Telomerase is a ribonucleoprotein enzyme that catalyzes the addition of the telomeric repeats (TTAGGG)n onto the 3′-end of the human chromosomes by using reverse transcription and its intrinsic RNA as a template.1 This process prevents eukaryotic cells from telomere shortening during cell division and has thus been hypothesized to account for a cancer cell’s infinite life.2 Having been found in over 85% of all known human tumors, but not in neighboring normal cells, telomerase has been regarded as a biomarker for cancer diagnosis as well as a therapeutic target.3,4 To date, the most widely used and most sensitive telomerase detection assay is the polymerase chain reaction (PCR)-based telomeric repeat amplification protocol (TRAP).4 Although quite powerful, TRAP requires the use of DNA polymerases and is therefore susceptible to PCR-derived artifacts,4,5 especially when screening compounds for telomerase inhibition. A number of PCR-free assays for telomerase activity, based on direct probing6 or forming a sandwiched structure to the elongated strands,7 have been developed over the past decade; however, due to the lack of an amplification mechanism, most do not achieve a sensitivity comparable to TRAP.
The recently developed biobarcode assay,8 based on polyvalent oligonucleotide-functionalized gold nanoparticles (AuNPs),9,10 has exhibited advantages over conventional detection methods in detecting a variety of biomolecule targets including nucleic acids and proteins. A key to its extraordinary sensitivity is the use of AuNP probes, modified with hundreds of thiolated single-stranded barcode oligonucleotides, and magnetic microparticle probes (MMPs) to first efficiently capture and isolate targets. Signal amplification is then achieved through the release and detection of the barcode strands. Typically, a high sensitivity scanometric detection method8 is used to quantify the concentration of the barcode strands originally in solution after their release from the AuNPs. More traditional methods (e.g., fluorescence) can be used, as well, albeit with lower sensitivity. In this paper, we report a new assay for human telomerase that is fundamentally different but inspired, in part, by the concepts that underlie the barcode assay. The use of polyvalent oligonucleotide AuNPs in our new assay provides a novel means of signal amplification, with the limit of detection comparable to the PCR-based TRAP method.
Telomerase detection with this assay involves three basic steps (Scheme 1 and Supporting Information). First, telomerases extracted from HeLa cells are complexed to their substrate oligonucleotides, which were preconjugated to the surface of AuNPs (30 nm diameter). These oligonucleotide sequences serve as both the telomerase binding substrates (designated as TS) and the strands for subsequent signal amplification. Using their intrinsic RNAs as templates, the bound telomerases can catalytically elongate the DNA strands in TTAGGG repeats in the presence of deoxyribonucleotide triphosphate (dNTP) monomers from a solution of the four dNTPs (i.e., dATP, dCTP, dGTP, and dTTP). After the bound telomerases are removed with a sodium dodecyl sulfate (SDS) solution, MMPs bearing a sequence complementary to the telomerase-elongated sequence on the AuNP are added to the assay solution. The AuNPs hybridize with the MMPs to form structures that can be magnetically separated from the solution. After isolation, the AuNPs are dissolved with I2 to release the bound oligonucleotides, including the elongated and unmodified strands originally bound to the AuNPs. A microarray consisting of oligonucleotide spots on a glass surface is used to isolate and detect both the elongated and unmodified oligonucleotide strands. Subsequent silver development and amplification via the scanometric method allows one to detect the different strands using a Verigene ID light scattering reader system.8
Scheme 1.
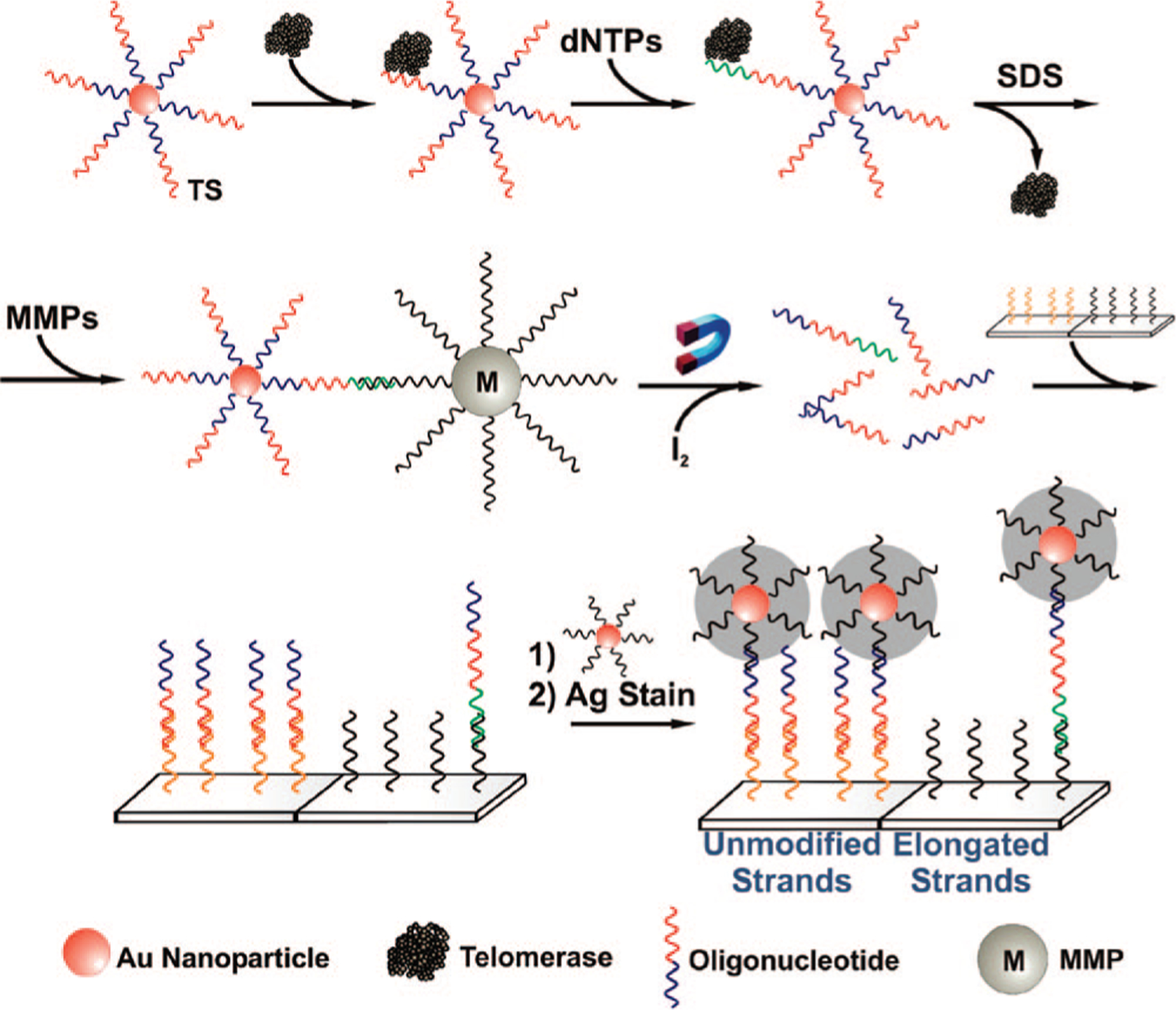
Polyvalent Oligonucleotide–AuNP Assay for Telomerase Enzymatic Activity: “TS” (= 5′ HS T20 AAT CCG TCG AGC AGA GTT 3′) Represents the Oligonucleotide Sequences Serving As the Telomerase Binding Substrate
The elongation of oligonucleotides on the AuNP surface by the enzymatic telomerase reaction was first characterized by fluorescence (Figure S1). The AuNPs with oligonucleotides complexed with telomerases (extracted from 104 HeLa cells) were incubated with biotin-labeled dATP and the other three types of dNTP monomers for various elongation times. Afterward, these oligonucleotide–AuNP conjugates were centrifuged and subsequently labeled with Cy5-modified streptavidin. The fluorescence spectra of the resulting conjugates showed a fluorescence band centered at 670 nm. The intensity of this band increased as a function of oligonucleotide elongation time (Figure S1), a consequence of an increasing amount of bound Cy5-streptavidin and therefore the number of elongated strands. In contrast, control experiments measured under otherwise identical conditions but in the absence of telomerase did not show any observable fluorescence at 670 nm, consistent with the conclusion that telomerase catalytically elongates oligonucleotides on the AuNP surfaces.
In a proof-of-concept detection experiment, telomerase samples extracted from 105, 104, 103, 102, and 10 HeLa cells were incubated with 200 pM oligonucleotide-functionalized AuNP probes (Supporting Information). In addition, a sample of cell extract from 105 normal human fibroblast cells was used as a control. Using a literature assay,11 we determined that the 30 nm diameter AuNPs have approximately 220 ± 30 strands. Because there is a large excess of AuNPs in the solution compared to the telomerase targets, only a small number of particles have oligonucleotides that become bound to telomerase and elongated. Addition of the MMPs leads to complexation of only the AuNPs with the elongated strands, which are subsequently separated with a magnetic field. After the release of bound oligonucleotides from AuNPs, both the elongated and unmodified strands were captured on a glass slide prespotted with their complementary sequences, and the signal was amplified by hybridization of a secondary AuNP probe and subsequent silver staining, as in a previously reported scanometric protocol.8 The resulting false color images allow one to quantify the number and type of captured oligonucleotide (Figure 1A,B). In each sample well, the left row of dots represents captured unmodified strands, while the right row of dots represents the elongated strands. The lowest concentration of telomerase detected using the elongated strands was extracted from 103 HeLa cells, while the lowest concentration detected by the unmodified strands was extracted from 10 cells, demonstrating that the sensitivity of this assay is comparable to the PCR-based detection for telomerase.4 The increased sensitivity, which arises from detecting the unmodified strands, is consistent with our expectation that the concentration of unmodified strands is much higher than elongated strands. Adding more cell extract to the AuNP solution was expected to increase the ratio of elongated to unmodified oligonucleotides, which was observed in the concentration range where both oligonucleotide strands could be detected (103 to 105 cell range). These complementary signals from both types of strands can serve as internal positive controls for each other and be useful for discriminating signal artifacts, which may arise from nonspecific binding, cross reactivity, or contamination.
Figure 1.

(A) Scanometric data (false color image) showing the detection of telomerase enzymatic activity. Telomerase samples were extracted from 10, 102, 103, 104, and 105 HeLa cells. The control sample was extracted from 105 normal human fibroblast cells. In each well, the dots in the left and right rows represent the signal resulting from the unmodified and elongated strands, respectively. (B) Bar graphs of the scanometric signal from A. (C) Bar graphs of a telomerase inhibition measurement from Figure S2. The quantities of AZT used are specified for each sample.
Finally, the inhibition of telomerase activity was measured by adding 3′-azido-3′-deoxythymidine (AZT), a known inhibitor of telomerase enzymatic activity,12 to the solution during the elongation step (Supporting Information, Figure S2). Scanometric analysis of the oligonucleotide concentrations shows decreasing signal intensity for both the elongated and unmodified oligonucleotide strands as a function of AZT concentration (Figure 1C). These results suggest the potential use of our assay for studies of telomerase inhibition.
In summary, we have developed a new PCR-free assay for telomerase activity that relies on polyvalent oligonucleotide–nanoparticle conjugates as probes and the concept of elongated and unmodified oligonucleotides on one particle for amplification. The assay can detect telomerase activity with as few as 10 HeLa cells, with on-chip positive and negative controls.
Supplementary Material
Acknowledgment.
C.A.M. acknowledges the AFOSR, DARPA, ONR, and NSF for the support of this research. C.A.M. is also grateful for the NIH Director’s Pioneer Award.
Footnotes
Supporting Information Available: Additional experimental details and supporting figures. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).(a) Morin GB Cell 1989, 59, 521–529. [DOI] [PubMed] [Google Scholar]; (b) Cohen SB; Graham ME; Lovrecz GO; Bache N; Robinson PJ; Reddel RR Science 2007, 315, 1850–1853. [DOI] [PubMed] [Google Scholar]; (c) Stone MD; Mihalusova M; O’Connor CM; Prathapam R; Collins K; Zhuang X Nature 2007, 446, 458–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).(a) Feng J; et al. Science 1995, 269, 1236–1241. [DOI] [PubMed] [Google Scholar]; (b) Finkel T; Serrano M; Blasco MA Nature 2007, 448, 767–774. [DOI] [PubMed] [Google Scholar]
- (3).(a) Shay JW; Bacchetti S Eur. J. Cancer 1997, 33, 787–791. [DOI] [PubMed] [Google Scholar]; (b) Hiyama E; Hiyama K Oncogene 2002, 21, 643–649. [DOI] [PubMed] [Google Scholar]
- (4).(a) Kim NW; Piatyszek MA; Prowse KR; Harley CB; West MD; Ho PLC; Coviello GM; Wright WE; Weinrich SL; Shay JW Science 1994, 266, 2011–2015. [DOI] [PubMed] [Google Scholar]; (b) Savoysky E; Akamatsu K; Tsuchiya M; Yamazaki T Nucleic Acids Res. 1996, 24, 1175–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Herbert B-S; Hochreiter AE; Wright WE; Shay JW Nat. Protocols 2006, 1, 1583–1590. [DOI] [PubMed] [Google Scholar]
- (5).(a) Niemeyer CM; Adler M; Wacker R Trends Biotechnol. 2005, 23, 208–216. [DOI] [PubMed] [Google Scholar]; (b) Lackey DB Anal. Biochem 1998, 263, 57–61. [DOI] [PubMed] [Google Scholar]
- (6).(a) Maesawa C; et al. Nucleic Acids Res. 2003, 31, e4. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Weizmann Y; Patolsky F; Lioubashevski O; Willner IJ Am. Chem. Soc 2004, 126, 1073–1080. [DOI] [PubMed] [Google Scholar]; (c) Zheng G; Patolsky F; Cui Y; Wang WU; Lieber CM Nat. Biotechnol 2005, 23, 1294–1301. [DOI] [PubMed] [Google Scholar]; (d) Eskiocak U; Ozkan-Ariksoysal D; Ozsoz M; Oktem HA Anal. Chem 2007, 79, 8807–8811. [DOI] [PubMed] [Google Scholar]
- (7).(a) Schmidt PM; Lehmann C; Matthes E; Bier FF Biosens. Bioelectron 2002, 17, 1081–1087. [DOI] [PubMed] [Google Scholar]; (b) Sato S; Kondo H; Nojima T; Takenaka S Anal. Chem 2005, 77, 7304–7309. [DOI] [PubMed] [Google Scholar]
- (8).(a) Nam JM; Thaxton CS; Mirkin CA Science 2003, 301, 1884–1886. [DOI] [PubMed] [Google Scholar]; (b) Hill HD; Mirkin CA Nat. Protocols 2006, 1, 324–336. [DOI] [PubMed] [Google Scholar]; (c) Stoeva SI; Lee JS; Thaxton CS; Mirkin CA Angew. Chem., Int. Ed 2006, 45, 3303–3306. [DOI] [PubMed] [Google Scholar]
- (9).(a) Mirkin CA; Letsinger RL; Mucic RC; Storhoff JJ Nature 1996, 382, 607–609. [DOI] [PubMed] [Google Scholar]; (b) Cao YC; Jin R; Mirkin CA Science 2002, 297, 1536–1540. [DOI] [PubMed] [Google Scholar]
- (10).(a) Nicewarner Peña SR; Raina S; Goodrich GP; Fedoroff NV; Keating CD J. Am. Chem. Soc 2002, 124, 7314–7323. [DOI] [PubMed] [Google Scholar]; (b) Liu J; Lu Y J. Am. Chem. Soc 2005, 127, 12677–12683. [DOI] [PubMed] [Google Scholar]; (c) Medley CD; Smith JE; Tang Z; Wu Y; Bamrungsap S; Tan W Anal. Chem 2008, 80, 1067–1072. [DOI] [PubMed] [Google Scholar]
- (11).Hurst SJ; Lytton-Jean AKR; Mirkin CA Anal. Chem 2006, 78, 8313–8318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).(a) Melana SM; Holland JF; Pogo BG-T Clin. Cancer Res 1998, 4, 693–696. [PubMed] [Google Scholar]; (b) White LK; Wright WE; Shay JW Trends Biotechnol. 2001, 19, 114–120. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


