Abstract
The Cdk8 kinase module (CKM) is a dissociable part of the coactivator complex mediator, which regulates gene transcription by RNA polymerase II. The CKM has both negative and positive functions in gene transcription that remain poorly understood at the mechanistic level. In order to reconstitute the role of the CKM in transcription initiation, we prepared recombinant CKM from the yeast Saccharomyces cerevisiae. We showed that CKM bound to the core mediator (cMed) complex, sterically inhibiting cMed from binding to the polymerase II preinitiation complex (PIC) in vitro. We further showed that the Cdk8 kinase activity of the CKM weakened CKM–cMed interaction, thereby facilitating dissociation of the CKM and enabling mediator to bind the PIC in order to stimulate transcription initiation. Finally, we report that the kinase activity of Cdk8 is required for gene activation during the stressful condition of heat shock in vivo but not under steady-state growth conditions. Based on these results, we propose a model in which the CKM negatively regulates mediator function at upstream-activating sequences by preventing mediator binding to the PIC at the gene promoter. However, during gene activation in response to stress, the Cdk8 kinase activity of the CKM may release mediator and allow its binding to the PIC, thereby accounting for the positive function of CKM. This may impart improved adaptability to stress by allowing a rapid transcriptional response to environmental changes, and we speculate that a similar mechanism in metazoans may allow the precise timing of developmental transcription programs.
Keywords: mediator, Cdk8 kinase module, transcription initiation, gene activation, gene regulation
Abbreviations: 1-NA-PP1, 1-Naphthyl-PP1; 4tU, 4-thiouracil; β-ME, β-mecaptoethanol; BS3, bis(sulfo)succinimidyl suberate; CKM, Cdk8 kinase module; cMed, core mediator; CTD, C-terminal repeat domain; DMSO, dimethyl sulfoxide; FDR, false discovery rate; ITA, immobilized template assay; LDS, lithium dodecyl sulfate; MBP, maltose binding protein; PI, protease inhibitor cocktail; PIC, preinitiation complex; pol II, RNA polymerase II; TBP, TATA box–binding protein; TF, transcription factor; TSS, transcription start site; UAS, upstream activation sequence
Transcription of protein-coding genes begins when RNA polymerase II (pol II) and the general transcription factors (TFs)—IIA, IIB, IID (or the TATA box–binding protein [TBP]), IIE, IIF, and IIH—assemble at gene promoters to form a preinitiation complex (PIC). The PIC bends and unwinds promoter DNA and positions the pol II active center at the transcription start site (TSS) for initiation of DNA-templated RNA synthesis. Whereas PICs across different gene promoters are likely to be similar, diversification of transcriptional outputs is mainly achieved by binding of gene-specific TFs to upstream activation sequences (UASs) in yeast or to enhancers in metazoan cells.
TFs that are bound to UASs or enhancers communicate their signals through coactivator complexes such as mediator and the Spt-Ada-Gcn5 acetyltransferase complex. These coactivators are large and modular complexes providing a plethora of possibilities to accommodate interactions with hundreds of different gene-specific TFs (1). Coactivators may facilitate chromatin accessibility, modulate the frequency of PIC formation events, or stabilize PICs to favor their formation and/or dissociation and release of pol II into productive transcription.
The mediator complex is a general coactivator and has a molecular weight of over 1 MDa. Mediator is composed of 25 subunits in yeast and 30 subunits in human (2). Early studies of mediator structure identified four structural modules called the head, middle, tail, and Cdk8 kinase module (CKM) (3, 4). The essential head and middle modules form the core mediator (cMed). Structures of cMed in isolation (5, 6) and interacting with pol II complexes (6, 7, 8) have been determined. Recently, the structure of mediator including the head, middle, and tail modules from mouse was also reported (9). cMed is composed of a head module with two jaws, with a neck, spine, and arm, linking it to the middle module, which extends in an arch forming a knob and hook. cMed interacts with pol II in the PIC, forming three contacts (7). First, the arm/spine of the cMed head module contacts the pol II Rpb4–Rpb7 stalk. Second, the moveable jaw of the cMed head module contacts the pol II dock. Third, there is a transient interaction between the mobile plank domain of the cMed middle module and the pol II foot region. Recently, structures of human mediator–PIC complexes became available showing similar contacts (10, 11).
The CKM is present in a subpopulation of mediator complexes in cells (12, 13), consistent with its dissociable nature. The CKM comprises the four subunits Med12, Med13, Cdk8, and cyclin C and is very large, roughly the same size as cMed (14). Whereas the head, middle, and tail modules of mediator are present at both promoters and upstream regulatory sequences (15, 16, 17, 18, 19), the CKM is present only at upstream sequences but not at promoters (18, 19). The Cdk8 kinase phosphorylates the pol II C-terminal repeat domain (CTD) (20) and various gene-specific TFs resulting in both negative and positive effects on gene transcription (21, 22, 23, 24, 25, 26, 27, 28, 29). Cdk8 phosphorylation increases the turnover of some TFs (30, 31, 32) but is also required for the activity of other factors (22, 25, 26, 27, 33, 34). CKM activity stimulates transcription of genes in stress and signal response networks (21, 35, 36), and its dysregulation has been implicated in over 100 different human cancers (37, 38, 39).
The CKM was proposed to sterically inhibit mediator from interacting with pol II at promoters (40, 41), independently of its kinase activity (42), but this has not been shown directly in a highly defined biochemical system. Also, it is not understood how the effects of Cdk8 kinase activity on gene expression relate to changes in the interactions between mediator, CKM, and the PIC. Here, we reconstitute an in vitro system to show that the CKM sterically inhibits cMed binding to the PIC, and that CKM-dependent phosphorylation releases this steric inhibition. Our data show that while CKM sterically represses transcription, its Cdk8 kinase activity releases this repression and enables gene activation.
Results
Preparation of recombinant yeast CKM
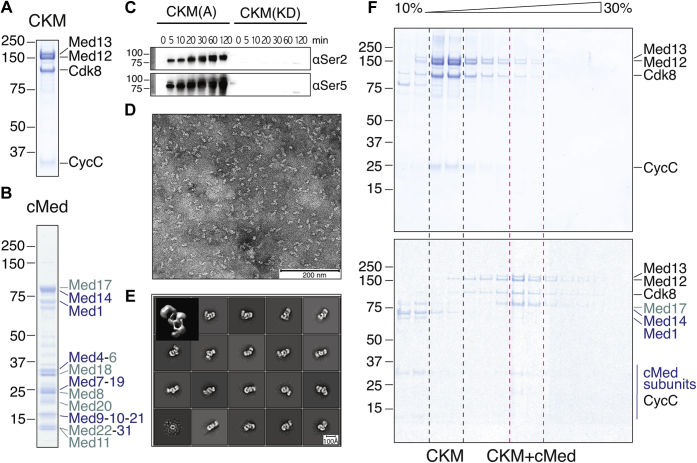
A hurdle toward the biochemical characterization of the CKM was obtaining the complex in sufficient quality and quantity. We established a method for heterologous coexpression of the complete yeast CKM (Fig. 1A) from a single vector in insect cells using a baculovirus-based expression system and its subsequent affinity purification (see Experimental procedures section). We prepared two variants of the CKM module. The first variant contains the catalytically active Cdk8 subunit (CKM(A)), and the second variant contains the kinase-dead Cdk8 mutation D286A (CKM(KD)). Both variants behaved similarly in biochemical purification (Fig. S1). Purified CKM(A) readily phosphorylated the pol II CTD on both serine-2 and serine-5 residues (Fig. 1C). These results show that our recombinant CKM module was active as a kinase on a natural substrate.
Figure 1.
Purification of recombinant, catalytically active, and structurally homogeneous CKM that binds cMed.A, purified recombinant complete CKM containing its four subunits Med13, Med12, cyclin C, and N-terminally Hexahistidine (His)-maltose binding protein (MBP)–tagged Cdk8 used for affinity capture. B, purified recombinant 16-subunit cMed. C, recombinant CKM is catalytically active as indicated by phosphorylation of one of its known substrates, the pol II C-terminal repeat domain (CTD) at serine 2 and serine 5. Conversely, kinase dead CKM (in which we mutated a critical aspartate in the active site to an alanine D286A) is unable to phosphorylate the pol II CTD under the same experimental conditions. D, purified recombinant CKM according to our newly established method distributes evenly on carbon-foil–coated copper grids stained with uranyl formate and shows particles of the expected size, ranging between 150 and 170 Å along the longest dimension. E, 2D classes and a 3D ab initio reconstruction (top left corner) from negatively stained particle images of approximately 60,000 particles show that purified recombinant CKM is homogeneous and forms a three-lobed overall architecture. F, sucrose density gradient ultracentrifugation of purified CKM (top) and CKM–cMed (bottom) shows a shift in the complex-containing fractions to higher density fractions in the CKM–cMed sample compared with CKM alone, indicating complex formation. CKM, Cdk8 kinase module; cMed, core mediator.
Recombinant CKM is structurally defined
We next performed negative stain EM on CKM(KD). We fixed the complex with glutaraldehyde during sucrose gradient ultracentrifugation according to the GraFix protocol (43) and incubated complex-containing fractions on carbon foil–coated EM grids, stained with uranyl formate. EM analysis showed homogeneous particles, evenly distributed over the surface of the grid (Fig. 1D). 2D classes calculated from roughly 60,000 particles showed that the majority of the particles were intact, confirming the homogeneity and quality of the sample. An ab initio 3D model generated using CryoSparc SPA Cryo-EM Software Systems (44) showed an elongated volume with three prominent wedges forming an overall shape resembling the capital letter “E” (Fig. 1E), which is consistent with the low-resolution cryo-EM densities previously reported for endogenously purified CKM (41, 45). While this article was under revision, a higher resolution CKM structure was also reported (46).
Recombinant CKM binds cMed
We next tested whether the CKM can bind to the 16-subunit cMed, which we produced in recombinant form and at high homogeneity as described (7, 8). Binding of the CKM to cMed was evident by a shift in the complex-containing fractions to higher density fractions upon sucrose density gradient ultracentrifugation (Fig. 1F). These results show that recombinant CKM binds cMed in vitro.
CKM binds neither PIC nor PIC–cMed
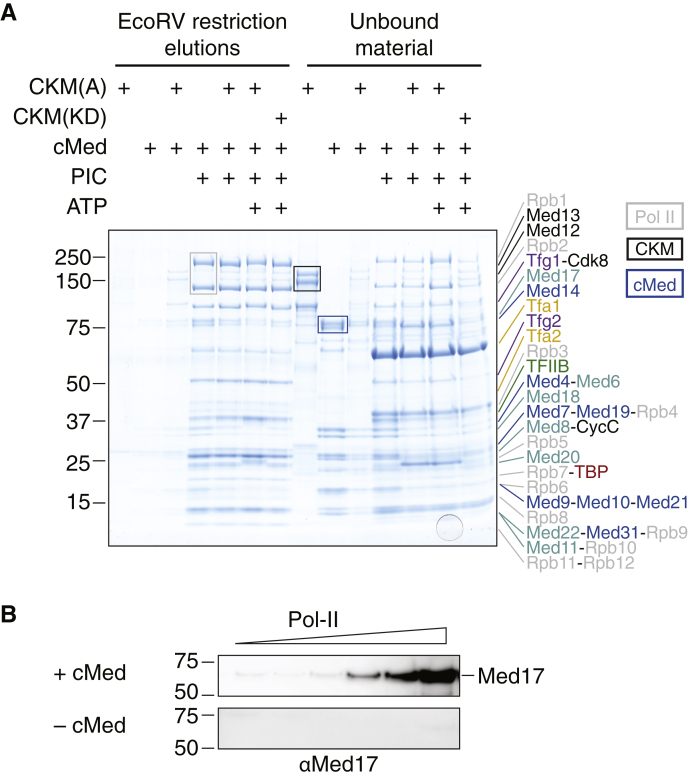
Having purified CKM and cMed at hand, we asked whether these complexes could bind to the pol II PIC. To test this, we set up an immobilized template assay (ITA) using a DNA scaffold containing a promoter sequence modeled on the yeast HIS4 promoter, with a TATA box and a TSS, as a platform for PIC assembly. Biotinylation on the DNA 3’ end allowed immobilization of the template on streptavidin beads, and an EcoRV restriction site downstream of the promoter sequence allowed elution by endonuclease cleavage. We could readily form PICs on the immobilized promoter template (Fig. 2A) providing a positive control.
Figure 2.
CKM is excluded from cMed–PIC and competes with pol II for cMed binding.A, to investigate whether the CKM binds to the PIC together with cMed, core PIC components were added as well as cMed and CKM to biotinylated promoter DNA immobilized on streptavidin beads with a downstream EcoRV restriction digestion site, to allow specific elution. CKM(A) alone, cMed alone, and CKM(A)–cMed alone had only minute background binding to promoter DNA (lanes 1, 2, and 3). Pol II, initiation factors, and cMed bound to promoter DNA forming a cMed–PIC complex (lane 4). CKM did not bind to cMed–PIC above background levels, and this was irrespective of whether CKM(A) (lane 5), CKM(A) in the presence of ATP (lane 6), or CKM(KD) in the presence of ATP (lane 7) was used, indicating that the exclusion of the CKM from cMed–PIC is a steric structural effect, rather than a kinase-dependent one. The unbound components corresponding to each lane (1–7) are shown in the same order in lanes 8 to 14. Characteristic complex bands are indicated with rectangles of the appropriate color for clarity. B, CKM(KD) immobilized on amylose beads was saturated with an excess of cMed (top) and then washed with increasing concentrations of pol II (left to right), and the washes were probed with an anti-Med17 (a cMed subunit) antibody. Pol II competed cMed away from CKM binding as indicated by an increase in the anti-Med17 signal in the washes with increasing added pol II. The same experiment but with no added cMed (bottom) showed no change in the anti-Med17 signal with increasing added pol II. CKM, Cdk8 kinase module; cMed, core mediator; PIC, preinitiation complex; pol II, RNA polymerase II.
We then incubated CKM(A) or CKM(KD) with the immobilized template, purified cMed, and purified PIC components pol II, TFIIA, TFIIB, TFIIE, TFIIF, and TBP. We excluded TFIIH to reduce the complexity of the system to a single catalytic activity being tested. The restriction digestion elutions were analyzed by SDS-PAGE to investigate what was bound to promoter DNA together with the PIC (Fig. 2A). In the presence of PIC components, cMed bound stoichiometrically to the PIC, as expected, but CKM did not bind, indicating that CKM was excluded from the PIC–cMed complex. This was true irrespective of whether CKM(A) alone, CKM(A) in the presence of ATP, or CKM(KD) in the presence of ATP was used. This demonstrates that CKM exclusion from the PIC–cMed complex is predominantly a steric effect, rather than an effect mediated by Cdk8 kinase activity.
CKM–cMed and pol II–cMed are alternative mutually exclusive complexes
To corroborate these findings, we performed an in vitro competition assay with purified CKM(KD), cMed, and pol II. In this assay, we took advantage of the maltose binding protein (MBP) tag on the CKM. After binding of the CKM to beads, overstoichiometric amounts of cMed were added to ensure saturated binding. The beads were then copiously washed to remove any unbound cMed. Finally, wash buffer with increasing concentrations of pol II was added, and the washes were analyzed by Western blot analysis using an antibody against Med17, one of the subunits of cMed. We observed the presence of increasing amounts of Med17 with increasingly added pol II (Fig. 2B). This experiment showed that pol II competes with the CKM for cMed binding and implicated pol II as the PIC component responsible for preventing the incorporation of the CKM into the cMed–PIC complex. Thus, we provide direct biochemical evidence that CKM–cMed and PIC–cMed are mutually exclusive complexes.
Crosslinking MS analysis of CKM–cMed
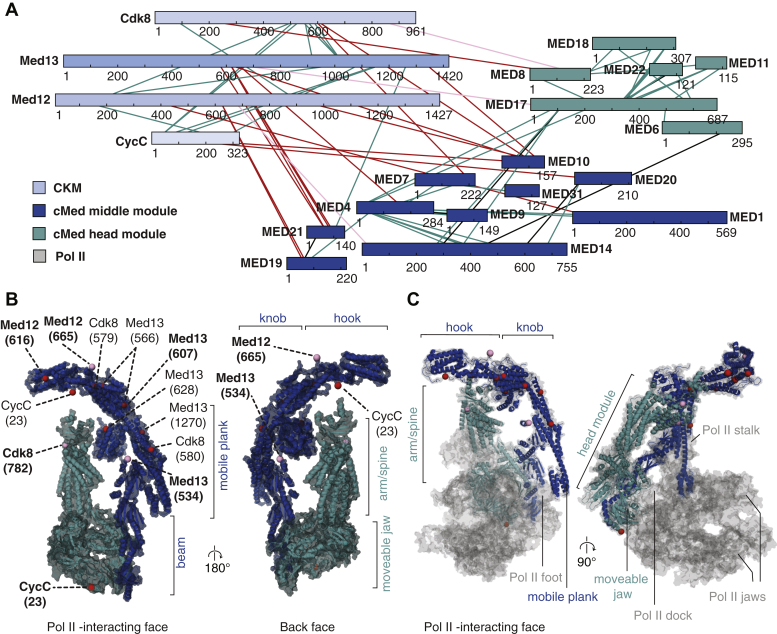
To investigate where on the cMed surface CKM binds, we subjected the CKM–cMed complex to chemical crosslinking coupled to MS (XL–MS). CKM and cMed were mixed at a 1:1 ratio, dialyzed into lower salt, and then crosslinked with 1 mM bis(sulfo)succinimidyl suberate (BS3), after having titrated the crosslinker concentration to a concentration just below complete crosslinking (see Experimental procedures section). The crosslinked complex was then analyzed on a native gel to separate the CKM–cMed complex from the two-component subcomplexes. CKM–cMed was extracted from the gel, trypsin digested, enriched for crosslinked peptides, and subjected to mass spectrometric analysis. The analysis was carried out with a false discovery rate (FDR) cutoff of 1%.
The obtained crosslinking network (Fig. 3A) showed not only extensive crosslinking within the CKM and cMed subcomplexes but also several high confidence crosslinks between CKM and cMed. To validate the interaction map, crosslinks between cMed subunits, for which the structure is already known, were mapped onto the structure (Fig. S2C). Crosslinks used for validation are shown on the interaction map in black, together with the measured atomic distances (Fig. S2C). The appearance of many crosslinks within the expected distance constraints confirmed the validity of the crosslinking network.
Figure 3.
An overlapping binding surface on mediator suggests steric competition between pol II and the CKM for cMed binding.A, bis-Sulfosuccinimidyl suberate (BS3) chemical crosslinking of the CKM–cMed complex shows extensive interactions within and between the CKM and cMed complexes. B, crosslinks between CKM and cMed are mapped on the cMed structure (Protein Data Bank ID: 5OQMSaccharomyces cerevisiae cMed–PIC with all other factors hidden). Exact crosslinking locations are indicated with red spheres. Crosslinks that occurred on residues not seen in the structure are shown on the most proximal residue that is seen in the structure with pink spheres. (These crosslinks are indicated in panel A in the same color as the respective spheres). The complementary lysine residues on the CKM are indicated with dashed lines as “subunit (residue number).” Residues for which more than one unique match were found are emphasized in bold face. C, mapping the crosslinks between CKM and cMed on the cMed–pol II structure (Protein Data Bank ID: 5OQMS. cerevisiae cMed–PIC with only cMed–pol II shown and other factors hidden) shows that some crosslinking appear at or close to the cMed–pol II interaction interface, indicating that the CKM and pol II share an overlapping binding interface on cMed. CKM, Cdk8 kinase module; cMed, core mediator; pol II, RNA polymerase II.
Mapping of CKM-binding site on cMed
The high confidence crosslinks between the CKM and cMed were mapped onto the cMed structure and are indicated by red spheres and corresponding red lines on the interaction network (Fig. 3B). Crosslinks that could not be mapped because of their presence in flexible regions were assigned to the closest structured residue and are indicated by pink spheres and corresponding pink lines on the interaction network. Thus, pink spheres indicate approximate crosslinking locations, and red spheres indicate exact locations. Dashed lines connected to the spheres indicate the complementary residues on the CKM. Crosslinks for which more than one unique match was found are emphasized in bold face.
We found that the vast majority of crosslinks between the CKM and cMed fall on the pol II-binding face of cMed, with a larger number of crosslinks to the cMed middle module than to the head. In particular, a clustering of crosslinking sites to the CKM subunits Med12 and Med13 is seen on the knob and hook domains of cMed. On the other hand, Cdk8 and CycC crosslink to the head module spine and moveable jaw, respectively. These results show that CKM binds on one side of cMed, the side that faces pol II in the PIC–cMed complex, suggesting that CKM and pol II sterically exclude each other in cMed binding.
In Figure 3C, the pol II–cMed interaction within the PIC is shown. Pol II is rendered with low opacity to allow seeing through pol II into the pol II–cMed interface. The presence of crosslinks directly on, and in close spatial proximity to, this interface, suggests that pol II and the CKM share an overlapping binding surface on cMed, indicating that CKM sterically excludes cMed from binding the PIC.
CKM phosphorylates PIC and cMed in vitro
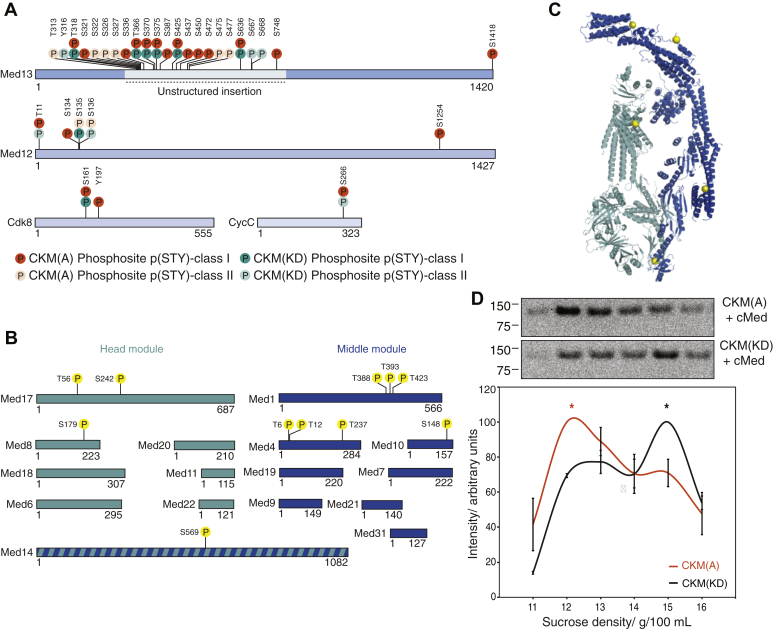
We next investigated the role of the kinase activity of the CKM. We set up a series of exploratory studies to identify novel phosphorylation targets of the CKM within transcription initiation complexes. We used phosphopeptide enrichment and MS to map and compare phosphorylation sites on CKM(KD) and CKM(A). We found that the CKM undergoes extensive intra-CKM phosphorylation in vitro. We mapped these sites onto the CKM subunits (Fig. 4A). Phosphorylation sites found only in the CKM(A) sample, but not in the CKM(KD) sample, and therefore attributed to intra-CKM phosphorylation by Cdk8, are represented in orange. Phosphorylation sites found on the CKM(KD) sample are represented in teal and are likely deposited by the insect cell expression host as they appear on both CKM(A) and CKM(KD).
Figure 4.
The CKM phosphorylates itself and cMed on their respective interaction interfaces.A, CKM undergoes extensive intra-CKM phosphorylation by Cdk8 (orange circles) on all its subunits, but particularly on Med13. Phosphorylation sites are shown that have a localization probability greater than or equal to 0.75 (p(STY) class I) or between 0.25 and 0.75 (p(STY) class II) in phosphopeptide enrichment mass spectrometry. B, CKM phosphorylates cMed on head and middle module subunits (yellow circles). Phosphorylation sites are shown that have a localization probability greater than or equal to 0.75. C, phosphorylation sites deposited by the CKM on cMed plotted on the cMed structure (Protein Data Bank ID: 5OQMSaccharomyces cerevisiae cMed–PIC with all other factors hidden) as yellow spheres lie on the CKM–cMed interaction interface. D, analytical sucrose density gradient ultracentrifugation showing that the peak presence of CKM (represented by anti-MBP antibody signal) occurs in lower density fractions when CKM(A) is used under phosphorylation reaction conditions (top), than when CKM(KD) is used under the same conditions (bottom). The mean band intensities from two replicates were quantified and plotted, and the respective peak positions are indicated with an asterisk of the corresponding color. This demonstrates that active phosphorylation by the CKM favors dissociation of the CKM–cMed complex. CKM, Cdk8 kinase module; cMed, core mediator.
A large number of intra-CKM phosphorylation sites occur on the ∼500-residue unstructured insertion in the Med13 subunit (Fig. 4A). This insertion is conserved from yeast to human and is located near the interface involved in binding cMed (41). To explore the phosphorylation targets of CKM within the PIC, we assembled the PIC on an immobilized template as described previously and incubated these complexes with cMed, CKM(A), and ATP. Only phosphosites with a localization probability p(STY) greater than or equal to 0.75 were considered. We found new phosphorylation sites on cMed (Fig. 4B) and mapped them onto cMed within the known PIC structure (8). This revealed sites on the hook and knob domains of the middle module (Fig. 4C). In addition, phosphorylation sites were found on the arm domain of the head module (Med8 and Med17), and on the scaffolding subunit Med14, close to the head module jaws. Phosphorylation sites were also found on the mobile plank domain of the mediator. All these sites lie on the CKM-interacting face of cMed, which we have previously identified by XL–MS.
CKM phosphorylation weakens CKM–cMed interaction
Because we observed phosphorylation deposited by Cdk8 on both the CKM and cMed on their respective interaction interfaces, we hypothesized that the Cdk8 kinase activity is involved in CKM release from cMed. To test this hypothesis, we mixed cMed with either CKM(A) or CKM(KD) together with ATP under conditions conducive to complete phosphorylation and then applied these complexes to sucrose density gradient ultracentrifugation (Fig. 4D). We took advantage of the MBP tag present on the CKM complex (but not on cMed) to locate CKM in the gradient fractions by Western blot analysis using an anti-MBP antibody. By quantifying the intensities of these bands in each gradient, we could locate the peak fractions in which the CKM was enriched. We performed this experiment in two replicates (Fig. S3C), quantified independently, and normalized against the respective highest intensity band. We found that CKM(A) is enriched in early fractions, corresponding to unbound CKM, whereas CKM(KD) is further enriched in later fractions corresponding to CKM–cMed (Fig. 4D). This indicates that phosphorylation by CKM weakens the CKM–cMed interaction.
Cdk8 kinase activity is required for transcription activation during heat shock
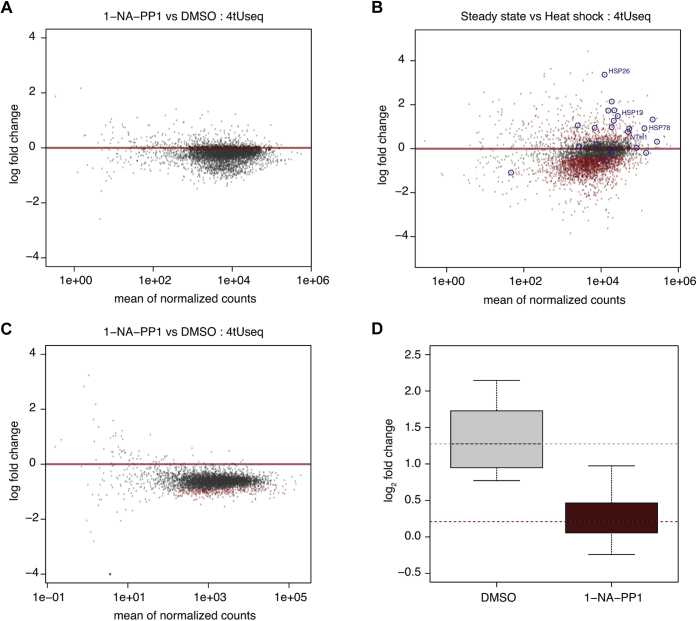
Thus far, our analysis was carried out exclusively in vitro. To investigate the role of the Cdk8 kinase activity in vivo, we used a Cdk8 analog-sensitive yeast strain (24) allowing us to specifically inhibit Cdk8 using the ATP analog 1-Naphthyl-PP1 (1-NA-PP1) (see Experimental procedures section). We applied either 1-NA-PP1 at a final concentration of 6 μM or dimethyl sulfoxide (DMSO) for 12 min, followed by 5 min of RNA labeling with 4-thiouracil (4tU). This allowed us to isolate newly transcribed RNA following the perturbation. We then sequenced the newly synthesized RNA and mapped it to the Saccharomyces cerevisiae genome (sacCer3). We performed each condition in two biological replicates and added spike-ins before RNA extraction to normalize RNA counts for quantification. The biological replicates correlated well (Fig. S3A). We found no statistically significant changes resulting from Cdk8 inhibition under steady-state growth conditions, as demonstrated by genome-wide differential gene expression analysis comparing the inhibited sample to the DMSO control (Fig. 5A).
Figure 5.
Cdk8 activity is required for transcription activation during heat shock.A, minus average (MA) plot shows differential gene expression between the Cdk8 inhibited sample and DMSO control for all protein coding genes (n = 4928) under steady-state conditions. Log2 fold changes are plotted against the mean of spike-in normalized counts over all samples. No significantly changed genes were detected (adjusted p value < 0.1). Whether a gene's expression is called significantly changed depends on both its log2 fold change and its dispersion estimate (see Experimental procedures section for details). B, MA plot shows differential gene expression between 12-min heat shock DMSO samples and the steady-state DMSO samples. Significantly changed genes are shown in red (adjusted p value < 0.1). Temperature response genes (Gene Ontology: 0009266) are encircled in blue. C, MA plot shows differential gene expression between the Cdk8 inhibited sample and DMSO control under heat shock conditions. Significantly changed genes are shown in red (adjusted p value < 0.1). D, box plots showing log2 fold changes in expression of induced heat shock response genes (see B) comparing 12-min heat shock DMSO samples to the steady-state DMSO samples (left) and comparing the 12-min heat shock Cdk8 inhibited samples to the steady state Cdk8 inhibited samples (right) (Wilcoxon rank sum test, p value = 0.0002). DMSO, dimethyl sulfoxide.
We then performed a second experiment, where we once again applied 1-NA-PP1 or DMSO for 12 min and then raised the temperature of the yeast cultures to 37 °C to impose heat shock for 12 min. The effect of heat shock treatment was validated by genome-wide differential gene expression analysis comparing the heat shock DMSO sample to the steady-state DMSO sample (Fig. 5B). Differential gene expression analysis identified significant upregulation of 7.7% and downregulation of 37% of genes, matching the expected pattern for the heat shock response in yeast. Gene ontology analysis showed that heat shock genes (encircled in blue) are enriched within the induced group (Fig. S3B). Under this condition, we found that Cdk8 inhibition resulted in a global downregulation of gene transcription by ∼1.5-fold (Fig. 5C), as indicated by differential expression analysis of the inhibited sample compared with the DMSO control. More specifically, the group of genes induced during the heat shock response in yeast fails to be induced upon Cdk8 inhibition (Fig. 5D). These observations show that Cdk8 kinase activity is required for heat shock gene activation during the heat shock response.
Discussion
In this study, we prepared recombinant yeast CKM in high quality and quantity and used a purified in vitro system to show directly that the CKM binds to cMed but not to the PIC–cMed complex. This biochemical observation is consistent with binding of CKM to UASs but not to promoters in vivo (18, 19, 47). It also indicates that no additional factors are required for exclusion of the CKM from promoters other than the assembled PIC. We mapped the location of CKM binding to cMed using XL–MS and found overlap between the CKM-binding and pol II-binding surfaces on cMed. These results directly demonstrate the mutual exclusivity that was suggested previously from low-resolution EM studies (40, 41) and in vivo interactomics (48, 49). Consistent with our findings, the human CKM can impair activated transcription from a template in vitro, even when its kinase activity is absent (42).
The catalytic subunit of the CKM, the Cdk8 kinase, has previously been shown to phosphorylate a host of targets. Cdk8 phosphorylates the pol II CTD in yeast (14, 20, 24) and human (50). In addition, Cdk8 phosphorylates various activators in both organisms. Cdk8 phosphorylation of the activator Gal4 is necessary for galactose-inducible transcription (22, 25, 26, 27, 33, 34). Cdk8 phosphorylation of activators involved in gluconeogenesis and stress response have also been shown to result in gene activation (23, 29). Conversely, Cdk8 phosphorylation of other activators involved in pathways of starvation and stress response has been found to increase their turnover or nuclear exclusion (30, 31, 32). We identified 11 phosphorylation sites deposited by Cdk8 on cMed subunits, of which two had been reported previously as kinase targets (24, 51, 52, 53). The sites of phosphorylation were predominantly located within mediator's pol II-binding (or CKM-binding) face. We also added new sites within the CKM itself on all its subunits but concentrated on Med13 and the PIC components TBP and TFIIF to the repertoire of known targets. Further regulatory phosphorylation sites had also been known within the mediator tail module (54), which we did not have in our preparations. Although in vitro phosphorylation experiments can sometimes result in spurious activity because of the presence of high concentrations of enzymes and substrates, five sites within the Med13 phosphorylation cluster have also been reported previously in large-scale in vivo phosphoproteomics studies, increasing their likelihood of physiological relevance (52, 53, 55). In a comprehensive analysis of the CKM phosphoproteome in human, sites on the CKM and mediator were identified among tens of other targets, indicating that our observed phosphorylation of these targets is likely conserved and functionally relevant (56).
It has remained unclear how the presence and phosphorylation activity of the CKM regulates transcription. The CKM is generally construed as a repressive molecule, but a large body of evidence has affirmed a positive role of its Cdk8 kinase on gene transcription in both yeast (22, 23, 24, 25, 26, 27, 29) and human (21, 28, 36, 57). Indeed, we found that Cdk8 inhibition impaired induction of heat shock genes in yeast, corroborating a positive role in gene activation. This apparent paradox is mitigated by our biochemical observation that the Cdk8 kinase activity facilitates the release of the CKM from mediator, which would free up its pol II-interacting surface needed for gene activation. As such, active Cdk8 ensures that mediator is free to establish its interaction with the PIC to stimulate transcription initiation.
In summary, the CKM plays a dual function—a repressive structural function and an activating Cdk8-dependent function that counteracts repression during stress response. The Cdk8 phosphorylation sites we identified on Med13 and the cMed surface may explain the sensitivity of the CKM–cMed interaction to Cdk8 activity, likely resulting from electrostatic repulsion.
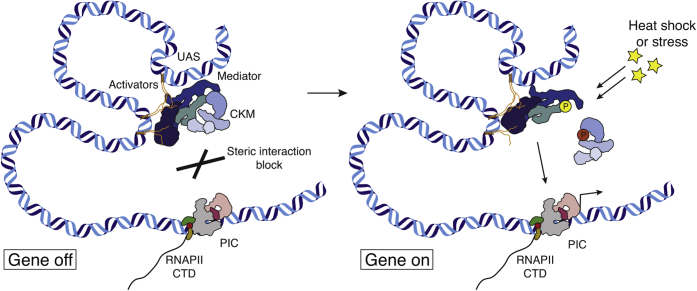
Our results converge with previous literature on a simple model for the role of the CKM module in gene activation (Fig. 6) (58). A stable complex of activators, mediator and the CKM is present at UASs (or at enhancers in metazoan cells). This complex may represent an inactive form of mediator that is unable to activate genes because of masking of its pol II-interacting surface by the CKM. This may explain the presence of the mediator complex on UASs or enhancers, irrespective of the activity status of their target genes, in both yeast (12) and human (59). In our model, the CKM serves to uncouple mediator presence from mediator activity at target genes by steric obstruction. This may be important to retain mediator near target genes and enable their rapid activation upon an external signal.
Figure 6.
Model of CKM steric repression and Cdk8-mediated release during stress response. CKM occludes mediator's PIC interacting interface resulting in a “gene off” status (left). Upon heat shock, Cdk8-dependent phosphorylation frees up the mediator–PIC interaction block, resulting in a “gene on” status (right). CKM, Cdk8 kinase module; PIC, preinitiation complex.
Finally, we suggest that gene activation may involve activation of the Cdk8 kinase, which weakens CKM–mediator interaction, thereby liberating mediator and enabling its binding to the PIC. Our observation that Cdk8 inhibition produces more pronounced transcriptomic effects under conditions of heat shock is consistent with similar observations during nutrient stress (60), oxidative stress (61), and in human cells (21, 35, 36) and argues that the Cdk8 kinase is not constitutively active. Indeed, stress and developmental signaling cascades culminate in phosphorylating various mediator or CKM complex subunits (62, 63, 64). Signaling-dependent exchange of activator-bound mediator from its CKM bound to its free form has previously been reported (64). The mediator complex and the rest of the CKM may thus act as a conduit of signals to the Cdk8. Cdk8 activity is sensitive to the location of a Med12 activation helix (65), an observation suggestive of allosteric signal transduction translated into an output of Cdk8 activity. This may allow Cdk8 to support rapid activation of genes within specific transcriptional programs, imparting improved adaptability to stress, and accounting for its early occurrence and high degree of evolutionary conservation (66).
Our model describes an important facet of the complex events taking place at the UAS-promoter axis. We have distilled a general functional principle of the CKM from the yeast system, in which no additional homologs of this complex are present. However, more of the Cdk8 functions in metazoans still remain unaccounted for, in congruence with the added regulatory complexity underpinning multicellularity and development (58). The evolution of homologs may have allowed a lateral diversification of regulatory programs, as evidenced by the observation that Cdk8 and its homolog Cdk19 control different gene sets (28). It also remains unclear what role the other phosphorylation targets of Cdk8 in metazoans (56) including histone proteins and combinatorial effects on the TFIIH kinase Cdk7 (50) and the PIC (this study) may play. In addition, Cdk8/19 inhibition was found to further activate gene expression at superenhancers in pluripotent and embryonic stem cells in mouse and human (67, 68), implying a different function in the superenhancer context.
In summary, our observation of Cdk8-dependent release of the CKM–mediator occlusion provides a crucial missing link that explains how the CKM's steric repression and the observed activating effects of its Cdk8 kinase on target gene transcription can be reconciled. Although it is likely that the same mode of steric occlusion and release has been purposed in metazoans, proof of this awaits structural and functional elucidation of the metazoan CKM–mediator complex.
Experimental procedures
CKM recombinant expression and purification
All CKM subunits were cloned from the yeast genome and inserted serially into Macrolab 438-series insect cell expression vectors to construct one large plasmid containing the complete CKM with a His(x6)-MBP-fusion tag on the N terminus of the Cdk8 subunit with all other subunits untagged. CKM(A) contained all the wildtype sequences, whereas CKM(KD) contained a kinase dead D286A mutation in the Cdk8 subunit. D286 is the highly conserved CDK active site residue, and mutation of the homologous residue in Cdk9 has previously been reported to abolish kinase activity (69). V0 and V1 viruses were made in Sf9 cells following standard protocol, and protein was expressed in Hi5 cells. Pellets from 1.2 to 2.4 l of Hi5 insect cell culture, frozen in lysis buffer (20 mM Hepes [pH 7.6], 400 mM KOAc, 10% [v/v] glycerol, 5 mM β-mecaptoethanol (β-ME), 1× protease inhibitor [PI] cocktail) were lysed by sonication, and the lysate was clarified by centrifugation. 100× PI contains 0.028 mg/ml leupeptin, 0.137 mg/ml pepstatin A, 17 mg/ml phenylmethylsulfonyl fluoride, and 33 mg/ml benzamidine. The lysate was bound to pre-equilibrated amylose beads in batch. The beads were washed three times with three resin volumes of lysis buffer. Protein was eluted with two resin volumes of elution buffer (20 mM Hepes [pH 7.6], 400 mM KOAc, 10% [v/v] glycerol, 5 mM β-ME, 100 mM maltose, and 1× PI). The protein was then loaded onto a HiTrapQ HP (1 ml) column (GE Healthcare), equilibrated with 30% ion exchange buffer B (20 mM Hepes [pH 7.6], 10% [v/v] glycerol, 1 M KOAc, and 5 mM β-ME), and eluted with a salt gradient from 30% to 100% ion exchange buffer B over 20 column volumes (20 ml). Complex-containing fractions were pooled and concentrated using an Amicon Ultra-15 centrifugal filter (Merck). Finally, complex was dialyzed overnight into gel filtration buffer (20 mM Hepes [pH 7.6], 400 mM KOAc, 10% [v/v] glycerol, and 5 mM β-ME) and then run on size exclusion chromatography on a Superose 6 Increase (10/300) column (GE Healthcare), or a sucrose gradient, or the final sizing step omitted, depending on the subsequent application. Purification of 16-subunit cMed and the rest of the recombinant initiation factors and endogenous pol II was performed (8).
CKM kinase activity assays
Two reactions containing either CKM(A) or CKM(KD) were set up in a total volume of 20 μl. The appropriate volume of components were pipetted to have 10 μM glutathione-S-transferase—histidine-tagged CTD (purified as in (70)), 0.4 μM CKM(A or KD), 1× of 5× buffer (100 mM Hepes [pH 7.5], 1.5 M KOAc, and 25 mM β-ME), 10 mM MgCl2, and (10 mM ATP—added later) in the final reaction. The volume was brought up to 20 μl with water. All components except for ATP were mixed together on ice and then transferred to a 30 °C heating block. About 1 μl for time point 0 and 2 μl for each subsequent time point (5, 10, 20, 30, 60, and 120 min) were quenched by mixing with 8 μl 4× lithium dodecyl sulfate (LDS) sample buffer. ATP was added after taking the sample for time point 0. Western blot analysis was performed using the CTD Ser5 (3E8) and Ser2 (3E10) phosphosite-specific antibodies (a kind gift from D. Eick, previously validated and peer reviewed for epitope binding on synthetic peptides (71) and further characterized in multiple peer-reviewed publications (70, 72, 73)).
Negative stain EM
About 5 μl of sample were pipetted onto the surface of a carbon-coated copper grid and incubated for 30 s to 2 min, depending on the sample concentration. The sample was blotted away using a Whatman filter paper (GE Life Sciences). Grids were washed twice with water and incubated face down on three 20 μl drops of 2% uranyl formate solution for 20 s each (a total of 60 s), and the excess stain blotted away. Negative stain grids were imaged on a CM120 (Phillips) electron microscope with a lanthanum hexaboride (LaB6) cathode and a spherical aberration of 6.7 mm. 4K × 4K TIFF micrographs were taken at a magnification of selected area magnification range 52,000×. Negative stain images were binned by a factor of 2 (4.4 Å/pixel), and particle picking was carried out in EMAN2 (74), with a box size of 88 pixel and a particle size of 60 pixel. Picked particle coordinates were exported. Particle extraction, contrast transfer function estimation, and 2D classification were carried out in Relion 1.4. The particle mask diameter used was 300 Å. 2D classification was run for 50 iterations to sort the particles into 100 2D classes, with a regularization parameter (T) of 2. 3D ab initio model building was carried out using CryoSparc (44), using picked particles from all the good 2D classes.
ITA
About 40 μl of Dynabeads MyOne Streptavidin T1 beads (Life Technologies) per planned reaction (x) = Y μl were transferred to a 2 ml tube (eppendorf). Using a DynaMag magnetic rack (Life Technologies), buffer was removed. Beads were washed three times with Y μl of streptavidin-binding buffer (20 mM Hepes [pH 7.5], 150 mM KOAc, 5–10% glycerol, 5 mM β-ME, 10 mM MgCl2, and 0.05% nonyl phenoxypolyethoxylethanol) and then resuspended in Y–x μl buffer, with x μl of biotinylated scaffold DNA (75) (5′-CGATATAGAAGGTAAGAAAAGGATAATGAACAGTAGCACGCTGTGTATATAATAGCTATGGAACGTTCGATTCACCTCCGATGTGTGTTGTACATACATAAAAATATCATAGCACAACTGCGCTGTGTAATAGTAATACATAGTGGAAACCCATACACAGGGAAGATATCCGGTCCGTAGG-3′) at 40 μM. Beads were incubated with DNA for 25 min at 25 °C and washed once with Y μl of streptavidin-binding buffer to remove excess scaffold and resuspended in Y μl. Resuspended beads were divided into 40 μl per reaction tube.
Seven reactions were set up (1) scaffold + CKM(KD); (2) scaffold + cMed; (3) scaffold + cMed–CKM(KD); (4) scaffold + PIC factors + cMed; (5) scaffold + PIC factors + cMed–CKM(A); (6) scaffold + PIC factors + cMed–CKM(A) +ATP; (7) scaffold + PIC factors + cMed–CKM(KD) + ATP. For PIC-containing reactions, the PIC factors were mixed such that 40 pmol each of TFIIA, TFIIB, and TBP, 10 pmol of pol II, 20 pmol of TFIIF, and 60 pmol of TFIIE are present per reaction. The volumes required for four reactions were mixed together, made up to 40 μl with buffer, or with buffer minus the volume of additional components. For reactions (4–7), 10 μl of PIC mix was added. To reaction (1), 15 pmol of CKM(KD) were added. To reaction (2) and (4), 22.5 pmol of cMed were added. To reactions (3) and (5–7), 15 pmol CKM(A or KD) as indicated and 22.5 pmol cMed, dialyzed to lower salt buffer (20 mM Hepes [pH 7.6], 250 mM KOAc, 10% [v/v] glycerol, and 5 mM β-ME) to allow complex formation, were added. To reactions (6) and (7), ATP to a final concentration of 2.5 mM was added. Reactions were pipetted into the respective tubes, and the final volume was adjusted to 40 μl with streptavidin-binding buffer and incubated for 10 min at 25 °C. Buffer was removed from the beads, and the reactions added to the respective tubes, mixed, and incubated at 25 °C for 45 min. The flowthrough (unbound) was collected, and the beads were washed three times with streptavidin-binding buffer. About 24.5 μl of streptavidin-binding buffer + 0.5 μl of 100 U/μl EcoRV-HF (New England Biolabs) were added per tube and incubated at 28 °C for 75 min to allow the restriction enzyme to cleave off the DNA from the beads. About 10 and 5 μl of 4× LDS loading dye were added to the unbound and elution fractions, respectively. Fractions were analyzed by SDS-PAGE.
CKM phosphoproteomics of the PIC
Buffers and experimental setup were exactly as described for the ITA, with the only difference being that 15 pmol per reaction of cMed were added (in a 1:1 ratio with CKM), instead of 22.5 pmol. Three reactions were set up: (1) scaffold + cMed + PIC; (2) scaffold + cMed + PIC + [CKM(A) + ATP] later; and (3) cMed + PIC + CKM(A) + ATP + [scaffold] later. The difference between reactions (2) and (3) is that, in reaction (2), PIC–cMed was first assembled on the DNA scaffold, washed, and then CKM(A) was added, whereas in reaction (3), PIC components were incubated with CKM(A) and ATP first on ice for 30 min, before adding in the bead-bound scaffold for the assembly reaction. All unbound and elution fractions were run on an SDS-PAGE gel. Full lanes from the gel were cut out, the proteins were extracted, digested, phosphor-enriched, and subjected to mass spectrometric analysis to identify phosphorylation sites on PIC components. In-gel trypsin digestion and extraction (76), peptide enrichment by size exclusion, and analysis using an LTQ-Orbitrap Velos or Q-Exactive mass spectrometer (Thermo Fisher Scientific). For phosphosite mapping, samples were treated the same way but enriched for phosphopeptides after gel extraction. Enriched phosphopeptides were submitted to HPLC–MS/MS analysis on a Q-Exactive HF quadrupole-orbitrap mass spectrometer (Thermo Scientific) coupled to a Dionex Ultimate 3000 RSLC nano system. Peptides were loaded on a Pepmap 300 C18 column at a flow rate of 10 μl/min in buffer A (0.1% [v/v] formic acid), and the column was washed for 2 min with buffer A. Peptides were separated on an in-house–made 30 cm main column (ReproSil-Pur 120 Å, 1.9 μm, C18-AQ, 75 mm inner diameter). During chromatographic separation, a linear gradient was used with a buffer system of buffer A (0.1% [v/v] formic acid) and buffer B (80% [v/v] acetonitrile and 0.08% [v/v] formic acid). The chromatographic separation took 58 min. The column was equilibrated with 5% buffer B for 3 min, and peptides were eluted with a linear gradient between 8% and 46% buffer B over 46 min. The column was washed with 90% buffer B for 6 min at the end of the chromatographic run. The parameters used during mass spectrometric measurement were as follows: data-dependent top 15 acquisition methods were used, and MS1 and MS2 spectra were acquired with 120,000 and 30,000 resolution, respectively. Automatic gain control was set to 1E6 for MS1 and 1E5 for MS2. Precursors between 350 and 1600 m/z with charge state between +2 to +6 were chosen. For MS2 fragmentation, higher-energy collision-induced dissociation fragmentation with 28% normalized collision energy was used. Maximum injection time was set to 50 and 110 ms for MS1 and MS2 acquisition, respectively. Dynamic exclusion was set to 30 s. Database search for the phosphosite identification was performed with MaxQuant (version 1.5.2.8) (77). Yeast protein sequence data including TrEMBL sequences was downloaded from Uniprot (July 2014, 108,460 entries). During database search, the following parameters were used: MS1 tolerance 4.5 ppm and MS2 tolerance 20 ppm. Carbamidomethylation on cysteine was set as fix modification. Methionine oxidation, N-terminal acetylation, and phosphorylation on serine, threonine, and tyrosine were set as variable modifications. 1% peptide spectrum match and 1% protein FDR were applied. Datasets were analyzed with the Mascot software (Matrix Science) (78) against the National Center for Biotechnology Information nonredundant proteome database. Phosphorylation sites from all fractions were pooled into one dataset of Cdk8-dependent phosphorylation sites.
CKM–cMed–pol II competition assay
Storage buffer was removed from 400 μl of amylose magnetic beads (New England Biolabs) using a magnetic rack in a 2 ml eppendorf tube, and the beads were washed once with 400 μl of water and then blocked with 600 μl of 1 mg/ml bovine serum albumin solution in binding buffer 1 (20 mM Hepes, pH 7.5, 300 mM KOAc, 10% glycerol, and 5 mM β-ME) at 37 °C for 20 min with vigorous shaking. The blocking solution was then removed, and the beads were washed three times with binding buffer 1. About 440 μg of CKM(KD) in storage buffer (240 μl) were adjusted to a total volume of 800 μl and then added to the beads and shaken at 800 rpm for 1 h at 4 °C to bind MBP-tagged CKM to the blocked amylose beads. After binding, the flowthrough was discarded, and the beads were washed twice with binding buffer 1, then twice with binding buffer 2 (20 mM Hepes, pH 7.5, 250 mM KOAc, 10% glycerol, and 5 mM β-ME) to lower the salt concentration in preparation for the addition of cMed. About 880 μg (360 μl) of cMed (2× molar ratio compared with CKM) adjusted to 800 μl in binding buffer 2 were added and shaken together at 800 rpm with the amylose–CKM beads at 4 °C. After binding, the flowthrough was discarded, and the beads were washed 12 times with 400 μl of binding buffer 2 (the number of washes had been previously determined in a preliminary experiment, where all washes were blotted with the same antibody used in this experiment to determine the times after which equilibrium is reached—there was always a trace of cMed detected, but after about ten washes, the level of detected cMed became constant). All previous steps were carried out in a single tube. After the washes, the beads were then resuspended in 410 μl of binding buffer 2. The beads were then divided into 50 μl of resuspended beads per tube. Six solutions with varying concentrations of polymerase were prepared. About 0, 0.2, 0.4, 1, 2, and 10 μl, respectively, of purified pol II at 9.45 μM were adjusted to 200 μl in binding buffer 2. This corresponded to a molar excess of RNA pol II over bead-bound CKM–cMed of 0, 0.5, 2, 5, 10, and 50×, respectively, approximated from the theoretical binding capacity of the amylose beads based on their binding to MBP5–paromysinΔSal. CKM–cMed–amylose beads were incubated with polymerase at room temperature for 1 h with shaking. The beads were then removed, and 30 μl from the 200 μl wash volume were taken and mixed with 10 μl of 4× LDS sample buffer. About 18 μl were then loaded onto a 4 to 12% SDS-PAGE gel (NuPAGE) run in MES buffer. The gel was then blotted onto a nitrocellulose membrane (Biorad) and then probed with anti-Srb4 antibody.
Crosslinking/MS
About 80 μg of CKM–cMed in roughly 100 μl were dialyzed overnight into lower salt buffer (20 mM Hepes [pH 7.6], 250 mM KOAc, 10% [v/v] glycerol, and 5 mM β-ME) to allow complex formation, then crosslinked with BS3, stored in powder form (no-weight format; Thermo Fisher Scientific) and dissolved in buffer freshly before use to a 30 mM stock, to a final concentration of 0.1 mM BS3. Crosslinked samples were separated by Native-PAGE (20 μg per lane), and species corresponding to the full CKM–cMed complex were excised from the gel (two lanes) and treated by in-gel digestion with trypsin as described (76). Briefly, the excised gel bands were cut to 1 mm2 pieces, reduced with 10 mM DTT, alkylated with 55 mM iodoacetamide, and digested with trypsin (Sigma–Aldrich) overnight. Tryptic peptides were extracted, dried, and reconstituted in a solution containing 2% (v/v) acetonitrile and 0.05% (v/v) TFA and subjected to LC–tandem MS. LC–MS/MS analyses were performed on a Q Exactive HF-X hybrid quadrupole-orbitrap mass spectrometer (Thermo Scientific) coupled to a Dionex Ultimate 3000 RSLCnano system. The peptide mixture from each gel lane was loaded on a Pepmap 300 C18 column (Thermo Fisher) at a flow rate of 10 μl/min in buffer A (0.1% [v/v] formic acid) and washed for 3 min with buffer A. The sample was separated on an in-house–packed C18 column (30 cm; ReproSil-Pur 120 Å, 1.9 μm, C18-AQ; inner diameter, 75 μm) at a flow rate of 300 nl/min. Sample separation was performed over 120 min using a buffer system consisting of 0.1% (v/v) formic acid (buffer A) and 80% (v/v) acetonitrile, 0.08% (v/v) formic acid (buffer B). The main column was equilibrated with 5% B before application of the peptide mixture to the column, and the column was washed for 3 min with 5% B. Peptides were eluted with a linear gradient over 105 min from 10 to 45% B (replicates 1–3) or 15 to 50% B (replicate 4), followed by an 8 min gradient from 90 to 95% B and re-equilibration for 4 min at 5% B. Eluting peptides were analyzed in positive mode using a data-dependent top 20 acquisition methods. MS1 and MS2 resolution were set to 120,000 and 30,000 full width at half maximum, respectively, and automatic gain control targets were 5∗105 and 2∗105. Precursors selected for MS2 (scan range m/z 380–1580) were fragmented using 30% normalized higher-energy collision-induced dissociation fragmentation. Allowed charge states of selected precursors were +3 to +7 (replicates 1, 2, and 4) or +2 to +7 (replicate 3). Further MS/MS parameters were set as follows: isolation width, 1.4 m/z; dynamic exclusion, 10 s; and maximum injection time (MS1/MS2), 60 ms/200 ms. The lock mass option (m/z 445.12002) was used for internal calibration. The .raw files of all replicates from both lanes were searched by the pLink 2, version 2.3.1 software (79) against a customized protein database containing the expressed proteins. Protein–protein crosslinks were filtered with a 1% FDR and plotted using xiNET (80). Manual validation of identified crosslinks was performed by a selection of eight crosslinks between cMed subunits, mapping them onto the S. cerevisiae cMed homology model derived from the S. pombe crystal structure (Protein Data Bank ID: 5N9J), which was used to model cMed in the cMed–PIC structure (Protein Data Bank ID: 5OQJ). Atomic distances were measured between α-carbon atoms of crosslinked residues using PyMol.
Analytical sucrose gradient ultracentrifugation
About 25 μg of CKM(A) or CKM(KD) were mixed with 25 μg of cMed, and the volume was made up to 25 μl with binding buffer 2 (20 mM Hepes potassium, 300 mM KOAc, 10% glycerol, and 5 mM β-ME). The complex was incubated on ice for 30 min, 2 μl of 100 mM ATP and 2 μl of 100 mM MgCl2 were added, and the reaction was incubated at 30 °C for 2 h to ensure completion of the phosphorylation reaction. About 26 μl of each reaction were then gently pipetted to the top of a 10 to 30% sucrose density gradient in gradient buffer (20 mM Hepes potassium, 300 mM KOAc, and 5 mM β-ME) and subjected to ultracentrifugation for 16 h at 32,000 rpm. About 200 μl fractions were taken from the top (lowest density to highest density) of which 15 μl were loaded on a gel and blotted using an anti-MBP–horseradish peroxidase antibody (Abcam ab49923). Two technical replicates were performed and are shown in Fig. S3C to confirm the observed effect and exclude any errors of loading. The bands were quantified separately for each run (in total four runs; two replicates of each of CKM(A) or CKM(KD) with cMed) using the U.S. National Institutes of Health ImageJ suite (81). The value of the highest intensity band within each run was used for internal normalization. All other values were calculated as a percentage of the highest intensity. These values were then used to calculate the mean intensity in arbitrary units shown in Figure 4D as well as the corresponding standard error of mean (shown as error bars).
4tU-seq and bioinformatics analysis
The Srb10 analogue-sensitive strain (Mat α, Δ ade2::hisG, his3Δ200, leu2Δ0, lys2Δ0, met15Δ0, trp1Δ63, ura3Δ0, srb10Δ:: KanMX, pSH599 [ars cen TRP1 srb10as-1 (Y236G)]) carries an srb10 Shokat-altered specificity mutation (as-1 Y236G) derived by transformation of SHY475B, shared by S. Hahn, and was previously published (24). For the steady-state growth experiment, fresh cultures were inoculated from an overnight preculture at a starting absorbance at 600 nm of 0.1 and grown until an absorbance at 600 nm of 0.8 was reached in yeast extract–peptone–dextrose–Trp growth media at 30 °C. Cultures were then divided into four smaller cultures of 40 ml each and treated with either 1-NA-PP1 (40 mM in DMSO) to a final concentration of 6 μM (two replicates), or an equal volume of DMSO (two replicates), and incubated for 12 min, followed by 5 min of 4tU labeling. 4tU labeling and subsequent extraction of labeled RNA was performed as described (82). For the heat shock experiment, a 300 ml culture was grown in the same way. The culture was then divided into two 140 ml cultures, to which either 1-NA-PP1 or DMSO was added in the same way as described for the steady-state growth experiment and incubated for 12 min. About 100 ml from each culture was diluted with 100 ml of 37 °C warm media to exert heat shock. About 80 ml were extracted after 12 min of heat shock and labeled with 4tU for 5 min. The entire experiment was carried out in the same way twice to obtain two replicates. 4tU-Seq data analysis was performed (82) but with minor modifications. Briefly, the raw fastq files of paired-end 75 base reads (for steady-state experiment) and 42 base reads (for heat shock experiment) with additional six base reads of barcodes were obtained for each of the samples. Reads were demultiplexed, and low-quality bases (<Q20) removed using Cutadapt (version 1.9.1) with parameters −q 20,20 −o 12 −m 25 (83). Reads were then mapped to the S. cerevisiae genome (sacCer3, version 64.2.1) using STAR 2.5.2b (84) with parameters –outFilterMismatchNmax 2 –outFilterMultimapScoreRange 0. Samtools (85) was used to quality filter SAM files. Alignments with MAPQ smaller than 7 (−q 7) were skipped, and only proper pairs (−f 2) were selected. We used a spike-in (RNAs) normalization strategy (86) to allow for observation of global changes in the 4tU-Seq signal. Sequencing depth from spike-in RNAs was calculated for each sample j according to
with read counts kij for the labeled spike-ins i in sample j and li for the length of labeled spike-ins i for the 4tU-seq samples.
Read counts for protein-coding genes (using TIF-seq derived TSS and pA site annotations for 5578 protein-coding genes (87)) and spike-ins were calculated using HTSeq (88) after mapping. Further processing of the 4tU-Seq data was carried out using the R/Bioconductor environment. Differential gene expression analysis was performed with the DESeq2 package (89) using the spike-in–derived normalization factors. DESeq2 uses a negative binomial distribution to model the counts for each gene with a given set of parameters (i.e., normalization factor, dispersion) and fit the normalized count data to it. After model fitting, coefficients (log2 fold changes) are estimated for each sample group along with their standard error. By default, p values are obtained by the Wald test and adjusted for multiple testing using the Benjamini and Hochberg method. We used an adjusted p value cutoff of 0.1 to call significant changes. Whether a gene's expression is called significantly changed depends on both its log2 fold change and its dispersion estimate. If the dispersion estimates from replicates as quantified by within-group variability are higher, the significance of log2 fold changes decreases. This explains why some genes with similar log2 fold change can appear as nonsignificant and significant in Figure 5, A–C, respectively.
To examine the effects of Cdk8 inhibition during steady-state growth, the 1-NA-PP1 samples were compared with the DMSO samples. To verify induction of temperature response genes (Gene Ontology: 0009266) upon heat shock, the 12-min heat shock DMSO samples were compared with the steady-state DMSO samples. To examine the effect of Cdk8 inhibition on gene expression during heat shock, the 12 min heat shock 1-NA-PP1 samples were compared with the 12-min heat shock DMSO samples. To examine the effect of Cdk8 inhibition on induction of heat shock genes, the 12-min heat shock 1-NA-PP1 samples were compared with the steady-state 1-NA-PP1 samples.
Data availability
Supporting information includes three supporting tables (Tables S1–S3) listing the selected phosphopeptide and crosslinking MS data presented in the main figures according to the criteria detailed in Experimental procedures section and three supporting figures. All supporting information can be found with this article online. All raw MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (90) partner repository with the dataset identifier PXD023361. All sequencing data have been deposited in Gene Expression Omnibus under the accession number GSE161140. The data can be accessed with the following token: ofybgcuurlotlul.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank S. Hahn for providing the yeast Cdk8-analog sensitive strain and D. Eick for providing the pol II CTD phosphosite-specific antibodies. We also thank K. Zumer, T. Velychko, and L. Caizzi for input on the genomic sequencing experiments and other members of the Cramer laboratory, particularly F. Wagner, G. Kokic, and P. Seweryn for helpful discussions. We also thank M. Raabe and A. Kuhn from the bioanalytical MS facility for phosphopeptide enrichment MS measurements.
Author contributions
S. O. designed and performed experiments and evaluated data unless otherwise stated. A. S., F. L. B., and H. U. performed MS measurements. K. C. M. performed the 4tU-seq experiments. E. M. and M. L. performed the bioinformatics analysis of the 4tU-seq data. P. C. supervised research. S. O. and P. C. wrote the article, with input from all authors.
Funding and additional information
P. C. was supported by the Deutsche Forschungsgemeinschaft (EXC 2067/1-390729940) and the European Research Council Advanced Investigator grant CHROMATRANS (grant agreement no: 882357).
Edited by Roger Colbran
Supporting information
References
- 1.Hahn S., Young E.T. Transcriptional regulation in Saccharomyces cerevisiae: Transcription factor regulation and function, mechanisms of initiation, and roles of activators and coactivators. Genetics. 2011;189:705–736. doi: 10.1534/genetics.111.127019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Verger A., Monté D., Villeret V. Twenty years of mediator complex structural studies. Biochem. Soc. Trans. 2019;47:399–410. doi: 10.1042/BST20180608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asturias F.J., Jiang Y.W., Myers L.C., Gustafsson C.M., Kornberg R.D. Conserved structures of mediator and RNA polymerase II holoenzyme. Science. 1999;283:985–987. doi: 10.1126/science.283.5404.985. [DOI] [PubMed] [Google Scholar]
- 4.Dotson M.R., Yuan C.X., Roeder R.G., Myers L.C., Gustafsson C.M., Jiang Y.W., Li Y., Kornberg R.D., Asturias F.J. Structural organization of yeast and mammalian mediator complexes. Proc. Natl. Acad. Sci. U. S. A. 2000;97:14307–14310. doi: 10.1073/pnas.260489497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nozawa K., Schneider T.R., Cramer P. Core mediator structure at 3.4 Å extends model of transcription initiation complex. Nature. 2017;545:248. doi: 10.1038/nature22328. [DOI] [PubMed] [Google Scholar]
- 6.Tsai K.L., Yu X., Gopalan S., Chao T.C., Zhang Y., Florens L., Washburn M.P., Murakami K., Conaway R.C., Conaway J.W., Asturias F.J. Mediator structure and rearrangements required for holoenzyme formation. Nature. 2017;544:196–201. doi: 10.1038/nature21393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plaschka C., Larivière L., Wenzeck L., Seizl M., Hemann M., Tegunov D., Petrotchenko E.V., Borchers C.H., Baumeister W., Herzog F., Villa E., Cramer P. Architecture of the RNA polymerase II–mediator core initiation complex. Nature. 2015;518:376–380. doi: 10.1038/nature14229. [DOI] [PubMed] [Google Scholar]
- 8.Schilbach S., Hantsche M., Tegunov D., Dienemann C., Wigge C., Urlaub H., Cramer P. Structures of transcription pre-initiation complex with TFIIH and mediator. Nature. 2017;551:204–209. doi: 10.1038/nature24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao H., Young N., Kalchschmidt J., Lieberman J., El Khattabi L., Casellas R., Asturias F.J. Structure of mammalian mediator complex reveals tail module architecture and interaction with a conserved core. Nat. Commun. 2021;12:1–12. doi: 10.1038/s41467-021-21601-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abdella R., Talyzina A., Chen S., Inouye C., Tjian R., He Y. Structure of the human mediator-bound transcription preinitiation complex. Science. 2021;372:52–56. doi: 10.1126/science.abg3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rengachari S., Schilbach S., Aibara S., Dienemann C., Cramer P. Structure of human mediator-RNA polymerase II transcription pre-initiation complex. bioRxiv. 2021 doi: 10.1101/2021.03.11.435010. [preprint] [DOI] [PubMed] [Google Scholar]
- 12.Andrau J.-C., van de Pasch L., Lijnzaad P., Bijma T., Koerkamp M.G., van de Peppel J., Werner M., Holstege F.C.P. Genome-wide location of the coactivator mediator: Binding without activation and transient Cdk8 interaction on DNA. Mol. Cell. 2006;22:179–192. doi: 10.1016/j.molcel.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 13.Taatjes D.J., Näär A.M., Andel F., Nogales E., Tjian R. Structure, function, and activator-induced conformations of the CRSP coactivator. Science. 2002;295:1058–1062. doi: 10.1126/science.1065249. [DOI] [PubMed] [Google Scholar]
- 14.Borggrefe T., Davis R., Erdjument-Bromage H., Tempst P., Kornberg R.D. A complex of the Srb8,-9,-10, and-11 transcriptional regulatory proteins from yeast. J. Biol. Chem. 2002;277:44202–44207. doi: 10.1074/jbc.M207195200. [DOI] [PubMed] [Google Scholar]
- 15.El Khattabi L., Zhao H., Kalchschmidt J., Young N., Jung S., Van Blerkom P., Kieffer-Kwon P., Kieffer-Kwon K.-R., Park S., Wang X. A pliable mediator acts as a functional rather than an architectural bridge between promoters and enhancers. Cell. 2019;178:1145–1158.e1120. doi: 10.1016/j.cell.2019.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan X., Chou D.M., Struhl K. Activator-specific recruitment of mediator in vivo. Nat. Struct. Mol. Biol. 2006;13:117. doi: 10.1038/nsmb1049. [DOI] [PubMed] [Google Scholar]
- 17.Fan X., Struhl K. Where does mediator bind in vivo? PLoS One. 2009;4 doi: 10.1371/journal.pone.0005029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeronimo C., Langelier M.-F., Bataille A.R., Pascal J.M., Pugh B.F., Robert F. Tail and kinase modules differently regulate core mediator recruitment and function in vivo. Mol. Cell. 2016;64:455–466. doi: 10.1016/j.molcel.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petrenko N., Jin Y., Wong K.H., Struhl K. Mediator undergoes a compositional change during transcriptional activation. Mol. Cell. 2016;64:443–454. doi: 10.1016/j.molcel.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hengartner C.J., Myer V.E., Liao S.-M., Wilson C.J., Koh S.S., Young R.A. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol. Cell. 1998;2:43–53. doi: 10.1016/s1097-2765(00)80112-4. [DOI] [PubMed] [Google Scholar]
- 21.Chen M., Liang J., Ji H., Yang Z., Altilia S., Hu B., Schronce A., McDermott M.S.J., Schools G.P., Lim C.-U., Oliver D., Shtutman M.S., Lu T., Stark G.R., Porter D.C. CDK8/19 mediator kinases potentiate induction of transcription by NFκB. Proc. Natl. Acad. Sci. U. S. A. 2017;114:10208–10213. doi: 10.1073/pnas.1710467114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirst M., Kobor M.S., Kuriakose N., Greenblatt J., Sadowski I. GAL4 is regulated by the RNA polymerase II holoenzyme-associated cyclin-dependent protein kinase SRB10/CDK8. Mol. Cell. 1999;3:673–678. doi: 10.1016/s1097-2765(00)80360-3. [DOI] [PubMed] [Google Scholar]
- 23.Lenssen E., Azzouz N., Michel A., Landrieux E., Collart M.A. The Ccr4-not complex regulates Skn7 through Srb10 kinase. Eukaryot. Cell. 2007;6:2251–2259. doi: 10.1128/EC.00327-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y., Kung C., Fishburn J., Ansari A.Z., Shokat K.M., Hahn S. Two cyclin-dependent kinases promote RNA polymerase II transcription and formation of the scaffold complex. Mol. Cell. Biol. 2004;24:1721–1735. doi: 10.1128/MCB.24.4.1721-1735.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rohde J.R., Trinh J., Sadowski I. Multiple signals regulate GALTranscription in yeast. Mol. Cell. Biol. 2000;20:3880–3886. doi: 10.1128/mcb.20.11.3880-3886.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sadowski I., Costa C., Dhanawansa R. Phosphorylation of Ga14p at a single C-terminal residue is necessary for galactose-inducible transcription. Mol. Cell. Biol. 1996;16:4879–4887. doi: 10.1128/mcb.16.9.4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sadowski I., Niedbala D., Wood K., Ptashne M. GAL4 is phosphorylated as a consequence of transcriptional activation. Proc. Natl. Acad. Sci. U. S. A. 1991;88:10510–10514. doi: 10.1073/pnas.88.23.10510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steinparzer I., Sedlyarov V., Rubin J.D., Eislmayr K., Galbraith M.D., Levandowski C.B., Vcelkova T., Sneezum L., Wascher F., Amman F. Transcriptional responses to IFN-γ require mediator kinase-dependent pause release and mechanistically distinct CDK8 and CDK19 functions. Mol. Cell. 2019;76:485–499.e488. doi: 10.1016/j.molcel.2019.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vincent O., Kuchin S., Hong S.-P., Townley R., Vyas V.K., Carlson M. Interaction of the Srb10 kinase with Sip4, a transcriptional activator of gluconeogenic genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 2001;21:5790–5796. doi: 10.1128/MCB.21.17.5790-5796.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chi Y., Huddleston M.J., Zhang X., Young R.A., Annan R.S., Carr S.A., Deshaies R.J. Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev. 2001;15:1078–1092. doi: 10.1101/gad.867501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nelson C., Goto S., Lund K., Hung W., Sadowski I. Srb10/Cdk8 regulates yeast filamentous growth by phosphorylating the transcription factor Ste12. Nature. 2003;421:187. doi: 10.1038/nature01243. [DOI] [PubMed] [Google Scholar]
- 32.Raithatha S., Su T.-C., Lourenco P., Goto S., Sadowski I. Cdk8 regulates stability of the transcription factor Phd1 to control pseudohyphal differentiation of Saccharomyces cerevisiae. Mol. Cell. Biol. 2012;32:664–674. doi: 10.1128/MCB.05420-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ansari A.Z., Koh S.S., Zaman Z., Bongards C., Lehming N., Young R.A., Ptashne M. Transcriptional activating regions target a cyclin-dependent kinase. Proc. Natl. Acad. Sci. U. S. A. 2002;99:14706–14709. doi: 10.1073/pnas.232573899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liao S.-M., Zhang J., Jeffery D.A., Koleske A.J., Thompson C.M., Chao D.M., Viljoen M., van Vuuren H.J., Young R.A. A kinase–cyclin pair in the RNA polymerase II holoenzyme. Nature. 1995;374:193. doi: 10.1038/374193a0. [DOI] [PubMed] [Google Scholar]
- 35.Galbraith M.D., Allen M.A., Bensard C.L., Wang X., Schwinn M.K., Qin B., Long H.W., Daniels D.L., Hahn W.C., Dowell R.D. HIF1A employs CDK8-mediator to stimulate RNAPII elongation in response to hypoxia. Cell. 2013;153:1327–1339. doi: 10.1016/j.cell.2013.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galbraith M.D., Andrysik Z., Pandey A., Hoh M., Bonner E.A., Hill A.A., Sullivan K.D., Espinosa J.M. CDK8 kinase activity promotes glycolysis. Cell Rep. 2017;21:1495–1506. doi: 10.1016/j.celrep.2017.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clark A.D., Oldenbroek M., Boyer T.G. Mediator kinase module and human tumorigenesis. Crit. Rev. Biochem. Mol. Biol. 2015;50:393–426. doi: 10.3109/10409238.2015.1064854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Firestein R., Hahn W.C. Revving the throttle on an oncogene: CDK8 takes the driver seat. Cancer Res. 2009;69:7899–7901. doi: 10.1158/0008-5472.CAN-09-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Philip S., Kumarasiri M., Teo T., Yu M., Wang S. Cyclin-dependent kinase 8: A new hope in targeted cancer therapy? J. Med. Chem. 2018;61:5073–5092. doi: 10.1021/acs.jmedchem.7b00901. [DOI] [PubMed] [Google Scholar]
- 40.Elmlund H., Baraznenok V., Lindahl M., Samuelsen C.O., Koeck P.J., Holmberg S., Hebert H., Gustafsson C.M. The cyclin-dependent kinase 8 module sterically blocks mediator interactions with RNA polymerase II. Proc. Natl. Acad. Sci. U. S. A. 2006;103:15788–15793. doi: 10.1073/pnas.0607483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsai K.L., Sato S., Tomomori-Sato C., Conaway R.C., Conaway J.W., Asturias F.J. A conserved mediator-CDK8 kinase module association regulates mediator-RNA polymerase II interaction. Nat. Struct. Mol. Biol. 2013;20:611–619. doi: 10.1038/nsmb.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knuesel M.T., Meyer K.D., Bernecky C., Taatjes D.J. The human CDK8 subcomplex is a molecular switch that controls mediator coactivator function. Genes Dev. 2009;23:439–451. doi: 10.1101/gad.1767009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kastner B., Fischer N., Golas M.M., Sander B., Dube P., Boehringer D., Hartmuth K., Deckert J., Hauer F., Wolf E. GraFix: Sample preparation for single-particle electron cryomicroscopy. Nat. Methods. 2008;5:53. doi: 10.1038/nmeth1139. [DOI] [PubMed] [Google Scholar]
- 44.Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 45.Wang X., Wang J., Ding Z., Ji J., Sun Q., Cai G. Structural flexibility and functional interaction of mediator Cdk8 module. Protein Cell. 2013;4:911–920. doi: 10.1007/s13238-013-3069-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y.-C., Chao T.-C., Kim H.J., Cholko T., Chen S.-F., Li G., Snyder L., Nakanishi K., Chang C.-e., Murakami K. Structure and noncanonical Cdk8 activation mechanism within an Argonaute-containing mediator kinase module. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abd4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jeronimo C., Robert F. Kin28 regulates the transient association of mediator with core promoters. Nat. Struct. Mol. Biol. 2014;21:449. doi: 10.1038/nsmb.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uthe H., Vanselow J.T., Schlosser A. Proteomic analysis of the mediator complex interactome in Saccharomyces cerevisiae. Sci. Rep. 2017;7:43584. doi: 10.1038/srep43584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paoletti A.C., Parmely T.J., Tomomori-Sato C., Sato S., Zhu D., Conaway R.C., Conaway J.W., Florens L., Washburn M.P. Quantitative proteomic analysis of distinct mammalian mediator complexes using normalized spectral abundance factors. Proc. Natl. Acad. Sci. U. S. A. 2006;103:18928–18933. doi: 10.1073/pnas.0606379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Knuesel M.T., Meyer K.D., Donner A.J., Espinosa J.M., Taatjes D.J. The human CDK8 subcomplex is a histone kinase that requires Med12 for activity and can function independently of mediator. Mol. Cell. Biol. 2009;29:650–661. doi: 10.1128/MCB.00993-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chi A., Huttenhower C., Geer L.Y., Coon J.J., Syka J.E., Bai D.L., Shabanowitz J., Burke D.J., Troyanskaya O.G., Hunt D.F. Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 2007;104:2193–2198. doi: 10.1073/pnas.0607084104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Holt L.J., Tuch B.B., Villén J., Johnson A.D., Gygi S.P., Morgan D.O. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325:1682–1686. doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li X., Gerber S.A., Rudner A.D., Beausoleil S.A., Haas W., Villén J., Elias J.E., Gygi S.P. Large-scale phosphorylation analysis of α-factor-arrested Saccharomyces cerevisiae. J. Proteome Res. 2007;6:1190–1197. doi: 10.1021/pr060559j. [DOI] [PubMed] [Google Scholar]
- 54.Miller C., Matic I., Maier K.C., Schwalb B., Roether S., Strässer K., Tresch A., Mann M., Cramer P. Mediator phosphorylation prevents stress response transcription during non-stress conditions. J. Biol. Chem. 2012;287:44017–44026. doi: 10.1074/jbc.M112.430140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Albuquerque C.P., Smolka M.B., Payne S.H., Bafna V., Eng J., Zhou H. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol. Cell. Proteomics. 2008;7:1389–1396. doi: 10.1074/mcp.M700468-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Poss Z.C., Ebmeier C.C., Odell A.T., Tangpeerachaikul A., Lee T., Pelish H.E., Shair M.D., Dowell R.D., Old W.M., Taatjes D.J. Identification of mediator kinase substrates in human cells using cortistatin A and quantitative phosphoproteomics. Cell Rep. 2016;15:436–450. doi: 10.1016/j.celrep.2016.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Garibaldi A., Carranza F., Hertel K.J. Isolation of newly transcribed RNA using the metabolic label 4-thiouridine. Methods Mol. Biol. 2017;1648:169–176. doi: 10.1007/978-1-4939-7204-3_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Allen B.L., Taatjes D.J. The mediator complex: A central integrator of transcription. Nat. Rev. Mol. Cell Biol. 2015;16:155–166. doi: 10.1038/nrm3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grünberg S., Henikoff S., Hahn S., Zentner G.E. Mediator binding to UASs is broadly uncoupled from transcription and cooperative with TFIID recruitment to promoters. EMBO J. 2016;35:2435–2446. doi: 10.15252/embj.201695020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Holstege F.C., Jennings E.G., Wyrick J.J., Lee T.I., Hengartner C.J., Green M.R., Golub T.R., Lander E.S., Young R.A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 61.Stieg D.C., Cooper K.F., Strich R. The extent of cyclin C promoter occupancy directs changes in stress-dependent transcription. J. Biol. Chem. 2020;295:16280–16291. doi: 10.1074/jbc.RA120.015215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chang Y.-W., Howard S.C., Herman P.K. The Ras/PKA signaling pathway directly targets the Srb9 protein, a component of the general RNA polymerase II transcription apparatus. Mol. Cell. 2004;15:107–116. doi: 10.1016/j.molcel.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 63.Kim S., Xu X., Hecht A., Boyer T.G. Mediator is a transducer of Wnt/β-catenin signaling. J. Biol. Chem. 2006;281:14066–14075. doi: 10.1074/jbc.M602696200. [DOI] [PubMed] [Google Scholar]
- 64.van de Peppel J., Kettelarij N., van Bakel H., Kockelkorn T.T., van Leenen D., Holstege F.C. Mediator expression profiling epistasis reveals a signal transduction pathway with antagonistic submodules and highly specific downstream targets. Mol. Cell. 2005;19:511–522. doi: 10.1016/j.molcel.2005.06.033. [DOI] [PubMed] [Google Scholar]
- 65.Klatt F., Leitner A., Kim I.V., Ho-Xuan H., Schneider E.V., Langhammer F., Weinmann R., Müller M.R., Huber R., Meister G. A precisely positioned MED12 activation helix stimulates CDK8 kinase activity. Proc. Natl. Acad. Sci. U. S. A. 2020;117:2894–2905. doi: 10.1073/pnas.1917635117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bourbon H.-M. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 2008;36:3993–4008. doi: 10.1093/nar/gkn349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lynch C.J., Bernad R., Martínez-Val A., Shahbazi M.N., Nóbrega-Pereira S., Calvo I., Blanco-Aparicio C., Tarantino C., Garreta E., Richart-Ginés L. Global hyperactivation of enhancers stabilizes human and mouse naive pluripotency through inhibition of CDK8/19 mediator kinases. Nat. Cell Biol. 2020;22:1223–1238. doi: 10.1038/s41556-020-0573-1. [DOI] [PubMed] [Google Scholar]
- 68.Pelish H.E., Liau B.B., Nitulescu I.I., Tangpeerachaikul A., Poss Z.C., Da Silva D.H., Caruso B.T., Arefolov A., Fadeyi O., Christie A.L. Mediator kinase inhibition further activates super-enhancer-associated genes in AML. Nature. 2015;526:273. doi: 10.1038/nature14904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vos S.M., Farnung L., Boehning M., Wigge C., Linden A., Urlaub H., Cramer P. Structure of activated transcription complex Pol II–DSIF–PAF–SPT6. Nature. 2018;560:607–612. doi: 10.1038/s41586-018-0440-4. [DOI] [PubMed] [Google Scholar]
- 70.Boehning M., Dugast-Darzacq C., Rankovic M., Hansen A.S., Yu T., Marie-Nelly H., McSwiggen D.T., Kokic G., Dailey G.M., Cramer P. RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat. Struct. Mol. Biol. 2018;25:833. doi: 10.1038/s41594-018-0112-y. [DOI] [PubMed] [Google Scholar]
- 71.Chapman R.D., Heidemann M., Albert T.K., Mailhammer R., Flatley A., Meisterernst M., Kremmer E., Eick D. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science. 2007;318:1780–1782. doi: 10.1126/science.1145977. [DOI] [PubMed] [Google Scholar]
- 72.Clemente-Blanco A., Sen N., Mayan-Santos M., Sacristán M.P., Graham B., Jarmuz A., Giess A., Webb E., Game L., Eick D. Cdc14 phosphatase promotes segregation of telomeres through repression of RNA polymerase II transcription. Nat. Cell Biol. 2011;13:1450–1456. doi: 10.1038/ncb2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Menafra R., Brinkman A.B., Matarese F., Franci G., Bartels S.J., Nguyen L., Shimbo T., Wade P.A., Hubner N.C., Stunnenberg H.G. Genome-wide binding of MBD2 reveals strong preference for highly methylated loci. PLoS One. 2014;9 doi: 10.1371/journal.pone.0099603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tang G., Peng L., Baldwin P.R., Mann D.S., Jiang W., Rees I., Ludtke S.J. EMAN2: An extensible image processing suite for electron microscopy. J. Struct. Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 75.Gradia S.D., Ishida J.P., Tsai M.S., Jeans C., Tainer J.A., Fuss J.O. MacroBac: new technologies for robust and efficient large-scale production of recombinant multiprotein complexes. Methods Enzymol. 2017;592:1–26. doi: 10.1016/bs.mie.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shevchenko A., Tomas H., Havli J., Olsen J.V., Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 77.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 78.Perkins D.N., Pappin D.J., Creasy D.M., Cottrell J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 79.Yang B., Wu Y.-J., Zhu M., Fan S.-B., Lin J., Zhang K., Li S., Chi H., Li Y.-X., Chen H.-F. Identification of cross-linked peptides from complex samples. Nat. Methods. 2012;9:904–906. doi: 10.1038/nmeth.2099. [DOI] [PubMed] [Google Scholar]
- 80.Combe C.W., Fischer L., Rappsilber J. xiNET: Cross-link network maps with residue resolution. Mol. Cell. Proteomics. 2015;14:1137–1147. doi: 10.1074/mcp.O114.042259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xu Y., Bernecky C., Lee C.-T., Maier K.C., Schwalb B., Tegunov D., Plitzko J.M., Urlaub H., Cramer P. Architecture of the RNA polymerase II-Paf1C-TFIIS transcription elongation complex. Nat. Commun. 2017;8:1–13. doi: 10.1038/ncomms15741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–12. [Google Scholar]
- 84.Dobin A., Gingeras T.R. Mapping RNA-seq reads with STAR. Curr. Protoc. Bioinformatics. 2015;51:11.14.11–11.14.19. doi: 10.1002/0471250953.bi1114s51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schwalb B., Michel M., Zacher B., Frühauf K., Demel C., Tresch A., Gagneur J., Cramer P. TT-seq maps the human transient transcriptome. Science. 2016;352:1225–1228. doi: 10.1126/science.aad9841. [DOI] [PubMed] [Google Scholar]
- 87.Pelechano V., Wei W., Steinmetz L.M. Extensive transcriptional heterogeneity revealed by isoform profiling. Nature. 2013;497:127–131. doi: 10.1038/nature12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Anders S., Pyl P.T., Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Anders S., Huber W. Differential expression analysis for sequence count data. Nat. Prec. 2010 doi: 10.1038/npre.2010.4282.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Supporting information includes three supporting tables (Tables S1–S3) listing the selected phosphopeptide and crosslinking MS data presented in the main figures according to the criteria detailed in Experimental procedures section and three supporting figures. All supporting information can be found with this article online. All raw MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (90) partner repository with the dataset identifier PXD023361. All sequencing data have been deposited in Gene Expression Omnibus under the accession number GSE161140. The data can be accessed with the following token: ofybgcuurlotlul.