Abstract
Pediatric cancers are generally characterized by low mutational burden and few recurrently mutated genes. Recent studies suggest that genomic alterations may help guide treatment decisions and clinical trial selection. Here, we describe genomic profiles from 1,215 pediatric tumors representing sarcomas, extracranial embryonal tumors, brain tumors, hematologic malignancies, carcinomas, and gonadal tumors. Comparable published datasets identified similar frequencies of clinically relevant alterations, validating this dataset as biologically relevant. We identified novel ALK fusions in a neuroblastoma (BEND5–ALK) and an astrocytoma (PPP1CB–ALK), novel BRAF fusions in an astrocytoma (BCAS1–BRAF) and a ganglioglioma (TMEM106B–BRAF), and a novel PAX3–GLI2 fusion in a rhabdomyosarcoma. Previously characterized ALK, NTRK1, and PAX3 fusions were observed in unexpected malignancies, challenging the “disease-specific” alterations paradigm. Finally, we identified recurrent variants of unknown significance in MLL3 and PRSS1 predicted to have functional impact. Data from these 1,215 tumors are publicly available for discovery and validation.
Introduction
Pediatric cancers are rare malignancies worldwide and represent ~1% of new cancer diagnoses in the United States (1). Unlike adult solid tumors that are predominantly carcinomas derived from epithelial cells, pediatric solid tumors are histologically diverse and include carcinomas from epithelial cells, embryonal tumors from developing tissues, gonadal tumors from sex-cord stromal cells, brain tumors from neural and glial cells, leukemias and lymphomas from hematopoietic cells, and sarcomas from mesenchymal cells (2). Advances in detection and treatment of childhood cancers have resulted in improved survival for many subtypes. However, over 1,900 pediatric patients in the United States succumb to disease each year, and survivors often face lifelong side effects from toxic chemo and/or radiotherapy treatments (1).
The genomic landscape of pediatric tumors is distinct from adult tumors due to low mutational burden, and relatively few, albeit highly recurrent, significantly mutated genes (3). Even within the same tumor type, mutation profiles in pediatric samples are distinct from their adult counterpart. For example, the BCR–ABL1 fusion protein is observed in 2% of childhood acute lymphoid leukemia (ALL) but up to 25% of adult ALL cases (4). Certain tumors that occur primarily in children and young adults are marked by characteristic alterations, such as BRAF alterations in low-grade gliomas, EWSR1 fusions in Ewing sarcoma, and ALK alterations in neuroblastoma (5–7). In some cases, the identification of genomic alterations has guided targeted therapy selection. Clinical efficacy has been demonstrated with crizotinib against ALK-driven neuroblastomas and anaplastic large cell lymphomas (8), and with imatinib against BCR–ABL1-driven ALL (9). Additionally, mutational signatures have assisted in the stratification of some disease, such as medulloblastoma, where alteration patterns can define clinically distinct subtypes within the same histology (10). Recent data suggest that the incorporation of genomic information can help inform therapeutic decisions in pediatric oncology (11–14).
While improvements in detection and treatment have led to a decline in pediatric cancer mortality, many survivors face decreased quality of life and an increased risk of secondary cancer development as side effects from efficacious, but toxic, treatments (15). Therefore, the development of less toxic regimens is needed urgently. Improved therapeutic strategies are also needed in cancers with the highest mortality rates, including CNS malignancies, high-risk neuroblastoma, metastatic bone cancers, and soft tissue sarcomas, where standard-of-care treatment has limited efficacy. An enhanced understanding of the genomic alterations contributing to tumorigenesis in this population may identify new targets and strategies for improved therapeutic intervention across childhood cancers.
Here, we describe a dataset of 1,215 pediatric tumors (ages 0–18) comprised of sarcomas (26.7%), extracranial embryonal tumors (22.8%), brain tumors (20.8%), hematologic malignancies (19.3%), carcinomas (9.1%), and gonadal tumors (1.4%). This collection represents tumor specimens referred for sequence analysis of all classes of somatic variation in cancer-associated genes to guide clinical management. While many studies have focused on the genomic analysis of more common tumors from which specimens are easy to obtain, this collection contains multiple rare entities that have not been profiled previously in large numbers. We describe the discovery of novel fusions, point mutations, and the spectrum of therapeutic targets across multiple tumor types. Collectively, this dataset represents one of the largest groups of genomically profiled pediatric tumors to date and can be used as a resource for discovery of novel alterations and validation of findings from other studies.
Materials and Methods
Samples were submitted to a CLIA-certified, New York State-accredited, and CAP-accredited laboratory (Foundation Medicine) for next-generation sequencing (NGS)-based genomic profiling. The pathologic diagnosis of each case was confirmed by review of hematoxylin and eosin (H&E)–stained slides or Wright-Giemsa stained blood/aspirate smears and all samples that advanced to DNA and/or RNA extraction contained a minimum of 20% tumor cells. DNA and RNA were extracted from formalin-fixed paraffin-embedded (FFPE) 10-μm sections or from fresh blood or bone marrow aspirates. For samples assayed on FoundationOne, DNA was adaptor ligated, and hybrid capture was performed for all coding exons of 182 (v1), 287 (v2), 323 (v3), or 395 (v5) cancer-related genes plus select introns from 14 (v1), 19 (v2), 24 (v3), or 31 (v5) genes frequently rearranged in cancer (Supplementary Tables S1–S5); samples assayed on FoundationOne Heme (v4) underwent DNA-based hybrid capture for all coding regions of 465 genes plus select introns from 31 genes frequently rearranged in cancer. For samples in which RNA was available, targeted RNA-seq was performed for rearrangement analysis in 333 genes (Supplementary Table S4; refs. 16, 17). Captured libraries were sequenced to a median exon coverage depth of >600× using Illumina sequencing, and resultant sequences were analyzed for base substitutions, insertions deletions, copy number alterations (focal amplifications and homozygous deletions) and select gene fusions, as previously described (16, 17). RNA sequences were analyzed for the presence of rearrangements only. Frequent germline variants from the 1000 Genomes Project (dbSNP142) were removed, and known confirmed somatic alterations deposited in the Catalog of Somatic Mutations in Cancer (COSMIC v62) were highlighted as biologically significant. Germline variants documented in the dbSNP database (http://www.ncbi.nlm.nih.gov/SNP/) and germline variants with two or more counts in the ExAC database (~0.0003% population frequency, http://exac.broadinstitute.org/ ) were removed, with the exception of known cancer driver germline events (e.g., documented hereditary BRCA1/2 and TP53 deleterious mutations). Additionally, recurrent variants of unknown significance that were predicted to be germline were removed using an internally developed algorithm (Sun and colleagues, in review 2016). In brief, a CGH-like profile was created based on coverage and allele frequencies (AF) of ~3,500 genome-wide SNPs. This profile incorporated tumor purity (p), copy number (C), and minor allele count (M). A variant’s measured frequency was compared with the expected frequency: AFgermline = (pM + 1 − p)/(pC + 2(1 − p)) versus AFsomatic = pM/(pC + 2(1 − p)) and a prediction was made with statistical confidence based on read depth and local variability of SNP allele frequencies (18). All truncations and deletions in known tumor suppressor genes were also called as significant. To maximize mutation-detection accuracy (sensitivity and specificity) in impure clinical specimens, the test was previously optimized and validated to detect base substitutions at a ≥5% mutant allele frequency (MAF), indels with a ≥10% MAF, and focal copy number alterations at ≥20% tumor fraction with high accuracy (16, 17). This study was reviewed and approved by the Western Institutional Review Board (WIRB). Sequence analysis results are available publicly in a browsable web portal at https://pediatric-data.foundationmedicine.com.
Results
Characteristics of the pediatric dataset
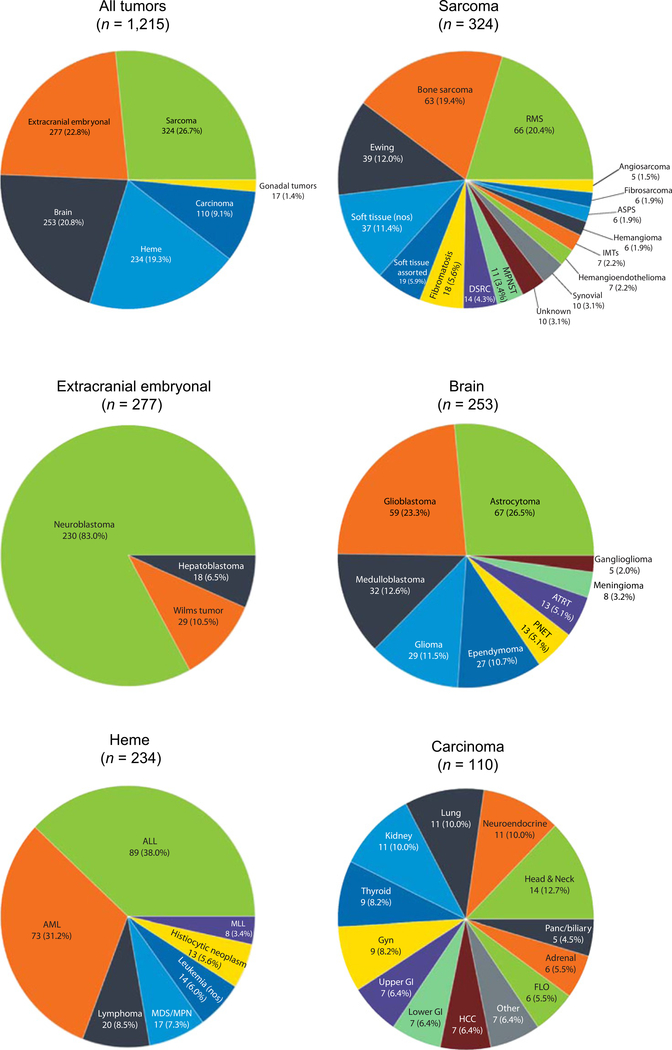
This dataset was composed of 1,215 unique samples from patients age 18 or younger that underwent genomic profiling as part of clinical care (Supplementary Table S6). To facilitate classification, tumors were grouped into one of six major categories and assigned a detailed tumor subtype that more accurately described the diagnosis at the time of genomic testing (Fig. 1). The samples represented sarcomas (26.7%; 16 subtypes), extracranial embryonal tumors (22.8%; 3 subtypes), brain tumors (20.8%; 9 subtypes), hematologic malignancies (19.3%; 7 subtypes), carcinomas (9.1%; 13 subtypes), and gonadal tumors (1.4%; 1 subtypes). The most common sarcoma subtypes included rhabdomyosarcomas (20.4%), bone sarcomas, including both osteosarcomas and other rare bone cancers (19.4%), and Ewing sarcomas (12.0%) followed by 13 other subtypes of varying frequency (Fig. 1). Extracranial embryonal tumors included neuroblastomas (83%), Wilms tumors (10.5%), and hepatoblastomas (6.5%). The brain tumors were astrocytomas (26.5%), glioblastomas (23.3%), medulloblastomas (12.6%), gliomas (11.5%), and five additional subtypes (Fig. 1). Hematologic malignancies included acute lymphoblastic leukemias (ALL; 38.0%), acute myeloid leukemias (AML; 31.2%), lymphomas (8.5%), as well as four subcategories with frequencies <10% (Fig. 1). Of the 13 carcinoma subtypes, the most common were head and neck cancers (12.7%), neuroendocrine tumors (10.0%), lung cancers (10.0%), and kidney cancers (10.0%). Finally, gonadal tumors were comprised entirely of ovarian/testis tumors.
Figure 1.
Distribution of sample types within the pediatric data cohort. Samples were grouped into one of six major categories (top left). Each major category was subsequently divided into multiple subcategories that contained detailed information about the tumor diagnosis, with the exception of gonadal tumors. Sarcomas contained 16 subtypes (top right), extracranial embryonal tumors contained 3 subtypes (middle left), brain tumors contained 9 subtypes (middle right), heme malignancies contained 7 subtypes (bottom left), and carcinomas contained 13 subtypes (bottom right). Gonadal tumors were composed entirely of this tumor type (not shown).
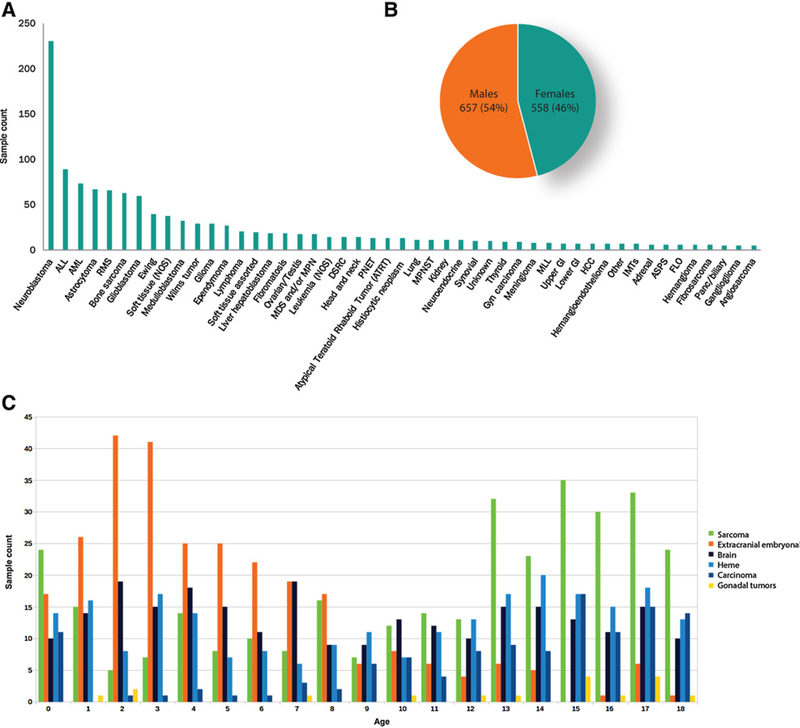
All sample subcategories contained at least five tumors; the most common tumors in this dataset were neuroblastoma, ALL, AML, astrocytoma, and rhabdomyosarcoma (Fig. 2A). The gender distribution showed a slight predominance of male patients (Fig. 2B). Age at testing showed a bimodal distribution with one peak in young patients (ages 0–8) and a second peak in teenage patients (ages 14–18). Extracranial embryonal tumors, including neuroblastomas, Wilms tumors, and hepatoblastomas, were the most common tumors in very young patients (<8 years old), while sarcomas were predominant in teenage patients (ages 14–18; Fig. 2C). It is unknown if the samples submitted for genomic profiling represented primary or recurrent tumors. Disease stage and prior treatment history were also unavailable.
Figure 2.
Characteristic of the pediatric data cohort. A, The sample cohort contained 1,215 tumors from 49 unique subtypes. B, Each subtype contained at least 5 samples. Slightly more males were presented. C, Age at the time of testing showed a bimodal distribution with a predominance of extracranial embryonal tumors at younger ages (<8 years old) and a peak of sarcomas at older ages (13–18 years old).
We next analyzed the most commonly altered genes within each broad disease category. Across the dataset, we observed a median of 2.5 alterations per sample. The most commonly affected genes in sarcomas were TP53 (18.8%), EWSR1 (15.4%), CDKN2A (9.4%), MYC (7.5%), and CDKN2B (6.6%; Supplementary Fig. S1). Extracranial embryonal tumors showed a different distribution of alterations with frequent events in MYCN (23.9%), ALK (14.9%), TP53 (5.4%), ATRX (5.4%), and CTNNB1 (4.0%; Supplementary Fig. S2). Brain tumors had frequent disrupting events in TP53 (25.3%), BRAF (19.4%), CDKN2A (12.6%), NF1 (12.3%), and H3F3A (9.9%; Supplementary Fig. S3). Hematologic malignancies harbored alterations in CDKN2A (18.9%), NRAS (15.5%), TP53 (14.7%), CDKN2B (12.9%), and KRAS (11.6%; Supplementary Fig. S4). Carcinomas were characterized by TP53 (18.2%), CTNNB1 (7.3%), CDKN2A (7.3%), SMARCA4 (6.4%), and KRAS (5.5%; Supplementary Fig. S5). Finally, gonadal tumors contained alterations in TP53 (29.4%), KRAS (17.6%), CHD2 (11.8%), ARID1A (11.8%), and DICER1 (11.8%; Supplementary Fig. S6).
Comparison of genomics with previously published pediatric tumors
We sought to compare how the genomic profiles in this clinical dataset compared with those from previously published studies. This analysis included tumors for which at least 30 samples were available in our dataset, and corresponding genomic landscape papers had been published for matched disease subtypes. Because our samples were analyzed for only focal copy number events, we omitted from this comparison disease subtypes that contained arm-level copy number events as distinct features (e.g., bone sarcomas). Samples within this dataset also lacked information about grade and stage, so we also excluded comparisons with datasets that were preselected for these features (e.g., low-grade glioma).
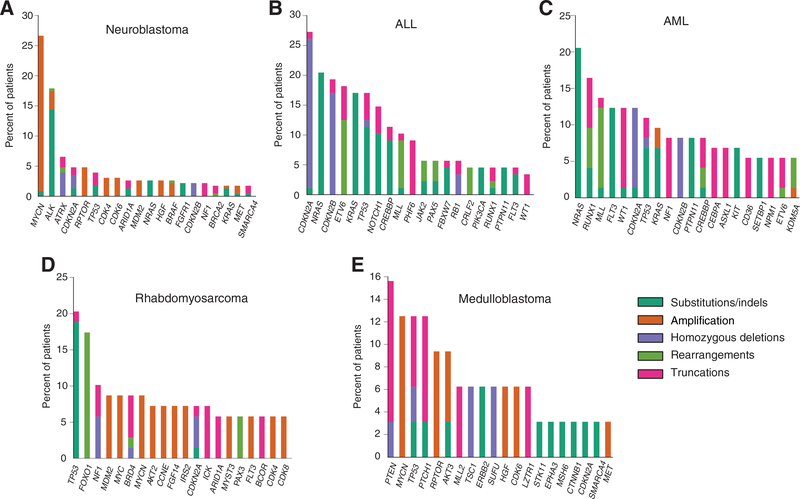
Neuroblastoma was the most common tumor in our database (n = 230). The most frequent alterations within this subtype were observed in MYCN (26.5%), ALK (17.8%), ATRX (6.5%), CDKN2A (4.8%), and RPTOR (4.8%; Fig. 3A). Previous integrative genome, exome, and transcriptome sequencing of a similarly sized high-risk neuroblastoma series (n = 240) identified five genes (ALK, ATRX, PTPN11, MYCN, NRAS) as biologically significant and statistically enriched in this disease (7). Comparing the frequency of alterations in these genes across the two datasets, no statistically significant differences were observed (Table 1).
Figure 3.
Long tail distributions across the five most common diseases. The top 20 altered genes in neuroblastoma (A), ALL (B), AML (C), rhabdomyosarcoma (D), and medulloblastoma (E). Types of alterations are color coded using the key to the right.
Table 1.
Comparison of alteration frequencies
| Tumor type and gene alteration | FM | Published frequency | Fisher exact test |
|---|---|---|---|
| Neuroblastoma | |||
| ALK point mutation | 14.3% (33/230) | 9.2% (22/240) | P = 0.0865 |
| PTPN11 point mutation | 1.3% (3/230) | 2.9% (7/240) | P = 0.3396 |
| ATRX point mutation | 1.8% (4/227)a | 2.5% (6/240) | P = 0.7522 |
| ATRX focal deletion | 4.0% (9/227)a | 7.1% (17/240) | P = 0.1610 |
| MYCN point mutation | 0.9% (2/230) | 1.7% (4/240) | P = 0.6859 |
| MYCN focal amplification | 25.7% (59/230) | 32.0% (77/240) | P = 0.1284 |
| NRAS point mutation | 2.6% (6/230) | 0.8% (2/240) | P = 0.1677 |
| ALL | |||
| ETV6-RUNX1 rearrangement | 12.0% (10/83) | 19.4% (47/242) | P = 0.1362 |
| TCF3-PBX1 rearrangement | 2.4% (2/83)a | 7.0% (17/242) | P = 0.1749 |
| BCR-ABL1 rearrangement | 1.1% (1/89) | 3.7% (9/242) | P = 0.2986 |
| MLL rearrangement | 4.8% (4/83)a | 11.4% (16/140) | P = 0.1442 |
| P2RY8-CRLF2 rearrangement | 4.8% (4/83)a | 7% (19/272) | P = 0.6151 |
| PAX5 rearrangement | 6.0% (5/83)a | 2.2% (10/446) | P = 0.0697 |
| PICALM-MLLT10 rearrangement | 2.4% (2/83)a | 9.5% (4/42) | P = 0.1778 |
| AML | |||
| FLT3 internal tandem duplication | 5.5% (4/73) | 16.5% (15/91) | P = 0.0472 |
| NPM1 point mutation | 4.1% (3/73) | 7.8% (23/295) | P = 0.4422 |
| CEBPA point mutation | 6.8% (5/73) | 4.5% (38/847) | P = 0.3785 |
| Fusion-negative RMS | |||
| NRAS point mutation | 6.7% (3/45) | 11.7% (11/94) | P = 0.5482 |
| FGFR4 point mutation | 4.4% (2/45) | 9.6% (9/94) | P = 0.5030 |
| PIK3CA point mutation | 2.2% (1/45) | 7.4% (7/94) | P = 0.4370 |
| BCOR point mutation | 6.7% (3/45) | 7.4% (7/94) | P = 1.00 |
| FBXW7 point mutation | 0% (0/45) | 7.4% (7/94) | P = 0.0960 |
| KRAS point mutation | 2.2% (1/45) | 6.4% (6/94) | P = 0.4279 |
| TP53 point mutation | 20% (9/45) | 5.3% (5/94) | P = 0.0132 |
| NF1 point mutation | 4.4% (2/45) | 5.3% (5/94) | P = 1.00 |
| HRAS point mutation | 2.2% (1/45) | 4.3% (4/94) | P = 1.00 |
| Medulloblastoma | |||
| CTNNB1 point mutation | 3.1% (1/32) | 6.5% (6/92) | P = 0.6760 |
| PTCH1 point mutation | 12.5% (4/32) | 7.6% (7/92) | P = 0.4722 |
| MLL2 point mutation | 6.3% (2/32) | 8.7% (8/92) | P = 1.00 |
| SMARCA4 point mutation | 3.1% (1/32) | 4.30% (4/92) | P = 1.00 |
| TP53 point mutation | 9.4% (3/32) | 3.3% (3/92) | P = 0.1775 |
| BCOR point mutation | 0% (0/31)a | 3.3% (3/92) | P = 0.5712 |
| DDX3X point mutation | 0% (0/5)a | 7.6% (7/92) | P = 1.00 |
| MYC amplification | 3.1% (1/32) | 1.1% (1/92) | P = 0.4511 |
| MYCN amplification | 12.5% (4/32) | 4.30% (4/92) | P = 0.2025 |
Abbreviation: FM, Foundation Medicine dataset.
Corrects for the gene not being assayed on all versions of the test.
We next investigated the frequency of common alterations in ALL. ALL is the most common childhood cancer (1), and the second most common tumor subtype in this series (n = 89). From our data, the most frequent alterations in this disease occurred in CDKN2A (28.1%), NRAS (21.3%), and CDKN2B (20.2%) (Fig. 3B). Multiple studies have previously investigated the genomic alterations underlying development of ALL and their association with treatment response or failure (19). Excluding chromosomal arm-level events, seven other genomic rearrangement events have been described as clinically significant in this disease (ETV6–RUNX1, TCF3–PBX1, BCR–ABL1, P2RY8–CRLF2, PICALM–MLLT10, and diverse rearrangements involving MLL and PAX5). Comparing the frequency of these alterations in our dataset to multiple published series (20–24), no significant differences were observed (Table 1).
Treatment of AML has incorporated mutations in three genes (FLT3, NPM1, and CEBPA) as markers of prognosis, therapeutic targets, and inclusion criteria for clinical trials. Our AML dataset contained 76 unique samples, and harbored frequent alterations in NRAS (20.5%), RUNX1 (16.4%), MLL (13.7%), FLT3 (12.3%), and WT1 (12.3%; Fig. 3C). A significantly lower rate of FLT3 ITD events was observed in our dataset (5.3% versus 16.5%, P = 0.0276; ref. 25). The frequencies of alterations in NPM1 and CEBPA were not statistically different between the Foundation Medicine cohort and published studies (Table 1; refs. 26, 27).
We next investigated the genomic landscape of rhabdomyosarcomas (RMS) in our dataset. The dataset presented herein (n = 66) harbored common alterations in TP53 (20.3%), FOXO1 (17.4%), NF1 (10.1%), MDM2 (8.7%), and MYC (8.7%; Fig. 3D). Information about alveolar or embryonal characterization was not available for these tumors. Within our dataset, 45 RMS tumors (68%) were tested for the presence of a PAX3/7 fusion by RNA-seq; because fusion information for the remaining 21 samples was unavailable, they were excluded from the subsequent analyses described below. Somatic alterations affecting the MAPK/PI3K signaling pathway, including point mutations in NRAS, FGFR4, PIK3CA, BCOR, FBXW7, KRAS, TP53, NF1, and HRAS, have been reported as potential driver alterations in tumors lacking PAX3/7 fusion events (28). Consistent with these data, we observed alterations exclusively in our collection of fusion negative tumors. Alteration frequencies in these genes were not significantly different in our cohort versus the published study (Table 1), except for the frequency of TP53 mutations, which was significantly higher in the Foundation Medicine dataset (20% versus 5.3%, P = 0.0132).
Finally, we undertook an analysis of medulloblastoma alterations. Within our series (n = 32), alterations were observed in PTEN (15.6%), MYCN (12.5%), TP53 (12.5%), PTCH1 (12.5%), and RPTOR (9.4%; Fig. 3E). We compared these data with a published series that interrogated the exomes of 92 primary medulloblastomas (10). Pugh and colleagues identified statistically significant rates of mutation in CTNNB1, PTCH1, MLL2, SMARCA4, and TP53 as well as recurrent mutations in DDX3X, GPS2, BCOR, and LDB1. Additionally, amplification of MYC and MYCN was shown previously to be important in subgroups of medulloblastoma (29). GPS2 and LDB1 were not included on any of our gene lists, and were therefore excluded from our comparison. Similar to the previous tumor types, no statistically significant differences were observed in mutation rates between our cohort and the published frequencies (Table 1).
Discovery of novel fusions in pediatric tumors
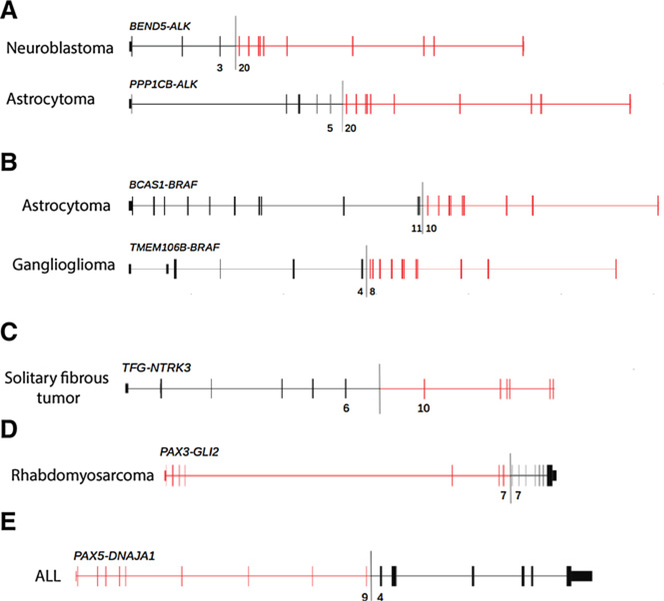
Because canonical fusion proteins involving kinases (e.g., ALK fusions in IMT) and transcription factors (e.g., EWSR1 fusions in Ewing sarcoma) are observed in multiple pediatric tumors, we next investigated the dataset for new and potentially oncogenic fusion proteins involving genes within these families. In total, we identified seven novel, but nonrecurrent, kinase fusions. Two novel ALK fusions were identified in a neuroblastoma (BEND5–ALK) and an astrocytoma (PPP1CB–ALK; Fig. 4A). In both cases, the breakpoints in ALK (intron 19) were similar to known fusions, such as EML4–ALK, with established oncogenic activity and therapeutic potential. We also identified two novel BRAF fusions in an astrocytoma (BCAS1–BRAF) and a ganglioglioma (TMEM106B–BRAF; Fig. 4B). The breakpoints in BRAF (introns 9 and 7) were also similar to those in other known oncogenic fusions. An analogous TMEM106B–ROS1 fusion that incorporated a similar region of TMEM106B was identified previously in an adult lung adenocarcinoma (30). The fusion protein identified in the lung cancer sample involved exons 1–3 of TMEM106B, while this pediatric ganglioglioma fusion involved exons 1–4 of the same gene. Finally, a novel TFG–NTRK3 fusion was identified in a solitary fibrous tumor (Fig. 4C); the breakpoint here kept the kinase domain intact and is predicted to produce a functional fusion protein. Interestingly, this solitary fibrous tumor lacked the canonical NAB2–STAT6 fusion that is ubiquitous in this disease (31); this fusion involving NTRK3 may suggest a differential diagnosis.
Figure 4.
Novel kinase and transcription factor fusions. Novel kinase fusions in ALK (A), BRAF (B), and NTRK3 (C) have similar breakpoints to known fusions involving these genes. Novel transcription factor fusions involving PAX3 (D) and PAX5 (E) were also identified. Exon numbers at the fusion boundary are depicted below each diagram.
In addition to the novel kinase fusions, we also identified a previously characterized fusion involving ALK and NTRK1 in different diseases from which they were originally reported. For example, an SQSTM1–NTRK1 fusion was identified in a fibrosarcoma (Supplementary Fig. S7A). This fusion protein was recently reported in a 45-year-old male with lung adenocarcinoma and associated with clinical sensitivity to the NTRK1 (TrkA) inhibitor entrectinib (32). However, this is the first report of this fusion in a soft tissue tumor. We also identified an STRN–ALK fusion protein in a pediatric kidney carcinoma (Supplementary Fig. S7B). This ALK fusion was reported previously as a recurrent genomic event in aggressive thyroid cancers from adults; the fusion was sensitive to ALK inhibitors in preclinical studies (33). EML4–ALK rearrangements are best known for their role in ~5% of lung adenocarcinomas and their clinical sensitivity to the ALK inhibitor crizotinib (34). Interestingly, we identified EML4–ALK fusion events in three non-lung cancers (thyroid cancer, histiocytic neoplasm, and a ganglioglioma). All three events were similar in structure to the variants that have been reported in lung adenocarcinoma and are predicted to be oncogenic (Supplementary Fig. S7C).
We next evaluated whether there were novel fusions involving transcription factors in the data. We focused specifically on PAX3/7 fusions as they define distinct subsets of rhabdomyosarcomas and ALL (28). In addition to known fusion events involving these genes, we identified a novel PAX3–GLI2 fusion in rhabdomyosarcoma (Fig. 4D) and confirmed a second occurrence of the rare PAX3–NCOA1 fusion in also in rhabdoymosarcoma (Supplementary Fig. S7D; ref. 6). The related protein PAX5 is fused to various partners in ~2.5% of ALL (23). This dataset adds to the growing list of PAX5 fusion partners in ALL with the identification of a novel PAX5-DNAJA1 rearrangement (Fig. 4E) that juxtaposes exons 1–9 of PAX5 with exons 4–9 of DNAJA1. Expression of this alteration was confirmed in RNA sequencing analysis.
Analysis of genomic alterations associated with clinical sensitivity to targeted therapies
Given the long-term side effects associated with many conventional therapies in pediatric cancer, we mined our data for alterations associated with sensitivity to potentially less toxic targeted therapies. This analysis was restricted to agents with established dosing regimens and documented clinical efficacy against specific alterations in pediatric populations. With these filters, we included vemurafenib for BRAF V600E-mutant tumors (35, 36), crizotinib for ALK driven cancers (8), various experimental TRK inhibitors for NTRK-rearranged cancers (NCT02637687; ref. 37), and imatinib in ABL1-rearranged cancers (9).
The canonical BRAF V600E alteration was observed in 27 samples, including astrocytomas (n = 8, 11.9%), glioblastomas (n = 5, 8.5%), gliomas (n = 4, 13.8%), histiocytic neoplasms (n = 3, 23.1%), gangliogliomas (n = 2, 40%), thyroid cancers (n = 2, 22.2%), meningioma (n = 1, 12.5%), rhabdomyosarcoma (n = 1, 1.5%), and AML (n = 1, 1.3%). Rearrangements involving ALK, including those novel events described above, were identified in 11 disease types including lymphoma (n = 6, 30%), IMTs (n = 3, 42.9%), neuroblastoma (n = 2, 0.9%), assorted soft tissue sarcomas (n = 2, 9.1%), and within a single specimen from lung (8.3%), unknown primary (7.1%), histiocytic neoplasms (7.7% thyroid (11.1%), astrocytoma (1.5%), ganglioglioma (20%), and kidney cancer (8.3%). NTRK fusions were identified in fibrosarcoma (n = 2, 28.6%), assorted soft tissue sarcoma (n = 1, 4.5%), glioblastoma (n = 1, 1.7%), hemangioma (n = 1, 16.7%), and bone sarcoma (n = 1, 1.6%). Finally, ABL1 rearrangements were identified in ALL (n = 2, 2.3%), lymphoma (n = 1, 5%), and myelodysplastic &/or MPN (n = 1, 5.9%).
Identification of potentially novel recurrent somatic variants with unknown clinical relevance
To identify potentially novel oncogenic alterations, we first investigated the list of single amino acid substitutions (i.e., point mutations) with unknown clinical relevance. These alterations were neither reported in COSMIC nor dbSNP databases and underwent additional filtering against the ExAc database (see Materials and Methods) to remove benign germline polymorphisms. Internal algorithms were also used to highlight those likely somatic alterations (see Materials and Methods). A complete list of variants meeting these criteria can be found in Supplementary Table S7. Recurrent point mutations (n ≥ 3) were evaluated further using MutationAssessor, an in silico analysis tool that predicts functional impact of base substitutions based on evolutionary conservation (38). Using this approach, we identified three likely somatic variants in two genes with potential functional consequences.
Two variants in KMT2C (A293V and P309L) predicted to have functional impact were each identified in five samples (Supplementary Table S7). KMT2C, also known as MLL3 (mixed-lineage leukemia protein 3), encodes a methyltransferase and is frequently rearranged in subsets of mixed lineage leukemias. Both A293V and P309L occur outside of annotated protein domains in KMT2C, but were conserved among species. KMT2C A293V was observed across multiple tumors types, including two sarcomas (synovial and DSRC tumors) and one each of PNET, ALL, and neuroblastoma. The alteration co-occurred with canonical fusions in synovial sarcoma (SS18-SSX1) and DSRC (EWSR1-WT1; Supplementary Fig. S8A). This mutation has been reported once in a gastric adenocarcinoma sequenced as part of The Cancer Genome Atlas (TCGA) project (39). In contrast, the P309L mutation occurred in brain tumors (2 glioblastomas and 1 astrocytoma) and neuroblastomas (n = 2). This alteration co-occurred with the canonical KIAA1549-BRAF in the astrocytoma sample (Supplementary Fig. S8B). A similar P309R mutation has been reported in a single clear-cell renal cell carcinoma (40).
We also identified four PRSS1 G191R mutations in 3 brain tumors (2 medulloblastomas and 1 glioblastoma) and a Wilms tumor. PRSS1 encodes the trypsin-1 protein in humans. Germline variants in this gene are implicated in hereditary pancreatitis and an increased risk of pancreatic ductal adenocarcinoma (41). The G191R alteration occurs within the peptidase domain and has been reported in a single primary central nervous system lymphoma (42). Notably, no other known alterations were found in two of the four samples harboring the PRSS1 mutation (Supplementary Fig. S9).
While in silico functional analysis of small insertions and deletions (indels) was not possible with available tools, we searched for recurrent indels (n ≥ 3) with potential functional significance based on domain structure. Using this approach, we observed alterations around DKC1 K505, including small indels (Supplementary Fig. S10), in eight tumors from hematologic malignancies (2 ALL and 2 AML), neuroblastoma (n = 2), a bone sarcoma, and a single tumor classified as other. This event occurs within the nuclear and nucleolar localization region (uniprot. org). Interestingly, six additional events around this region have been observed in sarcoma (n = 2; TCGA provisional data), breast cancer (n = 2; ref. 43), gastric cancer (n = 1; ref. 39), and clear cell renal cell carcinoma (n = 1; ref. 44).
Discussion
Pediatric cancers are diverse histological entities that have a distinct clinical course and genomic landscape compared with adult tumors. Increasing evidence suggests that an enhanced understanding of genomic alterations in pediatric patients may help to guide clinical decisions and the design of clinical trials (11–14). We describe a collection of 1,215 samples that underwent Genomic profiling. This dataset represents 49 tumor subtypes across sarcomas, extracranial embryonal tumors, brain tumors, hematologic malignancies, carcinomas, and gonadal tumors. To our knowledge, this cohort represents one of the largest sets of genomically characterized pediatric cancers published to date.
Compared with other large published datasets in neuroblastoma (7), ALL (20–24), AML (25–27), rhabdomyosarcoma (28), and medulloblastoma (10), few significant differences were observed in the frequencies of alterations in most genes that were deemed biologically significant in these disease types. The two exceptions were a decreased rate of FLT3 ITD mutations (5.3% vs. 16.5%, P = 0.0276) in AML (Table 1) and an increased rate of TP53 mutations (20% vs. 5.3%, P = 0.0132) in rhabdomyosarcomas (Table 1) within the Foundation Medicine samples. A lower frequency of FLT3 ITD events (5%) has been observed in patients younger than 10 years of age (45). Interestingly, the mean age of pediatric AML patients within this dataset was 9.1 years old. It is also unknown if these samples represent specific subtypes of AML associated with a low frequency of FLT3 ITD events, such as non-promyelocytic AML (6.6%–8.8%; ref. 46). Some reports have noted that a fraction of samples positive for FLT3 ITD at diagnosis are negative for this alteration at the time of relapse (47), suggesting a possible enrichment of relapsed samples within this cohort. Finally, variable frequencies of FLT3 ITD in pediatric patients have been reported in the literature and range from 15% (48) down to 4% (49) and highlight the bioinformatics challenges of correctly calling these events. The previous study in rhabdomyosarcoma (28) examined somatic events only and did not investigate germline alterations that may have contributed to disease development. However, germline TP53 mutations have been implicated in this tumor (50). Although mutations were not distinguished as somatic or germline in our data due to tumor-only testing, the observed increase in TP53 mutations may be explained in part by germline events that would have been reported here due to their established role in carcinogenesis. Earlier studies focusing on somatic events in this disease may have focused on somatic events only. Additionally, inadequate depth of coverage may have hindered calling of subclonal or rare events.
To demonstrate the discovery potential of this dataset, we mined for novel fusion and mutational events across pediatric cancers. This search resulted in the discovery of five novel kinase fusions involving ALK, BRAF, and NTRK3 (Fig. 4A–C). Based on structural similarities to similar characterized fusions, we hypothesize that these events are oncogenic and contribute to the expanding list of fusions observed in solid tumors. We also identified novel and rare transcription factor fusions involving PAX3 and PAX5 (Fig. 4D and E). While these events are not targetable directly, they have implications for diagnosis, prognosis, and risk stratification. In silico analysis of recurrent variants of unknown significance (VUSes) identified four alterations in three genes with potential functional significance. The MLL3 A293V mutation was observed across solid and heme tumors whereas the MLL3 P309L alteration was enriched in gliomas. A mutation within PRSS1 (G191R) was observed in medulloblastomas, a glioblastoma, and a Wilms tumor. Interestingly, two samples with this alteration lacked other known alterations in cancer genes. Finally, we report deletions around DKC1 K505 within the nucleolar localization region. Functional experiments are ultimately needed to confirm the role of these novel alterations in tumorigenesis.
A dataset this large challenges the paradigm of “disease-specific” alterations. For example, although EML4–ALK is observed primarily in lung cancer, we identified this fusion in three non-lung tumors. We also report other ALK and NTRK1 fusions in diseases other than the tumor types in which they were originally reported. Although rare, these data support the notion that so-called “disease-specific” events can be promiscuous and occur outside of their primary tissues. Therapeutically relevant alterations, such as BRAF V600E alterations, are also observed across a wide variety of tumor types. Prospective identification of such alterations can have potentially significant impact on consideration of treatment options and clinical trial selection (11–14).
We also sought to identify genomic alterations that may be sensitive to targeted therapies. Specially, vemurafenib, imatinib, and experimental NTRK inhibitors have demonstrated promising results against molecularly matched pediatric tumors (8, 9, 35–37; NCT02637687). For example, BRAF V600E has been documented in small percentages of central nervous system tumors, and recent data have demonstrated anecdotal, but durable, clinical responses to vemurafenib (35, 36). We observed proven therapeutically actionable alterations, including ALK, NTRK, and ABL1 fusions as well as oncogenic BRAF V600E mutations, across a variety of diseases, suggesting that targeted therapies may have a broader role in the treatment of some pediatric cancers than previously appreciated, and clinical trials investigating their efficacy outside of their approved indications are warranted. This and recently published datasets could be utilized for the rational design of biomarker-driven trials in pediatric oncology (11–14).
This tumor collection is not without limitations. Unfortunately, corresponding clinical data are unavailable for these specimens, and it is unknown from where in the clinical course the tumor tissue was obtained for genomic profiling. There is also no information about previous treatments or tumor grade/stage, making direct comparison with publicly available datasets challenging. Due to the design of the genomic assay, only focal copy-number events are reported and arm-level amplifications and deletions cannot be assessed. This is a significant void, especially for hematologic samples where such information is crucial for risk stratification and treatment selection. Despite these limitations, we believe that this dataset represents a valuable resource.
While many highly recurrent events have already been described in pediatric cancers, scientists and physicians are becoming increasingly aware of rare, yet equally important, clinically relevant genomic alterations in pediatric malignancies. Novel therapeutic strategies are needed to improve survival and spare patients from long-term side effects of toxic treatments. Large collections of genomically profiled tumors are ripe with discovery potential, and can be used to generate hypotheses, validate rare findings, and investigate the genomic landscape of rare tumors for which only small studies exist. To facilitate exploration of this dataset set by the research community, we have made it available publicly (http://pediatric-data.foundation-medicine.com), with the goal that these data will be incorporated into future experiments that will ultimately improve the treatment and prognosis for children with cancer.
Supplementary Material
Acknowledgments
The authors wish to thank colleagues at Foundation Medicine, including Catherine Winfield, Pauline Glushko, Kate Cook, Mark Freeman, Benjamin Newell, Jared White, Susan Hager, and Allen Nunnally, for their contributions to this project. We also thank the team at Extension Engine, including Eduardo Cortejoso, Ofri Markus, Furqan Nazeeri, Scout Stevenson, Marin Bareta, Antonio Zemunik, Jessica Pang, and Roya Rakhshan, for their assistance with developing the data portal.
Disclosure of Potential Conflicts of Interest
J. Chmielecki, M. Bailey, and J.A. Elvin have ownership interest (including patents) in Foundation Medicine. S. Ramkissoon is a consultant/advisory board member for Bristol-Meyers Squibb. J. Suh has received speakers bureau honoraria from Genentech. G.M. Frampton is a scientist at and has ownership interest (including patents) in Foundation Medicine. S.M. Ali has ownership interest (including patents) in Foundation Medicine. L. Gay is a senior scientist at and has ownership interest (including patents) in Foundation Medicine. R. Erlich is Senior Director, at Biomedical Informatics and has ownership interest (including patents) in Foundation Medicine. J.S. Ross is Medical Director at and reports receiving a commercial research grant from Foundation Medicine. J. Buxhaku is client services representative at Joana Buxhaku. V. Miller is Chief Medical Officer at Foundation Medicine, Inc. P.J. Stephens is CSO at and has ownership interest (including patents) in Foundation Medicine. D. Lipson is vice president at and has ownership interest (including patents) in Foundation Medicine. No potential conflicts of interest were disclosed by the other authors.
Footnotes
Note: Supplementary data[dummy_suppl data] for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
References
- 1.Ward E, DeSantis C, Robbins A, Kohler B, Jemal A. Childhood and adolescent cancer statistics, 2014. CA Cancer J Clin 2014;64:83–103. [DOI] [PubMed] [Google Scholar]
- 2.Stiller CA. Epidemiology and genetics of childhood cancer. Oncogene 2004;23:6429–44. [DOI] [PubMed] [Google Scholar]
- 3.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013;499:214–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernstein ML. Targeted therapy in pediatric and adolescent oncology. Cancer 2011;117:2268–74. [DOI] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Research N, Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med 2015;372:2481–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cantile M, Marra L, Franco R, Ascierto P, Liguori G, De Chiara A, et al. Molecular detection and targeting of EWSR1 fusion transcripts in soft tissue tumors. Med Oncol 2013;30:412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pugh TJ, Morozova O, Attiyeh EF, Asgharzadeh S, Wei JS, Auclair D, et al. The genetic landscape of high-risk neuroblastoma. Nat Genet 2013;45: 279–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mosse YP, Lim MS, Voss SD, Wilner K, Ruffner K, Laliberte J, et al. Safety and activity of crizotinib for paediatric patients with refractory solid tumours or anaplastic large-cell lymphoma: a Children’s Oncology Group phase 1 consortium study. Lancet Oncol 2013;14:472–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schultz KR, Carroll A, Heerema NA, Bowman WP, Aledo A, Slayton WB, et al. Long-term follow-up of imatinib in pediatric Philadelphia chromosome-positive acute lymphoblastic leukemia: Children’s Oncology Group study AALL0031. Leukemia 2014;28:1467–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pugh TJ, Weeraratne SD, Archer TC, Pomeranz Krummel DA, Auclair D, Bochicchio J, et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature 2012;488:106–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris MH, DuBois SG, Glade Bender JL, Kim A, Crompton BD, Parker E, et al. Multicenter feasibility study of tumor molecular profiling to inform therapeutic decisions in advanced pediatric solid tumors: The Individualized Cancer Therapy (iCat) Study. JAMA Oncol 2016. January 28. doi: 10.1001/jamaoncol.2015.5689. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 12.Mody RJ, Wu YM, Lonigro RJ, Cao X, Roychowdhury S, Vats P, et al. Integrative clinical sequencing in the management of refractory or relapsed cancer in youth. JAMA 2015;314:913–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khan J, Helman LJ. Precision therapy for pediatric cancers. JAMA Oncol 2016;2:575–7. [DOI] [PubMed] [Google Scholar]
- 14.Parsons DW, Roy A, Yang Y, Wang T, Scollon S, Bergstrom K, et al. Diagnostic yield of clinical tumor and germline whole-exome sequencing for children with solid tumors. JAMA Oncol 2016. January 28. doi: 10.1001/jamaoncol.2015.5699. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith MA, Altekruse SF, Adamson PC, Reaman GH, Seibel NL. Declining childhood and adolescent cancer mortality. Cancer 2014;120: 2497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, He J, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 2013;31: 1023–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He J, Abdel-Wahab O, Nahas MK, Wang K, Rampal RK, Intlekofer AM, et al. Integrated genomic DNA/RNA profiling of hematologic malignancies in the clinical setting. Blood 2016;127:3004–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun JX, Frampton GM, Wang K, Ross JS, Miller VA, Stephens PJ, et al. A computational method for somatic versus germline variant status determination from targeted next-generation sequencing of clinical cancer specimens without a matched normal control. American Association for Cancer Research Annual Meeting; 2014; San Diego, CA. [Google Scholar]
- 19.Roberts KG, Mullighan CG. Genomics in acute lymphoblastic leukaemia: insights and treatment implications. Nat Rev Clin Oncol 2015;12: 344–57. [DOI] [PubMed] [Google Scholar]
- 20.Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 2007;446:758–64. [DOI] [PubMed] [Google Scholar]
- 21.Rubnitz JE, Camitta BM, Mahmoud H, Raimondi SC, Carroll AJ, Borowitz MJ, et al. Childhood acute lymphoblastic leukemia with the MLL-ENL fusion and t(11;19)(q23;p13.3) translocation. J Clin Oncol 1999;17:191–6. [DOI] [PubMed] [Google Scholar]
- 22.Mullighan CG, Collins-Underwood JR, Phillips LA, Loudin MG, Liu W, Zhang J, et al. Rearrangement of CRLF2 in B-progenitor- and Down syndrome-associated acute lymphoblastic leukemia. Nat Genet 2009;41: 1243–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nebral K, Denk D, Attarbaschi A, Konig M, Mann G, Haas OA, et al. Incidence and diversity of PAX5 fusion genes in childhood acute lymphoblastic leukemia. Leukemia 2009;23:134–43. [DOI] [PubMed] [Google Scholar]
- 24.Brandimarte L, Pierini V, Di Giacomo D, Borga C, Nozza F, Gorello P, et al. New MLLT10 gene recombinations in pediatric T-acute lymphoblastic leukemia. Blood 2013;121:5064–7. [DOI] [PubMed] [Google Scholar]
- 25.Meshinchi S, Woods WG, Stirewalt DL, Sweetser DA, Buckley JD, Tjoa TK, et al. Prevalence and prognostic significance of Flt3 internal tandem duplication in pediatric acute myeloid leukemia. Blood 2001;97:89–94. [DOI] [PubMed] [Google Scholar]
- 26.Brown P, McIntyre E, Rau R, Meshinchi S, Lacayo N, Dahl G, et al. The incidence and clinical significance of nucleophosmin mutations in childhood AML. Blood 2007;110:979–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ho PA, Alonzo TA, Gerbing RB, Pollard J, Stirewalt DL, Hurwitz C, et al. Prevalence and prognostic implications of CEBPA mutations in pediatric acute myeloid leukemia (AML): a report from the Children’s Oncology Group. Blood 2009;113:6558–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shern JF, Chen L, Chmielecki J, Wei JS, Patidar R, Rosenberg M, et al. Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov 2014;4:216–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho YJ, Tsherniak A, Tamayo P, Santagata S, Ligon A, Greulich H, et al. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J Clin Oncol 2011;29:1424–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ou SH, Chalmers ZR, Azada MC, Ross JS, Stephens PJ, Ali SM, et al. Identification of a novel TMEM106B-ROS1 fusion variant in lung adenocarcinoma by comprehensive genomic profiling. Lung Cancer 2015;88: 352–4. [DOI] [PubMed] [Google Scholar]
- 31.Chmielecki J, Crago AM, Rosenberg M, O’Connor R, Walker SR, Ambrogio L, et al. Whole-exome sequencing identifies a recurrent NAB2-STAT6 fusion in solitary fibrous tumors. Nat Genet 2013;45:131–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Farago AF, Le LP, Zheng Z, Muzikansky A, Drilon A, Patel M, et al. Durable clinical response to entrectinib in NTRK1-rearranged non-small cell lung cancer. J Thorac Oncol 2015;10:1670–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelly LM, Barila G, Liu P, Evdokimova VN, Trivedi S, Panebianco F, et al. Identification of the transforming STRN-ALK fusion as a potential therapeutic target in the aggressive forms of thyroid cancer. Proc Natl Acad Sci U S A 2014;111:4233–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shaw AT, Kim DW, Nakagawa K, Seto T, Crino L, Ahn MJ, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med 2013;368:2385–94. [DOI] [PubMed] [Google Scholar]
- 35.Bautista F, Paci A, Minard-Colin V, Dufour C, Grill J, Lacroix L, et al. Vemurafenib in pediatric patients with BRAFV600E mutated high-grade gliomas. Pediatr Blood Cancer 2014;61:1101–3. [DOI] [PubMed] [Google Scholar]
- 36.Robinson GW, Orr BA, Gajjar A. Complete clinical regression of a BRAF V600E-mutant pediatric glioblastoma multiforme after BRAF inhibitor therapy. BMC Cancer 2014;14:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doebele RC, Davis LE, Vaishnavi A, Le AT, Estrada-Bernal A, Keysar S, et al. An oncogenic NTRK fusion in a patient with soft-tissue sarcoma with response to the tropomyosin-related kinase inhibitor LOXO-101. Cancer Discov 2015;5:1049–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res 2011;39: e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cancer Genome Atlas Research N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sato Y, Yoshizato T, Shiraishi Y, Maekawa S, Okuno Y, Kamura T, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet 2013;45:860–7. [DOI] [PubMed] [Google Scholar]
- 41.Rustgi AK. Familial pancreatic cancer: genetic advances. Genes Dev 2014;28:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Braggio E, Van Wier S, Ojha J, McPhail E, Asmann YW, Egan J, et al. Genome-wide analysis uncovers novel recurrent alterations in primary central nervous system lymphomas. Clin Cancer Res 2015;21: 3986–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cancer Genome Atlas N Comprehensive molecular portraits of human breast tumours. Nature 2012;490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cancer Genome Atlas Research N. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013;499:43–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liang DC, Shih LY, Hung IJ, Yang CP, Chen SH, Jaing TH, et al. Clinical relevance of internal tandem duplication of the FLT3 gene in childhood acute myeloid leukemia. Cancer 2002;94:3292–8. [DOI] [PubMed] [Google Scholar]
- 46.Kang HJ, Hong SH, Kim IH, Park BK, Han KS, Cho HI, et al. Prognostic significance of FLT3 mutations in pediatric non-promyelocytic acute myeloid leukemia. Leuk Res 2005;29:617–23. [DOI] [PubMed] [Google Scholar]
- 47.Levis M, Small D. FLT3: ITDoes matter in leukemia. Leukemia 2003;17:1738–52. [DOI] [PubMed] [Google Scholar]
- 48.Liang DC, Shih LY, Hung IJ, Yang CP, Chen SH, Jaing TH, et al. FLT3-TKD mutation in childhood acute myeloid leukemia. Leukemia 2003;17:883–6. [DOI] [PubMed] [Google Scholar]
- 49.Shimada A, Taki T, Tabuchi K, Tawa A, Horibe K, Tsuchida M, et al. KIT mutations, and not FLT3 internal tandem duplication, are strongly associated with a poor prognosis in pediatric acute myeloid leukemia with t(8;21): a study of the Japanese Childhood AML Cooperative Study Group. Blood 2006;107:1806–9. [DOI] [PubMed] [Google Scholar]
- 50.Hettmer S, Archer NM, Somers GR, Novokmet A, Wagers AJ, Diller L, et al. Anaplastic rhabdomyosarcoma in TP53 germline mutation carriers. Cancer 2014;120:1068–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.