same CNV effects in GM12878, we impose the K562 copy number profiles to GM12878 in silico. For any contact with virtual copy number (a, b), we multiply the value by either or . We found the resulted Hi-C map could reproduce aberrant signals in K562 within the same regions.
Rearranged fragment detection, SV filtering and complex SV assembling
Cancer genomes are shaped with both simple SVs and complex SVs. In this work, the complex SVs are defined as local assemblies that are made up of multiple (≥3) DNA fragments (or ≥2 junctions) from different genomic locations on the reference genome, while the simple SV assemblies are defined as assemblies that only contain two DNA fragments (or one junction). In our previous work20, we have developed an algorithm to identify various kinds of simple SVs using Hi-C. Here we demonstrate that Hi-C can also be used in assembling complex SVs.
The input to the algorithm is a list of simple SV breakpoints and the genome-wide Hi-C matrix (Extended Data Fig. 3b). Only large SVs are supported, including intra-chromosomal rearrangements greater than 1Mb and inter-chromosomal translocations, which could be identified from various platforms such as Hi-C, WGS and optical mapping. We should note that the input SV coordinates are just the coordinates of the breakpoints with strand information; the coordinates of the other end of junction fragments are still unknown. To determine whether a chain of SVs should be called complex SVs, we begin with the detection of the coordinates of the whole chromatin fragments that are rearranged by each SV. To do so, we first extend the interaction region from breakpoints by 5Mb (this parameter is configurable) in appropriate orientations (the gray box in Extended Data Figs. 3c and 3d). Then we calculate two correlation matrices by rows and columns of , respectively. As shown in Extended Data Figure 3d, the stretch of the rearranged fragments can be recognized by locating the corner square block on the correlation matrix. Therefore, we perform the principal component analysis, and the boundary loci are identified where the sign of the first eigenvector / principal component changes. Within , we denote the region encompassed by the detected boundaries as (the green box in Extended Data Fig. 3c).
We next exclude all SVs where no significant contact-distance decay can be detected within . Here we use a similar rationale used in Hi-C guided genome assembly: Hi-C signals should be continuous and attenuated along with the increasing genomic distance (contact-distance decay) across all regions on a valid SV assembly. Specifically, we first calculate the global average contact frequencies among all contact pairs with the same genomic distance throughout the whole genome:
where denotes the whole-genome contact matrix, and n(d) denotes the number of valid data points at the genomic distance d. Similarly, we calculate the local distance averaged contact frequencies within as follows:
Then a linear regression model is fitted between and for each SV, and only SVs with R2 > 0.6 will be considered in this study (Extended Data Fig. 3e-f).
Then the potential complex SVs are detected by checking the overlap of rearranged fragments between simple SVs. A directed graph is built with each node representing a simple SV, and each edge representing an overlap of rearranged fragments in consistent orientations. We calculate the shortest paths between any two nodes of the graph by the Dijkstra’s algorithm and define each such path as a candidate complex SV. During this procedure, we re-map Hi-C signals to each candidate assembly and apply the same linear regression-based method described above to determine the assembly continuity. In other words, we ensure each region of an assembly displays a significant distance-decay trend. We also remove the candidates if the whole path or part of the path form a circular assembly. Finally, we remove redundant candidates if their paths are a subset or a reverse of other paths.
In this study, we identified neo-loops and neo-TADs for both non-redundant complex SVs and simple SVs, and non-redundant neo-loops and neo-TADs were reported for each sample.
Allele normalization
Due to heterogeneity of patient samples and heterozygosity of SVs, different regions in an assembly could have different Hi-C “visibility”, that is, SVs that are less frequent in a sample and only occur within few alleles have a lower chance to be sequenced in the Hi-C experiment, which makes it difficult to accurately detect loops or TADs in these regions (Extended Data Fig. 5a). We found such biases can be captured by the distance decay curve between the contact frequencies and the genomic distance between the contact pairs. To remove the biases across different regions, we proposed a linear regression-based method to minimize the differences between local distance decay curves and the whole-genome distance decay curve.
We first calculated the global average contact frequencies using the same formula defined in the previous section. The maximum genomic distance we considered was limited to 2Mb. Similarly, for an assembly composed of N chromatin fragments, we calculated the local average contact frequencies for each of the contact regions between the fragment a (a ≤ N) and fragment b (b ≤ N). We ignored any genomic distances with less than 3 non-zero data points and applied an isotonic regression method to ensure the average contact frequencies are monotonically nonincreasing with the increasing genomic distance. Then the linear regression model was built as follows:
where β(a,b) denotes the estimated scaling factor. To make the model robust to outliers, we utilized the Huber loss function instead of the traditional least-squares. Finally, we rescaled the contact frequencies by applying:
which potentially accounts for both sample heterogeneity and allelic effects.
Machine-learning based loop detection
For loop detection, we applied Peakachu, a machine-learning based framework recently developed by our lab28. Briefly, Peakachu can learn the loop pattern on a genome-wide contact map from a set of known chromatin loops and then use the trained model to predict loops on other maps generated by the same experimental protocol.
We noticed that the collected cancer Hi-C datasets in this study were generated by different protocols, with varying sequencing depths and data quality (Supplementary Table 1). To build a general framework, we trained Peakachu models on both in-situ22 and dilution18 Hi-C map of the GM12878 cell. Both maps were down-sampled to contain ~60 million intra-chromosomal reads to improve the model sensitivity for a wider range of sequencing depths. For in-situ Hi-C, the model was trained for three resolutions: 10K, 20K, and 25K; for dilution Hi-C, the model was trained for five resolutions: 10K, 20K, 25K, 40K and 50K. The positive training sets were defined from the published CTCF ChIA-PET and H3K27ac HiChIP interactions, representing chromatin structural loops and regulatory loops, respectively.
In the prediction stage, the higher probability score computed by the CTCF or the H3K27ac model was recorded for each pixel of the reconstructed Hi-C map. After filtering with a pre-defined probability threshold (Supplementary Table 1, 0.9 and 0.8 for in situ Hi-C and dilution Hi-C datasets, respectively), the same pooling algorithm used in Peakachu was applied to each SV region independently to select the best-scored loop contacts from each cluster. Loop lists from different resolutions were combined in a way that excluded redundant lower resolution loops. That is, if a pixel was detected as a loop in both resolutions, we recorded the more precise location in higher resolution and discarded the lower resolution one.
Neo-TAD identification
Our neo-TAD detection algorithm is based on the directionality index (DI) and takes two steps:
The first step corresponds to the training stage of a traditional DI method, where DI is calculated along the reference genome (hg38) and used as input to learn a global HMM model. The DI is defined as a t-statistic as follows46:
where Uk denotes the upstream contact frequency between bin i and bin i − k, Dk denotes the downstream contact frequency between bin i and bin i + k and W denotes the fixed window size which is set to 2Mb in this study. A four-state (start, upstream bias, downstream bias and end) GMM-HMM model is defined by the pomegranate package and each state is emitted from a three-component Gaussian mixture. Again, the Baum-Welch algorithm is used for training.
In the second step, we recalculate DI on the allele normalized Hi-C map of each local assembly and use the model trained above and the Viterbi algorithm to predict the state of each bin. The TADs are then defined as intervals between a “start” state and the next “end” state. And if a TAD has SV breakpoints located within its interval, it is called a neo-TAD.
Visualization of reconstructed Hi-C map and genome browser tracks
NeoLoopFinder introduces a standalone browser tool, specifically designed for visualizing shuffled cancer genome in the correct order. The tool supports plotting most kinds of tracks in a modern web browser, including genes, 1D peaks (.bed), 1D signals of RNA-Seq, ChIP-Seq, ATAC-Seq or DNase-Seq (.bigwig), 2D contact matrix (.cool), loops (.bedpe) and TADs (.bedpe), etc.
When initializing a plotting object, the required input information is a cool URI and an individual line of the file outputted by the complex SV assembler, and both the number of tracks and the height of each track can be configured flexibly. For additional data types, we provide miscellaneous methods, which take regular bigwig/bed/bedpe files as input and automatically perform the coordinate conversion from the reference genome (hg38) to the local assembly in the correct orientations.
Internally, we use the cooler package for fast Hi-C data retrieval47, the pyensembl package (https://github.com/openvax/pyensembl) to extract the gene information of any given regions, the CoolBox to draw genes in different styles48, and the pyBigWig for bigwig file loading49. Most elements in each track are configurable, such as the colormap, value range of the Hi-C heatmap, the line color and style of the breakpoints, loop size and color, color and width of chromosome bars, the font and positions of gene names. The module is able to generate publication-quality figures, which can be conveniently saved to various image formats such as PNG, PDF, and SVG.
Loop detection using FitHiC2
To investigate whether existing loop detection tools can be directly used to detect neo-loops without considering SV information, we tested FitHiC250 (fithic 2.0.7) (Extended Data Fig. 6), the only software that is able to detect inter-chromosomal interactions (Supplementary Table 10), as inter-chromosomal translocations happen frequently in cancer. We ran FitHiC2 in all in situ Hi-C datasets analyzed in this study at 10kb resolution. For intra-chromosomal interactions, the results of the 2nd spline pass were first filtered with the q-value cutoff < 10−10 and then the nearby significant interactions were pooled using the script “CombineNearbyInteraction.py” (https://github.com/ay-lab/fithic/tree/master/fithic/utils) with default parameters. For inter-chromosomal interactions, we ran FitHiC2 in “-x interOnly” mode and applied a q-value cutoff of 10−10 for filtering. Both inter-chromosomal and intra-chromosomal interactions identified by FitHiC2 were mapped to SV assemblies reconstructed by NeoLoopFinder for comparison.
Statistics and reproducibility
The 4C-Seq data we generated for the MYC gene promoter is highly similar to the virtual 4C from an independent Hi-C dataset of the same cell line. For CRISPR editing experiments, we generated 6 and 4 independent clones for M1-del and M2-del, respectively. We showed that the ETV1 gene expressions were consistently reduced in all the clones. No statistical method was used to predetermine the sample size.
Data Availability
The cancer Hi-C datasets analyzed in this study are summarized in Supplementary Table 1. Details of all the other datasets collected for the validation and downstream analysis are summarized in Supplementary Table 7. Data used for survival analysis in leukemia and gastric cancer were downloaded from the cBioPortal for Cancer Genomics (https://www.cbioportal.org)45. The list of cancer-related genes was obtained from the Bushman Lab (http://www.bushmanlab.org/assets/doc/allOnco_May2018.tsv). The 4C-Seq data for the MYC gene promoter in SK-N-MC cells, and the Hi-C data generated for wild-type and enhancer-deleted LNCaP cells have been uploaded to the Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession code GSE161493. Source data are provided with this paper.
Code Availability
The NeoLoopFinder source code is publicly available in GitHub at https://github.com/XiaoTaoWang/NeoLoopFinder. The NeoLoopFinder code is also available at Code Ocean (DOI: 10.24433/CO.1323561.v1).
Extended Data
Extended Data Fig. 1. Evaluation of the CNV segmentation and CNV normalization module in NeoLoopFinder.
a-g, We implemented the same generalized additive model (GAM) used by HiNT-CNV to estimate the copy number profile directly from Hi-C. For CNV segmentation, we applied a different algorithm based on Hidden Markov Model (HMM). a, We compared the copy number profiles estimated by NeoLoopFinder with the CNV profiles computed by Control-FREEC with whole genome sequencing (WGS) data. Each dot represents a 25kb bin. Bins with zero reads were excluded from the calculation. b, Similar to a, but for the comparison between NeoLoopFinder and HiNT-CNV. c, The number of CNV segments identified by NeoLoopFinder and HiNT-CNV, with or without WGS support. Only segments with a copy number ratio larger than 1.5 or smaller than 0.3 were considered in the calculation. d, The fraction of WGS-detected CNV segments that are recalled by NeoLoopFinder or HiNT-CNV. e, Comparison of F1 scores between NeoLoopFinder and HiNT-CNV in eight cancer cell lines. f, Comparisons of CNV segments inferred from HiNT-CNV and NeoLoopFinder based on Hi-C data in K562 cells. We compared their results with the CNV segments computed by Control-FREEC with whole genome sequencing (WGS) data. Results from both HiNT-CNV and NeoLoopFinder are similar to Control-FREEC. However, for more fragmented regions (green circles), NeoLoopFinder’s performance is better. g, A similar example in SK-N-MC cells. h-i, Comparison of different Hi-C normalization methods in K562 (Resolution: 10kb). Hi-C contact heatmaps and copy number variation profiles are shown for two example regions: “chr22: 22,340,000 – 24,200,000” (h) and “chr9: 130,000,000 – 131,280,000” (i). The CAIC method is excluded from the analysis due to memory error (Supplementary Table 2).
Extended Data Fig. 2. Evaluating the performance of CNV normalization of Hi-C data by simulation.
The inputs in our simulation are Hi-C data in GM12878 (normal lymphoblastic cells) and K562 (chronic myeloid leukemia) cells, and the CNV profiles in K562 cells. a, Our algorithm learns the trans-/cis-scaling factors separately for all possible copy number pairs from K562 Hi-C data. The CNV effects of K562 are then imposed on GM12878 Hi-C by linearly transforming the signals with the factor of corresponding copy number pairs. The resulting simulated GM12878 Hi-C matrix with K562 CNV is highly similar to the original K562 Hi-C matrix. b, We applied ICE and the newly designed CNV normalization method in this project to the simulated matrix from Supplementary Fig. 2a (GM12878 Hi-C matrix with CNVs in K562). By visual inspection, the CNV normalized Hi-C is more similar to the original GM12878. c, We used HiCRep to calculate the Stratum-adjusted Correlation Coefficients (SCCs) between ICE normalized and CNV normalized matrix to the original GM12878 Hi-C data. The distributions of SCC scores are presented in box-and-whisker plots, where the box represents the interquartile range (IQR, Q3-Q1), the horizontal thick line represents the median, the upper whisker extends to the last datum less than Q3+1.5×IQR, and the lower whisker extends to the first datum greater than Q1-1.5×IQR. Each dot represents an individual chromosome.
Extended Data Fig. 3. Complex SV detection based on Hi-C maps.
a, Illustration of how we re-construct Hi-C map surrounding breakpoints. There are four orientation types of inter-chromosomal translocations. b, Overall workflow of the complex SV assembling module in NeoLoopFinder. c, In the first step of the pipeline, we determine the whole rearranged fragments (green box) of the input SV breakpoints within the checking window (gray box, by default 5Mb extended from the breakpoints). d, The algorithm for determining rearranged fragments. First, correlation matrices are calculated by rows (top) and columns (bottom) of the contact matrix separately within the checking window; then the rearranged fragment boundaries are determined by checking the first principal component profile (PC1) of the correlation matrices. e-f, Determination of the R2 cutoff for SV filtering. e, The distribution of R2 across all large SVs (intra-chromosomal rearrangements larger than 1Mb and inter-chromosomal translocations). The number was summarized from all 50 cancer samples. f, The fraction of SVs after filtering as a function of R2 cutoff. Data were merged from eight cancer cell lines: A549, Caki2, K562, LNCaP, NCI-H460, PANC-1, SK-N-MC, and T47D.
Extended Data Fig. 4. Examples of complex SVs assembled by NeoLoopFinder using Hi-C data.
All these complex SVs were also reported in our previous work, where the structure of each assembly was manually reconstructed by combining optical mapping, Hi-C and WGS. a-c, Assembly of a complex SV in K562 cells. a, The abnormal signals on the original Hi-C map indicate the complex SV structures between chromosome 13 and chromosome 9 in K562 cells. b, The correct lengths and orders of the rearranged regions B2 (chr13: 92.3M-92.66M), B3 (chr13: 93.2M-93.37M), C (chr13: 107.84M-108M) and D (chr9: 130.72M-131.28M) can be automatically identified and assembled by NeoLoopFinder solely based on Hi-C data. c, Linear regression of the global distance averaged contact frequencies and local distance averaged contact frequencies within the indicated contact regions, for example, “D-C” represents the contacts between region D and region C in b. d-f, Assembly of another complex SV in K562 cells. g-i, Reconstruction of a complex SV in T47D cells.
Extended Data Fig. 5. Cancer-specific allele normalization and neo-TAD identification.
a-c, We show an example here in SK-N-MC cells. As shown in a, there is a 1.5 Mb deletion event (chr8: 127.88M, +; chr8: 129.37M, −). Since this is a heterozygous deletion, the chromatin interactions in area A3 are between loci (127.28M – 127.88M) and loci (129.37M – 129.97M) on one allele where the deletion happens. On the contrary, chromatin interactions within A1 (or A2) area are from all alleles and therefore, the overall intensities in area A3 are much weaker than A1 or A2. We need to normalize the signals so that we can predict neo-TADs and neo-Loops. a, Hi-C matrix without normalizing cancer-specific allele. The neo-TAD is undetectable as shown by the directionality index (DI) track. b, Reconstructed Hi-C map after cancer-specific allele normalization. Now the neo-TAD becomes detectable, and the number of detected neo-loops is also enhanced. c, Linear regression of the local distance averaged contact frequencies and the global distance averaged contact frequencies in different regions (A1, A2 and A3) of the Hi-C map in a. d-e, Detection of neo-TADs in 50 cancer cell lines or patient samples. d, The number of neo-TADs detected in each sample. e, Aggregate analysis of neo-TADs and distribution of breakpoint locations. Hi-C signals were distance-normalized, averaged, and centered at neo-TAD midpoints.
Extended Data Fig. 6. Comparison of loops predicted by FitHiC2 and NeoLoopFinder in SV regions.
a-c, Neo-loops predicted by NeoLoopFinder and FitHiC2 in LNCaP cells. a, Global view of the Hi-C map of chromosomes 7 and 14 shows that there is an inter-chromosomal translocation (marked by arrow). b, High-resolution Hi-C map showing contact frequencies between ETV1 and its hijacked enhancers on chr14. Blue circles indicate neo-loops identified by NeoLoopFinder. c, Significant interactions (blue circles) identified by FitHiC2. d-g, For this analysis, we used all the loops from ten cancer cell lines with DNase-Seq data available in ENCODE data portal (MCF7, A549, LNCaP, T47D, HL-60, KBM7, RPMI 8226, SK-N-MC, SW480 and K562). d, Upset plot of chromatin loops detected by NeoLoopFinder and FitHiC2 within SV regions. e, Chromatin accessibility around anchors of NeoLoopFinder-unique loops and FitHiC2-unique loops. f-g, Aggregate Peak Analysis (APA) for NeoLoopFinder-unique loops (f) or FitHiC2-unique loops (g).
Extended Data Fig. 7. Reconstruction of complex SVs in 50 cancer samples.
a, Number of assembled complex SVs in each sample. Only samples with complex SVs detected are shown. b, Size distributions of complex SV fragments, simple inversions and simple deletions/duplications. Data were merged from eight cancer cell lines: A549, Caki2, K562, LNCaP, NCI-H460, PANC-1, SK-N-MC, and T47D. For the boxplot, the box represents the interquartile range (IQR, Q3-Q1), the white dot represents the median, the upper whisker extends to the last datum less than Q3+1.5×IQR, and the lower whisker extends to the first datum greater than Q1-1.5×IQR. c, Percentage of transition between different SV types in a complex SV assembly, averaged from our analysis in 50 cancer cell lines/tissues.
Extended Data Fig. 8. Examples of neo-loop-involved genes.
a, (left) Reconstructed Hi-C map for the translocation (chr19: 6.62M, -; chr11: 118.91M, -) in a T-ALL (T-cell Acute Lymphoblastic Leukemia) patient. Blue circles indicate the predicted neo-loops. (middle) Kaplan-Meier survival analysis of TCGA leukemia patients with high (top 35%) and low (bottom 35%) expressions of the UPK2 gene. The means and the 95% confidential intervals are shown for each group of patients. The p-value was calculated from two-sided log-rank test. (right) Log2 converted copy number ratios of the UPK2 gene with high (top 35%) or low (bottom 35%) expressions in patients. The horizontal bar in each violin plot represents the median. Genes with higher expression levels do not have higher copies. The p-value was computed from two-sided Mann-Whitney U test. b-c, Similar examples in a gastric cancer patient T2000877. The Kaplan-Meier survival and copy-number analysis were performed using TCGA stomach adenocarcinoma patient data.
Extended Data Fig. 9. Genes with hijacked enhancers are up-regulated in cancer.
Quantile normalized gene expression signals (Transcripts Per Kilobase Million, TPM) are compared between cancer cells and corresponding normal cells. Here the genes with hijacked enhancers are defined as expressed neo-loop-involved genes (TPM > 1 in cancer cells) when there are at least one DNase-Seq peaks in the other anchor of the neo-loop. Each dot represents an individual gene. The p-values were computed using the two-sided Wilcoxon signed-rank test.
Extended Data Fig. 10. Application of the NeoLoopFinder framework to developmental diseases with genomic rearrangements.
a, CHi-C map reconstruction and neo-loop detection for an inversion event (inv1) in the mouse forelimb at embryonic day 11.5 (E11.5). Data were downloaded from Kragesteen BK et al. Nature Genetics 2018. (left) CHi-C map of the wild-type forelimb. The Pitx1 gene shows weak interactions with the Pen enhancer. Blue circles indicate the predicted chromatin loops by Peakachu. (middle) Original CHi-C map of the forelimb that contains a homozygous inversion of a 113-kb fragment containing Pen. (right) Reconstructed CHi-C map for the inversion. Note the neo-loop between Pitx1 and the Pen enhancer was correctly detected by NeoLoopFinder. b, CHi-C map reconstruction and neo-loop detection for a duplication event (Dup-C) in the mouse limb buds at E12.5. Data were downloaded from Franke M et al. Nature 2016. The duplicated region (blue and green arrows) contains both the Sox9 enhancers (marked by H3K27ac peaks) and the Kcnj2 gene. The rightmost panel shows the reconstructed chromatin interaction map near the duplication breakpoints. Yellow circles highlight the Kcnj2-involved neo-loops. There are also two more predicted neo-loops in this region (blue circles). c, CHi-C map reconstruction and neo-loop detection for an inversion event (InvC) in E12.5 limb buds. Data were downloaded from Despang A et al. Nature Genetics 2019. The inverted region (green bar in the middle panel) contains Sox9 enhancers (marked by H3K27ac peaks) and the TAD boundary separating the Sox9 enhancers and the Kcnj2 gene. The rightmost panel shows the reconstructed map for the whole region. The blue circles indicate the detected neo-loops, and yellow circles highlight the Kcnj2-involved neo-loops.
Supplementary Material
Supplementary Table 7 Details of the datasets collected for the validation and downstream analysis, including WGS, ChIP-Seq, DNase-Seq, RNA-Seq and Capture Hi-C datasets in various cell lines or tissues.
Supplementary Table 5 List of neo-loop-involved genes identified in each sample
Supplementary Table 8 List of the annotated enhancer hijacking events in 11 cancer cell lines: A549, K562, LNCaP, MCF7, T47D, HepG2, SK-MEL-5, NCI-H460, PANC-1, HT-1080, and C4-2B
Supplementary Table 1 Details of the 50 cancer Hi-C datasets analyzed in this study.
Supplementary Table 3 List of large SVs detected in each sample.
Supplementary Table 4 Genomic coordinates of the detected neo-loops in each sample.
Acknowledgements
F.Y. is supported by NIH grants R35GM124820 and R01HG009906. We thank Dr. Hongbo Yang and the rest of Yue lab for discussion.
Footnotes
Competing Interests
F.Y. and X.W. are listed as inventors of a provisional patent titled “Detection of chromatin interactions in re-arranged genomes.” F.Y. is a co-founder of Sariant Therapeutic, Inc.
References
- 1.Futreal PA et al. A census of human cancer genes. Nat Rev Cancer 4, 177–183, doi: 10.1038/nrc1299 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Consortium EP An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74, doi: 10.1038/nature11247 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernstein BE et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol 28, 1045–1048, doi: 10.1038/nbt1010-1045 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weischenfeldt J et al. Pan-cancer analysis of somatic copy-number alterations implicates IRS4 and IGF2 in enhancer hijacking. Nat Genet 49, 65–74, doi: 10.1038/ng.3722 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spielmann M, Lupianez DG & Mundlos S Structural variation in the 3D genome. Nat Rev Genet 19, 453–467, doi: 10.1038/s41576-018-0007-0 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Groschel S et al. A Single Oncogenic Enhancer Rearrangement Causes Concomitant EVI1 and GATA2 Deregulation in Leukemia. Cell 157, 369–381, doi: 10.1016/j.cell.2014.02.019 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Drier Y et al. An oncogenic MYB feedback loop drives alternate cell fates in adenoid cystic carcinoma. Nat Genet 48, 265–272, doi: 10.1038/ng.3502 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franke M et al. Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature 538, 265–269, doi: 10.1038/nature19800 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Northcott PA et al. The whole-genome landscape of medulloblastoma subtypes. Nature 547, 311–317, doi: 10.1038/nature22973 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang M et al. 13q12.2 deletions in acute lymphoblastic leukemia lead to upregulation of FLT3 through enhancer hijacking. Blood, doi: 10.1182/blood.2019004684 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ooi WF et al. Integrated paired-end enhancer profiling and whole-genome sequencing reveals recurrent CCNE1 and IGF2 enhancer hijacking in primary gastric adenocarcinoma. Gut 69, 1039–1052, doi: 10.1136/gutjnl-2018-317612 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Martin-Garcia D et al. CCND2 and CCND3 hijack immunoglobulin light-chain enhancers in cyclin D1(−) mantle cell lymphoma. Blood 133, 940–951, doi: 10.1182/blood-2018-07-862151 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haller F et al. Enhancer hijacking activates oncogenic transcription factor NR4A3 in acinic cell carcinomas of the salivary glands. Nat Commun 10, 368, doi: 10.1038/s41467-018-08069-x (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zimmerman MW et al. MYC Drives a Subset of High-Risk Pediatric Neuroblastomas and Is Activated through Mechanisms Including Enhancer Hijacking and Focal Enhancer Amplification. Cancer Discov 8, 320–335, doi: 10.1158/2159-8290.CD-17-0993 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ryan RJ et al. Detection of Enhancer-Associated Rearrangements Reveals Mechanisms of Oncogene Dysregulation in B-cell Lymphoma. Cancer Discov 5, 1058–1071, doi: 10.1158/2159-8290.CD-15-0370 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Northcott PA et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature 511, 428–434, doi: 10.1038/nature13379 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He B et al. Diverse noncoding mutations contribute to deregulation of cis-regulatory landscape in pediatric cancers. Sci Adv 6, eaba3064, doi: 10.1126/sciadv.aba3064 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lieberman-Aiden E et al. Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science 326, 289–293, doi: 10.1126/science.1181369 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang S et al. HiNT: a computational method for detecting copy number variations and translocations from Hi-C data. Genome Biol 21, 73, doi: 10.1186/s13059-020-01986-5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dixon JR et al. Integrative detection and analysis of structural variation in cancer genomes. Nat Genet 50, 1388-+, doi: 10.1038/s41588-018-0195-8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chakraborty A & Ay F Identification of copy number variations and translocations in cancer cells from Hi-C data. Bioinformatics 34, 338–345, doi: 10.1093/bioinformatics/btx664 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rao SSP et al. A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell 159, 1665–1680, doi: 10.1016/j.cell.2014.11.021 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imakaev M et al. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat Methods 9, 999–1003, doi: 10.1038/nmeth.2148 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang T et al. HiCRep: assessing the reproducibility of Hi-C data using a stratum-adjusted correlation coefficient. Genome Res 27, 1939–1949, doi: 10.1101/gr.220640.117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu HJ & Michor F A computational strategy to adjust for copy number in tumor Hi-C data. Bioinformatics 32, 3695–3701, doi: 10.1093/bioinformatics/btw540 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vidal E et al. OneD: increasing reproducibility of Hi-C samples with abnormal karyotypes. Nucleic Acids Res 46, e49, doi: 10.1093/nar/gky064 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Servant N, Varoquaux N, Heard E, Barillot E & Vert JP Effective normalization for copy number variation in Hi-C data. BMC Bioinformatics 19, 313, doi: 10.1186/s12859-018-2256-5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salameh TJ et al. A supervised learning framework for chromatin loop detection in genome-wide contact maps. Nat Commun 11, 3428, doi: 10.1038/s41467-020-17239-9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fudenberg G et al. Formation of Chromosomal Domains by Loop Extrusion. Cell Rep 15, 2038–2049, doi: 10.1016/j.celrep.2016.04.085 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanborn AL et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci U S A 112, E6456–6465, doi: 10.1073/pnas.1518552112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crane E et al. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature 523, 240–244, doi: 10.1038/nature14450 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dixon JR et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380, doi: 10.1038/nature11082 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Derderian C, Orunmuyi AT, Olapade-Olaopa EO & Ogunwobi OO PVT1 Signaling Is a Mediator of Cancer Progression. Front Oncol 9, 502, doi: 10.3389/fonc.2019.00502 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quereda V et al. Therapeutic Targeting of CDK12/CDK13 in Triple-Negative Breast Cancer. Cancer Cell 36, 545–558 e547, doi: 10.1016/j.ccell.2019.09.004 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Parolia A et al. Distinct structural classes of activating FOXA1 alterations in advanced prostate cancer. Nature 571, 413–418, doi: 10.1038/s41586-019-1347-4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuleshov MV et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44, W90–97, doi: 10.1093/nar/gkw377 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spangle JM et al. PI3K/AKT Signaling Regulates H3K4 Methylation in Breast Cancer. Cell Rep 15, 2692–2704, doi: 10.1016/j.celrep.2016.05.046 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baena E et al. ETV1 directs androgen metabolism and confers aggressive prostate cancer in targeted mice and patients. Gene Dev 27, 683–698, doi: 10.1101/gad.211011.112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gasi D et al. Overexpression of full-length ETV1 transcripts in clinical prostate cancer due to gene translocation. Plos One 6, e16332, doi: 10.1371/journal.pone.0016332 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kragesteen BK et al. Dynamic 3D chromatin architecture contributes to enhancer specificity and limb morphogenesis. Nat Genet 50, 1463–1473, doi: 10.1038/s41588-018-0221-x (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Despang A et al. Functional dissection of the Sox9-Kcnj2 locus identifies nonessential and instructive roles of TAD architecture. Nat Genet 51, 1263–1271, doi: 10.1038/s41588-019-0466-z (2019). [DOI] [PubMed] [Google Scholar]
- 42.Ay F, Bailey TL & Noble WS Statistical confidence estimation for Hi-C data reveals regulatory chromatin contacts. Genome Res 24, 999–1011, doi: 10.1101/gr.160374.113 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y et al. The 3D Genome Browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biol 19, 151, doi: 10.1186/s13059-018-1519-9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boeva V et al. Control-FREEC: a tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics 28, 423–425, doi: 10.1093/bioinformatics/btr670 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu J et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 173, 400–416 e411, doi: 10.1016/j.cell.2018.02.052 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang XT, Cui W & Peng C HiTAD: detecting the structural and functional hierarchies of topologically associating domains from chromatin interactions. Nucleic Acids Res 45, e163, doi: 10.1093/nar/gkx735 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abdennur N & Mirny LA Cooler: scalable storage for Hi-C data and other genomically labeled arrays. Bioinformatics 36, 311–316, doi: 10.1093/bioinformatics/btz540 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu W et al. CoolBox: a interactive genomic data explorer for Jupyter Notebook. bioRxiv, 614222, doi: 10.1101/614222 (2019). [DOI] [Google Scholar]
- 49.Ramirez F et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44, W160–165, doi: 10.1093/nar/gkw257 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaul A, Bhattacharyya S & Ay F Identifying statistically significant chromatin contacts from Hi-C data with FitHiC2. Nat Protoc 15, 991–1012, doi: 10.1038/s41596-019-0273-0 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 7 Details of the datasets collected for the validation and downstream analysis, including WGS, ChIP-Seq, DNase-Seq, RNA-Seq and Capture Hi-C datasets in various cell lines or tissues.
Supplementary Table 5 List of neo-loop-involved genes identified in each sample
Supplementary Table 8 List of the annotated enhancer hijacking events in 11 cancer cell lines: A549, K562, LNCaP, MCF7, T47D, HepG2, SK-MEL-5, NCI-H460, PANC-1, HT-1080, and C4-2B
Supplementary Table 1 Details of the 50 cancer Hi-C datasets analyzed in this study.
Supplementary Table 3 List of large SVs detected in each sample.
Supplementary Table 4 Genomic coordinates of the detected neo-loops in each sample.
Data Availability Statement
The cancer Hi-C datasets analyzed in this study are summarized in Supplementary Table 1. Details of all the other datasets collected for the validation and downstream analysis are summarized in Supplementary Table 7. Data used for survival analysis in leukemia and gastric cancer were downloaded from the cBioPortal for Cancer Genomics (https://www.cbioportal.org)45. The list of cancer-related genes was obtained from the Bushman Lab (http://www.bushmanlab.org/assets/doc/allOnco_May2018.tsv). The 4C-Seq data for the MYC gene promoter in SK-N-MC cells, and the Hi-C data generated for wild-type and enhancer-deleted LNCaP cells have been uploaded to the Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession code GSE161493. Source data are provided with this paper.