Abstract
Purpose:
Blepharoptosis is a known cause of reversible vision loss. Accurate assessment can be difficult, especially amongst non-specialists. Existing automated techniques disrupt clinical workflow by requiring user input, or placement of reference markers. Neural networks are known to be effective in image classification tasks. We aim to develop an algorithm that can accurately identify blepharoptosis from a clinical photo.
Methods:
A total of 500 clinical photographs from patients with and without blepharoptosis were sourced from a tertiary ophthalmic center in Taiwan. Images were labeled by two oeuloplastie surgeons, with an independent third oeuloplastie surgeon to adjudicate disagreements. These images were used to train a series of convolutional neural networks (CNNs) to ascertain the best CNN architecture for this particular task.
Results:
Of the models that trained on the dataset, most were able to identify ptosis images with reasonable accuracy. We found the best performing model to use the DenseNet121 architecture without pre-training which achieved a sensitivity of 90.1 % with a specificity of 82.4 %, compared to the worst performing model which was used a Resnet34 architecture with pre-training, achieving a sensitivity of 74.1 %, and specificity of 63.6 %. Models with and without pre-training performed similarly (mean accuracy 82.6 % vs. 85.8 % respectively, p = 0.06), though models with pre-training took less time to train (1-minute vs. 16 min, p < 0.01).
Conclusions:
We report the use of AI to accurately diagnose blepharoptosis from a clinical photograph with no external reference markers or user input requirement. Most current-generation CNN architectures performed reasonably on this task, with the DenseNet121, and Resnet18 architectures without pre-training performing best in our dataset.
Keywords: Artificial intelligence, Deep learning models, Automated identification, Blepharoptosis, Novel medical image dataset, High accuracy
1. Introduction
Blepharoptosis, commonly referred to as ptosis, is the inferior displacement or drooping of the upper eyelid. In addition to its cosmetic implications, it is a known cause of reversible vision loss in adults [1] and amblyopia in children. Ptosis can also be a subtle clinical sign of genetic, neurologic, myogenic, or systemic disease. Therefore, it is important for primary care physicians and non-specialist healthcare workers to accurately diagnose ptosis for a proper referral to an ophthalmologist. Ptosis is diagnosed by using a ruler and light source to measure important landmarks such as the distance between the pupillary light reflex and the upper lid margin (margin reflex distance 1, or MRD1), together with clinical photographs. Commercial devices are also available which can make these measurements with varying degrees of accuracy [2]. While the reliability of these measurements is excellent amongst experienced experts, there is a learning curve [3,4]. Accurate diagnosis is particularly challenging for non-specialist healthcare workers who are referring patients and insurance company reviewers who often base authorization on MRD1 measurements [5]. This can lead to missed diagnoses, difficulties triaging referrals to a tertiary oculoplastic clinic, as well as challenges with insurance denials.
Recently, there has been a surge of interest in computer-based image recognition tasks due to advances in the field of deep learning, a subset of machine learning where multiple layers of a neural network aim to generate predictions from input data by modeling complex non-linear data patterns [6]. This approach led to a series of rapid advancements, with convolutional neural networks (CNNs) architectures such as AlexNet [7], ResNet [8], VGG [9], SqueezeNet [10], and DenseNet [11] showing excellent performance in image recognition, as demonstrated by their success in the ImageNet Large Scale Visual Recognition Competition [12]. These advances are now being translated into clinical tasks, with many successful examples demonstrating the use of artificial intelligence (AI) models in diabetic retinopathy, glaucoma [13–16], AMD [17,18], and numerous other areas of medicine [19–24]. Typically, CNN’s used for specialized purposes such as ours are initialized using pretrained weights from large general purposes imaging datasets to help them learn detection of basic shapes. The final layers of these models are then modified to suit the specialized needs of the task at hand. While this approach of transfer learning is successful in many domains, it is unclear whether relatively homogenous oculoplastic photos would benefit from this process.
We propose the use of an automated photo-based technique that will allow for a convenient, inexpensive alternative to manual assessment of ptosis with the potential for improving the reliability of measurements amongst non-experts. Given the recent advances in AI-assisted image recognition techniques and the lack of literature on this application to oculoplastics, we explore various AI architectures to create a highly sensitive and specific AI model for blepharoptosis. By using a CNN architecture, we aim to create a model that allows for greater flexibility on the input image. Herein, we report the first published CNN based AI model which can accurately diagnose blepharoptosis from a clinical photograph with no external reference markers or user input.
2. Methods
2.1. Dataset - Inclusion/Exclusion criteria
Full face images, collected as part of the oculoplastic clinic evaluation at a tertiary ophthalmology clinic in Taiwan, were de-identified and cropped to include one eye from each patient (data processing details provided below).
From this databank, 250 patients with blepharoptosis were chosen at random, and individual eye images were extracted. From these 500 images, 66 photographs with severe dermatochalasis, upper eyelid retraction, and photographs with poor image quality were removed, leaving 434 images used in this study (Fig. 1). The brow region is not included in the photos, so brow ptosis cannot be excluded. IRB approval was granted for this retrospective study by both Stanford University and National Taiwan University Hospital, and the research was conducted following the data use agreement signed by these two institutions. Pediatric patients (<18 years old) were excluded from this study.
Fig. 1.
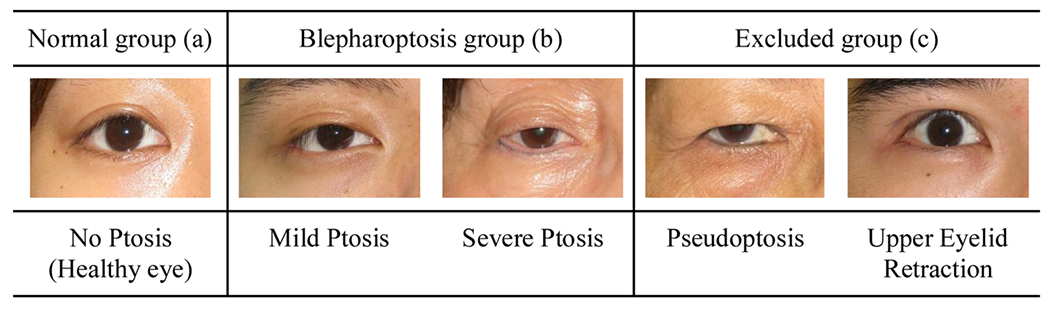
Normal (a), blepharoptosis (b), and excluded (c) examples. (Right eyes).
2.2. Dataset - pre-processing
The photos were taken with a regular digital camera together with a flashlight as a standard full-face photo. In order to crop a standardized image of a single eye, we used the OpenFace [25] open-source package to identify key facial landmarks in each photo. With these landmark positions, we can then crop single eye images as shown in Fig. 2. The resulting single eye images were 400 × 600 pixels each, which were then resized to 224 × 224 pixels (to match the input size most CNN architectures are optimized to use). These images were then used for further evaluation.
Fig. 2.
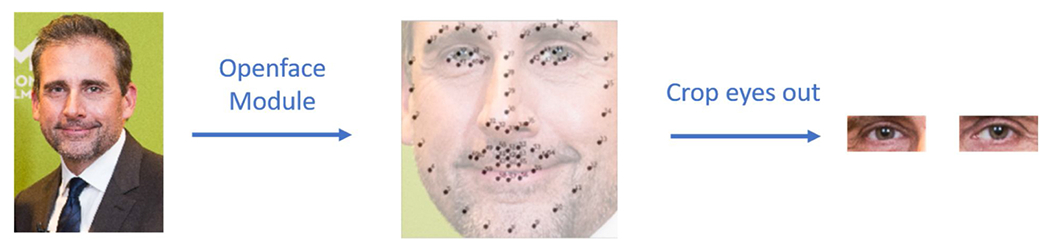
Automated extraction of single eye images from a full-face photo using the OpenFace package. Key facial landmarks were identified (black dots in the center image) and then used to crop each eye for further evaluation.
2.3. Dataset - ground truth labelling
The ground truth of the blepharoptosis was decided by three oculoplastic surgeons, through a voting system. Two of the three labelers both reviewed all 500 photographs in the first round of the labeling process. Where there was disagreement in the first round, a consensus meeting was then held by the same two oculoplastic surgeons to attempt to come to a consensus decision. For images where the two surgeons were unable to come to a consensus after the meeting, these images were adjudicated by an independent third oculoplastic surgeon. Their decision was then used to cast the deciding vote. Only single-eye regional photographs were provided to the labelers. Other information, such as the condition of the fellow eye, medical or surgical history, the measurements including margin to reflex distance 1,2,3 are not supplemented. The final labels assigned to the images designated them in a binary fashion as ‘blepharoptosis present’ or ‘blepharoptosis absent’. We consider the ‘blepharoptosis absent’ patients to be healthy, normal patients.
2.4. Dataset - subdivision
To train the AI model, the data was split randomly into a training set, validation set, and test set (Table 1). This enables the AI model to train using the “Training Dataset” while optimizing model weights by checking against the “Validation dataset”. Once all model parameters were finalized, the “Test Dataset” (which the model had not previously seen) was used to evaluate the accuracy of the trained model. Given the relatively small size of our dataset, we chose to perform 5-fold cross-validation on our dataset, reducing the probability that our findings are due to an aberrant split of data through chance. Results presented in the manuscript are the average results across all 5 folds unless specified.
Table 1.
The number of images in Training, Validation, and Test datasets.
| Number of Normal Eyelid Images | Number of Blepharoptosis Images | |
|---|---|---|
| Training Set | 136 | 177 |
| Validation Set | 34 | 45 |
| Test Set | 18 | 24 |
2.5. Model selection
We tested the eleven leading CNN architectures with Fast.AI implementations due to their excellent performance in image recognition when tested on the ImageNet challenge. Whilst it is possible to create a custom CNN architecture, many well-established CNN architectures are known to perform well in image classification problems and are unlikely to be beaten by a new custom architecture. By choosing models already available via Fast.AI, we can limit variations in implementation which may adversely affect experiment repeatability. These models have also been trained on large databases of images and can be initialized with pre-trained weights. This technique of transfer learning allows our models to utilize the ‘knowledge’ learned from training on these large image databases to optimize their performance. Given the lack of oculoplastic AI-specific research, we chose to perform a series of experiments using different model architectures with and without pretrained weights to determine which approach would provide the most accurate results, and to see the variance in outcomes from different models.
Each model’s last fully connected layer is modified to generate a vector of size two, followed by a SoftMax layer to predict a probability output value for our blepharoptosis binary classification problem. For example, our customized DenseNet121’s architecture can be seen in Table 2.
Table 2.
DenseNet121 [11] architecture.
| Layers | Architecture | Output Size |
|---|---|---|
| Convolution | 7 × 7 conv, stride 2 | 112 × 112 |
| Pooling | 3 × 3 max pool, stride 2 | 56 × 56 |
| Dense Block (1) | [1 × 1 conv, 3 × 3 conv] x 6 | 56 × 56 |
| Transition Layer (1) | 1 × 1 conv, 2 × 2 avg pool, stride 2 | 28 × 28 |
| Dense Block (2) | [1 × 1 conv, 3 × 3 conv] x 12 | 28 × 28 |
| Transition Layer (2) | 1 × 1 conv, 2 × 2 avg pool, stride 2 | 14 × 14 |
| Dense Block (3) | [1 × 1 conv, 3 × 3 conv] x 24 | 14 × 14 |
| Transition Layer (3) | 1 × 1 conv, 2 × 2 avg pool, stride 2 | 7 × 7 |
| Dense Block (4) | [1 × 1 conv, 3 × 3 conv] x 16 | 7 × 7 |
| Classification Layer | global avg pool, 2D fully connected, softmax | 1 × 1 |
And we trained our model with the binary cross entropy loss as follows:
Where N is the total number of training samples, i is the index of one training sample. The binary variable gi is i-th training sample’s groundtruth labeling, with value 1’s for positive samples (with blepharoptosis) and value 0’s for negative samples (without blepharoptosis). The variable si is our model’s SoftMax layer output’s first component, representing the probability that our model believes the i-th input training image has blepharoptosis.
2.6. Data augmentation
The dataset used consisted of a relatively small number of images for training a robust AI network. If a model is trained only using these images, the results usually generalize poorly to images the network has not previously seen. As such, we augmented our data to create a larger dataset by generating multiple variations of our images by randomly applying transformations:
Flip images horizontally
Rotate images up to 10 degrees
Zoom in on image up to 10 %
Adjust brightness/contrast by 20 %
Perspective warping images by 20 %
By applying these transformations, our model was better able to generalize to images it had not seen before.
2.7. AI training pipeline
The Fast.AI library was used for AI model development in this study. It is a high-level API built on top of PyTorch, which facilitates the incorporation of best practices to maximize performance. The analysis was performed on a machine with Intel Xeon CPU @ 2.2Ghz, 13GB RAM, and a Tesla P100 (16GB VRAM) GPU.
Images were loaded into the training and validation sets with data transformations applied to the training data set, and normalization was applied to all images. The chosen CNN architecture was then initialized either with or without pretrained weights depending on the experiment parameters. The model was then trained till convergence using a one-cycle policy [26]. This allows us to incorporate cyclical learning rates in the model where the learning rate continuously oscillates between reasonable minimum and maximum bounds, helping models converge much faster than using a constant learning rate. To calculate the optimal learning rate range – this was calculated using a function within the FastAI library which helps identify the optimal learning rate range for each model/dataset. We found in our datasets, the learning rate bounds tended to be between 3 × 10−6, and 3 × 10−4. The Adam optimizer was used in all models for consistency.
Where pretrained ImageNet weights were used, the classification head of the model is removed and substituted with a blank classification head, and the rest of the model was frozen. This was then trained for 4 epochs using a one-cycle policy as described above. Then the rest of the model was unfrozen and retrained for another 2 epochs using the one-cycle policy while adjusting weights in a graded manner where earlier layers of the model are affected to a lesser extent than later ones. We found in all cases of pretrained models, further epochs of training did not improve performance. For models where pretrained weights were not used, the entire model is kept unfrozen, and the optimal learning rate range for each model was calculated. These were then trained using the one-cycle policy for 100 epochs, as they took longer to reach convergence – as expected without the pretraining weights. Training loss, validation loss, and accuracy were monitored to minimize overfitting, which was ultimately verified by checking model performance against the unseen test dataset.
Each model was then created and tested 5 times using 5-fold cross-validation for each training/evaluation loop. This allows us to generate averaged evaluation statistics for the models across each of the 5 folds.
2.8. Statistical analysis
Metrics were calculated, and graphs plotted using Python (version 3.8, Python Software Foundation). A combination of packages was used to generate these, including fast.ai, PyTorch, matplotlib, and scikit-learn.
A variety of model performance metrics were calculated based on the number of true positive samples (TP), number of false positive samples (FP), number of true negative samples (TN), and number of false negative samples (FN). These were averaged across the 5 folds of cross-validation. The derivation of these performance metrics is shown below:
Where statistical significance was assessed, p values were calculated using Fisher’s exact test for binary variables and one way ANOVA for continuous variables with a p-value <0.05 considered significant.
3. Results
After the first round of independent labeling by the two oculoplastic surgeons, the agreement rate was 82 %. The remaining images with disagreement were then resolved as per the study protocol.
Following the test protocol, a total of 22 separate CNNs was trained. For each of these models, the accuracy score was calculated by testing the model against the Test image dataset. As seen in Table 3, there is a significant variation in accuracy seen between the models. The pre-trained models had an average accuracy of 82.6 %, and the models without pre-training had an average accuracy of 85.8 %, with no significant difference between the two groups (p = 0.06). A notable advantage of utilizing models with pre-training was that the time taken to take each model was significantly lower at 1 min vs. 16 min (p < 0.01).
Table 3.
Comparative Accuracy scores for a variety of CNN architectures both initialized with pretrained weights and without pretrained weights.
| Accuracy of Pretrained Version | Accuracy of Not Pretrained Version | |
|---|---|---|
| VGG11 | 80.5% | 83.8 % |
| VGG13 | 86.2% | 83.8 % |
| VGG16 | 85.7% | 85.2 % |
| VGG19 | 87.1 % | 84.7 % |
| ResNet18 | 81.0% | 88.6 % |
| ResNet34 | 71.4 % | 86.7 % |
| ResNet50 | 81.9% | 83.3 % |
| AlexNet | 85.2 % | 87.1 % |
| SqueezeNet | 84.7 % | 84.8 % |
| DenseNet121 | 88.0% | 88.6 % |
| DenseNet201 | 76.7% | 87.1 % |
Rather than present detailed scores for all 22 models, we chose to calculate scores for the best (DenseNet121 without pre-training) and worst (Resnet34 with pre-training) performing models. This was done to provide a contrasting overview of the range of model performances (Table 4). ResNet18 without pre-training also had the same results as DenseNet121 without pre-training. We chose to include only one ‘best performing model’ for simplicity and clarity of findings. DenseNet121 with pre-training was chosen as the best model, as larger models tend to generalize to unseen data more accurately. The best performing model (DenseNet121 without pre-training) achieved a sensitivity of 90.3 % with a specificity of 82.4 %, which was significantly better than the worst performing model (Resnet34 pretrained), which achieved a sensitivity of 74.1 %, and specificity of 63.6 %. This is demonstrated well in the confusion matrices (Fig. 3) revealing a far higher rate of false positives and false negatives in the worst-performing model. The AUROC (0.95 vs 0.84) and FI scores (88.2 % vs 72.6 %) also reflect these differences in model performance.
Table 4.
Comparative performance metrics for the best performing model (DenseNet121 - Not Pretrained) vs. a poorly performing model (Resnet34 - Pretrained).
| DenseNet121 - Not Pretrained (High Accuracy) | Resnet34 - Pretrained (Poorer Accuracy) | |
|---|---|---|
| Positive Predictive Value | 93.3 % | 85.1 % |
| Negative Predictive Value | 75.5 % | 46.6 % |
| Accuracy | 88.6 % | 71.4 % |
| F1 Score | 88.2 % | 72.6 % |
| ROC AUC | 0.95 | 0.84 |
| Sensitivity | 90.1 % | 74.1 % |
| Specificity | 82.4 % | 63.6 % |
Fig. 3.
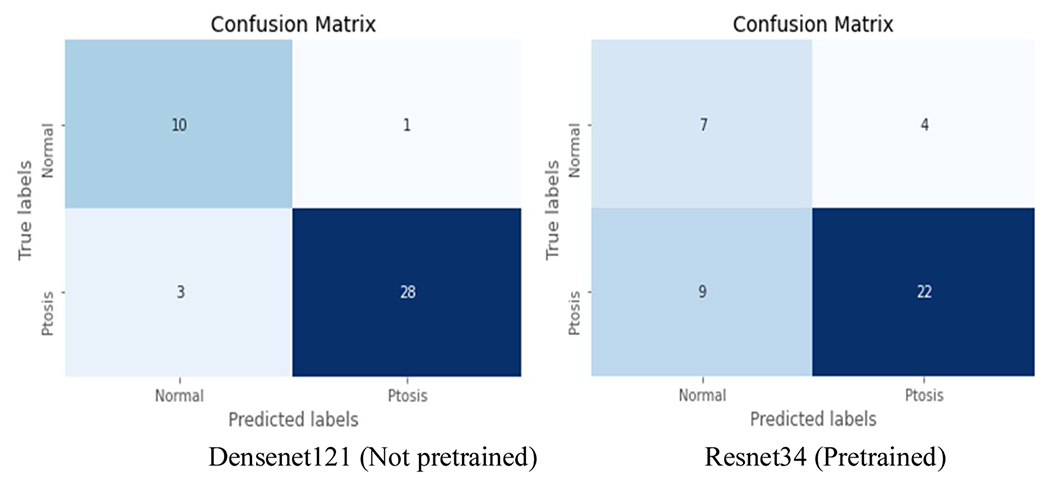
Confusion matrices showing results of test set for the best performing model (Densenet121 Not pretrained), and poorest performing model (Resnet34 Pretrained). Taken from a single representative fold during cross validation.
4. Discussion
Blepharoptosis is a frequent cause of peripheral vision loss which, typically requires surgical intervention when significant. The accurate diagnosis of blepharoptosis is complex and relies heavily on accurate MRD1 measurement in traditional clinical assessment. Studies have found MRD1 to be the most predictive measurement of visual field loss [27]. However accurate measurement of MRD1 by non-expert clinicians is known to be sub-optimal [3,4]. To facilitate accurate diagnosis by non-experts, we present the first publication demonstrating an AI algorithm to accurately and reliably diagnose blepharoptosis from a single photo without the need for external reference markers or manual annotation. We also explored various CNN architectures for this task and found many models that had reasonable performance, with the best models reaching a sensitivity of 90.1 % and specificity of 82.4 % on a previously unseen test dataset.
Several experiments were performed during the construction of our AI algorithm to help determine which architectures would lead to the best results in our particular task [28]. Transfer learning and large models are known to be effective in many medical image recognition tasks [29–31], however, our dataset differs in that there is limited variation in our images, and our data set is relatively small. Transfer learning is a technique that is designed to enhance the performance of a neural network by ‘pre-training’ the network on a large dataset that may not be related to the intended use (e.g. in this case on the 14 million images contained in the ImageNet database). This allows the model to learn general features of images, which can then be applied to the target image set. We tested some of the leading CNN architectures with and without pre-training and found that the DenseNet [11] and ResNet [8] architectures without pre-training provided the best performance. A number of the other architectures also performed quite well. In our study, we found that models without pre-training and those with pre-training had similar performance. Typically, pretrained models have better performance as they have already learned many common shapes and image patterns which makes it easier for them to deal with a wide variety of image recognition tasks. However, the authors hypothesize the limited benefit in this scenario may be due to limited variation in ocular photos used for this. The use of the pretrained networks did however significantly reduce the training time for the models. The complexity of the architectures also had a limited impact, with the 18-layer ResNet18 performing similarly to the 121-layer DenseNet121 architecture. The authors believe this is also due to the relative lack of variation in eyelid images, and thus the number of layers and weights needed for an accurate model is fewer for this particular task.
4.1. Discussion - comparison to previous techniques
Previous reports investigating automated blepharoptosis detection from facial photos have relied on traditional computer vision processing methodologies. The relative contrast between the eyelid vs sclera/iris does make this task possible under most circumstances as it is possible to handcraft these features. However, these techniques tend to fail when lighting is suboptimal, or the lid contours do not fit the expected parameters, thus leading to failure in challenging situations. Furthermore, specifically created computer vision techniques are limited to exactly what features are hand-coded initially, which limits the scope of their abilities. For example, cases of severe dermatochalasis could not be differentiated from true blepharoptosis through these algorithms (where the apparent lower lid margin may actually be overhanging skin rather than the true lid margin). Some techniques require hand annotation of images by the user [32,33], and others require a reference marker [34, 35] to be placed on the patient before the image is taken. This usually takes the form of a sticker of pre-specified dimensions being placed on the patient’s forehead before photos are taken. Requiring hand annotation of images takes a significant amount of time, and it is not practical to expect a busy clinician to annotate these photos when the software is being used at the point of care. For images where a reference marker is required – this also poses issues to clinical workflow, as clinicians must obtain the reference marker of appropriate size for that particular software, then have the reference markers available at all times – which can be limiting. For example, when reviewing a patient who is unwell in a hospital bed and unable to come to the eye clinic. Both of these methodologies thus have challenges to their implementation in a real-world clinical context. By utilizing a CNN architecture, we have shown that it is possible to identify ptosis in a variety of clinical settings, without any extra steps required from clinicians which may disrupt existing clinical workflows.
4.2. Discussion - limitations
It is important to note some limitations to this study. Firstly, our dataset has a relatively small number of images compared to the typical numbers of images used in AI imaging studies. This however reflects the difficulty of acquiring images in a subspecialty field, especially considering the sensitivity of the original clinical images - full face images. For this study, we chose to exclude patients with severe dermatochalasis, upper eyelid retraction, and poor image quality. The source images were from patients who were given a ptosis diagnosis at some stage, though the images for each eye of all patients were separated, and independently assessed by a consensus of oculoplastic surgeons to ascertain our ground truth labels. This early study was designed to establish the possibility of creating an AI algorithm to identify ptosis, and as such the image set was relatively simple. Furthermore, the image set consists of patients only from an Asian ethnic background, which limits the applicability of these findings to other ethnic groups given differences in eyelid features between populations. These are both issues we aim to address in future studies utilizing a larger dataset. We believe that a larger dataset including a more diverse range of photos will allow a more accurate model to be developed. Firstly, by having a larger dataset, we will be able to further optimize model hyperparameters. In our paper, we chose not to attempt further hyperparameter optimization beyond what has been described in the methodology of this paper, due to the relatively small size of our dataset. Having a larger range of clinical examples from different populations will also help the model be able to train across a larger set of examples, and ultimately perform better when generalizing to other patients.
Even amongst experts, there can be some subjectivity in the diagnosis of ptosis, especially when making judgments from an isolated clinical photo. As such we sought to have a consensus agreement between oculoplastic surgeons to try and increase the accuracy of our ground truth labels and feel that expert opinion is a reasonable method of classification in this situation. Finally, with regards to the choice of model, it is often impossible to predict which model will perform best on a particular dataset. As such we experimented with numerous different model types, potentially introducing an element of chance into model selection. We hope that future studies will be able to reference this study to have a more in-depth understanding of how different models perform on oculoplastic images and guide their model choices for study.
4.3. Discussion - future directions
To further validate our findings and fine-tune our algorithm, a multi-center prospective trial is underway to build a larger dataset with a broader set of eyelid pathologies and validate our algorithm against external datasets, and the diagnosis of other non-expert physicians. Finally, all photos taken for this study are formal clinical photos taken as part of clinical care. As such the quality of these images is likely to be of a higher caliber than images taken by primary care physicians or patients. To help account for this, our model was trained with data augmentation to vary the lighting and image quality which helps improve the generalizability of our model results. In future studies, we aim to capture imaging from a wider variety of devices.
Further studies are also warranted to explore additional applications of AI in oculoplastic and eyelid evaluation. By utilizing a larger dataset, we propose moving beyond binary classification to a multilabel classification of eyelid position, exploring degrees of blepharoptosis, and diagnosing other eyelid pathologies such as dermatochalasis, eyelid retraction, or eyelid malposition. Our long-term vision is to incorporate these tools into a clinically validated decision support tool that can be used in a variety of settings. For primary care physicians and ER physicians, it can offer a convenient way to way to assess patients with suspected ptosis either in the context of suspected systemic/neurologic disease or to facilitate referral to an oculoplastic surgeon. For ophthalmologists, it allows conveniently labeled photo documentation, with the potential to replace manual MRD1 measurements and visual field testing for insurance approvals.
5. Conclusion
This paper demonstrates that an AI algorithm utilizing CNN’s can detect patients with blepharoptosis with a high degree of accuracy, without manual user annotation, or the need for reference markers on the patient. We explored a variety of CNN architectures and found that most leading architectures had reasonable performance on this task, with the DenseNet121 and ResNet18 architectures achieving the highest accuracy. Pre-trained models had similar performance to non-pretrained models, though model training time was significantly lower for pre-trained models. Our findings form a foundation for future research applying AI in oculoplastics by demonstrating the possibility of accurate model creation and guiding CNN architecture selection in this subspecialty. We believe that through the use of a larger more diverse dataset, future studies will be able to create an AI algorithm with increased accuracy.
Acknowledgements
This work was supported by the LEAP Program (a Stanford-Taiwan Technology Fellows program sponsored by the Taiwan Ministry of Science and Technology) and departmental core grants from the National Eye Institute (P30 EY026877) and Research to Prevent Blindness (RPB) to the Byers Eye Institute at Stanford. Computational and storage resources were sponsored by Taiwan National Center for High-performance Computing (NCHC).
Footnotes
Declaration of Competing Interest
The authors declare no conflict of interest.
Author statement
We confirm that this work is original and has not been published elsewhere, nor is it currently under consideration for publication elsewhere. All authors have read and approved the manuscript. There are no financial or other relationships which might lead to conflicts of interest. In consideration of the journal “International Journal of Medical Informatics” taking action in reviewing and editing our submission, which represents an original article, the authors undersigned hereby transfers, assigns, or otherwise conveys all copyright ownership to “International Journal of Medical Informatics” in the event that such work is published in the journal “International Journal of Medical Informatics”
References
- [1].Ho SF, Morawski A, Sampath R, Burns J, Modified visual field test for ptosis surgery (Leicester Peripheral Field Test), Eye Lond. Engl 25 (3) (2011) 365–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Sinha KR, Yeganeh A, Goldberg RA, Rootman DB, Assessing the accuracy of eyelid measurements utilizing the volk eye check system and dinical measurements, Ophthal. Plast. Reconstr. Surg 34 (July (4)) (2018) 346–350. [DOI] [PubMed] [Google Scholar]
- [3].Boboridis K, Repeatability and reproducibility of upper eyelid measurements, Br. J. Ophthalmol 85 (January (1)) (2001) 99–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Nemet AY, Accuracy of marginal reflex distance measurements in eyelid surgery, J. Craniofac. Surg 26 (October (7)) (2015) e569–e571. [DOI] [PubMed] [Google Scholar]
- [5].Inc A, Eyelid Surgery - Medical Clinical Policy Bulletins [Internet]. [cited 2020 May 23]. Available from:, 2020, p. 2 http://www.aetna.com/cpb/medical/data/1_99/0084.html.
- [6].LeCun Y, Bengio Y, Hinton G, Deep learning, Nature. 521 (7553) (2015) 436–444. [DOI] [PubMed] [Google Scholar]
- [7].Krizhevsky A, Sutskever I, Hinton GE, ImageNet classification with deep convolutional neural networks, Commun. ACM 60 (6) (2017) 84–90. [Google Scholar]
- [8].He Kaiming, Zhang Xiangyu, Ren Shaoqing, Sun Jian, Deep residual learning for image recognition, arXiv.org (2016). [Google Scholar]
- [9].Simonyan K, Zisserman A, Very deep convolutional networks for large-scale image recognition, Arxiv. (2014). [Google Scholar]
- [10].Iandola FN, Han S, Moskewicz MW, Ashraf K, Dally WJ, Keutzer K, SqueezeNet: AlexNet-level accuracy with 50x fewer parameters and <0.5MB model size, Arxiv. (2016). [Google Scholar]
- [11].Huang G, Liu Z, Maaten LVD, Weinberger KQ, Densely connected convolutional networks, Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (2017) 4700–4708. [Google Scholar]
- [12].Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, et al. ImageNet large scale visual recognition challenge, Int. J. Comput. Vision. 115 (3) (2015) 211–252. [Google Scholar]
- [13].Yousefi PS, Elze PT, Pasquale MDLR, Saeedi MMO, Wang PM, Shen MDLR, et al. Monitoring glaucomatous functional loss using an artificial intelligence-enabled dashboard, Ophtha. 10 (March) (2020) 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Christopher M, Belghith A, Weinreb RN, Bowd C, Goldbaum MH, Saunders LJ, et al. Retinal nerve Fiber layer features identified by unsupervised machine learning on optical coherence tomography scans predict Glaucoma progression, Investig. Opthalmology Vis. Sci 59 (June (7)) (2018) 2748–2749. Available from: [Internet]. https://iovs.arvojournals.org/article.aspx?articleid=2683627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Baxter SL, Marks C, Kuo T-T, Ohno-Machado L, Weinreb RN, Machine learning-based predictive modeling of surgical intervention in Glaucoma Using systemic data from electronic health records, Ajopht. 1 (December 208) (2019) 30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Yang HK, Kim YJ, Sung JY, Kim DH, Kim KG, Hwang J-M, Efficacy for differentiating nonglaucomatous versus glaucomatous optic neuropathy using deep learning systems, Ajopht. 1 (April) (2020) 1–21. [DOI] [PubMed] [Google Scholar]
- [17].Treder M, Lauermann JL, Eter N, Automated detection of exudative age-related macular degeneration in spectral domain optical coherence tomography using deep learning, Graefes Arch. Clin. Exp. Ophthalmol 256 (2) (2017) 259–265. [DOI] [PubMed] [Google Scholar]
- [18].Bogunovic H, Waldstein SM, Schlegl T, Langs G, Sadeghipour A, Liu X, et al. Prediction of Anti-VEGF treatment requirements in Neovascular AMD using a machine learning approach, Investigat. Opthalmol. Vis. Sci 58 (7) (2017) 3240. [DOI] [PubMed] [Google Scholar]
- [19].Kermany D, Goldbaum M, Cai W, Valentim C, Liang H, Baxter S, et al. Identifying medical diagnoses and treatable diseases by image-based deep learning, Cell. 172 (5) (2018) 1122–1131, e9.n. [DOI] [PubMed] [Google Scholar]
- [20].Poplin R, Varadarajan A, Blumer K, Liu Y, McConnell M, Corrado G, et al. Prediction of cardiovascular risk factors from retinal fundus photographs via deep learning, Nat. Biomed. Eng 2 (3) (2018) 158–164. [DOI] [PubMed] [Google Scholar]
- [21].Ong Y, Hilal S, Cheung C, Venketasubramanian N, Niessen W, Vrooman H, et al. Retinal neurodegeneration on optical coherence tomography and cerebral atrophy, Neurosci. Lett 584 (2015) 12–16. [DOI] [PubMed] [Google Scholar]
- [22].Zeng N, Zhang H, Song B, Liu W, Li Y, Dobaie AM, Facial expression recognition via learning deep sparse autoencoders, Neurocomputing. 273 (2018) 643–649. [Google Scholar]
- [23].Zeng N, Li H, Wang Z, Liu W, Liu S, Alsaadi FE, et al. Deep-reinforcement-Learning-Based images segmentation for quantitative analysis of gold immunochromatographic strip, Neurocomputing. (2020). [Google Scholar]
- [24].Zeng N, Wang Z, Zineddin B, Li Y, Du M, Xiao L, et al. Image-based quantitative analysis of gold immunochromatographic strip via cellular neural network approach, Ieee T Med. Imaging 33 (5) (2014) 1129–1136. [DOI] [PubMed] [Google Scholar]
- [25].Amos B, Ludwiczuk B, Satyanarayanan M, Openface: a general-purpose face recognition library with mobile applications. CMU-CS-16-118, CMU School of Computer Science, Tech Rep (2016). [Google Scholar]
- [26].Smith LN, A disciplined approach to neural network hyper-parameters: part 1 – learning rate, batch size, momentum, and weight decay, Arxiv. (2018). [Google Scholar]
- [27].Cahill KV, Burns JA, Weber PA, The effect of Blepharoptosis on the field of vision, Ophth. Plastic Rec. Surg 3 (3) (1987) 121–126. [DOI] [PubMed] [Google Scholar]
- [28].Baker B, Gupta O, Naik N, Raskar R, Designing neural network architectures using reinforcement learning, arXiv preprint arXiv (2016) 161102167. [Google Scholar]
- [29].Yu Y, Lin H, Meng J, Wei X, Guo H, Zhao Z, Deep transfer learning for modality classification of medical images, Information 8 (3) (2017) 91. [Google Scholar]
- [30].. Khalifa N, Loey M, Taha M, Mohamed H, Deep transfer learning models for medical diabetic retinopathy detection, Acta Inform. Med 27 (5) (2019), 327–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Christopher M, Belghith A, Bowd C, Proudfoot JA, Goldbaum MH, Weinreb RN, et al. Performance of deep learning architectures and transfer learning for detecting glaucomatous optic neuropathy in fundus photographs, Sci. Rep. uk 8 (1) (2018) 16685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Nishihira T, Ohjimi H, Eto A, A new digital image analysis system for measuring blepharoptosis patients’ upper eyelid and eyebrow positions, Ophth. Plastic Rec. Surg 72 (February (2)) (2014) 209–213. [DOI] [PubMed] [Google Scholar]
- [33].Chun YS, Park HH, Park IK, Moon NJ, Park SJ, Lee JK, Topographic analysis of eyelid position using digital image processing software, Acta. Ophthalmol. (Copenh) 95 (November (7)) (2017) e625–32. [DOI] [PubMed] [Google Scholar]
- [34].Bodnar ZM, Neimkin M, Holds JB, Automated ptosis measurements from facial photographs, JAMA Ophthalmol. 134 (February (2)) (2016), 146–5. [Internet]. Available from: https://jamanetwork.com/journals/jamaophthalmology/fullarticle/2473361. [DOI] [PubMed] [Google Scholar]
- [35].Lou L, Yang L, Ye X, Zhu Y, Wang S, Sun L, et al. A novel approach for automated eyelid measurements in blepharoptosis using digital image analysis, Curr. Eye Res 44 (September (10)) (2019) 1075–1079. [DOI] [PubMed] [Google Scholar]


