Abstract
ATAC-seq, the assay for transposase-accessible chromatin using sequencing, is a quick and efficient approach to investigating the chromatin accessibility landscape. Investigating chromatin accessibility has broad utility for answering many biological questions, such as mapping nucleosomes, identifying transcription factor binding sites, and measuring differential activity of DNA regulatory elements. Because the ATAC-seq protocol is both simple and relatively inexpensive, there has been a rapid increase in the availability of chromatin accessibility data. Furthermore, advances in ATAC-seq protocols are rapidly extending its breadth to additional experimental conditions, cell types, and species. Accompanying the increase in data, there has also been an explosion of new tools and analytical approaches for analyzing it. Here, we explain the fundamentals of ATAC-seq data processing, summarize common analysis approaches, and review computational tools to provide recommendations for different research questions. This primer provides a starting point and a reference for analysis of ATAC-seq data.
Keywords: data analysis, ATAC-seq, chromatin accessibility, open chromatin, pipelines, bioinformatics tools
INTRODUCTION
As our understanding of gene regulation has improved, so has our awareness of the increasingly complex chromatin landscape that governs that regulation. Assays to better evaluate this landscape have been rapidly developed and improved, and the Assay for Transpose Accessible Chromatin using sequencing (ATAC-seq) has become a common first step for studying gene regulation. ATAC-seq interrogates chromatin openness, or chromatin accessibility, similar to earlier assays such as DNase-seq, MNase-seq, or FAIRE-seq (Nordström et al., 2019; Sheffield & Furey, 2012). These assays identify DNA regions that are accessible to external factors, which have been shown to correspond to regulatory elements, including promoters, enhancers, and other types of elements (Klemm, Shipony, & Greenleaf, 2019; Pálfy, Schulze, Valen, & Vastenhouw, 2020; Sheffield et al., 2013; Song et al., 2011; Thurman et al., 2012). Activity of regulatory elements varies spatially, temporally, and among cell types to influence the binding of transcription factors and the expression of target genes (Sheffield et al., 2013; Song et al., 2011). Studying the activity of regulatory elements promises to not only increase understanding of the fundamental biology of gene regulation, but also its influence on human health and disease (Chan et al., 2018; Corces et al., 2016; Corces, et al., 2018; Hatzi et al., 2019; Lara-Astiaso et al., 2014; Polak et al., 2015; Spivakov & Fraser, 2016; Tewari et al., 2012; Wang et al., 2018).
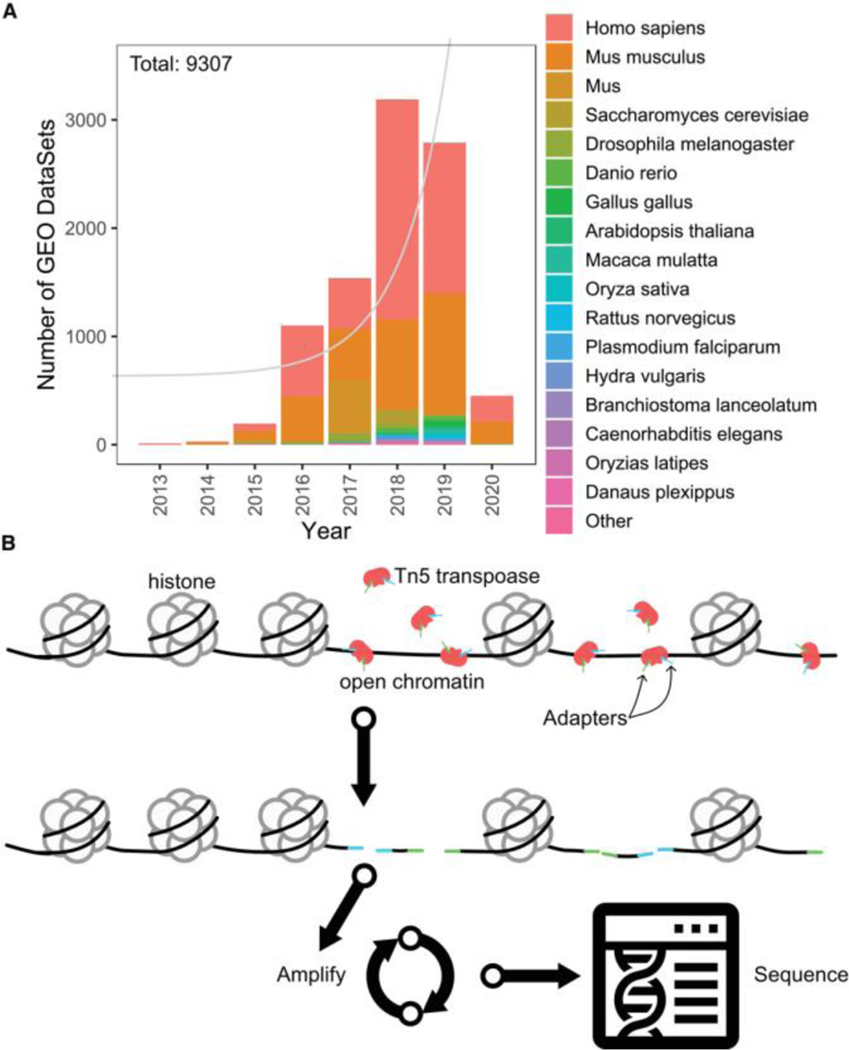
ATAC-seq has been adopted rapidly in the scientific community, with the number of studies using ATAC-seq approaching 10,000 in just a few years (Fig. 1A). The primary factor driving this adoption is efficiency, as ATAC-seq has dramatically improved the efficiency in cost, time, and required amount of sample over previous similar assays (Buenrostro, Giresi, Zaba, Chang, & Greenleaf, 2013). ATAC-seq relies on the activity of a hyperactive Tn5 transposase (Buenrostro et al., 2013; Reznikoff, 2008). This transposase is leveraged, through a process known as tagmentation (Adey et al., 2010), to simultaneously fragment the genome while inserting sequencing adapters (Buenrostro et al., 2013). These sequences can be PCR amplified and then sequenced using 2–4 orders of magnitude fewer cells, fewer protocol steps, and less time than analogous assays (Fig. 1B; Buenrostro et al., 2013; Chang, Gohain, Yen, & Chen, 2018). Protocols for ATAC-seq have improved since it was first introduced in 2013 (Buenrostro et al., 2013; Buenrostro, Wu, Chang, & Greenleaf, 2015), for example, with improved removal of contaminating mitochondrial DNA (Corces et al., 2017; Montefiori et al., 2017) and extension to single cells (Buenrostro, Wu, & Litzenburger, et al., 2015; Cusanovich et al., 2015; Cusanovich et al., 2018). As the protocol has developed and increased in popularity, analytical approaches have also been multiplying rapidly. Here, we provide guidance for both novice and experienced analysts on the advantages and limitations of ATAC-seq analysis pipelines, methods, and tools.
Figure 1.
(A) Increasing prevalence of ‘ATAC-seq’ DataSets in the Gene Expression Omnibus (GEO). Color = species; gray line = fitted exponential growth model. (B) Generalized ATAC-seq library preparation protocol.
FUNDAMENTALS OF ATAC-SEQ DATA ANALYSIS
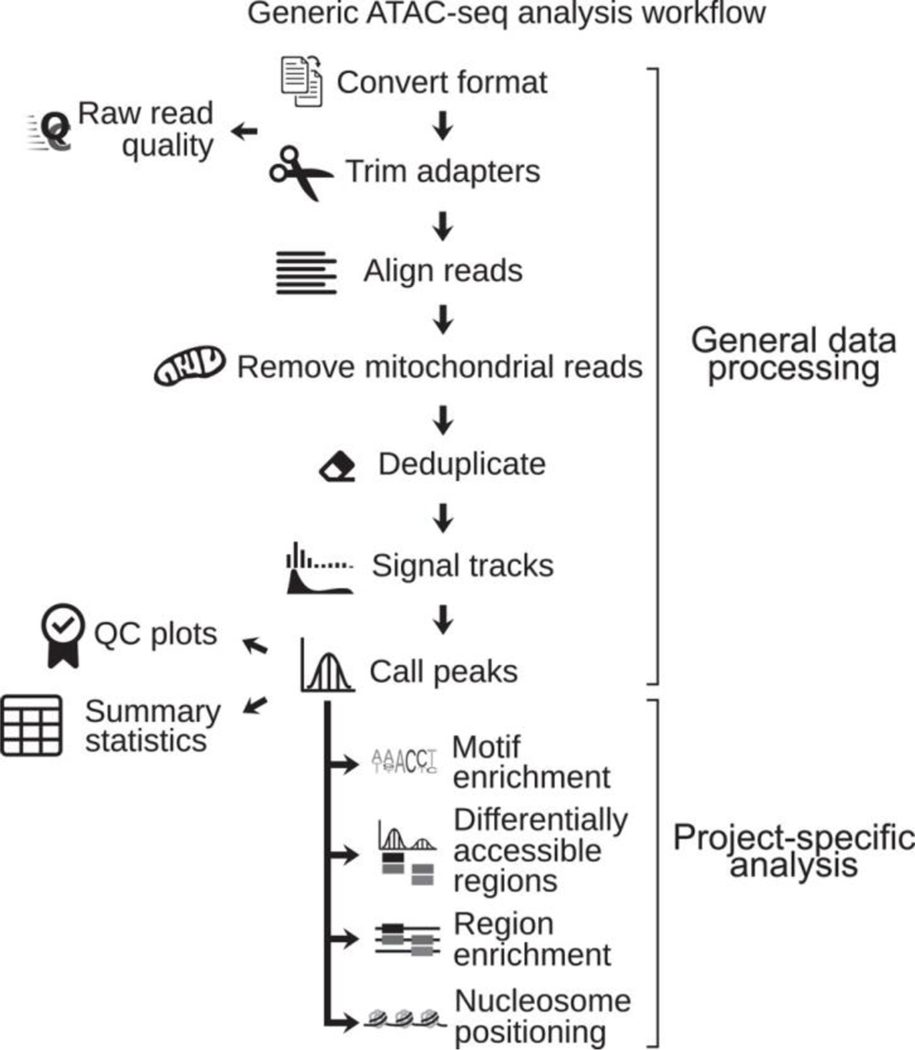
A typical ATAC-seq analysis can be divided into two major components: (1) general processing of raw sequencing reads, which produces intermediate outputs like annotated peak calls; and (2) detailed downstream analysis, which is more specific to a particular biological question (Fig. 2). In general, the first step is universal to all downstream analysis types, whereas the second step then requires more specialized software.
Figure 2.
ATAC-seq general workflow. Raw reads are processed through a series of steps to produce uniform intermediate results, which can then be further analyzed with more specific analyses relevant to a biological research question.
Alignment, Adapters, and Mitochondrial Reads
Analysis of ATAC data typically starts by processing raw sequences through a series of pipeline steps into outputs relevant to detailed biological questions (Fig. 2). A generalized workflow includes the following: first, reads are screened for quality, then adapter sequences are removed, and finally the reads are aligned to a reference assembly. After alignment, many pipelines are equipped to handle high mitochondrial DNA content, because ATAC-seq libraries are prone to high levels of mitochondrial DNA, which is typically considered undesirable. While recent protocol adaptations have succeeded in reducing mitochondrial DNA using optimized reagents (Corces et al., 2017; Rickner, Niu, & Cheng, 2019) or molecular biology techniques (Montefiori et al., 2017), many pipelines address this computationally by filtering out mitochondrial sequences. These sequences are removed through sequential alignments to mitochondrial DNA before genomic DNA, through removal of mitochondrial DNA from genome-wide genomic indices, or through blacklists of mitochondrial DNA after alignment. In our work, sequential alignment is the most accurate and computationally efficient way to eliminate mitochondrial contaminants–and also allows for later analysis of mitochondrial reads (Smith et al., 2020).
Removing Duplicates
Following adapter removal and alignment, pipelines remove read duplicates, although typical computational strategies may be overzealous in this approach if using only single-end sequencing data, since there is only a single end to compare. Single-end sequencing also provides less information, as it reduces the ability to identify PCR duplicates, which are typically removed. It also eliminates the ability to determine fragment lengths and whether identified fragments are therefore subnucleosomal or nucleosomal, which are important considerations if nucleosome positioning is of interest to the analyst. For these reasons, it is recommended to use paired-end ATAC-seq data when possible. After alignment and duplicate removal, low-quality, multi-mapping, or unmapped paired reads also typically get removed from downstream analyses.
Generating Signal Tracks
Once reads are aligned and filtered, they are shifted to accommodate the mechanics of transposase Tn5 activity (Adey et al., 2010; Buenrostro et al., 2013; Reznikoff, 2008). When the Tn5 transposase interacts with DNA, it effectively occupies about 9 bp of DNA and introduces the sequencing adapter at the 5' end of the interaction site. The Tn5 adapters are inserted in a staggered manner into the 5' ends of target sequence strands with a 9-bp gap between them (Adey et al., 2010; Buenrostro et al., 2013; Reznikoff, 2008). This means that the center of the Tn5 binding is actually 4 bp to the right of the edge on positive-strand reads, or 5 bp to the left on negative-strand reads. This shifting is intended to identify the center of the locus where Tn5 interaction occurred. An alternative approach is to account for the 9-bp size of the transposase binding event by mapping the reads as 9-bp insertion events instead of at nucleotide resolution. In either case, mapped reads are then transformed into signal tracks for visualization and further data analysis.
Peak Calling
As the goal of ATAC-seq is the identification of regions of accessible chromatin, and, by proxy, regulatory elements and sites of transcription factor binding, we must next identify those regions of interest. To do this, we identify areas of the genome that are enriched for aligned reads. These regions are identified and visualized as peaks. Calling peaks therefore represents the identification of regions of concentrated ATAC-seq signal that indicate regions of open chromatin. Peak calling necessitates choosing an appropriate peak-calling algorithm or tool that balances sensitivity and specificity of called peaks. User-defined settings can widely influence the number, width, and confidence of identified peaks (Bailey et al., 2013). Following the identification of peaks, they are typically broadly annotated into genomic partitions including known features such as promoters, exons, introns, or 3′ and 5′ UTR, among others.
Peak calling is typically the end of the general data processing pipeline that considers each sample independently. With signal tracks and called peaks for each sample, analysts are prepared for downstream analyses using more specialized analysis approaches that depend on specific user-defined biological questions.
Downstream Analysis
For detailed downstream analysis, the data is generally integrated across samples. These analyses include differential accessibility analysis, motif analysis, footprinting, and peak and region enrichment analysis. Because these analyses are more specific to particular biological questions, they are not typically performed by general-purpose ATAC-seq pipelines and must be manually performed for each study. Therefore, only a subset of these analyses will be relevant for a particular analysis, which should be determined before investing significant effort in a particular tool. We describe these analysis types in more detail in the next section.
SURVEY OF TOOLS FOR ATAC-SEQ ANALYSIS
Here, we present a survey of tools divided into classes based on their primary goal. This includes four classes geared toward general ATAC-seq data processing: step-by-step analysis guides, raw sequence pipelines and workflows, quality control, and peak calling tools. The remaining tools are for more detailed downstream analyses, which we divide into five additional categories: differential accessibility, motif enrichment and footprinting, nucleosome positioning, region enrichment, and single-cell analysis. The advantages and disadvantages of the tools vary widely, and some are targeted for novices while others require an experienced analyst. Our survey provides an overview of each analysis type, along with a table of some characteristics of relevant tools, such as mode of operation, language, and update frequency, along with a link to more information.
Step-by-Step Analysis Guides
For users who would prefer following a manual, stepwise procedure, several tutorials are available to walk a user through ATAC-seq data analysis (Table 1). These guides are a great starting point for an inexperienced user, as they explain how each step is manipulating raw data toward the goal of called peaks and further analyses. Users are required only to be able to work at the command line and have experience installing prerequisites. Examples include either formal classes available publicly (Steve Parker, Rockefeller University), training guides from public platforms (Delisle, Doyle, & Heyl, 2020), or guides from individual researchers sharing their own experiences (e.g., Yiwei Niu and John M. Gaspar). These step-by-step guides are primarily educational tools and are not intended to be automatic, re-usable pipelines that can be easily deployed on many samples across multiple projects; for this application, users will be more interested in the reusable pipelines described next.
Table 1.
Step by Step Guides to Performing ATAC-seq Data Analysis
| Title and author | Notes and link |
|---|---|
| ATAC-seq data analysis: from FASTQ to peaks | Blog style walkthrough of generalized ATAC-seq data analysis. |
| Yiwei Niu | |
| Last updated: 2019 | https://yiweiniu.github.io/blog/2019/03/ATAC-seq-data-analysis-from-FASTQ-to-peaks/ |
| BIOINF525 Lab 3.2 | Minimal standard ATAC-seq analysis walkthrough. |
| Steve Parker | |
| Last updated: 2016 | https://github.com/ParkerLab/ |
| Analysis of ATAC-seq data in R and Bioconductor | Bioconductor ATAC-seq analysis course. |
| Rockefeller Bioinformatics Resource | |
| Last updated: 2018 | https://rockefelleruniversity.github.io/RU_ATACseq/ |
| ATAC-seq | Generalized ATAC-seq analysis walkthrough with included custom scripts. |
| John M. Gaspar | |
| Last updated: 2019 | https://github.com/harvardinformatics/ATAC-seq |
| ATAC-seq data analysis | Galaxy training walkthrough of generalized ATAC-seq analysis. |
| Delisle L; Doyle M; & Heyl F | |
| Last updated: 2020 | https://galaxyproject.github.io/training-material/topics/epigenetics/tutorials/atac-seq/tutorial.html |
Raw Sequence Pipelines and Workflows
A more common need is a standardized pipeline to process raw data through fastq processing, alignment, peak calling, and signal track generation (Fig. 2). A number of raw data processing pipelines are available (Table 2). Many comprehensive pipelines now exist, with different target audiences. Some pipelines are geared toward the bench biologist, with graphical user interfaces (GUIs), including both open-source (I-ATAC, GUAVA) and commercial options (Basepair). While the GUI may simplify things for some users, these tools tend to have less documentation and also give less power to the user. The majority of raw data processing pipelines are executable at a command-line interface (CLI). Among these pipelines, there is a wide range of possible pipeline end-points. Some pipelines are geared toward doing only universal analysis, ending at annotated peaks to provide a starting point for more detailed downstream analysis. Other pipelines include substantial cross-sample analysis after peak calling. To delineate this distinction, we have categorized pipelines into two groups: entry-point pipelines provide a series of outputs intended as the beginning of a user-controlled downstream analysis, while end-point pipelines are intended as a complete analysis, running integrated analysis internally.
Table 2.
Raw ATAC-seq Data Processing Pipelines
| Language | Notes | Docs | Citation | |
|---|---|---|---|---|
| AIAP | Bash; R; Python | Optimized analysis with novel QC metrics | ++ |
Liu et al. (2019)
Last updated: 2019 |
| ATAC2GRN | Bash; Python | Parameter optimized ATAC-seq pipeline | + | Pranzatelli, Michael, & Chiorini (2018) |
| Last updated: 2018 | ||||
| ATAC-pipe | Python; R | Analysis pipeline for ATAC-seq data including TF footprinting; cell-type classification; and regulatory network creation | +++ | Zuo et al. (2019) |
| Last updated: 2019 | ||||
| ATACProc | Bash; Python; R | Complete pipeline with additional downstream analyses included | ++ | Unpublished Last updated: 2019 |
| Basepair | NA | Commercial. Web-based GUI for complete analysis | ? | Unpublished |
| CIPHER | R; Perl; Python | A data processing platform for ChIP-seq; RNA-seq; MNase-seq; DNase-seq; ATAC-seq; and GRO-seq datasets | + | Guzman & D’Orso (2017) |
| Last updated: 2017 | ||||
| ENCODE | Python; Bash | Complete pipeline following ENCODE standards for ATAC/DNase-seq analysis | ++ | Unpublished |
| Last updated: 2020 | ||||
| esATAC | R | Complete pipeline including downstream analyses | +++ | Wei, Zhang, Fang, Li, & Wang (2018) |
| Last updated: 2019 | ||||
| GUAVA | Java; Python; R | GUI based complete ATAC-seq pipeline | + |
Divate & Cheung (2018) Last updated: 2019 |
| I-ATAC | Java | GUI based interactive ATAC-seq pipeline | + |
Ahmed & Ucar (2017) Last updated: 2017 |
| nfcore/atacseq | Python; R | Complete pipeline build using Nextflow | +++ |
Ewels et al. (2019) Last updated: 2019 |
| PEPATAC | Python; R; Perl | Complete pipeline with unique analytical approaches and QC metrics | +++ | Unpublished Last updated: 2019 |
| pyflow-ATACseq | Bash; Python | ATAC-seq snakemake pipeline with included nucleosome positioning and TF footprinting | ++ | Unpublished |
| Last updated: 2019 | ||||
| snakePipes ATAC-seq | Python | Workflow system including ATAC-seq analysis | +++ |
Bhardwaj et al. (2019) Last updated: 2019 |
| Tobias Rausch | Bash; R; Python | Complete pipeline with emphasis on downstream analyses | ++ |
Rausch et al. (2019)
Last updated: 2020 |
Entry-point pipelines (AIAP, ENCODE, PEPATAC) are generally robust and reproducible, yielding consistent processing of few to many samples. This goal necessarily excludes some downstream steps–to improve efficiency and because not all researchers may wish to do all analyses all the time. This is particularly important if those additional procedures are not specific to the biological question being investigated. In that case, those additional procedures come at the increased cost of time and computational resources. All three of the entry-point pipelines include some level of shared and novel quality-control metrics to identify quality libraries with minimal project-specific analyses included.
The majority of the pipelines are end-point oriented, with substantial downstream processing following peak calling and signal track generation. The advantage of end-point pipelines is that they require the least additional effort for a complete analysis. These pipelines typically include the ability to incorporate sample structure (case versus control) for differential analysis of accessible regions, transcription factor binding sites, or motifs. However, the cost of this convenience is a lack of customizability, as the exact downstream analysis may or may not match the requirements of a particular study, and the exact settings and assumptions must be considered. Furthermore, the increased complexity of pipelines that include numerous downstream analyses may waste analysis time and computational resources if that analysis is irrelevant for the question under investigation.
Quality Control
Raw data processing pipelines have nearly universally adopted several standard quality control (QC) metrics. Briefly, these include QC of the raw and aligned sequence data, the distribution of aligned sequence fragments to confirm the presence of nucleosomes, measures of library complexity, the fraction of reads in peaks (FRiP), and the enrichment of reads at transcription start sites (TSS). Quality-control tools are dedicated tools that provide these and more advanced QC metrics (Table 3). Advanced metrics include the enrichment of promoter signal relative to gene body, measures of the proportion of nucleosome-free reads, and measures of signal to noise.
Table 3.
ATAC-seq Advanced Quality Control Metric Tools
| Languages | Notes | Docs | Citation | |
|---|---|---|---|---|
| ATAqC | Bash; Python | Generate ATAC-seq specific quality control metrics. | + | Unpublished Last updated: 2017 |
| ATACseqQC | R | Provides ATAC-seq specific quality control metrics and transcription factor footprinting. | +++ |
Ou et al. (2018) Last updated: 2018 |
| ataqv | C++; Bash | ATAC-seq QC and visualization. | +++ |
Orchard, Kyono, Hensley, Kitzman, & Parker (2020) Last updated: 2020 |
Peak Calling
Comprehensive ATAC-seq pipelines typically employ one of just a few widely adopted peak callers, which include tools originally developed for ChIP-seq or DNase-seq experiments, such as F-Seq (Boyle, Guinney, Crawford, & Furey, 2008), MACS (Zhang et al., 2008), or PeaKDEck (McCarthy & O’Callaghan, 2014). There are also other options built specifically for ATAC-seq data, including Genrich (Gaspar, 2018) and HMMRATAC (Tarbell & Liu, 2019; Table 4). The widely employed peak callers developed for ChIP-seq and DNase-seq experiments offer the advantage of years of demonstrated utility, support, and understanding of their strengths and weaknesses, but may neglect features of ATAC-seq data such as nucleosome positioning and transposase biases. Because ATAC-seq seeks to identify regions of open chromatin, the peak-calling step is critical, so there will likely continue to be effort dedicated to improving peak-calling tools and leveraging ATAC-specific data features to improve accuracy.
Table 4.
Peak Calling Tools
| Languages | Notes | Docs | Citation | |
|---|---|---|---|---|
| F-Seq | Java | Can be used as general peak caller to identify regions of open chromatin. | ++ |
Boyle et al. (2008) Last updated: 2016 |
| Genrich | C | Peak caller for genomic enrichment assays with specific ATAC-seq mode. | +++ | unpublished Last updated: 2020 |
| HMMRATAC | Java | Identify nucleosome positioning and leverage ATAC-seq specific read outs to call peaks. | +++ |
Tarbell & Liu (2019) Last updated: 2020 |
| Hotspot2 | C++ | Identify significantly enriched genomic regions. | ++ | Unpublished Last updated: 2019 |
| HOMER | Perl; C++ | Suite of tools that include the ability to call peaks from DNA enrichment assays. | +++ |
Heinz et al. (2010) Last updated: 2010 |
| MACS2 | Python | Specifically designed for CHiP-seq but broadly applicable to any DNA enrichment assay to call peaks. | +++ |
Zhang et al. (2020) Last updated: 2020 |
| PeaKDEck | Perl | Peak calling program for DNase-seq data. | +++ |
McCarthy & O’Callaghan (2014) Last updated: 2014 |
Differential Accessibility
ATAC-seq peaks correspond to regions of open chromatin, which have been shown to identify regulatory regions. One of the most common analyses is to identify differentially accessible regions. Analagous to identifying differential expression between two sample types, differential accessibility can demonstrate how gene regulation is governed in different biological settings. Typically, differential regions are identified by counting sequencing reads in individual peaks and then using mainstream count-based statistical tests to assess for statistical differences. Most analysis uses popular R packages for count-based data, such as edgeR (McCarthy, Chen, & Smyth, 2012; Robinson, McCarthy, & Smyth, 2010), DESeq2 (Love, Huber, & Anders, 2014), or DiffBind (Stark & Brown, 2011). While designed for other data types, e.g., RNA-seq, because ATAC-seq data is count-based, the statistical assumptions are often transferable.
After identifying differentially accessible regions, we typically want to better understand what factors are acting at these regions. A common follow-up is to identify which transcription factors are also differentially active between scenarios (Table 5). To accomplish this, there are at least two tools optimized to work with ATAC-seq data to identify differential transcription factor activity. By incorporating chromatin accessibility information and reported transcription factor binding sites, it becomes possible to identify differential TF activity (DAStk, Tripodi, Allen, & Dowell, 2018; diffTF, Berest et al., 2019). Should an experiment also include corresponding gene expression information, it is possible to then classify differential transcription factors as activators or repressors (Berest et al., 2019).
Table 5.
Tools to Investigate Differentially Accessible Regions
| Languages | Notes | Docs | Citation | |
|---|---|---|---|---|
| DAStk | Python | Identifies changes in transcription factor activity by looking at changes in chromatin accessibility | +++ |
Tripodi et al. (2018) Last updated: 2020 |
| diffTF | Python; R | Identifies differential transcription factors. Can operate in basic mode with just chromatin accessibility or in classification mode where it integrates RNA-seq. | +++ |
Berest et al. (2019) Last updated: 2020 |
Motif Enrichment and TF Footprinting
Another common analysis of differentially accessible regions is de novo motif analysis, which entails looking for an overrepresentation of transcription factor motifs in regions of interest relative to some background set. Motif discovery is typically used in analysis of ChIP-seq data, but is also relevant for accessible chromatin peaks with some specificity, such as for a particular cell type or treatment. Motif discovery has been an ongoing field of study for decades, and there are many tools to identify enriched motifs (Bailey et al., 2009; Berest et al., 2019; Galas & Schmitz, 1978; Heinz et al., 2010; Tripodi et al., 2018). Tools initially designed for ChIP-seq or DNase-seq experiments have been widely applied to ATAC-seq data as well (MEME Suite, Bailey et al., 2009; HOMER, Heinz et al., 2010). There are now dozens or hundreds of individual motif-finding tools (Hashim, Mabrouk, & Al-Atabany, 2019).
A related approach called footprinting explores the microarchitecture of reads within peaks to identify physical evidence of bound transcription factors that decrease the accessibility at small binding sites (typically under 20 bp) within an overall area of higher accessibility (Table 6; Vierstra & Stamatoyannopoulos, 2016). Following the introduction and rapid adoption of DNase-seq, the number of tools to perform TF footprinting rapidly expanded. A number of these were designed for DNase-seq, but have often been employed using ATAC-seq data successfully (CENTIPEDE, Pique-Regi et al., 2011; PIQ, Sherwood et al., 2014; DNase2TF, Sung, Guertin, Baek, & Hager, 2014; BinDNase, Kähärä & Lähdesmäki, 2015; Wellington, Piper et al., 2013; Piper, Elze, et al., 2013; TRACE, Ouyang & Boyle, 2019). One advantage of using tools designed for DNase-seq simply lies in their demonstrated utility, even when applied to ATAC-seq data. Yet, there are unique features of ATAC-seq data including nucleosome positioning information and transposase cleavage biases that can be used to inform on TF footprinting. Research has shown that biases and transcription factor dynamics must be carefully considered when interpreting results of footprinting analysis, whether from DNase-seq or ATAC-seq assays (Calviello et al., 2019; Martins, Walavalkar, Anderson, Zang, & Guertin, 2017; Sung, Baek, & Hager, 2016). Newer tools either have specific settings to work with ATAC-seq data, or were designed specifically for ATAC-seq and may be more appropriate going forward (DeFCoM, Quach & Furey, 2017; TOBIAS, Bentsen et al., 2019; HINT-ATAC, Li, Schulz, et al., 2019; BiFET, Youn, Marquez, Lawlor, Stitzel, & Ucar, 2019).
Table 6.
Motif Enrichment and Transcription Factor Footprinting Tools
| Languages | Notes | Docs | Citation | |
|---|---|---|---|---|
| BiFET | R | Identify overrepresented transcription factor footprints. | ++ |
Youn et al. (2019) Last updated: 2019 |
| BinDNase | R | Transcription factor binding prediction using DNase-seq. | + |
Kähärä & Lähdesmäki (2015) Last updated: 2015 |
| CENTIPEDE | R | Transcription factor footprinting and binding site prediction. | ++ |
Pique-Regi et al. (2011) Last updated: 2010 |
| DeFCoM | Python | Detecting transcription factor footprints and underlying motifs using supervised learning. | +++ |
Quach & Furey (2017) Last updated: 2017 |
| DNase2TF | R | Identify footprint candidates from DNase-seq data on user-specified regions. | + |
Sung et al. (2014) Last updated: 2017 |
| HINT-ATAC | Python | Use open chromatin data to identify transcription factor footprints with modifications specific to ATAC-seq data. | +++ |
Li et al. (2019) Last updated: 2019 |
| HOMER | Perl; C++ | A suite of tools for motif discovery and enrichment. | +++ |
Heinz et al. (2010) Last updated: 2019 |
| MEME Suite | Perl; Python | Suite of tools for motif discovery; enrichment; and GO term analyses. | +++ |
Bailey et al. (2009) Last updated: 2020 |
| PIQ | Bash; R | Models genome-wide DNase profiles to identify transcription factor binding sites. | ++ |
Sherwood et al. (2014) Last updated: 2016 |
| TOBIAS | Python | Identify transcription factor footprints. | ++ |
Bentsen et al. (2019) Last updated: 2020 |
| TRACE | Python | Transcription factor footprinting. | ++ |
Ouyang & Boyle (2019) Last updated: 2020 |
| Wellington | Python | Identify TF footprints using DNase-seq data. | +++ |
Piper et al. (2013) Last updated: 2019 |
Nucleosome Positioning
Nucleosome positioning is crucial in a number of DNA regulatory processes, particularly gene expression, and may be directly interrogated using ATAC-seq data (Radman-Livaja & Rando, 2010; Schep et al., 2015; Struhl & Segal, 2013). ATAC-seq is designed to assay regions of open chromatin–in other words, to identify regions not currently packaged into nucleosomes. As a consequence of this, sequenced fragment lengths and alignments occur in structured patterns that inform on the presence and positioning of nucleosomes (Table 7). Essentially, short ATAC-seq fragments represent nucleosome-free regions, and longer fragments represent nucleosome-associated DNA (Buenrostro et al., 2013). The earliest tool, NucleoATAC (Schep et al., 2015) reports the position and occupancy of nucleosomes. Building on the fact that this information is inherent in ATAC-seq data, later tools have extended the biological information that can be obtained from a more thorough understanding of nucleosome positioning. The use of nucleosome positioning information may now be easily compared between sample conditions, which ultimately allows for concurrent identification of transcription factor binding sites alongside additional epigenetic marks (NucTools, Vainshtein, Rippe, & Teif, 2017). Furthermore, this information may be leveraged to improve peak calling by incorporating nucleosome positioning and enrichment to more accurately predict true positive open chromatin (HMMRATAC, Tarbell & Liu, 2019).
Table 7.
Tools to Investigate Nucleosome Positioning
| Languages | Notes | Docs | Citation | |
|---|---|---|---|---|
| HMMRATAC | Java | Identify nucleosome positioning and leverage ATAC-seq specific read outs to call peaks. | +++ |
Tarbell & Liu (2019) Last updated: 2020 |
| NucleoATAC | Python; R | Call nucleosomes using ATAC-seq data. | +++ |
Schep et al. (2015) Last updated: 2019 |
| NucTools | Perl; R | Calculate nucleosome occupancy profiles on chromatin accessibility data. | +++ |
Vainshtein et al. (2017) Last updated: 2019 |
Region Enrichment
A widely successful analysis type for gene expression data is gene ontology analysis or gene set enrichment analysis, which can be extended to region-based enrichments. In this context, instead of genes as the units of interest, the analysis is done on non-coding regions corresponding to regulatory elements. As chromatin accessibility has increased, so has interest in assigning biological meaning to non-coding loci. Region-set enrichment analyses are one approach to this problem. Generally, these tools compare a set of regions of interest (i.e., called peaks) to regions with known biological function. The tools then assess similarity to determine whether there are significant enrichments of overlap between the region sets. This approach can function by identifying significantly enriched GO terms (GREAT, McLean et al., 2010) and/or by comparing any previously annotated region set with your unknown peak set (regioneR, Gel et al., 2016; LOLA, Sheffield & Bock, 2016; annotatr, Cavalcante & Sartor, 2017; GIGGLE, Layer et al., 2018). Therefore, to assign more meaningful biological relationships to annotated ATAC-seq peaks, one can investigate what specific biological features are correlated or enriched in your peak set (Table 8). These tools and other related tools have been reviewed elsewhere in detail (Dozmorov, 2017; Simovski et al., 2018).
Table 8.
Tools to Investigate Region Enrichement
| Languages | Notes | Docs | Citation | |
|---|---|---|---|---|
| Annotatr | R | Annotate summarize and visualize genomic regions. | +++ |
Cavalcante & Sartor (2017) Last updated: 2019 |
| BART/BARTweb | Python | Predict factors that bind at cis-regulatory regions. | +++ |
Wang et al. (2018) Last updated: 2020 |
| chipenrich | R | Perform gene set enrichment testing using genomic regions. | +++ |
Welch et al. (2014) Last updated: 2020 |
| coloc-stats | Python | Perform co-localization analysis of genomic regions. | +++ |
Simovski et al. (2018) Last updated: 2019 |
| COLO | JSP | Identify genomic features in close proximity to user-submitted genomic regions. | ++ |
Kim et al. (2015) Last updated: 2015 |
| FEATnotator | Perl; R | Annotate genomic regions. | ++ |
Podicheti & Mockaitis (2015) Last updated: 2018 |
| GenomeRunner | .NET | Perform annotation and enrichment of genomic regions against default or custom regulatory regions. | ++ |
Dozmorov et al. (2016) Last updated: 2016 |
| GenometriCorr | R | Determine spatial correlation between region sets. | ++ |
Favorov et al. (2012) Last updated: 2020 |
| Genomic Association Tester | Python | Calculate the significance of overlaps between multiple genomic region sets. | +++ |
Heger et al. (2013) Last updated: 2019 |
| GIGGLE | C | Genomics search engine to uncover significantly shared genomic loci (regions) between data. | +++ |
Layer et al. (2018) Last updated: 2019 |
| GLANET | Java; Perl | Genomic loci annotation and enrichment tool between sets of genomic regions. | +++ |
Otlu et al. (2017) Last updated: 2019 |
| GREAT | C | Annotate genomic regions. | +++ |
McLean et al. (2010) Last updated: 2019 |
| LOLA/LOLAweb | R | Determine significant enrichment between region sets to inform on biological meaning. | +++ |
Sheffield & Bock (2016) Last updated: 2019 |
| regioneR | R | Evaluate significant associations between region sets using permutation testing. | +++ |
Gel et al. (2016) Last updated: 2020 |
| StereoGene | C++; R | Estimate genome-wide correlation between pairs of genomic features. | ++ |
Stavrovskaya et al. (2017) Last updated: 2019 |
Single-Cell
Although single-cell ATAC-seq (scATAC-seq) is only a few years old (Buenrostro, Wu, & Litzenburger et al., 2015; Cusanovich et al., 2015), the number of available analysis tools has proliferated rapidly (Table 9). A primary challenge to any single-cell sequencing assay is the sparsity of data. For that reason, modifications to general ATAC-seq data processing are necessary. Tools specific to single-cell ATAC-seq analysis include both raw processing pipelines (Cell Ranger ATAC; BROCKMAN, de Boer & Regev, 2018; Scasat, Baker et al., 2019; SnapATAC, Fang et al., 2019; scATAC-pro, Yu, Uzun, Zhu, Chen, & Tan, 2019) and downstream analysis tools, particularly for clustering individual cells into separate cell-type populations (BAP, Lareau et al., 2019; scABC, Zamanighomi et al., 2018; SCALE, Xiong et al., 2019) and identifying transcription factor accessibility (SCRAT, Ji, Zhou, & Ji, 2017; chromVAR, Schep, Wu, Buenrostro, & Greenleaf, 2017; Cicero, Pliner et al., 2018; cisTopic, Bravo González-Blas et al., 2019; scOpen, Li & Kuppe, et al., 2019). Single-cell ATAC-seq analysis is a rapidly changing area, with many of these tools published only within the past year.
Table 9.
Available Tools for Single-Cell ATAC-seq Data Processing
| Languages | Notes | Docs | Citation | |
|---|---|---|---|---|
| BAP | R; Python | Bead-based scATAC-seq data processing. | ++ |
Lareau et al. (2019) Last updated: 2019 |
| BROCKMAN | R; Bash; Ruby | Convert genomics data into K-mer words associated with chromatin marks used to compare and identify changes across samples. | ++ |
de Boer & Regev (2018) Last updated: 2018 |
| Cell Ranger ATAC | NA | Commercial. Set of analysis pipelines for Chromium single cell ATAC-seq. | +++ | Unpublished |
| chromVAR | R | Identify transcription factor accessibility in single-cell data. Enables clustering of single-cell ATAC-seq data. | +++ |
Schep et al. (2017) Last updated: 2019 |
| Cicero | R | Predict cis-regulatory DNA interactions using single-cell chromatin accessibility data. | +++ |
Pliner et al. (2018) Last updated: 2019 |
| cisTopic | R | Identify cell states and cis-regulatory topics from single-cell data. | +++ |
Bravo González-Blas et al.(2019) Last updated: 2019 |
| scABC | R | Classify single-cell ATAC using unsupervised clustering and identify chromatin regions specific to cell identity. | + |
Zamanighomi et al. (2018) Last updated: 2019 |
| SCALE | Python | Clustering and visualization of single-cell ATAC-seq data into interpretable cell populations. | ++ |
Xiong et al. (2019) Last updated: 2019 |
| Scasat | Bash; Python; R | Complete pipeline to process scATAC-seq data with simple steps. | +++ |
Baker et al. (2019) Last updated: 2019 |
| scATAC-pro | R; Python | Comprehensive pipeline for single cell ATAC-seq analysis. | +++ |
Yu et al. (2019) Last updated: 2020 |
| scOpen | Python | Chromatin-accessibility estimation of single-cell ATAC data. | + |
Li et al. (2019) Last updated: 2020 |
| SCRAT | R | Useful for studying single cell heterogeneity. Can identify changes in gene sets or transcription factor binding sites. Includes GUI and web-based service. | +++ |
Ji et al. (2017) Last updated: 2018 |
| SnapATAC | R; Python | Single Nucleus Analysis Pipeline for ATAC-seq. | +++ |
Fang et al. (2019) Last updated: 2019 |
CONCLUSION
Chromatin accessibility analysis is becoming increasingly relevant for a range of biological research areas. As scientists realize the richness of chromatin accessibility data, new analytical approaches and tools are being developed. At the same time, chromatin accessibility analysis is now approachable by individuals with a wider range of perspective and experience. This has led to a wide increase in biological results, tools, and analytical approaches.
In our survey of ATAC-seq analysis tools, we identified more than 50 tools employed specifically for ATAC-seq data analysis. In assessing this diverse range of tools, we have found it useful to categorize them by primary aim. Because the diversity and number of available tools and approaches is likely only to increase as ATAC-seq analysis becomes mainstream, we believe it will be important to continue to revisit such tool surveys as the field develops. To address this, we maintain an expanding list of ATAC-seq tools at https://github.com/databio/awesome-atac-analysis. These summaries provide novices with a basic understanding and starting point, and also give experienced analysts a reference resource to provide ideas for more detailed analysis.
ACKNOWLEDGMENTS
J.P.S. was supported by the institutional training grant GM008136 and by National Institute of General Medical Sciences grant GM128636 (NCS).
LITERATURE CITED
- Adey A, Morrison HG, Asan Xun, X., Kitzman JO, Turner EH, …Shendure J(2010). Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biology, 11, R119. doi: 10.1186/gb-2010-11-12-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed Z, & Ucar D. (2017). I-ATAC: Interactive pipeline for the management and pre-processing of ATAC-seq samples. PeerJ, 5, e4040. doi: 10.7717/peerj.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, … Noble WS (2009). MEME suite: Tools for motif discovery and searching. Nucleic Acids Research, 37, W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T, Krajewski P, Ladunga I, Lefebvre C, Li Q, Liu T, … Zhang J. (2013). Practical guidelines for the comprehensive analysis of chip-seq data. PLoS Computational Biology, 9, e1003326. doi: 10.1371/journal.pcbi.1003326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SM, Rogerson C, Hayes A, Sharrocks AD, … Rattray M. (2019) Classifying cells with scasat, a single-cell ATAC-seq analysis tool. Nucleic Acids Research, 47, e10–e10. doi: 10.1093/nar/gky950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj V, Heyne S, Sikora K, Rabbani L, Rauer M, Kilpert F, … Manke T. (2019). snakePipes: facilitating flexible, scalable and integrative epigenomic analysis. Bioinformatics, 35, 4757–4759. doi: 10.1093/bioinformatics/btz436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsen M, Goymann P, Schultheis H, Klee K, Petrova A, Wiegandt R, … Looso M. (2019). Beyond accessibility: ATAC-seq footprinting unravels kinetics of transcription factor binding during zygotic genome activation. bioRxiv, 869560.doi: 10.1101/869560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berest I, Arnold C, Reyes-Palomares A, Palla G, Rasmussen KD, Giles H, … Zaugg JB (2019). Quantification of differential transcription factor activity and multiomics-based classification into activators and repressors: DiffTF. Cell Reports, 29, 3147–3159.e12. doi: 10.1016/j.celrep.2019.10.106. [DOI] [PubMed] [Google Scholar]
- Boyle AP, Guinney J, Crawford GE, & Furey TS (2008). F-seq: A feature density estimator for high-throughput sequence tags. Bioinformatics, 24, 2537–2538. doi: 10.1093/bioinformatics/btn480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo González-Blas C, Minnoye L, Papasokrati D, Aibar S, Hulselmans G, Christiaens V, … Aerts S. (2019). CisTopic: Cis-regulatory topic modeling on single-cell ATAC-seq data. Nature Methods, 16, 397–400. doi: 10.1038/s41592-019-0367-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Chang HY, & Greenleaf WJ (2015). ATAC-seq: A method for assaying chromatin accessibility genomewide. Current Protocols in Molecular Biology, 109, 21.29.1–21.29.9. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu W, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, … Greenleaf WJ (2015). Single-cell chromatin accessibility reveals principles of regulatory variation. Nature, 523, 486–490. doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, & Greenleaf WJ (2013). Transposition of native chromatin for multimodal regulatory analysis and personal epigenomics. Nature Methods, 10, 1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calviello AK, Hirsekorn A, Wurmus R, Yusuf D, & Ohler U. (2019). Reproducible inference of transcription factor footprints in ATAC-seq and DNase-seq datasets using protocol-specific bias modeling. Genome Biology, 20, 42. doi: 10.1186/s13059-019-1654-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalcante RG, & Sartor MA (2017). Annotatr: Genomic regions in context. Bioinformatics, 33, 2381–2383. doi: 10.1093/bioinformatics/btx183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan HL, Beckedorff F, Zhang Y, Garcia-Huidobro J, Jiang H, Colaprico A, … Morey L. (2018). Polycomb complexes associate with enhancers and promote oncogenic transcriptional programs in cancer through multiple mechanisms. Nature Communications, 9, 3377. doi: 10.1038/s41467-018-05728-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P, Gohain M, Yen M-R, & Chen P-Y (2018). Computational methods for assessing chromatin hierarchy. Computational and Structural Biotechnology Journal, 16, 43–53. doi: 10.1016/j.csbj.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces MR, Buenrostro JD, Wu B, Greenside PG, Chan SM, Koenig JL, … Chang HY (2016). Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nature Genetics, 48, 1193–1203. doi: 10.1038/ng.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna S, … Chang HY (2017). An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Scientific Reports, 14, 959–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces MR, Granja JM, Shams S, Louie BH, Seoane JA, Zhou W, … Chang HY (2018). The chromatin accessibility landscape of primary human cancers. Science, 362, eaav1898. doi: 10.1126/science.aav1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusanovich DA, Daza R, Adey A, Pliner HA, Christiansen L, Gunderson KL, … Shendure J. (2015). Multiplex single-cell profiling of chromatin accessibility by combinatorial cellular indexing. Science, 348, 910–914. doi: 10.1126/science.aab1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusanovich DA, Hill AJ, Aghamirzaie D, Daza RM, Pliner HA, Berletch JB, & Shendure J. (2018). A single-cell atlas of in vivo mammalian chromatin accessibility. Cell, 174(5), 1309–1324.e18. doi: 10.1016/j.cell.2018.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer CG, & Regev A. (2018). BROCKMAN: Deciphering variance in epigenomic regulators by k-mer factorization. BMC Bioinformatics, 19(1), 253. doi: 10.1186/s12859-018-2255-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delisle L, Doyle M, & Heyl F. (2020). ATACseq data analysis (galaxy training materials). Available at: https://galaxyproject.github.io/training-material/topics/epigenetics/tutorials/atac-seq/tutorial.html. [Google Scholar]
- Divate M, & Cheung E. (2018). GUAVA: A graphical user interface for the analysis and visualization of ATAC-seq data. Frontiers in Genetics, 9, 250. doi: 10.3389/fgene.2018. 00250eCollection2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dozmorov MG (2017). Epigenomic annotation-based interpretation of genomic data: From enrichment analysis to machine learning. Bioinformatics, 33, 3323–3330. doi: 10.1093/bioinformatics/btx414. [DOI] [PubMed] [Google Scholar]
- Dozmorov MG, Cara LR, Giles CB, & Wren JB (2016). GenomeRunner web server: Regulatory similarity and differences define the functional impact of SNP sets. Bioinformatics, 32, 2256–2263. doi: 10.1093/bioinformatics/btw169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewels PA, Peltzer A, Fillinger S, Alneberg JA, Patel H, Wilm A, … Nahnsen S. (2019). nf-core: Community curated bioinformatics pipelines. bioRxiv, 610741. doi: 10.1101/610741. [DOI] [PubMed] [Google Scholar]
- Fang R, Preissl S, Hou X, Lucero J, Wang X, Motamedi A, … Ren B. (2019). Fast and accurate clustering of single cell epigenomes reveals cis-regulatory elements in rare cell types. bioRxiv, 615179. [Google Scholar]
- Favorov A, Mularoni L, Cope LM, Medvedeva Y, Mironov AA, Makeev VJ, & Wheelan SJ (2012). Exploring massive, genome scale datasets with the GenometriCorr package. PloS Computational Biology, 8(5), e1002529. doi: 10.1371/journal.pcbi.1002529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas DJ, & Schmitz A. (1978). DNAse footprinting: A simple method for the detection of protein-DNA binding specificity. Nucleic Acids Research, 5, 3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar JM (2018). Genrich: Detecting sites of genomic enrichment. Available at https://github.com/jsh58/Genrich. [Google Scholar]
- Gel B, Díez-Villanueva A, Serra E, Buschbeck M, Peinado MA, & Malinverni R. (2016). RegioneR: An R/bioconductor package for the association analysis of genomic regions based on permutation tests. Bioinformatics, 32, 289–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman C, & D’Orso I. (2017). CIPHER: A flexible and extensive workflow platform for integrative next-generation sequencing data analysis and genomic regulatory element prediction. BMC Bioinformatics, 18(1), 363. doi: 10.1186/s12859-017-1770-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashim FA, Mabrouk MS, & Al-Atabany W. (2019). Review of different sequence motif finding algorithms. Avicenna Journal of Medical Biotechnology, 11, 130. [PMC free article] [PubMed] [Google Scholar]
- Hatzi K, Geng H, Doane AS, Meydan C, LaRiviere R, Cardenas M, … Melnick AM (2019). Histone demethylase LSD1 is required for germinal center formation and BCL6-driven lymphomagenesis. Nature Immunology, 20, 86–96. doi: 10.1038/s41590-018-0273-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heger A, Webber C, Goodson M, Ponting CP, & Lunter G. (2013). GAT: A simulation framework for testing the association of genomic intervals, Bioinformatics, 29(16), 2046–2048. doi: 10.1093/bioinformatics/btt343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, … Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular Cell, 38, 576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Zhou W, & Ji H. (2017). Single-cell regulome data analysis by SCRAT. Bioinformatics, 33, 2930–2932. doi: 10.1093/bioinformatics/btx315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kähärä J, & Lähdesmäki H. (2015). BinDNase: A discriminatory approach for transcription factor binding prediction using DNase I hypersensitivity data. Bioinformatics, 31, 2852–2859. doi: 10.1093/bioinformatics/btv294. [DOI] [PubMed] [Google Scholar]
- Kim R, Smith OK, Wong WC, Ryan AM, Ryan MC, & Aladjem MI (2015). ColoWeb: A resource for analysis of colocalization of 1genomic features. BMC Genomics, 16, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm SL, Shipony Z, & Greenleaf WJ (2019). Chromatin accessibility and the regulatory epigenome. Nature Reviews Genetics, 20, 207–220. doi: 10.1038/s41576-018-0089-8. [DOI] [PubMed] [Google Scholar]
- Lara-Astiaso D, Weiner A, Lorenzo-Vivas E, Zaretsky I, Jaitin DA, David E, … Amit I. (2014). Chromatin state dynamics during blood formation. Science, 345, 943–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau CA, Duarte FM, Chew JG, Kartha VK, Burkett ZD, Kohlway AS, … Buenrostro JD (2019). Droplet-based combinatorial indexing for massive-scale single-cell chromatin accessibility. Nature Biotechnology, 37, 916–924. doi: 10.1038/s41587-019-0147-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layer RM, Pedersen BS, DiSera T, Marth GT, Gertz J, & Quinlan AR (2018). GIGGLE: A search engine for large-scale integrated genome analysis. Nature Methods, 15, 123–126. doi: 10.1038/nmeth.4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Kuppe C, Cheng M, Menzel S, Zenke M, Kramann R, & Costa IG (2019). ScOpen: Chromatin-accessibility estimation of single-cell atac data. bioRxiv, 865931. doi: 10.1101/865931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Schulz MH, Look T, Begemann M, Zenke M, & Costa IG (2019). Identification of transcription factor binding sites using ATAC-seq. Genome Biology, 20, 45. doi: 10.1186/s13059-019-1642-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Li D, Lyu C, Gontarz P, Miao B, Madden P, … Zhang B. (2019). Improving ATAC-seq data analysis with AIAP, a quality control and integrative analysis package. BioRxiv. Available at: 10.1101/686808v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, & Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology, 15, 550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins AL, Walavalkar NM, Anderson WD, Zang C, & Guertin MJ (2017). Universal correction of enzymatic sequence bias reveals molecular signatures of protein/DNA interactions. Nucleic Acids Research, 46(2), e9. doi: 10.1093/nar/gkx1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy DJ, Chen Y, & Smyth GK (2012). Differential expression analysis of multifactor RNA-seq experiments with respect to biological variation. Nucleic Acids Research, 40, 4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy MT, & O’Callaghan CA (2014). PeaKDEck: A kernel density estimator−based peak calling program for DNaseI-seq data. Bioinformatics, 30, 1302–1304. doi: 10.1093/bioinformatics/btt774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, … Bejerano G. (2010). GREAT improves functional interpretation of cis-regulatory regions. Nature Biotechnology, 28, 495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montefiori L, Hernandez L, Zhang Z, Gilad Y, Ober C, Crawford G, & Sakabe NJ (2017). Reducing mitochondrial reads in ATAC-seq using crispr/cas9. Scientific Reports, 7, 2451. doi: 10.1038/s41598-017-02547-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström KJV, Schmidt F, Gasparoni N, Salhab A, Gasparoni G, Kattler K, … Walter J. (2019). Unique and assay specific features of NOMe-, ATAC- and DNase I-seq data. Nucleic Acids Research, 47, 10580–10596. doi: 10.1093/nar/gkz799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orchard P, Kyono Y, Hensley J, Kitzman JO, & Parker SCJ (2020). Quantification, dynamic visualization, and validation of bias in ATAC-Seq Data With Ataqv. Cell Systems, 10, 298–306. doi: 10.1016/j.cels.2020.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otlu B, Firtina C, Keles S, & Tastan O. (2017). GLANET: Genomic loci annotation and enrichment tool. Bioinformatics, 33, 2818–2828. doi: 10.1093/bioinformatics/btx326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang N, & Boyle AP (2019). TRACE: Transcription factor footprinting using DNase I hypersensitivity data and DNA sequence. bioRxiv, 801001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou J, Liu H, Yu J, Kelliher MA, Castilla LH, Lawson ND, & Zhu LJ (2018). ATACseqQC: A Bioconductor package for postalignment quality assessment of ATAC-seq data. BMC Genomics, 19(1), 169. doi: 10.1186/s12864-018-4559-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pálfy M, Schulze G, Valen E, & Vastenhouw NL (2020). Chromatin accessibility established by Pou5f3, Sox19b and Nanog primes genes for activity during zebrafish genome activation. PLoS Genetics, 16, e1008546. doi: 10.1371/journal.pgen.1008546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper J, Assi SA, Cauchy P, Ladroue C, Cockerill PN, Bonifer C, & Ott S. (2015). Wellington-bootstrap: Differential DNase-seq footprinting identifies cell-type determining transcription factors. BMC Genomics, 16, 1000. doi: 10.1186/s12864-015-2081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper J, Elze MC, Cauchy P, Cockerill PN, Bonifer C, & Ott S. (2013). Wellington: A novel method for the accurate identification of digital genomic footprints from DNase-seq data. Nucleic Acids Research, 41, e201. doi: 10.1093/nar/gkt850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pique-Regi R, Degner JF, Pai AA, Gaffney DJ, Gilad Y, & Pritchard JK (2011). Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Research, 21, 447–455. doi: 10.1101/gr.112623.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pliner HA, Packer JS, McFaline-Figueroa JL, Cusanovich DA, Daza RM, Aghamirzaie D, … Trapnell C. (2018). Cicero predicts cis-regulatory DNA interactions from single-cell chromatin accessibility data. Molecular Cell, 71, 858–871.e8. doi: 10.1016/j.molcel.2018.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podicheti R, & Mockaitis K. (2015). FEATnotator: A tool for integrated annotation of sequence features and variation, facilitating interpretation in genomics experiments. Methods, 79-80, 11–17. doi: 10.1016/j.ymeth.2015.04.028. [DOI] [PubMed] [Google Scholar]
- Polak P, Karlić R, Koren A, Thurman R, Sandstrom R, Lawrence MS, … Sunyaev SR (2015). Cell-of-origin chromatin organization shapes the mutational landscape of cancer. Nature, 518, 360–364. doi: 10.1038/nature14221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pranzatelli TJF, Michael DG, & Chiorini JA (2018). ATAC2GRN: Optimized ATAC-seq and DNase1-seq pipelines for rapid and accurate genome regulatory network inference. BMC Genomics, 19(1), 563. doi: 10.1186/s12864-018-4943-z. [Erratum in: BMC Genomics. 2019 Jan 15;20(1):44]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quach B, & Furey TS (2017). DeFCoM: Analysis and modeling of transcription factor binding sites using a motif-centric genomic footprinter. Bioinformatics, 33, 956–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radman-Livaja M, & Rando OJ (2010). Nucleosome positioning: How is it established, and why does it matter? Developmental Biology, 339, 258–266. doi: 10.1016/j.ydbio.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch T, Fritz M, Korbel JO, & Benes A. (2019). Alfred: Interactive multi-sample BAM alignment statistics, feature counting and feature annotation for long- and short-read sequencing. Bioinformatics, 35, 2489–2491. doi: 10.1093/bioinformatics/bty1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznikoff WS (2008). Transposon Tn5. Annual Review of Genetics, 42, 269–286. doi: 10.1146/annurev.genet.42.110807.091656. [DOI] [PubMed] [Google Scholar]
- Rickner HD, Niu S-Y, & Cheng CS (2019). ATAC-seq assay with low mitochondrial DNA contamination from primary human cd4+ T lymphocytes. JoVE, 145, e59120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, & Smyth GK (2010). EdgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26, 139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schep AN, Buenrostro JD, Denny SK, Schwartz K, Sherlock G, & Greenleaf WJ (2015). Structured nucleosome fingerprints enable high-resolution mapping of chromatin architecture within regulatory regions. Genome Research, 25, 1757–1770. doi: 10.1101/gr.192294.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schep AN, Wu B, Buenrostro JD, & Greenleaf WJ (2017). ChromVAR: Inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nature Methods, 14, 975. doi: 10.1038/nmeth.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield NC, & Bock C. (2016). LOLA: Enrichment analysis for genomic region sets and regulatory elements in R and bioconductor. Bioinformatics, 32, 587–589. doi: 10.1093/bioinformatics/btv612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield NC, Thurman RE, Song L, Safi A, Stamatoyannopoulos JA, Lenhard B, … Furey TS (2013). Patterns of regulatory activity across diverse human cell types predict tissue identity, transcription factor binding, and longrange interactions. Genome Research, 23, 777–788. doi: 10.1101/gr.152140.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield N, & Furey T. (2012). Identifying and characterizing regulatory sequences in the human genome with chromatin accessibility assays. Genes, 3, 651–670. doi: 10.3390/genes3040651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood RI, Hashimoto T, O’Donnell CW, Lewis S, Barkal AA, van Hoff JP, … Gifford DK (2014). Discovery of directional and nondirectional pioneer transcription factors by modeling DNase profile magnitude and shape. Nature Biotechnology, 32, 171–178. doi: 10.1038/nbt.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simovski B, Kanduri C, Gundersen S, Titov D, Domanska D, Bock C, … Sandve GK (2018). Coloc-stats: A unified web interface to perform colocalization analysis of genomic features. Nucleic Acids Research, 46, W186–W193. doi: 10.1093/nar/gky474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JP, Corces R, Xu J, Wei Y, Reuter VP, Chang HY, & Sheffield NC (2020). PEPATAC: A portable, optimized ATAC-seq pipeline.Available at http://pepatac.databio.org/en/latest/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Zhang Z, Grasfeder LL, Boyle AP, Giresi PG, Lee B-K, … Furey TS (2011). Open chromatin defined by DNaseI and faire identifies regulatory elements that shape cell-type identity. Genome Research, 21, 1757–1767. doi: 10.1101/gr.121541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spivakov M, & Fraser P. (2016). Defining cell type with chromatin profiling. Nature Biotechnology, 34, 1126–1128. doi: 10.1038/nbt.3724. [DOI] [PubMed] [Google Scholar]
- Stark R, & Brown G. (2011). DiffBind: Differential binding analysis of chip-seq peak data. Bioconductor, http://bioconductor.org/packages/release/bioc/vignettes/DiffBind/inst/doc/DiffBind.pdf. [Google Scholar]
- Stavrovskaya ED, Niranjan T, Fertig EJ, Wheelan SJ, Favorov AV, & Mironov AA (2017). StereoGene: Rapid estimation of genome-wide correlation of continuous or interval feature data. Bioinformatics, 33, 3158–3165. doi: 10.1093/bioinformatics/btx379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K, & Segal E. (2013). Determinants of nucleosome positioning. Nature Structural and Molecular Biology, 20, 267–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung M-H, Baek S, & Hager GL (2016). Genome-wide footprinting: Ready for prime time? Nature Methods, 13, 222–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung M-H, Guertin MJ, Baek S, & Hager GL (2014). DNase footprint signatures are dictated by factor dynamics and DNA sequence. Molecular Cell, 56, 275–285. doi: 10.1016/j.molcel.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarbell ED, & Liu T. (2019). HMMRATAC: A hidden markov modeleR for ATAC-seq. Nucleic Acids Research, 47, e91. doi: 10.1093/nar/gkz533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewari AK, Yardimci G, Shibata Y, Sheffield NC, Song L, Taylor BS, … Febbo PG (2012). Chromatin accessibility reveals insights into androgen receptor activation and transcriptional specificity. Genome Biology, 13, R88. doi: 10.1186/gb-2012-13-10-r88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, … Stamatoyannopoulos JA (2012). The accessible chromatin landscape of the human genome. Nature, 489, 75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripodi I, Allen M, & Dowell R. (2018). Detecting differential transcription factor activity from ATAC-seq data. Molecules, 23(5), 1136. doi: 10.3390/molecules23051136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainshtein Y, Rippe K, & Teif VB (2017). NucTools: Analysis of chromatin feature occupancy profiles from high-throughput sequencing data. BMC Genomics, 18, 158. doi: 10.1186/s12864-017-3580-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra J, & Stamatoyannopoulos JA (2016). Genomic footprinting. Nature Methods, 13, 213–221. doi: 10.1038/nmeth.3768. [DOI] [PubMed] [Google Scholar]
- Wang J, Zibetti C, Shang P, Sripathi SR, Zhang P, Cano M, … Qian J. (2018). ATAC-seq analysis reveals a widespread decrease of chromatin accessibility in age-related macular degeneration. Nature Communication, 9, 1364. doi: 10.1038/s41467-018-03856-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Zhang W, Fang H, Li Y, Wang X. (2018). esATAC: An easy-to-use systematic pipeline for ATAC-seq data analysis. Bioinformatics, 34(15), 2664–2665. doi: 10.1093/bioinformatics/bty141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch RP, Lee C, Imbriano PM, Patil S, Weymouth TE, Smith RA, Scott LJ, & Sartor MA (2014). ChIP-Enrich: Gene set enrichment testing for ChIP-seq data. Nucleic Acids Research, 42, e105. doi: 10.1093/nar/gku463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Xu K, Tian K, Shao Y, Tang L, Gao G, … Zhang QC (2019). SCALE method for single-cell ATAC-seq analysis via latent feature extraction. Nature Communications, 10, 4576. doi: 10.1038/s41467-019-12630-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn A, Marquez EJ, Lawlor N, Stitzel ML, & Ucar D. (2019). BiFET: Sequencing bias-free transcription factor footprint enrichment test. Nucleic Acids Research, 47, e11. doi: 10.1093/nar/gky1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Uzun Y, Zhu Q, Chen C, & Tan K. (2019). ScATAC-pro: A comprehensive workbench for single-cell chromatin accessibility sequencing data. bioRxiv, 824326. doi: 10.1101/824326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamanighomi M, Lin Z, Daley T, Chen X, Duren Z, Schep A, … Wong WH (2018). Unsupervised clustering and epigenetic classification of single cells. Nature Communications, 9, 2410. doi: 10.1038/s41467-018-04629-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, … Liu XS (2008). Model-based analysis of ChIP-seq (macs). Genome Biology, 9, R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Z, Jin Y, Zhang W, Lu Y, Li B, & Qu K. (2019). ATAC-pipe: general analysis of genome-wide chromatin accessibility. Briefing in Bioinformatics, 20(5), 1934–1943. doi: 10.1093/bib/bby056. [DOI] [PubMed] [Google Scholar]