Abstract
The spectrum of tumors arising in the salivary glands is wide and has recently been shown to harbor a network of tumor-specific fusion genes. Acinic cell carcinoma (AciCC) is one of the more frequently encountered types of salivary gland carcinoma, but it has remained a genetic orphan until recently when a fusion between the HTN3 and MSANTD3 genes was described in one case. Neither of these 2 genes is known to be implicated in any other malignancy. This study was undertaken to investigate whether the HTN3-MSANTD3 fusion is a recurrent genetic event in AciCC and whether it is a characteristic of one of its histological variants. Of the 273 AciCCs screened, 9 cases showed rearrangement of MSANTD3 by break-apart fluorescence in situ hybridization, 2 had 1 to 2 extra signals, and 1 had gain, giving a total of 4.4% with MSANTD3 aberrations. In 6 of 7 available cases with MSANTD3 rearrangement, the HTN3-MSANTD3 fusion transcript was demonstrated with real-time polymerase chain reaction. Histologically, all fusion-positive cases were predominantly composed of serous tumor cells growing in solid sheets, with serous tumor cells expressing DOG-1 and the intercalated duct-like cell component being CK7 positive and S-100 positive in 6/9 cases. All but one case arose in the parotid gland, and none of the patients experienced a recurrence during follow-up. In contrast, the case with MSANTD3 gain metastasized to the cervical lymph nodes and lungs. In conclusion, we find the HTN3-MSANTD3 gene fusion to be a recurrent event in AciCC with prominent serous differentiation and an indolent clinical course.
Keywords: carcinoma, acinic cell, salivary gland, salivary gland neoplasms, genes, HTN3-MSANTD3, gene fusion, genetics
Salivary gland acinic cell carcinoma (AciCC) constitutes ~10% of primary salivary gland carcinomas and arises in female individuals in two-thirds of cases.1 Although typically a low-grade malignancy with a favorable long-term prognosis, the high-grade transformation is well described and is associated with an aggressive clinical course.2,3 With the exception of frequent DOG-1 (ANO1) expression, the immunohistochemical profile of AciCC is nonspecific, and the diagnosis is based on the characteristic acinar differentiation, which is aided by demonstration of cytoplasmic PAS-positive zymogen granules, which are resistant to diastase digestion.3,4 During the last decade, molecular studies have revealed numerous type-specific translocations in salivary gland tumors, which have become valuable diagnostic tools in equivocal cases and in cases of high-grade transformation, small biopsy specimens, so-called hybrid tumors, and even in fine-needle aspiration cytologies.5–7
In 2010, Skálová and colleagues separated a morphologically and genetically distinct subset of “zymogen granule-poor AciCC” from conventional AciCC, and they found this subset to be histologically and genetically identical to secretory carcinoma of the breast.3,8–12 However, only recently was “classic” AciCC shown to belong to the group of salivary gland carcinomas driven by fusion oncogenes with the demonstration of a gene fusion between the Histatin 3 (HTN3) and Myb/SANT-like DNA-binding domain containing 3 (MSANTD3) genes in an index case, and rearrangement of MSANTD3 in 2/19 additional AciCCs by fluorescence in situ hybridization (FISH).13 Interestingly, rearrangement of the poorly characterized MSANTD3 gene was found to be unique for AciCC among all other types of salivary gland tumors.13 However, the HTN3-MSANTD3 fusion was demonstrated in the index case only, and the prevalence of this novel gene fusion in AciCC has been uncertain due to the relatively small sample size in this initial report. Moreover, MSANTD3 expression was not sensitive or specific for AciCC.13 Hence, the diagnostic, as well as prognostic, value of this gene fusion has remained unclarified.
In this study, we interrogated a large set of salivary gland AciCCs for the presence of MSANTD3 rearrangement and characterized cases with aberrations for expression of the HTN3-MSANTD3 fusion transcript. Moreover, we present the clinicopathologic data on the MSANTD3-rearranged AciCCs in order to ascertain the clinical course of this genetically distinct subset of AciCC.
MATERIALS AND METHODS
Patient Material
AciCC were retrieved from the surgical pathology archives from the authors’ institutions, including the material previously included by Barasch et al.13 Formalin-fixed paraffin-embedded (FFPE) blocks from 273 cases, including 5 cases with high-grade transformation, were retrieved and tissue microarrays constructed for the majority of cases after different protocols, either including large single cores or duplicate/triplicate 0.6 mm cores.13,14 Whole slides were evaluated for the remaining cases. Clinical information was collected from patient files. The study was approved by the Danish Data Protection Agency (J.no. REG-94–2014) and the Ethics Committee of the Capital Region of Denmark (J.no. H-6–2014-086 add. 58080).
Histochemistry and Immunohistochemistry
From MSANTD3-rearranged cases and 20 nonrearranged controls, whole sections were cut to 4 μm slides and stained with hematoxylin and eosin (H&E), periodic acid-Schiff with (PAS+D) and without (PAS-D) diastase, phosphotungstic acid hematoxylin (PTAH), and Perls’ prussian blue using standard protocols. Immunohistochemistry was performed as previously described using the antibodies listed in Table 1.15,16 Briefly, sections were mounted on coated slides, deparaffinized using EZ-prep, and the antigens retrieved with CC1 in Tris buffer (pH 8.5) (all Ventana Medical Systems, Tucson, AZ). The ultraView DAB Detection kit (Ventana) was used for visualization, and slides were counterstained with Mayer’s hematoxylin. Positive controls as suggested on datasheets were used on each slide, and the expected reaction and subcellular site were confirmed. Positive and negative controls were used on all slides.
TABLE 1.
Antibodies Used for Immunohistochemical Characterization
| Antibody | Clone | Dilution | Source |
|---|---|---|---|
| Alpha-amylase | EPR19605 | 1:10,000 | AbCam (Cambridge, UK) |
| CK7 | OV-TL 12/30 | 1:1000 | Dako (Glostrup, Denmark) |
| DOG-1 | SP31 | Ready-to-use | Roche (Hvidovre, Denmark) |
| Ki-67 | MIB1 | 1:100 | Dako |
| Mammaglobin | 304–1A5 | Ready-to-use | Dako |
| S-100 | Polyclonal | 1:4,000 | Dako |
Fluorescence In Situ Hybridization
In order to screen for MSANTD3 rearrangement, the material was characterized using either a custom break-apart FISH probe for MSANTD3 generated from bacterial artificial chromosomes flanking MSANTD3, CTD-3186I20 Cy5 (telomeric), and CTD-2363K7 Cy3 (centromeric) (BACPAC Resources Center, Children’s Hospital Oakland Research Institute) labeled with either Cy5 or Cy3 (GE Healthcare Life Sciences, Chicago, IL) or a commercially available break-apart probe (Empire Genomics, Buffalo, NY), which was hybridized using Vysis (Abbott Molecular, Des Plaines, IL) reagents and protocols. After hybridization, nuclei were counterstained with DAPI II (ZytoVision, Bremerhaven, Germany), and the images were captured using Ariol software (Applied Imaging, San Jose, CA). For each case, 60 to 100 nuclei were evaluated and rearrangement defined as ≥ 1 split signal in ≥ 10% of nuclei.8,17 The gain was defined as > 2 of one or both signals in ≥ 10% of nuclei.
Ribonuclease Acid Sequencing
Total ribonuclease acid (RNA) was prepared for RNA sequencing in accordance with the manufacturer’s instructions (Illumina, San Diego, CA). RNA sequencing was performed using Illumina’s TruSeq Stranded Total RNA Library Prep Kit, and paired-end sequencing was performed to gain an output of 100 M reads according to the manufacturer’s instructions. FusionMap was used for the screening of fusion transcripts.18
Real-time Polymerase Chain Reaction and Nucleotide Sequence Analysis
Total RNA was isolated from FFPE sections using Maxwell RSC FFPE kit (Promega, Madison, WI) according to the manufacturer’s instructions. RNA was converted to complementary deoxyribonucleic acid (cDNA) using the Superscript VILO cDNA synthesis kit (Thermo Fisher Scientific, Waltham, MA) and performed according to the manufacturer’s instructions. RNA integrity was tested using primer pairs for the phosphoglycerate kinase gene (PGK1) in each case, as described previously.19,20 The cDNA was subjected to real-time polymerase chain reaction (RT-PCR) using the primers TGAGACTTCACTTCAGCTTCAC (NM_000200) and CAACGAAATTATAAAGCCTGCCA (NM_080655) spanning the fusion site between exon 1 and exon 2 of HTN3 and MSANTD3, respectively. PCR conditions were as follows: 94°C for 90 seconds followed by 40 cycles of 94°C for 45 seconds, 60°C for 1 minute, 72°C for 1 minute, and a final extension of 72°C for 10 minutes. T3 and T7 tails were attached to the forward and reverse primers, respectively, which were used for later sequencing of the amplified products. The amplified fusion products were visualized using Qiaxcel (Qiagen GmbH, Hilden, Germany), and the 160 bp amplicons were subsequently sequenced using BigDye Terminator 3.1 Cycle Sequencing Kit, T3/T7-primers, and an ABI 3500 Dx DNA sequenator according to the manufacturer’s instructions (Thermo Fisher Scientific). Ten nonrearranged AciCCs were used as controls.
RESULTS
MSANTD3 Aberrations in AciCC
Among the 273 AciCCs comprising the cohort, 9 cases (3.3%) showed split signals with the MSANTD3 probe in 17% to 48% percent of tumor cells, one case each showed 1 to 2 extra green and red signals, respectively, giving a total of 4% with MSANTD3 aberrations (Table 2 and Fig. 1). In addition to the index case from the study by Barasch et al13 (case 1, Table 2), RT-PCR identified the fusion between HTN3 exon 1 and MSANTD3 exon 2 in 6/7 rearranged cases with available tissue (lanes 1 to 6, Table 2, Fig. 2). Despite showing rearrangement of MSANTD3 by FISH and having sufficient RNA quality for RT-PCR, no fusion transcript was identified in case 7, and RNA did not meet quality requirements for RNA sequencing. One case that showed a gain of one MSANTD3 signal did not express a fusion transcript, which was also the case for 10 nonrearranged AciCCs (Fig. 3).
TABLE 2.
Demographics, Presentation, Treatment, and Outcome of Patients With MSANTD3 Aberrated Acinic Cell Carcinoma
| Case# | MSANTD3 FISH | RT-PCR | Age (y), Sex | Site | Size (mm) | Presentation | Stage | Treatment | Outcome, follow-up (mo) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Break | HTN3-MSANTD3 | 59, M | Parotid | 30×35×30 | Parotid mass | T2N0M0 | Surgery | DOC 98 |
| 2 | Break | HTN3-MSANTD3 | 52, F | Parotid | 80×110 | Parotid mass | T3N0M0 | Surgery+RT | NED 269 |
| 3 | Break | HTN3-MSANTD3 | 51, F | Parotid | NA | NA | NA | NA | NA |
| 4 | Break | HTN3-MSANTD3 | 75, F | Parotid | 25 | Parotid mass | T2cN0M0 | Surgery+RT | NED 120 |
| 5 | Break | HTN3-MSANTD3 | 65, F | Parotid, accesssory | 18 | Buccal mass | T1cN0M0 | Surgery | NED 36 |
| 6 | Break | HTN3-MSANTD3 | 28, F | Parotid | 32 | Incidental finding on MRI | T2cN0M0 | Surgery | NED 24 |
| 7 | Break | —* | 59, M | Submandibular | 25×20×25 | Submandibular mass | T2N0M0 | Surgery+RT | NED 58 |
| 8† | Break | HTN3-MSANTD3‡ | 76, F | Parotid | 25 | NA | NA | Surgery | NA |
| 9† | Break | NA | 49, F | Parotid | 12 | NA | NA | Surgery | NA |
| 10† | Break | NA | 30, M | Parotid | 12 (recurrence) | NA | NA | Surgery | NA |
| 11 | Break | NA | NA | Parotid | NA | NA | NA | NA | NA |
Absence of HTN3-MSANTD3 fusion transcript by RT-PCR and uninformative RNA sequencing.
Cases previously reported by Barasch et al13 “Complex pattern, 1 to 2 extra green signals.” “Complex pattern, 1 to 2 extra red signals.”
Fusion identified by RNA sequencing.
DOC indicates died of other causes; MRI, magnetic resonance imaging; NA, not available; NED, no evidence of disease; RT, radiotherapy.
FIGURE 1.
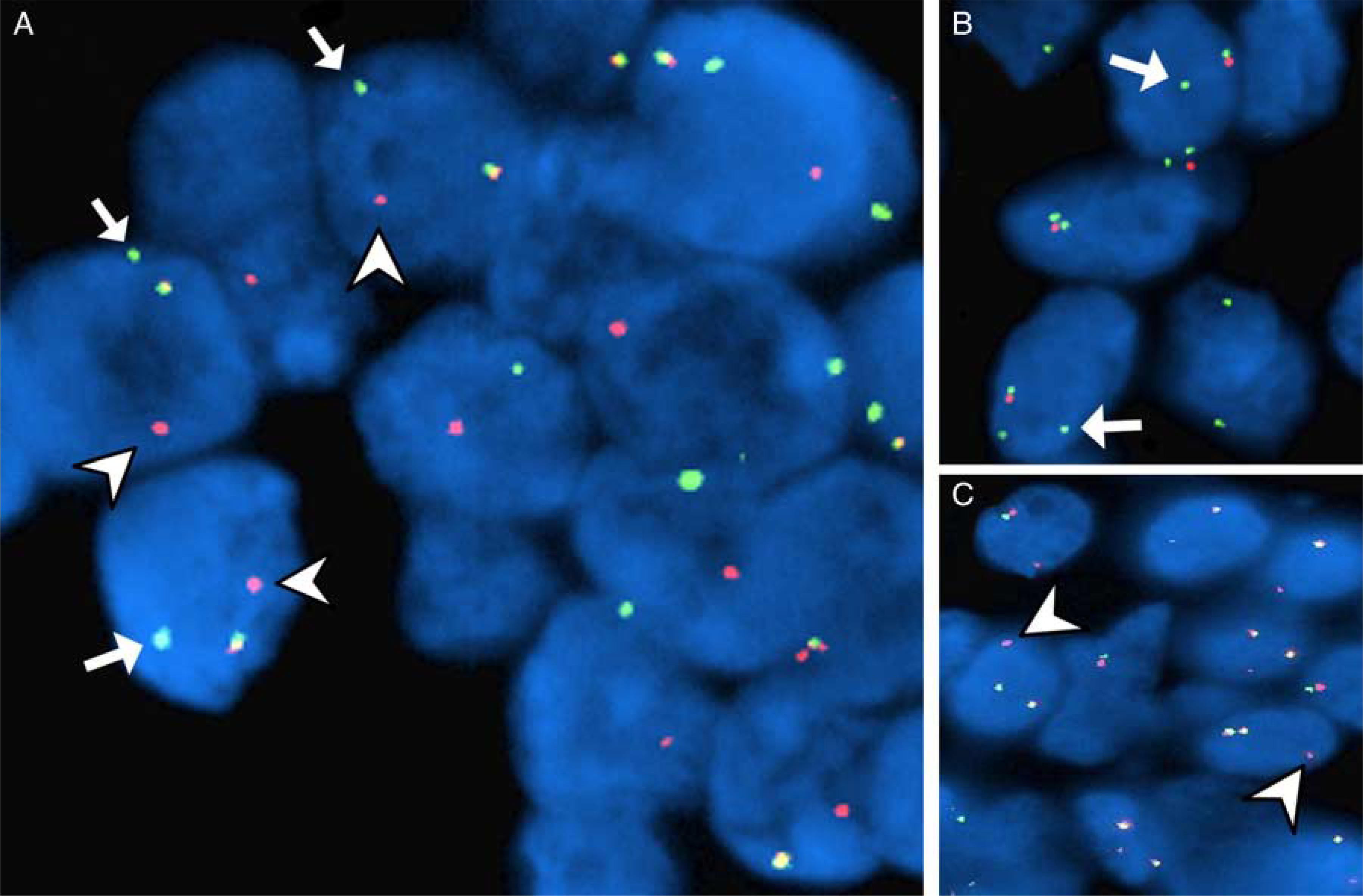
Patterns of MSANTD3 aberrations in acinic cell carcinoma of the salivary gland. A, FISH demonstrating separate green (arrows) and red (arrowheads) signals, consistent with fusion gene formation. The gain of one green signal (arrows) (B) and one red signal (arrowheads) (C) was present in a subset of cases.
FIGURE 2. HTN3-MSANTD3.
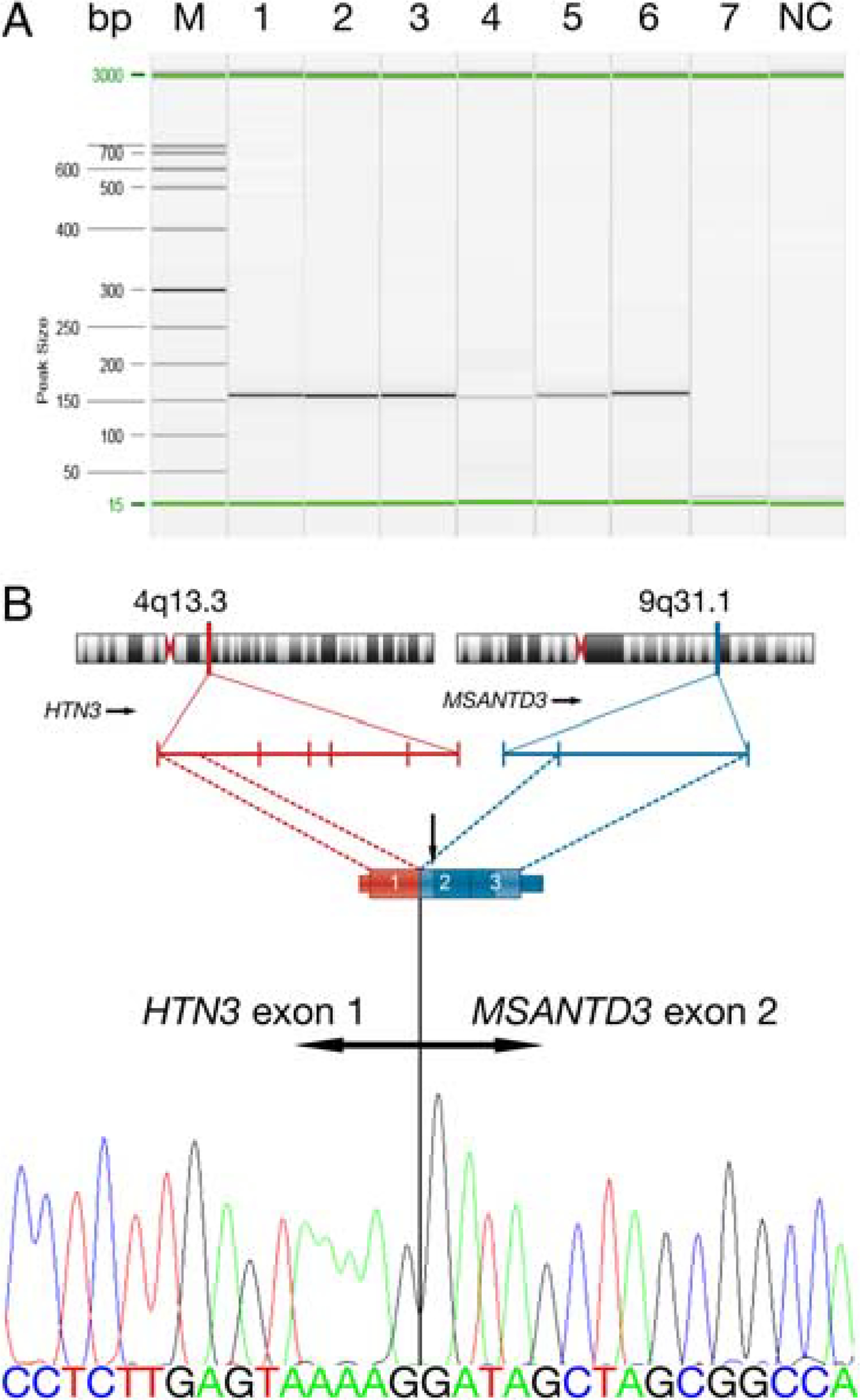
HTN3-MSANTD3 fusion transcripts in acinic cell carcinoma of the salivary gland. A, RT-PCR revealed a 160 bp fragment in cases 1 to 6 but not in case 7. RT-PCR did not yield an amplicon in any of the cases with complex MSANTD3 FISH patterns (not shown). All cases had PGK1 products, ensuring sufficient RNA quality (not shown). B, The breakpoints within the genomic location of the HTN3 gene on chromosome 4 and MSANTD3 gene on chromosome 9 with exons denoted by vertical bars schematically presented. An illustrative part of the fusion transcript demonstrating the fusion of exon 1 of HTN3 and exon 2 of MSANTD3, which was shown to be the same in all 6 cases with amplified PCR fragments by nucleotide sequencing. Note that the start codon (vertical arrow) lies within exon 2 of MSANTD3, causing for full-length MSANTD3 transcripts as marked by dark blue. Bp indicates base pair; M, marker; NC, negative control.
FIGURE 3.
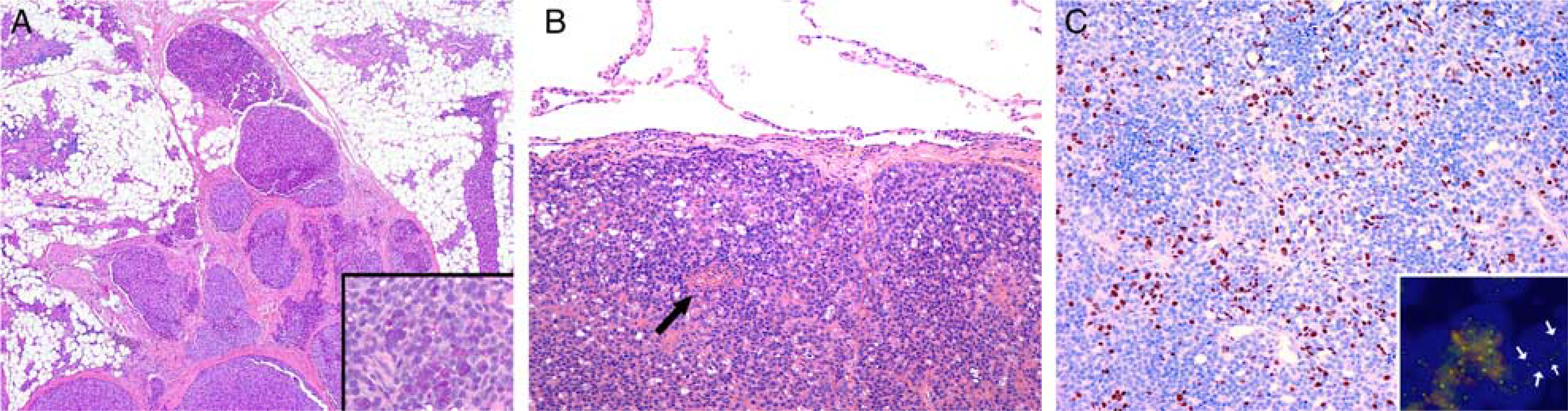
Invasive growth of salivary gland acinic cell carcinoma with MSANTD3 gain. A, A locally invasive tumor growing along interlobular fibrous septae in an atrophic parotid gland (H&E). Note the preservation of cytoplasmic PAS+D-positive granulation (inset). B, A microcystic pulmonary metastasis from the tumor shown in (A) surrounded by a pseudocapsule with a small necrotic area (arrow, H&E). C, The proliferative activity as ascertained with Ki-67 was 10%, up to 25% in hotspots (Ki-67, IHC). The gain of the green signal in MSANTD3 was found in the primary tumor as well as the metastasis (inset, arrows).
Clinical and Histologic Findings in AciCC With MSANTD3 Aberrations
The clinical and pathologic findings of the cases are summarized in Table 2. Sex was known in 10/11 cases with 3 male patients and 7 female patients, with ages ranging from 28 to 76 years (mean: 54 y). All except one arose in the parotid gland and varied in size from 12 to 110 mm in largest dimension (mean: 33 mm). Stage information was available for 6/11 patients, and all presented with localized disease. Treatment information was available for 9/11 patients, and all underwent surgical resection of the primary site and 3 received adjuvant radiotherapy. Follow-up was available for 6/11 patients and ranged from 24 to 269 months (median: 78 mo). None of the patients experienced a recurrence during follow-up, although case 10 itself was a recurrent tumor for which no additional information was available on the primary tumor. The case with MSANTD3 gain arose in a 75-year-old woman who presented with cervical lymph node metastases and subsequently developed pulmonary metastases. She was lost to follow-up 3 months after pulmonary resection.
Only cores embedded in tissue microarrays were available in 4 cases, but the remaining cases ranged from well circumscribed (3/7) to locally invasive (4/7). All were lobulated lesions showing hallmark AciCC features with widespread to almost universally dominant neoplastic serous cells growing in solid sheets with pronounced cytoplasmic granulation (Figs. 3, 4A–D). Accordingly, the intercalated duct-like cell component was present in variable proportions ranging from minuscule to more conspicuous (Figs. 4C–F). Intercalated duct-like cells were variable in size and shape with eosinophilic to pale cytoplasm arranged in cysts, microcysts, and cribriform formations filled with a homogenous eosinophilic material (Figs. 4D–F). No areas of papillary or follicular growth or clear cell or oncocytic change were observed in any of the primary tumors, and none had lymphoid stroma or areas of high-grade transformation. Immunohistochemically, all available cases showed intense positivity for CK7 and S-100 in the intercalated duct-like component and intense expression of DOG-1 in the luminal membrane of serous tumor cells (Figs. 5A, B and Table 3). Alpha-amylase was expressed in the cytoplasmic granules of 6/10 cases (Fig. 5B, inset). Mammaglobin was consistently negative and Ki-67 index varied from 1% to 5% (mean: 2.2) (Table 3). This immunohistochemical profile was similar to what was found in 20 AciCCs without MSANTD3 aberrations (Table 3).
FIGURE 4.
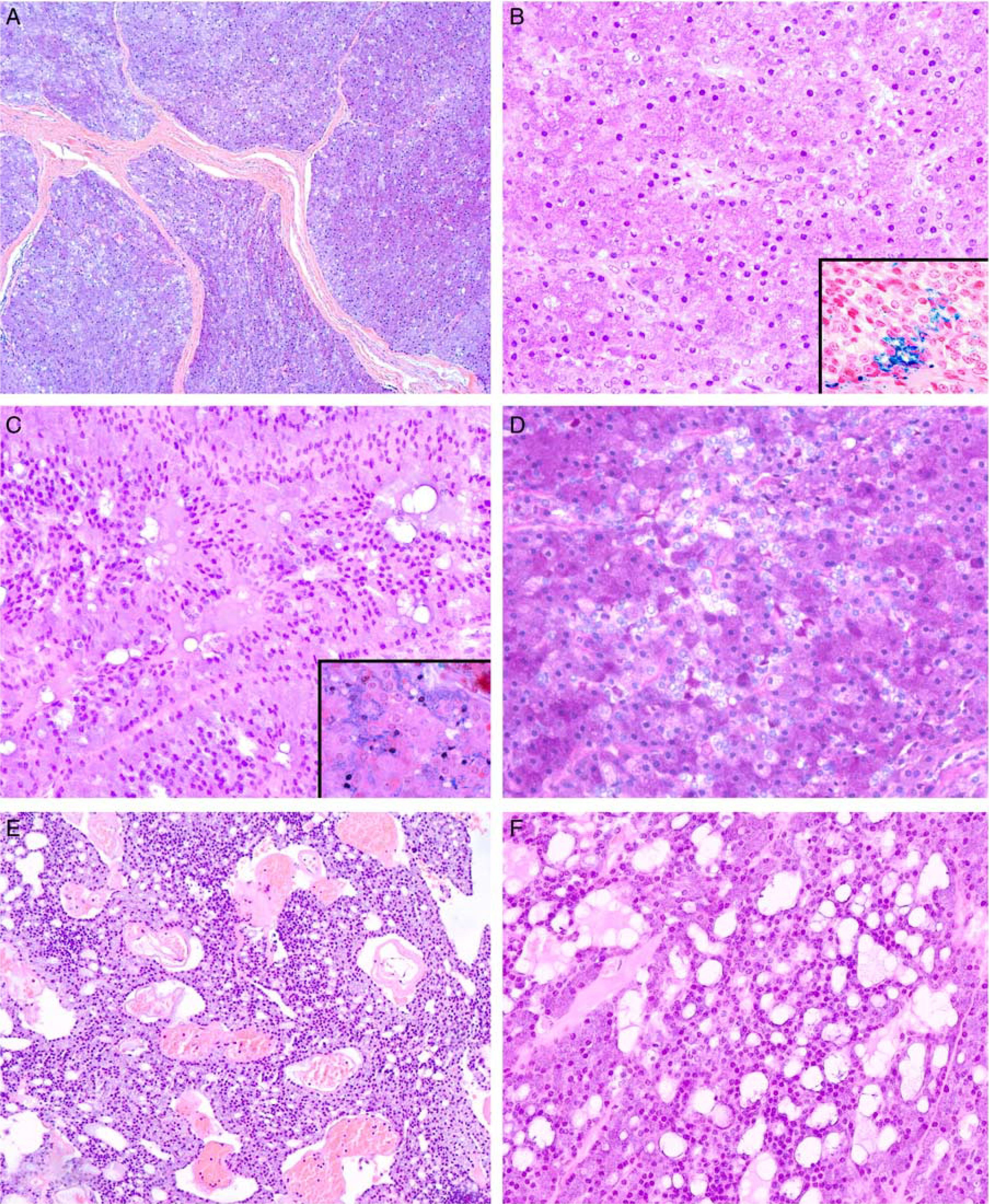
Histologic spectrum of salivary gland acinic cell carcinoma with MSANTD3 aberrations. A, Low-power magnification of a lobulated lesion composed of serous tumor cells in a solid growth pattern (H&E). B, High-power magnification of this cellular lesion showing these serous tumor cells with poorly defined borders, membrane-bound nuclei, and minimal intervening stroma (H&E). Minute cytoplasmic vacuoles are widespread, and hemosiderin granules are only rarely encountered (inset). C, Tumor cells showed occasional remnants of acinar arrangements and discrete cytoplasmic granulation readily discernable with PTAH (inset). Intercalated duct-like tumor cells line small luminae with eosinophilic secretion. Note the basally located nuclei in tumor cells growing along thin fibrovascular septae (H&E). D, Cytoplasmic granulation was present in all cases to a variable extent but was pronounced in most cases. The intercalated duct-like cell component had eosinophilic to pale cytoplasm, as exemplified by the central portion of this image (H&E). E, Low-power view showing larger cysts and cribriform growth mainly lined by intercalated duct-like cells with interspersed neoplastic serous cells (H&E). F, Most cases had areas of microcystic overgrowth with evenly distributed serous and intercalated duct-like tumor cells. Microcysts were filled with variable amounts of bubbly eosinophilic secretion (H&E).
FIGURE 5.
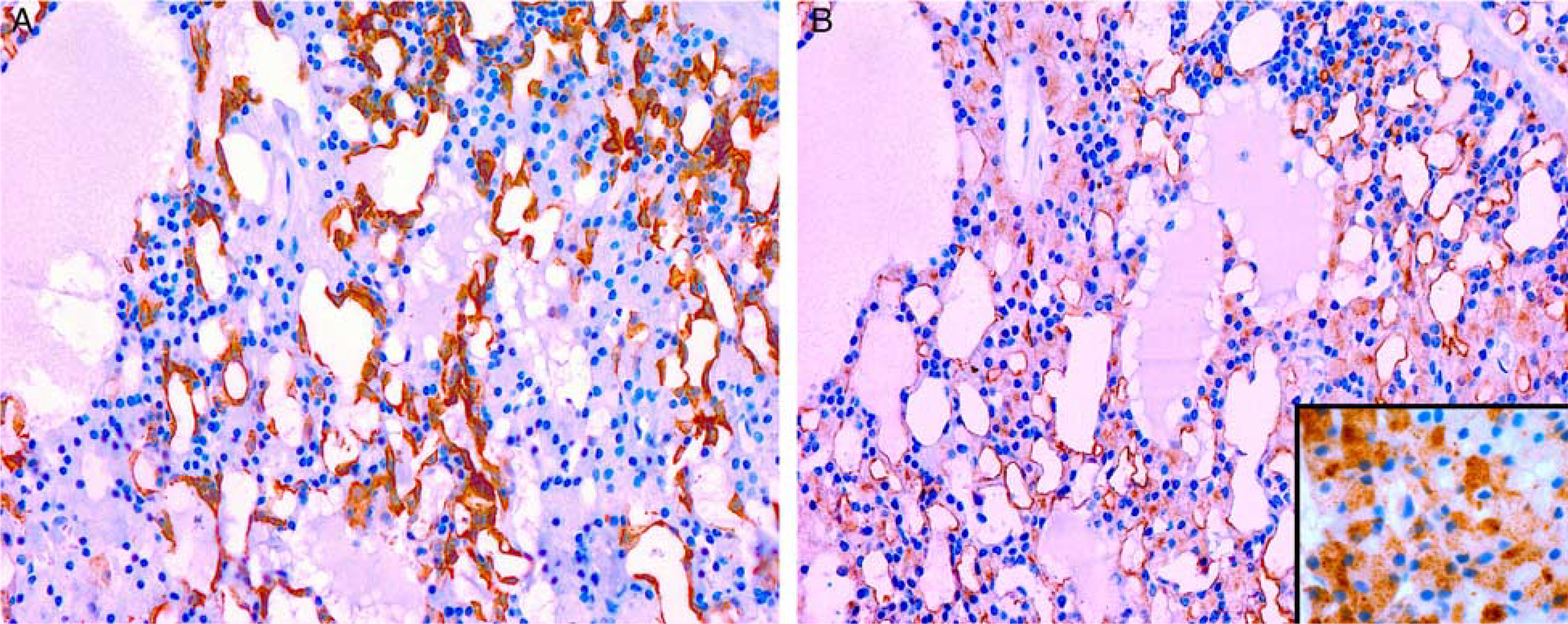
Immunohistochemical profile of salivary gland acinic cell carcinoma with MSANTD3 aberrations. A, S-100 highlights the cytoplasm of intercalated duct-like cells with serous cells being negative. B, DOG-1 highlight the luminal membrane of serous tumor cells with only a few, scattered intercalated duct-like cells showing faint cytoplasmic staining. Alpha-amylase highlights the cytoplasmic granules of the serous tumor cells (inset).
TABLE 3.
Immunohistochemical Profile of Acinic Cell Carcinoma With and Without MSANTD3 Aberrations
| Case# | Alpha amylase | CK7 | DOG-1 | Ki-67 | Mammaglobin | S-100 |
|---|---|---|---|---|---|---|
| 1 | + | + | + | 2% | − | + |
| 2 | + | + | + | 1% | − | + |
| 3 | NA | NA | + | NA | NA | NA |
| 4 | + | + | + | 2% | − | − |
| 5 | − | + | + | 1% | − | − |
| 6 | − | + | + | 2% | − | − |
| 7 | − | + | + | 4% | − | + |
| 8 | + | + | + | 1% | − | + |
| 9 | − | + | + | 2% | − | + |
| 10 | + | + | + | 5% | − | + |
| Total | 6/9 | 9/9 | 10/10 | Mean 2.2 | 0/9 | 6/9 |
| Non-rearranged AciCC (n = 20) | 11/20 | 20/20 | 20/20 | Mean 2 | 0/20 | 20/20 |
No material from case 11 was available for immunohistochemistry.
NA indicates not available.
DISCUSSION
The landscape of tumor-specific gene fusions in salivary gland tumors has grown increasingly complex during the last decade and was recently expanded to include AciCC with the description of the HTN3-MSANTD3 fusion in one single case.5,21–23 The HTN3 gene encodes the histatin 3 peptide, which is secreted exclusively in saliva and has antimicrobial activity important in the maintenance of oral health.24,25 As a consequence of the fusion, the highly active promoter region of HTN3 is positioned to control the expression of the poorly characterized MSANTD3 gene (Fig. 2B). Neither of these 2 genes has previously been reported to be recurrently involved in any type of malignancy, and aberrations in either of these genes are rare across the > 65,000 samples of various human malignancies listed in cBioPortal.26 The oncogenic properties of the highly conserved MSANTD3 are not well understood, but the gene shows some degree of homology with the MYB gene known from adenoid cystic carcinoma of the salivary gland and various other sites.27–29 Furthermore, the oncogenic properties of MSANTD3 are strongly supported by our finding of MSANTD3 gain in one case with an unusually aggressive clinical course with marked local invasion and metastatic spread to cervical lymph nodes and lungs (Fig. 3). To our knowledge, MSANTD3 gain has not previously been reported in any type of malignancy.
We show the HTN3 gene to be the partner gene in the majority of salivary gland AciCC with MSANTD3 aberrations (Table 2). Considering the function of HTN3 in saliva, it is not unexpected that the HTN3 gene is expressed exclusively in the serous acinar cell, and this cell, therefore, is the dominant one in HTN3-MSANTD3–driven tumors. Yet, although in a minor proportion, the intercalated duct-like cells were present in all cases, thereby suggesting an ability of the serous cell to differentiate into this particular cell type. However, the one case arising outside the parotid gland (case 8) did not harbor this fusion transcript by RT-PCR, and RNA sequencing failed to be informative about the fusion status of this particular case due to insufficient RNA quality. Hence, we can only speculate whether the fusion partner, in this case, was fused to a different exon of HTN3 or an entirely different gene, most likely a gene with a highly active promoter region.
In our expanded material of 273 cases, including the material from Barasch et al,13 we found MSANTD3 aberrations in a low proportion of AciCCs (4.4%), as compared with the 15% originally reported (Table 2). The histologic hallmark of AciCC is the serous cell component, which is present in variable proportions, together with the intercalated duct-like component growing in one or more different patterns, which include solid, microcystic, papillary-cystic, and follicular. Interestingly, with the exception of the case with isolated MSANTD3 gain, the AciCCs with MSANTD3 aberrations were all highly differentiated “classic” AciCCs dominated by serous tumor cells in a solid growth pattern. Moreover, none of the cases had significant lymphoid infiltrates, an otherwise common finding in AciCC and associated with a favorable outcome.30
We found an identical immunohistochemical profile between our 12 cases and wild-type controls, distinctly different from that of secretory carcinoma, which was recently separated from AciCC (Table 3).8,31 Therefore, the archetypical features of AciCC with MSANTD3 rearrangement is solid growth, serous cell domination, and absence of lymphoid stroma. However, these characteristics are not unique to AciCC with MSANTD3 aberrations, and the clinical impact of identifying these patients is unlikely to be of significance, as the clinical course of the present material did not differ from the generally indolent nature of AciCC. Collectively, these findings do not merit the separation of MSANTD3-rearranged AciCC as a diagnostic entity, but rather as a genetic characteristic of a subset of classic AciCC.
Similarities between tumors of the salivary gland and breast are well known and include several entities in which identical fusion genes are found in histologically identical tumors of both sites, including adenoid cystic carcinoma and secretory carcinoma.29,32 Among the most intensely debated of these is breast AciCC, as it has a genetic profile similar to other triple-negative breast carcinomas with a high proportion of TP53 mutations, whereas the mutational burden in salivary AciCC is low.32–35 However, in contrast to most triple-negative breast carcinomas, AciCC has a favorable prognosis similar to other salivary-type breast carcinomas found in the breast.32 The identification of a gene fusion involving the promoter region of the HTN3 gene, a gene not expressed in the breast, lends further support to AciCC of the salivary gland and that of the breast as being distinctly different entities.24
In conclusion, the HTN3-MSANTD3 fusion is a recurrent fusion event in the salivary gland AciCC but is found in <5% of cases, in particular, those dominated by serous tumor cells in a solid growth pattern without lymphoid stroma. Although we report on relatively few cases of which one case with MSANTD3 gain metastasized, the clinical course of patients with MSANTD3 aberrations is not distinct from that of AciCC in general. The molecular underpinnings for the vast majority of AciCC remain unknown and await further investigation.
ACKNOWLEDGMENT
The authors express their gratitude to Camilla Cordua Mortensen for invaluable assistance with RT-PCR
Conflicts of Interest and Source of Funding: The authors have disclosed that they have no significant relationships with, or financial interest in, any commercial companies pertaining to this article.
REFERENCES
- 1.Bjø rndal K, Krogdahl A, Therkildsen MH, et al. Salivary gland carcinoma in Denmark 1990–2005: a national study of incidence, site and histology. Results of the Danish Head and Neck Cancer Group (DAHANCA). Oral Oncol. 2011;47:677–682. [DOI] [PubMed] [Google Scholar]
- 2.Thompson LD, Aslam MN, Stall JN, et al. Clinicopathologic and immunophenotypic characterization of 25 cases of acinic cell carcinoma with high-grade transformation. Head Neck Pathol. 2016;10:152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiosea SI, Griffith C, Assaad A, et al. The profile of acinic cell carcinoma after recognition of mammary analog secretory carcinoma. Am J Surg Pathol. 2012;36:343–350. [DOI] [PubMed] [Google Scholar]
- 4.Wenig BM. Neoplasms of the salivary glands. In: Wenig BM, ed. Atlas of Head and Neck Pathology, 3rd ed. Philadelphia, PA: Elsevier; 2015:499–504. [Google Scholar]
- 5.Andersson MK, Stenman G. The landscape of gene fusions and somatic mutations in salivary gland neoplasms—implications for diagnosis and therapy. Oral Oncol. 2016;57:63–69. [DOI] [PubMed] [Google Scholar]
- 6.Hudson JB, Collins BT. MYB gene abnormalities t(6;9) in adenoid cystic carcinoma fine-needle aspiration biopsy using fluorescence in situ hybridization. Arch Pathol Lab Med. 2014;138:403–409. [DOI] [PubMed] [Google Scholar]
- 7.Bishop J, Westra W. MYB translocation status in salivary gland epithelial-myoepithelial carcinoma: evaluation of classic, variant, and hybrid forms. Am J Surg Pathol. 2017;42:319–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skálová A, Vanecek T, Sima R, et al. Mammary analogue secretory carcinoma of salivary glands, containing the ETV6-NTRK3 fusion gene: a hitherto undescribed salivary gland tumor entity. Am J Surg Pathol. 2010;34:599–608. [DOI] [PubMed] [Google Scholar]
- 9.Skálová A, Bell D, Bishop J, et al. Secretory carcinoma. In: El-Naggar A, Chan J, Grandis J, Takata T, Slootweg P, eds WHO Classification of Head and Neck Tumours, 4th ed. Lyon: IARC Press; 2017: 177–178. [Google Scholar]
- 10.Bishop JA. Unmasking MASC: bringing to light the unique morphologic, immunohistochemical and genetic features of the newly recognized mammary analogue secretory carcinoma of salivary glands. Head Neck Pathol. 2013;7:35–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiosea SI, Peel R, Barnes EL, et al. Salivary type tumors seen in consultation. Virchows Arch. 2009;454:457–466. [DOI] [PubMed] [Google Scholar]
- 12.Griffith C, Seethala R, Chiosea SI. Mammary analogue secretory carcinoma: a new twist to the diagnostic dilemma of zymogen granule poor acinic cell carcinoma. Virchows Arch. 2011;459:117–118. [DOI] [PubMed] [Google Scholar]
- 13.Barasch N, Gong X, Kwei KA, et al. Recurrent rearrangements of the Myb/SANT-like DNA-binding domain containing 3 gene (MSANTD3) in salivary gland acinic cell carcinoma. PLoS One. 2017;12:e0171265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miettinen M A simple method for generating multitissue blocks without special equipment. Appl Immunohistochem Mol Morphol. 2012;20:410–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andreasen S, Therkildsen MH, Grauslund M, et al. Activation of the interleukin-6/Janus kinase/STAT3 pathway in pleomorphic adenoma of the parotid gland. APMIS. 2015;123:706–715. [DOI] [PubMed] [Google Scholar]
- 16.Andreasen S, Persson M, Kiss K, et al. Genomic profiling of a rare case of combined large-cell neuroendocrine carcinoma of the submandibular gland. Oncol Rep. 2016;35:2177–2182. [DOI] [PubMed] [Google Scholar]
- 17.Andreasen S, Bishop JA, Hansen TV, et al. Human papillomavirus-related carcinoma with adenoid cystic-like features of the sinonasal tract: clinical and morphological characterization of 6 new cases. Histopathology. 2016;70:880–888. [DOI] [PubMed] [Google Scholar]
- 18.Ge H, Liu K, Juan T, et al. FusionMap: detecting fusion genes from next-generation sequencing data at base-pair resolution. Bioinformatics. 2011;27:1922–1928. [DOI] [PubMed] [Google Scholar]
- 19.Argani P, Perez-Ordonez B, Xiao H, et al. Olfactory neuroblastoma is not related to the Ewing family of tumors: absence of EWS/FLI1 gene fusion and MIC2 expression. Am J Surg Pathol. 1998;22: 391–398. [DOI] [PubMed] [Google Scholar]
- 20.Andreasen S, Skálová A, Agaimy A, et al. ETV6 gene rearrangements characterize a morphologically distinct subset of sinonasal low-grade non-intestinal-type adenocarcinoma: a novel translocation-associated carcinoma restricted to the sinonasal tract. Am J Surg Pathol. 2017;41:1552–1560. [DOI] [PubMed] [Google Scholar]
- 21.Weinreb I, Piscuoglio S, Martelotto LG, et al. Hotspot activating PRKD1 somatic mutations in polymorphous low-grade adenocarcinomas of the salivary glands. Nat Genet. 2014;46:1166–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawahara A, Harada H, Abe H, et al. Nuclear β-catenin expression in basal cell adenomas of salivary gland. J Oral Pathol Med. 2011;40:460–466. [DOI] [PubMed] [Google Scholar]
- 23.Chiosea SI, Miller M, Sethala RR. HRAS mutations in epithelial-myoepithelial carcinoma. Head Neck Pathol. 2014;8:146–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uhlén M, Fagerberg L, Hallström BM, et al. Proteomics: tissue-based map of the human proteome. Science. 2015;347:1260419. [DOI] [PubMed] [Google Scholar]
- 25.Khurshid Z, Najeeb S, Mali M, et al. Histatin peptides: pharmacological functions and their applications in dentistry. Saudi Pharm J. 2017;25:25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cerami E, Gao J, Dogrusoz U, et al. The cBio Cancer Genomics Portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Persson M, Andrén Y, Mark J, et al. Recurrent fusion of MYB and NFIB transcription factor genes in carcinomas of the breast and head and neck. Proc Natl Acad Sci USA. 2009;106:18740–18744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brill LB II, Kanner WA, Fehr A, et al. Analysis of MYB expression and MYB-NFIB gene fusions in adenoid cystic carcinoma and other salivary neoplasms. Mod Pathol. 2011;24:1169–1176. [DOI] [PubMed] [Google Scholar]
- 29.Andreasen S, Tan Q, Agander TK, et al. Adenoid cystic carcinomas of the salivary gland, lacrimal gland, and breast are morphologically and genetically similar but have distinct microRNA expression profiles. Mod Pathol. 2018;31:1211–1225. [DOI] [PubMed] [Google Scholar]
- 30.Michal M, Skalová A, Simpson RH, et al. Well-differentiated acinic cell carcinoma of salivary glands associated with lymphoid stroma. Hum Pathol. 1997;28:595–600. [DOI] [PubMed] [Google Scholar]
- 31.Bishop JA, Yonescu R, Batista D, et al. Utility of mammaglobin immunohistochemistry as a proxy marker for the ETV6-NTRK3 translocation in the diagnosis of salivary mammary analogue secretory carcinoma. Hum Pathol. 2013;44:1982–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geyer FC, Pareja F, Weigelt B, et al. The spectrum of triple-negative breast disease. Am J Pathol. 2017;187:2139–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piscuoglio S, Hodi Z, Katabi N, et al. Are acinic cell carcinomas of the breast and salivary glands distinct diseases? Histopathology. 2015;67:529–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guerini-Rocco E, Hodi Z, Piscuoglio S, et al. The repertoire of somatic genetic alterations of acinic cell carcinomas of the breast: an exploratory, hypothesis-generating study. J Pathol. 2015;237:166–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grünewald I, Vollbrecht C, Meinrath J, et al. Targeted next generation sequencing of parotid gland cancer uncovers genetic heterogeneity. Oncotarget. 2015;6:18224–182237. [DOI] [PMC free article] [PubMed] [Google Scholar]


