Abstract
In the stem-cell niche, the extracellular matrix (ECM) serves as a structural support that additionally provides stem cells with signals that contribute to the regulation of stem-cell function, via reciprocal interactions between cells and components of the ECM. Recently, cell-derived ECMs have emerged as in vitro cell culture substrates to better recapitulate the native stem-cell microenvironment outside the body. Significant changes in cell number, morphology and function have been observed when mesenchymal stem cells (MSC) were cultured on ECM substrates as compared to standard tissue-culture polystyrene (TCPS). As select ECM components are known to regulate specific stem-cell functions, a robust characterization of cell-derived ECM proteomic composition is critical to better comprehend the role of the ECM in directing cellular processes. Here, we characterized and compared the protein composition of ECM produced in vitro by bone marrow-derived MSC, adipose-derived MSC and neonatal fibroblasts from different donors, employing quantitative proteomic methods. Each cell-derived ECM displayed a specific and unique matrisome signature, yet they all shared a common set of proteins. We evaluated the biological response of cells cultured on the different matrices and compared them to cells on standard TCPS. The matrices lead to differential survival and gene-expression profiles among the cell types and as compared to TCPS, indicating that the cell-derived ECMs influence each cell type in a different manner. This general approach to understanding the protein composition of different tissue-specific and cell-derived ECM will inform the rational design of defined systems and biomaterials that recapitulate critical ECM signals for stem-cell culture and tissue engineering.
Keywords: extracellular matrix, mesenchymal stem cell, proteomic analysis, stem-cell derived matrices, ECM cell-culture substrate, tissue engineering
1. Introduction
In the body, cells are surrounded by a complex three-dimensional microenvironment, termed the extracellular matrix (ECM), which provides cells with many chemical and biophysical signals required for cell function. Both specific components and biophysical properties of the ECM coordinate intracellular signaling and downstream biological responses through bidirectional interactions with the cells, regulating numerous physiological processes such as cell survival, migration, proliferation and differentiation [1-3]. In the stem-cell niche, cell-matrix interactions influence and modulate stem-cell self-renewal and differentiation. That is, the ECM operates in vivo not only as a cellular support but also directs cell fate through coordinated physical and biochemical cues [4-6].
During mammalian stem-cell culture, stem cells are removed from their native microenvironment (e.g., the bone-marrow niche for bone-marrow-derived mesenchymal stem cells (MSC), adipose tissue for adipose-derived mesenchymal stem cells) and need to adapt to a relatively foreign environment, that is, tissue-culture polystyrene (TCPS), which is fully synthetic and does not present standard ECM signals. Evidence in the literature has shown that TCPS biases MSC function resulting in lower proliferation rates and a loss of stemness over sequential passages [7-9]. Therefore, ECM-based cell-culture substrates have been developed in an attempt to better recapitulate in vitro the native cellular microenvironment [10-14]. MSC are able to deposit an ECM on TCPS over the course of two weeks and this ECM can be used, after decellularization, as a culture substrate for a new batch of MSC. It has been reported that the culture of MSC on in vitro cell-derived ECM induces significant biological changes in MSC function compared to standard culture conditions [10,11,15,16]. As ECM components are key players in the regulation of cellular processes, it is critical to gain a better knowledge of ECM composition to decipher how ECM components regulate cell function. In addition, a comprehensive understanding of cell-matrix interactions will provide further insight into the rational design of ECM-mimicking substrates for tissue engineering and regenerative medicine [17,18].
Despite several reports on the development of cell-derived ECMs for MSC culture, detailed data about their molecular composition is limited. Traditional biochemical analysis of ECM is challenging on account of the insolubility and complexity of ECM components [19]. To address these issues, we have applied a proteomic approach initially described for the analysis of tumoral ECM [20]. The method consists of a sequential digestion of the ECM proteins followed by tandem mass spectrometry and bioinformatic analyses, yielding a detailed inventory of the ECM and ECM-associated proteins (termed the “matrisome”). By coupling this method to label-based quantitative proteomics, we were able to characterize and compare the molecular composition of cell-derived ECM produced by different cell types in vitro, specifically bone-marrow-derived human mesenchymal stem cells (Bm MSC), adipose-derived MSC (Ad MSC) and human neonatal dermal fibroblasts (NHDF), as well as to evaluate the ECM produced by cells from different donors. Proliferation and mRNA transcriptomic profiling of the cells cultured on the different ECM were performed and compared to standard culture conditions.
We observe that, in addition to a set of common proteins, each cell-derived ECM contains cell-type-specific proteins. Quantitative proteomic analysis reveals a specific matrisome signature for each type of ECM. The matrices lead to differential cell growth and gene expression among the cells as compared to TCPS culture, indicating that the ECM signatures influence each cell type in a differential fashion.
2. Materials and methods
2.1. Preparation of in vitro cell-derived ECM
ECM plates were provided by StemBioSys (San Antonio, Texas) and prepared according to a published protocol [16]. Briefly, human mesenchymal stem cells derived from bone marrow (Bm MSC, Lonza) or from adipose tissue (Ad MSC, Life Technologies), or neonatal dermal fibroblasts (NHDF, Life Technologies) were seeded onto a 75 cm2-cell culture flask coated with human fibronectin (1 h at 37°C) at a cell density of 3,500 cells/cm2 and cultured in α-MEM medium supplemented with 15% fetal bovine serum (FBS) and 1% penicillin-streptomycin for 14 days. The medium was refreshed the day after initial seeding and then every 3 days. At day 7, ascorbic acid 2-phosphate (A2P, Sigma) was added to the medium at a final concentration of 50 μM, and A2P-supplemented medium was used until the end of ECM production, with medium changes every other day. At day 14, plates were decellularized using 0.5% Triton in 20 mM ammonium hydroxide for 5 min, rinsed two times with Hank's Balanced Salt Solution containing both calcium and magnesium (HBSS +/+), and once with ultra-pure H2O. Plates coated with cell-derived ECM were stored dry at 4°C until use for cell culture. In vitro ECM produced by commercially available Ad MSC (two different 35- and 45-year-old female donors), Bm MSC (six different 19- to 22-year-old males and female donors) and NHDF (two different new born male donors) are designated as Ad ECM, Bm ECM and Der ECM, respectively. The donor characteristics listed above were provided by the companies.
2.2. Proteomic analysis of ECM samples
The ECM was mechanically detached from the 75 cm2-cell culture flask using a cell scraper in 2 ml of HBSS +/+, centrifuged at 16,000x g for 5 min, washed with 1 ml of HBSS +/+, centrifuged, and dried in a Speed-Vac (Savant) for 15 min. The ECM pellet was then processed as described previously [20,21]. Briefly, the ECM pellet was resuspended and reduced in a solution of 8 M urea, 100 mM ammonium bicarbonate, and 10 mM dithiothreitol at pH 8 under agitation at 37°C for 2 h. After cooling, cysteines were alkylated by adding iodoacetamide at a final concentration of 25 mM for 30 min. The ECM sample was then diluted to 2 M urea, 100 mM ammonium bicarbonate (pH 8), and deglycosylated with PNGaseF (2000 U, New England BioLabs, Ipswich, MA) for 2 h under agitation at 37°C, followed by digestion with Lys-C (Wako Chemicals USA, Richmond, VA), at a ratio of 1:100 enzyme:substrate, under agitation at 37°C for 2 h. Final digestion was done using trypsin (Sequencing Grade, Promega, Madison, WI), at a ratio of 1:50 enzyme:substrate, under agitation at 37°C overnight, followed by a second aliquot of trypsin, at a ratio of 1:100 enzyme:substrate, and an additional 2 h of incubation. Digests were acidified and desalted using 30mg HLB Oasis Cartridges (Waters Corp., Milford, MA) eluted with 50% acetonitrile with 0.1% trifluoroacetic acid (TFA), followed by concentration in a Speed-Vac.
2.3. Analysis by mass spectrometry (LC-MS/MS)
Each sample was separated by reverse-phase HPLC using an EASY-nLC1000 liquid chromatograph (Thermo Fisher Scientific, Waltham, MA) over a 140-minute gradient before nanoelectrospray using a Q Exactive mass spectrometer (Thermo Fisher Scientific). The mass spectrometer was operated in a data-dependent mode. The parameters for the full-scan MS were: resolution of 70,000 across 350-2000 m/z; AGC 3e6; and maximum IT 50 ms. The full MS scan was followed by MS/MS for the top 10 precursor ions in each cycle with a normalized collision energy (NCE) of 28 (unlabeled samples) or 32 (labeled samples) and dynamic exclusion of 30 s. Raw mass spectral data files (.raw) were searched using Proteome Discoverer (Thermo Fisher Scientific) and Mascot version 2.4.1 (Matrix Science) using the SwissProt Homo sapiens database (SwissProt_2016_02, Homo sapiens 20199 sequences) containing 20,199 entries. Mascot search parameters were: 10 ppm mass tolerance for precursor ions; 0.8 Da for fragment-ion mass tolerance; 2 missed cleavages of trypsin; fixed modifications were carbamidomethylation of cysteines and for the quantitative experiments: Tandem Mass Tag (TMT) 6-plex modifications of lysines and peptide N-termini; variable modifications were oxidized methionines, deamidation of asparagines, pyro-glutamic acid modification at N-terminal glutamines; and hydroxylation of prolines and lysines. Only peptides with a Mascot score greater than or equal to 25 and an isolation interference less than or equal to 30 were included in the quantitative data analysis. The average false discovery rate was 0.0080 (ranging from 0.0033-0.0107). Proteins were identified as being ECM-derived or not using the human matrisome annotations as previously described [22,23].
2.4. Label-based quantitative proteomics
Peptide labeling with Tandem Mass Tag (TMT) reagents (Thermo Fisher Scientific) was performed according to the manufacturer’s instructions. Lyophilized samples were dissolved in 70 μl ethanol and 30 μl of 500 mM triethylammonium bicarbonate (pH 8.5) and the TMT reagent was dissolved in 30 μl of anhydrous acetonitrile. The solutions containing peptides and TMT reagents were vortexed and incubated at room temperature for 1 h. Samples were labeled using the TMT 6-plex channels as follows:
Study of ECMs produced by bone marrow-derived MSC from different donors:
Replicate 1: donor 1: TMT-129N, donor 2: TMT-127N, donor 3: TMT-126, donor 4: TMT-130C, donor 5: TMT-128C, donor 6: TMT-128N. Replicate 2: donor 1: TMT-126, donor 2: TMT-127, donor 3: TMT-128, donor 4: TMT-129, donor 5: TMT-130, donor 6: TMT-131.
Study of ECMs produced by different cell types:
Replicate 1: Bm ECM: TMT-126, Der ECM: TMT-127, Ad ECM: TMT-128. Replicate 2: Bm ECM: TMT-126, Der ECM: TMT-128, Ad ECM: TMT-127.
An initial LC-MS/MS analysis was performed on each sample, and the total number of peptides identified and the sum of the intensity of the precursor ions were used as initial metric to determine equivalent peptide amount and normalize the samples. Equal amounts of samples labeled with the different isotopic TMT reagents were combined and concentrated to completion in a vacuum centrifuge, resuspended in 100 μl of 0.1% formic acid, and analyzed by LC-MS/MS as described above. TMT quantification was isotopically corrected according to manufacturer instructions, and the values were normalized to the median of each channel.
The protein expression data were analyzed as described in [24]. Fold changes were expressed in the log2 scale relative to the average of all samples. To account for biological variation, observed proteins with TMT log2 fold change ratios below −1 were considered reduced in expression and proteins with TMT log2 fold change ratios above 1 were considered increased in expression. The coefficients of variation across the biological replicates were calculated for all proteins as the standard deviation divided by the mean.
The raw mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD005521 and 10.6019/PXD005521 [25,26].
2.5. Cell culture
Bone-marrow-derived human mesenchymal stem cells (Bm MSC, Texas A&M University System Health Science Center), adipose-derived human MSC (Ad MSC, Thermo Fisher Scientific) and human neonatal dermal fibroblasts (NHDF, Lonza) were seeded on each substrate (i.e., Bm ECM, Ad ECM, Der ECM or TCPS) at a cell density of 3,500 cells/cm2. Cells were cultured in α–MEM medium supplemented with 20% FBS and 1% penicillin-streptomycin. The medium was changed the day after seeding and every 3 days thereafter. For the cell growth experiment, cells were harvested and counted using a hemocytometer 72 h after seeding. After fixation with 3.2% paraformaldehyde (PFA) solution, actin filaments were stained with Alexa Fluor® 488 Phalloidin (Life Technologies, Carlsbad, CA) for 30 min and cell nuclei was stained with DAPI for 15 min. Samples were imaged using an Evos fluorescent microscope.
2.6. Immunostaining of the ECM
For confocal imaging, each ECM was prepared on glass-bottom dishes using the same protocol as above to generate ECM-coated plates. ECM was fixed with 3.2% PFA for 15 min at room temperature and then incubated with donkey serum (0.5% in PBS) at 4°C for 1 h. Anti-Collagen I antibody (Abcam, 1:100 in 1% BSA) was then added to the sample. After three rinses with 1% BSA solution, Alexa Fluor® 555 donkey anti-rabbit secondary antibody was added for 1 h at 4°C (1:1000 in 0.5% donkey serum). The sample was washed 5 times with 1% BSA and incubated with an anti-fibronectin antibody (Rabbit monoclonal F1 Alexa Fluor® 647, Abcam, 1:100 1 h 4°C). Cell nuclei were stained with DAPI for 15 min. Olympus FV1200 confocal microscope was used for imaging and image reconstruction was done using Image J.
2.7. Electron microscopy
The protocol for electron microscopy was adapted from [16]. Briefly, ECM samples were hydrated for 1 h with PBS at 37°C and fixed with 2% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) at room temperature for 1 h. Following 30 min incubation with 0.1 M sodium cacodylate buffer at room temperature, the samples were dehydrated in ascending concentrations of ethanol (35%, 70%, 95%, 100%), and coated with 30 nm gold-palladium. After coating, ECM samples were imaged using a scanning electron microscope (Zeiss Merlin High-resolution SEM).
2.8. Transcriptomic analysis
Total RNA was extracted using RNeasy Mini Kit (Qiagen, Hilden, Germany) from NHDF, Ad MSC, and Bm MSC after 3 days of culture on TCPS, Ad ECM, Bm ECM or Der ECM, in triplicate. Total RNA was amplified and labeled using Gene Chip 3’ IVT Express Kit (Affymetrix) and hybridized to Human Genome Microarrays (Affymetrix). Microarray data were analyzed using Spotfire. 310 genes with differential expression (abs log2FC≥1, p-value≤0.05) in at least one of the 3 comparisons (Der ECM versus TCPS, Ad ECM versus TCPS, Bm ECM versus TCPS) for at least one of the 3 cell types (NHDF, Ad MSC, Bm MSC) were selected for self-organizing map (SOM) clustering. Replicates were averaged and row normalized with the GenePattern module PreprocessDataset so that the average of each row is set to 0 with a variance of 1 [27]. These row-centered data were then clustered using the GenePattern module SOMclustering. A range of cluster numbers was tested and the final number of clusters to consider was identified using elbow analysis where the last increase in cluster number explaining an additional 1% of the variance in the data was selected. In this experiment, this corresponded to 13 clusters that explained about 81% of the variance in the data. Gene Set Enrichment Analysis (GSEA) was performed to analyze the gene sets and obtain the enrichment sets [28,29]. All microarray data are publicly available through the Gene Expression Omnibus archives under accession number GSE94667.
The same RNA was also used for real time Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR) to confirm microarray results. Real-time RT-PCR was performed with validated primers and Cells-to-CT Taqman reagents (Applied Biosystems, Foster City, CA) on a StepOnePlus thermocycler (Applied Biosystems). GAPDH was used as a loading control for each sample.
2.9. Statistical analyses
Data are presented as mean ± standard deviation (SD) and were analyzed using GraphPad Prism 6 (GraphPad Software Inc, La Jolla). Statistically significant differences were assessed by unpaired t-test or, for multiple comparisons, by an analysis of variance (ANOVA) at a 0.05 significance level and Tukey's post test, with *p<0.05, **p<0.01, and ***p<0.001.
3. Results
3.1. Bm ECM improves the growth of mesenchymal stem cells in vitro
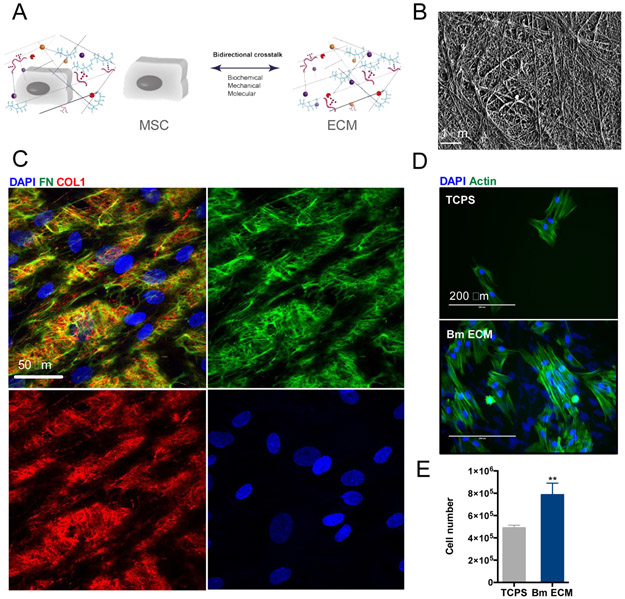
MSC produced an insoluble ECM after 14 days in culture in ascorbic acid-supplemented medium (Fig 1B-C). This cell-derived ECM obtained after decellularization was used as a substrate for the culture of a new batch of MSC. The ECM produced by bone-marrow-derived MSC (Bm MSC) displayed a dense fibrillar structure as imaged by electron microscopy (Fig 1B). Immunostaining showed that the cells were embedded in complex networks of fibronectin and collagen I (Fig 1C).
Figure 1: ECM produced by Bm MSC in vitro.
(A): Bidirectional interactions between MSC and ECM regulate cell function. (B): Electron microscopy of Bm ECM reveals a fibrillar structure. Scale bar is 1 μm. (C): Immunostaining of Bm MSC on their ECM after 14 days using fibronectin antibody (green, top right picture), collagen I antibody (red, bottom left picture) and DAPI (cell nuclei, blue, bottom right picture). The overlay is shown in top left picture. Scale bar is 50 μm. (D): Fluorescence microsopy images of Bm MSC on TCPS (top) or Bm ECM (bottom) after 3 days of culture. Actin cytoskeleton was stained with Alexa Fluor® 488 Phalloidin and cell nuclei with DAPI; scale bar is 200 μm. (E): Number of Bm MSC after 3 days of culture on Bm ECM (blue bar) or TCPS (grey bar). Results show mean ± SD of three independent experiments performed in triplicate. Statistics were done using unpaired t-test (**p<0.01).
We compared the growth of Bm MSC on TCPS and on their own matrix, Bm ECM, 72 h after seeding. MSC on Bm ECM grew significantly more than those on TCPS, indicated by an increase in total cell number (Fig 1D and E).
3.2. Proteomic characterization of in vitro cell-derived Bm ECM
As we and others have observed changes in MSC cell behavior on cell-derived ECM as compared to standard TCPS culture, we were motivated to develop a comprehensive characterization of the Bm ECM molecular composition.
3.2.1. Proteomic approach to characterize in vitro cell-derived ECM
Proteomic characterization of ECM is challenging on account of the complexity and biochemical properties of ECM components, which are mainly large, insoluble, highly cross-linked and glycosylated proteins. The primary technique for proteomic characterization, mass spectrometric analysis, requires proteins to be solubilized and digested into peptides. Traditional approaches to overcome ECM biochemical intractability employ solubilizing agents in concentrations that are incompatible with mass spectrometers [19]. This technical issue, along with the difficulty to functionally annotate ECM components accurately, have impeded the development of a global characterization of stem-cell ECM and explain why only partial characterizations of stem-cell matrices have been reported so far. Recently, Naba et al. have overcome this hurdle by developing a global proteomic approach that consists of the sequential solubilization and digestion of ECM proteins into peptides, without the need for centrifugation, minimizing material loss. This multistep process is comprised of ECM denaturation, reduction, alkylation, deglycosylation and protease digestion, followed by liquid chromatography combined with tandem mass spectrometry (LC-MS/MS) [20,21], bioinformatic analysis and filtering through MatrisomeDB, an inventory of all human matrisome genes and proteins, to specifically identify matrisome proteins (i.e., the proteins that constitute the ECM) [22,23,30].
Here, Bm ECM produced in vitro by bone marrow-derived MSC was characterized using this proteomic approach. The total number of proteins detected in Bm ECM across the donors was 420 on average (range 340 to 499 proteins), of which 20% were expressly identified as ECM or ECM-associated proteins (corresponding to between 64 and 84 proteins). The remaining proteins, designated as non-ECM proteins, consisted of insoluble intracellular components that remained after the decellularization step and were, for the most part, proteins associated with the actin cytoskeleton or with intermediate filaments (Supplementary Table III). Despite their number, the non-ECM-associated proteins represented only a small fraction of the proteomic content, i.e., approximately 25% of the precursor-ion MS signal intensity, which is representative of peptide abundance and the matrisome proteins were detected in large abundance (Supplementary Fig 1).
Fibronectin pre-coating was used in this study to help prevent ECM detachment from the culture dish during ECM production, most likely by providing a more adhesive surface for the cells [11]. To verify that the fibronectin pre-coating did not affect the final ECM composition, and in particular, did not contribute to an overestimation of the fibronectin amount in the ECM, we performed mass spectrometry analysis of ECM deposited on pre-coated TCPS and compared this ECM deposited on uncoated TCPS. Results showed that fibronectin was the most abundant protein, and was detected in similar amounts, in both conditions. In addition, the pre-coating did not affect ECM composition, as shown by the distribution of the 10 most abundant ECM proteins (Supplementary Fig 2). Finally, to estimate the contribution of bovine serum proteins to the composition of cell-derived ECM, mass spectrometry data were queried for bovine proteins and results showed that they represent less than 2% of the total protein content (Supplementary Fig 3). Some peptide sequences are common to both human and bovine species, and were considered as human in this study.
3.2.2. ECM produced by bone marrow-derived MSC displays a specific and consistent matrisome signature
To gain a thorough understanding of the ECM produced by Bm MSC in vitro, we characterized the ECM produced by MSC from six different donors. All proteins, even those detected with only one peptide, were included in the analysis to provide an exhaustive proteomic composition of Bm ECM. The detailed individual composition for each donor is listed in Supplementary Table I.
The ECM produced by Bm MSC in culture over the course of 14 days presents a relatively complex composition. Indeed, across the six donors, 108 ECM and ECM-associated proteins were identified, of which 62 proteins were present in more than five donors (listed in Table I) and 49 proteins were consistently found in all six donors (Fig 2B). The matrisome proteins of the common set were present in large amounts, as indicated by the precursor-ion MS intensity and peptide number, while the proteins found in fewer than three donors were detected with only a few peptides, indicative of their low abundance (Supplementary Table I). As the most abundant peptides are preferentially selected for fragmentation and identified with LC-MS/MS, proteins in low amounts are thus less likely to be consistently detected, partly explaining why some proteins are only detected in some of the donors. The common proteins were detected consistently across donors, as indicated by the similar numbers of peptides and peptide intensities, suggesting that ECM produced by Bm MSC isolated from different donors present a relatively consistent composition.
Table I: Bm ECM signature.
The list shows the proteins identified in at least 5 donors or in at least 3 donors for the secreted factors.
| CORE MATRISOME PROTEINS | |||||||
|---|---|---|---|---|---|---|---|
| COLLAGENS | |||||||
| Fibril forming | Network forming | FACIT | Beaded filament forming | ||||
| Gene symbol | Protein | Gene symbol | Protein | Gene symbol | Protein | Gene symbol | Protein |
| COL1A1/2 | Collagen I α1/2 | COL4A1/2/5 | Collagen IV α1/2/5 | COL12A1 | Collagen XII α1 | COL6A1/2/3 | Collagen VI α1/2/3 |
| COL2A1 | Collagen II α1 | COL8A1 | Collagen VIII α1 | COL14A1 | Collagen XIV α1 | ||
| COL3A1 | Collagen III α1 | COL16A1 | Collagen XVI α1 | ||||
| COL5A1/2 | Collagen V α1/2 | ||||||
| COL11A1 | Collagen XI α1 | ||||||
| GLYCOPROTEINS | |||||||
| Major known | Growth factor binding protein | Vascular ECM proteins | Other | ||||
| Gene symbol | Protein | Gene symbol | Protein | Gene symbol | Protein | Gene symbol | Protein |
| EFEMP2 | Fibulin 4 | LTPB1 | Latent TGF-β binding protein 1 | FGB | Fibrinogen β | AEBP1 | AE-binding protein 1 |
| EMILIN1 | Emilin 1 | FGG | Fibrinogen γ | EDIL3 | EGF-like repeats and discoidin I-like domains 3 | ||
| FBN1/2 | Fibulin 1/2 | VTN | Vitronectin | ||||
| FBLN1/2 | Fibrillin 1/2 | MFGE8 | Milk fat globule-EGF factor 8 protein | ||||
| FN1 | Fibronectin | MXRA5 | Matrix-remodeling associated 5 | ||||
| MFAP2/5 | Microfibrillar-associated proteins | PXDN | Peroxidasin homolog | ||||
| POSTN | Periostin | TGFBI | TGF β-induced protein | ||||
| THBS1/2 | Thrombospondin 1/2 | VWA1 | von Willebrand factor A domain containing protein 1 | ||||
| TNC | Tenascin C | ||||||
| PROTEOGLYCANS | |||||||
| Leucine-rich repeat (LRR) | LINK | Complex | Hyalectans (LINK/CLEC/CCP) | ||||
| Gene symbol | Protein | Gene symbol | Protein | Gene symbol | Protein | Gene symbol | Protein |
| BGN | Biglycan | HAPLN1 | Hyaluronan and proteoglycan link protein 1 | HSPG2 | Perlecan | VCAN | Versican |
| DCN | Decorin | ||||||
| MATRISOME ASSOCIATED PROTEINS | |||||
|---|---|---|---|---|---|
| REGULATORS | |||||
| Proteases | Protease inhibitors | Crosslinkers | |||
| Gene symbol | Protein | Gene symbol | Protein | Gene symbol | Protein |
| CTSB | Cathepsin B | SERPINH1 | Serpin peptidase inhibitor, H1 | LOXL1/2 | Lysyl oxidase-like 1/2 |
| HTRA1 | HtrA serin peptidase 1 | PZP | Pregnancy zone protein | TGM2 | Transglutaminase 2 |
| MMP2 | Matrix metallopeptidase 2 | ||||
| ECM-AFFILIATED PROTEINS | |||||
| Gene symbol | Protein | ||||
| ANXA2/5 | Annexin A2/A5 | ||||
| LGALS1/3 | Lectin galactoside-binding soluble 1/3 (Galectins) | ||||
| SECRETED FACTORS | |||||
| Gene symbol | Protein | ||||
| S100A6/10 | S100 calcium binding protein A6/10 | ||||
| CXCL12 | Chemoxin C-X-C motif ligand 12 | ||||
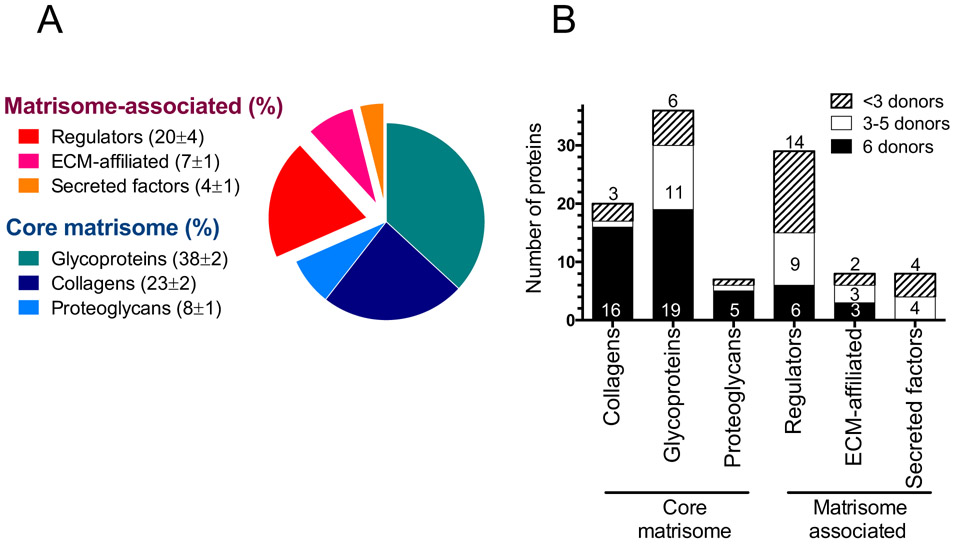
Figure 2: Proteomic characterization of ECM produced by Bm MSC from different donors.
(A): Matrisome signature of Bm ECM. Pie chart represents the distribution of Bm ECM proteins by percentage of total number for each matrisome protein sub-category. The average percentage ± SD for each sub-category was calculated for the 6 donors and is presented in the parentheses. (B): Protein distribution among donors. Columns represent the number of proteins for each matrisome sub-category detected in all donors (black bars), 3 to 5 donors (white bars) or fewer than 3 donors (hatched bars). Proteins represented by at least 1 peptide are included. Results were collected from two technical replicates of two independent experiments.
Matrisome proteins can be classified in two distinct categories: those proteins that constitute the core matrisome (i.e., collagens, glycoproteins, and proteoglycans) and those proteins that are associated and interact with the core matrisome proteins, designated as matrisome-associated proteins (i.e., regulators, secreted factors, and other ECM-affiliated proteins). Figure 2A shows the matrisome signature of Bm ECM as a segmentation of the matrisome proteins into each sub-category. Core matrisome proteins were dominant; they represented around 60% of the total number of Bm ECM matrisome proteins and were also very abundant (75% of the precursor-ion MS signal intensity and 50% of the number of peptides, Supplementary Fig 1). The proteins of the core matrisome are structural proteins that confer mechanical properties to the ECM, and also play roles in cell adhesion and signaling predominantly via integrin binding [31]. Some of the most abundant glycoproteins detected by LC/MS-MS were fibronectin, tenascin-C, fibulin-1, emilin-1 and thrombospondin-1. Collagens I, VI and XII as well as perlecan were identified as the most abundant collagens and proteoglycans, respectively (Table I). Components of the core matrisome were detected with a similar number of peptides and in comparable abundance across the 6 donors, suggesting that Bm ECM composition is consistent across biological replicates (Supplementary Table I).
In addition to the core matrisome proteins, the ECM serves as a reservoir for regulators (i.e., ECM-remodeling enzymes, cross-linkers and protease inhibitors), secreted factors and other ECM-affiliated proteins. These proteins fall into the category of matrisome-associated proteins and are less abundant than the core matrisome proteins (Fig 2A). Although more variation in the qualitative composition of matrisome-associated proteins was observed among the donors, a common set of matrisome-associated proteins was consistently found and includes cross-linkers such as transglutaminase-2 and LOXL1/2, the ECM-remodeling enzymes cathepsin-B and matrix metalloproteinase MMP-2, as well as the protease inhibitor serpine H1. These enzymes participate in ECM arrangement by controlling the fine balance between stability and remodeling [32,33]. In addition to these regulators, annexin A2 and galectins 1 and 3 were identified as ECM-affiliated proteins. These proteins are associated with structural ECM proteins and regulate diverse cell functions through downstream signaling. For example, the binding of annexin A2 to the glycoprotein tenascin-C is believed to promote cell migration and mitogenesis through the loss of cell adhesion [34].
Finally, the ECM serves as a reservoir for numerous secreted factors that bind to core matrisome proteins, including proteoglycans as well as ECM-associated proteins, and trigger various signaling pathways [1]. On account of their small size, low abundance and higher solubility, they may be partially lost during decellularization and their detection is more variable among the donors. Thus, we have chosen to include the secreted factors detected in more than 3 donors in Table I and they include the chemokine CXCL12 that is constitutively expressed in the bone marrow, and responsible for bone-marrow stem-cell homeostasis regulation [35], as well as the calcium-binding proteins S100A6/A10 implicated in various cellular processes such as cell proliferation and differentiation through interactions with annexin A2 [36].
3.2.3. Quantitative proteomic characterization of ECM produced by Bm MSC from different donors
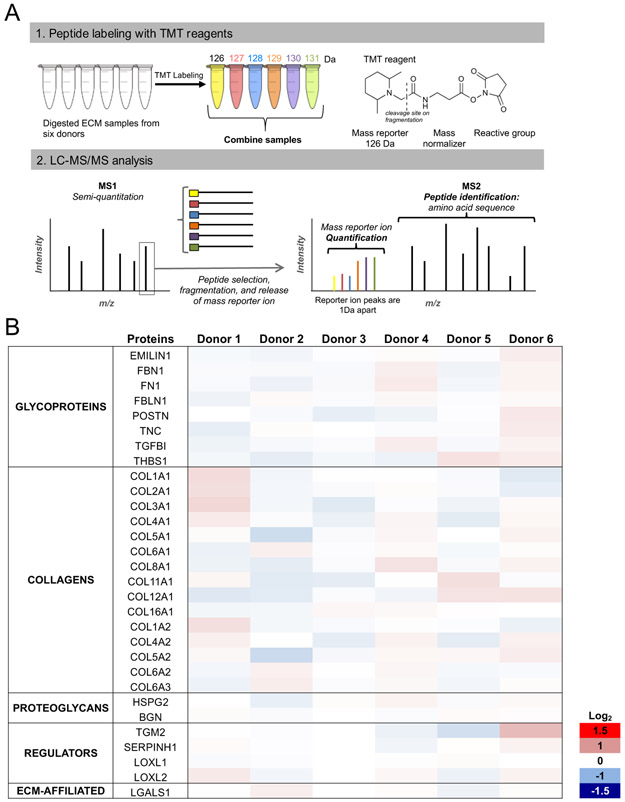
Quantitative proteomic approaches enable an accurate measure of relative protein amounts between different samples [37,38]. Isobaric mass tag labeling or Tandem Mass Tag (TMT) labeling was performed to evaluate the relative abundance of ECM proteins produced by Bm MSC from different donors and thus gain a better understanding of Bm ECM composition consistency. This method allows the measurement of relative expression levels of proteins in ECM derived from different donors after labeling the peptides of each sample with a different chemical tag [39]. On fragmentation during MS2, each tag releases a different mass fragment (see color code in Fig 3A), and quantification of these released fragments is exploited as the basis of the relative quantification approach. Results are presented in Figure 3B in the format of a heat map, where the colors indicate the relative protein expression level for each donor relative to the average across the six donors (red are increased; blue are decreased). The coefficients of variation for protein expression level across the different donors were 5.7%, 5.8%, 6.6%, 7.6%, 6.5% and 7.1% for donor 1 to donor 6, respectively (average of 6.5%). All protein ratios were within a log2 fold change range of −0.6 to 0.65 indicative of a relatively consistent protein profile across the six donors. Thus, the TMT labeling suggests that Bm MSC isolated from different donors produce similar ECMs, both qualitatively and quantitatively.
Figure 3: Quantitative proteomic analysis of the ECM produced by Bm MSC from six donors.
(A): TMT labeling experimental workflow. Each peptide solution is labeled with a specific TMT reagent, samples are combined, and peptides are separated based on their mass-to-charge m/z ratios (MS1). On MS2, peptides of particular m/z are selected, fragmented, and the different mass fragments are released, enabling relative quantification. (B): Relative quantitative comparison among the six donors. Proteins represented by at least 2 peptides are presented. Results show the fold-change (FC) in protein detection levels normalized to the mean of all samples in log2 scale. Data were collected from two technical replicates of two independent experiments.
3.3. Comparison of the composition of ECMs produced in vitro by different cell types
As Bm MSC from different donors produce an ECM with a defined and consistent proteomic composition, we investigated whether this ECM composition was unique to MSC originating from the bone marrow or universal among different cell types. Adipose-derived MSC (Ad MSC) were chosen in order to compare ECM produced by mesenchymal stem cells from a different tissue of origin. Neonatal dermal fibroblasts (NHDF) were chosen as a different primary cell type from a different tissue of origin (Fig 4A). ECM from Ad MSC and NHDF (Ad ECM and Der ECM, respectively) were produced using the same experimental procedure as for Bm ECM and their protein composition was compared, first qualitatively, then quantitatively, following the same approach as described above.
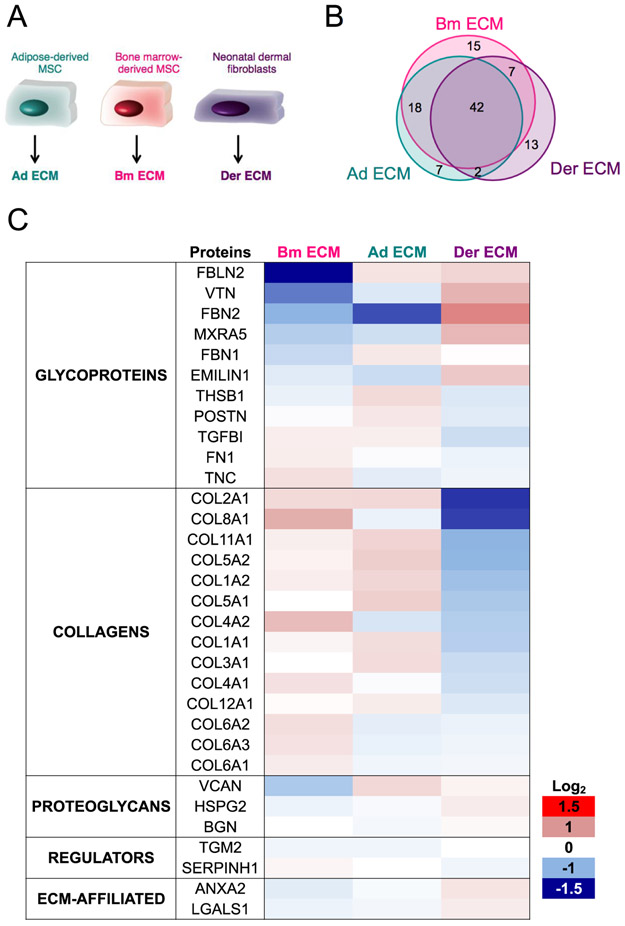
Figure 4: Characterization of ECM produced by different cell types.
(A): ECM from Bm MSC (Bm ECM), Ad MSC (Ad ECM) and NHDF (Der ECM) were produced and characterized. (B): Venn diagram of the overlap of protein numbers. (C): Quantitative proteomics using TMT labeling. Results show the fold-change (FC) in protein detection levels normalized to the mean of all samples in log2 scale. Results were collected from two technical replicates of two independent experiments and proteins identified with at least 2 peptides were considered.
The number of matrisome proteins detected for each sample was in the same range, despite a lower peptide intensity signal for Der ECM compared to Ad ECM and Bm ECM, that might be explained by the lower total mass of insoluble matrix produced by NHDF after 14 days (Supplementary Fig 3). Qualitative analysis based on matrisome protein lists showed that ECM from the cells originating from different tissue types contain a large set of common proteins that represents 40% of the total protein number (Fig 4B). The overlap includes all collagens previously found in Bm ECM, except for collagen 14 α1. The predominant glycoproteins detected in Bm ECM were also identified in Ad ECM and Der ECM, as well as most of the proteoglycans.
Quantitative TMT labeling approach was applied to compare the relative protein abundance in the three ECM samples, and significant tissue-type variations in the composition of the ECM were observed (Fig 4C). Only proteins represented with at least two peptides were taken into account for the analysis to guarantee more reliability, and this explained why 32 proteins were represented on the TMT heat map (Fig 4C) while 42 proteins were qualitatively detected in all three matrices (Fig 4B).
Der ECM was the most diverse of the three ECMs and a key observation was that Der ECM possessed the lowest abundance of collagens, compared to ECM from both types of MSC. More precisely, the amounts of collagens II and VIII were more than two times lower (log2 FC of approximately −1.35), as well as for collagens XI and V whose quantities were halved (log2 FC of approximately −1). Another observation was the differential levels of fibrillin-2 (FBN2) between the three samples: high in Der ECM and low or very low in Bm ECM and Ad ECM, respectively. In addition, fibulin-2 (FBLN2) and vitronectin (VTN) were three and two times less abundant in Bm ECM. Some variations in FBLN2, COL8A1, COL4A1 and, to a lesser extent, VCAN expression were also noted between Ad ECM and Bm ECM. In addition to the quantitative differences in the common proteins, some proteins were detected in one ECM and not in the others (Table II). For example, tenascin-X, a glycoprotein that is primarily expressed in loose connective tissues was solely detected in Ad ECM [34]. The detailed composition of Ad ECM and Der ECM is available in Supplementary Table II.
Table II: Source-specific protein lists for ECM derived from the three cell types.
Proteins presented here were identified in the ECM of one cell-type and not detected in the others. For Bm ECM proteins, only proteins present in at least 5 donors were considered (or in at least 3 donors for the secreted factors).
| Ad ECM | ||
|---|---|---|
| Gene Symbol | Protein | |
| Collagens | COL15A1 | Collagen XV α1 |
| Glycoproteins | FGL2 | Fibrinogen-like 2 |
| TNXB | Tenascin XB | |
| Regulators | ITIH3 | Inter-alpha (globulin) inhibitor H3 |
| SERPINA5 | Serpin peptidase inhibitor, clade A, member 5 | |
| Secreted factors | SCUBE3 | Signal peptide, CUB domain, EGF-like 3 |
| CTGF | Connective tissue growth factor | |
| Der ECM | ||
| Glycoproteins | LTBP4 | Latent transforming growth factor beta binding protein 4 |
| MATN2 | Matrillin 2 | |
| THBS3 | Thrombospondin 3 | |
| THSD4 | Thrombospondin, type 1 domain containing 4 | |
| Regulators | ADAMTS1 | ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| ADAMTSL4 | ADAMTS-like 4 | |
| CD109 | CD109 molecule | |
| P4HA1 | Prolyl 4-hydroxylase, alpha polypeptide I | |
| PRSS12 | Neurotrypsin | |
| SERPINC1 | Serpin peptidase inhibitor, clade C, member 1 | |
| Secreted factors | MDK | Midkine |
| SFRP1 | Secreted frizzled-related protein 1 | |
| WNT5A | Wingless-type MMTV integration site family, member 5A | |
| Bm ECM | ||
| Glycoproteins | AEBP1 | AE-bindin prot. 1 |
| EDIL3 | EGF-like repeats and discoidin I-like domains 3 | |
| EFEMP2 | Fibulin 3 | |
| FGB | Fibrinogen β | |
| FGG | Fibrinogen γ | |
| MFGE8 | Milk fat globule-EGF factor 8 protein | |
| VWA1 | Von Willebrand factor A domain containing prot. 1 | |
| Collagens | COL14A1 | Collagen XIV α1 |
| Regulators | CTSB | Cathepsin B |
| LOXL1 | Lysyl oxidase-like 1 | |
| MMP2 | Matrix metallopeptidase 2 | |
| ECM-affiliated proteins | ANXA5 | Annexin A5 |
| LGALS3 | Lectin galactoside-binding soluble 3 | |
| Secreted factors | S100A6/10 | S100 calcium binding protein A6/10 |
These data highlight that ECMs from different cell types are not identical but not totally discrete: while sharing a large set of common proteins, each ECM presents some unique components that may relate to the specialized functions of the tissue of origin.
3.4. Cell response to ECM substrates
Proteomic analyses revealed that ECM from Ad MSC, Bm MSC and NHDF shared a large set of common proteins, although significant variations in levels were detected for some proteins. Also, each matrix presents specific proteins that were not detected in others. We hypothesized that the differential ECM compositions might affect cell responses when cultured on each of the three cell-derived ECMs.
3.4.1. Effect of the different cell-derived ECMs on cell survival
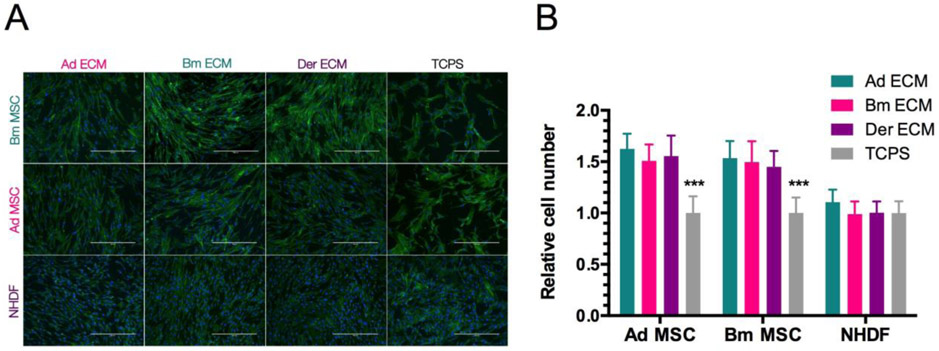
To investigate how cells respond to the different matrices, we evaluated and compared the cell number and cell morphology 72 h after seeding on each of the substrates, i.e., Ad ECM, Bm ECM, Der ECM and TCPS (Fig 5).
Figure 5: Cell survival on the different matrices.
(A): Fluorescence microscopy pictures with actin cytoskeleton staining (Alexa Fluor® 488 Phalloidin) and cell nuclei (DAPI). Scale bar is 400 μm. (B): Cell number relative to TCPS after 3 days in culture. Results show mean ± SD of two independent experiments (n=6). Statistics were done using two-way ANOVA and Tukey’s post test (***p<0.001).
All cell types plated on any of the ECM substrates showed a more aligned arrangement than on TCPS that can be explained by the fibrillar ECM architecture (Fig 1B), which likely guides cell morphology and migration [1]. Both types of MSC showed improved survival on cell-derived ECMs compared to TCPS, as indicated by the 1.5 increase in cell number (Fig 5B). In addition, MSC exhibited a different morphology on ECM compared to TCPS, with a more elongated shape and a smaller size, which may be related to the higher cell density (Fig 5A). On the other hand, NHDF survival was similar on TCPS as to the ECM-based substrates and NHDF do not exhibit significant morphologic changes beyond collective alignment.
These data indicate that ECM impacts cells differentially depending on the cell type. While the type of substrate does not affect NHDF survival, MSC number was significantly increased on ECM-based substrates compared to TCPS, regardless of the cellular origin of the ECM.
3.4.2. Cell-derived ECMs alter the cellular transcriptome
To investigate the effect of the substrates on the cell transcriptomes, RNA from the three cell types (i.e., Ad MSC, Bm MSC and NHDF) was extracted after seeding the cells on each of the four substrates (i.e., Ad ECM, Bm ECM, Der ECM, and TCPS) and the differential gene expression was evaluated across the different samples.
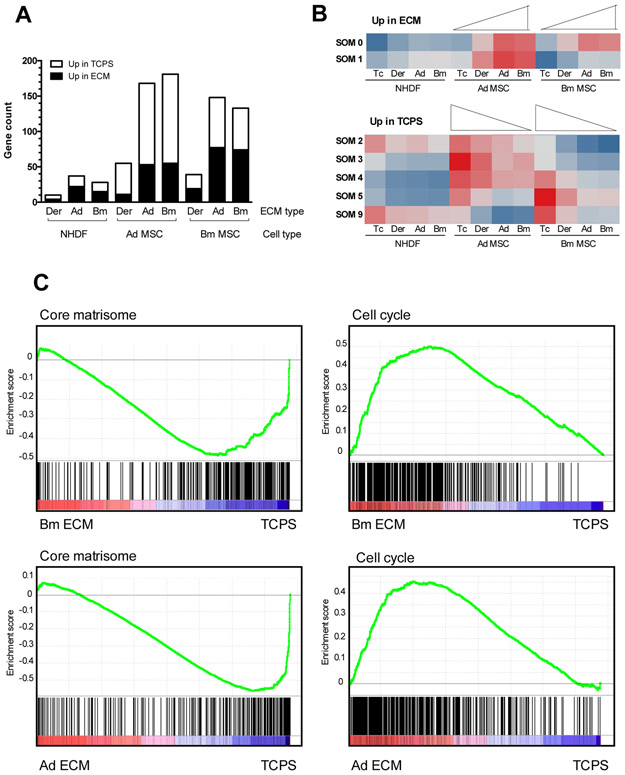
Few genes were differentially expressed when NHDF were seeded on TCPS as compared to cell-derived ECM, indicating a limited impact of the substrate on NHDF transcriptome (Fig 6A). This corroborates what was observed for cell growth and morphology. On the other hand, extensive significant differential gene expression was observed when MSC (both Ad MSC and Bm MSC) were plated on TCPS compared to ECM, suggesting that the signals from the substrate significantly alter the overall transcriptome of both types of MSC. The differentially expressed genes are detailed in Supplementary Table IV.
Figure 6: Influence of the substrate on the cell transcriptome.
(A): Differentially expressed genes when Ad MSC, Bm MSC and NHDF were cultured on the different ECM compared to TCPS, with the threshold of log2 FC≥1 and p<0.05. (B): Gene response clustering with self-organizing maps (SOM). Each row corresponds to a different gene cluster. Red represents up-regulation and blue represents down-regulation (C): Enrichment plots for core matrisome gene set (left) and cell cycle KEGG pathway (right) in MSC on their own ECM versus TCPS (Bm MSC top, Ad MSC bottom). Green curve indicates the enrichment score. p-values <0.002.
310 genes with differential expression (abs log2FC≥1, p-value≤0.05) in at least one of the three comparisons (Ad ECM versus TCPS, Bm ECM versus TCPS, or Der ECM versus TCPS) for at least one of the three cell types were selected for self-organizing map (SOM) clustering in order to characterize the response of these genes in all comparisons. Clustering algorithms are designed to elucidate general patterns in large data sets by grouping similar elements together [40]. Here, the differentially expressed genes were grouped into 13 clusters that explained 81% of the variance of the data. We focused our attention on the clusters in which differential responses for Bm MSC on disparate matrices were observed. This corresponds to 7 different clusters that displayed two main patterns: i) genes up-regulated in Bm MSC on MSC-derived ECM (Fig 6B upper panel); and ii) genes expressed at a lower level in Bm MSC on MSC-derived ECM (Fig 6B lower panel). The genes assigned to each of the clusters represented in Fig 6B are listed in Supplementary Table V. For both Bm MSC and Ad MSC, the responses to MSC-derived ECM (i.e., Bm ECM or Ad ECM) were similar to one another, while the response to Der ECM was intermediate between MSC-derived ECM and TCPS. As seen in Fig 6B, little variation across the NHDF samples was observed using SOM clustering.
We then looked at the genes differentially expressed by MSC (both Ad MSC and Bm MSC) on their own ECM compared to TCPS, and similar gene-set enrichments were observed for both cell types (Fig 6C and Supplementary Table VI). The leading edge genes responsible for the enrichment are listed in Supplementary Table VII. Of the genes more highly expressed on TCPS, a large set encode ECM and ECM-associated proteins, as well as proteins involved in cell adhesion (e.g., ITGAV, ITGB8, CD36 or FGFR2). Increase in mRNA expression of ECM proteins was validated by real time PCR for a subset of ECM proteins (Supplementary Figure 5). On the other hand, genes that regulate cell division were over-expressed by MSC on cell-derived ECM as compared to TCPS (Fig 6C and Supplementary Table V). In addition to genes involved in cell cycle, genes involved in MSC mobility and migration (e.g., genes coding for the CXCL chemokine family members, matrix proteases, PREX-1, GNG2) were up-regulated when cells were on MSC-derived ECM versus TCPS.
4. Discussion
In the body, stem cells are in constant and intimate contact with the ECM. This matrix serves not only as a structural support, but as a reservoir of many biochemical and mechanical signals that are transduced via cell surface receptor-protein interactions and influence cell signaling pathways and dictate cell functions [1]. ECM components are at the center of this complex interplay and have a vital role in regulating physiological processes. During standard cell culture, stem cells are isolated from their native microenvironment (i.e., the stem-cell niche), and need to adapt to a polymeric surface where ECM signals are limited to the serum proteins in the medium or adsorbed proteins on the plastic surface. This environment has been shown to affect cell biology; MSC cultured on ECM substrates compared to those on TCPS displayed different biologic features that impact cell morphology, motility and proliferation as well as, in other reports, differentiation [10,11]. In the context of stem cell biology, it is critical to decipher how individual ECM components regulate specific stem cell functions to better comprehend the natural role of the ECM. Understanding cellular processes requires a robust characterization of the ECM’s molecular composition. In addition, this understanding will provide further insight for the rational design of ECM mimicking biomaterials for tissue engineering and regenerative medicine [18,41,42].
By employing a proteomic approach, we were able to provide a comprehensive characterization of the molecular composition of ECM produced in vitro by bone marrow-derived MSC (Bm ECM), adipose-derived MSC (Ad ECM) and human neonatal dermal fibroblasts (Der ECM). The analysis of Bm ECM of MSC from 6 different donors revealed a complex yet consistent protein composition. Specifically, the composition of these cell-derived ECMs (approximately 80 unique ECM and ECM-associated proteins) is rather complex as compared to protein-based substrates, which often comprise a surface coating of one or two proteins, and are occasionally employed for stem cell culture as an alternative to TCPS. The variance of protein expression across the Bm ECM formed by cells from the 6 different donors was relatively low (log2 fold change range of −0.6 to 0.65) indicative of a consistent matrix formed by Bm MSC from different donors. However, when the proteomic composition of Bm ECM was compared to Ad ECM and Der ECM, a distinct signature was observed for each of the three cell-derived matrices. Despite the sensitivity and precision of TMT labeling techniques, underestimation of protein abundance differences by a factor of 20 to 30% can occur due to ratio compression phenomena [43,44] and real effect sizes might be larger than what was measured. High-resolution techniques as well as correction algorithms have been developed to minimize the ratio compression and help detect subtle variations between samples.
The major proportion of Bm ECM proteins belongs to the core matrisome that includes collagens, glycoproteins and proteoglycans. A large variety of collagens were detected in Bm ECM, of which the fibrillar family was the most abundant (i.e., collagens I, II, III, V and XI) that are known to contribute to the molecular architecture and mechanical properties of many tissues [45-47]. In addition, Fibril Associated Collagens with Interrupted Triple helices (FACIT) collagens that associate to the surface of fibrillar collagens were identified (collagens XII and XIV that associate with fibrillar collagen I to facilitate fibril thickening), along with network-forming collagens (collagen IV) and beaded-filament forming collagens (collagen VI) [45]. The presence of collagens in the ECM was associated with proteinases that specifically degrade collagens (MMP-2 that lyses collagens I, IV, V) as well as collagen cross-linkers such as the LOX-like family members [48]. While Bm ECM and Ad ECM present a similar collagen composition, the expression level of collagens in Der ECM was overall lower. Dermal tissues are elastic, while collagens confer stiffness and strength to tissues, as the collagen fibers have a high capacity for energy storage but minimal elasticity as a consequence of their semi-crystalline packing [45,49,50]. In addition, overall glycoprotein levels were found to be increased in Der ECM compared to Ad ECM and Bm ECM, although differences in the levels of specific glycoproteins (e.g., fibulin-2) were observed between both MSC-derived ECMs. These glycoproteins are implicated in the ECM structural organization and have somewhat restricted tissue distribution [51]. For example, a major function of fibulin-2 is the formation of large proteoglycan networks and its overexpression in Ad ECM and Der ECM might be responsible for different ECM macromolecular architectures, compared to Bm ECM. Similarly, fibrillins form microfibrils that provide tissue with long-range extensibility and associate with elastic fibers [52,53]. The different ratios of core matrisome proteins lead to different supramolecular assemblies that might ultimately provide the matrices with different mechanical properties [49,50,54].
In addition to the core matrisome ECM proteins, numerous other proteins that contribute to ECM function and dynamics were detected. Regulators maintain a balance between ECM stability and dynamic remodeling by degrading proteins (e.g., cathepsin B, Adamalysin ADAMTS that degrade fibronectin and collagen IV, or sulfatase SULF1 that cleaves proteoglycans and removes GAG chains) or by modifying ECM topography through protein crosslinking (e.g., TGM2 and LOX). ECM remodeling promotes cell movement and migration, and the released proteolytic fragments may also play a role in cell signaling [32]. These proteases are tightly regulated at the transcriptome level, and some of them are secreted as pro-enzymes that require extracellular activation. This is the case for the matrix metalloproteinase MMP-2 that needs to be activated by other proteinases to be functional, such as the transmembrane type MMP-14, also identified in Bm ECM [33]. Proteinases can be inactivated by protease inhibitors such as TIMP3 and A2M found in Bm ECM. While several proteases were identified in Bm ECM, Ad ECM and Der ECM, it is not clear from these data what their relative activity is and on-going work is characterizing their role in ECM remodeling during stem cell culture.
Interestingly, ECM impacts different cells in different ways. Human neonatal dermal fibroblasts survive as well on TCPS as on cell-derived ECM, and minimal changes at the RNA levels were observed, suggesting that this cell type is negligibly impacted by the substrate over 72 h. In contrast, MSC number was significantly increased on ECM compared to TCPS as well as to a fibronectin-coated substrate (unpublished data), indicating that a synergy of multiple proteins is advantageous. Interestingly, the differences among the compositions of ECMs from different tissue types do not seem to affect MSC survival, as the cell number was similarly increased on each ECM. This suggests that MSC survival depends more on the common core of proteins than on the unique proteins expressed in each of the cell-derived ECM. However, the different ECMs might affect other cell functions that were not investigated in this study. Future work will focus on the secretome, another major regulator of MSC fate and paracrine effects.
Transcriptomic analysis revealed that the culture of MSC on TCPS induces an up-regulation of genes that code for ECM and ECM-associated proteins as well as proteins implicated in cell adhesion as compared to MSC on cell-derived ECM. On the other hand, NHDF seem less affected by the type of substrate they are seeded on. One hypothesis is that MSC on TCPS are more sensitive to certain ECM signals than other cell types, such as NHDF. In addition, MSC population was increased on ECM and the cells over-express several genes implicated in cell cycle regulation and migration. Interactions with ECM and its components impact MSC function compared to TCPS where MSC receive restricted molecular cues. Perhaps, the lack of pertinent ECM signals at early time points during TCPS culture or the relative metabolic cost of construction of a new ECM is responsible for the lower number of MSC on TCPS as compared to cell-derived ECM.
5. Conclusions
In conclusion, we characterized the molecular composition of ECM deposited in vitro by bone marrow-derived MSC as well as adipose-derived MSC and neonatal dermal fibroblasts. While the three matrices share a set of common proteins, each ECM displayed distinct features specific to the tissue of origin that might be related to specialized tissue functions. MSC derived from bone marrow or adipose tissue both showed increased cell number on the cell-derived ECMs versus standard TCPS; however, the unique ECM signatures had a minimal impact on MSC growth as cell number was similarly increased across the different cell-derived ECMs. In addition, significant changes in MSC transcriptome profiles were observed between both culture conditions; MSC on TCPS over-expressed genes coding for matrisome proteins and MSC on cell-derived ECM over-expressed genes implicated in cell cycle regulation and migration. On the other hand, the type of substrate had a limited effect on NHDF behavior. These data corroborate the importance of ECM signals for the in vitro culture of MSC. Utilizing these methods to further characterize ECM protein composition as well as understanding ECM-stem cell interactions can provide insight into stem cell biology and inform the design of next-generation ECM-mimicking substrates for in vitro stem cell culture.
Supplementary Material
Acknowledgements
The authors gratefully acknowledge J. Wyckoff for expert advice with confocal imaging of the cell-derived ECM as well as M.W. Tibbitt and O.S. Fenton for helpful feedback on the manuscript. This work was supported in part by StemBioSys, the Helmsley foundation, grants from the National Cancer Institute – Tumor Microenvironment Network (U54 CA126515/CA163109), the Howard Hughes Medical Institute, of which ROH is an Investigator, and in part by Support Grant to the Koch Institute (P30-CA14051 from the National Cancer Institute).
References
- [1].Hynes RO, The extracellular matrix: not just pretty fibrils., Science. 326 (2009) 1216–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Discher DE, Janmey P, Wang Y-L, Tissue cells feel and respond to the stiffness of their substrate., Science. 310 (2005) 1139–1143. [DOI] [PubMed] [Google Scholar]
- [3].Discher DE, Mooney DJ, Zandstra PW, Growth factors, matrices, and forces combine and control stem cells., Science. 324 (2009) 1673–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lane SW, Williams DA, Watt FM, Modulating the stem cell niche for tissue regeneration., Nat. Biotechnol 32 (2014) 795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Engler AJ, Sen S, Sweeney HL, Discher DE, Matrix elasticity directs stem cell lineage specification., Cell. 126 (2006) 677–689. [DOI] [PubMed] [Google Scholar]
- [6].Watt FM, Huck WTS, Role of the extracellular matrix in regulating stem cell fate., Nat. Rev. Mol. Cell Biol 14 (2013) 467–473. [DOI] [PubMed] [Google Scholar]
- [7].Banfi A, Muraglia A, Dozin B, Mastrogiacomo M, Cancedda R, Quarto R, Proliferation kinetics and differentiation potential of ex vivo expanded human bone marrow stromal cells: Implications for their use in cell therapy., Exp. Hematol 28 (2000) 707–715. [DOI] [PubMed] [Google Scholar]
- [8].Bruder SP, Jaiswal N, Haynesworth SE, Growth kinetics, self-renewal, and the osteogenic potential of purified human mesenchymal stem cells during extensive subcultivation and following cryopreservation., J. Cell. Biochem 64 (1997) 278–294. [DOI] [PubMed] [Google Scholar]
- [9].Stenderup K, Justesen J, Clausen C, Kassem M, Aging is associated with decreased maximal life span and accelerated senescence of bone marrow stromal cells., Bone. 33 (2003) 919–926. [DOI] [PubMed] [Google Scholar]
- [10].Ng CP, Sharif ARM, Heath DE, Chow JW, Zhang CBY, Chan-Park MB, et al. , Enhanced ex vivo expansion of adult mesenchymal stem cells by fetal mesenchymal stem cell ECM., Biomaterials. 35 (2014) 4046–4057. [DOI] [PubMed] [Google Scholar]
- [11].Prewitz MC, Seib FP, von Bonin M, Friedrichs J, Stissel A, Niehage C, et al. , Tightly anchored tissue-mimetic matrices as instructive stem cell microenvironments., Nat. Methods 10 (2013) 788–794. [DOI] [PubMed] [Google Scholar]
- [12].Abraham S, Riggs MJ, Nelson K, Lee V, Rao RR, Characterization of human fibroblast-derived extracellular matrix components for human pluripotent stem cell propagation., Acta Biomater. 6 (2010) 4622–4633. [DOI] [PubMed] [Google Scholar]
- [13].Chen X-D, Extracellular matrix provides an optimal niche for the maintenance and propagation of mesenchymal stem cells., Birth Defects Res. C. Embryo Today 90 (2010) 45–54. [DOI] [PubMed] [Google Scholar]
- [14].Li J, Hansen KC, Zhang Y, Dong C, Dinu CZ, Dzieciatkowska M, et al. , Rejuvenation of chondrogenic potential in a young stem cell microenvironment., Biomaterials. 35 (2014) 642–653. [DOI] [PubMed] [Google Scholar]
- [15].Antebi B, Zhang Z, Wang Y, Lu Z, Chen X-D, Ling J, Stromal-cell-derived extracellular matrix promotes the proliferation and retains the osteogenic differentiation capacity of mesenchymal stem cells on three-dimensional scaffolds., Tissue Eng. Part C. Methods 21 (2015) 171–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lai Y, Sun Y, Skinner CM, Son EL, Lu Z, Tuan RS, et al. , Reconstitution of marrow-derived extracellular matrix ex vivo: a robust culture system for expanding large-scale highly functional human mesenchymal stem cells., Stem Cells Dev. 19 (2010) 1095–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gobaa S, Hoehnel S, Roccio M, Negro A, Kobel S, Lutolf MP, Artificial niche microarrays for probing single stem cell fate in high throughput., Nat. Methods 8 (2011) 949–955. [DOI] [PubMed] [Google Scholar]
- [18].Gattazzo F, Urciuolo A, Bonaldo P, Extracellular matrix: A dynamic microenvironment for stem cell niche, Biochim. Biophys. Acta - Gen. Subj 1840 (2014) 2506–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Byron A, Humphries JD, Humphries MJ, Defining the extracellular matrix using proteomics., Int. J. Exp. Pathol 94 (2013) 75–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Naba A, Clauser KR, Hoersch S, Liu H, Carr SA, Hynes RO, The matrisome: in silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices., Mol. Cell. Proteomics 11 (2012) M111.014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Naba A, Clauser KR, Hynes RO, Enrichment of Extracellular Matrix Proteins from Tissues and Digestion into Peptides for Mass Spectrometry Analysis., J. Vis. Exp (2015) e53057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Naba A, Clauser KR, Ding H, Whittaker CA, Carr SA, Hynes RO, The extracellular matrix: Tools and insights for the “omics” era., Matrix Biol. 49 (2016) 10–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Naba A, Hoersch S, Hynes RO, Towards definition of an ECM parts list: an advance on GO categories., Matrix Biol. 31 (2012) 371–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Johnson H, Del Rosario AM, Bryson BD, Schroeder MA, Sarkaria JN, White FM, Molecular characterization of EGFR and EGFRvIII signaling networks in human glioblastoma tumor xenografts., Mol. Cell. Proteomics 11 (2012) 1724–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ternent T, Csordas A, Qi D, Gomez-Baena G, Beynon RJ, Jones AR, et al. , How to submit MS proteomics data to ProteomeXchange via the PRIDE database., Proteomics. 14 (2014) 2233–2241. [DOI] [PubMed] [Google Scholar]
- [26].Vizcaino JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, et al. , 2016 update of the PRIDE database and its related tools., Nucleic Acids Res. 44 (2016) D447–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP, GenePattern 2.0., Nat. Genet 38 (2006) 500–501. [DOI] [PubMed] [Google Scholar]
- [28].Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, et al. , PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes., Nat. Genet 34 (2003) 267–273. [DOI] [PubMed] [Google Scholar]
- [29].Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. , Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles., Proc. Natl. Acad. Sci. U. S. A 102 (2005) 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].http://matrisomeproject.mit.edu, The Matrisome Project, (n.d.).
- [31].Hynes RO, Naba A, Overview of the matrisome-An inventory of extracellular matrix constituents and functions, Cold Spring Harb. Perspect. Biol 4 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lu P, Takai K, Weaver VM, Werb Z, Extracellular matrix degradation and remodeling in development and disease., Cold Spring Harb. Perspect. Biol 3 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Loffek S, Schilling O, Franzke C-W, Series “matrix metalloproteinases in lung health and disease”: Biological role of matrix metalloproteinases: a critical balance., Eur. Respir. J 38 (2011) 191–208. [DOI] [PubMed] [Google Scholar]
- [34].Chiquet-Ehrismann R, Tucker RP, Tenascins and the importance of adhesion modulation., Cold Spring Harb. Perspect. Biol 3 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Karin N, The multiple faces of CXCL12 (SDF-1alpha) in the regulation of immunity during health and disease., J. Leukoc. Biol 88 (2010) 463–473. [DOI] [PubMed] [Google Scholar]
- [36].Donato R, Functional roles of S100 proteins, calcium-binding proteins of the EF-hand type., Biochim. Biophys. Acta 1450 (1999) 191–231. [DOI] [PubMed] [Google Scholar]
- [37].Steen H, Mann M, The ABC’s (and XYZ's) of peptide sequencing., Nat. Rev. Mol. Cell Biol 5 (2004) 699–711. [DOI] [PubMed] [Google Scholar]
- [38].Ong S-E, Mann M, Mass spectrometry-based proteomics turns quantitative., Nat. Chem. Biol 1 (2005) 252–262. [DOI] [PubMed] [Google Scholar]
- [39].Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, et al. , Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents., Mol. Cell. Proteomics 3 (2004) 1154–1169. [DOI] [PubMed] [Google Scholar]
- [40].Garrigues GE, Cho DR, Rubash HE, Goldring SR, Herndon JH, Shanbhag AS, Gene expression clustering using self-organizing maps: analysis of the macrophage response to particulate biomaterials., Biomaterials. 26 (2005) 2933–2945. [DOI] [PubMed] [Google Scholar]
- [41].Murphy WL, McDevitt TC, Engler AJ, Materials as stem cell regulators., Nat. Mater 13 (2014) 547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Peerani R, Zandstra PW, Enabling stem cell therapies through synthetic stem cell-niche engineering., J. Clin. Invest 120 (2010) 60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ow SY, Salim M, Noirel J, Evans C, Rehman I, Wright PC, iTRAQ underestimation in simple and complex mixtures: “the good, the bad and the ugly”., J. Proteome Res 8 (2009) 5347–5355. [DOI] [PubMed] [Google Scholar]
- [44].Ow SY, Salim M, Noirel J, Evans C, Wright PC, Minimising iTRAQ ratio compression through understanding LC-MS elution dependence and high-resolution HILIC fractionation., Proteomics. 11 (2011) 2341–2346. [DOI] [PubMed] [Google Scholar]
- [45].Ricard-Blum S, The collagen family., Cold Spring Harb. Perspect. Biol 3 (2011) a004978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Canty EG, Kadler KE, Procollagen trafficking, processing and fibrillogenesis., J. Cell Sci 118 (2005) 1341–1353. [DOI] [PubMed] [Google Scholar]
- [47].Kadler KE, Baldock C, Bella J, Boot-Handford RP, Collagens at a glance., J. Cell Sci 120 (2007) 1955–1958. [DOI] [PubMed] [Google Scholar]
- [48].Bonnans C, Chou J, Werb Z, Remodelling the extracellular matrix in development and disease., Nat. Rev. Mol. Cell Biol 15 (2014) 786–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Mouw JK, Ou G, Weaver VM, Extracellular matrix assembly: a multiscale deconstruction., Nat. Rev. Mol. Cell Biol 15 (2014) 771–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Muiznieks LD, Keeley FW, Molecular assembly and mechanical properties of the extracellular matrix: A fibrous protein perspective., Biochim. Biophys. Acta 1832 (2013) 866–875. [DOI] [PubMed] [Google Scholar]
- [51].Timpl R, Sasaki T, Kostka G, Chu M-L, Fibulins: a versatile family of extracellular matrix proteins., Nat. Rev. Mol. Cell Biol 4 (2003) 479–489. [DOI] [PubMed] [Google Scholar]
- [52].Kielty CM, Baldock C, Lee D, Rock MJ, Ashworth JL, Shuttleworth CA, Fibrillin: from microfibril assembly to biomechanical function., Philos. Trans. R. Soc. Lond. B. Biol. Sci 357 (2002) 207–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Kielty CM, Wess TJ, Haston L, Ashworth JL, Sherratt MJ, Shuttleworth CA, Fibrillin-rich microfibrils: elastic biopolymers of the extracellular matrix., J. Muscle Res. Cell Motil 23 (2002) 581–596. [PubMed] [Google Scholar]
- [54].Daley WP, Peters SB, Larsen M, Extracellular matrix dynamics in development and regenerative medicine., J. Cell Sci 121 (2008) 255–264. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.