Correction to: Genome Biol 22, 163 (2021)
https://doi.org/10.1186/s13059-021-02367-2
Following publication of the original paper [1], it was noticed that a typesetting error occurred.
The HTML version of this article erroneously contained the incorrect version of Table 1. It was also reported that “SSPsimSeq” should be “SPsimSeq” in the 12th row of Table 1. The correct Table 1 is given below.
Table 1.
Summary of 16 simulators (including our proposed scDesign2) in six properties
| Property | 1 protocol adaptive | 2 gene preserved | 3 gene cor. captured | 4 cell num. seq. dep. flexible | 5 transparent | 6 comp. and sample efficient |
|---|---|---|---|---|---|---|
| Simulator | ||||||
| dyngen [77] | ✗ | ✗ | ✓ | ✓ | ||
| Lun2 [78] | ✓ | ✗ | ✓ | ✓ | ✓ | |
| powsimR [75] | ✓ | ✓ | ✗ | ✓ | ✓ | ✓ |
| PROSST [68] | ✓ | ✗ | ✓ | ✓ | ||
| scDD [74] | ✓ | ✗ | ✗ | ✓ | ✓ | |
| scDesign[35] | ✓ | ✗ | ✓ | ✓ | ✓ | |
| scGAN [72] | ✓ | ✓ | ✗ | ✗ | ||
| splat simple[69] | ✓ | ✗ | ✗ | ✗ | ✓ | ✓ |
| splat [69] | ✓ | ✗ | ✗ | ✗ | ✓ | ✓ |
| kersplat [69] | ✓ | ✗ | ✗ | ✓ | ✓ | |
| SPARSim [71] | ✓ | ✓ | ✗ | ✓ | ✓ | |
| SPsimSeq [79] | ✓ | ✓ | ✓ |
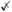 ** **
|
✓ | ✓ |
| SymSim[70] | ✓ | ✗ | ✗ | ✗ | ✓ | ✓ |
| ZINB-WaVe[39] | ✓ | ✗ | ✓ | ✓ | ||
| SERGIO [76] | ✓ | ✗* | ✓ | ✓ | ✓ | |
| scDesign2 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
Property 1: protocol adaptiveness
Property 2: gene preservation
Property 3: gene correlation capture
Property 4: flexible cell number and sequencing depth choices
Property 5: transparency
Property 6: computational and sample efficiency
For each simulator and each property, a checkmark, checkcross, or cross means that the simulator satisfies, partially satisfies, or does not satisfy the property, respectively
*SERGIO requires a user-specified gene regulatory network, and it does not capture/estimate gene correlations from a real dataset
**SPsimSeq can vary cell number but not sequencing depth
The original article [1] has been updated.
Contributor Information
Wei Vivian Li, Email: li@rutgers.edu.
Jingyi Jessica Li, Email: jli@stat.ucla.edu, Email: lijy03@g.ucla.edu.
Reference
- 1.Sun T, Song D, Li WV, et al. scDesign2: a transparent simulator that generates high-fidelity single-cell gene expression count data with gene correlations captured. Genome Biol. 2021;22:163. doi: 10.1186/s13059-021-02367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]


