Abstract
The intestinal invasion of pathogenic microorganisms can have serious health consequences. Recent evidence has shown that the N6-methyladenosine (m6A) mRNA modification is closely associated with innate immunity; however, the underlying mechanism is poorly understood. Here, we examined the function and mechanism of m6A mRNA modification and the YTH domain-containing protein YTHDF1 (YTH N6-methyladenosine RNA-binding protein 1) in the innate immune response against bacterial pathogens in the intestine. Ribo-seq and m6A-seq analyses revealed that YTHDF1 directs the translation of Traf6 mRNA, which encodes tumor necrosis factor receptor-associated factor 6, thereby regulating the immune response via the m6A modification near the transcript's stop codon. Furthermore, we identified a unique mechanism by which the P/Q/N-rich domain in YTHDF1 interacts with the DEAD domain in the host factor DDX60, thereby regulating the intestinal immune response to bacterial infection by recognizing the target Traf6 transcript. These results provide novel insights into the mechanism by which YTHDF1 recognizes its target and reveal YTHDF1 as an important driver of the intestinal immune response, opening new avenues for developing therapeutic strategies designed to modulate the intestinal immune response to bacterial infection.
INTRODUCTION
Bacterial infections are a major cause of both morbidity and mortality, with high costs both in the healthcare field and to society (1). Bacteria can pass the intestinal epithelium relatively easily, leading to various gastrointestinal disorders such as Crohn's disease, celiac disease, duodenal ulcers, acute pancreatitis, infectious diarrhea, and inflammatory bowel disease (2). For instance, enterotoxigenic Escherichia coli (ETEC) causes diarrhea by colonizing the small intestine, leading to morbidity and mortality in children and infants (3). Intestinal epithelial cells (IECs) are an essential component of a highly regulated communication network that senses microbial and environmental stimuli, as well as endogenous danger signals (4), and activation of an IEC-specific immune response induces the production of a myriad of cytokines, chemokines and acute phase proteins (5). Moreover, signaling via toll-like receptors (TLRs) and nuclear factor-κB (NF-κB) plays important roles in the production of cytokines and chemokines by inducing the epithelial immune response, mobilizing immune effector cells, and activating the adaptive immune system (6).
Recently, a growing body of evidence suggests that RNA modification can provide a powerful system for dynamically regulating gene expression (7). Methylation of adenosine nucleotides at the N6 position (m6A) is the most prevalent posttranscriptional modification in mammalian mRNA (8). Similar to DNA methylation, the reversible m6A modification is catalyzed by a specific set of enzymes; the methylation step is catalyzed by a stable core catalytic complex (METTL3/METTL14/WTAP) (9–11) and is reversed by the demethylases FTO and ALKBH5 (12,13). Thus, m6A nucleotides function as a ‘readable marker’ in the mRNA molecule (14). To date, three major ‘reader’ proteins (YTHDF1, YTHDF2 and YTHDF3) have been shown to recognize m6A nucleotides via their YTH (YT521-B homology) domain (15–19). YTHDF1 is believed to interact with translation initiation factors to mediate the translation of m6A-modified transcripts (17), whereas YTHDF2 promotes the degradation of target transcripts by recruiting the CCR4-NOT deadenylase complex (20). YTHDF3 coordinates with YTHDF1 and YTHDF2 to regulate the translation and decay, respectively, of methylated transcripts (21). Although the function of m6A modifications and the role of YTHDF proteins in the intestinal immune response are clinically relevant, they have not been investigated in detail.
To address these questions, we screened the three members of the YTHDF family and found that YTHDF1 is essential for mediating the intestinal immune response to bacterial infection both in vitro and in vivo. Specifically, loss of Ythdf1 in mice resulted in a substantially reduced immune response by downregulating the translation of Traf6 transcripts. We also found that the METTL3-mediated generation of m6A nucleotides either within the 3′ UTR or the coding sequence (CDS) plays a role in regulating translation. Traf6 encodes tumor necrosis factor receptor−associated factor 6, a key regulator of TLRs and subsequent NF-κB signaling, and is triggered by complex stimuli, including bacterial infection (22); however, the mechanisms that regulate the intestinal immune response are poorly understood.
Importantly, we also found that an interaction between DDX60 (DEAD/H-box helicase 60) and YTHDF1 is required in order to ‘read’ the m6A nucleotides in Traf6 transcripts and direct their translation. Members of the DEAD-box (DDX) family of helicases participate broadly as coregulators in the innate immune response. DDX60 was previously identified in virus-infected human dendritic cells (23) and is involved in a subset of innate immune pathways in a variety of cell types (24).
Taken together, our findings indicate that YTHDF1 plays an important role in protecting against intestinal bacterial infection by maintaining the intestinal immune system.
MATERIALS AND METHODS
Cell culture
The porcine intestinal epithelial cell line IPEC-J2 and the human intestinal epithelial cell line Caco-2 were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and were cultured as described previously (25,26).
Pathogens
Escherichia coli strain O111:B4 LPS (Sigma-Aldrich) and enterotoxigenic E. coli K88 (ETEC) were generously provided by Dr W. Fang (Zhejiang University, China) and cultured in LB broth (Aoboxing, China) or on LB agar plates (27).
Antibodies
The following antibodies were used in this study: anti-m6A (Synaptic Systems, 202003 for m6A-seq; Abcam, ab208577 for m6A-IP qPCR), anti-YTHDF1 (Proteintech, 66745–1-lg), anti-TRAF6 (Abcam, ab181622), anti-TRAF3 (Proteintech, 18099-1-AP), anti-β-actin (HuaAn, R1207–1), anti-Flag (Sigma-Aldrich, F3165), anti-MYC (Abcam, ab32), anti-DDX60 (Abcam, ab139807), anti-MyD88 (Proteintech, 23230-1-AP), anti-phosphor-p65 (Cell Signaling Technology, 3033), anti-phosphor-IκBα (Cell Signaling Technology, 2859), anti-TRIF (Proteintech, 23288-1-AP), anti-p65 (Proteintech, 10745-1-AP), anti-IκBα (Proteintech, 10268-1-AP), anti-BCLAF1 (Proteintech, 26809-1-AP), anti-THRAP3 (Proteintech, 19744-1-AP), anti-TRAF3 (Proteintech, 18099-1-AP), anti-phospho-IKKα/β (Cell Signaling Technology, 2078), anti-IKKα (HuaAn, ER30911), anti-IKKβ (HuaAn, ER1706-13), anti-p38 (HuaAn, ET1602-26), phospho-p38 (Cell Signaling Technology, 9211), anti-phospho-JNK (HuaAn, RT1488), anti-JNK (HuaAn, RT1550), anti-ERK1/2 (Cell Signaling Technology, 9102), phospho-ERK1/2 (HuaAn, ET1610–13), normal rabbit IgG (Cell Signaling Technology, 2729) and normal mouse IgG (Cell Signaling Technology, 5946).
Mice and treatments
C57BL/6J mice (6–8 weeks old) were obtained from the Laboratory Animal Center of the Chinese Academy of Sciences (Shanghai, China). Ythdf1-knockout (Ythdf1−/−) mice on a C57BL/6J background were generated using CRISPR-Cas9 obtained from the Bin Shen Laboratory at Nanjing Medical University (Nanjing, China) (28). The oligonucleotides for gRNAs were as follows: TACCTGTCCAGTTACTATCC and GGCACCATGGTCCACTGGAG. For ETEC infection, Ythdf1+/+ and Ythdf1−/− mice received 100 μL of the prepared bacterial suspension at a dose equivalent to 108 CFU/mouse by oral gavage. Histological and anatomical analyses of the intestinal tract were performed 3 days after ETEC infection. All animal procedures were performed in accordance with the Guide for the Care and Use of Laboratory Animals and were approved by Zhejiang University (Hangzhou, China).
Lentiviral short-hairpin RNAs, siRNAs and transfection
IPEC-J2 cells in which Ythdf1 or Ddx60 were knocked down were generated by transfecting a lentivirus expressing the Ythdf1 or Ddx60 short-hairpin RNA (shRNA), respectively; the shRNA targeting sequences and siRNA sequences are listed in Supplementary Table S2. Plasmids and siRNAs were transfected into cells using Lipofectamine 2000 (Invitrogen) in accordance with the manufacturer's instructions.
Generation of knockout cell lines using CRISPR-Cas9
Knockout (KO) cell lines were generated by co-transfecting IPEC-J2 cells with a pLentiCRISPRv2 vector carrying the targeting sequence and pGFP-Puro plasmids at a ratio of 10:1 (Cyagen, Hangzhou, China) as previously described (29). The gRNA sequences used to generate the KO cells are listed in Supplementary Table S2.
Gene cloning
Total RNA was extracted from IPEC-J2 cells, and genomic cDNA was prepared using RT-PCR. FLAG-tagged Ythdf1 was cloned into the pCMV-Tag 4 vector (Agilent, 211174), and Myc-tagged Ddx60 was cloned into the pRK-5 vector (BD Pharmingen, 556104). We generated truncation plasmids based on the corresponding full-length cDNAs. All other plasmids, including full-length, coding sequence, and mutation plasmids, were generated using GenScript and cloned into the FLAG-pcDNA3.1 vector. All plasmids were confirmed by DNA sequencing. The primers used for cloning are listed in Supplementary Table S3.
RNA extraction, cDNA synthesis, and qPCR
Total RNA was isolated using TRIzol reagent (Invitrogen), and poly(A)+ mRNA was enriched from total RNA using the GenElute mRNA Miniprep Kit (Sigma-Aldrich). The cDNA was reverse-transcribed using M-MLV reverse transcriptase (Thermo Fisher Scientific) with 2 μg of RNA from each sample, followed by quantitative PCR (qPCR) as previously described (25). Gapdh or β-actin mRNA was used as an endogenous control, and each reaction was run in triplicate. The primer sequences used for qPCR are listed in Supplementary Table S4.
RNA immunoprecipitation (RIP) assay
RIP was performed using the native RIP protocol as described previously (30,31). In brief, cells were lysed on ice in polysome lysis buffer containing 100 mM KCl, 5 mM MgCl2, 10 mM HEPES (pH 7.0), 0.5% NP40, DTT, protease inhibitor cocktail (ApexBio Technology) and RNase inhibitor (Promega). The cell lysates were then incubated with the appropriate antibody and protein A/G magnetic beads (Sigma) ranging from 3 h to overnight at 4°C on a rotator; 10% percent of the starting cell lysate supernatant was reserved for input analysis. After incubation, the beads were washed four times, and the RNA was released using proteinase K incubation for 30 min at 55°C. Precipitated RNA was then extracted and analyzed using qPCR with the primers listed in Supplementary Table S4.
RNA m6A quantification using dot blot assay and LC/MS/MS
RNA m6A dot blot assays were conducted as previously described (16,25). Equal amounts of poly(A)+ mRNA were spotted on an Amersham Hybond-N+ membrane (GE Healthcare), followed by UV crosslinking. After blocking, the membrane was incubated with anti-m6A antibody overnight at 4°C. The dots were visualized using a ChemiScope series 3400 Mini Imaging System. Methylene blue (Sigma-Aldrich) staining was used to visualize the amount of total RNA on the membrane.
LC/MS/MS was performed as described previously (32), and the total contents of m6A and A were quantified based on corresponding standard curves generated using pure standards, after which the m6A/A ratio was calculated. After digestion, the nucleosides were separated using reverse-phase ultra-performance liquid chromatography on a C18 column with online MS detection using an Agilent 6410 QQQ triple-quadrupole LC mass spectrometer in positive electrospray ionization mode.
Western blot analysis
Cells were lysed on ice in SDS-PAGE sample buffer containing 50 mM Tris (pH 6.8), 100 mM DTT, 2% SDS, 0.1% bromophenol blue, and 10% glycerol, and western blot analysis was performed and quantified as described previously (25).
Luciferase reporter assays
IPEC-J2 cells were plated in 24-well plates and transfected with the pGL4.21 firefly luciferase reporter plasmid and the Renilla luciferase plasmid together with various amounts of the appropriate control or protein-expressing plasmid(s). Luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega), and reporter gene activity was determined by normalizing the firefly luciferase activity to Renilla luciferase activity.
Protein co-immunoprecipitation (co-IP)
Cell pellets were resuspended in 2× volume of IP lysis buffer (Pierce) supplemented with cocktail protease inhibitor (ApexBio Technology), incubated on ice for 10 min, and then incubated with the primary antibody and Protein A/G magnetic beads at 4°C overnight. The beads were then washed and resuspended in SDS sample buffer and analyzed using western blot analysis or tandem mass spectrometry.
Tandem mass spectrometry analysis
Peptide samples were analyzed by Novogene (Beijing, China) using an Orbitrap Elite hybrid mass spectrometer (Thermo Fisher). In brief, the purified protein samples were separated by SDS-PAGE and visualized using silver staining. The proteins were then reduced, alkylated, digested and extracted as previously described (33). The extracted samples were dried, resuspended in 0.1% trifluoroacetic acid, desalted with C18 Zip Tips, dried again, and dissolved in 0.1% formic acid. The samples were then separated on a C18 analytical column using a two-solvent system.
Cytokine multiplex assay
Porcine cytokine antibody arrays (QAP-CYT-1) were performed by RayBiotech (Guangzhou, China). Serum-free media from IPEC-J2 cultures (or an equivalent amount of mouse intestinal tissue homogenate) were used, and intensity was normalized to an internal positive control.
Cell treatment and ELISA
The cells were cultured in six-well plates until they reached approximately 80% confluence, and then treated with LPS (50 μg/ml) or ETEC (multiplicity of infection: 10:1) for various durations; untreated/uninfected cells were used as controls. The concentration of cytokines in the supernatant or in the intestines was measured using a Quantikine ELISA Kit (Proteintech).
Histology and immunohistochemistry
The mice were euthanized, and the intestines were removed and fixed in 4% paraformaldehyde overnight at 4°C. The samples were then sent to Servicebio (Wuhan, China) for histological and immunohistochemical analyses. In brief, paraffin-embedded samples were deparaffinized and processed through a graded series of alcohol concentrations. The sections were then incubated with the appropriate primary antibody followed by HRP-conjugated secondary antibody. The sections were scored by a blinded observer based on goblet cell depletion, leukocyte infiltration, and submucosal inflammation on a total scale of 0–3, with 0 representing no pathology and 3 indicating the most severe pathology.
Ribosome-protected fraction isolation, Ribo-seq and analysis
Isolation of the ribosome-protected fraction, library construction, high-throughput sequencing, and analysis were performed by Novogene (Beijing, China). In brief, ribosome-protected mRNA was obtained using MNase digestion and RNA purification (34,35). In total, 3 μg of ribosome-protected mRNA was used to prepare a library with the NEBNext Small RNA Library Prep Set (New England Biolabs) in accordance with the manufacturer's instructions. After PCR amplification, the samples were used for quality control and deep sequencing with an Illumina HiSeq 2000. The Sequence Read Archive (SRA) accession number for the Ribo-Seq data reported in this study is PRJNA645024.
m6A sequencing and analysis
The library preparation for m6A sequencing and high-throughput sequencing were performed by Novogene (Beijing, China). m6A immunoprecipitation and library preparation were performed as previously described (36). Purified RNA fragments were used for library construction with the NEBNext Ultra RNA Library Prep Kit and were sequenced using an Illumina HiSeq 2000. Sequencing reads were aligned to Sscrofa11.1 using the Burrows-Wheeler Alignment tool, and m6A peak calling was performed using MACS2. The SRA accession number for the m6A-Seq data reported in this study is PRJNA645290.
Isolation of bone marrow–derived macrophages (BMDMs) and primary small intestinal epithelial cells from ETEC-infected mice
BMDMs were isolated from bone marrow samples as described previously (37). To study the maturation of BMDMs, 2 × 106 cells were cultured for 6 days in 35-mm Petri dishes in RPMI-1640 containing 30% L929 cell supernatant, 20% FCS, l-glutamine, and 1× penicillin/streptomycin.
Primary small intestinal epithelial cells were isolated as previously described (38). In brief, the entire small intestine was dissected from the mice, sectioned longitudinally, and any residual feces and mucus were removed. After shaking gently in 15 ml HBSS (Invitrogen) containing 2 mM EDTA (pH 8.0) for 30 min at 37°C, the tissue was transferred to a fresh 50-ml tube containing 10 ml phosphate-buffered saline (PBS) and vortexed until the supernatant was almost clear. The crypts were allowed to settle for up to 5 min at room temperature, and single cells were collected by centrifugation at 1500 rpm for 5 min at room temperature.
Statistical analysis
All summary results are expressed as the mean ± the s.e.m. Multiple groups were analyzed using a one-way ANOVA, and differences between two groups were analyzed using the Student's t-test; *P < 0.05, **P < 0.01, and n.s. indicates not significant (P > 0.05). Data were analyzed using GraphPad Prism software, and each experiment was performed at least in triplicate unless otherwise stated.
RESULTS
YTHDF1 is required by intestinal epithelial cells for host defense against bacterial infection
To examine the role of m6A RNA methylation in IECs in response to bacterial infection, we first measured m6A methylation levels in the intestines of mice infected with ETEC. We found that ETEC infection increased global m6A levels in the colon and jejunum measured using both an m6A dot blot assay with poly(A)+ RNA (Supplementary Figure S1A) and LC/MS/MS (Supplementary Figure S1B). In separate experiments, we used siRNA (small interfering RNA) to silence various m6A methylation−related genes in cultured IPEC-J2 cells and found that silencing Mettl3, which encodes an m6A methyltransferase, increased cell survival following both ETEC infection and LPS stimulation (Figure 1A), indicating that m6A modification plays a role in bacterial infection. With respect to genes that encode m6A reader proteins, we found that silencing Ythdf1 significantly increased cell survival, whereas silencing either Ythdf2 or Ythdf3 mRNA had no effect on cell survival (Figure 1A).
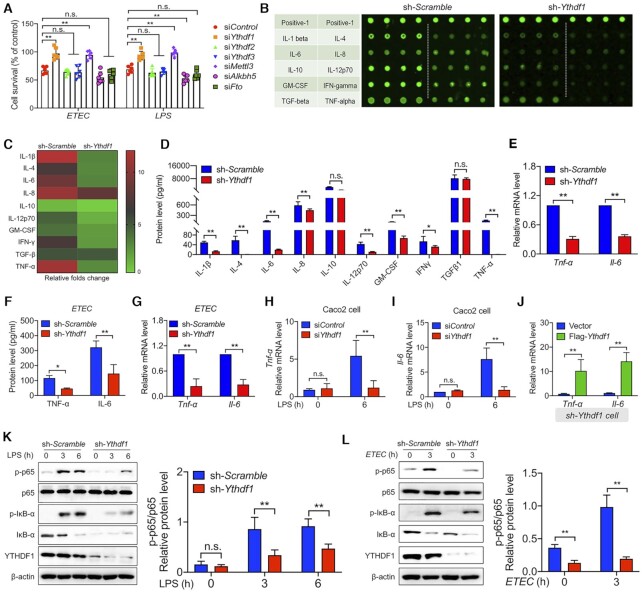
Figure 1.
YTHDF1 mediates the bacterial immune response in Intestinal epithelial cells. (A) Summary of the survival of IPEC-J2 cells transfected with a control siRNA or Ythdf1, Ythdf2, Ythdf3, Mettl3, Alkbh5, Fto siRNA and then either infected with ETEC (left) or stimulated with LPS (right). (B) Laser-scanning map of a porcine cytokine antibody array in LPS-treated cells expressing either a scrambled shRNA or the Ythdf1 shRNA. Each cytokine was measured in quadruplicate, and the table at the left indicates the cytokines. The top row is a positive control. (C) Heat map showing the fold change in signal intensity measured in (B), relative to cells expressing the scrambled shRNA. (D) The indicated cytokines were measured in the supernatants of IPEC-J2 cells expressing either the scrambled shRNA or Ythdf1 shRNA, 6 h after LPS stimulation. (E) Summary of Tnf-α and Il-6 mRNA measured 6 h after LPS stimulation in IPEC-J2 cells expressing either the scrambled shRNA or Ythdf1 shRNA, expressed relative to Gapdh mRNA. (F, G) Summary of TNF-α and IL-6 protein in the supernatant and Tnf-α and Il-6 mRNA levels (expressed relative to Gapdh mRNA) in ETEC-infected IPEC-J2 cells. (H, I) Summary of Tnf-α (H) and Il-6 (I) mRNA measured in Caco2 cells 6 h after LPS stimulation, expressed relative to Gapdh mRNA. (J) Summary of Tnf-α and Il-6 mRNA levels in IPEC-J2 cells co-expressing the Ythdf1 shRNA and an empty vector or FLAG-Ythdf1, expressed relative to Gapdh mRNA. (K, L) Western blot analysis and summary of the indicated proteins measured in IPEC-J2 cells expressing either the scrambled shRNA or Ythdf1 shRNA before and after LPS stimulation (K) or ETEC infection (L).
To examine the role of YTHDF1 in protecting against bacterial infection, we silenced Ythdf1 expression in IPEC-J2 cells using lentiviral expression of short hairpin RNA (shRNA) (Supplementary Figure S2A and B). Strikingly, a cytokine multiplex assay revealed that the LPS-induced production of a wide range of cytokines, including TNF-α, IL-6, and IL 2 p70, were significantly reduced in cells expressing the Ythdf1 shRNA compared to cells expressing a scrambled control shRNA (Figure 1B and C).
Even in the absence of LPS stimulation—when cytokine levels are extremely low—silencing Ythdf1 expression still visibly reduced the levels of various cytokines (Supplementary Figure S2C and D). These results were confirmed at the protein and mRNA levels using ELISA and qPCR analysis, respectively (Figure 1D-E). Moreover, we generated Ythdf1 KO cells using CRISPR/Cas9 and obtained similar results (Supplementary Figure S2E and F), supporting the notion that YTHDF1 plays a role in the intestinal immune response. Similarly, ETEC-induced expression of the inflammatory cytokines TNF-α and IL-6 was reduced in cells expressing the Ythdf1 shRNA (Figure 1F and G). Similar results were obtained using Caco-2 cells, a human epithelial colorectal adenocarcinoma cell line, in which LPS-induced TNF-α and IL-6 levels were reduced in cells expressing the Ythdf1 siRNA (Figure 1H and I). Consistent with these results, we found that co-expressing the Ythdf1 shRNA together with a FLAG-tagged Ythdf1 construct in IPEC-J2 cells increased the expression of both TNF-α and IL-6 (Figure 1J). Finally, we found that knocking down Ythdf1 expression with the Ythdf1 shRNA reduced the LPS- and ETEC-induced activation of the transcription factor NF-κB (i.e., p65) (Figure 1K and L), suggesting that YTHDF1 functions upstream of NF-κB. Taken together, these data indicate that YTHDF1 plays an important role in mediating the immune response in IECs during bacterial infection, possibly by regulating the NF-κB signaling pathway.
YTHDF1 regulates the immune response in IECs by mediating the translation of Traf6 transcripts
The principal function of YTHDF1 is the ribosomal loading of m6A-modified mRNA in order to facilitate the initiation of translation (17). To identify the downstream targets of YTHDF1, we used ribosome profiling to compare translation efficiency in LPS-stimulated IPEC-J2 cells expressing either the Ythdf1 shRNA or the scrambled control shRNA. Consistent with its role in initiating translation, we found that cells expressing the Ythdf1 shRNA had decreased translation efficiency compared to control cells, particularly with respect to m6A-modified transcripts (Figure 2A and B). A total of 2030 differentially translated transcripts, including 947 upregulated genes and 1083 downregulated genes, were then selected for KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway enrichment analysis. We found that among the 1083 downregulated genes, the most enriched pathways were involved in the ‘TNF signaling pathway,’ the ‘NOD-like receptor signaling pathway,’ and the ‘NF-κB signaling pathway’ (Figure 2C). We then focused on the NF-κB signaling pathway and found that the translation efficiency of Traf6 transcripts was significantly decreased (Figure 2D and E). In addition to Traf6, other transcripts such as Traf3 were also downregulated in cells lacking YTHDF1, whereas IκBα translation was largely unaffected (Supplementary Figure S3A, B).
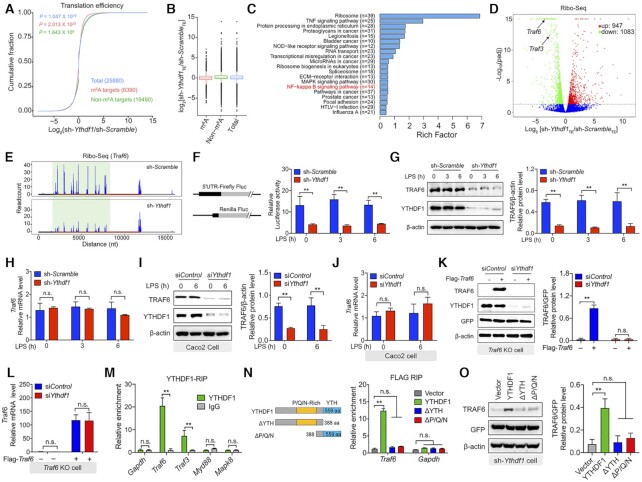
Figure 2.
YTHDF1 regulates TRAF6 expression by initiating Traf6 translation. (A) Cumulative distribution of the log fold change in translation efficiency between LPS-treated IPEC-J2 cells expressing the scrambled shRNA and cells expressing the Ythdf1 shRNA. m6A targets, non-m6A targets, and total mRNA are shown in red, green and blue, respectively. n = 2 independent biological replicates, and P-values were calculated using a two-sided Kolmogorov-Smirnov test. (B) Box plot summarizing the cumulative distributions shown in (A). The horizontal line indicates the median value, the upper and lower boxes represent the upper and lower quartiles, respectively, and the whiskers represent the 1%–99% range. (C) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of Ribo-seq data. Pathway analyses of significantly downregulated genes were conducted using ClusterFile and filtered using the Benjamini-Hochberg procedure. (D) Volcano plot of genes with differential translation efficiency derived from the same samples shown in (A). (E) Genome-wide ribosome profiling of ribosome-protected Traf6 mRNA fragments in LPS-treated IPEC-J2 cells expressing either scrambled shRNAs or Ythdf1 shRNAs. The x-axis represents the nucleotide numbers in the unedited RNA transcript. (F) Luciferase activity in IPEC-J2 cells expressing the indicated plasmids, relative to Renilla luciferase activity. (G) Western blot analysis and summary of TRAF6 protein in LPS-stimulated IPEC-J2 cells expressing the scrambled shRNA or Ythdf1 shRNA. (H) Summary of Traf6 mRNA measured in LPS-stimulated IPEC-J2 cells expressing the scrambled shRNA or Ythdf1 shRNA, relative to Gapdh mRNA. (I) Western blot analysis and summary of TRAF6 protein in LPS-stimulated Caco2 cells expressing a control siRNA or Ythdf1 siRNA. (J) Summary of Traf6 mRNA measured in LPS-stimulated Caco2 cells expressing a control siRNA or Ythdf1 siRNA, relative to Gapdh mRNA. (K, L) Rescue experiment in LPS-stimulated Traf6 KO IPEC-J2 expressing the control siRNA or Ythdf1 siRNA using plasmids expressing Traf6 CDS. TRAF6 protein was measured using western blot analysis and is normalized to GFP (K). Traf6 mRNA was measured using qPCR and is expressed relative to Gapdh mRNA (L). (M) RNA immunoprecipitation (RIP)-qPCR analysis, expressed relative to the input levels. (N) RIP-qPCR analysis of Traf6 and Gapdh immunoprecipitated using an anti-FLAG antibody in IPEC-J2 cells expressing the indicated Ythdf1 plasmids (shown schematically at the left). The data were normalized to the input levels. (O) Western blot analysis and summary of TRAF6 protein in LPS-stimulated expressing the Ythdf1 shRNA and the indicated Ythdf1 plasmids.
To examine directly the outcome of altering Traf6 translation, we performed a firefly luciferase reporter assay in IPEC-J2 cells expressing either the Ythdf1 shRNA or the scrambled shRNA. Consistent with our ribosome profiling results, we found significantly reduced reporter gene activity in cells expressing the Ythdf1 shRNA (Figure 2F). Next, we measured TRAF6 protein levels in cells expressing the Ythdf1 shRNA or the scrambled shRNA and found significantly reduced levels of TRAF6 protein in cells expressing the Ythdf1 shRNA (Figure 2G). This reduction in protein levels was likely due to reduced translation, as Traf6 mRNA levels were similar between cells expressing the Ythdf1 shRNA and cells expressing the scrambled shRNA (Figure 2H), similar to findings obtained with Ythdf1 KO cells (Supplementary Figure S3C and D). Similar results were obtained in both LPS-stimulated Caco-2 cells (Figure 2I and J) and ETEC-infected IPEC-J2 cells (Supplementary Figure S3E and F). As an independent control, we transfected Traf6 KO cells with a Traf6-expressing plasmid together with either the Ythdf1 siRNA or the control siRNA, yielding similar results (Figure 2K and L). Furtherly, we confirmed that TRAF6 plays a role in cell survival using Traf6 KO cells, and we found that the YTHDF1-mediates translation of TRAF6 is associated with cell survival (Supplementary Figure S3G).
Next, we measured the effect of knocking down Ythdf1 on TLR4 signaling. As expected, downstream targets of TRAF6, including IKK, JNK, and MAPK, were suppressed in cells expressing the Ythdf1 shRNA, while proteins upstream of TRAF6 such as MyD88 and TRIF were unaffected (Supplementary Figure S4A). Similar with LPS stimulation, we found that knocking down Ythdf1 did not affect the mRNA levels of Traf6 or Traf3, but markedly decreased their proteins levels upon treatment with the TLR3 ligand poly(I:C) (Supplementary Figure S4B, C). Furthermore, loss of Ythdf1 expression significantly reduced the expression of Tnf-α and Il-6 in cells stimulated with poly(I:C), but had little effect on cells treated with the dectin-1 agonist zymosan (Supplementary Figure S4D-E). Interestingly, the reduction in TRAF6 protein levels was specific to cells lacking Ythdf1 expression, as knocking down either Ythdf2 or Ythdf3 had no effect on TRAF6 protein levels (Supplementary Figure S4F).
To regulate translation, YTHDF1 must bind to its target RNA. Thus, we performed RNA immunoprecipitation (RIP)-qPCR analysis in order to confirm binding between YTHDF1 and the Traf6 transcript. Using an anti-YTHDF1 antibody for immunoprecipitation, we detected robust levels of Traf6 and Traf3 mRNA, but virtually no Myd88 or Mapk8 mRNA (Figure 2M). These results suggest that the inactivation of downstream MAPK signaling is due to reduced TRAF6 protein levels, and is not simply a direct effect of knocking down Ythdf1. Thus, the Traf6 transcript may serve as a key target of YTHDF1 in the immune response.
Next, we generated plasmids expressing truncated versions of YTHDF1 (Supplementary Figure S5A) and found that deleting either the YTH domain (ΔYTH) or the Pro/Gln/Asn-rich domain (ΔP/Q/N) in YTHDF1 eliminated the protein's ability to bind to the Traf6 transcript (Figure 2N). Moreover, TRAF6 protein levels were virtually eliminated in cells co-expressing the Ythdf1 shRNA and the truncated Ythdf1-truncated plasmids (Figure 2O), and expressing the truncated Ythdf1 plasmids failed to rescue the decreased cytokine expression in IPEC-J2 cells transfected with the Ythdf1 shRNA (Supplementary Figure S5B). These data suggest that the YTHDF1 protein binds to and initiates the translation of the Traf6 transcript, thereby regulating the expression of various cytokines; these data also indicate that both the YTH and P/Q/N-rich domains in YTHDF1 are required for mediating the interaction between the YTHDF1 protein and its target RNA.
The YTHDF1-mediated translation of Traf6 mRNA requires m6A methylation of the transcript
As shown above, we found that the YTHDF1 protein can ‘read’ m6A-containing RNA. To test whether changes in YTHDF1-mediated gene expression are indeed associated with m6A modifications, we mapped the m6A methylome in IPEC-J2 cells before and after ETEC infection using m6A sequencing (m6A-Seq). We then searched for consensus motifs and identified the GACU sequence (Figure 3A). In addition, the distribution of m6A nucleotides was similar between uninfected and ETEC-infected cells, regardless of whether the transcripts were upregulated or downregulated, with the majority of m6A modifications enriched near the stop codon and within the 3′ UTR (Figure 3B and Supplementary Figure S6A). Among the 230 differently methylated transcripts, 141 (including Traf6) were upregulated and 89 were downregulated (Figure 3C). To support this finding, we performed m6A-IP qPCR and found altered m6A levels in the Traf6 transcripts (Supplementary Figure S6B). Notably, we identified three statistically significant m6A peaks in the Traf6 transcript, with two peaks in the CDS near the stop codon and one peak within the 3′ UTR; moreover, all three peaks were increased after infection (Figure 3D). We found a similar increase in m6A in the Traf3 transcript (Supplementary Figure S6C). As a negative control, we found that the level of m6A in the Myd88 transcript was similar between uninfected and infected cells (Supplementary Figure S6D). Taken together, these results suggest that Traf6 transcripts undergo m6A modification upon bacterial infection, thereby playing a role in their translation.
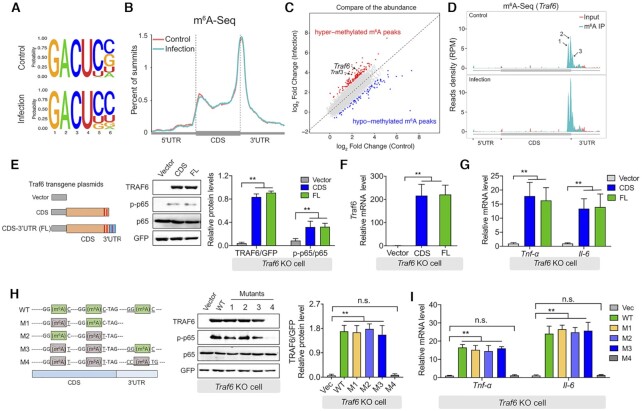
Figure 3.
Translation of the Traf6 transcript requires m6A methylation. (A) Sequence motifs within the m6A peaks were identified using Homer software. (B) Metagene plot showing virtually identical m6A peak distributions in control IPEC-J2 cells and IPEC-J2 cells 3 h after ETEC infection. The 5′ UTR, coding sequence (CDS), and 3′ UTR in the Traf6 transcript are indicated. (C) Summary of differentially expressed m6A-modified transcripts measured by comparing ETEC-infected IPEC-J2 cells and uninfected cells. (D) RNA-Seq analysis of input RNA and immunoprecipitated (IP) m6A RNA in Traf6 transcript. The three m6A motif sequences that correspond to an immunoprecipitation-enriched region are shown in green. (E) Rescue experiment in LPS-stimulated Traf6 KO IPEC-J2 cells expressing the indicated FLAG-tagged Traf6 plasmids. (F, G) Summary of Traf6 (F) and Tnf-α and Il-6 (G) mRNA levels measured in the cells show in (E), expressed relative to Gapdh mRNA. (H) Western blot analysis and summary of TRAF6, p65, and p-p65 in LPS-stimulated Traf6 KO IPEC-J2 cells expressing the indicated Traf6 plasmids. The left panel shows the various plasmid with mutations in the indicated m6A sites in the Traf6 transcript. (I) Summary of Tnf-α and Il-6 mRNA levels measured in the cells shown in (H), expressed relative to Gapdh mRNA.
To further analyze the functional role of m6A modifications in the transcript, we generated constructs expressing either the FLAG-tagged full-length Traf6 cDNA or a FLAG-tagged Traf6 cDNA containing the coding sequence but lacking the 3′ UTR (Figure 3E, left). We found that expressing either the full-length construct or the 3′ UTR−lacking construct increased both TRAF6 protein and Traf6 mRNA levels in Traf6 KO IPEC-J2 cells following LPS stimulation (Figure 3E and F), suggesting that methylation of the 3′ UTR is not required for the regulation of Traf6 translation via YTHDF1, a notion supported by our finding that the expression of both Tnf-α and Il-6 was increased in cells expressing either of the two FLAG-tagged Traf6 constructs (Figure 3G). Next, we introduced mutations at one, two, or three m6A consensus sites in the FLAG-Traf6 constructs (Figure 3H); these constructs were transfected into Traf6 KO IPEC-J2 cells, which were then stimulated with LPS. We found that LPS stimulation induced robust levels of TRAF6 protein in cells transfected with plasmids containing either one or two m6A mutations, including cells transfected with plasmid M3, which contains only the m6A site in the 3′ UTR (Figure 3H). In contrast, cells expressing plasmid M4—in which all three m6A sites are mutated—had virtually no TRAF6 protein, suggesting impaired translation (Figure 3H). Consistent with these results, Tnf-α and Il-6 expression in cells expressing plasmid M1, M2, or M3 was similar to cells expressing wild-type Traf6, but was undetectable in cells expressing plasmid M4 (Figure 3I). These results indicate that during the immune response to bacterial infection, the m6A sites near the stop codon—either in the CDS or in the 3′ UTR—mediate the translation of Traf6 transcripts.
The binding of YTHDF1 to the Traf6 transcript requires DDX60
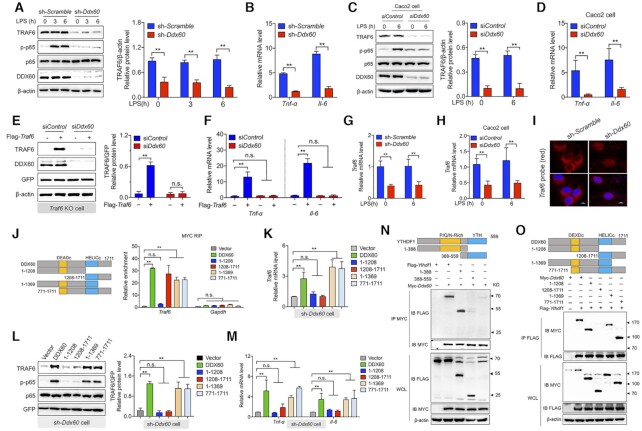
To investigate further the mechanism by which YTHDF1 reads the methylation status of Traf6 transcripts, we investigated whether a coregulatory protein plays a role in the ability of YTHDF1 to accurately recognize its target. We therefore isolated YTHDF1-binding proteins using an anti-YTHDF1 antibody in lysates obtained from LPS-stimulated IPEC-J2 cells. We then used tandem mass spectrometry (MS/MS) to identify and quantify the associated protein components. In total, 466 proteins were identified using this approach, with DDX60, THRAP3, and BCLAF1 as the most enriched proteins (Figure 4A and Supplementary Table S1). Gene Ontology (GO) pathway analysis revealed that the top pathways included proteins involved in RNA processing, translation, ribosomes, RNA binding, and structural constituents of ribosomes (Figure 4B). We then tested whether any of these proteins interact with the Traf6 transcript using RNA immunoprecipitation and found that DDX60, but not THRAP3 or BCLAF1, interacts with the Traf6 transcript (Figure 4C). As a negative control, no binding was observed between the Gapdh transcript and DDX60, THRAP3, or BCLAF1 (Figure 4C). Binding between the Traf6 transcript and DDX60 was confirmed by performing fluorescence in situ hybridization (FISH) and immunostaining, respectively, in LPS-stimulated cells (Figure 4D). Next, we performed co-IP experiments in LPS-stimulated cells and confirmed that YTHDF1 interacts with DDX60 (Figure 4E); this interaction remained even after the lysates were treated with RNase to digest the RNA transcripts (Figure 4E).
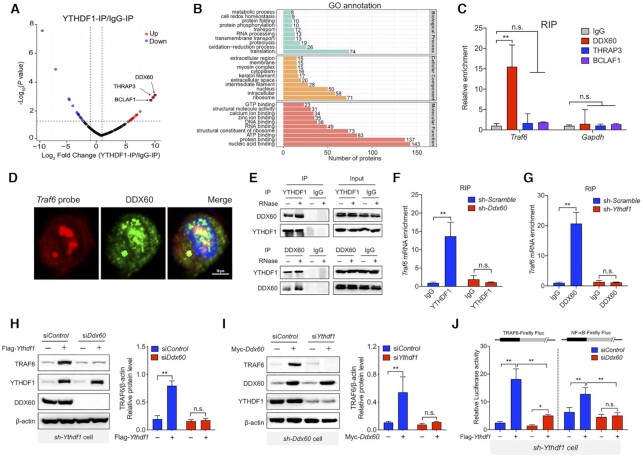
Figure 4.
Traf6 transcripts recognized by YTHDF1 depends on DDX60. (A) Volcano plot depicting the Log10 (P-value) versus the Log2 fold change (YTHDF1-IP/IgG-IP) in MS-identified proteins obtained from two biological replicates. Intensities were obtained from a MaxQuant database search. (B) Pathway analyses (with Gene Ontology direct terms) of proteins measured using co-IP of YTHDF1 in LPS-stimulated IPEC-J2 cells. (C) Summary of the interaction between the DDX60, THRAP3, and BCLAF1 proteins and the Traf6 transcript measured using RIP q-PCR, normalized to the input levels. (D) DDX60 immunofluorescence (green) and FISH analysis of Traf6 mRNA (red) in an IPEC-J2 cell 6 h after LPS stimulation. The nucleus was counterstained with DAPI (blue). (E) Co-IP experiment using anti-YTHDF1 (upper) or DDX60 (lower) and control IgG. After immunoprecipitation, the samples were washed and incubated with RNase where indicated. YTHDF1 and DDX60 proteins were then detected using western blot analysis. (F) RIP-qPCR analysis of the interaction between YTHDF1 and the Traf6 transcript in IPEC-J2 cells expressing a scrambled shRNA or the Ddx60 shRNA. (G) RIP-qPCR analysis of the interaction between DDX60 and the Traf6 transcript in IPEC-J2 cells expressing a scrambled shRNA or the Ythdf1 shRNA. (H, I) Western blot analysis and summary of TRAF6 in IPEC-J2 cells co-expressing the Ythdf1 shRNA, the FLAG-Ythdf1 plasmid or empty vector, and either a scrambled siRNA or Ddx60 siRNA (H). The reciprocal experiment is shown in (I). (J) Luciferase assay to measure TRAF6 and NF-κB expression in IPEC-J2 cells expressing the Ythdf1 shRNA, the FLAG-Ythdf1 plasmid or empty vector, and a scrambled siRNA or the Ddx60 siRNA. Luciferase activity is expressed relative to Renilla luciferase activity.
To investigate whether DDX60 is required for mediating the interaction between the Traf6 transcript and the YTHDF1 protein, we used RIP analysis to measure the binding between YTHDF1 and Traf6 mRNA in cells expressing either a scrambled shRNA or a Ddx60 shRNA. We found that knocking down Ddx60 virtually eliminated the binding between YTHDF1 and Traf6 mRNA (Figure 4F); conversely, knocking down Ythdf1 eliminated the interaction between DDX60 and Traf6 mRNA (Figure 4G). To confirm these results, we co-transfected cells with the Ythdf1 shRNA and the FLAG-tagged Ythdf1 construct (or an empty vector) together with either the scrambled siRNA or the Ddx60 siRNA; we then measured TRAF6 protein. We found that expressing Ythdf1 was unable to rescue TRAF6 expression in cells lacking DDX60 (Figure 4H). Conversely, expressing a Myc-tagged Ddx60 construct in cells expressing the Ddx60 shRNA failed to rescue TRAF6 expression in cells lacking YTHDF1 (Figure 4I). Similar results were obtained using TRAF6 and NF-κB luciferase reporter assays (Figure 4J). These results indicate that the ability of YTHDF1 to bind the Traf6 transcript requires DDX60.
DDX60 serves as a coregulator with YTHDF1 in regulating the translation of Traf6 mRNA and downstream signaling
The above results suggest that DDX60 may serve as a functional coregulator with YTHDF1 in order to mediate the translation of Traf6 mRNA. To test this hypothesis, we measured TRAF6 protein levels in cells expressing either the scrambled shRNA or the Ddx60 shRNA. Following LPS stimulation, cells lacking DDX60 had significantly reduced TRAF6 protein and significantly reduced NF-κB signaling (Figure 5A), as well as significantly reduced decreased Tnf-α and Il-6 expression (Figure 5B), compared to LPS-stimulated cells expressing the scrambled shRNA. To test this hypothesis further, we generated a Ddx60 KO IPEC-J2 cell line using CRISPR/Cas9 and found similar results (Supplementary Figure S7A-B). Moreover, similar results were obtained using both LPS-stimulated Caco-2 cells (Figure 5C and D) and ETEC-infected IPEC-J2 cells (Supplementary Figure S7C and D). Next, we expressed the FLAG-tagged Traf6 construct (or an empty vector) in Traf6 KO IPEC-J2 cells together with the Ddx60 shRNA or scrambled control shRNA and found that DDX60 was required for driving Traf6 translation (Figure 5E) and downstream Tnf-α and Il-6 expression (Figure 5F). Interestingly, knocking down Ddx60 also significantly reduced Traf6 mRNA levels in IPEC-J2, independent of LPS stimulation (Figure 5G) and ETEC infection (Supplementary Figure S7E and F). Knocking down Ddx60 had a similar effect in Caco-2 cells with respect to reducing Traf6 mRNA levels (Figure 5H). Moreover, FISH analysis revealed that knocking down Ddx60 decreased the number of Traf6 transcripts, particularly in the cytoplasm (Figure 5I).
Figure 5.
The interaction between DDX60 and YTHDF1 is required for initiating translation of the Traf6 transcript. (A) Western blot analysis and summary of TRAF6 protein in IPEC-J2 cells expressing the scrambled shRNA or theDdx60 shRNA and stimulated with LPS for the indicated times. (B) Total RNA was extracted from the same samples in (A), and Tnf-α and Il-6 mRNA was measured using qPCR and expressed relative to Gapdh mRNA. (C) Western blot analysis and summary of TRAF6 protein in LPS-stimulated Caco2 cells expressing a scrambled siRNA or Ddx60 siRNA. (D) Total RNA was extracted from the same samples in (C), and Tnf-α and Il-6 mRNA was measured and expressed relative to Gapdh mRNA. (E) Western blot analysis and summary of TRAF6 protein in LPS-stimulated Traf6 KO IPEC-J2 cells expressing the FLAG-Traf6 plasmid or an empty vector together with either a control siRNA or Ddx60 siRNA. (F) Total RNA was extracted from the same samples in (E), and Tnf-α and Il-6 mRNA was measured and expressed relative to Gapdh mRNA. (G-H) Total RNA was extracted from the same samples in (A), and Traf6 mRNA was measured and expressed relative to Gapdh mRNA. (I) FISH analysis of Traf6 (red) transcripts in LPS-stimulated IPEC-J2 cells expressing the scrambled shRNA or Ddx60 shRNA; the nuclei were counterstained with DAPI (blue). Scale bars, 10 μm. (J) The fold enrichment of Traf6 and Gapdh mRNA was measured using RIP-qPCR in IPEC-J2 cells expressing the indicated plasmids. The structure of the full-length and truncated DDX60 proteins is shown schematically at the left; the DEADc and HELICc domains are indicated. (K) Total RNA was extracted from IPEC-J2 expressing the Ddx60 shRNA and the indicated Myc-Ddx60 plasmids, and Traf6 mRNA was measured and expressed relative to Gapdh mRNA. (L) Western blot analysis and summary of TRAF6 protein measured in the same samples in (K). (M) Total RNA was extracted from the same samples in (K), and Tnf-α and Il-6 mRNA was measured and expressed relative Gapdh mRNA. (N, O) Co-IP experiments with LPS-stimulated IPEC-J2 cells co-expressing the indicated FLAG-Ythdf1 plasmids together with Myc-Ddx60 or an empty vector (N) or co-expressing the indicated Myc-Ddx60 plasmids together with FLAG-Ythdf1 or an empty vector (O). For reference, western blot analyses using whole-cell lysate (WCL) samples are shown at the bottom.
To identify the functional region(s) in DDX60 that underlie its coregulatory activity, we generated plasmids expressing various truncated forms of DDX60 (Figure 5J, left). RIP-qPCR revealed that the DDX60 construct lacking the HELICc domain was unable to interact with the Traf6 transcript (Figure 5J and Supplementary Figure S8A), indicating that this domain is required for the protein's RNA-binding activity. Moreover, neither the DEADc domain nor the HELICc domain alone was sufficient to rescue TRAF6 expression or p65 activation in cells expressing the Ddx60 shRNA (Figure 5K and L), suggesting that both domains are functionally important; these results were supported by overexpression experiments using Ddx60-truncated plasmids (Supplementary Figure S8B and C). Additional functional experiments revealed that the expression of Tnf-α and Il-6 was rescued in Ddx60 shRNA−transfected cells expressing truncated Ddx60 plasmids containing both the DEADc and HELICc domains, but not in cells transfected with plasmids containing only the DEADc domain or the HELICc domain (Figure 5M); similar results were obtained with respect to cells overexpressing various plasmids containing specific Ddx60 domains (Supplementary Figure S8D). Taken together, these results indicate that the HELICc domain in DDX60 is required for binding to the Traf6 transcript, whereas both the DEADc and HELICc domains are required for driving the translation of Traf6 mRNA. Finally, we generated plasmids expressing various truncated fragments of tagged YTHDF1 and DDX60 proteins and found that the binding between YTHDF1 and DDX60 requires the P/Q/N-rich domain in YTHDF1 (Figure 5N) and the DEADc domain in DDX60 (Figure 5O).
Mice lacking YTHDF1 have an impaired intestinal immune response
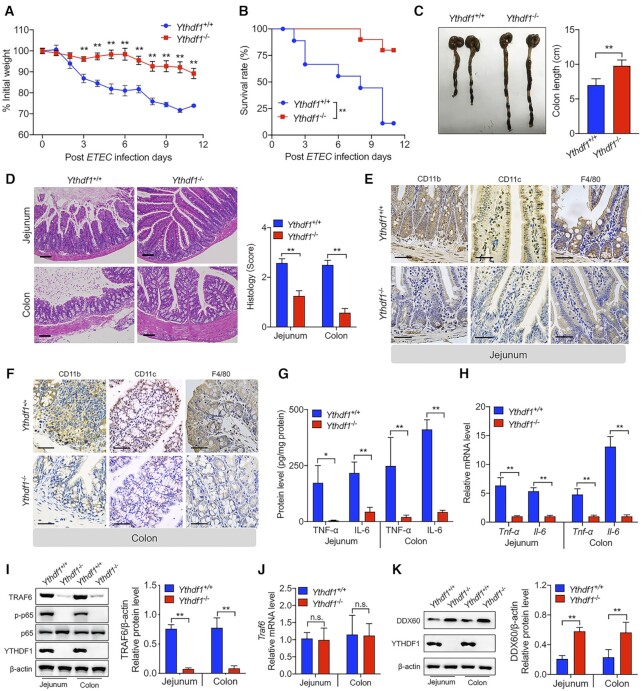
To investigate the clinical relevance of our in vitro results with respect to the role of YTHDF1 in mediating the intestinal immune response to bacterial infection, we created a targeted deletion in the Ythdf1 gene in mice using CRISPR/Cas9, yielding Ythdf1−/− mice (28) (Supplementary Figure S9A). Western blot analysis confirmed the loss of YTHDF1 protein in the jejunum of Ythdf1−/− mice (Supplementary Figure S9B). We then challenged weight-matched Ythdf1+/+ and Ythdf1−/− mice with an oral dose of ETEC (108 CFU). We found that ETEC-infected Ythdf1−/− mice had significantly less weight loss (Figure 6A), higher survival (Figure 6B), and reduced colon atrophy (Figure 6C) compared to ETEC-infected Ythdf1+/+ mice.
Figure 6.
Loss of Ythdf1 in mice reduces the intestinal immune response to bacterial infection. (A, B) Time course of relative body weight (A) and survival (B) measured in Ythdf1+/+ and Ythdf1−/− littermates following ETEC infection (n = 10 mice/group). (C) Representative gross images of colons removed from Ythdf1+/+ and Ythdf1−/− littermates 3 days after ETEC infection. Colon length is summarized at the right (n = 10 mice/group). (D) Representative histological images of jejunum and colon sections obtained from Ythdf1+/+ and Ythdf1−/− mice 3 days after ETEC infection. The right panel shows the histology scores (n = 6 mice/group), scale bar: 50 mm. (E, F) Immunohistochemical staining of the indicated proteins in jejunum (E) and colon (F) segments obtained from Ythdf1+/+ and Ythdf1−/− mice 3 days after ETEC infection; scale bars, 50 μm. (G) Summary of TNF-α and IL-6 proteins measured in the jejunum and colon of Ythdf1+/+ and Ythdf1−/− mice using ELISA (n = 6 mice/group). (H) Total RNA was extracted from the same samples in (G), and Tnf-α and Il-6 mRNA was measured and expressed relative to Gapdh mRNA (n = 6 mice/group). (I) Western blot analysis and summary of TRAF6 protein in the jejunum and colon of Ythdf1+/+ and Ythdf1−/− mice (n = 6 mice/group). (J) Total RNA was extracted from the same samples in (I), and Traf6 mRNA was measured and expressed relative to Gapdh mRNA (n = 6 mice/group). (K) Western blot analysis and summary of DDX60 protein in the jejunum and colon of Ythdf1+/+ and Ythdf1−/− mice (n = 6 mice/group).
Histological analyses of the jejunum and colon revealed a reduced inflammatory response (quantified as reduced histology scores) in ETEC-infect Ythdf1−/− mice compared to ETEC-infected Ythdf1+/+ mice (Figure 6D). This reduced response was accompanied by reduced infiltration of inflammatory cells, including CD11b+, CD11c+ and F4/80+ cells, in both the jejunum (Figure 6E) and colon (Figure 6F). Moreover, ETEC-infected Ythdf1−/− mice had drastically reduced levels of TNF-α and IL-6 in the jejunum and colon, measured at both the protein (Figure 6G) and mRNA (Figure 6H) levels. Interestingly, however, circulating levels of TNF-α and IL-6 were similar between ETEC-infected Ythdf1+/+ and Ythdf1−/− mice (Supplementary Figure S9C and D), confirming that the effects of knocking out YTHDF1 were specific to the intestinal tract. These results might therefore be useful for performing a differential analysis between myeloid cells and epithelial cells.
Next, we measured TRAF6 expression in vivo and found significantly lower levels of TRAF6 protein in the jejunum and colon of Ythdf1−/− mice compared to Ythdf1+/+ mice (Figure 6I), despite similar levels of Traf6 mRNA (Figure 6J), suggesting that YTHDF1 plays a role in regulating the translation—but not the transcription—of Traf6. Finally, we found that Ythdf1−/− mice have significantly higher levels of DDX60 protein in the jejunum and colon compared to Ythdf1−/− mice (Figure 6K), consistent with our in vitro results. Taken together, these findings support the notion that YTHDF1 plays an essential role in mediating the intestinal immune response to bacterial infection.
DISCUSSION
The study of epigenetic factors has yielded new insights into the pathogenesis of the immune response. Among the many epigenetic modifications studied, the m6A modification in RNA has been shown to play a major role in innate immunity (21,24,39,40). However, the specific functional role of m6A ‘reader’ proteins with respect to the immune response in intestinal epithelial cells, and whether m6A is involved in this process, remain unclear. Bacteria activate the immune response in IECs via LPS, which can pass through the intestinal barrier. Here, we established models in which ETEC infection and LPS stimulation recapitulate the immune response observed in IECs following bacterial infection. We found that YTHDF1 regulates the immune response in IECs by directing the translation of Traf6 transcripts in an m6A-dependent manner. In addition, we identified DDX60 as a necessary coregulator in facilitating the binding of YTHDF1 to m6A-containing transcripts during bacterial infection.
At the biological level, m6A-containing nucleotides are functionally mediated by the coordinated activity of methyltransferases, demethylases, and reader proteins (41). Using an established mouse model, we found that ETEC infection significantly increased the number of m6A-containing transcripts in the colon and jejunum. Moreover, knocking down Mettl3, which encodes an m6A methyltransferase, increased the survival of ETEC-infected IECs, suggesting that m6A nucleotides play a role in bacterial infection. Interestingly, we found that knocking down Ythdf1—but not knocking down either Ythdf2 or Ythdf3—increased the survival of ETEC-infected IECs. This may have clinical relevance, given the recent report that knocking out Ythdf1 in mice with colorectal cancer leads to tumor shrinkage and prolonged survival (42). Moreover, knocking out Ythdf1 in classic dendritic cells increased the cross-presentation of tumor antigens and the cross-priming of CD8+ T cells, suggesting that YTHDF1 may be a potential therapeutic target in anticancer immunotherapy (43). Using a series of in vivo and in vitro experiments, we found that deleting YTHDF1 suppresses the intestinal immune response by blocking the NF-κB signaling pathway. YTHDF1 is believed to increase the translation of its target transcripts by interacting with translation initiation factors and facilitating ribosome loading (17). Using Ribo-seq and m6A-seq analyses, we found that knocking down Ythdf1 in IPEC-J2 cells drastically reduced the translation efficiency of m6A-containing transcripts. We then focused on the NF-κB signaling pathway and found that the translation efficiency of Traf6 mRNA—but not IκBα mRNA—was significantly reduced, leading us to speculate that the Traf6 is an important target of YTHDF1 in mediating the immune response. In addition to Traf6, we found that other transcripts such as Traf3 are decreased in Ythdf1 KO cells, albeit to a lesser extent than Traf6. Thus, the Traf6 transcript is likely not the sole target of YTHDF1. Interestingly, a recent study by Rubio et al. found that human cytomegalovirus (HCMV) and double-stranded DNA (dsDNA) can trigger the m6A-dependent production of IFN-β via the m6A methyltransferase METTL3 and the m6A demethylase ALKBH5 (44). In the context of these results, we hypothesize that YTHDF1 may utilize two distinct mechanisms to regulate the immune response in IECs. In one mechanism, YTHDF1 drives the translation of m6A modified transcripts such as Traf6 to activate the NF-κB signaling pathway; in the second mechanism, YTHDF1 initiates the translation of m6A-containing effector molecule mRNAs such as the IFNB1 transcript in order to regulate their production directly.
We also confirmed and characterized the putative m6A sites in the Traf6 transcript using m6A-seq analysis. Consistent with the classic distribution of m6A nucleotides within genes (36), we found that the m6A modifications in the Traf6 transcript are concentrated in the vicinity of the stop codon at the conserved ‘RACU’ motif. However, the use of m6A mapping is controversial (45); we therefore performed m6A-IP qPCR and confirmed the altered m6A levels in the Traf6 transcript using a separate antibody. To examine the mechanism by which YTHDF1 binding to m6A-containing transcripts participates in the IEC immune response, we generated Traf6 plasmids lacking the 3′ UTR and found robust Traf6 translation regardless of whether the 3′ UTR was present or absent, suggesting that m6A within the 3′ UTR is not required for the YTHDF1-mediated translation of Traf6 transcripts. This finding is similar to TGF-β−induced SNAI1 expression, in which an m6A nucleotide in the 3′ UTR was not required for translation (46). Recently, Mao et al. reported that removing the m6A nucleotides in the coding sequence of transcripts further decreased translation (47). Together, these results clarify whether m6A nucleotides in the 3′ UTR mediate the regulatory role of YTHDF1 with respect to translation (17,48). By systematically mutating the m6A consensus sites in Traf6, we found that the m6A modification is required for translation but is not required for transcription. Given our findings and the previous report that YTHDF1 interacts with the eIF3 (eukaryotic initiation factor 3) complex (17), we propose that formation of a mediated loop structure is likely a key step in the YTHDF1-m6A−mediated initiation of translation, regardless of whether the m6A nucleotide is located within the 3′ UTR or in the CDS near the stop codon.
Structural and binding studies have suggested that the YTH domain serves as the specific m6A ‘reader’ (49,50). Consistent with this notion, we found that YTHDF1 functions by binding to the Traf6 transcript via its YTH domain. On the other hand, we found that expressing a YTHDF1 construct lacking the P/Q/N-rich domain failed to rescue cytokine expression, indicating that the entire YTHDF1 protein is required for driving translation of the Traf6 transcript. Although YTHDF family members have a common domain structure, YTHDF1 and YTHDF2 target specific transcripts in endometrial cancer cells, regulating the translation and decay, respectively, of their targets (15). Precisely how YTHDF1 and YTHDF2 recognize their specific m6A-containing targets using the same domain is currently unknown, although a coregulator may interact with the P/Q/N-rich domain in order to mediate the specific functional role of YTHDF1 when interacting with the Traf6 transcript.
Importantly, we identified DDX60 as a coregulator of YTHDF1 in mediating the translation of Traf6 transcripts. DEAD-box (DDX) proteins contain a conserved Asp-Glu-Ala-Asp motif and are the largest family of RNA helicases (51). DDX proteins interact with both rRNA and mRNA to regulate a wide range of biological functions, including mRNA synthesis, RNA splicing, and translation (52). Interestingly, Zheng et al. recently showed that DDX46 binds to specific antiviral transcripts in order to negatively regulate the innate antiviral response (53). We confirmed the putative interaction between YTHDF1, DDX60, and the Traf6 transcript using a series of rescue experiments in IPEC-J2 cells lacking both Ddx60 and Ythdf1. Mechanistically, our data suggest that DDX60 binds to the Traf6 transcript via its HELICc domain and interacts with the P/Q/N-rich domain in YTHDF1 via its DEAD helicase domain, which may explain—at least in part—how YTHDF1 can specifically recognize unique targets using the same domain present in other YTHDF family members.
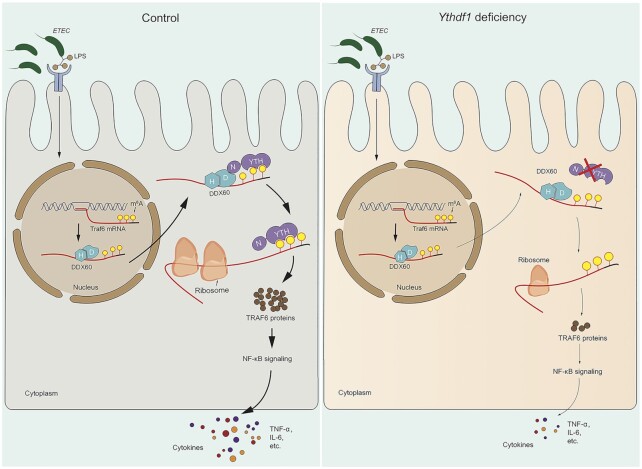
In summary, our results support a model in which m6A modifications in the Traf6 transcript regulate the intestinal immune response to bacterial infection via a complex consisting of YTHDF1, DDX60, and the Traf6 transcript (Figure 7). These findings provide novel insights into the molecular mechanisms underlying the immune response in the intestinal epithelium and suggest new therapeutic strategies for treating bacterial infections in the intestine.
Figure 7.
Model depicting the proposed mechanism by which YTHDF1 modulates the intestinal immune response to bacterial infection. Left: Upon bacterial infection (e.g. with ETEC) of an intestinal epithelial cell, DDX60 binds to the Traf6 transcript via its HELICc domain, transporting the transcript to the cytoplasm, where DDX60 interacts with YTHDF1 by recognizing the m6A-modified Traf6 transcript, driving the translation of Traf6 and upregulating the production of cytokines such as TNF-α and IL-6. Right panel: Loss of YTHDF1 significantly decreases the translation efficiency of Traf6 transcripts, leading to reduced cytokine production, increased survival of intestinal epithelial cells, and a reduced immune response.
DATA AVAILABILITY
The data used and/or analyzed to support the findings of this study are available in the main paper or in the Supplementary Information. Any other raw data that support the findings of this study are available upon reasonable request from the corresponding author.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the Electronic Microscopy Center and the Agricultural, Biological, and Environmental Test Center of Zhejiang University for assistance with the microscopy work. The authors also thank Dr Shu Zhu at Zhejiang University for assistance with ETEC infection, and Dr Sufen Zhang at the Institute of Nuclear Agricultural Sciences, Zhejiang University, for help with the translation efficiency experiments.
Contributor Information
Xin Zong, Key Laboratory of Molecular Animal Nutrition, Ministry of Education, College of Animal Sciences, Zhejiang University, Hangzhou, China; The First Affiliated Hospital, School of Public Health, Institute of Translational Medicine, Zhejiang University School of Medicine, Hangzhou, China; Key Laboratory of Animal Nutrition and Feed Science in Eastern China, Ministry of Agriculture, College of Animal Sciences, Zhejiang University, Hangzhou, China.
Xiao Xiao, Key Laboratory of Molecular Animal Nutrition, Ministry of Education, College of Animal Sciences, Zhejiang University, Hangzhou, China.
Bin Shen, State Key Laboratory of Reproductive Medicine, Department of Histology and Embryology, Nanjing Medical University, Nanjing, China.
Qin Jiang, Key Laboratory of Molecular Animal Nutrition, Ministry of Education, College of Animal Sciences, Zhejiang University, Hangzhou, China.
Hong Wang, Key Laboratory of Molecular Animal Nutrition, Ministry of Education, College of Animal Sciences, Zhejiang University, Hangzhou, China.
Zeqing Lu, Key Laboratory of Molecular Animal Nutrition, Ministry of Education, College of Animal Sciences, Zhejiang University, Hangzhou, China; Key Laboratory of Animal Nutrition and Feed Science in Eastern China, Ministry of Agriculture, College of Animal Sciences, Zhejiang University, Hangzhou, China.
Fengqin Wang, Key Laboratory of Molecular Animal Nutrition, Ministry of Education, College of Animal Sciences, Zhejiang University, Hangzhou, China; Key Laboratory of Animal Nutrition and Feed Science in Eastern China, Ministry of Agriculture, College of Animal Sciences, Zhejiang University, Hangzhou, China.
Mingliang Jin, Key Laboratory of Molecular Animal Nutrition, Ministry of Education, College of Animal Sciences, Zhejiang University, Hangzhou, China; Key Laboratory of Animal Nutrition and Feed Science in Eastern China, Ministry of Agriculture, College of Animal Sciences, Zhejiang University, Hangzhou, China.
Junxia Min, The First Affiliated Hospital, School of Public Health, Institute of Translational Medicine, Zhejiang University School of Medicine, Hangzhou, China.
Fudi Wang, The First Affiliated Hospital, School of Public Health, Institute of Translational Medicine, Zhejiang University School of Medicine, Hangzhou, China; Hengyang Medical School, University of South China, Hengyang, China.
Yizhen Wang, Key Laboratory of Molecular Animal Nutrition, Ministry of Education, College of Animal Sciences, Zhejiang University, Hangzhou, China; Key Laboratory of Animal Nutrition and Feed Science in Eastern China, Ministry of Agriculture, College of Animal Sciences, Zhejiang University, Hangzhou, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31630075, 32002185, 31930057, 31970689]; Zhejiang Provincial Natural Science Foundation of China [LQ21C170002]; China Postdoctoral Science Foundation [2020M671741]. Funding for open access charge: National Natural Science Foundation of China [31630075].
Conflict of interest statement. None declared.
REFERENCES
- 1. Hwang I.Y., Koh E., Wong A., March J.C., Bentley W.E., Lee Y.S., Chang M.W.. Engineered probiotic Escherichia coli can eliminate and prevent Pseudomonas aeruginosa gut infection in animal models. Nat. Commun. 2017; 8:15028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Balzan S., Quadros C.D., de Cleva R., Zilberstein B., Cecconello I.. Bacterial translocation: Overview of mechanisms and clinical impact. J Gastroen Hepatol. 2007; 22:464–471. [DOI] [PubMed] [Google Scholar]
- 3. Qadri F., Akhtar M., Bhuiyan T.R., Chowdhury M.I., Ahmed T., Rafique T.A., Khan A., Rahman S.I.A., Khanam F., Lundgren A.et al.. Safety and immunogenicity of the oral, inactivated, enterotoxigenic Escherichia coli vaccine ETVAX in Bangladeshi children and infants: a double-blind, randomised, placebo-controlled phase 1/2 trial. Lancet Infect. Dis. 2019; 20:208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Peterson L.W., Artis D.. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat. Rev. Immunol. 2014; 14:141–153. [DOI] [PubMed] [Google Scholar]
- 5. Henderson P., van Limbergen J.E., Schwarze J., Wilson D.C.. Function of the intestinal epithelium and its dysregulation in inflammatory bowel disease. Inflamm. Bowel Dis. 2011; 17:382–395. [DOI] [PubMed] [Google Scholar]
- 6. Price A.E., Shamardani K., Lugo K.A., Deguine J., Roberts A.W., Lee B.L., Barton G.M.. A map of toll-like receptor expression in the intestinal epithelium reveals distinct spatial, cell type-specific, and temporal patterns. Immunity. 2018; 49:560–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhao B.S., Roundtree I.A., He C.. Post-transcriptional gene regulation by mRNA modifications (vol 18, pg 31, 2017). Nat Rev Mol Cell Bio. 2018; 19:808–808. [DOI] [PubMed] [Google Scholar]
- 8. Roundtree I.A., Evans M.E., Pan T., He C.. Dynamic RNA modifications in gene expression regulation. Cell. 2017; 169:1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bokar J.A., Shambaugh M.E., Polayes D., Matera A.G., Rottman F.M.. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA. 1997; 3:1233–1247. [PMC free article] [PubMed] [Google Scholar]
- 10. Liu J., Yue Y., Han D., Wang X., Fu Y., Zhang L., Jia G., Yu M., Lu Z., Deng X.et al.. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014; 10:93–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ping X.L., Sun B.F., Wang L., Xiao W., Yang X., Wang W.J., Adhikari S., Shi Y., Lv Y., Chen Y.S.et al.. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014; 24:177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jia G.F., Fu Y., Zhao X., Dai Q., Zheng G.Q., Yang Y., Yi C.Q., Lindahl T., Pan T., Yang Y.G.et al.. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011; 7:885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zheng G.Q., Dahl J.A., Niu Y.M., Fedorcsak P., Huang C.M., Li C.J., Vagbo C.B., Shi Y., Wang W.L., Song S.H.et al.. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 2013; 49:18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang C.Y., Fu J.R., Zhou Y.F.. A review in research progress concerning m6A methylation and immunoregulation. Front. Immunol. 2019; 10:00922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shi H., Wei J., He C.. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell. 2019; 74:640–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhou J., Wan J., Gao X., Zhang X., Jaffrey S.R., Qian S.B.. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature. 2015; 526:591–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang X., Zhao B.S., Roundtree I.A., Lu Z., Han D., Ma H., Weng X., Chen K., Shi H., He C.. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015; 161:1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang X., Lu Z., Gomez A., Hon G.C., Yue Y., Han D., Fu Y., Parisien M., Dai Q., Jia G.et al.. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014; 505:117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Meyer K.D., Patil D.P., Zhou J., Zinoviev A., Skabkin M.A., Elemento O., Pestova T.V., Qian S.B., Jaffrey S.R.. 5′ UTR m(6)A promotes cap-independent translation. Cell. 2015; 163:999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Du H., Zhao Y., He J.Q., Zhang Y., Xi H.R., Liu M.F., Ma J.B., Wu L.G.. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 2016; 7:12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shi H.L., Wang X., Lu Z.K., Zhao B.X.S., Ma H.H., Hsu P.J., Liu C., He C.. YTHDF3 facilitates translation and decay of N-6-methyladenosine-modified RNA. Cell Res. 2017; 27:315–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Walsh M.C., Lee J., Choi Y.. Tumor necrosis factor receptor- associated factor 6 (TRAF6) regulation of development, function, and homeostasis of the immune system. Immunol. Rev. 2015; 266:72–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Miyashita M., Oshiumi H., Matsumoto M., Seya T.. DDX60, a DEXD/H box helicase, is a novel antiviral factor promoting RIG-I-like receptor-mediated signaling. Mol. Cell. Biol. 2011; 31:3802–3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goubau D., van der Veen A.G., Chakravarty P., Lin R.T., Rogers N., Rehwinkel J., Deddouche S., Rosewell I., Hiscott J., Sousa C.R.E.. Mouse superkiller-2-like helicase DDX60 is dispensable for type I IFN induction and immunity to multiple viruses. Eur. J. Immunol. 2015; 45:3386–3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zong X., Zhao J., Wang H., Lu Z.Q., Wang F.Q., Du H.H., Wang Y.Z.. Mettl3 deficiency sustains long-chain fatty acid absorption through suppressing Traf6-dependent inflammation response. J. Immunol. 2019; 202:567–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jiang Q., Sun B.F., Liu Q., Cai M., Wu R.F., Wang F.Q., Yao Y.X., Wang Y.Z., Wang X.X.. MTCH2 promotes adipogenesis in intramuscular preadipocytes via an m(6)A-YTHDF1-dependent mechanism. FASEB J. 2019; 33:2971–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gao Y., Rong Y., Wang Y., Xiong H., Huang X., Han F., Feng J., Wang Y.. Expression pattern of porcine antimicrobial peptide PR-39 and its induction by enterotoxigenic Escherichia coli (ETEC) F4ac. Vet. Immunol. Immunopathol. 2014; 160:260–265. [DOI] [PubMed] [Google Scholar]
- 28. Shi H.L., Zhang X.L., Weng Y.L., Lu Z.Y., Liu Y.J., Lu Z.K., Li J.N., Hao P.L., Zhang Y., Zhang F.et al.. m(6)A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature. 2018; 563:249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. de Castro R.O., Previato L., Goitea V., Felberg A., Guiraldelli M.F., Filiberti A., Pezza R.J.. The chromatin-remodeling subunit Baf200 promotes homology-directed DNA repair and regulates distinct chromatin-remodeling complexes. J. Biol. Chem. 2017; 292:8459–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shen Q.C., Zhang Q., Shi Y., Shi Q.Z., Jiang Y.Y., Gu Y., Li Z.Q., Li X., Zhao K., Wang C.M.et al.. Tet2 promotes pathogen infection-induced myelopoiesis through mRNA oxidation. Nature. 2018; 554:123–127. [DOI] [PubMed] [Google Scholar]
- 31. Gagliardi M., Matarazzo M.R.. RIP: RNA immunoprecipitation. Methods Mol. Biol. 2016; 1480:73–86. [DOI] [PubMed] [Google Scholar]
- 32. Jia G.F., Fu Y., Zhao X., Dai Q., Zheng G.Q., Yang Y., Yi C.Q., Lindahl T., Pan T., Yang Y.G.et al.. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2012; 8:1008–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shevchenko A., Tomas H., Havlis J., Olsen J.V., Mann M.. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006; 1:2856–2860. [DOI] [PubMed] [Google Scholar]
- 34. Reid D.W., Shenolikar S., Nicchitta C.V.. Simple and inexpensive ribosome profiling analysis of mRNA translation. Methods. 2015; 91:69–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ingolia N.T., Brar G.A., Rouskin S., McGeachy A.M., Weissman J.S.. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 2012; 7:1534–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M.et al.. Topology of the human and mouse m(6)A RNA methylomes revealed by m(6)A-seq. Nature. 2012; 485:201–206. [DOI] [PubMed] [Google Scholar]
- 37. Canetti C., Aronoff D.M., Choe M., Flamand N., Wettlaufer S., Toews G.B., Chen G.H., Peters-Golden M.. Differential regulation by leukotrienes and calcium of Fc gamma receptor-induced phagocytosis and Syk activation in dendritic cells versus macrophages. J. Leukocyte Biol. 2006; 79:1234–1241. [DOI] [PubMed] [Google Scholar]
- 38. Labi V., Erlacher M., Kiessling S., Manzl C., Frenzel A., O’Reilly L., Strasser A., Villunger A.. Loss of the BH3-only protein Bmf impairs B cell homeostasis and accelerates gamma irradiation-induced thymic lymphoma development. J. Exp. Med. 2008; 205:641–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hao H.J., Hao S.J., Chen H.H., Chen Z., Zhang Y.F., Wang J., Wang H.Z., Zhang B., Qiu J.M., Deng F.et al.. N-6-methyladenosine modification and METTL3 modulate enterovirus 71 replication. Nucleic. Acids. Res. 2019; 47:362–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li H.B., Tong J.Y., Zhu S., Batista P.J., Duffy E.E., Zhao J., Bailis W., Cao G.C., Kroehling L., Chen Y.Y.et al.. m(6)A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature. 2017; 548:338–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hsu P.J., Shi H., He C.. Epitranscriptomic influences on development and disease. Genome Biol. 2017; 18:197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Han B., Yan S.J., Wei S.S., Xiang J., Liu K.L., Chen Z.H., Bai R.P., Sheng J.H., Xu Z.P., Gao X.W.. YTHDF1-mediated translation amplifies Wnt-driven intestinal stemness. EMBO Rep. 2020; 21:e49229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Han D., Liu J., Chen C., Dong L., Liu Y., Chang R., Huang X., Liu Y., Wang J., Dougherty U.et al.. Anti-tumour immunity controlled through mRNA m(6)A methylation and YTHDF1 in dendritic cells. Nature. 2019; 566:270–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rubio R.M., Depledge D.P., Bianco C., Thompson L., Mohr I.. RNA m(6)A modification enzymes shape innate responses to DNA by regulating interferon beta. Gene Dev. 2018; 32:1472–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. McIntyre A.B.R., Gokhale N.S., Cerchietti L., Jaffrey S.R., Horner S.M., Mason C.E.. Limits in the detection of m(6)A changes using MeRIP/m(6)A-seq. Sci Rep-Uk. 2020; 10:e49229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lin X.Y., Chai G.S., Wu Y.M., Li J.X., Chen F., Liu J.Z., Luo G.Z., Tauler J., Du J., Lin S.B.et al.. RNA m(6)A methylation regulates the epithelial mesenchymal transition of cancer cells and translation of Snail. Nat. Commun. 2019; 10:2065. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47. Mao Y.H., Dong L.M., Liu X.M., Guo J.Y., Ma H.H., Shen B., Qian S.B.. m(6)A in mRNA coding regions promotes translation via the RNA helicase-containing YTHDC2. Nat. Commun. 2019; 10:5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang S.C., Zhao B.S., Zhou A.D., Lin K.Y., Zheng S.P., Lu Z.K., Chen Y.H., Sulman E.P., Xie K.P., Bogler O.et al.. m(6)A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017; 31:591–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhu T.T., Roundtree I.A., Wang P., Wang X., Wang L., Sun C., Tian Y., Li J., He C., Xu Y.H.. Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine. Cell Res. 2014; 24:1493–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Xu C., Wang X., Liu K., Roundtree I.A., Tempel W., Li Y.J., Lu Z.K., He C., Min J.R.. Structural basis for selective binding of m(6)A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 2014; 10:927–929. [DOI] [PubMed] [Google Scholar]
- 51. Banroques J., Cordin O., Doere M., Linder P., Tanner N.K.. Analyses of the functional regions of DEAD-box RNA “Helicases” with deletion and chimera constructs tested in vivo and in vitro. J. Mol. Biol. 2011; 413:451–472. [DOI] [PubMed] [Google Scholar]
- 52. Fu T.Y., Wu C.N., Sie H.C., Cheng J.T., Lin Y.S., Liou H.H., Tseng Y.K., Shu C.W., Tsai K.W., Yen L.M.et al.. Subsite-specific association of DEAD box RNA helicase DDX60 with the development and prognosis of oral squamous cell carcinoma. Oncotarget. 2016; 7:85097–85108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zheng Q.L., Hou J., Zhou Y., Li Z.Y., Cao X.T.. The RNA helicase DDX46 inhibits innate immunity by entrapping m(6)A-demethylated antiviral transcripts in the nucleus. Nat. Immunol. 2017; 18:1094–1103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used and/or analyzed to support the findings of this study are available in the main paper or in the Supplementary Information. Any other raw data that support the findings of this study are available upon reasonable request from the corresponding author.