Abstract
Background:
Hairy cell leukemia (HCL) and hairy cell leukemia variant (HCLv) are rare diseases with overlapping clinicopathological features. Features distinguishing HCL from HCLv include expression of CD25, CD123, CD200, annexin-A1, and the presence of BRAF V600E mutation. HCLv typically lacks these markers, but they may occur in a subgroup of HCL patients with an aggressive clinical course. We examined CD43, CD81, CD79b, and CD200 expression in HCL and HCLv.
Methods:
Multiparametric flow cytometry (FCM) was performed on blood from 59 HCL and 15 HCLv patients for protocol entry. Mean fluorescent intensity (MFI) of CD43, CD79b, CD81, and CD200 was determined (for CD200, n = 17 and 7, respectively).
Results:
Median MFI of HCL vs HCLv was 545 vs 272 for CD43, 602 vs 2,450 for CD81, 4,962 vs 1,969 for CD79b, and 11,652 vs 1,405 for CD200, respectively. Analysis of the median differences, HCL minus HCLv (and their 95% confidence intervals and P-values) indicated that CD43 MFI (estimated median difference (95% CI): 212 [72–413; P = 0.0027) and CD200 MFI (9,883 [3,514–13,434]; P < 0.0001) were higher in HCL than in HCLv, while CD81 MFI (−1,858 [−2,604 to −1,365]; P < 0.0001) was lower in HCL than in HCLv. CD79b MFI HCL median was more than double that of HCLv, but the observed difference (1,571 [−739 to 4,417]) was consistent with the null hypothesis of no difference (P = 0.13).
Conclusions:
CD200, CD43, and CD81 are likely differentially expressed between HCL and HCLv, reflecting their differing disease biology. Inclusion of these markers in FCM is potentially informative. © 2019 International Clinical Cytometry Society
Keywords: hairy cell leukemia, hairy cell leukemia variant, CD43, CD81, CD79b, CD200, flow cytometry, immunophenotype, B-cell lymphoma, B-cell lymphoproliferative disorder, splenic lymphoma
INTRODUCTION
Hairy cell leukemia (HCL) is a rare, indolent neoplasm of mature B-cells representing approximately 2% of lymphoid leukemias. HCL variant (HCLv) is an even rarer B-cell lymphoproliferative disorder that may resemble HCL morphologically but is clinically and biologically distinct from classic HCL. As such, the 2017 WHO classification of hematological malignancies classifies HCLv under “splenic B-cell lymphoma/leukemia, unclassifiable” (1). HCLv is a more aggressive disease, with significantly shorter median survival than HCL (2). Since HCL and HCLv may show overlapping clinicopathologic features, distinguishing these diseases from each other is critical for appropriate treatment and prognostic assessment (2,3). Multiparameter flow cytometry (FCM) is necessary in the diagnosis and differentiation of HCL from HCLv, has the sensitivity for detecting minimal residual disease (MRD) posttherapy (4–6), and is appropriate in the evaluation of patients with suspected HCL or HCLv (3). HCL classically expresses bright CD20, bright CD22, bright CD11c, bright CD25, CD103, and bright CD123, while HCLv is typically negative for CD25, with diminished or negative CD123 (2,7–11).
The clinical and diagnostic utility of CD43, CD79b, CD81, and CD200 has been shown in a variety of B-cell neoplasms by FCM. The expression patterns of CD43, CD79b, CD81, and CD200 are described in chronic lymphocytic leukemia (CLL) (12,13), CD10-expressing lymphomas (14), and other B-cell lymphoproliferative disorders and in plasma cell neoplasms (15–18). CD81 is also described in B-cell precursors, and aberrantly decreased CD81 expression is a helpful feature in the diagnosis of B-lymphoblastic leukemia (19). Distinct CD200 expression patterns are also described in HCL (18,20) and HCLv (21), as well as in CLL and mantle cell lymphoma (MCL) (22). Although there are reports of CD43 (23) and CD79b (16,24) expression in HCL and/or HCLv by FCM, these markers have not been extensively assessed in this context. Our study explores the potential utility of the expression patterns of CD43, CD79b, CD81, and CD200 in a substantial number of these two rare diseases.
MATERIALS AND METHODS
Patients
FCM of 74 cases (59 HCL and 15 HCLv), evaluated for initial protocol eligibility from 2011 to 2014, was included in this study, and included both treated and newly diagnosed patients (NCT01087333, NCT01059786, NCT00923013). The HCL group included 45 males and 14 females, ranging from 32 to 92 years of age, with a median age of 53 years. The HCLv group included 12 males and 5 females, ranging from 41 to 83 years of age, with a median age of 70 years. CD43, CD81, and CD79b were assessed in all cases. CD200 was evaluated in a subset of these cases (17/59 HCL, 7/15 HCLv), because this testing was not available in our laboratory prior to 2014 and not all samples had sufficient numbers of cells to include CD200. All patients signed institutional review board-approved informed consent to be screened.
Flow Cytometric Immunophenotyping
Peripheral blood specimens were stained within 24 h of collection with a panel of antibodies. Erythrocytes were lysed by incubating with lysing solution (150 mmol/L NH4Cl, 10 mmol/L KHCO3, and 0.1 mmol/L EDTA) for 10 min at room temperature at a ratio of 1:9 (volume of sample/volume of lysing solution). Specimens were subsequently washed with phosphate-buffered saline, and stained for 30 min at room temperature with antibody combinations in eight-color cocktails (Supporting Information Table S1) according to Clinical Laboratory Standards Institute document H43-A2 recommendations (25). All cells were fixed in 1.0% paraformaldehyde after staining and stored at 4C for up to 12 h before acquisition. Specimens were acquired on an FACSCanto II (BD Biosciences, San Jose, CA). A target of at least 500,000 to 1 million cells were acquired per tube. Data collected in list mode were analyzed with FCS Express v4 (De Novo Software, Glendale, CA). For analysis, cell populations were gated by forward and side light scatter properties consistent with mononuclear cells, in conjunction with antigen backgating to ensure that relevant cell populations were included. FCM analysis and diagnostic interpretation were performed as previously described (10) and rendered in accordance with WHO 2017 guidelines (1). Normal hematopoietic elements within the specimen served as internal controls for interpretation of expression intensity; furthermore, in accordance with recommended guidelines (26), antigen intensity of abnormal/disease populations (e.g., HCL or HCL-v cells) was described as “dim” or “bright” relative to the fluorescence of relevant normal populations within the sample (e.g., normal B-cells).
Assessment and Statistical Analysis of MFI
The mean fluorescence intensity (MFI) of CD43, CD79b, CD81, and CD200 was determined on HCL and HCLv cells and on normal B-cells (when present). The Wilcoxon rank sum test was performed to compare distributions of MFI of CD43, CD79b, CD81, and CD200 between the HCL and HCLv groups with the null hypothesis of zero difference between medians. The Hodges–Lehmann estimate of the median difference and its 95% confidence interval (CI) are reported as well. A small P-value indicates that the data (or more extreme data) may be inconsistent with the null hypothesis; a large P-value indicates that the data (or less extreme data) may be consistent with the null hypothesis. All reported P-values are two-tailed and unadjusted for multiple comparisons. Thus, the results should be viewed as exploratory. Since all the MFI distributions were positively skewed, the data were base 10 log-transformed for graphical presentation.
RESULTS
CD43 expression was variable in HCL and predominantly negative in HCLv. In HCL, CD43 was dimly positive in 20/59 cases (34%) and negative in the remaining 39 cases (66%). CD43 was dimly positive in 1 HCLv case, and negative in the remaining 14/15 (93%) cases. The median value for MFI of CD43 was double that of HCL (545 [95% CI: 439–670]) compared with HCLv (272 [95% CI, 200–523]) (Table 1). The estimated median difference between the CD43 MFI of the HCL and HCLv groups was 212 (95% CI, 72–413) (Table 2). CD43 MFI showed a wide distribution in HCL, as well as complete overlap with the CD43 MFI of the HCLv group (Fig. 1). Nevertheless, there was evidence that the median difference between the two groups was inconsistent with the null hypothesis (P = 0.0027).
Table 1.
MFI for CD43, CD81, CD79b, and CD200 in HCL and HCLv Groups
| HCL |
HCLv |
|||||
|---|---|---|---|---|---|---|
| Antigen | n | Median MFI | Distribution-free 95% CI | n | Median MFI | Distribution-free 95% CI |
| CD43 | 59 | 545 | 439–670 | 15 | 272 | 200–523 |
| CD81 | 59 | 602 | 514–791 | 15 | 2,450 | 1,866–4,183 |
| CD79b | 59 | 4,962 | 3,391–7,453 | 15 | 1,969 | 367–7,008 |
| CD200 | 17 | 11,652 | 5,687–14,227 | 7 | 1,405 | 438–4,543 |
Table 2.
The Median Differences of MFI for CD43, CD81, CD79b, and CD200 Between HCL and HCLv Groups
| Antigen MFI | Median difference* (95% CI) | P-value** |
|---|---|---|
| CD43 MFI | 212 (72–413) | 0.0027 |
| CD81 MFI | −1,858 (−2604 to −1,365) | <0.0001 |
| CD79b MFI | 1,571 (−739 to 4,417) | 0.13 |
| CD200 MFI | 9,883 (3,514–13,434) | <0.0001 |
Hodges–Lehmann estimate, HCL group minus HCLv group.
Wilcoxon rank sum test.
Fig. 1.
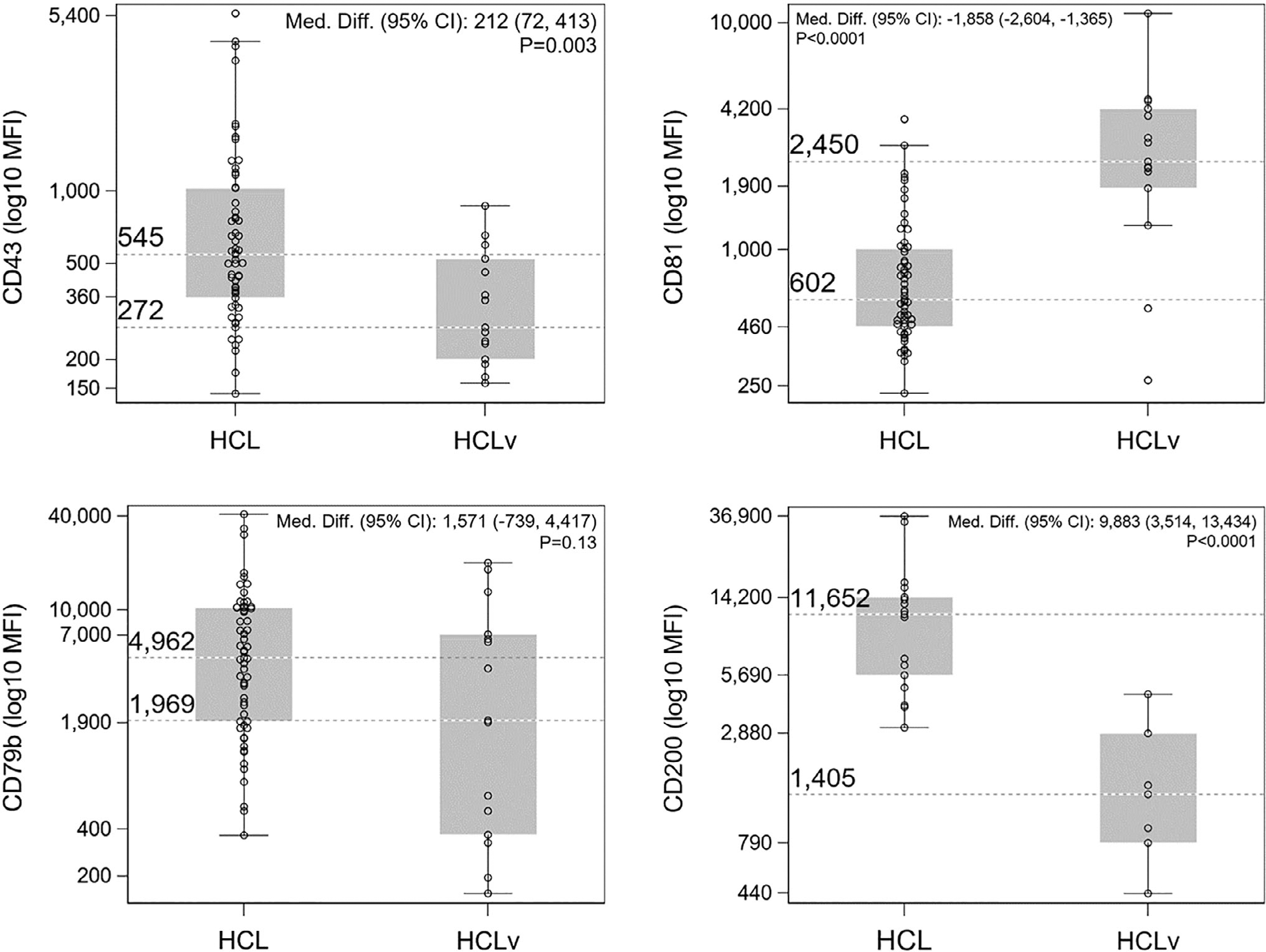
Box-and-whisker plots representing the MFI distribution of CD43, CD81, CD79b, and CD200 in HCL and HCLv. For CD43, CD81, and CD79b, n = 59 (HCL) and n = 15 (HCLv); for CD200, n = 17 (HCL) and n = 7 (HCLv). Values of y-axis represent approximate minimum/maximum values as well as rounded upper/lower quartiles for the box plots. Median values are shown as reference lines.
CD81 was expressed in both HCL and HCLv, but bright CD81 expression was a notable feature of HCLv. In HCL, 57/59 (97%) specimens were positive for CD81, with bright CD81 expression observed in 6/57 cases (11%). In HCLv, all specimens expressed CD81, with bright expression observed in most cases (10/15 cases, 67%). The median CD81 MFI for HCLv was 2,450 (95% CI: 1,866–4,183) which was about fourfold the median MFI for HCL (602 [95% CI: 514–791]). There was strong evidence that the median difference between the two groups (−1,858 [95% CI: −2,608 to −1,365]) was inconsistent with the null hypothesis (P < 0.0001) with less overlap between the two disease entities, especially when compared with the overlap of CD43 MFI (Fig. 1).
CD200 was expressed in all HCL specimens (17/17, 100%), and bright CD200 expression (15/17; 88%) was a characteristic feature. In contrast, CD200 expression was lower in HCLv specimens; expression was predominantly dim (5/7), with moderate expression noted in 2/7 cases (29%). The MFI of CD200 in HCL (11,652 [95% CI: 5,687–14,227]) was more than eight times that in HCLv (1,405 [95% CI: 438–4,543]), and there was strong evidence that the median difference between the two groups (9,883 [95% CI: 3,514–13,434]) was inconsistent with the null hypothesis (P < 0.0001). The distribution of CD200 MFI showed the least degree of overlap between HCL and HCLv among all the antigens studied (Fig. 1). However, given the small sample sizes (especially for HCLv, n = 7), these results should be viewed cautiously.
CD79b was expressed on all HCL specimens (59/59, 100%), with a single case (1/59, 2%) showing variable/partial expression, and five cases (5/59, 8%) showing dim expression. CD79b was also expressed in 12/15 (80%) cases of HCLv. Although the median MFI of CD79b in HCL (4,962 [95% CI: 3,391–7,453]) was more than double that of HCLv (1,969 [95% CI: 367–7,008]), the median difference between the groups (1,571 [95% CI: −739 to 4,417]) was consistent with the null hypothesis (P = 0.13). The expression of CD79b tended to be more variable than the other three antigens and the distribution of CD79b MFI widely overlapped between HCL and HCLv (Fig. 1).
Figure 2 illustrates the above findings by demonstrating the flow cytometric expression patterns of CD43, CD81, CD200, and CD79b in a representative case of HCL (Fig. 2A) and HCLv (Fig. 2B). The contrasting expression patterns of CD43, CD81, and CD200 are shown. Although CD79b expression was observed for all HCL cases and most (80%) HCLv cases, expression of CD79b was variable, and lack of CD79b was noted in a minority of HCLv cases (Fig. 2B).
Fig. 2.
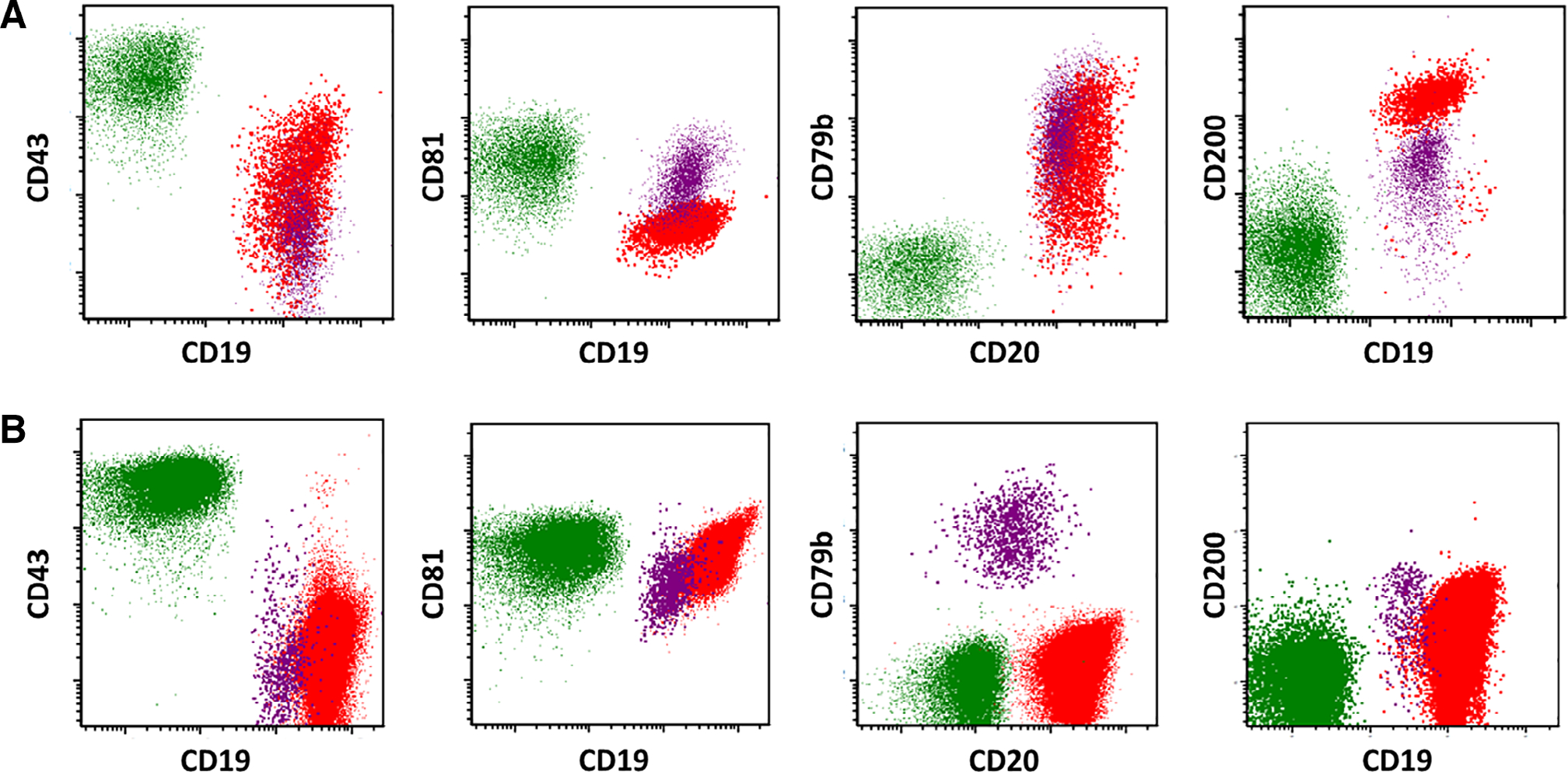
Expression patterns of CD43, CD81, CD79b, and CD200 in a representative case of HCL (A, top row) and HCLv (B, bottom row). Cell populations are designated as follows: T-cells (green), normal B-cells (purple), and HCL or HCLv cells (red).
DISCUSSION
HCL and HCLv are biologically distinct entities, with different treatment and prognosis. The simultaneous expression of bright CD25, bright CD11c, and bright CD123 is valuable in differentiating HCL from HCLv (1,7,9–11,27). Annexin-1, tartrate-resistant acid phosphatase staining, and BRAF V600E mutations also distinguish HCL from HCLv (28,29). The BRAF mutation is an important disease-defining molecular genetic event in HCL, resulting in constitutive MAPK activation. HCLv typically lacks Annexin-1 and exhibits wild-type BRAF; however, a minor but significant subset of immunophenotypically classic HCL cases has been identified with these features by molecular studies (30), and biologically behave more aggressively than the typically indolent HCL (31). The rarity of HCL, and especially HCLv, limits the number of cases available for evaluation and reporting in the literature. Our institution is in the unique position to evaluate both HCL and HCLv on a routine basis, and the size of this study is noteworthy. Additionally, BRAF and MEK inhibitor therapies for HCL (32,33) may downregulate expression of CD25 and alter other HCL-defining immunophenotypic features (34). Therapy-related immunophenotypic alterations may pose a challenge in detecting MRD, if additional immunophenotypic markers are not also carefully evaluated. Identifying and incorporating new, clinically useful markers into FCM panels may be helpful in this regard, as new therapies continue to be developed.
CD43 (leukosialin) is a cell-surface sialoglycoprotein involved in cell adhesion, differentiation, and activation. It is predominantly expressed on hematopoietic cells (T-cells, NK cells, granulocytes, monocytes, macrophages, hematopoietic stem cells, platelets) and nonhematopoietic tumors (35), but not on erythrocytes (36). Interestingly, CD43 is able to mediate seemingly opposing functions. Due to the inherent negative charge of its sialic acid-rich extracellular domain, CD43 facilitates cellular repulsion; conversely, posttranslational modification and various resulting glycoforms promote cellular adhesion (37). In T-cells, intracellular processing of CD43 and its nuclear translocation regulate cellular pathways that protect a cell from apoptosis (38). Consequently, CD43 expression may facilitate oncogenic cell survival in both nonhematopoietic and hematopoietic malignancies (35,39).
Most circulating B-cells lack CD43, with the exception of a minor subset of activated B-cells, and its expression is observed to varying degrees in a range of B-cell lymphomas (40,41). Flow cytometric assessment of CD43, in combination with CD20, shows a reproducible expression pattern that is reminiscent of the expression pattern of CD10 and CD20 in the progressive maturation of marrow B-cell precursors/hematogones (42). Bright CD43 expression is a notable feature for MRD detection of CLL, when evaluated in the context of an appropriate immunophenotypic marker combination (13). Though a report including CD43 assessment by FCM exists (23) in a case of CD27(+) HCLv/japanese variant, FCM assessment of CD43 in HCL and HCLv has not been extensively evaluated. In our series, we demonstrate that HCL exhibits a median difference of 212 (95% CI: 72–413) in CD43 MFI over HCLv (Table 2) with a twofold ratio of the medians (Table 1). However, overlapping distributions of CD43 MFI values suggest that CD43 may have limited clinical utility to distinguish between HCL and HCLv, especially at lower levels of CD43 expression (Fig. 1). Nevertheless, bright expression of CD43, when used in combination with other traditional immunophenotypic markers (i.e., CD20, CD22, CD11c, CD25, CD103, CD123), is a helpful confirmatory feature of HCL.
CD81 is a tetraspanin molecule expressed as part of a B-cell receptor (BCR) complex with a multitude of functions. CD81 is involved in lowering the threshold for B-cell activation, appropriate trafficking, and cell surface expression of CD19, signaling through downstream kinases, cellular proliferation, adhesion, and migration (43,44). Hepatitis C virus enters and infects B-cells via CD81, leading to subsequent B-cell stimulation and IGHV mutation (45).
Patterns of tetraspanin molecule expression, including CD81, have been described in stages of normal B-cell maturation, as well as in various B-cell lymphoproliferative disorders and plasma cell myeloma (46). Bright CD81 is a feature of Burkitt lymphoma and distinguishes it from other CD10-expressing lymphomas (14). CD81 is expressed on B-cell precursors, and its expression is aberrantly decreased in B-lymphoblastic leukemia (19). FCM assessment of CD81 in HCL and HCLv, however, has not been extensively evaluated. In our study, CD81 was expressed in all HCLv (100%) and almost all HCL (97%) specimens. Bright CD81 expression was a notable feature of HCLv; a median difference of 1,858 (95% CI: 1,365–2,604) in CD81 MFI between HCLv and HCL was observed, with a roughly fourfold ratio of median MFI of HCLv over that of HCL (Table 1 and Fig. 1). Furthermore, the expression of both CD43 and CD81 tended to show an inverse pattern when comparing HCL (CD43+, CD81dim+) with HCLv (CD43 dim+, CD81bright+) (Figs. 1 and 2).
CD79b is the beta chain of a heterodimeric protein that associates with the BCR complex. It appears early in B-cell development and its surface expression persists until the plasma cell stage; it plays an important role in B-cell development and drives receptor signal transduction (47). It is ubiquitously expressed among B-cell lymphomas and lymphoproliferative disorders. CD79b detection and quantitation by FCM demonstrates decreased expression in CLL/small lymphocytic lymphoma (CLL/SLL), increased expression in HCL and MCL, and marked overexpression in splenic lymphoma with villous lymphocytes and B-cell prolymphocytic leukemia, when compared with normal B-cells (16,48,49), and CD79b was shown to be expressed on HCL (16). Previously, CD79b exhibited moderate/weak expression by FCM in 4/7 cases (57%) of HCLv, compared with 23/24 cases (96%) of HCL with strong expression (27). In HCLv, CD79b is reportedly expressed in just under one-third of cases (3/11 cases, 27%) (24). Regarding HCL, our CD79b expression data are consistent with the preceding studies. Interestingly, we observed a slightly higher percentage of CD79b positivity (80%) in HCLv than that reported in the preceding studies, but still less than in HCL (100%). We observed a median difference of 1,571 (95% CI: −739 to 4,417) between CD79b MFI HCL values (Fig. 2A far left, HCL representative case) compared with HCLv (Fig. 2B far left, HCLv representative case) with a roughly 2.5-fold ratio of the medians (Table 1). However, the distributions of CD79b MFI overlapped considerably (Table 1 and Fig. 1), and so the data are consistent with no difference between the two groups. (Table 2). Based on these results, CD79b may not discriminate between HCL and HCLv. However, from a treatment standpoint, therapeutic modalities targeting CD79b may potentially be applicable to all cases of HCL and most cases of HCLv. Two examples of CD79b-targeted therapy, Polatuzumab vedotin, and Iladatuzumab vedotin (DCDS0780A), are antibody-drug conjugates comprised of an anti-CD79b antibody linked to monomethylauristatin E, and are currently under investigation in clinical trials for potential therapeutic utility in B-cell malignancies (50).
CD200 (OX-2) is an immunoglobulin superfamily membrane glycoprotein that is expressed by various cell types including B-cells, thymocytes, activated T-cells, and neuronal and endothelial cells. It is believed to exert an immunosuppressive effect via interaction with its receptor (CD200R) (51,52). CD200 demonstrates different expression patterns in a various B-cell lymphoproliferative disorders and B-cell lymphomas, such as B-lymphoblastic leukemia/lymphoma (CD200+), Burkitt’s lymphoma (CD200-), diffuse large B-cell lymphoma (CD200-), and mediastinal large B-cell lymphoma (CD200+) (53). CD200 expression distinguishes CLL from MCL and marginal zone lymphoma; the latter two entities are both typically negative for CD200 (17). CD200 is expressed on plasma cell myeloma and is associated with poor prognosis (54). CD200 assessment by FCM is well documented for HCL and has also been described for HCLv. Brunetti et al identified CD200 expression in 10/10 cases of HCL, with levels significantly higher than CD200 levels in normal B-cells (20). Sandes et al. identified bright CD200 expression in 13/13 cases of HCL, in contrast to dim CD200 expression observed in six cases of splenic marginal zone lymphoma (18). Other studies have also demonstrated similar findings (21,22,55–57). In contrast to HCL, HCLv demonstrates diminished or lack of CD200 expression. In a series including mature B-cell lymphoproliferative disorders, Mason et al. identified CD200 expression in 34/34 HCL cases and 0/3 HCLv (56). Pillai et al. demonstrate bright CD200 in 10 cases of HCL (with a mean MFI of 15,411) that was significantly higher than in four cases of HCLv (mean MFI of 742) (21). Similar observations were reported by Rahman et al.; CD200 was negative in 1 case of HCLv, whereas 6/6 cases of HCL showed bright CD200 (median MFI 5,050, MFI range 3,400–13,101) (57). In our study, the median difference between HCL and HCLv CD200 MFI values was 9,883 (95% CI: 3,514–13,434) with a roughly eightfold ratio of the medians (Table 1), which confirms the findings of the preceding studies, with bright expression of CD200 in HCL, and diminished CD200 in HCLv. However, given the small sample sizes (especially for HCLv, n = 7), the results for CD200 should be viewed cautiously.
HCL and HCLv, despite their nomenclature, represent two biologically divergent entities, and their different expression patterns of CD43, CD81, and CD200 reflect that divergence. Expression of CD200 induces immune tolerance and suppresses antitumor response. Engagement of CD81 results in phosphorylation and activation of multiple downstream kinases that facilitate cytoskeletal reorganization and induce increased cell proliferation. CD43 modulates both cellular repulsion and adhesion, and regulates apoptosis. In this study, the majority of HCL were positive for CD43, consistently brightly positive for CD200, but with lower CD81 expression. The expression of CD43 and CD200 reflects the features of HCL as an indolent malignancy, with protection from apoptosis and cell survival conferred by the effect of CD43, along with immune tolerance granted to the HCL cells due to high levels of CD200. In this respect, HCL resembles another indolent B-cell lymphoproliferative disorder, CLL, which shows characteristic bright CD43, dim CD81, and CD200 expressions (13,17). On the other hand, HCLv exhibits high levels of CD81, with lower expression of CD43 and CD200. Because CD81 plays a role in cytoskeletal reorganization and cell proliferation, the CD81 expression pattern in HCLv is more compatible with that of a more actively proliferating neoplasm and may reflect the characteristic lymphocytosis presentation of HCLv.
The therapeutic implications of these findings are also an interesting consideration. Due to its critical role in regulating immune tolerance, OX-2/CD200 is emerging as a potentially interesting target for immune checkpoint blockade (58). Differential expression of CD43 glycoforms in various malignancies also makes it an attractive target for cancer immunotherapy (35). The role of CD81 as a therapeutic target is currently being explored (59). Currently, interest in these molecular targets lies predominantly in the treatment of solid tumors; however, CD79b targeted therapy is already under investigation in clinical trials for B-cell lymphoma (50).
In summary, multiparametric FCM is a critical test in the differentiation of HCL from HCLv. Newer treatment modalities may alter and/or mask the distinguishing immunophenotypic features unique to each of these disease entities; therefore, searching for other newer, potentially useful markers is important. Our study examines CD200, CD43, CD79b, and CD81 in HCL and HCLv. We identify potentially distinguishing differences in CD43 and CD81 expression in HCL from HCLv. We demonstrate CD79b expression in all the cases of HCL and most cases of HCLv, a finding that may be of therapeutic interest. We demonstrate differences in expression of CD200, corroborating results of previous studies. These markers may be informative if added to the clinical FCM arsenal and may have therapeutically useful implications.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the intramural program of the Center for Cancer Research, National Cancer Institute, and National Institutes of Health. The authors have no conflicts of interest to disclose.
Grant sponsor: National Institutes of Health
Grant sponsor: National Cancer Institute
Grant sponsor: Center for Cancer Research
Footnotes
Additional supporting information may be found online in the Supporting Information section at the end of the article.
LITERATURE CITED
- 1.Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J (Eds). WHO classification of tumours of haematopoietic and lymphoid tissues. Lyon: IARC; 2017. [Google Scholar]
- 2.Kreitman RJ, Arons E. Update on hairy cell leukemia. Clin Adv Hematol Oncol. 2018;16:205–215. [PMC free article] [PubMed] [Google Scholar]
- 3.Grever MR, Abdel-Wahab O, Andritsos LA, Banerji V, Barrientos J, Blachly JS, Call TG, Catovsky D, Dearden C, Demeter J, et al. Consensus guidelines for the diagnosis and management of patients with classic hairy cell leukemia. Blood. 2017;129:553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ravandi F, O’Brien S, Jorgensen J, Pierce S, Faderl S, Ferrajoli A, Koller C, Challagundla P, York S, Brandt M, et al. Phase 2 study of cladribine followed by rituximab in patients with hairy cell leukemia. Blood. 2011;118:3818–3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sausville JE, Salloum RG, Sorbara L, Kingma DW, Raffeld M, Kreitman RJ, Imus PD, Venzon D, Stetler-Stevenson M. Minimal residual disease detection in hairy cell leukemia. Comparison of flow cytometric immunophenotyping with clonal analysis using consensus primer polymerase chain reaction for the heavy chain gene. Am J Clin Pathol. 2003;119:213–217. [DOI] [PubMed] [Google Scholar]
- 6.Kreitman RJ, Tallman MS, Robak T, Coutre S, Wilson WH, Stetler-Stevenson M, FitzGerald DJ, Santiago L, Gao G, Lanasa MC, et al. Minimal residual hairy cell leukemia eradication with moxetumomab pasudotox: Phase 1 results and long-term followup. Blood. 2018;131:2331–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Totero D, Tazzari PL, Lauria F, Raspadori D, di Celle PF, Carbone A, Gobbi M, Foa R. Phenotypic analysis of hairy cell leukemia: “variant” cases express the interleukin-2 receptor beta chain, but not the alpha chain (CD25). Blood. 1993;82: 528–535. [PubMed] [Google Scholar]
- 8.Hassan IB, Hagberg H, Sundstrom C. Immunophenotype of hairy-cell leukemia. Eur J Haematol. 1990;45:172–176. [DOI] [PubMed] [Google Scholar]
- 9.Matutes E, Martinez-Trillos A, Campo E. Hairy cell leukaemia-variant: Disease features and treatment. Best Pract Res Clin Haematol. 2015;28:253–263. [DOI] [PubMed] [Google Scholar]
- 10.Shao H, Calvo KR, Gronborg M, Tembhare PR, Kreitman RJ, Stetler-Stevenson M, Yuan CM. Distinguishing hairy cell leukemia variant from hairy cell leukemia: Development and validation of diagnostic criteria. Leuk Res. 2013;37:401–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Venkataraman G, Aguhar C, Kreitman RJ, Yuan CM, Stetler-Stevenson M. Characteristic CD103 and CD123 expression pattern defines hairy cell leukemia: Usefulness of CD123 and CD103 in the diagnosis of mature B-cell lymphoproliferative disorders. Am J Clin Pathol. 2011;136:625–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jung G, Eisenmann JC, Thiebault S, Henon P. Cell surface CD43 determination improves diagnostic precision in late B-cell diseases. Br J Haematol. 2003;120:496–499. [DOI] [PubMed] [Google Scholar]
- 13.Rawstron AC, Villamor N, Ritgen M, Bottcher S, Ghia P, Zehnder JL, Lozanski G, Colomer D, Moreno C, Geuna M, et al. International standardized approach for flow cytometric residual disease monitoring in chronic lymphocytic leukaemia. Leukemia. 2007;21:956–964. [DOI] [PubMed] [Google Scholar]
- 14.Cardoso CC, Auat M, Santos-Pirath IM, Rudolf-Oliveira RCM, da Silva JP, Lange BG, Siegel D, de Moraes ACR, Del Moral JAG, Santos-Silva MC. The importance of CD39, CD43, CD81, and CD95 expression for differentiating B cell lymphoma by flow cytometry. Cytometry B Clin Cytom. 2018;94:451–458. [DOI] [PubMed] [Google Scholar]
- 15.Alapat D, Coviello-Malle J, Owens R, Qu P, Barlogie B, Shaughnessy JD, Lorsbach RB. Diagnostic usefulness and prognostic impact of CD200 expression in lymphoid malignancies and plasma cell myeloma. Am J Clin Pathol. 2012;137:93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olejniczak SH, Stewart CC, Donohue K, Czuczman MS. A quantitative exploration of surface antigen expression in common B-cell malignancies using flow cytometry. Immunol Invest. 2006;35: 93–114. [DOI] [PubMed] [Google Scholar]
- 17.Palumbo GA, Parrinello N, Fargione G, Cardillo K, Chiarenza A, Berretta S, Conticello C, Villari L, Di Raimondo F. CD200 expression may help in differential diagnosis between mantle cell lymphoma and B-cell chronic lymphocytic leukemia. Leuk Res. 2009; 33:1212–1216. [DOI] [PubMed] [Google Scholar]
- 18.Sandes AF, Chauffaille MD, Oliveira C, Maekawa Y, Tamashiro N, Takao TT, Ritter EC, Rizzatti EG. CD200 has an important role in the differential diagnosis of mature B-cell neoplasms by multiparameter flow cytometry. Cytometry B Clin Cytom. 2014;86: 98–105. [DOI] [PubMed] [Google Scholar]
- 19.Muzzafar T, Medeiros LJ, Wang SA, Brahmandam A, Thomas DA, Jorgensen JL. Aberrant underexpression of CD81 in precursor B-cell acute lymphoblastic leukemia: Utility in detection of minimal residual disease by flow cytometry. Am J Clin Pathol. 2009;132: 692–698. [DOI] [PubMed] [Google Scholar]
- 20.Brunetti L, Di Noto R, Abate G, Gorrese M, Gravetti A, Raia M, Scalia G, Pascariello C, Camera A, Del Vecchio L. CD200/OX2, a cell surface molecule with immuno-regulatory function, is consistently expressed on hairy cell leukaemia neoplastic cells. Br J Haematol. 2009;145:665–667. [DOI] [PubMed] [Google Scholar]
- 21.Pillai V, Pozdnyakova O, Charest K, Li B, Shahsafaei A, Dorfman DM. CD200 flow cytometric assessment and semiquantitative immunohistochemical staining distinguishes hairy cell leukemia from hairy cell leukemia-variant and other B-cell lymphoproliferative disorders. Am J Clin Pathol. 2013;140: 536–543. [DOI] [PubMed] [Google Scholar]
- 22.Challagundla P, Medeiros LJ, Kanagal-Shamanna R, Miranda RN, Jorgensen JL. Differential expression of CD200 in B-cell neoplasms by flow cytometry can assist in diagnosis, subclassification, and bone marrow staging. Am J Clin Pathol. 2014;142:837–844. [DOI] [PubMed] [Google Scholar]
- 23.Tabata R, Tabata C, Iwama H, Yasumizu R, Kojima M. CD27-positive hairy cell leukemia-Japanese variant. Virchows Arch. 2016;468:375–379. [DOI] [PubMed] [Google Scholar]
- 24.Matutes E, Wotherspoon A, Catovsky D. The variant form of hairy-cell leukaemia. Best Pract Res Clin Haematol. 2003;16:41–56. [DOI] [PubMed] [Google Scholar]
- 25.Wayne P, Clinical and Laboratory Standards Institute. Clinical flow cytometric analysis of neoplastic hematolymphoid cells; approved guideline. 2nd ed. USA: Clinical and Laboratory Standards Institute. CLSI document H43-A2, 2007. [Google Scholar]
- 26.Davis BH, Holden JT, Bene MC, Borowitz MJ, Braylan RC, Cornfield D, Gorczyca W, Lee R, Maiese R, Orfao A, et al. 2006 Bethesda International Consensus recommendations on the flow cytometric immunophenotypic analysis of hematolymphoid neoplasia: Medical indications. Cytometry B Clin Cytom. 2007;72(Suppl 1):S5–S13. [DOI] [PubMed] [Google Scholar]
- 27.Del Giudice I, Matutes E, Morilla R, Morilla A, Owusu-Ankomah K, Rafiq F, A’Hern R, Delgado J, Bazerbashi MB, Catovsky D. The diagnostic value of CD123 in B-cell disorders with hairy or villous lymphocytes. Haematologica. 2004;89:303–308. [PubMed] [Google Scholar]
- 28.Falini B, Tiacci E, Liso A, Basso K, Sabattini E, Pacini R, Foa R, Pulsoni A, Dalla Favera R, Pileri S. Simple diagnostic assay for hairy cell leukaemia by immunocytochemical detection of annexin A1 (ANXA1). Lancet. 2004;363:1869–1870. [DOI] [PubMed] [Google Scholar]
- 29.Tiacci E, Trifonov V, Schiavoni G, Holmes A, Kern W, Martelli MP, Pucciarini A, Bigerna B, Pacini R, Wells VA, et al. BRAF mutations in hairy-cell leukemia. N Engl J Med. 2011;364:2305–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xi L, Arons E, Navarro W, Calvo KR, Stetler-Stevenson M, Raffeld M, Kreitman RJ. Both variant and IGHV4–34-expressing hairy cell leukemia lack the BRAF V600E mutation. Blood. 2012; 119:3330–3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arons E, Suntum T, Stetler-Stevenson M, Kreitman RJ. VH4–34+ hairy cell leukemia, a new variant with poor prognosis despite standard therapy. Blood. 2009;114:4687–4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Follows GA, Sims H, Bloxham DM, Zenz T, Hopper MA, Liu H,Bench A, Wright P, Van’t Veer MB, Scott MA. Rapid response of biallelic BRAF V600E mutated hairy cell leukaemia to low dose vemurafenib. Br J Haematol. 2013;161:150–153. [DOI] [PubMed] [Google Scholar]
- 33.Kreitman RJ. Hairy cell leukemia-new genes, new targets. Curr Hematol Malig Rep. 2013;8:184–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pettirossi V, Santi A, Imperi E, Russo G, Pucciarini A, Bigerna B, Schiavoni G, Fortini E, Spanhol-Rosseto A, Sportoletti P, et al. BRAF inhibitors reverse the unique molecular signature and phenotype of hairy cell leukemia and exert potent antileukemic activity. Blood. 2015;125:1207–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tuccillo FM, Palmieri C, Fiume G, de Laurentiis A, Schiavone M, Falcone C, Iaccino E, Galandrini R, Capuano C, Santoni A, et al. Cancer-associated CD43 glycoforms as target of immunotherapy. Mol Cancer Ther. 2014;13:752–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brown WRA, Barclay AN, Sunderland CA, Williams AF. Identification of a glycophorin-like molecule at the cell surface of rat thymocytes. Nature. 1981;289:456–460. [DOI] [PubMed] [Google Scholar]
- 37.Ostberg JR, Barth RK, Frelinger JG. The Roman god Janus: A paradigm for the function of CD43. Immunol Today. 1998;19:546–550. [DOI] [PubMed] [Google Scholar]
- 38.Seo W, Ziltener HJ. CD43 processing and nuclear translocation of CD43 cytoplasmic tail are required for cell homeostasis. Blood. 2009;114:3567–3577. [DOI] [PubMed] [Google Scholar]
- 39.Ma XB, Zheng Y, Yuan HP, Jiang J, Wang YP. CD43 expression in diffuse large B-cell lymphoma, not otherwise specified: CD43 is a marker of adverse prognosis. Hum Pathol. 2015;46:593–599. [DOI] [PubMed] [Google Scholar]
- 40.Treasure J, Lane A, Jones DB, Wright DH. CD43 expression in B cell lymphoma. J Clin Pathol. 1992;45:1018–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lai R, Weiss LM, Chang KL, Arber DA. Frequency of CD43 expression in non-Hodgkin lymphoma. A survey of 742 cases and further characterization of rare CD43+ follicular lymphomas. Am J Clin Pathol. 1999;111:488–494. [DOI] [PubMed] [Google Scholar]
- 42.Tsao L, Colovai AI, Jiang JG, Bhagat G, Alobeid B. CharacterizingCD43 expression in haematogones using multicolor flow cytometric analysis. Br J Haematol. 2005;128:820–823. [DOI] [PubMed] [Google Scholar]
- 43.Feneant L, Levy S, Cocquerel L. CD81 and hepatitis C virus (HCV) infection. Viruses. 2014;6:535–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levy S Function of the tetraspanin molecule CD81 in B and T cells. Immunol Res. 2014;58:179–185. [DOI] [PubMed] [Google Scholar]
- 45.Machida K, Cheng KT, Pavio N, Sung VM, Lai MM. Hepatitis C virus E2-CD81 interaction induces hypermutation of the immunoglobulin gene in B cells. J Virol. 2005;79:8079–8089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barrena S, Almeida J, Yunta M, Lopez A, Fernandez-Mosteirin N, Giralt M. Aberrant expression of tetraspanin molecules in B-cell chronic lymphoproliferative disorders and its correlation with normal B-cell maturation. Leukemia. 2005;19:1376–1383. [DOI] [PubMed] [Google Scholar]
- 47.Seda V, Mraz M. B-cell receptor signalling and its crosstalk with other pathways in normal and malignant cells. Eur J Haematol. 2015;94:193–205. [DOI] [PubMed] [Google Scholar]
- 48.Cabezudo E, Carrara P, Morilla R, Matutes E. Quantitative analysis of CD79b, CD5 and CD19 in mature B-cell lymphoproliferative disorders. Haematologica. 1999;84:413–418. [PubMed] [Google Scholar]
- 49.D’Arena G, Musto P, Cascavilla N, Dell’Olio M, Di Renzo N, Carotenuto R. Quantitative flow cytometry for the differential diagnosis of leukemic B-cell chronic lymphoproliferative disorders. Am J Hematol. 2000;64:275–281. [DOI] [PubMed] [Google Scholar]
- 50.Herrera AF, Molina A. Investigational antibody-drug conjugates for treatment of B-lineage malignancies. Clin Lymphoma Myeloma Leuk. 2018;18:452–468. e4. [DOI] [PubMed] [Google Scholar]
- 51.Gorczynski RM, Cattral MS, Chen Z, Hu J, Lei J, Min WP, Yu G, Ni J. An immunoadhesin incorporating the molecule OX-2 is a potent immunosuppressant that prolongs Allo- and xenograft survival. J Immunol. 1999;163:1654–1660. [PubMed] [Google Scholar]
- 52.Hoek RM, Ruuls SR, Murphy CA, Wright GJ, Goddard R, Zurawski SM, Blom B, Homola ME, Streit WJ, Brown MH, et al. Down-regulation of the macrophage lineage through interaction with OX2 (CD200). Science. 2000;290:1768–1771. [DOI] [PubMed] [Google Scholar]
- 53.Dorfman DM, Shahsafaei A. CD200 (OX-2 membrane glycoprotein) expression in b cell-derived neoplasms. Am J Clin Pathol. 2010;134:726–733. [DOI] [PubMed] [Google Scholar]
- 54.Moreaux J, Hose D, Reme T, Jourdan E, Hundemer M, Legouffe E, Moine P, Bourin P, Moos M, Corre J, et al. CD200 is a new prognostic factor in multiple myeloma. Blood. 2006;108:4194–4197. [DOI] [PubMed] [Google Scholar]
- 55.El Desoukey NA, Afify RA, Amin DG, Mohammed RF. CD200 expression in B-cell chronic lymphoproliferative disorders. J Investig Med. 2012;60:56–61. [DOI] [PubMed] [Google Scholar]
- 56.Mason EF, Pozdnyakova O, Li B, Dudley G, Dorfman DM. Flow cytometric patterns of CD200 and CD1d expression distinguish CD10-negative, CD5-negative mature B-cell lymphoproliferative disorders. Am J Clin Pathol. 2017;148:33–41. [DOI] [PubMed] [Google Scholar]
- 57.Rahman K, Kumar P, Gupta R, Singh MK, Nityanand S. Role ofCD200 in differential diagnosis of mature B-cell neoplasm. Int J Lab Hematol. 2017;39:384–391. [DOI] [PubMed] [Google Scholar]
- 58.Ring EK, Markert JM, Gillespie GY, Friedman GK. Checkpoint proteins in pediatric brain and extracranial solid tumors: Opportunities for immunotherapy. Clin Cancer Res. 2017;23:342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vences-Catalan F, Duault C, Kuo CC, Rajapaksa R, Levy R, Levy S.CD81 as a tumor target. Biochem Soc Trans. 2017;45:531–535. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


