Abstract
Marburg virus (MARV) is a lipid-enveloped virus harboring a negative-sense RNA genome, which has caused sporadic outbreaks of viral hemorrhagic fever in sub-Saharan Africa. MARV assembles and buds from the host cell plasma membrane where MARV matrix protein (mVP40) dimers associate with anionic lipids at the plasma membrane inner leaflet and undergo a dynamic and extensive self-oligomerization into the structural matrix layer. The MARV matrix layer confers the virion filamentous shape and stability but how host lipids modulate mVP40 oligomerization is mostly unknown. Using in vitro and cellular techniques, we present a mVP40 assembly model highlighting two distinct oligomerization interfaces: the (N-terminal domain [NTD] and C-terminal domain [CTD]) in mVP40. Cellular studies of NTD and CTD oligomerization interface mutants demonstrate the importance of each interface in matrix assembly. The assembly steps include protein trafficking to the plasma membrane, homo-multimerization that induced protein enrichment, plasma membrane fluidity changes, and elongations at the plasma membrane. An ascorbate peroxidase derivative (APEX)-transmission electron microscopy method was employed to closely assess the ultrastructural localization and formation of viral particles for wildtype mVP40 and NTD and CTD oligomerization interface mutants. Taken together, these studies present a mechanistic model of mVP40 oligomerization and assembly at the plasma membrane during virion assembly that requires interactions with phosphatidylserine for NTD–NTD interactions and phosphatidylinositol-4,5-bisphosphate for proper CTD–CTD interactions. These findings have broader implications in understanding budding of lipid-enveloped viruses from the host cell plasma membrane and potential strategies to target protein–protein or lipid–protein interactions to inhibit virus budding.
Keywords: lipid bilayer; lipid-binding protein; lipid–protein interaction; Marburg virus; phosphatidylinositol-4,5-bisphosphate; phosphatidylserine; phospholipid; plasma membrane; virus assembly; VP40
Abbreviations: APEX, ascorbate peroxidase tagging; CTD, C-terminal domain; DAB, 3,3’-diaminobenzidine; EBOV, Ebola virus; EGFP, enhanced green fluorescent protein; eVP40, Ebola virus VP40; GBP, GFP-binding protein; GP, generalized polarization; GUV, giant unilamellar vesicle; HDX-MS, hydrogen–deuterium exchange mass spectrometry; HRP, horse radish peroxidase; LUV, large unilamellar vesicle; MARV, Marburg virus; mGP, Marburg virus glycoprotein; mVP40, Marburg virus VP40; N&B, number and brightness; NTD, N-terminal domain; PI(4,5)P2, phosphatidylinositol-4,5-bisphosphate; PIP, phosphoinositide; PMT, photo multiplier tube; PS, phosphatidylserine; SEC, size exclusion chromatography; TEM, transmission electron microscopy; VLP, virus-like particle; WGA, wheat germ agglutinin; WNL-mVP40, W83R/N148A/L226R Marburg virus VP40
The Filoviridae family of viruses, which includes Marburg virus (MARV) and its cousin Ebola virus (EBOV), has been responsible for several highly fatal outbreaks since the late 1960s (1, 2, 3, 4, 5). Filoviruses are lipid-enveloped viruses harboring a negative-sense RNA genome, which bud and release new filamentous viral particles from the host cell plasma membrane (1, 6, 7, 8, 9, 10, 11). The viral matrix protein VP40 of MARV and EBOV (mVP40 and eVP40, respectively) is the primary viral component responsible for directing the assembly and budding of viral particles from the host cell plasma membrane inner leaflet (7, 9, 12, 13, 14). Indeed, VP40 expression alone is sufficient to produce virus-like particles (VLPs) with similar morphology to virions when expressed in mammalian cells (7, 9, 13, 14). Understanding the mechanism by which filoviruses assemble to form new virions is tightly related to understanding VP40 structure–function properties upon protein binding to target lipid that may induce or stabilize VP40 oligomers.
VP40 is a peripheral protein, and mVP40 lipid binding was first speculated when the protein was shown to accumulate at intracellular membranes, mostly multivesicular bodies and late endosomes early after its synthesis in cells (9, 10, 15). Later, the critical role of anionic lipids, phosphatidylserine (PS) and phosphoinositides (PIPs), for both mVP40 and eVP40 trafficking and interactions with the plasma membrane inner leaflet has been more well established (16, 17, 18, 19, 20, 21, 22).
VP40 forms a dimer (16, 23) involving interaction at the amino-terminal domain (NTD). In addition, this NTD is involved in large oligomer formation. The carboxy-terminal domain (CTD) containing the lipid-binding loops is important for VP40–membrane interaction (16, 17, 23, 24); it is also involved in proper protein oligomerization at the lipid membrane (18, 23). Homo-oligomerization of the filovirus matrix protein is a key and required step for budding of virions (21, 22, 23, 25, 26, 27, 28). mVP40 and eVP40 are 34% identical in their amino acid sequence but only 16% identical in their CTDs, which gives rise to their different lipid binding selectivity. Differences in their CTDs may also contribute to differences in their oligomerization at the plasma membrane and within the cell. Indeed, mVP40 was previously described as forming large structures in cells (29, 30) and an octamer ring was observed when only the NTD (1–186 aa) was purified (29). Timmins et al. (29) hypothesized that the paucity of distinct higher-ordered mVP40 oligomeric structures was a result of the extremely high propensity of mVP40 (1–186 aa) to oligomerize, indicated by the presence of extensive stacked rings (29). The same investigation successfully captured four distinct eVP40 oligomeric states, suggesting that mVP40 and eVP40 oligomerization may have fundamental differences (29). Furthermore, the dimeric and hexameric eVP40 crystal structures were resolved in 2013 lending significant insight to the origins of eVP40 lipid binding and oligomerization (23).
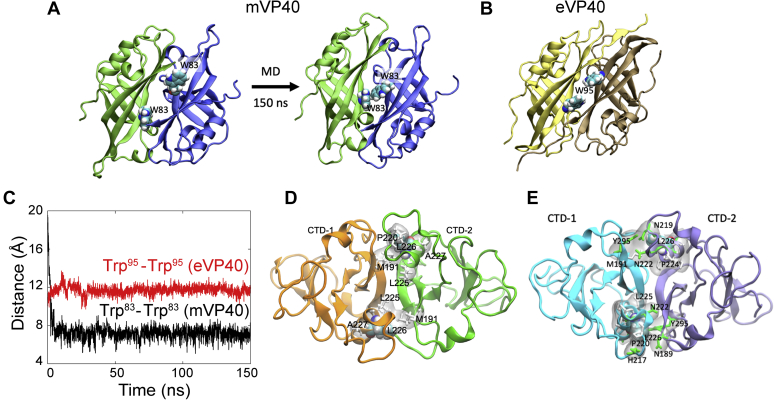
The first proposed model of eVP40 oligomerization and arrangement in 2013, VP40 was shown to form hexamers as building blocks of the matrix layer, postulating that electrostatic interactions facilitate the disengagement of the VP40 CTD from the NTD during matrix assembly. This disengagement sets up a conformational change that exposes two key residues within the NTD, Trp95 and Glu160, as part of an oligomeric interface. In 2016, the dimeric structure of mVP40 was resolved (16) revealing a conserved Trp (Trp83) and Asn (Asn148) in mVP40 that align with eVP40-Trp95 and Glu160 (Fig. 1A, NTD panel), respectively. In a previous study, we reported that the Trp83 residue was in a region that exhibited significant shielding during mVP40 membrane association using hydrogen–deuterium exchange-mass spectrometry (HDX-MS) analysis (18), suggesting it may be important for mVP40 oligomerization following binding to anionic lipids. Furthermore, the previous work demonstrated a reduction of deuterium exchange at the CTD involving residues Leu226 and Ser229 when mVP40 was bound to anionic membranes ((18), Fig. 1A). Of note, this region, dubbed α-helix 4 (α4 helix), just underlies the lipid-binding surface and is distinct in residue composition and in structure when compared with eVP40. Therefore, we postulated two separate oligomerization interfaces within dimeric mVP40, one involving the CTD α4 helix and a conserved interface within the NTD, as key regulators of mVP40 oligomerization (18). In this study, the initial model of VP40 oligomerization was used. This model involved the formation of VP40 hexamer at the membrane as a building block of the viral matrix assembly. However, a new model of linear arrangement of dimeric VP40 forming a 2D lattice that assembles across the virion membrane has been reported (31). In this model, the linear arrangement of VP40 was shown to be mediated by CTD–CTD interactions (31).
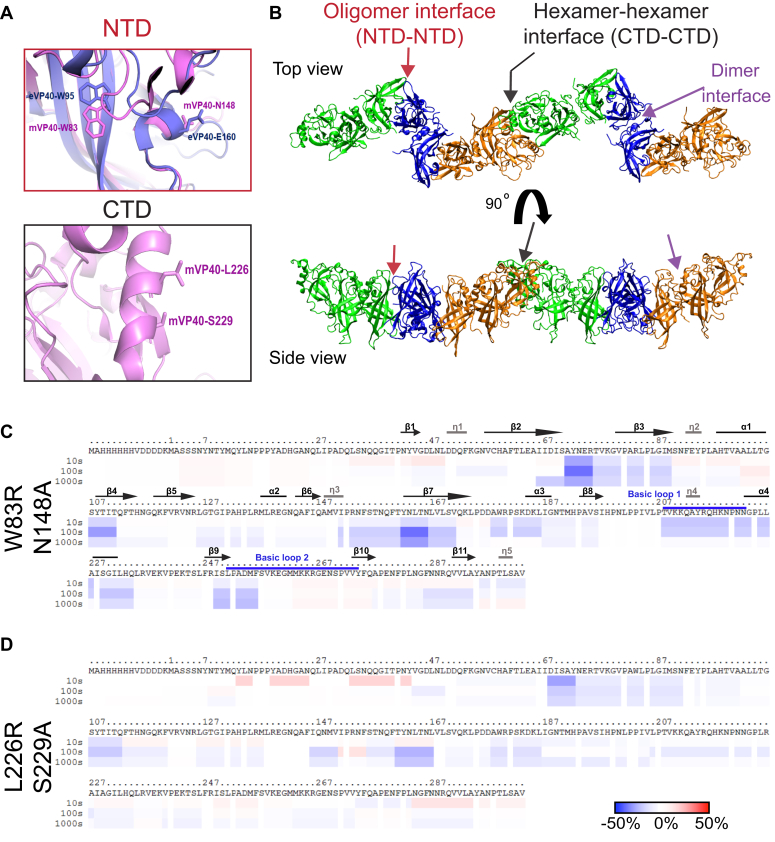
Figure 1.
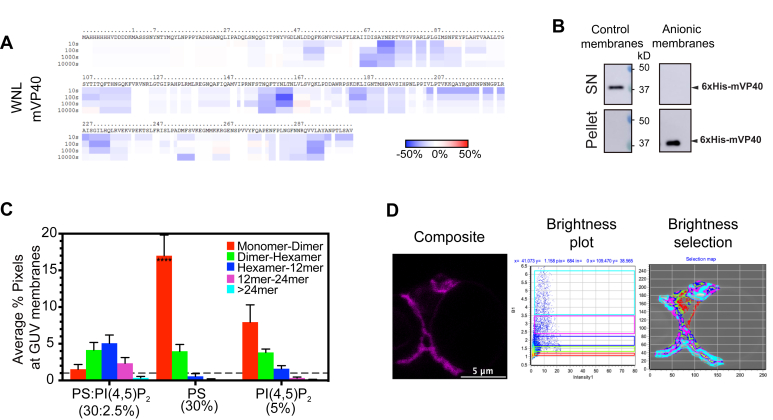
mVP40 potential oligomerization interfaces at NTD and CTD regions.A, zoomed in views of the structure of mVP40 at the NTD oligomer interface (top) indicating Trp83 and Asn148 residues (pink) involved in the oligomerization with an overlay of Ebola virus VP40 (eVP40) structure with corresponding residues Trp95 and Glu160 (purple), and at the CTD interface (bottom) showing the potential residues Leu226 and Ser229 involved in dimer–dimer interactions. Modeled using PyMOL (mVP40 Protein Data Bank ID: 5B0V) and (eVP40 Protein Data Bank ID: 4LDB). B, top and side views of a mVP40 filament (two hexamers formed through the NTD–NTD interface, Fig. S1). C and D, ribbon maps of W83R/N148A and L226R/S229A mutants, respectively, indicating the difference in deuteration percentage of mVP40 in the presence of PS-containing liposomes. Each row corresponds to each time point collected (10–1000 s). Color coding: blue indicates the regions that exchange slower and red indicates the regions that exchange faster in the presence of liposomes. CTD, C-terminal domain; NTD, N-terminal domain.
To determine the mechanism of lipid-induced mVP40 oligomerization, we assessed different in vitro lipid binding assays with HDX-MS analysis to study the effect of mutations at potential NTD and/or CTD oligomerization interfaces in mVP40 conformational changes upon binding membranes. Then, we conducted cellular studies to rationally investigate how the NTD and CTD oligomerization interfaces coordinate the matrix of mVP40 at the plasma membrane. Findings described here demonstrate that each oligomerization interface mutant displays a significant defect in VLP budding, impairment in overall and proper mVP40 trafficking, and oligomerization at the plasma membrane.
Results
Effects of phospholipid membrane interaction on mVP40 oligomerization interface mutants
In order to better understand the origins of mVP40 oligomers, we constructed the mVP40 hexamer–hexamer interface using the eVP40 hexamer–hexamer interface as the template (Protein Data Bank ID: 4LDD) and performed a 100-ns molecular dynamics simulation. Figure 1B shows the section of the mVP40 filament composed of two hexamers next to each other involving CTD–CTD interactions (Fig. S1A). To test our hypothesis that both the conserved NTD and newly identified residues in the CTD are involved in mVP40 oligomerization, we first generated several mutant constructs. These included the NTD oligomerization interface double mutant W83R/N148A and a CTD oligomerization interface double mutant L226R/S229A. Size exclusion chromatography (SEC) of purified proteins indicated that all proteins formed dimers in solution (Fig. S2).
To dissect changes in mVP40 residue solvent accessibility and oligomerization in the absence and presence of membranes, HDX-MS experiments were performed on mVP40 mutants incubated with liposomes containing 45% phosphatidylserine (% molar ratio) as described previously (18). In Figure 1C, the ribbon map of the double mutant W83R/N148A indicates the differences in deuterium incorporation (%D) of the protein in presence of PS-containing liposomes compared with the protein alone. Overall, this double mutant showed little detectable changes in HD exchange pattern in both the NTD (from residue Met1 to Lys47) and CTD (from residue Met263 to Ala284). Similarly, residues Lys96-Gly106 on the helix α1 and residues Gln112-Phe120 on the β4-β5 strands exhibited slightly more rapid HD exchanges. Helix α1 is involved in the dimerization of mVP40, and it had been shown previously that the HD exchange at this region is slower in the presence of anionic lipid-containing liposomes (18). The HDX-MS profile of W83R/N148A also showed an increase of HD exchange at the β6 strand (residues Glu140-Gln146) as well as in the region Met261 to Gln276, which is in basic loop 2 and the β10 strand. Oda et al. (16) showed that residues in this region are involved in the efficient binding of mVP40 to PS-containing liposomes. All together, these results suggest that the residues Trp83 and Asn148 are involved in the formation of oligomers that shields these specific regions from exposure to the aqueous environment resulting in slow deuterium incorporation/exchange rates upon binding to PS-containing lipid vesicles. Furthermore, the double mutant W83R/N148A exhibited an intermediate change in deuteration level compared with wildtype mVP40 (WT-mVP40) in the presence or absence of zwitterionic phospholipid (Fig. S1B adapted from (18)).
Next, we analyzed the solvent accessibility of the CTD double mutant L226R/S229A upon binding to PS-containing lipid vesicles. Similar to W83R/N148A, L226R/S229A exhibited an overall increase of the HD exchange profile compared with WT-mVP40 (Fig. S1B), except in the region including residues Ile88-Asn91. Furthermore, no changes of the deuteration level of the β6 strand (residues Glu140-Phe145) were observed for L226R/S229A compared with WT-mVP40, which showed very slow HD exchange in the presence of PS-containing vesicles within the same region (18). As mentioned above, L226R/S229A-mVP40 showed a faster HD exchange profile than WT-mVP40, including the following regions: i) in the NTD from residue Tyr13 to Tyr44 (which contains the β1 strand), residues Glu73-Gly87 (unstructured loop between β2-β3 strands and the N terminus of β3 strand), Phe113-Phe120 (β4-β5 unstructured loop), and residues Ile146-Asp177 (which includes the unstructured loop between helix η3-β7 strand and the entire β7 strand (Fig. 1D); ii) in the CTD of mVP40, the entire region including residues Thr208-Lys264, which contains helices η4 and α4, the unstructured loops between these two helices, β9 strand as well as the unstructured loop β9-β10 harboring basic loop 2, and finally the region containing the β11 strand until the C terminus (Fig. 1D). Altogether, this analysis suggests that mutation of the hypothesized CTD oligomerization interface reduces oligomerization of mVP40 in the presence of PS-containing vesicles resulting in an exposure of residues at or close to the CTD of the protein.
Mutations of key residues in mVP40-NTD or CTD oligomerization interfaces alter mVP40 plasma membrane localization and proper protein oligomerization at the plasma membrane
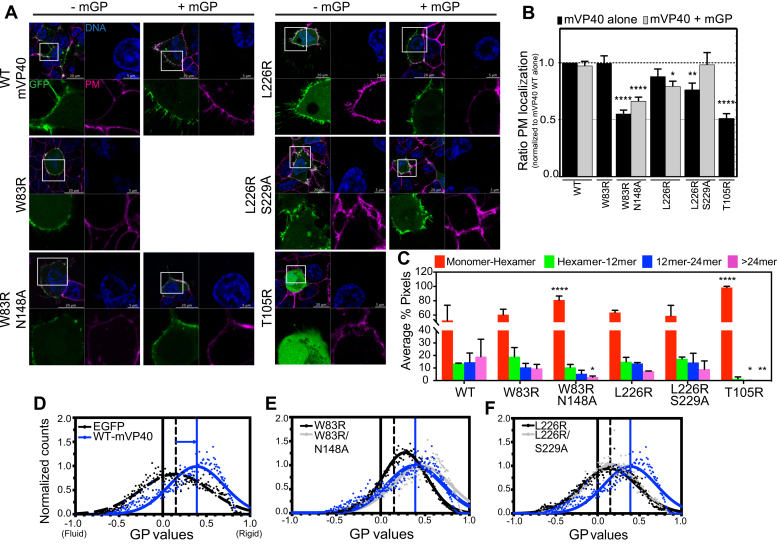
To investigate the biological role of the NTD and CTD oligomerization interfaces of mVP40 on the protein trafficking and binding to the plasma membrane, we performed live cell imaging of EGFP-tagged WT mVP40, single mutants of each oligomerization interface, W83R and L226R, as well as the double mutants W83R/N148A and L226R/S229A (Fig. 2, A and B). Confocal imaging showed the ability of the mutant EGFP-W83R-mVP40 to traffic and localize properly to the plasma membrane (Fig. 2A) to a level comparable with WT-mVP40 (Fig. 2B). In addition, W83R exhibited elongated structures at the plasma membrane similar to WT, which corresponds to assembled VLPs. The double mutant EGFP-W83R/N148A-mVP40 exhibited a similar membrane localization deficiency as the monomeric mutant T105R (Fig. 2, A and B). This result is consistent with previous data described in Oda et al. (16) and Koehler et al. (32). However, unlike WT-mVP40, no significant intracellular aggregations were observed in W83R/N148A-expressing cells 14 h post transfection (Fig. 2A). Coexpression of mVP40 or the mutants with mGP increased the plasma membrane localization of W83R/N148A by 11% (Fig. 2, A and B). Despite this modest increase in plasma membrane localization, no elongated tubulations were detected on the surface of transfected cells (Fig. 2A), which are abundant on cells expressing WT-mVP40 (in absence or presence of mGP, see Fig. 2A, top left panel). These observations may indicate the requirement of an interaction with both Trp83 and Asn148 residues within the NTD oligomerization interface for efficient membrane localization of the protein and assembly into VLPs, consistent with previous studies (16, 32, 33).
Figure 2.
N-terminal domain and C-terminal domain oligomerization interfaces required for efficient mVP40 trafficking and oligomerization at the plasma membrane.A, confocal live images of cells expressing EGFP constructs (green) +/− glycoprotein mGP, stained for DNA (blue) and plasma membrane (PM, pink). B, ratio of PM retention from A quantified by calculating the integrated density of pixels at PM to total pixels within the cell and normalized to WT. C, average % pixels with each estimated oligomerization form from number and brightness analysis performed 24 h.p.t of HEK293 cells with EGFP-mVP40 constructs. Functional budding assays were performed to assess the capacity of WT-mVP40 and mutants to produce virus-like particles. Experimental and fitted normalized generalized polarization (GP) distribution curves of Laurdan dye across PM of HEK293 cells with EGFP (black dashed line) (D), mVP40 mutants of N-terminal domain (E), and C-terminal domain (F) oligomerization interfaces, compared with WT (blue line). GP values range from −1 (very fluid lipid domains) to +1 (very rigid lipid domains). The fitting procedure was performed using a nonlinear Gaussian curve. Values are reported as mean ± SEM (B) or SD (C) of three independent means. One-way ANOVA with multiple comparisons were performed (∗p < 0.05, ∗∗∗p < 0.0005, ∗∗∗∗p < 0.0001).
In contrast, the single mutant EGFP-L226R-mVP40 showed a nonsignificant decrease in plasma membrane localization in both the presence and absence of mGP coexpression (Fig. 2, A and B). However, the double mutant EGFP-L226R/S229A-mVP40 had a more pronounced and significant reduction in localization at the plasma membrane compared with WT-mVP40 (25% reduction) (Fig. 2, A and B). These observations may suggest collaborative interactions at the CTD between L226 and S229 to ensure proper plasma membrane enrichment of mVP40. Both the L226R and L226R/S229A mutants were still able to form filamentous protrusions at the plasma membrane. Furthermore, coexpression of the CTD oligomerization interface double mutant L226R/S229A with mGP appeared to fully recover the wildtype phenotype (Fig. 2, A and B). Taken together, these results indicate that the residues involved in NTD oligomerization interface are essential to matrix assembly at the plasma membrane for the elongation of VLPs while the CTD oligomerization interfaces may be required for efficient trafficking and binding of mVP40 to the plasma membrane of mammalian cells. This was further supported by the lack of reduction in deuterium exchange for L226R/S229A in regions of membrane binding previously mapped for mVP40 (16, 18). As a control, we also analyzed the monomeric mVP40 mutant T105R that had been shown to exhibit a diffuse signal in the cytosol (16). As expected, EGFP-T105R-mVP40 failed to translocate to the plasma membrane (Fig. 2, A and B).
Next, to assess the effect of NTD and CTD oligomerization interface mutations on mVP40 ability to assemble into a matrix in cells, we performed cellular protein oligomerization analysis through number and brightness (N&B) analysis. N&B is a method used to analyze the assembly state of proteins in real time based on the variance of the intensity within single pixels over time (34). Moreover, this technique has been used to evaluate viral matrix protein oligomerization (21, 22). To determine the brightness value for a monomer, GFP was expressed in HEK293 cells, whereas to determine the brightness value of higher-ordered oligomeric states of GFP-mVP40 constructs expressed in HEK293 cells, multiples of the EGFP monomer brightness value were extrapolated to the corresponding oligomeric states. Pixel intensities correlating to monomer–hexamer (red), hexamer–12mer (green), 12mer–24mer (blue), and >24mer (pink) oligomeric states of mVP40 were analyzed, mapped onto the original composite image of the cell, and plotted as a percent of total pixels in the image (see Fig. S3A). The oligomerization profile of EGFP-WT-mVP40 revealed that the largest population of mVP40 was in the monomer–hexamer assembly state (~52.6% total pixels, Fig. 2C, Table S2, Fig. S3A). Of importance, each higher-ordered oligomeric state was roughly equally represented (from ~13% to 19.1% total pixels, Fig. 2C, Table S2, Fig. S3A).
Analysis of the oligomerization profiles of the EGFP-mVP40 mutants differed from the WT oligomerization profile. In the NTD oligomerization interface mutant W83R, we noted ~10% reduction in large oligomers >24mer (from 19.1% to 9.68%) with a nonsignificant but proportional increase of 8% in monomer–hexamer (from 52.62% to 60.7%) and ~6% in hexamer–12mer (from 13.58% to 19.04%) (Fig. 2C, Table S2, Fig. S3A). A similar but more significant pattern was observed for the double mutant W83R/N148A, where a significant increase in monomer–hexamer was observed (~29% increase from 52.6% to 81.2%) concomitantly with a notable decrease of ~16% in oligomers >24mer (decreased from 19.1% to 2.8%) (Fig. 2C, Table S2, Fig. S3A). Given these findings, this analysis demonstrated that the mutants have an impaired ability to form large oligomers and accumulated at the plasma membrane in small oligomers.
In contrast, CTD oligomerization interface mutants did not exhibit a drastic change in their oligomerization profile compared with WT except for a slight decrease in oligomers >24mer (~12% and ~10% reduction for L226R and L226R/S229A, respectively) and modest increase of monomer–hexamer structures (~11% and ~7.5% increase for L226R and L226R/S229A, respectively) (Fig. 2C, Table S2, Fig. S3A). In brief, CTD oligomerization interface mutants have a smaller effect on the ability of the protein to form large oligomers >24mer that may involve other residues and may have a compensatory effect. The monomeric and nonfunctional T105R mutant was used as control and did not show any detectable oligomerization (Fig. 2C, Table S2, Fig. S3A). Altogether, these results support our hypothesis of a potential oligomerization interface in the NTD required for proper and efficient mVP40 matrix assembly at the plasma membrane.
mVP40 matrix assembly through CTD–CTD oligomerization interface modifies plasma membrane fluidity
Membrane reorganization and dynamic remodeling are common processes observed during viral infection and are important for successful viral replication and budding. These membrane changes include the alteration of membrane fluidity and plasticity (35, 36, 37). We speculated that mVP40 lipid-specific binding and oligomerization at the host cell plasma membrane may induce changes in membrane fluidity required for proper matrix assembly and virus particle elongation. To test this, we employed a Laurdan fluidity imaging assay of cells expressing the different EGFP-mVP40 variants (or EGFP plasmid as a control) (Fig. S3B). Laurdan is a fluorescent hydrophobic probe that penetrates cell membranes and aligns parallel to the phospholipid tails (38). In ordered or rigid membranes with a highly hydrophobic environment, the probe has a peak emission wavelength of ~440 nm. However, in fluid membranes water molecules adjacent to the glycerol backbone induce dipolar relaxation of Laurdan, resulting in a spectral shift in the emission wavelength to ~500 nm (39). Changes in membrane fluidity can then be measured by a normalized ratio of the two emission regions and is called the generalized polarization (GP) index (which ranges between −1 and 1, for fluid to rigid membranes, respectively) (38).
The analysis of Laurdan fluorescence was performed under a two-photon confocal microscope and we focused the analysis on cells with the largest enrichment of mVP40 at the plasma membrane (Fig. S3B). The GP index shifted from 0.15 at the plasma membrane of HEK293 cells expressing EGFP to ~0.4 at the plasma membrane of EGFP-WT-mVP40 expressing cells (Fig. 2D). This observation suggests that the binding and proper assembly of mVP40 at the plasma membrane increases membrane rigidity. Next, to investigate if oligomerization mutants have a deficient effect on plasma membrane fluidity, we analyzed the GP index of NTD and CTD oligomerization interface mutants compared with WT-mVP40 (Fig. 2, E and F). Expression of W83R in cells did increase membrane rigidity compared with EGFP alone (GP index ~0.25 in W83R-expressing cells), albeit to a lesser extent than WT (Fig. 2E). Of interest, the double mutant W83R/N148A had a comparable effect as WT on the plasma membrane rigidity, with a GP index of ~0.45 in W83R/N148A-expressing cells (Fig. 2E). Conversely, L226R and L226R/S229A mutants exhibited exactly the same GP index (~0.15) as EGFP-expressing cells (Fig. 2F), indicating that their association with membranes does not change membrane fluidity. All together, these data suggest that the membrane rigidity observed in the wildtype is a result of CTD–CTD oligomerization of the virus matrix at the plasma membrane.
NTD oligomerization-deficient mVP40 mutants fail to produce VLPs
For a clearer understanding of the role of each oligomerization interface in mVP40 multimerization and assembly at the host plasma membrane, we performed transmission electron microscopy (TEM) imaging of the W83R/N148A double mutant and L226R single mutant. We chose these two mutants because of their altered phenotypes in cells and because we observed highly similar VLP structures in the L226R and L226R/S229A-expressing cells from live cell imaging (Fig. 2A). To ensure that we only evaluated cells expressing mVP40, we performed a novel ascorbate peroxidase-tagging (APEX) TEM method that utilizes the coexpression of EGFP-tagged proteins with GFP-binding protein (GBP) fused to APEX2 (GBP-APEX2) (40). Upon coexpression of GBP-APEX2 with GFP-mVP40 proteins, GBP-APEX2 binds to the EGFP tag on mVP40. During TEM processing, APEX2 catalyzes the conversion of diaminobenzidine (DAB) into a precipitate that deposits at the site of the GBP-APEX2:GFP interaction. Following chemical fixation, the precipitate allows a specific and localized signal of EGFP-mVP40 localization with high contrast for TEM imaging.
First, we tested the ability of WT-mVP40 to translocate to the plasma membrane and form the typical elongated structure of VLPs in cells cotransfected with EGFP-WT-mVP40 and GBP-APEX2. As shown in Fig. S4, B and C, the coexpression of the two constructs resulted in normal VLP protrusions from the plasma membrane. To validate this observation, we compared post-stained cells expressing EGFP-WT-mVP40 alone (Fig. S4, A and D) with cells expressing both EGFP-WT-mVP40 and GBP-APEX2 (Fig. S4, B and E) and found that VLP structures between both conditions were morphologically indistinguishable. We also assessed if post-staining APEX2-expressing cells could enrich the contrast detected for TEM, as post-stain enhances membrane staining for organelle identification; therefore, we compared cells expressing GBP-APEX2 and EGFP-WT-mVP40 with and without post-stain. Cells that did not receive post-stain yielded superior APEX2 signal at the membrane of the cell and the VLP membranes where mVP40 is enriched (Fig. S4, B and E) compared with post-stained APEX2 cells (Fig. S3, C and F). The post-stain appeared to introduce artifacts at the plasma membrane that could alter our observations and analysis. Therefore, we decided to continue our investigations of the structures of VLPs in cells expressing mVP40 mutants and GBP-APEX2 without any post-stain processing.
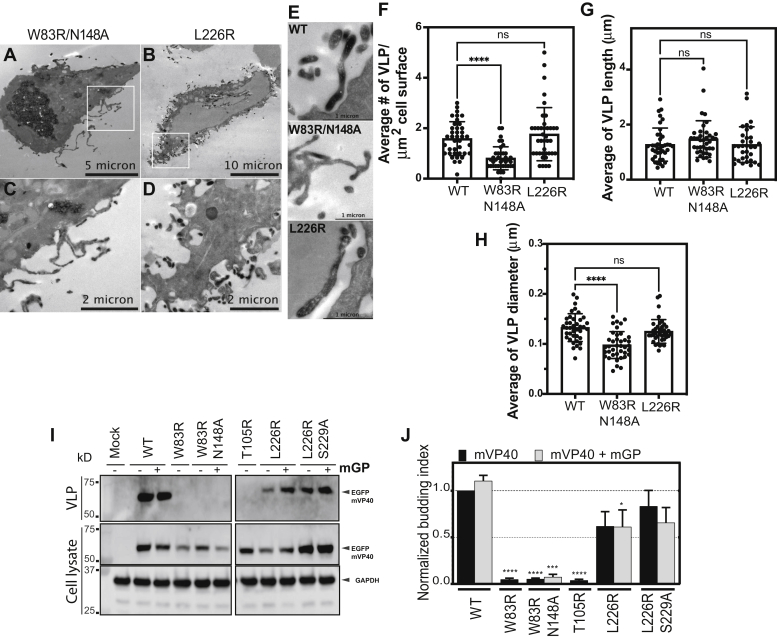
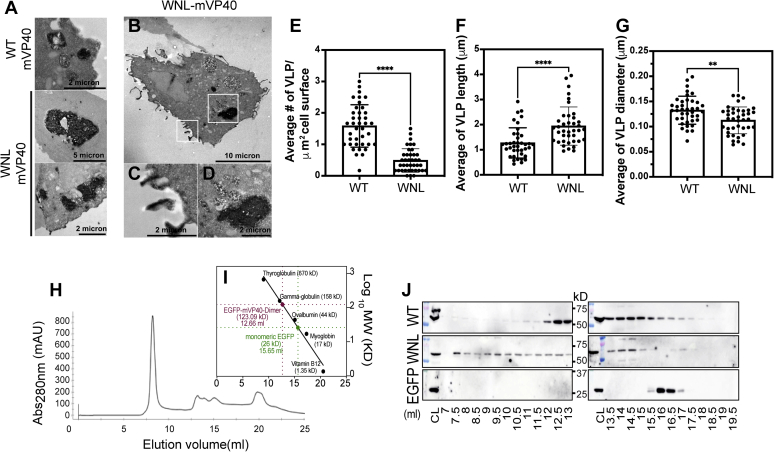
Figure 3, A and B are representative micrographs of cells coexpressing GBP-APEX2 and EGFP-W83R/N148A or EGFP-L226R-mVP40, respectively. The APEX2 signal from EGFP-W83R/N148A mutant was more distributed within the cell in APEX-enriched intracellular structures (Fig. 3A) with some distinct puncta at the membrane and across some tubulations. On the other hand, APEX2 signal from the EGFP-L226R mutant was located at the cell periphery (Fig. 3B) and was detected inside VLP structures (Fig. 3B). In contrast, EGFP-W83R/N148A did not show characteristic VLP structures found at the plasma membrane of WT-mVP40-expressing cells (Fig. 3, C–E); instead, moderate APEX2 signal was detected in ruffled membranes (Fig. 3, C and E). Next, we quantified the abundancy of VLP structures at the surface of cells, indicating a reduction by 50% of the number of VLP or APEX2-positive tubulations per μm2 of cell surface in EGFP-W83R/N148A, whereas the EGFP-L226R mutant had VLP abundancy comparable with WT-mVP40 (Fig. 3F). The analysis of the VLP structures (length and diameter) also highlighted a difference in VLP diameters of EGFP-W83R/N148A but not of EGFP-L226R, whereas both mutants exhibited VLP length similar to that of WT-mVP40 (Fig. 3, G and H). Taken together, the data from these analyses corroborated our confocal imaging results where the mutations of residues within the NTD oligomerization interface impaired mVP40 accumulation at the plasma membrane and the protein ability to assemble and form proper VLPs unlike mutation within the CTD oligomerization interface.
Figure 3.
Cellular oligomerization defects of N-terminal domain and C-terminal domain interface mutants reduce VLP budding.A and B, are representative TEM micrographs of HEK293 cells coexpressing GBP-APEX2 and EGFP-mVP40 W83R/N148A and L226R, respectively. C and D, zoomed insets in (A) and (B), respectively. E, TEM micrographs of potential VLPs at cell surfaces when expressing EGFP-mVP40 indicated constructs. Quantification of the average number of VLP per μm2 of cell surface and VLP length and diameter measurements were performed on TEM micrographs are shown in (F–H), respectively. For VLP length analysis, only full and non-bud VLP were measured. I, representative Western blot assays performed on VLPs (top) and cell samples (middle and bottom) from cells 24 h.p.t in the presence and absence of MARV glycoprotein (mGP). J, quantification of the budding index for each mVP40 protein (normalized to mVP40 WT) was determined by densitometry analysis. Values are reported as mean ± SD of three independent means. One-way ANOVA with multiple comparisons were performed (∗∗∗p < 0.0005, ∗∗∗∗p < 0.0001). VLP, virus-like particle.
To understand the functional significance of mVP40 oligomerization-deficient mutants, functional budding assays of HEK293 cells expressing EGFP-mVP40 or mutants were performed. We hypothesized that mVP40 mutants with aberrant oligomerization would fail to produce VLPs. In addition, an interaction between mGP and mVP40 has been previously reported (41); therefore, coexpression of mGP and mVP40 was performed for some of the functional budding assays. Robust VLP production was observed for cells expressing WT-mVP40, with a slight increase in VLP production when WT-mVP40 was coexpressed with mGP (Fig. 3, I and J). Both NTD oligomerization interface mutants lost their ability to release VLPs as described previously in (16, 32), even in the presence of mGP (Fig. 3, I and J). These results demonstrated that despite the ability of W83R and W83R/N148A mutants to bind and form small oligomers at the plasma membrane, their deficient ability to form large oligomers affects proper VLP release. Furthermore, the mutations at the NTD oligomerization interface may reduce or block interaction of mVP40 with host partners that could be important for proper VLP budding (42, 43).
The CTD oligomerization interface mutants, on the other hand, had a reduction of VLP release of ~40% for L226R (in the presence and absence of mGP) and of ~25% for the double mutant L226R/S229A in the absence of mGP. Furthermore, coexpression of L226R/S229A with mGP resulted in an even more profound reduction in VLP production compared with WT (~40% reduction) than when L226R/S229A was expressed alone (Fig. 3, I and J). The APEX2 TEM analysis of VLP structures at the plasma membrane of the L226R mutant did not show a significant morphological defect (Fig. 3E); however, the functional budding assay suggests a defect in efficient scission from the plasma membrane to form VLPs. Taken together, these observations highlight the importance of the L226 and S229 residues in the CTD oligomerization interface to ensure a functional mVP40 matrix layer, despite the ability of these mutants to multimerize and elongate at the plasma membrane, albeit to a lesser extent than WT. This also underscores the importance of CTD-CTD–mediated changes in membrane rigidity, which may be an important step in the proper matrix layer formation for effective scission. The monomeric nonfunctional T105R mutant was used as control of budding deficiency and failed to produce VLPs (Fig. 3, I and J).
Oligomerization profiles of wildtype mVP40 at the surface of GUVs depends on the anionic lipid compositions
The oligomerization profile in cells of mVP40 variants did not provide adequate details concerning the profile of small oligomers at the plasma membrane. This may be due to nonbound proteins in the cytosol that may make distinction of pixel intensities correlating to monomer–dimer from ones correlating to dimer–hexamer more difficult, in addition to intracellular factors that could also promote protein oligomerization. In order to address this point, we performed N&B analysis on purified 6xHis-mVP40 proteins incubated with giant unilamellar vesicles (GUVs), and this analysis required a fluorescent protein. For this purpose, we used (Ni)-NTA-Atto550 conjugate probe that is specific for poly-histidine tags with minimal cross-reactivity (44).
Previously, Wijesinghe and Stahelin (17) demonstrated that mVP40 associated nonspecifically with the anionic lipids within the plasma membrane (e.g., PS and PIPs). Here, we aimed to understand the oligomerization profile of mVP40 during virus matrix assembly at the plasma membrane, the building block of the virus particles. For this reason, we used the well-established GUV assay, which allows tailored lipid compositions with the ability to incorporate small amounts of fluorescent lipids for visualization. Thus, using the GUVs, we are able to selectively determine binding and oligomerization of mVP40 in the presence of PS, PI(4,5)P2, or both (Fig. 4, A and B, Fig. S5A). Only pixels detected at mVP40-enriched GUV membranes were analyzed and normalized to the total amount of pixels detected to estimate the oligomeric distribution across the in vitro membrane.
Figure 4.
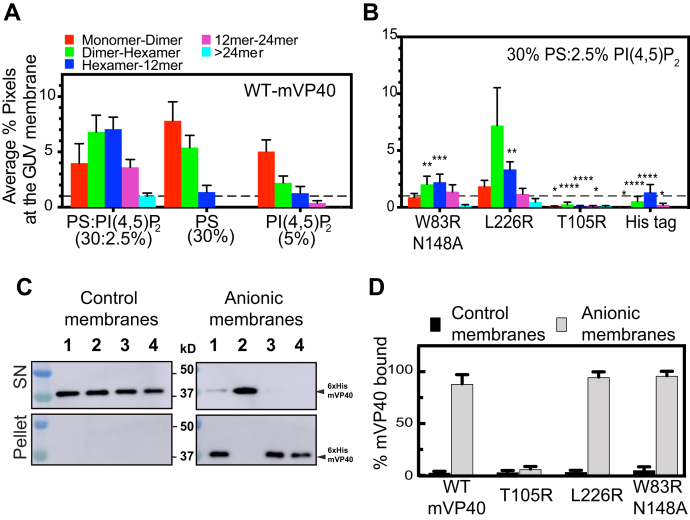
N-terminal domain and C-terminal domain oligomerization interface mutants bind anionic membranes efficiently and display lipid-specific oligomerization profiles.A, plotted average % pixel from number and brightness analysis of WT-mVP40 enriched at giant unilamellar vesicle membranes indicating the oligomerization profile of mVP40. B, oligomerization profiles of W83R/N148A, L226R, the monomeric mutant T105R and His-tag alone at the PS:PI(4,5)P2-containing membranes. C, binding efficiency of WT-mVP40 (lane 1) and mutants (lane 2, T105R; lane 3, L226R; lane 4, W83R/N148A) to anionic membrane (30% PS:2.5% PI(4,5)P2) assessed by liposome sedimentation assay and quantified in (D). Values are reported as mean ± SEM (A and B) or ± SD (D) of three independent means. Two-way ANOVA with multiple comparisons were performed. (∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.0005, ∗∗∗∗p < 0.0001).
This analysis demonstrated for the first time that mVP40 protein oligomerization profiles depend on the lipid composition of the membrane. Indeed, WT-mVP40 is able to bind PS:PI(4,5)P2-containing GUVs, where ~25% of the total pixel counts corresponded to membrane-bound mVP40 (Fig. 4A) (i.e., the total pixel count was taken as 100% bound mVP40 to GUV membrane). Equal population of dimer–hexamer and hexamer–12mer detected at the vesicle membrane corresponded to 60% of membrane-bound protein (30.3 and 31.5%, respectively, Table S3). For the remaining fraction of mVP40 membrane bound, ~17.8% was monomer–dimer and ~16% was 12mer–24mer. Finally, 4.4% of total bound mVP40 were very large oligomers, >24mer. Furthermore, in PS only-containing GUVs, WT-mVP40 was detected mostly as small oligomers with an abundance of monomer–dimer (~53% total bound protein, Table S3) and dimer–hexamer (37% total bound protein). However, only a small population of hexamer–12mer was detected (9.4%) and no larger oligomers could be detected (>12mer) without PI(4,5)P2 in the GUVs.
This first analysis suggests that both PS and PI(4,5)P2 are required for mVP40 to form larger oligomers and assemble the viral matrix. Moreover, in PI(4,5)P2-containing GUVs, a small population of pixels at the membrane of the GUV were detected; this may explain the low abundance of anionic charge at the surface of the membrane (20% of total charge) compared with the previous liposome compositions employed, 50% and 30%, respectively (17). Overall, 56.5% of the bound protein was monomer–dimer, 24.7% dimer–hexamer, 14.3% as hexamer–12mer, and 4.2% 12mer–24mer, whereas no >24mer could be detected (Fig. 4A, Table S3). This result indicates that PI(4,5)P2 promotes mVP40 to form larger oligomers (over 12mer), which requires the presence of PS in the GUVs. Thus, both PS and PI(4,5)P2 are required for large VP40 oligomers suggesting that PS is sufficient to promote small VP40 oligomers such as hexamers, whereas PI(4,5)P2 is likely involved in promoting or stabilizing interactions among dimers and larger aggregates.
In vitro oligomerization of mVP40 is altered upon mutation of key residues
To investigate the effect of NTD and CTD oligomerization interface mutations on in vitro oligomerization, we performed N&B analysis using 6xHis-W83R/N148A-mVP40 and 6xHis-L226R-mVP40 purified proteins. We decided to continue our investigations using only these mutants as W83R/N148A had a more profound phenotype in cells compared with W83R and owing to L226R and L226R/S229A displaying similar phenotypes in cells (aside from impaired plasma membrane binding of L226R/S229A). First, we confirmed by size exclusion that these two mutants formed the dimer (Fig. S2) indicating that the mutations had no effect on the dimerization of the protein. Next, we compared the oligomerization profiles of both W83R/N148A and L226R on GUVs that contained both PS and PI(4,5)P2 (Fig. 4B, Table S3). W83R/N148A showed efficient binding to the GUV membranes; however, large oligomer formation was significantly reduced (Fig. 4B, Table S3). In this analysis, a small population of pixels were detected at mVP40-enriched GUV membranes (6.6% total pixels). Because we focused this analysis on mVP40 enriched GUVs, it is important to note that this small population of detected pixels did not suggest a defect of binding to the GUV membrane. Instead, this result clearly demonstrates the inability of the mutants to enrich properly at the membranes that could be translated by a defect of oligomerization. The distribution of mVP40 consisted of 13.3% of the total bound protein profile as monomer–dimer, 30.3% dimer–hexamer, 33.3% hexamer–12mer, 20.7% 12–24mers, and 2.2% oligomers >24mer (Fig. 4B, Table S3). The major differences between this NTD mutant and the WT oligomerization profile is the increase of the hexamer–12mer and 12mer–24mer population in the W83R/N148A mutant compared with the WT (Fig. 4, A and B, Table S3). In addition, L226R displayed a unique oligomerization profile where the most abundant structures were dimer–hexamer, with over 51% of total bound protein (14% total pixels) at the membrane of GUV containing PS:PI(4,5)P2 (Fig. 4B, Table S3). The other oligomers, in contrast to WT and the NTD mutant, exhibited a decrease in their abundancy with 13.1% monomer–dimer, 23.8% hexamer–12mer, and 8.4% of 12mer–24mer. This result strongly supports our hypothesis that the α4 helix and residue Leu226 plays a critical role in oligomerization by facilitating CTD–CTD interaction to expand the matrix from a hexamer (of dimers) to larger filaments in vitro.
We next extended our investigations into the role of specific lipids in facilitating mVP40 oligomerization at both the NTD and CTD oligomerization interfaces by performing N&B using W83R/N148A and L226R on GUVs that contained only PS or only PI(4,5)P2 (Fig. S5, B and C). First, we observed that both mutants displayed a high abundance at the PS only-containing membrane as a monomer–dimer, 79.5% and 69.1% of total bound protein, respectively. In contrast, a reduction of other oligomers was observed for W83R/N148A and L226R with dimer–hexamer (18.8% and 25.1%, respectively) and hexamer–12mer (1.7% and 5.7%, respectively) (Fig. S5B, Table S3). None of the WT-mVP40 or either mutant was able to form larger oligomers on PS-only GUVs (Fig. 4A, Table S3, Fig. S5B); however, the oligomerization profile of W83R/N148A was notably defective compared with both WT and L226R on PS-only GUVs.
We next performed N&B using GUVs with only PI(4,5)P2. Using PI(4,5)P2 vesicles we demonstrated that the W83R/N148A mutant had an oligomerization profile comparable with that of WT, with a small increase of hexamer–12mer population from 14.3% in the WT to 16.3% in the mutant. W83R/N148A also exhibited a depletion of the >24mer population with less than 1% of total bound protein (Fig. S5C, Table S3). The single mutant, L226R, showed a high enrichment at the membrane of PI(4,5)P2-containing GUVs, where ~22% of total pixels corresponded to bound mVP40, compared with 8.9% for the WT and 11.7% for W83R/N148A, and a higher population of dimer–hexamer with 28.9% of total bound protein (24.7% and 26.4% for WT and W83R/N148A, respectively) (Fig. S5C, Table S3). The other oligomers detected for the L226R mutant were 45.5% of monomer–dimer, 17.5% of hexamer–12mer, 6.0% of 12mer–24mer, and 2.1% of >24mer of total bound protein (Fig. S5C, Table S3). The monomeric mVP40 mutant T105R and 6xHis-tag were used as controls for no binding and oligomerization on GUV membranes (Fig. S5, B and C, Table S3). Taken together, we demonstrated that both W83R/N148A and L226R mutants exhibited oligomerization profiles that are consistent with the role of the mutated residues in mVP40 matrix assembly, where the CTD oligomerization interface mutant L226R displayed an accumulation of dimer-hexamer population in both PI(4,5)P2 and PS:PI(4,5)P2-containing vesicles; on the other hand, NTD oligomerization interface double mutant W83R/N148A accumulated mostly as monomer–dimer at PS only-containing membranes with a deficiency to form other oligomers.
Association with anionic lipids is not altered in mVP40 oligomerization interface mutants
To assess the effect of NTD and CTD interface mutations on the ability of mVP40 to bind PS:PI(4,5)P2-containing membranes in vitro, a liposome sedimentation assay was performed. Large unilamellar vesicles (LUVs) were prepared containing either no anionic lipids (control membranes) or with 30% PS and 2.5% PI(4,5)P2 (anionic membranes). This assay showed a clear ability of all proteins to efficiently bind anionic membranes with no detectable binding to control membranes (Fig. 4, C and D). The monomeric T105R mutant was used as a control and lacked detectable binding to membranes (Fig. 4, C and D) demonstrating the necessity of the intact dimer in binding anionic membranes as previously reported (16). This suggests that neither the NTD nor the CTD oligomerization interfaces are involved in mVP40 binding to anionic phospholipid-containing membranes and that observations from orthogonal experiments are not a result of an inability of the protein to associate with PS or PI(4,5)P2-containing membranes or the plasma membrane.
NTD/CTD oligomerization interfaces triple mutant displayed a unique profile
To deepen our understanding of the oligomerization process of mVP40 and the role of both NTD and CTD oligomerization interfaces in this process as well as in the viral matrix assembly, we generated a 6xHis-mVP40 triple mutant of both the NTD and CTD oligomerization interfaces (W83R, N148A and L226R (WNL-mVP40)). SEC of the purified protein indicated that the triple mutant formed a dimer in solution (Fig. S2). HDX-MS analysis was performed with membranes as described above (Fig. 5A) and demonstrated that WNL displayed an overall decrease of HD exchange compared with WT-mVP40 except for four regions: C-terminal region of β2 strand (Ile66-Ser70), β6 strand (residues Glu140-Phe145), N-terminal region of β7 strand (residues Leu167-Val171), and basic loop-2 with the β10 strand (residues Lys265-Gln276). However, the two last regions had an increased rate of HD exchange at longer time points. On the other hand, some regions showed a slower HD exchange than WT, which included residues Ala71-Arg75 in the loop region between the β2 and β3 strands, residues Tyr162-Asn166 within the β7 strand, the residues constituting helix α3 (Lys183-Ile187), residues Ile249-Val259 found in the β9 strand and the N-terminal region of the basic loop-2, and residues Asn280-Tyr295 in the unstructured loop between the β10 and β11 strands (Fig. 5A). Other regions displaying low HD exchange at longer time points included residues in unstructured loops, Tyr157-Asn166 (unstructured loop between η3 and the N terminus of β7 strand), Tyr208-Arg226 (unstructured loop between η4 and helix α4), and Leu235-Lys239 (unstructured loop helix α4-β9 strand). WNL-mVP40 had a comparable ability to bind PS-PI(4,5)P2-containing membranes akin to WT-mVP40 (Fig. 5B). As expected, WNL-mVP40 did not bind control membranes (neutral) indicating that the triple mutation did not affect the lipid binding efficiency of the protein. Taken together, these analyses provide insight into a potential stable structure rearrangement or oligomerization of WNL-mVP40 upon binding to anionic membranes.
Figure 5.
In vitro study of N-terminal domain/C-terminal domain oligomerization interfaces triple mutant WNL-mVP40.A, ribbon maps of W83R/N148A/L226R (WNL) mutant, indicating the difference in deuteration percentage in the presence of PC:PS (55%:45%) liposomes over the entire exchange period. Each row corresponds to each time point from 10 to 1000 s. Color coding: blue indicates the regions that exchange slower and red indicates the regions that exchange faster in the presence of liposomes. B, liposome sedimentation assay of WNL-mVP40 was performed using control membranes (no anionic lipids) or anionic membranes (30% PS:2.5% PI(4,5)P2). C, oligomerization profile of WNL according to different anionic membranes 30%PS:2.5%PI(4,5)P2 (molar ratio), 30% PS only, and 5% PI(4,5)P2 only, determined from number and brightness analysis. D, representative original composite of the time-lapsed images (left), the number of pixels versus intensity plot (middle), and brightness selection plot of the 30%PS:2.5%PI(4,5)P2-containing giant unilamellar vesicle (GUV) (right). Two-way ANOVA with multiple comparisons were performed compared with WT-mVP40. (∗∗∗∗p < 0.0001).
To test this hypothesis and assess the ability of the triple mutant to oligomerize in the presence of anionic membranes, we performed in vitro N&B analysis with GUVs as described above. The data are summarized in Figure 5C and Table S3 with respect to the oligomerization profile of WNL-mVP40 in the presence of different anionic membranes, whereas Figure 5D shows the ability of the mutant to bind and enrich efficiently at PS:PI(4,5)P2 membranes (composite panel). In these analyses, we demonstrated that ~13% of total pixels detected were enriched protein at the membrane of the GUVs containing either PS:PI(4,5)P2 or PI(4,5)P2 only. However, the oligomerization profiles of the protein were different in the two lipid membranes. In short, PS:PI(4,5)P2-bound protein formed mostly hexamer–12mer (~37.8% of total bound protein), 30.9% dimer–hexamer, 17.4% 12–24mer, 11.3% monomer–dimer, and about 2.6% were larger oligomers (>24mer, Fig. 5C). On the other hand, and using PS only-containing membranes, the mutant displayed mostly a monomer-dimer profile at the GUV membrane with more than 78% of total bound protein, 18.4% were dimer–hexamer, and 2.6% were hexamer–12mer (Fig. 5C). Finally, on PI(4,5)P2-containing membranes, WNL-mVP40 was mostly abundant as monomer–dimer with about 57.7% of total bound protein, 27.6% dimer–hexamer, 11.6% hexamer–12mer, ~2.4% 12mer–24mer, and a very small population could be detected as >24mer (less than 1%). All together, these analyses indicate that WNL-mVP40 exhibited an oligomerization profile comparable with but more exaggerated than the NTD oligomerization interface mutant W83R/N148A-mVP40 (Figs. 4B and 5C). Altogether, in vitro analysis of NTD/CTD oligomerization interfaces demonstrate that they are not involved in the ability of mVP40 to bind anionic membranes; however, both the NTD and CTD interfaces are required for efficient mVP40 oligomerization at the membrane and matrix assembly.
NTD/CTD oligomerization interfaces triple mutant is unable to localize and oligomerize at the plasma membrane
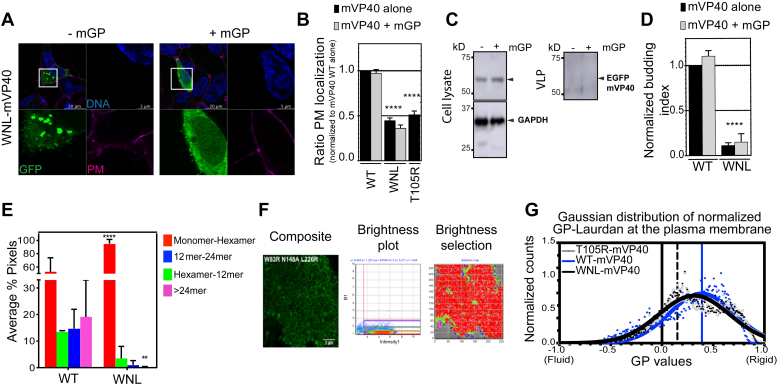
Next, we generated an EGFP-tagged triple mutant of both NTD and CTD oligomerization interfaces (EGFP-WNL-mVP40) to expand on the involvement of these two interfaces in plasma membrane localization. As shown in Figure 6A and B, the triple mutant was unable to localize to the plasma membrane similar to the T105R monomeric mutant (Fig. 2, A and B). Furthermore, the VLP budding efficiency was tested by functional budding assays and indicated that WNL-mVP40 was unable to bud from the plasma membrane (Fig. 6, C and D). In both assays, coexpressing the mutant with mGP failed to rescue the WT phenotype (Fig. 6, A, C and D) indicating that the trafficking and stabilization at the plasma membrane interface was highly dependent on the oligomerization efficiency of mVP40. To confirm that WNL-mVP40 is unable to oligomerize at the plasma membrane, we performed N&B analysis in living cells. Figure 6, E and F revealed that oligomerization of WNL was abrogated, where hexamer–12mer represented only 3.5%, 12mer–24mer 1.1%, and >24mer 0.21% of total pixels detected. Likewise, a significant increase in monomer–hexamer was observed with up to 95% of the total pixels (Fig. 6F, Table S2). Taken together, this analysis supports the requirement of both NTD and CTD oligomerization interfaces for the correct and efficient binding of mVP40 to the plasma membrane of host cells and productive homo-oligomerization to form the viral matrix needed for VLP budding. Based on this phenotype, we were interested to know if W/N/L mutations had an effect on plasma membrane fluidity. Laurdan imaging analysis described in Figure 6G (images in Fig. S3) highlighted the ability of WNL-mVP40 to induce a mild increase of rigidity at the plasma membrane, albeit slightly less than WT-mVP40. Of interest, the phenotype was identical to the monomer mutant T105R-mVP40 with a GP index about 0.3. These data suggest that cellular expression of mVP40 may affect the plasma membrane lipid composition or distribution even before mVP40 resides there.
Figure 6.
WNL-mVP40 mutant had impaired trafficking, oligomerization, and budding from the plasma membrane.A, HEK293 cells, expressing EGFP-constructs ± mGP, stained for DNA (blue) and plasma membrane (PM; pink). B, ratios of PM retention represented as averages ± SEM of three independent means. WT-mVP40 data are extracted from Figure 1B. Statistical analysis was performed as described in Figure 2 (∗∗∗∗p < 0.0001). C, Western blot assay performed on cells and virus-like particle (VLP) quantified in (D) as described in Figure 3. E, average % pixels of estimated oligomerization forms of EGFP- WT and WNL-mVP40 from number and brightness analysis of cellular EGFP-WNL-mVP40 24 h.p.t. (F). G, Gaussian fitted normalized GP index distribution curves of Laurdan across PM of cells expressing EGFP-WNL-mVP40 (black) compared with WT (blue) and T105R-mVP40 (gray) as described in Figure 2.
When performing confocal imaging analysis of WNL-mVP40, we observed GFP-enriched intracellular vesicular structures within transfected cells that probably hindered the protein from reaching the plasma membrane (Fig. 6A, -mGP). To investigate these observations further, TEM analysis on cells coexpressing EGFP-WNL-VP40 with GBP-APEX2 was performed. Figure 7, A–D are representative micrographs of cells coexpressing EGFP-WNL-mVP40 and GBP-APEX2. Trace levels of APEX2 signal were detected at the cell periphery (Fig. 7, A–D). However, a large accumulation of APEX2 signal was observed in the cytosol (Fig. 7, A, B and D). A similar accumulation was observed previously for WT-mVP40 (32) in addition to our TEM experiments (Fig. 7A) and confocal imaging (Fig. 2A). In Figure 7A, we compared the structure of the intracellular accumulations in both WT (top panel) and WNL-mVP40 (middle and bottom panels). We noted that these protein accumulations were more abundant, larger, and less structured in the triple mutant compared with WT. Furthermore, WNL-mVP40 failed to efficiently concentrate and form proper VLP structures at the surface of transfected cells (Fig. 7, E–G).
Figure 7.
WNL-mVP40 mutant accumulates in intracellular structures. TEM micrographs of zoomed intracellular structures EGFP-mVP40 (WT or WNL) coexpressed with GBP-APEX2 in (A), while (B) and insets (C and D) show intracellular accumulations of WNL protein in cells. Quantification of the average number of virus-like particles (VLP) per μm2 of cell surface and VLP length and diameter measurements performed on TEM micrographs are shown in (E–G), respectively. These analyses were performed as described in Figure 3. Values are reported as mean ± SD of three independent means. One-way ANOVA with multiple comparisons were performed (∗∗p < 0.005, ∗∗∗∗p < 0.0001). H, the chromatogram of gel filtration analysis of protein extract from HEK293 cells transfected with EGFP-WT-mVP40 shown as absorbance (280 nm) versus elution volume. Molecular mass standard curve is plotted in (I) as log values of molecular weights versus elution volume. J, Western blot analyses of each protein are indicated. EGFP empty vector served as a negative control. CL, cell lysate.
At this point it was necessary to examine whether the triple mutant displayed a specific oligomerization profile in cells and not only at the plasma membrane. For this purpose, we performed SEC on protein extract from cells expressing either EGFP-WT-mVP40 or EGFP-WNL-mVP40 as described previously (30). Monomeric EGFP alone was used as a control for a protein that does not oligomerize. In brief, cells transiently expressed different constructs for 24 h and proteins were extracted in 1% Triton X-100 prior to separation by SEC (Fig. 7, H–J). Internal molecular weight standards were also used for molecular weight estimation (Fig. 7I). EGFP-WT-mVP40 was detected in different fractions that correspond to the peaks at elution volumes 12.5 to 13.5 ml and 14 to 15 ml (Fig. 7J). These peaks likely correspond to dimeric and monomeric forms, respectively. In addition, a very small amount of protein was detected at elution volumes 10.5 to 11.5 ml that should correspond to larger oligomers. In contrast, EGFP-WNL-mVP40 was detected predominantly in fractions at elution volumes 8.5 to 10 ml along with elution volumes from 12.5 to 13.5 ml and 14 to 15 ml, corresponding most likely to larger homo-oligomeric, dimeric, and monomeric forms (Fig. 7J). Of interest, the protein was also detected in the void volume (elution volume 7 ml), which may indicate the presence of very large oligomers or aggregates (Fig. 7J). These data suggest that the NTD/CTD oligomerization interface triple mutant in the cell is forming the dimer as shown in Figure 6A, and also larger oligomers that block its trafficking to the plasma membrane. EGFP extract eluted from the fast-protein liquid chromatography column in one peak at elution volume 15.5 to 16.5 ml, which corresponded to a monomeric form as expected, supporting that the previous observations are a result of the homo-oligomerization of mVP40.
Molecular dynamics simulation of NTD and CTD oligomerization interfaces
To characterize the differences in the oligomer interfaces of eVP40 and mVP40 (Fig. 8, A and B, Movie S1), we calculated the distance between the tryptophan residues (Trp95–Trp95 for eVP40 and Trp83–Trp83 for mVP40) at the interface. Upon relaxation with molecular dynamics simulation, initially separated Trp83 residues in the modeled mVP40 oligomer interface get closer and interact with each other (Fig. 8A, right panel). This is shown by the significant decrease in the Trp83–Trp83 distance, whereas the Trp95 residues in eVP40 remain separated and the Trp95–Trp95 distance does not change during the simulation window (Fig. 8C). In addition, Asn148 is in close proximity to make occasional backbone hydrogen bonding with Ile88 at the interface.
Figure 8.
Molecular dynamics (MD) simulations of the oligomer interfaces ofmVP40.A, the mVP40 oligomer interface modeled based on eVP40 structure initially shows separated W83 residues as in eVP40 (Trp95) shown in (B). However, upon 150 ns MD simulation, the structure relaxes so that the interface residues W83 interact with each other. C, center of mass distance between Trp83 residues in mVP40 (black curve) and between Trp95 residues in eVP40 (red curve) as a function of time. D, dimer–dimer interface in the mVP40 filament (CTD from each monomer is shown in different colors). The hydrophobic residues within 3 Å of Leu226 at the mVP40 dimer–dimer interface are highlighted. The hydrophobic interaction at the dimer–dimer interface may provide an agile interface, giving flexibility to the filaments. E, zoom into dimer–dimer interface in the mVP40 filament formed through CTD–CTD linear oligomerization as proposed by Wan et al. (31). (CTD from each monomer is shown in different colors). CTD, C-terminal domain.
To investigate the interactions at the CTD–CTD interface formed between the two dimers shown in Figure 1A, we simulated the CTD–CTD complex (Fig. 8D) for 100 ns. Similar to eVP40, this interface consists of primarily hydrophobic residues including Leu226, Pro220, Met191, Ala227, and Leu225. Therefore, Leu226 is part of the hydrophobic core at the interface that provides stability as well as flexibility to the CTD dimer–dimer interface.
Discussion
mVP40 is described as an anionic charge sensor with lack of stereospecificity for PI(4,5)P2 at the plasma membrane inner leaflet. This is in contrast to eVP40, which requires PI(4,5)P2 for proper binding and matrix assembly and has been attributed to differences in sequence homology in the CTD of mVP40 and eVP40 (17, 22). However, it is still not known how the oligomerization of mVP40 occurs and what role anionic lipids play in promoting the proper mVP40-mVP40 oligomerization during the virus matrix assembly. HDX-MS analysis previously conducted on mVP40 in the presence and absence of anionic lipids revealed two potential oligomerization interfaces (18). The NTD oligomerization interface was proposed to include β2, β3 (Trp83 residue), and β7 antiparallel β sheet structures and the CTD oligomerization interface was proposed to include the α helix (Leu226 residue) similarly to higher-ordered oligomerization of eVP40 hexamers (via CTD end-to-end contacts as previously described (23)). These two regions of mVP40 exhibited a reduced deuteration level in the presence of anionic lipids (18). Furthermore, both NTD and the CTD oligomerization interfaces are hydrophobic suggesting multimerization driven by hydrophobic interactions. It is also possible that each interface is involved in a specific lipid-dependent oligomerization pattern of mVP40. To better understand the mechanism of these potential hydrophobic interactions at the NTD and CTD, we replaced the residues Trp83 in NTD and Leu226 in CTD with the charged amino acid arginine to repulse protein–protein interactions in these regions.
The cellular analysis of NTD oligomerization interface double mutant W83R/N148A indicated an inability to enrich at the plasma membrane compared with WT-mVP40. Furthermore, the double mutant had reduced higher-ordered oligomerization, significant increase of small oligomers (monomer–hexamer), and decreased budding efficiency. Similar results had been described previously for untagged or HA-tagged mVP40 (16, 32). W83R/N148A has been described to dimerize in solution (16), but no data were available on the oligomerization pattern of W83R/N148A at lipid membranes. Here, we demonstrated that the mutant is still able to multimerize (hexamer–12mer) but deficient to form higher-ordered oligomers at the plasma membrane. This inability of the NTD mutant can explain the deficiency in VLP formation and its low enrichment at the plasma membrane. This was also observed in vitro using PS:PI(4,5)P2-containing GUVs where only a small population (3.5-fold less than WT) could enrich at the vesicle’s membranes, suggesting that proper enrichment and assembly at these membranes requires proper mVP40 NTD-mediated oligomerization.
The single mutant W83R showed an oligomerization profile similar to WT but still was unable to form VLPs. This may suggest compensation in the Trp83 mutant by the adjacent residue (Asn148). However, the NTD–NTD oligomerization through Trp83 is a key process for VLP elongation and release. Moreover, the mutations at the NTD oligomerization interface had a comparable effect on the membrane rigidity increase compared with WT-mVP40 probably due to lipid rearrangement and/or clustering at the plasma membrane upon protein oligomerization. Lipid rearrangement and clustering (e.g., domain formation) is often required for virus particle budding (45, 46, 47, 48), and a similar phenomenon has been proposed at the inner leaflet of the plasma membrane where eVP40 oligomers significantly enhanced PI(4,5)P2 clustering (49). In this present study, we could not omit the ability of the NTD oligomerization interface mutant to form proper VP40 filament structures, but our data clearly demonstrate a significant deficiency in forming a functional viral matrix.
The CTD region of mVP40 contains two basic loops (1 and 2) involved in anionic lipid interactions. Previous HDX-MS studies highlighted the potential involvement of α helix at the CTD region, including residues Leu226 and potentially Ser229 in hexamer–hexamer interactions (18). The impacts of mutations on the CTD region were nonsignificant (L226R) or mild (L226R/S229A) with respect to cellular localization, mVP40 oligomerization, and VLP release. Of importance, in the presence of the MARV glycoprotein, L226R had reduced plasma membrane localization compared with WT-mVP40 and this consistently resulted in a reduction of VLPs. Furthermore, membrane fluidity analysis demonstrated that both L226R and L226R/S229A are unable to induce changes in plasma membrane rigidity compared with EGFP controls. Thus, we hypothesize that CTD–CTD oligomerization of mVP40 is important but not required to stabilize the matrix assembly that can result in lipid rearrangements at the plasma membrane. In eVP40, the residues Met241 and Ile307 in the center of the CTD–CTD interface are important for proper protein migration to the plasma membrane and VLP formation (23). The eVP40-M241R mutant displayed an exaggerated membrane-ruffling morphology at the surface of transfected cells suggesting that the mutant is able to reach the plasma membrane compared with eVP40-I307R (23). The structural analysis of eVP40-M241R showed that the mutation limited the assembly of dimers to larger filamentous structures and eVP40 interacts with a neighboring dimer via a twisted CTD–CTD interface, with Leu203 and Ile307 at the center rather than Met241 (23).
In the mVP40 dimer, Leu226 aligns with Met241 in the eVP40 dimer. Using both VP40 filament models (23, 31), the residue Leu226, along with Ser229 and Met191, is located outside the hydrophobic core of the CTD interface, whereas residues including Leu225 and Leu293 (aligns with Ile304 in eVP40) are buried inside the hydrophobic core. Although the mutation L226R does not completely block VLP budding, Leu226 may stabilize interactions within the hydrophobic core of the CTD–CTD interface. Our in vitro oligomerization assay of L226R supports this hypothesis. Indeed, L226R enriched 1.5-fold less than the WT-mVP40 at PS:PI(4,5)P2-containing membranes and more than 50% of enriched L226R were small oligomers (dimer to hexamer) with a consistent decrease of larger oligomers (hexamer–12mer, 12mer–24mer, and >24mer). Thus, larger mVP40 oligomers, which are attributed to CTD–CTD are more likely to alter the plasma membrane rigidity. Further analysis of other mutants of the CTD–CTD interface may provide a better understanding of this mechanism of matrix assembly.
Investigation of the role of each phospholipid in the in vitro oligomerization of WT-mVP40 and the mutants was in our opinion critical to better understand the involvement of each oligomerization interface in MARV matrix assembly. This simplified system with GUVs containing only PS, only PI(4,5)P2, or both PS and PI(4,5)P2 provided an important understanding of anionic lipid-dependent mVP40 oligomerization. First, in PS only-containing GUVs, mVP40 forms mostly small oligomers (from monomer to hexamer) with a very small population (9.4%) of hexamer–12mer. This clearly demonstrates that, upon binding to PS, mVP40 clusters at the membrane without further high-ordered oligomerization suggesting that NTD–NTD oligomerization is more prominent in the presence of PS alone. In PI(4,5)P2-containing GUVs, mVP40 was able to form higher-ordered oligomers up to 24mer compared with the previous conditions with a decrease of dimer-hexamer population. This indicated that, upon PI(4,5)P2 binding, mVP40 may undergo conformational changes that promote CTD–CTD interaction for high-ordered oligomerization. In our predictions on the role of Trp83 and Asn148 in mediating NTD–NTD oligomerization and of Leu226 in CTD–CTD oligomerization, the W83R/N148A mutant is most likely to exhibit a deficient oligomerization profile in PS-containing membranes, while L226R should have oligomerization defects in PI(4,5)P2-containing membranes. The relative decrease of oligomerization of W83R/N148A was indeed observed in PS-containing GUVs with a significant accumulation of monomeric–dimeric protein at the membrane. However, L226R showed a very pronounced increase of the dimer–hexamer population in PS:PI(4,5)P2 but not in PI(4,5)P2-containing membranes. This suggested that the role of CTD–CTD interactions is more important in membranes close to physiological compositions and helps explain the inability of L226R and L226R/S229A mutations to increase plasma membrane fluidity upon mVP40 binding and assembly.
In the present study, we provided insight on the potential role of NTD and CTD interfaces in mVP40 membrane enrichment, protein oligomerization and matrix assembly, and VLP budding. Our in vitro analyses using anionic lipid-containing vesicles highlighted the structural changes that CTD and NTD oligomerization interfaces undergo upon lipid binding and oligomerization. It is not completely clear to us how and when the CTD–CTD oligomerization occurs. CTD–CTD interactions may be required at a specific stage of the matrix assembly after protein associates with the plasma membrane and establishment of NTD–NTD interactions to initiate the higher-ordered oligomerization. A recent model proposed CTD–CTD linear oligomerization that is most likely in both MARV and EBOV virions and VLPs (31). However, our study suggests the importance of NTD-NTD oligomerization in cells and in vitro to establish the building blocks for higher-ordered oligomer formation and particle release. It is possible that NTD–NTD interactions are required to increase membrane bending, for elongation of tubule, and/or for host cell factor recruitment at assembly sites. Furthermore, using the recent model proposed in CTD–CTD linear oligomerization ((31), Fig. S6), we simulated the CTD–CTD complex that indicated this interface may involve Met191, Asn222, Tyr295, and Leu226 as we report here (Fig. 6E). Future studies aimed at mutations of this region should help to clarify the detailed interactions that take place at these CTD–CTD interaction sites. Finally, the ability of the triple mutant to bind lipids efficiently and oligomerize suggests an uncommon and uncharacterized homo-oligomerization in cells and with lipid membranes, involving nonstudied residues, which seemingly blocks the protein trafficking to the plasma membrane. Further analysis of structural rearrangement of this mutant can provide precious information on potential oligomerization of mVP40 required for cell signaling and/or trafficking.
In closing, this study demonstrated that mVP40 has two oligomerization interfaces at the NTD and the CTD. Each interface regulates specific protein oligomerization at the plasma membrane in a lipid-dependent manner, membrane fluidity changes, matrix assembly, VLP elongation, and budding. Thus, small molecule or other therapeutic agents can be considered to disrupt the inter- and intramolecular interactions of mVP40 to block the proper viral matrix assembly and prevent release of virus progeny.
Experimental procedures
Site-directed mutagenesis
Site-directed mutagenesis was performed using a Q5 Site-Directed Mutagenesis Kit (New England Biolabs) using primers listed in Table S1 according to the manufacturer’s protocol. The same primer sets were used to generate mutants with pcDNA3.1-EGFP-WT-mVP40 or a 6xHis-tag or EGFP tag in pET46 with the 6xHis-WT-mVP40 vector originally a kind gift from Dr E. Ollmann Saphire (La Jolla Institute for Immunology).
Cell culture and live cell imaging
COS-7 or HEK293 cells were maintained in Dulbecco's modified Eagle's medium (Corning) containing 10% fetal bovine serum and 1% penicillin/streptomycin at 37 °C in a 5% CO2 humidified incubator. Cells were grown until 70% confluency before transfection in 8-well Nunc Lab Tek II chambered slides with 0.16-mm cover glass thickness from Thermo Fisher Scientific. Transfections were performed using Lipofectamine 2000 or Lipofectamine LTX and PLUS reagents (supplied Thermo Fisher Scientific) according to the manufacturer’s protocol.
The enhanced green fluorescent protein (EGFP) signal was imaged 14 h post transfection (performed at 37 °C) on a Nikon Eclipse Ti Confocal inverted microscope (Nikon), using a Plan Apochromat 60x 1.4 numerical aperture oil objective or a 100x 1.45 numerical aperture oil objective, respectively. Cells were stained for 15 min at 37 °C with 5 μg/ml Hoechst 3342 and 5 μg/ml wheat germ agglutinin, Alexa Fluor 647 conjugate (WGA-Alexa Fluor 647, Molecular Probes) in growth media, for nucleus and plasma membrane staining, respectively. A minimum of ten cells per replicate were imaged using the 405-, 488-, and 647-nm argon lasers to excite Hoechst, EGFP, and WGA-Alexa Fluor 647, respectively. Plasma membrane localization ratios were calculated using the integrative density intensities at the plasma membrane determined using the WGA-Alexa Fluor 647 signal compared with the total intensities of the entire cell using ImageJ (50).
Functional budding assays and Western blotting
Functional budding assays were adapted from an established protocol (51). HEK293 cells at 1 to 1.5 × 106 density were transfected with EGFP-mVP40 constructs with or without coexpression of mGP using Lipofectamine LTX and PLUS reagent according to the manufacturer’s protocol. At 24 h post transfection, the media containing VLPs were harvested and collected as previously described (16). Total protein contents (5 μg) from cell lysates and VLP samples were resolved on a 12% SDS-PAGE gel (protein amount appropriate for 15 well gels) prior to transferring on nitrocellulose membrane. Target proteins were detected using the indicated primary antibody followed by the appropriate secondary antibodies horseradish peroxidase (HRP) conjugated. Antibody dilutions used were as follows: 1:200,000 dilution of Rabbit α-mVP40 (IBT BioServices), 1:2000 dilution of Mouse α-GFP (ThermoFisher), 1:10,000 dilution Mouse α-GAPDH (ThermoFisher), and 1:5000 dilution for both Goat α-Rabbit and Sheep α-Mouse (Abcam). HRP signal was detected using Amersham Prime ECL reagent (GE Lifesciences) and imaged on an Amersham Imager 600. VLP budding index of different mVP40 proteins was performed with densitometry analysis using ImageJ (50). The following equation was applied:
RPD is the relative pixel density. The budding index of each mutant was normalized to the WT-mVP40 budding index.
Transmission electron microscopy: Chemical fixation and APEX processing
A total of 5.2 × 105 of HEK293 cells were seeded on 25-mm-diameter poly-L-lysine-coated cover glass. On the next day, 2.5 μg of each APEX2-csGBP plasmid and mVP40 constructs were cotransfected using Lipofectamine LTX reagents. Cells were incubated at 37 °C, 5% CO2 for 6 h; then the transfection medium was changed for Dulbecco's modified Eagle's medium (Corning) containing 10% fetal bovine serum. Cells were then incubated at 37 °C, 5% CO2 for 8 h, after which time cells were rinsed with Dulbecco’s 1× PBS and were chemically fixed with 2.5% glutaraldehyde in 0.1 M cacodylate buffer for 30 min. Fixed cells were then rinsed 3× for 5 min each with 0.1 M cacodylate buffer and washed with 1 mg/ml of 3,3’-diaminobenzidine (DAB) (Sigma-Aldrich) in cacodylate buffer for 2 min. Following the wash, cells were incubated in a freshly made solution of 1 mg/ml of DAB and 5.88 mM of hydrogen peroxide in cacodylate buffer for 25 min on ice. Cells were washed 3× for 5 min each with cacodylate buffer, incubated in an aqueous solution of 1% osmium tetroxide for 10 min, and then washed with distilled water. Dehydration was conducted using increasing concentrations of ethanol (50%, 75%, 95%, and 100% made from 200 proof ethanol), transitioned using 100% acetonitrile, and followed by resin infiltration of the cells using increasing concentrations of Embed 812 Epoxy resin without the accelerator in acetonitrile (2:1 and then 1:2), and finally with Embed 812 containing the accelerator. Coverslips were then embedded on resin-filled beam capsules (cell-face-down) and incubated in an oven at 60 °C for 24 h. After polymerization, coverslips were removed by dipping the coverslip faced block in liquid nitrogen. Serial sections were then collected by sectioning the block samples en face, and ribbons were collected on formvar-coated slot grids.
Thin (90-nm) serial sections were obtained using a UC7 ultramicrotome (Leica) and collected onto formvar-coated copper slot grids (EMS). Glass knives were prepared for trimming, whereas an Ultra 35° diamond knife (Diatome) was used for sectioning the block samples. Sections were screened on a Tecnai T-12 80 kV transmission electron microscope and average 10 to 15 cells were visualized from each sample. VLP quantification and measurements were performed using ImageJ (50).
Number and brightness analysis on mammalian cells
N&B experiments were performed as described (22, 52, 53). HEK293 cells were seeded onto 1.5-mm poly-D-lysine-coated coverslips with 0.17 mm thickness in six-well plates at 70% confluency. Cells were transfected with either EGFP or EGFP-tagged mVP40 constructs as described. Cells were washed 24 h post transfection with 1× PBS, transferred to an Attofluor chamber (Invitrogen), and imaged in Live Cell Imaging Solution (Gibco, Life Technologies) using the Zeiss LSM 880 upright microscope (Carl Zeiss AG) and an LD “C-Apochromat” 40×/1.1 W Corr M27 objective and a 488-nm argon laser to excite EGFP. Each image was acquired using the same laser power (0.01), resolution (256 × 256), pixel dwell time (16 μs), frames (50), and zoom (pixel size of 50 nm). SimFCS Globals Software (Laboratory for Fluorescence Dynamics, University of California) was used for analysis.
On each experimental day, EGFP-expressing cells were imaged and SimFCS4 software (G-SOFT Inc) was used to determine the true brightness (B) of monomeric EGFP (0.058–0.13), which is consistent with previous analyses (54). To calculate the apparent brightness value of mVP40 oligomers, the Bmonomer value was multiplied by the corresponding oligomer value (i.e., dimer = 2, hexamer = 6). Using SimFCS, bins were placed in the brightness plot to correspond with the respective oligomer size. The number of pixels of monomer-hexamer, hexamer–12mer, 12mer–24mer, and 24mer+ bins were recorded. Average % pixels of each oligomeric state was ratiometrically determined by the total number of pixels in each bin versus the total number of pixels in the image.
Laurdan and membrane fluidity analysis
Membrane fluidity analysis was performed according to Owen et al. (55). In brief, 14 h post transfection of HEK293 cells with different mVP40 constructs or EGFP plasmid, cells were treated with 10 μM Laurdan (Invitrogen, stock made in dimethyl sulfoxide at final concentration of 5 mM) in culture media and incubated for 30 min at 37 °C in a humidified 5% CO2 atmosphere. A minimum of ten cells per replicate were then imaged with a Ti-E inverted microscope equipped with Nikon's A1R confocal and a Spectra Physics IR laser tunable to 800 nm for multiphoton confocal imaging of the Laurdan dye, and images were collected with photon multiplier tubes (PMTs) set at 400 to 460 nm and 470 to 530 nm for ordered (PMT1) and disordered (PMT2) membranes, respectively. Calibration images were acquired with 100 μM Laurdan in culture media to calculate the measured generalized polarization (GP) factor. Image processing was done using ImageJ, and GP index distribution was determined using the Laurdan_GP macro (55).
Gel filtration analysis of EGFP and EGFP-tagged WT and mutant mVP40 protein
Human HEK293 cells were transfected with EGFP constructs 24 h prior to protein extraction described in Liu et al. (30). In brief, cells were washed with PBS and lysed with PBS containing 1% Triton X-100, scraped, collected, and incubated on ice for 10 min. Lysates were cleared by centrifugation at 2500g for 10 min at 4 °C and filtrated through a 0.22-μm-pore-size filter. The cleared protein extract was then separated according to protein sizes on Superdex 200 Increase 10/300 GL, fast-protein liquid chromatography column using ÄKTA pure (GE healthcare). Eluted proteins were collected in 0.5-ml fractions and analyzed by SDS-PAGE and then by Western blotting with anti-EGFP antibody, as described above. The chromatogram plotting absorbance (280 nm) versus elution volume was generated with Unicorn 7.2 software.
Protein purification
Purification of mVP40 wildtype, mutants (W83R/N148A, L226R, W83R/N148A/L226R), and 6xHis-tag alone proteins was adapted from a previously established protocol (18). In brief, protein expression was performed overnight at 18 °C with 250 μM IPTG at an absorbance (A600nm) from 0.7 to 0.8. The bacterial pellets were lysed for 30 min on ice in lysis buffer consisting of 20 mM Tris pH 8.0, 500 mM NaCl, 1× halt protease inhibitors, 300 μg/ml lysozyme, 100 μg/ml RNAse, and 3 μM phenylmethylsulfonylfluoride (Thermo Fisher Scientific). The lysed solutions were then subjected to five sonication cycles at 38% (10 s ON, 59 s OFF). After 1 h centrifugation at 15,000g at 4 °C to clarify the lysate from cell debris and membranes, the protein solutions were incubated with Ni-NTA agarose for 30 min at 4 °C with continuous rocking. The proteins were washed with 20 mM Tris pH 8.0 containing 500 mM NaCl and 50 mM imidazole prior to three 5-min stepwise elution with 20 mM Tris pH 8.0 containing 500 mM NaCl and 300 mM imidazole. The mVP40 eluted fractions were washed and dialyzed against storage buffer 20 mM Tris pH 8.0 containing 500 mM NaCl and 20% glycerol using 30K MWCO concentration tubes (or 3K MWCO for His-tag alone purification). The protein purity and enrichment were confirmed by SDS-PAGE and size exclusion using a HiLoad 16/600 Superdex 200 pg column using an ÄKTA pure (GE Healthcare). However, for in vitro assays with lipids, the proteins were used post dialysis.
Liposome sedimentation assays
All lipids used here were purchased from Avanti Polar Lipids, Inc. LUVs were used for liposome sedimentation assays. Lipid mixtures were prepared at the indicated compositions, and chloroform-soluble lipids were dried to form lipid films under a continuous stream of N2. In each experiment, addition of anionic lipids was compensated with an equal mol% decrease in POPC, while POPE (9%) and dansylPE (1%) were held constant. Lipid films were then hydrated in liposome sedimentation buffer (260 μM raffinose pentahydrate in PBS, pH 7.4), vortexed vigorously, and extruded through a 200-nm Whatman polycarbonate filter (GE Healthcare) after incubation at 37 °C. Vesicle size was confirmed by dynamic light scattering using a DelsaNano S Particle Analyzer (Beckman Coulter). LUV solutions were diluted four times in PBS (pH 7.4) to reduce the raffinose pentahydrate concentration, and LUVs were pelleted at 50,000g (22 °C) for 15 min. The supernatant was discarded, and the raffinose-filled LUVs were resuspended in PBS (pH 7.4).
Liposome sedimentation assays were performed as described (56). In brief, protein and LUVs were mixed at final concentrations of 5 μg/ml and 2 mM, respectively, and incubated for 30 min on ice. Following incubation, protein-bound LUVs were pelleted (16,000g, 4 °C, 30 min), and the supernatants containing unbound proteins were transferred into new tubes. The protein-bound LUV pellet was washed in PBS and pelleted again (16,000g, 4 °C, 30 min). The supernatant was discarded, and the pellet was resuspended in an equal volume as the unbound protein supernatant sample. Equal volumes of supernatant and pellet samples were resolved on a 10% SDS-PAGE gel and transferred onto a nitrocellulose membrane. The proteins were detected using the primary antibody (Mouse α-His at 1:2500 dilution, Sigma Aldrich) followed by the HRP-conjugated secondary antibody (Sheep α-Mouse at 1:7000 dilution) The HRP signals were detected and analyzed as described above. To calculate %protein bound the following equation was used:
RPD is the relative pixel density, P corresponds to the pellet fraction, and SNT corresponds to the supernatant fraction.
Giant unilamellar vesicle preparation
GUVs were prepared by a gentle hydration method (57, 58, 59). Briefly, 1 mM lipid of lipid control mixture was made and contained POPC:POPE:POPS:Biotin-PC:fluorescent PC (TopFluor PC) at 59:10:30:1:0.2% molar ratio, or with 2.5% molar ratio brain phosphatidylinositol 4,5-bisphosphate PI(4,5)P2 added with the ratio of POPC adjusted accordingly. PI(4,5)P2-containing lipid mixtures were made by mixing POPC, POPE, PI(4,5)P2, Biotin-PC, and TopFluor PC at 84:10:5:1:0.2% molar ratio. The lipid mixtures were made in a 5-ml round-bottom glass flask, and the chloroform was removed with rotary movements under a continuous stream of N2. The lipid films were then hydrated overnight at 37 °C in an appropriate volume of GUV hydration buffer (10 mM Hepes, pH 7.4 containing 150 mM NaCl, and 0.5 M sucrose).
N&B analysis on GUVs
Freshly made GUVs were diluted ten times in GUV dilution buffer (10 mM Hepes, pH 7.4 containing 150 mM NaCl and 0.5 M glucose) and placed on 6-mm-diameter chambers made from a silicon sheet using a core sampling tool (EMS # 69039-60). The silicon chamber was mounted on a 1.5-mm clean coverglass (EMS # 72200-31) precoated with 1 mg/ml BSA:BSA-Biotin (9:1 molar ratio) for 20 min at room temperature, washed in a water bath, and then overnight at room temperature with 5 μg/ml Neutravidin in PBS. Extra Neutravidin was also washed with water. The setup was then assembled with an Attofluor chamber. GUVs were immobilized for 10 min on BSA:BSA-Biotin and Neutravidin-coated clean cover glasses. mVP40 proteins, 7.5 μM, or His-tag alone was incubated with 50 μg/ml Ni-NTA-Atto 550 dye (Millipore Sigma) in a final volume of 500 μl, overnight at 4 °C. Prior to incubation with GUVs, the proteins were concentrated to 100 μl using 30K MWCO concentration tubes (or 3K MWCO for His-tag alone purification). This step allowed the removal of excess Ni-NTA-Atto 550 not bound to the proteins. The GUVs and proteins were then incubated for 30 min at 37 °C with a final protein concentration of 1.5 μM with the GUVs. N&B analysis was performed with a similar setup as described above with some optimization on a minimum of 11 GUVs per assay. Briefly, at least 100 frames were imaged with Zeiss LSM 880 upright microscope using a Plan Apochromat 63x 1.4 numerical aperture oil objective, laser power: 0.1% using 561-nm laser, image size 256 × 256 pixel, pinhole: 4 μm, scan speed: 8.19 drop μsec, 16 bit depth.
On each experimental day, free NTA-Atto550 dye with GUVs was imaged and the true brightness (B) of a monomeric dye was determined (0.075–0.098). The apparent brightness value of mVP40 oligomers was calculated as described above using SimFCS software. Bins were placed in the brightness plot to correspond with the respective oligomer size. The number of pixels of monomer–dimer, dimer–hexamer, hexamer–12mer, 12mer–24mer, and 24mer+ bins were recorded. Average % pixels of each oligomeric state at the GUV membrane was ratiometrically determined by the total number of pixels in each bin versus the total number of pixels in the image.
HDX-MS analysis
HDX-MS analysis of W83R/N148A, L226R/S229A, and WNL mutants in the presence and absence of anionic lipid vesicles (PC:PS 55:45) was conducted as described in (18).
Molecular dynamics simulations
The mVP40 hexamer structure was modeled based on the eVP40 hexamer (Protein Data Bank ID: 4LDD) as the template. The modeled mVP40 hexamer was relaxed with all atom molecular dynamics simulation using NAMD2.12 (60). For this, an mVP40 hexamer system was set up using Charmm gui solvation builder (61). The system was solvated using TIP3 water molecules in 0.15 M KCl (61). The simulation was performed with Charmm36m force fields, and a SHAKE algorithm was used to treat covalent atoms (62), whereas pressure was maintained using the Nose-Hoover Langevin-piston method. Similarly, the particle mesh Ewald method was used for the long-range electrostatic interactions. After 10,000 steps of minimization and 200 ps equilibration, production simulation was performed for 100 ns at 300 K using 2 fs time step. In addition, two NTDs making up an oligomer interface was simulated for 150 ns. Visual molecular dynamics (63) was used to analyze the trajectories and protein images.
Data availability
The majority of the data are contained within the article. For data not included within the article, data can be shared by contacting the corresponding author.
Supporting information
This article contains supporting information (18).
Conflict of interest
The authors declare they have no conflicts of interest with the contents of the article.
Acknowledgments
S. A., M. L. H., and R. V. S. thank Nathan J. Dissinger for excellent technical support and editing. The authors acknowledge the use of the facilities of the Bindley Bioscience Center, a core facility of the NIH-funded Indiana Clinical and Translational Sciences institute and the use of the Purdue Life Science Electron Microscopy facility.
Author contributions
S. Amiar, M. L. H, K. J. W., and R. V. S. conceived and designed the study. S. Amiar, M. L. H., K. J. W., S. Angel, N. B., P. P. C., and S. L. performed investigation, methodology, and formal analysis. B. S. G., P. P. C., S. L., and R. V. S. provided supervision, project administration, and resources. M. L. H., K. J. W., S. L., and R. V. S. acquired funding for the project. S. Amiar and M. L. H. wrote the original draft with input from all authors. S. Amiar and R. V. S. reviewed and edited the manuscript.
Funding and additional information
These studies were supported by the NIH AI081077 to R. V. S., NIH GM020501, GM121964, AI117905, NS070899 to S. L., NIH T32 GM075762 to M. L. H. and K. J. W., and the Purdue Pharmacy Live Cell Imaging Facility (NIH OD027034 R. V. S.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Dennis Voelker
Supporting information
References
- 1.Leroy E.M., Gonzalez J.-P., Baize S. Ebola and Marburg haemorrhagic fever viruses: Major scientific advances, but a relatively minor public health threat for Africa. Clin. Microbiol. Infect. 2011;17:964–976. doi: 10.1111/j.1469-0691.2011.03535.x. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization . Elsevier; Amsterdam, Netherlands: 2019. Ebola Virus Disease: Democratic Republic of the Congo. [Google Scholar]
- 3.Suzuki Y., Gojobori T. The origin and evolution of Ebola and Marburg viruses. Mol. Biol. Evol. 1997;4:800–806. doi: 10.1093/oxfordjournals.molbev.a025820. [DOI] [PubMed] [Google Scholar]
- 4.Breman J.G., Heymann D.L., Lloyd G., McCormick J.B., Miatudila M., Murphy F.A., Muyembé-Tamfun J.-J., Piot P., Ruppol J.-F., Sureau P., van der Groen G., Johnson K.M. Discovery and description of Ebola Zaire virus in 1976 and relevance to the west African epidemic during 2013–2016. J. Infect. Dis. 2016;214:S93–S101. doi: 10.1093/infdis/jiw207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Slenczka W., Klenk H.D. Forty years of Marburg virus. J. Infect. Dis. 2007;196:S131–S135. doi: 10.1086/520551. [DOI] [PubMed] [Google Scholar]
- 6.Beer B., Kurth R., Bukreyev A. Characteristics of Filoviridae: Marburg and Ebola viruses. Naturwissenschaften. 1999;86:8–17. doi: 10.1007/s001140050562. [DOI] [PubMed] [Google Scholar]
- 7.Noda T., Sagara H., Suzuki E., Takada A., Kida H., Kawaoka Y. Ebola virus VP40 drives the formation of virus-like filamentous particles along with GP. J. Virol. 2002;76:4855–4865. doi: 10.1128/JVI.76.10.4855-4865.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bray M., Geisbert T.W. Ebola virus: The role of macrophages and dendritic cells in the pathogenesis of Ebola hemorrhagic fever. Int. J. Biochem. Cell Biol. 2005;37:1560–1566. doi: 10.1016/j.biocel.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 9.Kolesnikova L., Bugany H., Klenk H., Becker S. VP40, the matrix protein of Marburg virus, is associated with membranes of the late endosomal compartment. J. Virol. 2002;76:1825–1838. doi: 10.1128/JVI.76.4.1825-1838.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kolesnikova L., Bamberg S., Berghofer B., Becker S. The matrix protein of Marburg virus is transported to the plasma membrane along cellular membranes: Exploiting the retrograde late endosomal pathway. J. Virol. 2004;78:2382–2393. doi: 10.1128/JVI.78.5.2382-2393.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bharat T.A.M., Noda T., Riches J.D., Kraehling V., Kolesnikova L., Becker S., Kawaoka Y., Briggs J.A.G. Structural dissection of Ebola virus and its assembly determinants using cryo-electron tomography. Proc. Natl. Acad. Sci. U. S. A. 2012;109:4275–4280. doi: 10.1073/pnas.1120453109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feldmann H., Klenk H.D., Sanchez A. Molecular biology and evolution of filoviruses. Arch. Virol. Suppl. 1993;7:81–100. doi: 10.1007/978-3-7091-9300-6_8. [DOI] [PubMed] [Google Scholar]
- 13.Harty R.N., Brown M.E., Wang G., Huibregtse J., Hayes F.P. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: Implications for filovirus budding. Proc. Natl. Acad. Sci. U. S. A. 2000;97:13871–13876. doi: 10.1073/pnas.250277297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jasenosky L.D., Neumann G., Lukashevich I., Kawaoka Y. Ebola virus VP40-induced particle formation and association with the lipid bilayer. J. Virol. 2001;75:5205–5214. doi: 10.1128/JVI.75.11.5205-5214.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolesnikova L., Berghofer B., Bamberg S., Becker S. Multivesicular bodies as a platform for formation of the Marburg virus envelope. J. Virol. 2004;78:12277–12287. doi: 10.1128/JVI.78.22.12277-12287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oda S., Noda T., Wijesinghe K.J., Halfmann P., Bornholdt Z.A., Abelson D.M., Armbrust T., Stahelin R.V., Kawaoka Y., Saphire E.O. Crystal structure of Marburg virus VP40 reveals a broad, basic patch for matrix assembly and a requirement of the N-terminal domain for immunosuppression. J. Virol. 2016;90:1839–1848. doi: 10.1128/JVI.01597-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wijesinghe K.J., Stahelin V. Investigation of the lipid binding properties of the Marburg virus matrix protein VP40. J. Virol. 2016;90:3074–3085. doi: 10.1128/JVI.02607-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wijesinghe K.J., Urata S., Bhattarai N., Kooijman E.E., Gerstman B.S., Chapagain P.P., Li S., Stahelin R.V. Detection of lipid-induced structural changes of the Marburg virus matrix protein VP40 using hydrogen/deuterium exchange-mass spectrometry. J. Biol. Chem. 2017;292:6108–6122. doi: 10.1074/jbc.M116.758300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruigrok R.W.H., Schoehn G., Dessen A., Forest E., Volchkov V., Dolnik O., Klenk H.D., Weissenhorn W. Structural characterization and membrane binding properties of the matrix protein VP40 of Ebola virus. J. Mol. Biol. 2000;300:103–112. doi: 10.1006/jmbi.2000.3822. [DOI] [PubMed] [Google Scholar]
- 20.Adu-Gyamfi E., Soni S.P., Xue Y., Digman M.A., Gratton E., Stahelin R.V. The Ebola virus matrix protein penetrates into the plasma membrane: A key step in viral protein 40 (VP40) oligomerization and viral egress. J. Biol. Chem. 2013;288:5779–5789. doi: 10.1074/jbc.M112.443960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adu-Gyamfi E., Johnson K.A., Fraser M.E., Scott J.L., Soni S.P., Jones K.R., Digman M.A., Gratton E., Tessier C.R., Stahelin R.V. Host cell plasma membrane phosphatidylserine regulates the assembly and budding of Ebola virus. J. Virol. 2015;89:9440–9453. doi: 10.1128/JVI.01087-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson K.A., Taghon G.J.F., Scott J.L., Stahelin R.V. The Ebola virus matrix protein, VP40, requires phosphatidylinositol 4,5-bisphosphate (PI(4,5)P 2) for extensive oligomerization at the plasma membrane and viral egress. Sci. Rep. 2016;12:19125. doi: 10.1038/srep19125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bornholdt Z.A., Noda T., Abelson D.M., Halfmann P., Wood M.R., Kawaoka Y., Saphire E.O. Structural rearrangement of Ebola virus VP40 begets multiple functions in the virus life cycle. Cell. 2013;154:763–774. doi: 10.1016/j.cell.2013.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Del Vecchio K., Frick C.T., Gc J.B., Oda S.I., Gerstman B.S., Saphire E.O., Chapagain P.P., Stahelin R.V. A cationic, C-terminal patch and structural rearrangements in Ebola virus matrix VP40 protein control its interactions with phosphatidylserine. J. Biol. Chem. 2018;293:3335–3349. doi: 10.1074/jbc.M117.816280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adu-Gyamfi E., Digman M.A., Gratton E., Stahelin R.V. Single-particle tracking demonstrates that actin coordinates the movement of the Ebola virus matrix protein. Biophys. J. 2012;103:L41–L53. doi: 10.1016/j.bpj.2012.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakai K., Okamoto T., Kimura-Someya T., Ishii K., Lim C.K., Tani H., Matsuo E., Abe T., Mori Y., Suzuki T., Miyamura T., Nunberg J.H., Moriishi K., Matsuura Y. Oligomerization of Hepatitis C virus core protein is crucial for interaction with the cytoplasmic domain of E1 envelope protein. J. Virol. 2006;80:11265–11273. doi: 10.1128/JVI.01203-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hilsch M., Goldenbogen B., Sieben C., Höfer C.T., Rabe J.P., Klipp E., Herrmann A., Chiantia S. Influenza A matrix protein M1 multimerizes upon binding to lipid membranes. Biophys. J. 2014;107:912–923. doi: 10.1016/j.bpj.2014.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freed E.O. HIV-1 assembly, release and maturation. Nat. Rev. Microbiol. 2015;13:484–496. doi: 10.1038/nrmicro3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Timmins J., Schoehn G., Ricard-Blum S., Scianimanico S., Vernet T., Ruigrok R.W.H., Weissenhorn W. Ebola virus matrix protein VP40 interaction with human cellular factors Tsg101 and Nedd4. J. Mol. Biol. 2003;326:493–502. doi: 10.1016/s0022-2836(02)01406-7. [DOI] [PubMed] [Google Scholar]
- 30.Liu Y., Cocka L., Okumura A., Zhang Y.-A., Sunyer J.O., Harty R.N. Conserved motifs within Ebola and Marburg virus VP40 proteins are important for stability, localization, and subsequent budding of virus-like particles. J. Virol. 2010;84:2294–2303. doi: 10.1128/JVI.02034-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wan W., Clarke M., Norris M.J., Kolesnikova L., Koehler A., Bornholdt Z.A., Becker S., Saphire E.O., Briggs J.A. Ebola and Marburg virus matrix layers are locally ordered assemblies of VP40 dimers. Elife. 2020;9 doi: 10.7554/eLife.59225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koehler A., Pfeiffer S., Kolesnikova L., Becker S. Analysis of the multifunctionality of Marburg virus VP40. J. Gen. Virol. 2018;99:1614–1620. doi: 10.1099/jgv.0.001169. [DOI] [PubMed] [Google Scholar]
- 33.Hoenen T., Biedenkopf N., Zielecki F., Jung S., Groseth A., Feldmann H., Becker S. Oligomerization of Ebola virus VP40 is essential for particle morphogenesis and regulation of viral transcription. J. Virol. 2010;84:7053–7063. doi: 10.1128/JVI.00737-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Digman M.A., Dalal R., Horwitz A.F., Gratton E. Mapping the number of molecules and brightness in the laser scanning microscope. Biophys. J. 2008;94:2320–2332. doi: 10.1529/biophysj.107.114645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heaton N.S., Randall G. Multifaceted roles for lipids in viral infection. Trends Microbiol. 2011;19:368–375. doi: 10.1016/j.tim.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ketter E., Randall G. Virus impact on lipids and membranes. Annu. Rev. Virol. 2019;6:319–340. doi: 10.1146/annurev-virology-092818-015748. [DOI] [PubMed] [Google Scholar]
- 37.de Armas-Rillo L., Valera M.S., Marrero-Hernández S., Valenzuela-Fernández A. Membrane dynamics associated with viral infection. Rev. Med. Virol. 2016;26:146–160. doi: 10.1002/rmv.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bagatolli L.A., Sanchez S.A., Hazlett T., Gratton E. Giant vesicles, laurdan, and two-photon fluorescence microscopy: Evidence of lipid lateral separation in bilayers. Methods Enzymol. 2003;360:481–500. doi: 10.1016/s0076-6879(03)60124-2. [DOI] [PubMed] [Google Scholar]
- 39.Gaus K., Le Lay S., Balasubramanian N., Schwartz M.A. Integrin-mediated adhesion regulates membrane order. J. Cell Biol. 2006;174:725–734. doi: 10.1083/jcb.200603034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ariotti N., Hall T.E., Rae J., Ferguson C., McMahon K.A., Martel N., Webb R.E., Webb R.I., Teasdale R.D., Parton R.G. Modular detection of GFP-labeled proteins for rapid screening by electron microscopy in cells and organisms. Dev. Cell. 2015;35:513–525. doi: 10.1016/j.devcel.2015.10.016. [DOI] [PubMed] [Google Scholar]
- 41.Kolesnikova L., Ryabchikova E., Shestopalov A., Becker S. Basolateral budding of Marburg virus: VP40 retargets viral glycoprotein GP to the basolateral surface. J. Infect. Dis. 2007;196:S232–236. doi: 10.1086/520584. [DOI] [PubMed] [Google Scholar]
- 42.Lu J., Qu Y., Liu Y., Jambusaria R., Han Z., Ruthel G., Freedman B.D., Harty R.N. Host IQGAP1 and Ebola virus VP40 interactions facilitate virus-like particle egress. J. Virol. 2013;87:7777–7780. doi: 10.1128/JVI.00470-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Makino A., Yamayoshi S., Shinya K., Noda T., Kawaoka Y. Identification of amino acids in Marburg virus VP40 that are important for virus-like particle budding. J. Infect. Dis. 2011;204 Suppl 3:S871–S877. doi: 10.1093/infdis/jir309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.You C., Piehler J. Multivalent chelators for spatially and temporally controlled protein functionalization. Anal. Bioanal. Chem. 2014;406:3345–3357. doi: 10.1007/s00216-014-7803-y. [DOI] [PubMed] [Google Scholar]
- 45.Krementsov D.N., Rassam P., Margeat E., Roy N.H., Schneider-Schaulies J., Milhiet P.E., Thali M. HIV-1 assembly differentially alters dynamics and partitioning of tetraspanins and raft components. Traffic. 2010;11:1401–1414. doi: 10.1111/j.1600-0854.2010.01111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hogue I.B., Grover J.R., Soheilian F., Nagashima K., Ono A. Gag induces the coalescence of clustered lipid rafts and tetraspanin-enriched microdomains at HIV-1 assembly sites on the plasma membrane. J. Virol. 2011;85:9749–9766. doi: 10.1128/JVI.00743-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mücksch F., Laketa V., Müller B., Schultz C., Kräusslich H.G. Synchronized HIV assembly by tunable PIP2 changes reveals PIP2 requirement for stable Gag anchoring. Elife. 2017;6 doi: 10.7554/eLife.25287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Madsen J.J., Grime J.M.A., Rossman J.S., Voth G.A. Entropic forces drive clustering and spatial localization of influenza A M2 during viral budding. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E8595–E8603. doi: 10.1073/pnas.1805443115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gc J.B., Gerstman B.S., Stahelin R.V., Chapagain P.P. The Ebola virus protein VP40 hexamer enhances the clustering of PI(4,5)P 2 lipids in the plasma membrane. Phys. Chem. Chem. Phys. 2016;18:28409–28417. doi: 10.1039/c6cp03776c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rasband W. U. S. National Institutes of Health; Bethesda, MD: 2015. ImageJ [Software] [Google Scholar]
- 51.Harty R.N. Hemorrhagic fever virus budding studies. Methods Mol. Biol. 2018;1604:209–215. doi: 10.1007/978-1-4939-6981-4_15. [DOI] [PubMed] [Google Scholar]
- 52.Bobone S., Hilsch M., Storm J., Dunsing V., Herrmann A., Chiantia S. Phosphatidylserine lateral organization influences the interaction of influenza virus matrix protein 1 with lipid membranes. J. Virol. 2017;91:1–15. doi: 10.1128/JVI.00267-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adu-Gyamfi E., Digman M.A., Gratton E., Stahelin R.V. Investigation of Ebola VP40 assembly and oligomerization in live cells using number and brightness analysis. Biophys. J. 2012;102:2517–2525. doi: 10.1016/j.bpj.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Youker R.T., Teng H. Measuring protein dynamics in live cells: Protocols and practical considerations for fluorescence fluctuation microscopy. J. Biomed. Opt. 2014;19:90801–90810. doi: 10.1117/1.JBO.19.9.090801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Owen D.M., Rentero C., Magenau A., Abu-Siniyeh A., Gaus K. Quantitative imaging of membrane lipid order in cells and organisms. Nat. Protoc. 2012;7:24–35. doi: 10.1038/nprot.2011.419. [DOI] [PubMed] [Google Scholar]
- 56.Julkowska M.M., Rankenberg J.M., Testerink C. Liposome-binding assays to assess specificity and affinity of phospholipid--protein interactions. Methods Mol. Biol. 2013;1009:261–271. doi: 10.1007/978-1-62703-401-2_24. [DOI] [PubMed] [Google Scholar]
- 57.Reeves J.P., Dowben R.M. Formation and properties of thin-walled phospholipid vesicles. J. Cell. Physiol. 1969;73:49–60. doi: 10.1002/jcp.1040730108. [DOI] [PubMed] [Google Scholar]
- 58.Darszon A., Vandenberg C.A., Schönfeld M., Ellisman M.H., Spitzer N.C., Montal M. Reassembly of protein-lipid complexes into large bilayer vesicles: Perspectives for membrane reconstitution. Proc. Natl. Acad. Sci. U. S. A. 1980;77:239–243. doi: 10.1073/pnas.77.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamashita Y., Oka M., Tanaka T., Yamazaki M. A new method for the preparation of giant liposomes in high salt concentrations and growth of protein microcrystals in them. Biochim. Biophys. Acta. 2002;1561:129–134. doi: 10.1016/s0005-2736(02)00338-3. [DOI] [PubMed] [Google Scholar]
- 60.Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Chipot C., Skeel R.D., Kale L., Schulten K.J.J. Scalable molecular dynamics with AMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jo S., Kim T., Iyer V.G., Im W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 62.Huang J., MacKerell A.D., Jr. CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. J. Comput. Chem. 2013;34:2135–2145. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Humphrey W., Dalke A., Schulten K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The majority of the data are contained within the article. For data not included within the article, data can be shared by contacting the corresponding author.