Abstract
Deoxyribonucleic acid (DNA) has evolved to be a naturally selected, robust biomacromolecule for gene information storage, and biological evolution and various diseases can find their origin in uncertainties in DNA-related processes (e.g. replication and expression). Recently, synthetic DNA has emerged as a compelling molecular media for digital data storage, and it is superior to the conventional electronic memory devices in theoretical retention time, power consumption, storage density, and so forth. However, uncertainties in the in vitro DNA synthesis and sequencing, along with its conjugation chemistry and preservation conditions can lead to severe errors and data loss, which limit its practical application. To maintain data integrity, complicated error correction algorithms and substantial data redundancy are usually required, which can significantly limit the efficiency and scale-up of the technology. Herein, we summarize the general procedures of the state-of-the-art DNA-based digital data storage methods (e.g. write, read, and preservation), highlighting the uncertainties involved in each step as well as potential approaches to correct them. We also discuss challenges yet to overcome and research trends in the promising field of DNA-based data storage.
INTRODUCTION
Modern society is characterized by advanced information technologies, generating increasingly vast amounts of digital data (1,2). Therefore, a variety of data storage techniques have been developed, including those based on magnetic media (e.g. tape, hard disk drive), electronic media (e.g. flash memory), and optical media (e.g. digital video disc, blue-ray disc). However, the amount of data that can be stored on the conventional media will very soon be exceeded by one that humans will produce, not to mention for most data media the retention time is only about 10 years. It is estimated that the data generated worldwide in 2025 will reach 175 zettabytes (≈1021 bytes), and then 3 yottabytes (≈1024 bytes) in 2040, which will greatly challenge the storage capability of our digital memory if a new medium is still not available at that time (https://www.seagate.com/our-story/data-age-2025/, (3,4)). Therefore, novel data storage media with high performance in data density, retention time, energy cost and read-write speed are urgently needed.
Since billions of years ago, DNA has been naturally selected as the storage medium of gene information which serves as the blueprint to construct and maintain the most intricate biological system in the world. The naturally selected DNA is also an extraordinary candidate as a new data storage medium featuring high volumetric density, long retention time, and low energy cost (5). As a promising biological molecular data storage media, the most significant advantage would be its unprecedented data density (i.e. theoretical density up to 455 EB g–1 (6)), which is approximately three orders of magnitude greater than that of the state-of-the-art flash memories (5,7,8). Although reading this amount of data at a high speed is still challenging even with the well-established second generation high-throughput sequencing (9), it can still serve as the complementary to traditional storage media when high density and long retention time are more critical than short access time (e.g. for information archiving and backup), not to mention the promising third generation single-molecule DNA sequencing technique is on its way with long read lengths and real-time data acquisition (10).
DNA is also a remarkably stable biomacromolecule that can be preserved for thousands of years if kept away from high humidity, irradiation, and air. The molecule chain can accommodate any base sequence and form a double helix structure with specific Watson–Crick base pairing. The base pairs are formed based on the weak and reversible hydrogen bonding, which enables duplication and transcription of the gene information with small amounts of energy consumption. Taking advantage of these unique features, enormous replicas of DNA can be accurately prepared in a short time starting from a single copy (e.g. the polymerase chain reaction, PCR) (11,12). The advances in biotechnology also facilitate facile DNA synthesis and editing of the DNA sequence. Therefore, writing, copy, and paste of data into the DNA molecule can be rather efficient in the near future.
In the past few years, many efforts have been devoted to encoding non-biological information in the synthetic DNA sequences for digital data storage (13–16). Since current DNA synthesis and sequencing techniques inevitably lead to various uncertainties in data storage, many researchers have been trying to figure out and deal with the data errors, which can be one of the major challenges for large-scale DNA-based data storage. Therefore, in this review, we first give an overview of the mainstream procedures for synthetic DNA-based data storage and then discuss the uncertainties involved in each step, highlighting the mechanisms and potential methods for error correction. Other than data writing and reading, we also summarize the immobilization and preservation of DNA that are also highly relevant to the robustness of DNA-based data storage. At last, the challenges and future research trends toward the large-scale application of DNA-based data storage are also discussed.
OVERVIEW OF DNA-BASED DATA STORAGE
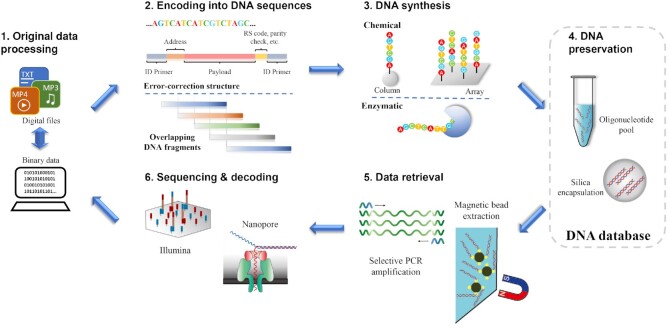
Akin to conventional electronic memory, the operation of the DNA-based data storage system generally involves five major steps: encoding, writing, preservation, retrieval and decoding, as illustrated in Figure 1.
Figure 1.
Schematic diagram of the DNA-based data storage and its operation. (1) The original data was transformed into binary data. (2) Binary data was encoded into corresponding DNA sequences. One usual strategy was to split the binary data into chunks, each of which was converted into a specific DNA base sequence. Additional base sequences including address information, error correction, and DNA amplification were also attached to the data sequence (16). Another approach was to split the data into chunks and transfer them to sequences having overlapping DNA fragments, which served as both address information and data redundancy for error correction (13). (3) All DNA strands with the specific sequences were synthesized, either chemically or enzymatically. The chemical synthesis included column-based and array-based phosphoramidite methods, while the enzymatical synthesis relied on polymerases such as template-independent terminal transferase. (4) The synthesized DNA samples were preserved in aqueous solutions or encapsulated in silica beads for preservation. (5) Data was retrieved by the selective extraction or amplification of relevant DNA strands in the pool. This could be achieved by using magnetic beads with specific ligands or polymerase chain reaction. (6) The relevant DNA sequences were obtained by a DNA sequencer, which was usually based on a sequencing-by-synthesis method or a nanopore. Then the DNA base sequences were decoded to obtain the original binary data.
Encoding
It is well known that the DNA sequence consists of four different bases (i.e. adenine (A), thymine (T), cytosine (C), guanine (G)) that are arranged in a specific order. The first step for DNA-based data storage is to transfer a binary data stream into quaternary DNA base sequences. Considering the sequence length of DNA that can be currently synthesized with relatively high fidelity, original binary data is usually broken into chunks and transferred into DNA sequences with lengths no more than two hundred nucleotides (nt). For accurate data recovery by the reassembly of these chunks, strategies based on data overlapping in adjacent chunks or adding addressable index in each chunk are often involved (13). Although the former is well established in gene assembly, the latter has proven to be more efficient for large databases (e.g. on the petabyte scale). Besides, error detection and correction algorithms, including those based on Fountain codes or Reed-Solomon codes (RS codes), are usually applied to deal with the errors in subsequent processes such as DNA synthesis and sequencing. After the encoding step, the DNA sequences are obtained for the next synthesis step.
Writing
Current nucleic acid synthesis methods are based on chemical or enzymatic methods. The chemical method usually relies on phosphoramidite chemistry developed by Caruthers and co-workers (17,18). To be specific, a nucleotide protected by a photolabile or acid-labile group at the 5′ end is deblocked under light or exposed to acid, then another nucleotide can react with the deblocked nucleotide, forming a phosphite internucleotide linkage. The array-based chemical synthesis, which can produce a large number of DNA strands in parallel, is usually preferred over column-based synthesis for DNA-based data storage. Enzymatic synthesis based on terminal deoxynucleotidyl transferase (TdT) has been of great interest owing to its advantages in speed, efficiency, and costs compared with the conventional methods (19,20), so it has been developed rapidly as a compelling candidate for DNA synthesis in data storage applications.
Preservation
Proper preservation condition is indispensable for the long-term integrity of DNA databases (21,22). It is well known that high humidity, light, and heat can induce irreversible damage to DNA molecules, leading to data errors or even complete loss of data. Meanwhile, in certain cases, DNA strands have to be immobilized on various carriers, so the linkage chemistry also directly determines the robustness of the system. Favourable preservation conditions in combination with proper immobilization promise the outstanding lifetime of DNA databases up to thousands of years.
Data retrieval
To selectively read certain data rather than sequentially read the whole database, specific chunks in the DNA pool need to be extracted and assembled, akin to random data access in the conventional electronic memories. However, unlike flash drivers or hard disks, locating the specific DNA strands with the desired data is difficult, especially for DNA that is freely dissolved in a solution. So base sequences for specific address information should be introduced in the encoding step. To retrieve the data, two approaches have been usually involved, which are based on DNA extraction (e.g. binding to a complementary probe immobilized on magnetic beads) and selective PCR amplification of required sequences using a specific primer, respectively. However, it is still very challenging to achieve random access for a very large database on the terabyte or even petabyte scale. The DNA sequence for address information will be prohibitively long, which can greatly affect the efficiency of encoding and accuracy of data locating (23).
Sequencing and decoding
Taking advantage of the high-throughput sequencing technique, the human genome sequence with a size of 3.2 gigabytes (GB) can be obtained within hours (24). The commercial DNA sequencers can be used to read enormous sequences in the DNA data storage system followed by using decoding algorithms to convert these sequences back to the original data. More compact third-generation single-molecule sequencers with greater reading lengths and ability of real-time sequencing (e.g. Oxford Nanopore Technology, ONT) can also be utilized in the future for the data reading (25), which can potentially reduce the reading time and the size of the DNA memory devices.
Note that the DNA-based database is essentially a chemical system governed by chemical thermodynamics and kinetics, so each step aforementioned, except the encoding and decoding steps, is error-prone like the conventional digital data storage system. Therefore, to establish a robust and efficient DNA database, uncertainties and their correcting methods should be concerned, as discussed in detail in the following sections.
UNCERTAINTIES IN DNA-BASED DATABASE
The origin of uncertainties in DNA-based data storage has been of great interest. It is believed that the main sources of errors are from DNA synthesis and sequencing. For example, Bornholt et al. (26) found that phosphoramidite-based synthesis and Illumina sequencing led to an error rate of about 1% in the final consequence (i.e. one error out of 100 bases), and the majority of the errors were from the sequencing process. A prior work from Organick et al. (14) reported a higher error rate of up to ∼10% from nanopore sequencing. To avoid data corruption, advanced error detection and correction algorithms are requisite for robust DNA-based data storage. In this section, uncertainties, their origins, and approaches for error correction are discussed. Key parameters in recent works are summarized in Table 1.
Table 1.
Summary of current works on DNA-based data storage. The table is presented chronologically with the earliest on top. The encoding techniques are illustrated in detail in Figure 2. The error correction methods refer to the approaches shown in Figure 3. Synthesis technologies include the phosphoramidite-based chemical synthesis and the emerging enzyme-based synthesis. Random access indicates the ability to acquire specific pieces of data from the whole data pool of the DNA storage system. Sequencing technologies include Illumina's high-throughput technology based on the sequencing-by-synthesis principle, and ONT’s nanopore technology with real-time data reading. The information density denotes the average number of binary information (bits) encoded in each nucleotide (nt). The binary information includes the data payload and the auxiliary sequences for error-correction, primers, and so forth. Table 1 is adapted from reference (14,16)
| Coding features | Data size | Error correction method | Synthesis technology | Random access | Sequencing technology | Coverage | Information density (bits/nt) | |
|---|---|---|---|---|---|---|---|---|
| Church et al. (15) | Conventional | 650KB | Repetition | Phosphoramidite (Agilent) | No | Illumina | 3000× | 0.60 |
| Goldman et al. (13) | Conventional | 750KB | Repetition (Overlapped segments) | Phosphoramidite (Agilent) | No | Illumina | 51× | 0.19 |
| Grass et al. (108) | Conventional | 80KB | RS codes | Phosphoramidite (CustomArray) | No | Illumina | 372× | 0.86 |
| Bornholt et al. (26) | Conventional | 150KB | Repetition (XOR operation) | Phosphoramidite | Yes | Illumina | 40× | 0.57 |
| Blawat et al. (109) | Conventional | 22MB | RS codes | Phosphoramidite (Agilent) | No | Illumina | 160× | 0.89 |
| Yazdi et al. (107) | Conventional | 3KB | Consensus estimation | Phosphoramidite (IDT) | Yes | Nanopore | 200× | 1.71 |
| Erlich and Zirlinsky (16) | Conventional | 2.15MB | Fountain codes | Phosphoramidite (Twist) | No | Illumina | 10.5× | 1.18 |
| Organick et al. (14) | Conventional | 200.2MB/32KB | Repetition (XOR operation) & RS codes | Phosphoramidite (Twist) | Yes | Illumina / Nanopore | 5×/36× | 0.81 |
| Lopez et al. (77) | Conventional | 1.67MB | RS codes & assembly | Phosphoramidite (Twist) | Yes | Nanopore | 43× | 0.84 |
| Tomek et al. (63) | Conventional | 94.3KB | Repetition (XOR operation) | Phosphoramidite | Yes | Illumina | 10× | 0.66 |
| Organick et al. (6) | Conventional | 19.8KB | Logical redundancy | Phosphoramidite | Yes | Illumina | 35× | 0.003 |
| Anavy et al. (53) | Degenerate bases | 8.5MB | RS codes | Phosphoramidite (Twist) | No | Illumina | 164× | 1.94 |
| Choi et al. (54) | Degenerate bases | 854 bytes | RS codes | Phosphoramidite (column) | No | Illumina | 250× | 1.78 |
| Lee et al. (61) | Transition coding | 18 bytes | Synchronization nucleotides | Enzymatic (column) | No | Nanopore | 175× | 1.57 |
Uncertainties in DNA synthesis
One of the key steps in DNA storage is to synthesize the DNA strands having data-encoded sequences. Current DNA synthesis can be realized by chemical and enzymatic methods. As early as the 1970s, Caruthers et al. reported the phosphoramidite-based oligonucleotide synthesis, which has been widely used for decades (17,18,27,28). In a typical synthesis, nucleotide monomers with hydroxyl groups at the 5′ terminal were blocked with a protecting group (e.g. dimethoxytrityl (DMT) group). The protecting group was labile, which could be detached under certain conditions (e.g. acid, light). Then the deblocked nucleotide was exposed to the next monomer for coupling. To generate a specific sequence, only one type of nucleotide was introduced to the reaction pool at a time for the coupling reaction, which was repeated until a DNA molecule of hundreds of nucleotides was obtained. After the introduction of each nucleotide into the DNA strand, the phosphite linkage was further converted to a more stable phosphate bond using moderate oxidant.
The chemical synthesis was usually accomplished on a glass substrate with controlled pores (29,30). Initial nucleotides were seeded on the glass via linking to the surface hydroxyl groups. The chemical reactions were accomplished by sequentially introducing the oxidizing agent, deblocking acid, and monomer solution, respectively, onto the substrate. These steps were repeated to synthesize DNA molecules with specific sequences. The synthesis could also be fully automated using a programmable microfluidic system to save time and reduce costs (31,32). After decades of development, various methods for the chemical synthesis of DNA have been reported, including those based on ink-jet printing (33,34), photolithography (35–37) and electrochemistry (38–40). The major difference between these methods is how the nucleotides are deblocked/activated. For example, Chow et al. reported the chemical DNA synthesis on a Ti array that was achieved on an undoped α-Si substrate. When the back of the substrate was exposed to light, the Ti array generated protons to trigger acidic deblocking of the nucleotide (41). Affymetrix reported another chemical DNA synthesis method based on the standard photolithography using phosphoramidite derivatives that were unstable to the light with a specific wavelength (42,43). In addition, the electrochemical microelectrode array has also been used for DNA synthesis, which is a competitive method because of its low cost and portability. The activation was usually induced by protons from an electrochemical reaction (44,45). For the construction of the DNA databases, a diverse group of DNA strands having different sequences needs to be synthesized in parallel. So, the high-throughput array-based synthesis is usually preferred over the conventional column-based synthesis.
Unfortunately, current chemical synthesis methods still have drawbacks such as low yield and synthetic errors (average error rate ∼0.7%) owing to substitution (0.5%), insertion (0.1%) and deletion (0.1%) (46,47). For instance, owing to incomplete coupling reaction, part of active sites remains on the substrate, which can subsequently react with the next nucleotide, leading to an erroneous insertion of a base. Likewise, incomplete deblocking of the protecting groups results in deletion errors. The residual acidic deprotection reagents may cause depurination or even cleavage of the backbone (48,49). Other problems can be associated with paralleled array-based synthesis. For example, protons produced at the reaction sites are likely to diffuse to the proximity, causing significant cross-talks between reaction spots, which is also called the edge effect (50). Therefore, array-based DNA synthesis possibly increases the error frequency and offsets the advantage of high density. Additionally, with the increasing length of the synthesized DNA, the uncertainty increased dramatically. For example, if the accuracy for adding one nucleotide is about 99.5%, the final accuracy for the synthesis of the whole DNA strand of 200 nt is only 36.7%. (0.995200 = 0.367).
Various methods have been proposed to solve these problems. For example, the remaining reaction sites can be deactivated by acetic anhydride to prevent insertion errors (30,51). LeProst et al. reported that unwanted depurination resulted from residual detritylation solution could be avoided by using relatively moderate reagents and by optimizing fluid introduction and washing steps (50). Error correction reaction (ECR) of the synthesized oligonucleotides via surveyor DNA endonuclease was also reported by Saaem et al. (52). In ECR, DNA with the mismatch was located and excised by the endonucleases, and then the fragments were reassembled by overlap extension PCR (OE-PCR) to obtain the correct strands.
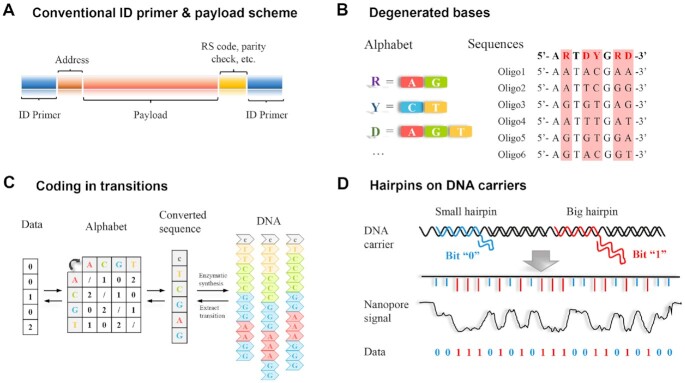
To address the problem from errors in the chemical DNA synthesis and pursue a high DNA storage density, composite encoding relied on base combinations have been previously reported, as an improvement based on conventional encoding structure (53,54). For the traditional coding framework (Figure 2A), four kinds of nucleotides at one site were capable to store 2 bits of data (log24 = 2 bits). In the degenerated base encoding method (Figure 2B), data was encoded not by specific type of nucleotides (A, T, C, G), but by their combinations/ratios. For example, the letter R could be encoded as equal amounts of A and G at the same site on different DNA strands (R = 50% A + 50% G), and the letter D could be encoded as equal amounts of A, G and T at the same site (D = 33% A + 33% G + 33% T) on different DNA strands. So, part of random errors could be eliminated during the data reading step. Note that, to obtain the statistic base ratio, each sequence must be of sufficient physical redundancy. For example, at least reading of several DNA strands at one site could determine whether R (A: G = 1:1) or D (A: G: T = 1:1:1) was encoded. The demanded redundancy was usually acquired in the DNA synthesis step and the PCR step. Also, increased sequencing depth and corresponding statistical algorithms were required to ensure accurate data reading. Anavy et al. put logical redundancy into sequences via fountain codes and introduced extra composite bases in the encoding (53). Apart from four basic nucleotides, extra composite bases K (K = 50% A + 50% C) and M (M = 50% G + 50% T) were also used to encode a file of 2.12 megabytes (MB). In this way, the storage density could be improved by 24% compared with Erlich's previous work (16). Furthermore, Choi et al. (54) utilized an alphabet consisting of 11 additional composite bases to encode 854 kilobytes (KB) files, which greatly shortened the DNA length and led to a dramatical leap of theoretical data coding density up to 3.9 bits per character (log215 ≈ 3.9 bits). The design of composite bases could store more data in a specific length but it required a greater number of DNA copies and sequencing depth for data reading. In other words, the application of composite bases was equivalent to reduce synthesis cost at the expense of increasing sequencing overhead. But under ideal conditions, the overall expected cost would be reduced greatly (∼52% as estimated, (53)).
Figure 2.
Encoding for DNA-based data storage. (A) Schematics of a DNA strand with the encoded address information, error-correction information (RS codes), data (payload), and adapters (ID primer). The address represented the relative location of the payload in the original data, which was required for data reassembly. The error-correction segment contained additional information that ensured error-free data recovery. Adapter sequences at both ends provided binding sites of complementary primers for PCR amplification and random access of certain data (16). (B) Schematics of the composite encoding strategy based on degenerated bases. Instead of directly transforming each binary data into four bases, the information was encoded by the ratio of A, T, C, G at the same sites on different DNA strands. For example, the letter R could be represented by A and G in a 1:1 ratio. This composite strategy improved the theoretical information density for DNA-based storage (54). (C) Schematics of an encoding method based on transitions of bases (e.g. C to G), which was applied in enzymatic synthesis having low synthesis accuracy but high speed. The data was represented by the transition of one nucleotide to another in the sequence. For instance, the transition of bases from C to G represented ‘1’ and A to T represented ‘2’ (61). (D) Schematics of an encoding method for nanopore-based DNA database. The data was encoded by oligonucleotide hairpins of two lengths so that two types of electronic signals could be recorded when they penetrated the nanopore. By monitoring the current signals during the DNA passed the nanopore, binary data could be obtained (80).
The emerging enzyme-based DNA synthesis is accomplished in aqueous solutions, which is more biocompatible than the organic solutions involved in the chemical synthesis. The length of the DNA strand that can be enzymatically synthesized was estimated to be ∼8000 nt, which is greater than that of the chemical approach (55–57). For the enzymatic synthesis, deoxynucleoside triphosphates (dNTPs) were coupled consecutively with the help of template-independent DNA polymerase, such as TdT. Since TdT could lead to random coupling of any type of dNTP in the solution, the controlled addition of a specific dNTP was crucial for the enzymatic synthesis, similar to the chemical methods. Besides, the blocking strategy for chemical synthesis could also be utilized by the enzymatic synthesis. The 3′ terminal of the nucleotide monomers could be blocked while the 5′ terminal was associated with triphosphate for the chain extension. The blocking group was removed under certain conditions to expose the hydroxyl group for the downstream reaction (58,59).
Palluk et al. (60) reported the use of a TdT-dNTP complex linked by a covalent disulfide bond for the enzymatic DNA synthesis. When the complex was coupled with the terminal of the DNA strand, the dNTP could not be detached from the TdT, thus other dNTPs or complexes could not react with it and the reaction stopped. When the disulfide bond between TdT and dNTP was cleaved by dithiothreitol (DTT), the DNA strand was ready for the next TdT-dNTP complex addition to the terminal. The averaged stepwise yield was estimated to be 97.7% from a total of 4861 reads based on the comparison between the sequencing results and the target sequence. Alternatively, Lee et al. (61) combined enzymatic synthesis with nanopore sequencing for DNA-based data storage. Different from the traditional encoding method, the data was encoded using a specific transition of one nucleotide to another in the sequence, as illustrated in Figure 2C. For instance, the transition of bases from C to G represented ‘1’ and that from A to T represented ‘2’. A competitive mechanism, in which the TdTs assisted the coupling of dNTPs with the strands, while apyrase served to inhibit this process, was used to controllably add several nucleotides to the DNA strand. Synchronous nucleotides were also included in the sequences to serve as fixed ‘spots’ between the information-encoding nucleotides, which facilitated the comparison between sequences, reconstruction as well as subsequent error correction. Despite the uncertainties in enzymatic synthesis and nanopore sequencing, this layout with the synchronous nucleotides enabled the reconstruction of template sequences. Although they contained all kinds of erroneous nucleotides, a statistical model could be used to evaluate the error probability and all the reconstructed sequences were scored. The final DNA sequences were obtained based on majority voting, in which the most probable nucleotide at each site was determined according to the statistics.
Current enzymatic synthesis is still less well-established than the chemical method, but it is a promising candidate for DNA-based data storage because of the use of less toxic reagents, mild reaction conditions, and a higher reaction rate. Moreover, longer DNA strands can be obtained using the enzymatic method, leading to a decrease of overheads for both address information and PCR primer in one strand, and consequently a remarkable increase in data density. Research in this area is now focused on optimizing the enzymatic DNA synthesis to further reduce errors.
Uncertainties in data retrieval
Random access has been well established in the computer-based data system to selectively read data from the whole database. Random access enhances data reading efficiency by eliminating unnecessary reading tasks. However, selective retrieval of data from a large DNA database is still challenging, especially when the DNA molecules are dissolved in an aqueous solution or preserved as a lyophilized powder with no fixed physical location for direct access. Several approaches, including physical separation and PCR amplification, have been usually employed to specifically collect the DNA strands for random access of data. For the physical separation, magnetic beads carrying complementary probes or chemical labels were introduced into the pool, then the target DNA strands bound to the probes or labels and were selectively separated by a magnet. Another method was to store DNA as isolated partitions (solved in individual droplets or dehydrated as powder spots). For the PCR amplification, the target DNA strands were selectively hybridized with primers and then amplified to a predominant amount by PCR.
In a large-scale DNA database, physical partitioning of the whole database is an efficient way, considering both storage scale and random accessibility. In other words, instead of managing all data in one bulky mixed pool, dividing them into several sub-pools and retrieving data can avoid the use of a long sequence occupied by address information. Newman et al. (62) recently reported the use of digital microfluidics for DNA data retrieval. Dehydrated DNA was stored in sub-pools on a glass plate, which was interfaced with a digital microfluidic chip. When a file was requested, a droplet of an aqueous solution was moved to the target site via electrowetting enabled by the embedded electrode array on the chip. DNA was then dissolved in the aqueous droplet and transported for subsequent data reading. Taking advantage of the high-throughput digital microfluidics, the proposed system was promising to build automatic large-scale DNA databases. To conclude, the physical distribution of several sub-pools addressed the adversity of the crowded molecular composition in a highly dense single DNA database and improved the practical scalability of the DNA storage system. However, physical separation decreased the storage density since isolated partitions occupied more volume than a mixed large database. Besides, more duplication was needed to eliminate retrieval failure due to insufficient DNA redissolution or low separation efficiency (e.g. 9× extra redundancy applied in the work of Newman et al. (62)).
For traditional PCR-based random access, its ability to acquire a specific file is limited to terabyte-scale in a single pool (14). It can be interpreted that the retrieval efficiency monotonically decreases with the increasing amount of background DNA because the accurate random access of data, which highly depends on proper hybridization of the probe DNA or PCR primer, can be affected by the nontarget strands. Therefore, many efforts have been devoted to improving the orthogonality and new coding structure as data addresses (63–67). For example, Yamamoto et al. (65) theoretically designed a file retrieval system based on nested PCR. The concatenated nested PCR of three layers resulted in 10 million individual addresses for data storage. When a certain file was requested, the system searched for the target sequences layer by layer using cascaded PCR. In addition, the orthogonality of primers was an important concern for successful file extraction, which has been designed and optimized in previous works (14,26). Recently, Song et al. (67) proposed a hybrid nested and semi-nested PCR address structure based on the previous work. A virtual 4-dimensional address structure was created, in which the DNA encoded with data was assigned to an address including row, column, higher layer, and block-level information. For the random access of data, forward/reverse primer pairs from the same or different address layers were leveraged to virtually locate and retrieve data in the form of rows, columns, layers, and blocks. For example, the forward and reverse primers close to the data payload indicated the data location in the columns and rows, respectively. An additional pair of primers indicated the data location in the other two dimensions (i.e. layer and block). The virtual data structure enabled multiple random-access modes and reduced the number of primers requested from a very large address space. Theoretically, k*n pairs of orthogonal primers were capable of covering nk addresses, where k referred to the number of virtual dimensions and n represented the number of data sequences stored in each dimension. Yazdi et al. (64) proposed several constraints for designing PCR primers with high orthogonality. Large mutual Hamming distance and irrelevance of the addresses were beneficial to construct a robust address system. The former reduced the probability of disordered address selection, while the latter prevented the prefixes of one address from acting as suffixes of the same or another address to minimize the incorrect hybridization of primer pairs. Based on these principles, the primers designed were highly reliable for random access to the data. Tomek et al. (63) first reported the experimental combination of physical file separation based on magnetic extraction and nested PCR structure, which led to selective retrieval of 9.15KB of data from a database as large as 5 terabytes. The required sequences were modified with chemical labels via emulsion PCR prior to the magnetic extraction. The combined approach retrieved little unrelated DNA strands so that high sequencing efficiency and accuracy were achieved compared with that by PCR amplification alone.
However, there are also several limitations with PCR-based data retrieval. Heckel et al. (47) reported that the initial physical abundance was not consistent for different DNA strands due to the natural deviation in synthesis. The difference in the initial copy numbers of these DNA strands would be further magnified by PCR. Furthermore, the affinity of DNA polymerases to each primer is slightly different probably due to secondary structure formation (68,69). Commercial DNA polymerases exhibit a bias on amplification of sequences with high GC content, which significantly change the coverage and distribution of the DNA strands after cycles of amplification (70). Ideally, in each PCR cycle, sequences are amplified by a factor of two. But the actual amplification factor can be slightly below two, which also differs between sequences (PCR bias). These features lead to issues in the DNA-based database. For example, if the factor for PCR amplification of sequence A is 1.8, while the factor of sequence B is 1.9, after 60 PCR cycles, the ratio of copy numbers between these two sequences will be (1.8/1.9)60 = 0.039. Therefore, multiple PCR cycles can result in remarkably unbalanced physical redundancy for different DNA strands, leading to a biased data recovery or even complete data loss.
For these problems associated with PCR-based random access of data, there are two general solutions: (i) ensure that every sequence has sufficient and similar physical abundance in the synthesis step and reduce the number of PCR cycles appropriately to minimize PCR bias; (ii) choose orthogonal PCR primers with large affinity differences, and improve the efficiency of primers from the kinetic perspective. In other words, the more the primers are different, the less the non-specific and erroneous amplification will usually expect (71,72).
Uncertainties in DNA sequencing
Reading digital information stored in DNA strands is started from DNA sequencing. For this purpose, high-throughput second-generation sequencing (e.g. Illumina, Roche) or real-time third-generation single-molecule sequencing (e.g. Pacific Biosciences, ONT) is often involved. For instance, Illumina sequencing relies on the principle of sequencing by synthesis (73,74). DNA fragments to be sequenced were first linked with adapters at both ends and then hybridized with primers in the flow cell. Distal ends of the DNA interacted with other nearby primers to form a ‘bridge’ structure. These DNA strands subsequently underwent several rounds of ‘bridge’ amplification to generate multiple individual clusters of DNA copies. Then the sequencing step started, in which four types of fluorophore-labelled and 3′ blocked dideoxynucleotides consecutively coupled with the anchored strands, respectively. After incorporation of each dNTP to the DNA strands, the unbound dNTP was washed and the fluorescence in the flow cell was imaged to identify where this type of dNTP was coupled at the cluster. Different dNTPs were characterized by their corresponding fluorophore labels. After the fluorescent imaging, the fluorophore and blocking groups were cleaved to initiate the next cycle.
Nanopore-based technology enables real-time single-molecule DNA sequencing (10,75,76). In this technique, solid-state (e.g. graphene) or biological nanopores such as α-hemolysin, MspA porin from Mycobacterium smegmatis, were sandwiched by two separate chambers filled with ionic solution. When a constant voltage bias was applied, an ionic current was generated through the nanopore. The DNA was driven to pass through the nanopore towards the other chamber. An enzyme (e.g. a polymerase or a helicase) could combine with the DNA strand tightly and ratcheted the DNA through the pore step by step. The narrowest part of the nanopore (i.e. the sensing region) was sensitive to the type of nucleotide passing through, which led to different levels of blockades on the ionic current. So, the DNA sequence could be inferred by analysing the profile of the current signals.
Second-generation sequencing has shown its higher throughput and accuracy, while third-generation sequencing exhibits promising features of long read lengths and real-time readouts. Besides, the readout for nanopore-based sequencing relies on relatively simple instrumentation, such as an integrated circuit for small current measurement. So, it can be as small as a memory stick (USB). The apparatus for the second-generation sequence is relatively bulky, which usually involves sophisticated fluidic and optical systems. For the second-generation Illumina sequencing, continuous fluorescence images need to be taken and analysed for sequence information. On the other hand, for the nanopore sequencing, only the current signals produced by the DNA translocation are recorded so that the DNA sequences can be obtained in real-time. The real-time reading ability makes it superior for digital data storage. Besides, when only a few files are requested by assessing a small part of the database, nanopore sequencing is also more practical and economical than high-throughput second-generation sequencing.
For instance, Lopez et al. (77) reported a method for assembly of multiple DNA fragments into longer DNA strands using OE-PCR. The spliced DNA strands with overlapping primers were sequenced using a MinION nanopore sequencer from ONT. Random access for demanded data relied on multiple sequence alignments of file addresses. Data payloads with an identical address were collected and sorted for a consensus to reconstruct the original information. In their work, small fragments with the length of hundreds of nucleotides were integrated into long DNA strands, which could readily be sequenced based on nanopore technology. It is worth mentioning that the consensus algorithm was modified. The mismatched sequences were not immediately discarded, but labelled as out of sync and further searched for possible alignments in subsequent steps. Therefore, fewer coverages (<30× for three files) were required for proper decoding.
The reading length of nanopore-based DNA sequencing can be a hundred kbps (a thousand base pairs) without sophisticated sample pre-treatments (e.g. ONT MinION platform), which is much greater than that of the second-generation sequencing. Taking advantage of this, nanopore-based DNA sequencing has also promoted the development of new data encoding strategies for the DNA databases (78,79). Chen et al. (80) attached short DNA hairpins to double-strand DNA with different stem lengths (Figure 2D). When such a structured DNA translocated the nanopore, the hairpins blocked the nanopore and generated a secondary current decrease corresponding to the hairpin length. In their experiments, the 8 and 16 bp represented bit ‘0’ and ‘1’, respectively. After optimization, data of 56 bits was encoded in a DNA carrier of 7228 nt. Recently, they reported the data rewriting, deleting, and encrypting via strand displacement reaction (81). Although the efficiency of this encoding method was relatively low, the data writing through direct strand hybridization was simple, and the reading using highly portable nanopore devices avoided the use of complex instruments or vulnerable enzymes. The data storage was based on larger structures rather than single bases, which increased the signal-to-noise ratio but at the expense of decreased data density. Automated data storage devices by a combination of chemical synthesis and nanopore sequencing have been previously reported (82).
However, errors are also inevitable in DNA sequencing. For most sequencing technologies, high GC content and long homopolymers lead to sequencing errors. To be specific, the probability of insertion and deletion errors rise significantly for homopolymers consisting of more than six repeated nucleotides, especially for polyG because they tend to form guanine tetraplex structures (83). These structures are of high thermal stability, thus difficult to sequence. Additionally, sequencing coverage of DNA fragments with GC content <20% or >75% is much lower in conventional sequencing methods (84–86). At optimal conditions, Illumina sequencing accounts for <1% of errors in the retrieved data (26,87,88). Any DNA strand with a broken primer sequence, which can result from DNA damage or erroneous DNA synthesis, also cannot be read properly either for most second-generation sequencing.
For the nanopore sequencing, current fluctuation, noises, nanopore defects, and uncontrolled DNA translocation speed may result in interference and subsequent base-calling failure (89,90). For example, the sensing region of α-hemolysin is relatively large, thus the ion current can be affected by multiple nucleotides simultaneously. So, convoluted signal profiles are obtained, and difficult to correlate with a specific base (91). Besides, the signals from biological protein nanopores are highly sensitive to chemical conditions such as pH, temperature, and ionic strength, which complicate the interpretation of data and reduce reproducibility (92). As for the solid-state nanopores, despite they have a comparable dimension with the size of the DNA nucleotide, unwanted interactions with DNA nucleotides like adsorption on the substrate affect the translocation of DNA strands through the nanopore (93). Furthermore, compared with biological nanopores, the solid-state nanopores are less sensitive to different nucleotides, leading to low specificity for base recognition (94,95). Therefore, despite many advantages of nanopore sequencing, it still suffers from a relatively high error rate (∼10%) even under optimal conditions (14,96). Extensive error correction and signal analysis algorithms are highly required to improve the accuracy for practical applications.
Therefore, to eliminate the mistakes and recover loss during the DNA sequencing, the DNA strands in the database should include an appropriate amount of data redundancy. Besides, enhancing sequencing coverage and depth is an effective way widely used to ensure correct data retrieval, despite the expense of high costs. Signal processing is also of great importance for nanopore-based sequencing and accurate data recovery. Approaches based on hidden Markov model (HMM) (97–100), deep neural network (Clair (101)), likelihood alignment (Samtools/Bcftools package (102)) and so forth have been reported for the sequencing data processing (103–106). For example, in the HMM-based method, the signal of six nucleotides was treated as an event. A series of emission distribution functions were used so that the event could be correlated to the corresponding identities of the 6 nucleotides. After repeated training, the model could be used to obtain an unknown base sequence from an electric signal.
Error corrections by encoding and decoding
Error correction is one of the crucial functions of the data encoding and decoding steps. Error-correction codes (ECCs) are initially utilized in error-free information transmission and communication technology. A general idea for error correction is adding data redundancy to original data so that the errors can be corrected based on a statistical voting principle. Many efforts have been devoted to using substantial data redundancy to achieve acceptable error tolerance (26,107–109). Data redundancy can be categorized into two types: physical redundancy and logical redundancy. Physical redundancy means the presence of multiple copies of identical DNA segments, akin to backups of computer files. Note that, the physical redundancy generated either in the synthesis or in the PCR amplification process is not sufficient to eliminate all potential errors in the DNA database. Church et al. (15) reported that even with abundant physical redundancy added in their encoding process and high sequencing depth (3000×), manual intervention and error correction algorithm were still required owing to inevitable mistakes. Therefore, considering both efficiency and robustness, logical redundancy is usually preferred. For the logical redundancy, extra information was added into the DNA sequences apart from the original data. Usually, the encoding algorithm converted binary data into DNA sequences, which were then split into many pieces (payload). Then, a piece of sequence for error correction, an index sequence indicating the relative location of the data (address), and two terminal sequences that served as the PCR primers were linked with the payload to form a complete DNA strand in the data pool. Such an encoding framework has been proven to exhibit ideal storage capacity (110). Note that, high GC contents, homopolymers, or self-complementary sequences may lead to low synthesis yields and difficulty in sequencing, and they should be avoided in the DNA sequence.
For the data encoding with the logical redundancy, various approaches including those based on Huffman codes (111), RS codes (112,113), logical exclusive-or (XOR) operation (14,26) and fountain codes (114,115) have been reported. Huffman codes are a kind of variable length code to compress data. In Huffman encoding, each symbol in data was sorted by frequency in increasing order and regarded as several nodes. Then, two nodes (child) with the lowest frequency (e.g. 0.02 and 0.1) were combined as a new node (parent) with a summated frequency (0.02 + 0.1 = 0.12). Two steps above were iterated until all the nodes were assembled to generate a binary tree of nodes, where each node had two directions (left as ‘0’ and right as ‘1’) and was assigned with a unique codeword determined by the path from the root to the node. In this way, more common symbols were represented using fewer bits than less common symbols, so that the length of data was reduced.
RS codes can detect and correct multiple errors. In RS encoding, the data matrix was multiplied with a pre-calculated encoding matrix, and the redundant information would be added as new rows or columns of the result. The added rows or columns of the matrix aided to detect errors in the message and correct them if the number of errors did not exceed the correction capability (Figure 3A).
Figure 3.
Error correction methods for the DNA database. (A) Reed-Solomon codes multiplied the data block with a pre-calculated encoding matrix, and the redundant information would be added at the end of the result (108). (B) Fountain codes separated data into many segments, selected them using a special distribution function and packaged them into many ‘droplets’. Unqualified sequences were excluded from the screening procedure. Droplets of good quality were used for oligonucleotide synthesis (16). (C) Overlapping was a simple but effective method to avoid errors. Repeated pieces of the sequence under a shifting pattern were included in different oligonucleotide strands to obtain available copies (For example, four-fold redundancy generated in the figure) (13). (D) Exclusive-or operations between any two information strands generated the third strand. Any two of them could restore the remaining strand (26).
Fountain codes are a kind of codeless erasure code, in which the original data can be recovered from any subset of the encoding data which is equal to or slightly larger than the number of the original information. In the encoding process, binary data was divided into multiple non-overlapping segments of a certain length. Then, Luby transformation selected a random subset of segments according to a special distribution function, applied bitwise operation to add them up under a binary field, and wrapped them up into desired numbers of small fragments called droplets. For further recovery, each of them was assigned with seeds, which was used to generate pseudorandom numbers related to the previous transformation (Figure 3B).
As a popular ECC, RS codes are widely used for information encoding in optical disks, quick response (QR) codes, and other storage media. In previous works, RS codes have also been applied in synthetic DNA-based databases (14,77,108,109,116). For example, Grass et al. (108) used inner and outer RS encoding in the 2D data matrix to eliminate single-base errors or sequence loss. It was calculated that inner RS codes could correct ∼0.7 errors per 100 bases, and the outer codes rectified the loss of 0.7% further in the raw result. The cascaded encoding and in silica encapsulation strategy exhibited good robustness as demonstrated in an aging experiment, in which the DNA samples were preserved at a high temperature to accelerate the generation of errors and data loss. Based on the estimation using the Arrhenius Equation, the robust DNA data system based on the hybrid RS codes and silica encapsulation was effective for error-free preservation of DNA-based data for 1000 years in Zurich (∼ 9.4°C), or over 2 million years at the Global Seed Vault (-18°C). In another work by Organick and co-workers (14), the unprecedented 200 MB data of audio, video, and text files were stored in DNA using the RS codes. With selective PCR amplification and the Illumina sequencing, only a sequencing depth of 5× was sufficient for high-quality data retrieval without error. For nanopore sequencing with a high error rate, increased sequencing depth (e.g. 36× or 80× according to the size of files) and extracting information from all sequences were critical for promoting data recovery using corresponding statistical algorithms.
Blawat et al. (109) developed an encoding method based on the RS codes for storing 22MB files in DNA. Every eight bits in the original data were transformed into five successive bases in the encoded DNA sequences. Such an encoding approach reduced the possibility of self-complementation in DNA sequences. Furthermore, robust RS codes and the additional parity-check process were collaborated to encode data in synthesized oligonucleotides. The raw readout from sequencing underwent a series of decoding steps including majority voting for the most possible sequences, incomplete oligonucleotide reconstruction, and RS decoding. The possibility of residual errors in the decoded data was relatively low, which was comparable with modern hard disk drivers (HDDs), or even less if more data redundancy was included in the encoding step.
In addition to the RS codes, other encoding methods used in information technology have also been applied in the DNA-based database. Based on fountain codes, Erlich et al. (16) reported the theoretically highest physical density for the data storage. To eliminate unqualified droplets for synthesis and sequencing, an additional screening step was designed to rule out those sequences which consisted of homopolymers or high GC base content. Considering the restriction of synthesis and sequencing, unqualified droplets with high GC contents and homopolymers were abandoned in the screening step. Finally, a file with the size of 2.14MB was converted to 72 000 strands of oligonucleotides for storage. Then the data was recovered with no mistake, even 2000 of the 72 000 DNA strands were lost after the whole storage process. Furthermore, with PCR amplification and dilution, researchers managed to retrieve the data from only 10 pg of DNA. Therefore, the physical density was astonishingly as large as 215PB/g.
Depending on the type of errors during DNA synthesis, a specific coding method can be more appropriate. RS codes are more efficient in rectifying substitution errors, while the fountain codes are preferred to deal with deletion or addition errors (117,118). RS codes can effectively detect and correct substitution errors when the length of DNA is unaltered (no addition or deletion errors). On the other hand, when part of the DNA pool is lost or other sequences are added, fountain codes may be the better option. Erlich et al. reported that the combination of these two codes could provide better results (16).
Other than fountain codes, other concepts were also applied in recent studies for error correction. Goldman et al. (13) used Huffman codes to transform each byte in original files into several ternary digits to eliminate homopolymers. The resulting information was divided into overlapping segments under a shifting pattern, as demonstrated in Figure 3C. For a certain segment, in addition to itself, encoded information could be identified from the other three ‘backup’ sequences, resulting in a fourfold redundancy. Segments were designed as reverse complement alternatively, which reduced the probability of data loss and failed recovery. Bornholt et al. (26) also reported a new way to add data redundancy. To be specific, two original strands produced a new strand via an XOR operation so that any two out of the three DNA strands were sufficient to recover the original data (Figure 3D). This method reduced the overhead, and the data redundancy could be flexibly tuned by the number of replicated XOR operation. Multiple XOR operations between strands provided higher redundancy and vice versa.
Adding error correction information and data redundancy in the encoding step is indispensable for the accurate recovery of data in DNA. Various encoding methods such as RS codes, fountain codes, Huffman codes, and XOR operation are introduced in this section. The choice of the most appropriate coding and their combination for a specific project highly depends on the application scenario and the type of data. Furthermore, there is always a trade-off between data redundancy for error correction and coding density (6), so the reduction of error rate from the origin (e.g. DNA synthesis and sequencing) is much preferred from this respect.
DNA IMMOBILIZATION AND PRESERVATION
DNA is a naturally selected molecule storing our essential gene information which can inherit from generation to generation. It serves as the blueprint to construct all kinds of biological creatures, and maintain homeostasis despite the environmental changes. Although DNA can be degraded when exposed to air, high humidity, or radiation, the half-life of DNA can readily exceed thousands of years when preserved under suitable conditions (119–121). For example, the complete genome of a Neanderthal from about 50 000 years ago was successfully obtained and sequenced (122). The sequences of the 16S ribosomal DNA identified as a spore-forming bacterium belonging to Bacillus species were acquired from a 250 million-year-old salt crystal from the Permian Salado Formation (123). Taking advantage of the long retention time of DNA, long-term archival storage of molecular digital data can be accomplished. To achieve the long-term stability of DNA, several critical factors need to be considered, such as the DNA immobilization and the preservation conditions, which we will discuss in this section.
DNA immobilization
A vital step in DNA-based data storage is the attachment of DNA molecules to a substrate. The stability of the DNA immobilization and the DNA density on the substrate have a dramatic influence on the robustness of the storage system and the data density. DNA is a polymer built up with four nucleotides covalently bonded by phosphate linkage. Due to the phosphate backbone, DNA is negatively charged under physiological conditions (pH 7.4). Physical and chemical methods are often used for DNA immobilization on substrates (124).
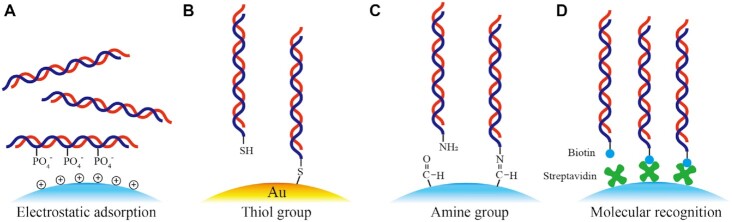
The physical immobilization is based on relatively weak interactions between DNA and substrates, such as the van der Waals force, hydrophobic interaction, and electrostatic adsorption. Among them, electrostatic adsorption based on the negative charges on the DNA backbone is widely used for DNA immobilization. The layer-by-layer structure is formed by alternatively stacking two oppositely charged polymers (Figure 4A) (116,125,126). For example, positively charged polymers such as poly ethylenimine and poly (dimethyl diallyl ammonium chloride) have been used to form condensed layer-by-layer structure with the negatively charged DNA on a substrate. Chen et al. (116) reported a layer-by-layer assembly method to preserve the DNA database on magnetic nanoparticles with large surface areas. An additional encapsulation layer of silica was deposited to protect the DNA molecules in the accelerated aging experiments. By this method, 83 KB data was encoded in DNA with a storage density of 160 ng/cm2 on the nanoparticles. Alternatively, Saurer et al. (127) assembled plasmid DNA with hydrolytically degradable cationic poly (β-amino ester) on microneedles via electrostatic adsorption. The microneedle could preserve bioactive DNA molecules, and controllably release the DNA molecules, which could be used for DNA data storage in the future.
Figure 4.
Schematics of various DNA immobilization methods. (A) DNA immobilized on the substrate using the electrostatic adsorption between polycations on the surface and negative phosphate groups of DNA molecules. (B) Immobilization of DNA with a thiol group on gold nanoparticles via Au–S bonding. (C) Immobilization of aminated DNA strands on the surface of the aldehyde group-modified substrate. The reaction between the amine group and the aldehyde group formed a Schiff base. (D) Immobilization of biotinylated DNA strands on the streptavidin-modified substrate.
For the chemical immobilization of DNA, the modified DNA reacts with the functional groups on the substrate surface. The most widely used reactions include Au-S interaction (128–131) and amine-based reactions (e.g. EDC (1-(3-dimethyl aminopropyl)-3-ethyl carbodiimide hydrochloride)-NHS (N-hydroxy succinimide) reaction and Schiff base reaction) (132–134). The immobilization of DNA on Au surfaces by forming Au-S bonds has been developed. For example, thiol-modified DNA covalently binds to Au after incubation at room temperature (Figure 4B). Cao et al. (128) reported the attachment of thiol-modified DNA to the surface of core/shell Ag/Au nanoparticles for colorimetric detecting. The DNA hybridization-induced nanoparticle aggregation led to visible colour change. Kim et al. (130) developed a programmable system based on DNA-linked colloidal gold nanoparticles building blocks termed nBLOCKs, in which gold nanoparticles were used as the nodes and thiol-modified DNA served as the scaffolds. By controlling the number, placement, and relative orientation of the DNA scaffolds on the surface of the gold nanoparticles, they could adjust the structures of the final self-assembled complexes, from simple linear dimers to complex 3D pyramidal and octahedral shapes. Although thiol-modified DNA may be detached from the Au substrate under high temperature or react with other competitive reagents like DTT (135,136), the biocompatible reaction conditions and relatively stable bonds make it appropriate for DNA-based data storage.
DNA immobilization using amino-based reactions is another widely used approach. The amino group is also highly reactive, which can be activated by EDC/NHS reaction and then react with a carboxylic group to form an amide bond (137–139). Another way to immobilize the amino-modified DNA is to react with an aldehyde group to form a Schiff base (Figure 4C). The resulting imine bond is unstable which can be further reduced by sodium borohydride to generate a stable product. For instance, Fuentes et al. (134) attached amino-modified DNA to the surface of superparamagnetic nanoparticles with a hetero-functional polymer (aldehyde-aspartic-dextran) to detect Hepatitis C Virus cDNA. Recently, Nguyen et al. (133) reported that the immobilization of DNA in microfluidic channels via the amino-based modification and the complementary fluorophore-labeled strands with controllable ON/OFF states helped to develop a new type of DNA storage system.
Recently, DNA immobilization by acrylate-based polymerization for data storage has been reported by Choi et al. (140). The acrylate moiety on the DNA could react and covalently link to poly (ethylene glycol) diacrylate (PEGDA) hydrogel. Auxiliary QR codes were printed on the DNA hydrogel based on photolithography. Details and related primer information for data retrieval could be obtained by scanning the QR codes. This covalent linking was also stable, which ensured reproducible reading with a long retention time (half-life >100 years under 10°C according to the decay kinetics estimation).
Except for the covalent immobilization above, another way that relies on specific recognition between molecules has also been used. For example, streptavidin and biotin exhibit strong specific interactions. Their binding strength is close to that of covalent bonding, as proven by its large affinity constant of ∼1015 mol/L (141,142). Therefore, the attachment of biotin-labeled DNA to the streptavidin-modified surface is an effective way for DNA immobilization (Figure 4D) (143). Song et al. (144) reported a hybridization system regulated by the electric field for DNA data storage, where the streptavidin-modified hydrogel matrix was attached to an electrode array. When a positive potential was applied to the electrodes, the biotinylated polyanionic DNA was allowed to combine with the hydrogel. Three hybridization regions were designed on the fixed DNA molecules so that three kinds of complementary DNA probes with different fluorescence labels could selectively hybridize with the DNA molecule on the hydrogel. The hybridization of three DNA probes on one fixed DNA strand led to a total of eight (23) combinations or one byte for data storage. Moreover, editing, rewriting, and bit shifting were realized via strand displacement reaction, and random access was also achieved on the individually addressable electrodes.
For two immobilization methods, the chemical method based on covalent bonding provides stable and localized DNA immobilization, which is compatible with array-based DNA processes. For frequent data reading and writing, the chemically immobilized DNA exhibits far better reliability against solvation, diffusion, and electric migration than physical adsorption. However, other than the long-term stability, the data storage density is also an important aspect. For the chemical method, only a monolayer of DNA can be deposited on the surface of the substrate, so the density is limited by the steric hindrance and surface area. In contrast, the physical method (i.e. layer-by-layer assembly) can accommodate large amounts of DNA on the surface, despite that the relatively weak physical interaction may lead to poor stability and robustness. To conclude, from the perspective of storage density, the layer-by-layer assembly method is usually preferred, but possible data loss and damage to DNA should be further studied.
Critical conditions for DNA preservation
Paleontological genome DNA stored in fossils, ambers, and salt crystals can retain their biological functions, which implies their incredible lifespan. Under favourable preservation conditions, DNA molecules can be stable for thousands of years, making it an ideal medium for non-volatile memory. However, as a biomacromolecule, DNA is also susceptible to degradation. For example, DNA can be irreversibly damaged when exposed to acids, light, high humidity, high temperature, or reactive oxygen species (ROS). So, the protection of a DNA-based database requires the elimination of these factors from the storage conditions.
DNA has good solubility in water, and most of the DNA-related biological processes are accomplished in an aqueous solution. Therefore, directly storing DNA in water is straightforward for subsequent reading operations. Unfortunately, the dissolved DNA in an aqueous solution is prone to depurination, deamination, depyrimidination, and hydrolytic cleavage of the phosphate backbone (145–149). Under alkaline conditions, the DNA degradation catalysed by acids is prevented (150), and the depurination of DNA can also be suppressed in solutions of high ionic strength (151). Under favourable alkaline conditions with high ionic strength, DNA stability is mainly affected by oxidation. Apart from dissolved oxygen, trace amounts of metals (e.g. Fe3+, Cu2+, etc.) notably enhance oxidative damage to DNA by producing hydroxyl radicals through the Fenton reaction (152–154). Unfortunately, the presence of transition metal ions in a DNA sample is almost inevitable. DNA purified at the highest standard still contains ∼30 ppb of Fe, and that common stabilizers and buffers lead to additional metal contamination (154). Lanthanide ions like CeIV ions have been proven to activate the phosphodiester linkages in DNA, which is susceptible to nucleophilic attack, leading to DNA hydrolysis (155). The addition of metal chelators such as ethylenediaminetetraacetic acid (EDTA) may mitigate the metal pollution, but it cannot avoid oxidation damage generated by the Fenton reaction (156,157). To prevent DNA from oxidation, antioxidants and radical scavengers can be added to the storing medium.
Another environmental factor affecting DNA preservation is temperature. Under high temperature, the probability of molecular collision increases so that a greater amount of molecules acquires the activation energy for unwanted reactions, as predicted by Arrhenius's Law (158). In a pyrolysis experiment, complete degradation of DNA was observed at 180°C in a few minutes (159). Although cryopreservation of DNA decreases the DNA degradation reactivity, cryolysis can also be induced by the formation of cracks within the ice at low temperatures (160). To address these problems, additives such as trehalose can improve the stability and integrity of DNA during prolonged preservation (161–163). Strong hydrogen bonds between trehalose molecules and DNA strands offset the electrostatic repulsion between DNA backbones (161). The effect of limited molecular mobility of DNA can be enhanced by the vitreous matrix provided by trehalose (164,165). Other additives such as resveratrol and DNAStable have also been examined as effective preservatives (166,167).
On the other hand, despite low temperature is beneficial for long-term preservation, it increases the cost remarkably. So, natural permafrost is concerned as a favourable choice for low-frequency archival storage to reduce the cost. However, low temperature attributes less to the robustness of DNA storage compared with keeping at the dry state (168,169). Although DNA can maintain its integrity in an aqueous solution for tens of thousands of years at 10°C, it is still three magnitudes lower than that stored in the dry state (8). The dehydrated DNA is more chemically inert owing to decreased molecular mobility and absence of water or dissolved oxygen that usually participated in the DNA damage. Consequently, for DNA-based data storage, keeping the DNA samples dry is superior to keeping them at a low temperature.
Ionising radiation, including β, γ and X rays, can also induce the damage of DNA molecules, such as chain break and chemical changes of bases. The damage can be attributed to a combination of direct and indirect reactions. For the direct reactions, ionising radiations indiscriminately transfer energy to electrons in the irradiated molecules, which are excited to react with other reagents (170). For example, a C-H bond in DNA can be activated to generate a highly reactive carbon-centred radical, leading to oxidization, reduction, and even strand breakage. Sanche et al. reported that transient radical anions in DNA molecules formed under the radical attacks (171), which might further induce bond breakage.
For the indirect reactions, ionising radiation activate water molecules to produce aggressive ⋅OH radicals (172), which can react with sugar and base moieties of DNA and cause subsequent side reactions such as ring-opening, pyrimidine oxidation, and bond breakage (173,174). Due to DNA repair mechanisms (175) and radical scavengers (176), the problem of radiation damage can be alleviated in vivo. As previously estimated, about one single-strand break occurred per 107 bp/Gy of radiation (177). However, the molecular damage is much more significant for in vitro DNA systems in absence of repair pathways. Besides, for certain conditions, such as in an space station, the intensity of cosmic rays is much higher than that at sea level (178), which lead to much more DNA damages. So, the effect of ionising radiation on a DNA database can also be a significant concern.
Considering all conditions mentioned above, a currently feasible solution for DNA database preservation is to encapsulate the DNA in a dehydrated state (Figure 5A) (116,179,180). Paunescu et al. (180) reported the preservation of DNA by adsorption on the surface of submicron-sized silica particles modified with positive-charged ammonium groups and subsequently depositing a silica layer on it. The encapsulation of the silica layer prevented the DNA from damages under high temperature, ROS, and UV irradiation. The dense silica layer acted as a hermetic diffusion barrier to external oxygen, ROS, and metal ions. Although the outmost amorphous silica was a UV-permeable material (>60% transparent at 170 nm wavelength), the geometric shape was able to scatter light of low wavelengths so that the DNA inside could maintain its integrity even under the UV radiation. Recently, Koch et al. reported the dispersion of as-synthesized microparticles in polycaprolactone (PCL) for three-dimensional (3D) printing (179). The files encoded in the DNA molecules were required for 3D modelling and printing. They iterated as high as six times of these procedures including extracting DNA from printed objects via PCR amplification, decoding DNA to obtain the data file, and 3D printing objects using PCL fibres mixed with microparticles according to the files. The hard silica shell was capable of protecting the DNA from the violent stress in filament extrusion (Figure 5B). To conclude, the encapsulation of DNA is a currently feasible and effective method for long-term preservation, which endows the DNA storage system with relatively high stability and considerable storage density.
Figure 5.
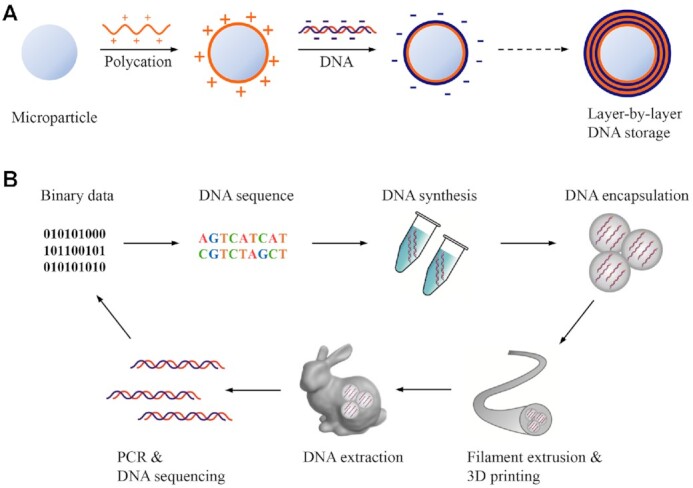
Schematics for long-term DNA preservation. (A) Preservation for layer-by-layer assembled DNA, in which cationic polymer and anionic DNA were organized into a layer-by-layer structure based on electrostatic interaction on the surface of microparticles (116). (B) DNA preservation by packaging in silica beads and then dispersed in polycaprolactone fibres. The DNA strands were encoded with data files that were used for 3D printing of a rabbit model. DNA molecules could be further extracted, amplified, and sequenced to acquire encoded files (179). Reproduced with permission (179). Copyright 2020, Springer Nature.
CHALLENGES BEFORE LARGE-SCALE APPLICATIONS
Although DNA-based data storage has theoretically higher data density, longer retention time, and lower power consumption than current electronic memory devices, the maximum storage capacity until now is less than 1GB, much lower than the state-of-the-art data storage devices. There remain several challenges to achieve the large-scale application of DNA-based data storage. Further improvements rely on the progress in all of the steps involved in the data storage, including data encoding, DNA synthesis, preservation, data retrieval, and DNA sequencing.
To ensure the correct retrieval of data from the DNA sequences, error-correction based on data redundancy is indispensable. Generally, more data redundancy leads to better correction of errors, either from DNA synthesis or sequencing. However, more data redundancy also results in decreased encoding efficiency, in other words, less information can be encoded in the DNA sequences. Thus, there is a trade-off between error-correction ability and logical data density. In addition, the performance of ECC differs when applied in different encoding frameworks shown in Figure 2. The choices of encoding framework and the ECC should be systematically considered for optimal error tolerance and coding efficiency.
The array-based DNA synthesis enables the paralleled synthesis of a large group of DNA molecules with different sequences, and the reagent consumption is less than the column-based synthesis method. However, it is still challenging to push the synthesis scale to the level that can be competitive against the scale of conventional electronic memory, in terms of encoded data. A larger synthesis scale relies on the fabrication of smaller features (e.g. electrodes) on the chip, leading to problems that affect the fidelity of DNA syntheses, such as the cross-talk between adjacent microelectrodes in the array and low reproducibility. The advance in this aspect probably relies on the advent of new enzymatic synthesis, which may considerably enhance the length, quality, and speed of the DNA synthesis (e.g. with the use of template-independent TdT). The greater synthesis length, higher accuracy, and speed will also reduce the synthesis cost, which is a crucial factor that restricts the applicability of DNA-based data storage. Akin to the transition from column-based to array-based chemical synthesis, the development of paralleled high-throughput enzymatic DNA synthesis will be probably the focus of research in the future.
Preservation of DNA molecules away from adverse factors (e.g. by encapsulation in silica beads) can ensure long-term stability for data retention. Proper environmental conditions and additives should be investigated to pursue a long retention time to the theoretical limit. Additionally, the preservation by encapsulation avoids direct access to the data files. For example, DNA encapsulated in silica cannot be directly amplified and retrieved by PCR unless released from the beads. Therefore, an optimized preservation method should balance the long-term stability and the accessibility of data.
In the past few decades, high-throughput sequencing technology has achieved remarkable progress, which is characterized by a significant reduction in sequencing costs and time. Nevertheless, the data reading speed is still several orders of magnitude lower than that on conventional data media (e.g. flash memory). Therefore, the application of DNA storage at present is usually limited to archival storage and is impractical for low-latency data memory units used in mobile phones, computers, or other consumer electronic devices. So far, third-generation sequencing based on nanopore technology has been a promising technique to address this problem. But many problems hinder its utilization in DNA data storage. The nanopore sequencing relies on the detection of weak electric signals, which can be easily interfered with by background noises caused by vibrations and electric circuits. Furthermore, the translocation speed of DNA is difficult to control. High speed usually results in poor resolution of the electronic signals, which makes it hard to distinguish both the type and length of nucleotides. So, a technical breakthrough is still for fast and real-time data reading in large-scale application of DNA-based storage systems.
Another issue is the cost to store data in DNA. Despite the theoretical advantages compared with conventional digital memory, DNA-based data storage is still much more expensive, and the majority of the cost comes from DNA synthesis. Currently, synthesizing millions of different DNA sequences still requires cutting-edge technology. The cost for synthesis and sequencing has been estimated to be at least ∼$3500/MB, as previously estimated by Erlich and Zielinski (16), which renders the large-scale DNA storage currently impractical. The technical development of DNA synthesis and sequencing is urgently demanded to reduce the expense of DNA data storage to an acceptable level.
Despite those challenges, we are still optimistic about the future of DNA-based data storage. It is always instructive to learn the wisdom of natural creatures, such as cells that have proofreading mechanisms for error correction of the DNA synthesized (181,182). This leads to in vivo storage of digital information or the use of proofreading enzymes for in vitro databases. In addition, genetic information can be exchanged between cells by plasmids, which are applied by bacteria to proliferate and overcome adversity (183–185). The plasmid can act as a carrier for information storage in living organisms. Emerging technologies, including the enzymatic synthesis of DNA, the development of computer and information technology, will lead to new robust encoding structures and error correction schemes. Other optimizations of DNA editing, encrypting and maintaining, immobilization, and storing approaches (e.g. clustered regularly interspaced short palindromic repeats (103,186,187), rewriting based on strand replacement reaction (66), and random number generation for encryption (188)) should help to reduce the uncertainties in large-scale DNA-based data storage and eventually makes the DNA a promising data storing medium for human beings.
CONCLUSION
With the rapid development of information technology, the worldwide explosion of data generation has increasingly challenged the production of conventional electronic memory based on silicon. Carbon-based DNA molecular data storage discussed in this review is probably one of the most promising technologies to address this issue considering its high data retention time, storage density, and relatively low energy consumption in data copying, pasting, editing, and reading. In this review, we introduced the general processes of data storage in DNA. Then we focused on the uncertainties involved in those steps and potential methods for error correction. Finally, we discussed the remaining challenges for the realization of large-scale DNA-based data storage systems and further research direction in this area. As a novel molecular media for data storage, DNA is still in its infancy. Indeed, substantial breakthroughs are still required to achieve the large-scale application. However, we believe it is a promising technology to solve the problem of data explosion, especially considering the rapid developments in DNA synthesis, sequencing, and other related technologies. As Grass et al. proposed (179), everything may act as a carrier of a DNA database in the future, which may have a profound influence on a range of fields, including data management, the internet of things (IoT), and blockchain.
Contributor Information
Chengtao Xu, State Key Laboratory of Bioelectronics, School of Biological Science and Medical Engineering, Southeast University, Nanjing, Jiangsu 210096, China.
Chao Zhao, State Key Laboratory of Bioelectronics, School of Biological Science and Medical Engineering, Southeast University, Nanjing, Jiangsu 210096, China.
Biao Ma, State Key Laboratory of Bioelectronics, School of Biological Science and Medical Engineering, Southeast University, Nanjing, Jiangsu 210096, China.
Hong Liu, State Key Laboratory of Bioelectronics, School of Biological Science and Medical Engineering, Southeast University, Nanjing, Jiangsu 210096, China.
FUNDING
National Natural Science Foundation of China [21635001 to H.L.]; State Key Project of Research and Development [2016YFF0100802 to H.L.]; Key Project and Open Research Fund of State Key Laboratory of Bioelectronics, Southeast University. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Goda K., Kitsuregawa M.. The history of storage systems. Proc. IEEE. 2012; 100:1433–1440. [Google Scholar]
- 2. Hilbert M., Lopez P.. The world's technological capacity to store, communicate, and compute information. Science. 2011; 332:60–65. [DOI] [PubMed] [Google Scholar]
- 3. Reisel D., Gantz J., Rydning J.. Data age 2025: the digitization of the world from edge to core. Seagate. 2018; https://www.seagate.com/in/en/our-story/data-age-2025/. [Google Scholar]
- 4. Xu Z.-W. Cloud-sea computing systems: Towards thousand-fold improvement in performance per watt for the coming zettabyte era. J. Comput. Sci. Technol. 2014; 29:177–181. [Google Scholar]
- 5. Extance A. Could the molecule known for storing genetic information also store the world's data. Nature. 2016; 537:22–24. [DOI] [PubMed] [Google Scholar]
- 6. Organick L., Chen Y.J., Dumas Ang S., Lopez R., Liu X., Strauss K., Ceze L.. Probing the physical limits of reliable DNA data retrieval. Nat. Commun. 2020; 11:616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gregory T.R., Nicol J.A., Tamm H., Kullman B., Kullman K., Leitch I.J., Murray B.G., Kapraun D.F., Greilhuber J., Bennett M.D.. Eukaryotic genome size databases. Nucleic Acids Res. 2007; 35:D332–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhirnov V., Zadegan R.M., Sandhu G.S., Church G.M., Hughes W.L.. Nucleic acid memory. Nat. Mater. 2016; 15:366–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goodwin S., McPherson J.D., McCombie W.R.. Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016; 17:333–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Branton D., Deamer D.W., Marziali A., Bayley H., Benner S.A., Butler T., Di Ventra M., Garaj S., Hibbs A., Huang X.et al.. The potential and challenges of nanopore sequencing. Nat. Biotechnol. 2008; 26:1146–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Garibyan L., Avashia N.. Polymerase chain reaction. J. Invest. Dermatol. 2013; 133:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ochman H., Gerber A.S., Hartl D.L.. Genetic applications of an inverse polymerase chain reaction. Genetics. 1988; 120:621–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goldman N., Bertone P., Chen S., Dessimoz C., LeProust E.M., Sipos B., Birney E.. Towards practical, high-capacity, low-maintenance information storage in synthesized DNA. Nature. 2013; 494:77–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Organick L., Ang S.D., Chen Y.J., Lopez R., Yekhanin S., Makarychev K., Racz M.Z., Kamath G., Gopalan P., Nguyen B.et al.. Random access in large-scale DNA data storage. Nat. Biotechnol. 2018; 36:242–248. [DOI] [PubMed] [Google Scholar]
- 15. Church G.M., Gao Y., Kosuri S.. Next-generation digital information storage in DNA. Science. 2012; 337:1628–1628. [DOI] [PubMed] [Google Scholar]
- 16. Erlich Y., Zielinski D.. DNA Fountain enables a robust and efficient storage architecture. Science. 2017; 355:950–953. [DOI] [PubMed] [Google Scholar]
- 17. Caruthers M.H. The chemical synthesis of DNA/RNA: our gift to science. J. Biol. Chem. 2013; 288:1420–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Beaucage S.L., Caruthers M.H.. Deoxynucleoside phosphoramidites - a new class of key intermediates for deoxypolynucleotide synthesis. Tetrahedron Lett. 1981; 22:1859–1862. [Google Scholar]
- 19. Mathews A.S., Yang H., Montemagno C.. Photo-cleavable nucleotides for primer free enzyme mediated DNA synthesis. Org. Biomol. Chem. 2016; 14:8278–8288. [DOI] [PubMed] [Google Scholar]
- 20. Motea E.A., Berdis A.J.. Terminal deoxynucleotidyl transferase: the story of a misguided DNA polymerase. Biochim. Biophys. Acta. 2010; 1804:1151–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cano R.J., Borucki M.K.. Revival and identification of bacterial spores in 25-to 40-million-year-old Dominican amber. Science. 1995; 268:1060–1064. [DOI] [PubMed] [Google Scholar]
- 22. Fish S.A., Shepherd T.J., McGenity T.J., Grant W.D.. Recovery of 16S ribosomal RNA gene fragments from ancient halite. Nature. 2002; 417:432–436. [DOI] [PubMed] [Google Scholar]
- 23. Chen Y.J., Takahashi C.N., Organick L., Bee C., Ang S.D., Weiss P., Peck B., Seelig G., Ceze L., Strauss K.. Quantifying molecular bias in DNA data storage. Nat. Commun. 2020; 11:3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shendure J., Balasubramanian S., Church G.M., Gilbert W., Rogers J., Schloss J.A., Waterston R.H.. DNA sequencing at 40: past, present and future. Nature. 2017; 550:345–353. [DOI] [PubMed] [Google Scholar]
- 25. Jain M., Olsen H.E., Paten B., Akeson M.. The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biol. 2016; 17:239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bornholt J., Lopez R., Carmean D.M., Ceze L., Seelig G., Strauss K.. A DNA-Based Archival Storage System. Proceedings of the Twenty-First International Conference on Architectural Support for Programming Languages and Operating Systems - ASPLOS '16. 2016; NY: ACM; 637–649. [Google Scholar]
- 27. Caruthers M.H. A brief review of DNA and RNA chemical synthesis. Biochem. Soc. Trans. 2011; 39:575–580. [DOI] [PubMed] [Google Scholar]
- 28. Hughes R.A., Ellington A.D.. Synthetic DNA synthesis and assembly: putting the synthetic in synthetic biology. Cold Spring Harbor Perspect. Biol. 2017; 9:a023812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Damha M.J., Giannaris P.A., Zabarylo S.V.. An improved procedure for derivatization of controlled-pore glass-beads for solid-phase oligonucleotide synthesis. Nucleic Acids Res. 1990; 18:3813–3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Matteucci M.D., Caruthers M.H.. Synthesis of deoxyoligonucleotides on a polymer support. J. Am. Chem. Soc. 1992; 24:92–98. [PubMed] [Google Scholar]
- 31. Moorcroft M.J., Meuleman W.R., Latham S.G., Nicholls T.J., Egeland R.D., Southern E.M.. In situ oligonucleotide synthesis on poly(dimethylsiloxane): a flexible substrate for microarray fabrication. Nucleic Acids Res. 2005; 33:e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee C.C., Snyder T.M., Quake S.R.. A microfluidic oligonucleotide synthesizer. Nucleic Acids Res. 2010; 38:2514–2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hughes T.R., Mao M., Jones A.R., Burchard J., Marton M.J., Shannon K.W., Lefkowitz S.M., Ziman M., Schelter J.M., Meyer M.R.et al.. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol. 2001; 19:342–347. [DOI] [PubMed] [Google Scholar]
- 34. Butler J.H., Cronin M., Anderson K.M., Biddison G.M., Chatelain F., Cummer M., Davi D.J., Fisher L., Frauendorf A.W., Frueh F.W.et al.. In situ synthesis of oligonucleotide arrays by using surface tension. J. Am. Chem. Soc. 2001; 123:8887–8894. [DOI] [PubMed] [Google Scholar]
- 35. Gao X.L., LeProust E., Zhang H., Srivannavit O., Gulari E., Yu P.L., Nishiguchi C., Xiang Q., Zhou X.C.. A flexible light-directed DNA chip synthesis gated by deprotection using solution photogenerated acids. Nucleic Acids Res. 2001; 29:4744–4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Singh-Gasson S., Green R.D., Yue Y.J., Nelson C., Blattner F., Sussman M.R., Cerrina F.. Maskless fabrication of light-directed oligonucleotide microarrays using a digital micromirror array. Nat. Biotechnol. 1999; 17:974–978. [DOI] [PubMed] [Google Scholar]
- 37. Fodor S.P.A., Read J.L., Pirrung M.C., Stryer L., Lu A.T., Solas D. Light-directed, spatially addressable parallel chemical synthesis. Science. 1991; 251:767–773. [DOI] [PubMed] [Google Scholar]
- 38. Egeland R.D., Southern E.M.. Electrochemically directed synthesis of oligonucleotides for DNA microarray fabrication. Nucleic Acids Res. 2005; 33:e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Egeland R.D., Marken F., Southern E.M.. An electrochemical redox couple activitated by microelectrodes for confined chemical patterning of surfaces. Anal. Chem. 2002; 74:1590–1596. [DOI] [PubMed] [Google Scholar]
- 40. Maurer K., Cooper J., Caraballo M., Crye J., Suciu D., Ghindilis A., Leonetti J.A., Wang W., Rossi F.M., Stover A.G.et al.. Electrochemically generated acid and its containment to 100 micron reaction areas for the production of DNA microarrays. PLoS One. 2006; 1:e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chow B.Y., Emig C.J., Jacobson J.M.. Photoelectrochemical synthesis of DNA microarrays. Proc. Natl. Acad. Sci. USA. 2009; 106:15219–15224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dalma-Weiszhausz D.D., Warrington J., Tanimoto E.Y., Miyada C.G.. The Affymetrix Genechip® platform: an overview. Methods Enzymol. 2006; 410:3–28. [DOI] [PubMed] [Google Scholar]
- 43. Irizarry R.A., Bolstad B.M., Collin F., Cope L.M., Hobbs B., Speed T.P.. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003; 31:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu R., Munro S., Nguyen T., Siuda T., Suciu D., Bizak M., Slota M., Fuji H., Danley D., McShea A.. Integrated microfluidic customarray device for bacterial genotyping and identification. J. Am. Chem. Soc. 2006; 11:360–367. [Google Scholar]
- 45. Roth K.M., Peyvan K., Schwarzkopf K.R., Ghindilis A.. Electrochemical detection of short DNA oligomer hybridization using the combimatrix electrasense microarray reader. Electroanalysis. 2006; 18:1982–1988. [Google Scholar]
- 46. Antkowiak P.L., Lietard J., Darestani M.Z., Somoza M.M., Stark W.J., Heckel R., Grass R.N.. Low cost DNA data storage using photolithographic synthesis and advanced information reconstruction and error correction. Nat. Commun. 2020; 11:5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Heckel R., Mikutis G., Grass R.N.. A characterization of the DNA data storage channel. Sci. Rep. 2019; 9:9663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Efcavitch J.W., Heiner C.. Depurination as a yield decreasing mechanism in oligodeoxynucleotide synthesis. Nucleosides. Nucleotides. 2006; 4:267–267. [Google Scholar]
- 49. Septak M. Kinetic studies on depurination and detritylation of CPG-bound intermediates during oligonucleotide synthesis. Nucleic Acids Res. 1996; 24:3053–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. LeProust E.M., Peck B.J., Spirin K., McCuen H.B., Moore B., Namsaraev E., Caruthers M.H.. Synthesis of high-quality libraries of long (150mer) oligonucleotides by a novel depurination controlled process. Nucleic Acids Res. 2010; 38:2522–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Andrus A., Efcavitch J.W., McBride L.J., Giusti B.. Novel activating and capping reagents for improved hydrogen-phosphonate DNA-synthesis. Tetrahedron Lett. 1988; 29:861–864. [Google Scholar]
- 52. Saaem I., Ma S., Quan J., Tian J.. Error correction of microchip synthesized genes using Surveyor nuclease. Nucleic Acids Res. 2012; 40:e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Anavy L., Vaknin I., Atar O., Amit R., Yakhini Z.. Data storage in DNA with fewer synthesis cycles using composite DNA letters. Nat. Biotechnol. 2019; 37:1229–1236. [DOI] [PubMed] [Google Scholar]
- 54. Choi Y., Ryu T., Lee A.C., Choi H., Lee H., Park J., Song S.H., Kim S., Kim H., Park W.et al.. High information capacity DNA-based data storage with augmented encoding characters using degenerate bases. Sci. Rep. 2019; 9:6582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Covo S., Blanco L., Livneh Z.. Lesion bypass by human DNA polymerase μ reveals a template-dependent, sequence-independent nucleotidyl transferase activity. J. Biol. Chem. 2004; 279:859–865. [DOI] [PubMed] [Google Scholar]
- 56. Ramadan K., Shevelev I., Hubscher U.. The DNA-polymerase-X family: controllers of DNA quality. Nat. Rev. Mol. Cell Biol. 2004; 5:1038–1043. [DOI] [PubMed] [Google Scholar]
- 57. Service R.F. DNA printers poised to jump from paragraphs to pages. Science. 2018; 362:143. [DOI] [PubMed] [Google Scholar]
- 58. Jensen M.A., Davis R.W.. Template-independent enzymatic oligonucleotide synthesis (TiEOS): its history, prospects, and challenges. Biochemistry. 2018; 57:1821–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Metzker M.L., Raghavachari R., Richards S., Jacutin S.E., Civitello A., Burgess K., Gibbs R.A.. Termination of DNA synthesis by novel 3′-modifieddeoxyribonucleoside 5′-triphosphates. Nucleic Acids Res. 1994; 22:4259–4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Palluk S., Arlow D.H., de Rond T., Barthel S., Kang J.S., Bector R., Baghdassarian H.M., Truong A.N., Kim P.W., Singh A.K.et al.. De novo DNA synthesis using polymerase-nucleotide conjugates. Nat. Biotechnol. 2018; 36:645–650. [DOI] [PubMed] [Google Scholar]
- 61. Lee H.H., Kalhor R., Goela N., Bolot J., Church G.M.. Terminator-free template-independent enzymatic DNA synthesis for digital information storage. Nat. Commun. 2019; 10:2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Newman S., Stephenson A.P., Willsey M., Nguyen B.H., Takahashi C.N., Strauss K., Ceze L.. High density DNA data storage library via dehydration with digital microfluidic retrieval. Nat. Commun. 2019; 10:1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tomek K.J., Volkel K., Simpson A., Hass A.G., Indermaur E.W., Tuck J.M., Keung A.J.. Driving the scalability of DNA-based information storage systems. ACS Synth. Biol. 2019; 8:1241–1248. [DOI] [PubMed] [Google Scholar]
- 64. Yazdi S.M., Yuan Y., Ma J., Zhao H., Milenkovic O.. A rewritable, random-access DNA-based storage system. Sci. Rep. 2015; 5:14138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yamamoto M., Kashiwamura S., Ohuchi A., Furukawa M.. Large-scale DNA memory based on the nested PCR. Nat. Comput. 2008; 7:335–346. [Google Scholar]
- 66. Lin K.N., Volkel K., Tuck J.M., Keung A.J.. Dynamic and scalable DNA-based information storage. Nat. Commun. 2020; 11:2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Song X., Shah S., Reif J.. Multidimensional data organization and random access in large-scale DNA storage systems. 2019; bioRxiv doi:22 August 2019, preprint: not peer reviewed 10.1101/743369. [DOI]
- 68. Ruijter J.M., Ramakers C., Hoogaars W.M., Karlen Y., Bakker O., van den Hoff M.J., Moorman A.F.. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009; 37:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Warnecke P.M., Stirzaker C., Melki J.R., Millar D.S., Paul C.L., Clark S.J.. Detection and measurement of PCR bias in quantitative methylation analysis of bisulphite-treated DNA. Nucleic Acids Res. 1997; 25:4422–4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Pan W., Byrne-Steele M., Wang C., Lu S., Clemmons S., Zahorchak R.J., Han J.. DNA polymerase preference determines PCR priming efficiency. BMC Biotechnol. 2014; 14:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Henry O.Y.F., O’Sullivan C.K.. Rapid DNA hybridization in microfluidics. TrAC, Trends Anal. Chem. 2012; 33:9–22. [Google Scholar]
- 72. Marimuthu K., Chakrabarti R.. Sequence-dependent theory of oligonucleotide hybridization kinetics. J. Chem. Phys. 2014; 140:175104. [DOI] [PubMed] [Google Scholar]
- 73. Guo J., Xu N., Li Z., Zhang S., Wu J., Kim D.H., Marma M.S., Meng Q., Cao H., Li X.et al.. Four-color DNA sequencing with 3′-O-modified nucleotide reversible terminators and chemically cleavable fluorescent dideoxynucleotides. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:9145–9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ju J., Kim D.H., Bi L., Meng Q., Bai X., Li Z., Li X., Marma M.S., Shi S., Wu J.et al.. Four-color DNA sequencing by synthesis using cleavable fluorescent nucleotide reversible terminators. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:19635–19640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Meller A., Nivon L., Brandin E., Golovchenko J.. Rapid nanopore discrimination between single polynucleotide molecules. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:1079–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Deamer D., Akeson M., Branton D.. Three decades of nanopore sequencing. Nat. Biotechnol. 2016; 34:518–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lopez R., Chen Y.J., Dumas Ang S., Yekhanin S., Makarychev K., Racz M.Z., Seelig G., Strauss K., Ceze L.. DNA assembly for nanopore data storage readout. Nat. Commun. 2019; 10:2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Clarke J., Wu H.C., Jayasinghe L., Patel A., Reid S., Bayley H.. Continuous base identification for single-molecule nanopore DNA sequencing. Nat. Nanotechnol. 2009; 4:265–270. [DOI] [PubMed] [Google Scholar]
- 79. Ip C.L.C., Loose M., Tyson J.R., de Cesare M., Brown B.L., Jain M., Leggett R.M, Eccles D.A., Zalunin V., Urban J.M.et al.. MinION analysis and reference consortium: phase 1 data release and analysis. F1000Research. 2015; 4:1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Chen K., Kong J., Zhu J., Ermann N., Predki P., Keyser U.F.. Digital data storage using DNA nanostructures and solid-state nanopores. Nano Lett. 2019; 19:1210–1215. [DOI] [PubMed] [Google Scholar]
- 81. Chen K., Zhu J., Boskovic F., Keyser U.F.. Secure data storage on DNA hard drives. 2019; bioRxiv doi:28 November 2019, preprint: not peer reviewed 10.1101/857748. [DOI] [PubMed]
- 82. Takahashi C.N., Nguyen B.H., Strauss K., Ceze L.. Demonstration of end-to-end automation of DNA data storage. Sci. Rep. 2019; 9:4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Poon K., Macgregor R.B. Jr. Unusual behavior exhibited by multistranded guanine-rich DNA complexes. Biopolymers: Orig. Res. Biomol. 1998; 45:427–434. [DOI] [PubMed] [Google Scholar]
- 84. Schwartz J.J., Lee C., Shendure J.. Accurate gene synthesis with tag-directed retrieval of sequence-verified DNA molecules. Nat. Methods. 2012; 9:913–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Bentley D.R., Balasubramanian S., Swerdlow H.P., Smith G.P., Milton J., Brown C.G., Hall K.P., Evers D.J., Barnes C.L., Bignell H.R.et al.. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008; 456:53–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Benjamini Y., Speed T.P.. Summarizing and correcting the GC content bias in high-throughput sequencing. Nucleic Acids Res. 2012; 40:e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Schirmer M., D’Amore R., Ijaz U.Z., Hall N., Quince C.. Illumina error profiles: resolving fine-scale variation in metagenomic sequencing data. BMC Bioinf. 2016; 17:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ross M.G., Russ C., Costello M., Hollinger A., Lennon N.J., Hegarty R., Nusbaum C., Jaffe D.B.. Characterizing and measuring bias in sequence data. Genome Biol. 2013; 14:R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Yusko E.C., Johnson J.M., Majd S., Prangkio P., Rollings R.C., Li J., Yang J., Mayer M.. Controlling protein translocation through nanopores with bio-inspired fluid walls. Nat. Nanotechnol. 2011; 6:253–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Fragasso A., Schmid S., Dekker C.. Comparing current noise in biological and solid-state nanopores. ACS Nano. 2020; 14:1338–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Wanunu M. Nanopores: a journey towards DNA sequencing. Phys. Life Rev. 2012; 9:125–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Dekker C. Solid-state nanopores. Nat. Biotechnol. 2007; 2:209–215. [DOI] [PubMed] [Google Scholar]
- 93. Schneider G.F., Xu Q., Hage S., Luik S., Spoor J.N., Malladi S., Zandbergen H., Dekker C.. Tailoring the hydrophobicity of graphene for its use as nanopores for DNA translocation. Nat. Commun. 2013; 4:2619. [DOI] [PubMed] [Google Scholar]
- 94. Agah S., Zheng M., Pasquali M., Kolomeisky A.B.. DNA sequencing by nanopores: advances and challenges. J. Phys. D: Appl. Phys. 2016; 49:413001. [Google Scholar]
- 95. Keyser U.F. Controlling molecular transport through nanopores. J. R. Soc., Interface. 2011; 8:1369–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Goodwin S., Gurtowski J., Ethe-Sayers S., Deshpande P., Schatz M.C., McCombie W.R.. Oxford Nanopore sequencing, hybrid error correction, and de novo assembly of a eukaryotic genome. Genome Res. 2015; 25:1750–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Timp W., Comer J., Aksimentiev A.. DNA base-calling from a nanopore using a Viterbi algorithm. Biophys. J. 2012; 102:L37–L39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Szalay T., Golovchenko J.A.. De novo sequencing and variant calling with nanopores using PoreSeq. Nat. Biotechnol. 2015; 33:1087–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Schreiber J., Karplus K.. Analysis of nanopore data using hidden Markov models. Bioinformatics. 2015; 31:1897–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Simpson J.T., Workman R.E., Zuzarte P.C., David M., Dursi L.J., Timp W.. Detecting DNA cytosine methylation using nanopore sequencing. Nat. Methods. 2017; 14:407–410. [DOI] [PubMed] [Google Scholar]
- 101. Luo R., Wong C.-L., Wong Y.-S., Tang C.-I., Liu C.-M., Leung C.-M., Lam T.-W.. Exploring the limit of using a deep neural network on pileup data for germline variant calling. Nat. Mach. Intell. 2020; 2:220–227. [Google Scholar]
- 102. Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011; 27:2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Shipman S.L., Nivala J., Macklis J.D., Church G.M.. CRISPR-Cas encoding of a digital movie into the genomes of a population of living bacteria. Nature. 2017; 547:345–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Gilpatrick T., Lee I., Graham J.E., Raimondeau E., Bowen R., Heron A., Downs B., Sukumar S., Sedlazeck F.J., Timp W.. Targeted nanopore sequencing with Cas9-guided adapter ligation. Nat. Biotechnol. 2020; 38:433–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Stranges P.B., Palla M., Kalachikov S., Nivala J., Dorwart M., Trans A., Kumar S., Porel M., Chien M., Tao C.et al.. Design and characterization of a nanopore-coupled polymerase for single-molecule DNA sequencing by synthesis on an electrode array. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E6749–E6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Loman N.J., Quick J., Simpson J.T.. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat. Methods. 2015; 12:733–735. [DOI] [PubMed] [Google Scholar]
- 107. Yazdi S., Gabrys R., Milenkovic O.. Portable and error-free DNA-based data storage. Sci. Rep. 2017; 7:5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Grass R.N., Heckel R., Puddu M., Paunescu D., Stark W.J.. Robust chemical preservation of digital information on DNA in silica with error-correcting codes. Angew. Chem., Int. Ed. Engl. 2015; 54:2552–2555. [DOI] [PubMed] [Google Scholar]
- 109. Blawat M., Gaedke K., Hütter I., Chen X.-M., Turczyk B., Inverso S., Pruitt B.W., Church G.M.. Forward error correction for DNA data storage. Procedia Comput. Sci. 2016; 80:1011–1022. [Google Scholar]
- 110. Heckel R., Shomorony I., Ramchandran K., Tse D.N.C. Fundamental limits of DNA storage systems. 2017 IEEE International Symposium on Information Theory (ISIT). 2017; NY: IEEE; 3130–3134. [Google Scholar]
- 111. Ailenberg M., Rotstein O.. An improved Huffman coding method for archiving text, images, and music characters in DNA. BioTechniques. 2009; 47:747–754. [DOI] [PubMed] [Google Scholar]
- 112. Guruswami V., Sudan M.. Improved decoding of Reed-Solomon and algebraic-geometry codes. IEEE Trans. Inf. Theory. 1999; 45:1757–1767. [Google Scholar]
- 113. Reed I.S., Solomon G.. Polynomial codes over certain finite fields. J. Soc. Ind. Appl. Math. 1960; 8:300–304. [Google Scholar]
- 114. MacKay D.J.C Fountain codes. IEEE Int. Sib. Conf. Control Commun., SIBCON-2005, Proc. 2005; 152:1062–1068. [Google Scholar]
- 115. Dimakis A.G., Prabhakaran V., Ramchandran K.. Distributed fountain codes for networked storage. 2006 IEEE International Conference on Acoustics, Speech and Signal Processing. 1–13:NY: IEEE; 6007–6010. [Google Scholar]
- 116. Chen W.D., Kohll A.X., Nguyen B.H., Koch J., Heckel R., Stark W.J., Ceze L., Strauss K., Grass R.N.. Combining data longevity with high storage capacity—layer-by-layer DNA encapsulated in magnetic nanoparticles. Adv. Funct. Mater. 2019; 29:1901672. [Google Scholar]
- 117. Luby M. LT codes. The 43rd Annual IEEE Symposium on Foundations of Computer Science, 2002. Proceedings. 2002; NY: IEEE; 271–280. [Google Scholar]
- 118. Dong Y., Sun F., Ping Z., Ouyang Q., Qian L.. DNA storage: research landscape and future prospects. Natl. Sci. Rev. 2020; 7:1092–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Paabo S., Gifford J.A., Wilson A.C.. Mitochondrial-DNA sequences from a 7000-year old brain. Nucleic Acids Res. 1988; 16:9775–9787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993; 362:709–715. [DOI] [PubMed] [Google Scholar]
- 121. Lindahl T. Recovery of antediluvian DNA. Nature. 1993; 365:700–700. [DOI] [PubMed] [Google Scholar]
- 122. Prufer K., Racimo F., Patterson N., Jay F., Sankararaman S., Sawyer S., Heinze A., Renaud G., Sudmant P.H., de Filippo C.et al.. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014; 505:43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Vreeland R.H., Rosenzweig W.D., Powers D.W.. Isolation of a 250 million-year-old halotolerant bacterium from a primary salt crystal. Nature. 2000; 407:897–900. [DOI] [PubMed] [Google Scholar]
- 124. Rashid J.I.A., Yusof N.A. The strategies of DNA immobilization and hybridization detection mechanism in the construction of electrochemical DNA sensor: a review. Sensing Bio-Sensing Res. 2017; 16:19–31. [Google Scholar]
- 125. Loan P.T., Zhang W., Lin C.T., Wei K.H., Li L.J., Chen C.H.. Graphene/MoS2 heterostructures for ultrasensitive detection of DNA hybridisation. Adv. Mater. 2014; 26:4838–4844. [DOI] [PubMed] [Google Scholar]
- 126. Pei R.J., Cui X.Q., Yang X.R., Wang E.K.. Assembly of alternating polycation and DNA multilayer films by electrostatic layer-by-layer adsorption. Biomacromolecules. 2001; 2:463–468. [DOI] [PubMed] [Google Scholar]
- 127. Saurer E.M., Flessner R.M., Sullivan S.P., Prausnitz M.R., Lynn D.M.. Layer-by-layer assembly of DNA- and protein-containing films on microneedles for drug delivery to the skin. Biomacromolecules. 2010; 11:3136–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Cao Y.W., Jin R., Mirkin C.A.. DNA-modified core-shell Ag/Au nanoparticles. J. Am. Chem. Soc. 2001; 123:7961–7962. [DOI] [PubMed] [Google Scholar]
- 129. Li F., Zhang H., Dever B., Li X.F., Le X.C.. Thermal stability of DNA functionalized gold nanoparticles. Bioconjugate Chem. 2013; 24:1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Kim J.W., Kim J.H., Deaton R.. DNA-linked nanoparticle building blocks for programmable matter. Angew. Chem., Int. Ed. Engl. 2011; 50:9185–9190. [DOI] [PubMed] [Google Scholar]
- 131. White S.P., Dorfman K.D., Frisbie C.D.. Label-free DNA sensing platform with low-voltage electrolyte-gated transistors. Anal. Chem. 2015; 87:1861–1866. [DOI] [PubMed] [Google Scholar]
- 132. Moreno-Hagelsieb L. Sensitive DNA electrical detection based on interdigitated Al/Al2O3 microelectrodes. Sens. Actuators B. 2004; 98:269–274. [Google Scholar]
- 133. Nguyen H.H., Park J., Hwang S., Kwon O.S., Lee C.S., Shin Y.B., Ha T.H., Kim M.. On-chip fluorescence switching system for constructing a rewritable random access data storage device. Sci. Rep. 2018; 8:337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Fuentes M., Mateo C., Rodriguez A., Casqueiro M., Tercero J.C., Riese H.H., Fernandez-Lafuente R., Guisan J.M.. Detecting minimal traces of DNA using DNA covalently attached to superparamagnetic nanoparticles and direct PCR-ELISA. Biosens. Bioelectron. 2006; 21:1574–1580. [DOI] [PubMed] [Google Scholar]
- 135. Li Z., Jin R.C., Mirkin C.A., Letsinger R.L.. Multiple thiol-anchor capped DNA-gold nanoparticle conjugates. Nucleic Acids Res. 2002; 30:1558–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Dougan J.A., Karlsson C., Smith W.E., Graham D.. Enhanced oligonucleotide-nanoparticle conjugate stability using thioctic acid modified oligonucleotides. Nucleic Acids Res. 2007; 35:3668–3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Fixe F., Dufva M., Telleman P., Christensen C.B.. Functionalization of poly(methyl methacrylate) (PMMA) as a substrate for DNA microarrays. Nucleic Acids Res. 2004; 32:e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Ahangar L.E., Mehrgardi M.A.. Nanoporous gold electrode as a platform for the construction of an electrochemical DNA hybridization biosensor. Biosens. Bioelectron. 2012; 38:252–257. [DOI] [PubMed] [Google Scholar]
- 139. Guo M., Chen J., Liu D., Nie L., Yao S.. Electrochemical characteristics of the immobilization of calf thymus DNA molecules on multi-walled carbon nanotubes. Bioelectrochemistry. 2004; 62:29–35. [DOI] [PubMed] [Google Scholar]
- 140. Choi Y., Bae H.J., Lee A.C., Choi H., Lee D., Ryu T., Hyun J., Kim S., Kim H., Song S.H.et al.. DNA micro-disks for the management of DNA-based data storage with index and write-once-read-many (WORM) memory features. Adv. Mater. 2020; 32:e2001249. [DOI] [PubMed] [Google Scholar]
- 141. Green N.M. Avidin .1. Use of 14c biotin for kinetic studies and for assay. Biochem. J. 1963; 89:585–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Piran U., Riordan W.J.. Dissociation rate-constant of the biotin-streptavidin complex. J. Immunol. Methods. 1990; 133:141–143. [DOI] [PubMed] [Google Scholar]
- 143. Jung Y.K., Kim T.W., Jung C., Cho D.Y., Park H.G.. A polydiacetylene microchip based on a biotin-streptavidin interaction for the diagnosis of pathogen infections. Small. 2008; 4:1778–1784. [DOI] [PubMed] [Google Scholar]
- 144. Song Y., Kim S., Heller M.J., Huang X.. DNA multi-bit non-volatile memory and bit-shifting operations using addressable electrode arrays and electric field-induced hybridization. Nat. Commun. 2018; 9:281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Zoltewicz J.A., Clark D.F., Sharpless T.W., Grahe G.. Kinetics and mechanism of acid-catalyzed hydrolysis of some purine nucleosides. J. Am. Chem. Soc. 1970; 92:1741–1750. [DOI] [PubMed] [Google Scholar]
- 146. Pruvost M., Schwarz R., Correia V.B., Champlot S., Braguier S., Morel N., Fernandez-Jalvo Y., Grange T., Geigl E.-M.. Freshly excavated fossil bones are best for amplification of ancient DNA. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Suzuki T., Ohsumi S., Makino K.. Mechanistic studies on depurination and apurinic site chain breakage in oligodeoxyribonucleotides. Nucleic Acids Res. 1994; 22:4997–5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Lindahl T., Nyberg B.. Heat-induced deamination of cytosine residues in deoxyribonucleic-acid. Biochemistry. 1974; 13:3405–3410. [DOI] [PubMed] [Google Scholar]
- 149. Lindahl T., Karlstro O.. Heat-induced depyrimidination of deoxyribonucleic acid in neutral solution. Biochemistry. 1973; 12:5151–5154. [DOI] [PubMed] [Google Scholar]
- 150. Evans R.K., Xu Z., Bohannon K.E., Wang B., Bruner M.W., Volkin D.B.. Evaluation of degradation pathways for plasmid DNA in pharmaceutical formulations via accelerated stability studies. J. Pharm. Sci. 2000; 89:76–87. [DOI] [PubMed] [Google Scholar]
- 151. Lindahl T., Nyberg B.. Rate of depurination of native deoxyribonucleic acid. Biochemistry. 1972; 11:3610–3618. [DOI] [PubMed] [Google Scholar]
- 152. Pogocki D., Schoneich C.. Chemical stability of nucleic acid-derived drugs. J. Pharm. Sci. 2000; 89:443–456. [DOI] [PubMed] [Google Scholar]
- 153. Hovorka S.W., Schoneich C.. Oxidative degradation of pharmaceuticals: theory, mechanisms and inhibition. J. Pharm. Sci. 2001; 90:253–269. [DOI] [PubMed] [Google Scholar]
- 154. Molina M., Anchordoquy T.J.. Metal contaminants promote degradation of lipid/DNA complexes during lyophilization. Biochim. Biophys. Acta. 2007; 1768:669–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Komiyama M., Takeda N., Shigekawa H.. Hydrolysis of DNA and RNA by lanthanide ions: mechanistic studies leading to new applications. Chem. Commun. 1999; 1999:1443–1451. [Google Scholar]
- 156. Graf E., Mahoney J.R., Bryant R.G., Eaton J.W.. Iron-catalyzed hydroxyl radical formation - stringent requirement for free iron coordination site. J. Biol. Chem. 1984; 259:3620–3624. [PubMed] [Google Scholar]
- 157. Bucak M.N., Tuncer P.B., Sariozkan S., Baspinar N., Taspinar M., Coyan K., Bilgili A., Akalin P.P., Buyukleblebici S., Aydos S.et al.. Effects of antioxidants on post-thawed bovine sperm and oxidative stress parameters: antioxidants protect DNA integrity against cryodamage. Cryobiology. 2010; 61:248–253. [DOI] [PubMed] [Google Scholar]
- 158. Vilenchik M.M. Studies of DNA damage and repair of thermal-induced and radiation-induced lesions in human-cells. Int. J. Radiat. Biol. 1989; 56:685–689. [DOI] [PubMed] [Google Scholar]
- 159. Bonnet J., Colotte M., Coudy D., Couallier V., Portier J., Morin B., Tuffet S.. Chain and conformation stability of solid-state DNA: implications for room temperature storage. Nucleic Acids Res. 2010; 38:1531–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Lyscov V.N., Moshkovsky Y.S.. DNA cryolysis. Biochim. Biophys. Acta. 1969; 190:101–110. [DOI] [PubMed] [Google Scholar]
- 161. Zhang M., Oldenhof H., Sydykov B., Bigalk J., Sieme H., Wolkers W.F.. Freeze-drying of mammalian cells using trehalose: preservation of DNA integrity. Sci. Rep. 2017; 7:6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Zhu B., Furuki T., Okuda T., Sakurai M.. Natural DNA mixed with trehalose persists in B-form double-stranding even in the dry state. J. Phys. Chem. B. 2007; 111:5542–5544. [DOI] [PubMed] [Google Scholar]
- 163. Emanuele E., Bertona M., Sanchis-Gomar F., Pareja-Galeano H., Lucia A.. Protective effect of trehalose-loaded liposomes against UVB-induced photodamage in human keratinocytes. Biomed. Rep. 2014; 2:755–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Yoshioka S., Aso Y.. Correlations between molecular mobility and chemical stability during storage of amorphous pharmaceuticals. J. Pharm. Sci. 2007; 96:960–981. [DOI] [PubMed] [Google Scholar]
- 165. Hancock B.C., Zograf G.. Characteristics and significance of the amorphous state in pharmaceutical systems. J. Pharm. Sci. 1997; 86:1–12. [DOI] [PubMed] [Google Scholar]
- 166. Branco C.S., Garcez M.E., Pasqualotto F.F., Erdtman B., Salvador M.. Resveratrol and ascorbic acid prevent DNA damage induced by cryopreservation in human semen. Cryobiology. 2010; 60:235–237. [DOI] [PubMed] [Google Scholar]
- 167. Howlett S.E., Castillo H.S., Gioeni L.J., Robertson J.M., Donfack J.. Evaluation of DNAstable for DNA storage at ambient temperature. Forensic Sci. Int.: Genet. 2014; 8:170–178. [DOI] [PubMed] [Google Scholar]
- 168. Ivanova N.V., Kuzmina M.L.. Protocols for dry DNA storage and shipment at room temperature. Mol. Ecol. Resour. 2013; 13:890–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Wan E., Akana M., Pons J., Chen J., Musone S., Kwok P.-Y., Liao W.. Green technologies for room temperature nucleic acid storage. Curr. Issues Mol. Biol. 2010; 12:135–141. [PMC free article] [PubMed] [Google Scholar]
- 170. Hutchinson F. Chemical changes induced in DNA by ionizing radiation. Prog. Nucleic Acid Res. Mol. Biol. 1985; 32:115–154. [DOI] [PubMed] [Google Scholar]
- 171. Boudaıffa B., Cloutier P., Hunting D., Huels M.A., Sanche L.. Resonant formation of DNA strand breaks by low-energy (3 to 20 eV) electrons. Science. 2000; 287:1658–1660. [DOI] [PubMed] [Google Scholar]
- 172. Ward J.F. DNA Damage Produced by Ionizing Radiation in Mammalian Cells: Identities, Mechanisms of Formation, and Reparability. Prog. Nucleic Acid Res. Mol. Biol. 1988; 35:95–125. [DOI] [PubMed] [Google Scholar]
- 173. Sutherland B.M., Bennett P.V., Sidorkina O., Laval J.. Clustered damages and total lesions induced in DNA by ionizing radiation: oxidized bases and strand breaks. Biochemistry. 2000; 39:8026–8031. [DOI] [PubMed] [Google Scholar]
- 174. Teoule R. Radiation-induced DNA damage and its repair. Int. J. Radiat. Biol. Relat. Stud. Phys., Chem. Med. 1987; 51:573–589. [DOI] [PubMed] [Google Scholar]
- 175. Wood R.D., Mitchell M., Lindahl T.. Human DNA repair genes, 2005. Mutat. Res. 2005; 577:275–283. [DOI] [PubMed] [Google Scholar]
- 176. Ward J. The yield of DNA double-strand breaks produced intracellularly by ionizing radiation: a review. Int. J. Radiat. Biol. 1990; 57:1141–1150. [DOI] [PubMed] [Google Scholar]
- 177. Elkind M., Redpath J.. Becker F.F. Molecular and cellular biology of radiation lethality. Radiotherapy, Surgery, and Immunotherapy. 1977; 6:Boston: Springer Press; 51–99. [Google Scholar]
- 178. Reitz G., Beaujean R., Benton E., Burmeister S., Dachev T., Deme S., Luszik-Bhadra M., Olko P.. Space radiation measurements on-board ISS–the DOSMAP experiment. Radiat. Prot. Dosimetry. 2005; 116:374–379. [DOI] [PubMed] [Google Scholar]
- 179. Koch J., Gantenbein S., Masania K., Stark W.J., Erlich Y., Grass R.N.. A DNA-of-things storage architecture to create materials with embedded memory. Nat. Biotechnol. 2020; 38:39–43. [DOI] [PubMed] [Google Scholar]
- 180. Paunescu D., Fuhrer R., Grass R.N.. Protection and deprotection of DNA–high-temperature stability of nucleic acid barcodes for polymer labeling. Angew. Chem., Int. Ed. Engl. 2013; 52:4269–4272. [DOI] [PubMed] [Google Scholar]
- 181. Schaaper R.M. Base selection, proofreading, and mismatch repair during DNA replication in Escherichia coli. J. Biol. Chem. 1993; 268:23762–23765. [PubMed] [Google Scholar]
- 182. Garmendia C., Bernad A., Esteban J.A., Blanco L., Salas M.. The bacteriophage phi 29 DNA polymerase, a proofreading enzyme. J. Biol. Chem. 1992; 267:2594–2599. [PubMed] [Google Scholar]
- 183. von Wintersdorff C.J., Penders J., van Niekerk J.M., Mills N.D., Majumder S., van Alphen L.B., Savelkoul P.H., Wolffs P.F.. Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 2016; 7:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Acman M., van Dorp L., Santini J.M., Balloux F.. Large-scale network analysis captures biological features of bacterial plasmids. Nat. Commun. 2020; 11:2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Davison J. Genetic exchange between bacteria in the environment. Plasmid. 1999; 42:73–91. [DOI] [PubMed] [Google Scholar]
- 186. Sheth R.U., Wang H.H.. DNA-based memory devices for recording cellular events. Nat. Rev. Genet. 2018; 19:718–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Ceze L., Nivala J., Strauss K.. Molecular digital data storage using DNA. Nat. Rev. Genet. 2019; 20:456–466. [DOI] [PubMed] [Google Scholar]
- 188. Meiser L.C., Koch J., Antkowiak P.L., Stark W.J., Heckel R., Grass R.N.. DNA synthesis for true random number generation. Nat. Commun. 2020; 11:5869. [DOI] [PMC free article] [PubMed] [Google Scholar]