Abstract
Polycomb group (PcG) proteins are widely utilized for transcriptional repression in eukaryotes. Here, we characterize, in the protist Tetrahymena thermophila, the EZL1 (E(z)-like 1) complex, with components conserved in metazoan Polycomb Repressive Complexes 1 and 2 (PRC1 and PRC2). The EZL1 complex is required for histone H3 K27 and K9 methylation, heterochromatin formation, transposable element control, and programmed genome rearrangement. The EZL1 complex interacts with EMA1, a helicase required for RNA interference (RNAi). This interaction is implicated in co-transcriptional recruitment of the EZL1 complex. Binding of H3K27 and H3K9 methylation by PDD1—another PcG protein interacting with the EZL1 complex—reinforces its chromatin association. The EZL1 complex is an integral part of Polycomb bodies, which exhibit dynamic distribution in Tetrahymena development: Their dispersion is driven by chromatin association, while their coalescence by PDD1, likely via phase separation. Our results provide a molecular mechanism connecting RNAi and Polycomb repression, which coordinately regulate nuclear bodies and reorganize the genome.
INTRODUCTION
Polycomb group (PcG) proteins are highly conserved in eukaryotes and widely involved in transcriptional repression: Among the best characterized are their roles in Hox gene repression in Drosophila and vertebrates, and in X chromosome inactivation in female mammals (reviewed in (1,2)). Biochemical analysis of PcG proteins has defined two major Polycomb Repressive Complexes (PRCs), PRC1 and PRC2: PRC1 drives nucleosome compaction and histone H2A ubiquitylation, while PRC2 is required for histone H3 Lys27 (H3K27) methylation (reviewed in (3–6)). In Drosophila, PcG proteins are targeted to specific DNA sequences, PcG Response Elements (PRE) (7). However, the poorly conserved nature of PRE strongly suggests alternative mechanisms for recruiting PcG proteins (8).
Growing evidence implicates long noncoding RNA (ncRNA) as well as small RNA (sRNA) in PcG protein-mediated transcriptional repression (reviewed in (6,9,10)). Many long ncRNA in mammalian cells, including Xist RNA involved in X inactivation, are associated with PRC2 and implicated in PRC2-mediated transcriptional repression (11–13). However, there are still controversies concerning the RNA-binding specificity of PRC2 (14,15), as well as the exact role played by long ncRNA in targeting or regulating PRC2 (16–19). X inactivation also intersects with nuclear RNA interference (RNAi)—a conserved pathway for co-transcriptional gene silencing (reviewed in (20–22)), though contrasting findings complicate their interpretation (23–25). In Drosophila, both RNAi components and PcG proteins are required for silencing induced by transgene repeats (26–28); RNAi components are also required for the higher-order nuclear organization mediated by PcG proteins (29). While these results implicate long ncRNA and sRNA in Polycomb repression, mechanistic details concerning their role in the recruitment of PcG proteins are still largely missing, and they may differ between species (30–32).
Studies in the unicellular eukaryote Tetrahymena thermophila have revealed a heterochromatin formation pathway requiring both nuclear RNAi and PcG proteins, providing a unique opportunity to dissect the interaction between them (reviewed in (33,34)). Like most ciliated protozoa, Tetrahymena contains in the same cytoplasmic compartment two types of nuclei: the germline micronucleus (MIC) and the somatic macronucleus (MAC) (reviewed in (35)). MIC differentiate into MAC during conjugation, the sexual phase of Tetrahymena life cycle (see Supplemental Figure S1 for a timeline of major events). The nuclear differentiation is accompanied by heterochromatinization and eventually removal of repetitive sequences—mostly derived from transposable elements (TEs) (reviewed in (34,36)). The development program starts with RNA polymerase II (Pol II)-catalyzed transcription of long ncRNA in the meiotic MIC (37–39). A special class of sRNA, referred to as scan RNA (scnRNA), accumulates in a manner dependent upon the nuclear RNAi machinery, which includes DCL1, a Dicer-like protein that processes long ncRNA into scnRNA (40,41), and TWI1, an Argonaute/piwi homologue that binds scnRNA (42–44). The sequence specificity of heterochromatinization and DNA elimination is determined by base-pairing between complementary scnRNA and nascent transcripts in the developing MAC, facilitated by a RNA helicase, EMA1 (39). Heterochromatin-specific histone modifications, H3K27 and H3K9 methylation, are deposited in a manner dependent upon the nuclear RNAi machinery as well as EZL1, a Tetrahymena homologue to the Drosophila PcG protein E(z) (45–48). These modifications are subsequently recognized by chromodomain-containing effectors like PDD1, which help to form heterochromatic structures containing DNA sequences eventually eliminated (46,48–50). Even as the case is being built for nuclear RNAi-dependent recruitment of EZL1, there remains a tantalizing gap in the molecular mechanism.
PcG proteins display dynamic distribution patterns in the nucleus of metazoan cells; the more discrete forms, previously variably described as speckles, foci, or puncta, are now recognized as a class of nuclear bodies and generally referred to as Polycomb bodies (reviewed in (51,52)). Emerging evidence connects the dynamic behavior of PcG proteins with protein phase separation (53,54), as a special manifestation of nuclear condensates (reviewed in (55,56)). The same set of mutations in CBX2, a chromodomain-containing component of PRC1, undermines nucleosome compaction (57) and organismal development (58), as well as nuclear condensate formation (53,54). More generally, nuclear condensates have been implicated in chromatin organization (59,60), including HP1-mediated heterochromatin formation (61,62). Intriguingly, similar dynamic behavior is also observed in developmentally programmed heterochromatin formation and genome rearrangement in Tetrahymena. PDD1 has long been known to exhibit dynamic distribution patterns during conjugation—from diffuse presence to large membraneless structures (49,63). This observation has since been extended to many other proteins in the same pathway (46,64–70). Like protein phase separation, the dynamic behavior is affected by a similar panel of factors, including post-translational modifications, RNA-binding, and prion-like low-complexity domains (66,67,71). Formation of Polycomb bodies (and nuclear condensates in general) therefore provides a theoretical framework to unify a wide range of phenomena involving cooperative organization of the genome and the chromatin at multiple levels.
Our previous characterization of the E(z) homologue EZL1 supports the presence of a functional Polycomb repression pathway in Tetrahymena (46,47). Here we characterize the EZL1 complex, with components conserved in Polycomb Repressive Complexes 1 and 2 (PRC1 and PRC2) of higher eukaryotes. The EZL1 complex is required for transcriptional repression of TE-related sequences and programmed genome rearrangement in Tetrahymena. It interacts with EMA1, and through scnRNA-mediated tethering to nascent ncRNA transcripts, is co-transcriptionally targeted to the chromatin. The chromatin recruitment of the EZL1 complex is reinforced by its association with PDD1. We also show that dynamic behavior of Polycomb bodies in Tetrahymena is regulated by the nuclear RNAi-dependent Polycomb repression pathway. Based upon conservation of key components and presence of similar pathways in eukaryotes, we propose that nuclear RNAi-dependent recruitment of PcG proteins may be widely implicated in transcriptional repression. We also argue that opposing forces exerted by chromatin tethering and phase separation may be generally utilized in regulating Polycomb bodies and other nuclear condensates, with critical roles in genome organization and rearrangement.
MATERIALS AND METHODS
Additional details are available in supplemental information.
Strains and culture conditions
Tetrahymena strains (Supplemental Table S2) were produced using fusion PCR generated constructs (Supplemental Table S3), as previously described (72). Tetrahymena cells were grown at 30°C in SPP medium (73). To initiate conjugation, log-phase growing cells (∼2 × 105/ml) of two different mating types were washed, starved, and mixed in 10 mM Tris (pH 7.4) or Dryl's buffer (2 mM sodium citrate, 1 mM NaH2PO4, 1 mM Na2HPO4, 1.5 mM CaCl2, pH 6.8) at 30°C (73,74).
Purification and characterization of the EZL1 complex
Conjugating Tetrahymena cells were homogenized in immunoprecipitation buffer (30 mM HEPES (pH 7.4), 150 mM NaCl, 20 mM KCl, 2 mM MgCl2, 0.1% Triton X-100, 10% glycerol, 1 mM PMSF, and the Complete® protease inhibitor cocktail (Roche)). The soluble fraction was separated by centrifugation and incubated with anti-HA agarose (Sigma) for 4 h at 4°C. Benzonase or other nucleases was used to limit interference from nucleic acids. After repeated wash, the associated proteins were eluted with HA peptide (500 ng/μl) in immunoprecipitation buffer.
For crosslinking immunoprecipitation, conjugating Tetrahymena cells were resuspended in fixation buffer (PBS, 0.1% paraformaldehyde) and incubated at room temperature for 5m. After washing (50 mM Tris (pH 8.0), 1 mM MgCl2, 10 mM KCl), cells were resuspended in ice-cold immunoprecipitation buffer and sonicated (Branson Sonifier 250, 90% duty cycle, output 4, 4 × 15 s burst). The soluble fraction was recovered after centrifugation and filtration, and incubated with anti-HA agarose (Sigma) for 4 h at 4°C.
EZL1-associated proteins were identified by Orbitrap tandem mass spectrometry after in-gel or in-solution trypsin digestion. The functionality of the EZL1 complex was confirmed by the histone methyltransferase assay. In 30 μl of the reaction buffer (50 mM Tris (pH 8.8), 4 mM DTT), 2 μg WT or mutant histone H3 was incubated for 30 m at 30°C in the presence of [3H-me] SAM and 1 μl of the purified EZL1 complex (∼50 ng). Reactions were analyzed by SDS-PAGE followed by fluorography.
Antibody generation and immunofluorescence staining
EZL1, RNF1, NUD1 and SUZ12 coding regions were synthesized after codon optimized for E. coli expression. They were inserted into the pGEX-4T1 vector for expression as GST-fusion proteins. The purified fusion proteins were used to immunize rabbits to generate specific polyclonal antibodies (Covance). For immunofluorescence staining, Tetrahymena cells were fixed in 2% paraformaldehyde (PBS) for 10 min, and permeabilized in 0.4% Triton X-100 (PBS) for 3 min, before incubation with appropriate antibodies.
Preparation of the developing MAC and Illumina sequencing of genomic DNA
The developing MAC were purified from WT, ΔEZL1, ΔRNF1 and ΔPDD1 cells at the end of conjugation (36 h post-mixing). The developing MAC were first separated from the parental MAC and the new MIC by differential centrifugation (75). Nuclear pellets collected at 2500g, containing a large proportion of the developing MAC, were used as input for fluorescence-activated cell sorting (FACS) (76). Briefly, nuclear pellets were resuspended in PBS, stained with propidium iodide, sorted with gates optimized for collecting the developing MAC. Illumina sequencing libraries for genomic DNA were prepared by NEBNext® kit (New England Biolabs) to provide even coverage across GC%.
Illumina sequencing data analysis
All sequencing data were cleaned by trim_galore with default parameters and then mapped to the Tetrahymena MIC genome (77), using Bowtie2 (v2–2.2.4) (78) with parameters ‘-q –phred33 –very-sensitive -p 10’. BAM files of mapping results were merged for the same sample using SAMtools by using BEDTools (79). Then, we removed duplicated reads using SAMtools (v1.5) (80). Bigwig files were generated from BAM files by using deepTools2 (v3.2.1) (81) with command ‘–outFileFormat bigwig –normalizeUsing RPKM –minMappingQuality 30 –binSize 20 –smoothLength 60 –numberOfProcessors 10 –extendReads 126’. The heatmap plots of signals centered on 177 well-defined CBSs and 6174 well-defined IESs (No sequencing gaps (Ns) in the regions for analysis) were generated by deepTools2 subcommand plotHeatmap with bigwig files. The signals in these selected regions were represented relative to the maximum value in each sample.
Chromatin immunoprecipitation (ChIP) and RNA immunoprecipitation (RIP)
For ChIP, 2 × 107 conjugating Tetrahymena cells were resuspended in 10 ml of fixation buffer I (PBS, 1 mg/ml disuccinimidyl glutarate (DSG, Thermo Scientific)) and incubated at room temperature for 30 m. Cells were washed with PBS twice, resuspended in fixation buffer II (PBS, 1% paraformaldehyde (w/v)), and incubated at room temperature for 10 m. After washing (50 mM Tris (pH 8.0), 1 mM MgCl2, 10 mM KCl), cells were resuspended in 10 ml of SDS lysis buffer (2% SDS, 10 mM EDTA, 50 mM Tris (pH8.0)) and sonicated (Branson Sonifier 250, 90% duty cycle, output 4, 20 × 15 s burst). After centrifugation and filtration, the soluble fraction was recovered and diluted in immunoprecipitation buffer (1:20) as the input for ChIP. ChIP was performed as previously described (46).
For RIP, 1 × 107 conjugating Tetrahymena cells were homogenized in 1 ml of immunoprecipitation buffer (see Purification and characterization of the EZL1 complex), supplemented with 10 mM Ribonucleoside Vanadyl Complex (New England Biolabs) and 0.4 U/ul of RNaseOUT (Invitrogen). After centrifugation and filtration, the supernatant was recovered and incubated with anti-HA agarose beads (Sigma) for 2 h at at 30°C. The beads were washed with immunoprecipitation buffer without Triton X-100, and treated with Turbo DNA-free™ (Ambion) for 30 m at 25°C. The beads were washed and extracted with Trizol® Reagent. The recovered RNA was analyzed by RT-PCR.
RESULTS
The EZL1 complex is a prototypical Polycomb repressive complex required for H3K27 and H3K9 methylation
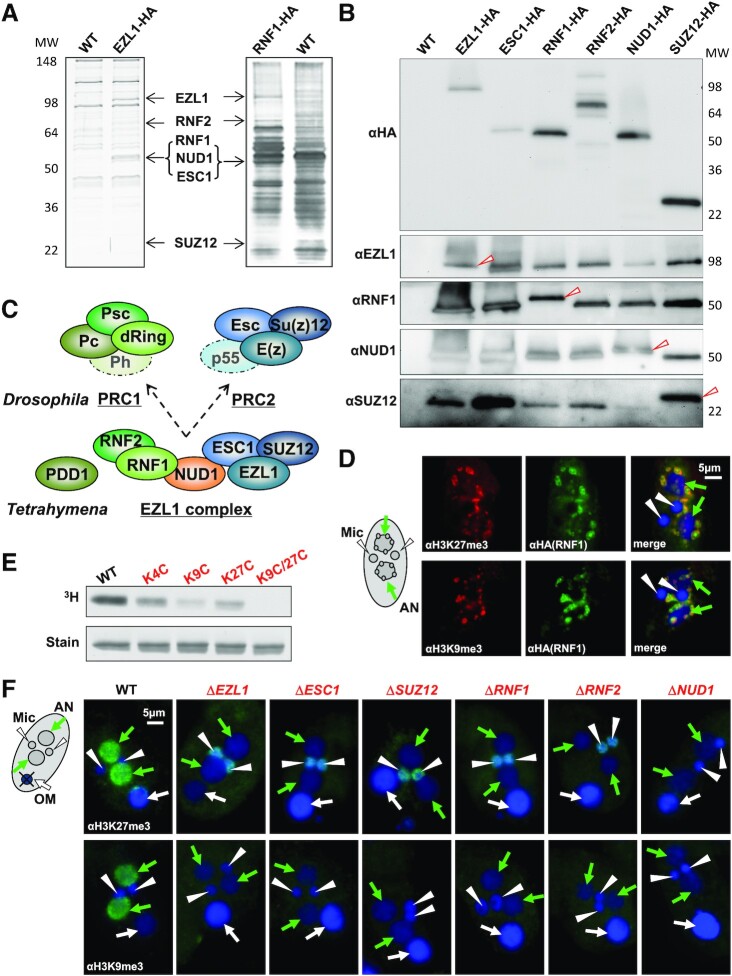
We identified EZL1-associated proteins by affinity purification and mass spectrometry (Figure 1A; Supplemental Figure S2; Supplemental Table S1). EZL1 was epitope-tagged at the C-terminus, which was fully functional as it could rescue the defect of ΔEZL1 cells (Supplemental Figure S2A-C). EZL1 and its associated proteins were affinity purified from conjugating cells (Figure 1A: left panel; Supplemental Figure S2D). Mass spectrometry analysis identified five EZL1-associated proteins: ESC1, SUZ12, RNF1, RNF2 and NUD1 (Figure 1A; Supplemental Table S1). EZL1 and the five associated proteins apparently co-fractionated during gel filtration (Supplemental Figure S2E), consistent with the formation of a stable complex. To verify their interactions, we also tagged all five EZL1-associated proteins and recovered by affinity purification essentially the same set of proteins (Figure 1A: right panel, Figure 1B; and Supplemental Table S1). Furthermore, these proteins were co-expressed (Supplemental Figure S3A) and co-localized (Supplemental Figure S3B); absence of any one of them destabilized at least some other components, if not the whole complex (Supplemental Figure S3C). Our results support that they are integral components of a complex—the EZL1 complex.
Figure 1.
The EZL1 complex is a prototypical Polycomb repressive complex. (A) Affinity purification of the EZL1 complex from conjugating Tetrahymena cells. IP samples from WT control, EZL1-HA, and RNF1-HA cells were resolved by SDS-PAGE and visualized by silver staining, with EZL1 and its associated proteins—RNF2, RNF1, NUD1, ESC1 and SUZ12, designated at proper positions. (B) Co-purification of the EZL1 complex components. All six EZL1 complex components were HA tagged and subjected to IP with the anti-HA antibody. EZL1, RNF1, NUD1 and SUZ12 were detected in IP samples by custom antibodies. Note the slight up-shift in the migration of the HA-tagged proteins compared with the endogenous proteins (arrowhead). Only the tagged form was purified even when the untagged form was also present in the input, supporting that the complex contains only one subunit for each of the components tested. (C) Conservation of the EZL1 complex. The Tetrahymena EZL1 complex shares components with the Drosophila Polycomb Repressive Complexes, PRC1 and PRC2 (indicated by the same colors). The schematic for the EZL1 complex does not indicate any spatial arrangement or interactions between its components. (D) Co-localization of the EZL1 complex with H3K27 and H3K9 methylation. RNF1-HA cells were stained with the anti-HA antibody (green) and specific antibodies against H3K27me3 or H3K9me3 (red); nuclei counterstained with DAPI (blue). Note the prominent DNA elimination bodies in the developing MAC. Developing MAC (AN, for anlagen): green arrow; new MIC (Mic): white arrowhead. (E) Histone methyltransferase (HMT) activity of the EZL1 complex. HMT activity was detected in the affinity purified EZL1 complex, with its substrate specificity pinpointed by recombinant H3 mutated at designated lysine residues. (F) The EZL1 complex-dependent H3K27 and H3K9 methylation in the developing MAC. Conjugating cells from WT and all the EZL1 complex KO mutants were stained with specific antibodies against H3K27 and H3K9 methylation (green), and counterstained with DAPI (blue). Both H3K27 and H3K9 methylation were missing from developing MAC in the mutants. Note the abnormal H3K27 methylation often found in the new MIC of the mutants, which may be attributed to occasional germline activation of transposable elements (Figure 2F) (47). Developing MAC (AN): green arrow; new MIC (Mic): white arrowhead; old MAC (OM): white arrow.
Biochemical analyses in Drosophila and mammalian cells have defined two major Polycomb Repressive Complexes (PRCs), PRC1 and PRC2 (reviewed in (82,83)). PRC1 contains RING finger (dRing and Psc in Drosophila) and chromo domain proteins (Pc) (84–86), while PRC2 features a SET domain-containing histone methyltransferase (E(z)) (87–90). Homologues to both PRC1 and PRC2 can be found in the EZL1 complex (Figure 1C; Supplemental Figure S4): EZL1, ESC1 and SUZ12 are homologous to Drosophila PRC2 components E(z), Esc and Su(z)12, respectively, while RNF1 and RNF2 feature the RING finger domain conserved in PRC1 components dRing and Psc. PDD1, sharing a chromodomain with the PRC1 component Pc (Figure 1C; Supplemental Figure S4), showed weak but functional interaction with the EZL1 complex (Supplemental Figure S5). We also found a NUDIX domain protein (putative pyrophosphohydrolase involved in metabolism of nucleotides and their derivatives), NUD1 (Figure 1C; Supplemental Figures S4 and S6), whose association with PcG proteins was not previously reported. RBBP4, the Tetrahymena homologue of Drosophila NURF55 and mammalian RBBP4/RBBP7 (auxiliary components in metazoan PRC2), was not detected in the EZL1 complex; reciprocally, the EZL1 complex was not pulled down with RBBP4 (Supplemental Figure S7). Tetrahymena SUZ12 is much shorter than its metazoan counterpart (Supplemental Figure S4), only containing the VEFS domain defining the minimal PRC2 with methyltransferase activity (87,91), but lacking the long N-terminal extension required to engage NURF55 (RBBP4/RBBP7) and other cofactors (92). As a composite with both PRC1 and PRC2 components, the EZL1 complex is noncanonical. It may represent the ancestral PRC, from which PRC1 and PRC2 split off during evolution; alternatively, PRC1 and PRC2 may have merged in ciliates. A wide survey of PRC (especially in early branching eukaryotes) is needed to distinguish these two possibilities.
Immunofluorescence (IF) staining showed that the EZL1 complex co-localized with H3K27 and H3K9 methylation as well as their reader protein PDD1 at DNA elimination bodies (Figure 1D; Supplemental Figure S5D), heterochromatic structures containing germline-specific sequences in the late developing MAC (49). The affinity purified EZL1 complex methylated recombinant histone H3 in vitro; the histone methyltransferase activity was significantly reduced for both the H3 K9C and H3 K27C substrates, and was completely abolished when both lysine residues were mutated (Figure 1E). In knockout (KO) strains lacking any one of the EZL1 complex components, both H3K27 and H3K9 methylation in the developing MAC were abolished (Figure 1F). Moreover, the H3K27 and H3K9 methylation reader PDD1 was mis-localized (Figure 6A, D). Even though it remains controversial whether Drosophila E(z) and its mammalian homologues can methylate H3K9 as well as H3K27 (87–90), our result strongly supports the dual substrate specificity for the EZL1 complex, a feature likely conserved in ciliates (93).
Figure 6.
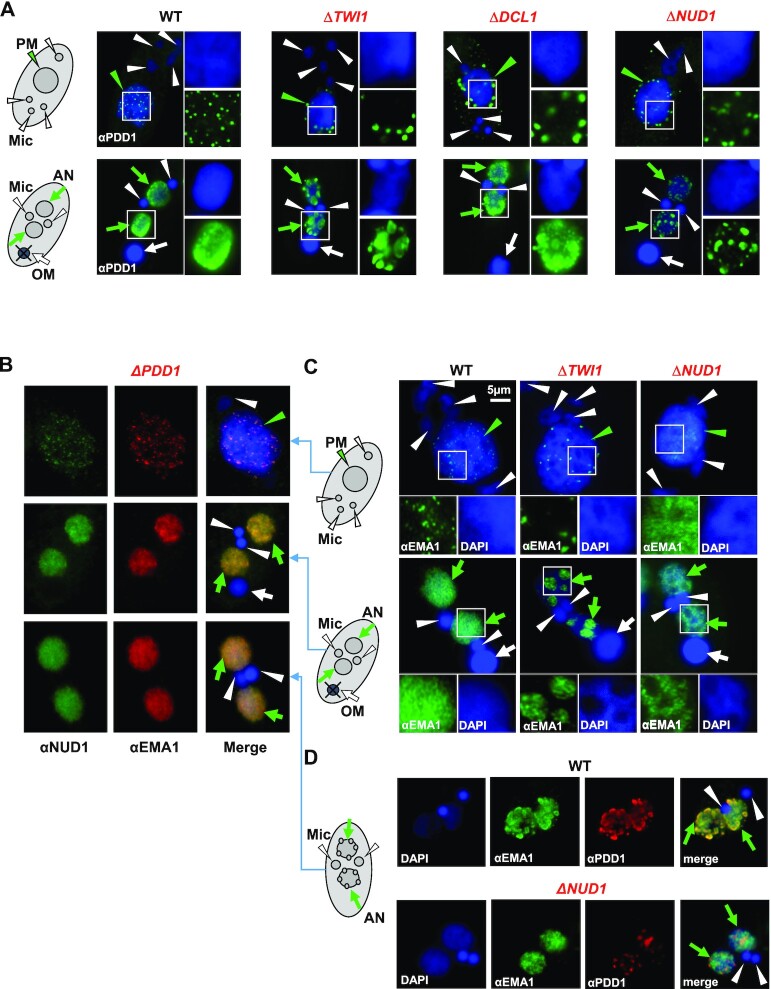
The EZL1 complex and PDD1 drive coalescence of Polycomb bodies. (A) RNAi-dependent localization of PDD1. WT and the designated RNAi mutants at early (top panels: 6 h post-mixing) and late conjugation (bottom panels: 10 h post-mixing) were stained with the anti-PDD1 antibody (green) and counterstained with DAPI (blue). Parental MAC (PM): green arrowhead; developing MAC (AN): green arrows; MIC (Mic): white arrowheads; old MAC (OM): white arrow. Granularity analysis of the PDD1 foci in the images was provided in Supplemental Figure S11C. (B) Co-localization of the EZL1 complex and EMA1 in ΔPDD1 cells. Cells at different conjugation stages (6, 10 and 14 h post-mixing; see schematics to the right) were co-stained with the anti-NUD1 (green) and anti-EMA1 (red) antibodies; counterstained with DAPI (blue). Note their abnormal dispersion, even at the late anlagen development stage. The EZL1 complex and EMA1 overlap even in this altered distribution, consistent with their co-IP in ΔPDD1 cells (Figure 3G). (C) Opposite EMA1 localization patterns in ΔTWI1 and ΔNUD1. WT and the mutants at early (top panels: 6 h post-mixing) and late conjugation (bottom panels: 10 h post-mixing; see schematics to the left) were stained with the anti-EMA1 antibody (green) and counterstained with DAPI (blue). Note abnormal aggregation of EMA1 in DAPI-poor regions in ΔTWI1, as well as its dispersion in DAPI-rich regions in ΔNUD1 cells (insets). (D) Co-localization of EMA1 and PDD1 disrupted in ΔNUD1 cells. WT and the mutant at late conjugation (14 h post-mixing; see schematics to the left) were sequentially stained with the anti-EMA1 (green) and anti-PDD1 (red) antibodies; counterstained with DAPI (blue). Note in the mutant abnormal dispersion of EMA1 in DAPI-rich regions, while PDD1 aggregated in DAPI-poor regions.
In mammalian cells, the heterodimer formed by the RING finger domain-containing Ring1B and Bmi-1 (PRC1 components homologous to dRing and Psc in Drosophila) can mono-ubiquitylate histone H2A (94). Despite the presence of the RING finger proteins RNF1 and RNF2, ubiquitin E3 ligase activity was not detected in the EZL1 complex using standard assay conditions (Supplementary Figure S8A–D). Furthermore, in ΔRNF1 and ΔRNF2 conjugating cells, H2A ubiquitylation level was not substantially affected (Supplementary Figure S8E). We note that H2A ubiquitylation activity is also not required for Polycomb repression in Drosophila and mammals (95,96). In addition to stabilizing the EZL1 complex, RNF1 and RNF2 may play a role for chromatin compaction, as reported for metazoan PRC1 (96–98).
The EZL1 complex is required for processing IESs and CBSs, and controlling TEs
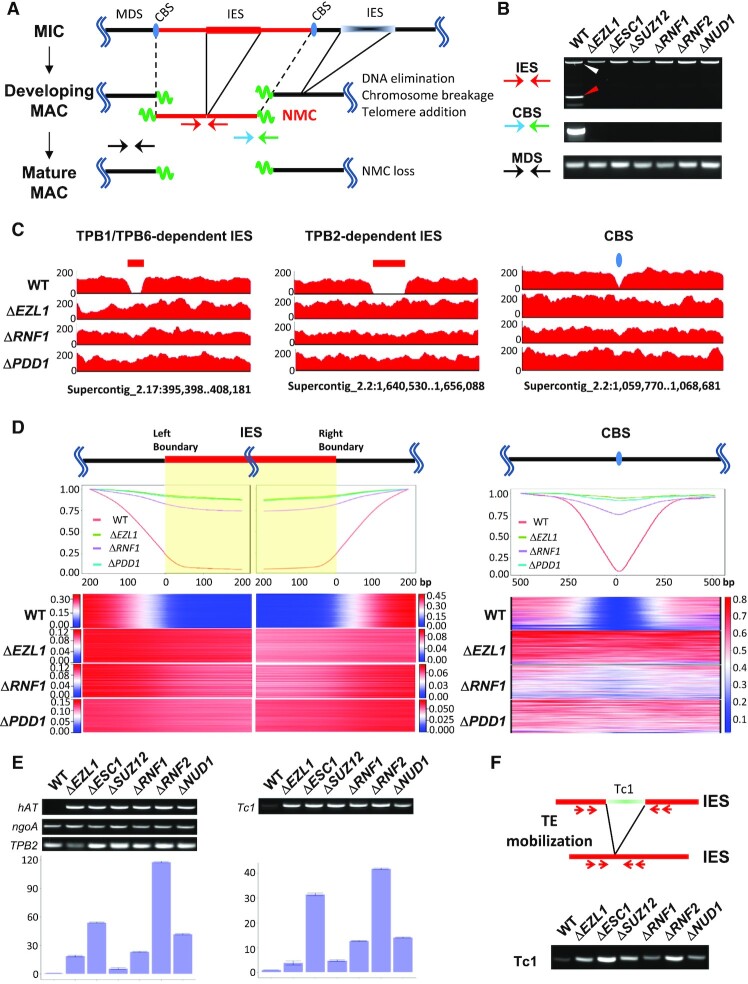
The loss of H3K27 and H3K9 methylation as well as altered PDD1 distribution in the developing MAC of the EZL1 complex mutants strongly suggests that heterochromatin formation is disrupted. In Tetrahymena, heterochromatinization in the developing MAC is followed by programmed genome rearrangement, which includes excision of internally eliminated sequences (IESs) and fragmentation at chromosome breakage sequences (CBSs) (Figure 2A). We performed standard PCR assays on a well-studied IES, the M element, revealing that elimination of the M element was completely abolished in KO strains of the EZL1 associated proteins (Supplemental Figure S9A–C). A standard PCR assay also revealed their deficiency in CBS processing (Supplemental Figure S9D, E). We corroborated defective IES and CBS processing in all EZL1 complex mutants by a new set of PCR assays (Figure 2A, B), taking advantage of the recently discovered non-maintained chromosomes (NMCs) (99,100). As NMCs are transiently present in the developing MAC and lost in the mature MAC, PCR products corresponding to successfully processed IESs and CBSs associated with NMCs are only detectable during late conjugation (99,100), eliminating the background from parental cells and increasing the assay sensitivity. This assay is readily applicable in any Tetrahymena strains with simple sample preparation.
Figure 2.
The EZL1 complex is required for controlling TEs and processing IESs and CBSs. (A) A schematic of programmed genome rearrangement in Tetrahymena. Several processes are involved: (i) excision of internally eliminated sequences (IESs, shaded boxes) and re-ligation of flanking MAC-destined sequences (MDSs, black solid lines); ii) chromosome fragmentation at chromosome breakage sequences (CBSs, blue ovals) and healing of double-strand breaks by de novo telomere addition (green wavy lines); iii) loss of non-maintained chromosomes (NMCs, red solid lines) from the mature MAC. Arrows represent PCR primers used in the IES and CBS processing assays (Figure 2B): IES (red), CBS (blue and green), MDS (black). (B) PCR assays for IES (top) and CBS processing (middle). PCR from a MDS region was included as the positive/loading control (bottom). PCR was performed on total genomic DNA purified at the end of conjugation (24 h post-mixing) from the specified cells. Two primers (red) flanking an IES regions will amplify a long PCR product if the IES is not excised (white arrowhead), or a short product upon IES excision (red arrowhead). As the IES resides in an NMC, the excision product is therefore only transiently present in the developing MAC. A NMC-specific primer (blue) and a telomere-specific primer (green) will amplify a chromosome end broken at a CBS and healed by telomere addition. This chromosome end belongs to an NMC, which is therefore only transiently present in the developing MAC. See the schematic (Figure 2A) for primer locations. (C) IES (left and middle panels) and CBS processing (right panel), visualized by sequencing coverage of selected genomic regions (coordinates in the Tetrahymena MIC genome assembly provided at the bottom). Genomic DNA was purified from the developing MAC of WT, ΔEZL1, ΔRNF1, and ΔPDD1 cells at the end of conjugation (36 h post-mixing), and subjected to Illumina sequencing. IES and CBS processing in the developing MAC of WT cells was revealed by a gap in sequencing coverage (left: TPB1/TPB6-dependent IES; middle: TPB2-dependent IES; right: CBS), which was absence in the mutants. Data are visualized by IGV (132). (D) Composite analysis of well-defined IESs (∼6000) and CBSs (∼200). Each IES was aligned at its left or right main boundary, and further extended in both directions for 200 bp (left panels). Each CBS was aligned at the left of its 15 bp consensus, and extended in both directions for 500 bp (right panel). On the top, normalized coverage (reads per million reads, RPM) of each IES or CBS regions were cumulated. IES and CBS processing in the developing MAC of WT cells was revealed by low coverage (close to 0) within the left and right boundaries of IESs or around CBSs. The coverage increased quickly (the gradient is due to heterogeneity in IES boundaries), and reached plateau values a few hundred bp away from IES boundaries and CBSs. The slightly reduced coverage within IESs and around CBSs in the mutants was attributable to the contaminating parental MAC. The coverage is plotted as ratios relative to the plateau values at specified positions. On the bottom, normalized coverage (RPM) for all well-defined IESs and CBSs is illustrated by the stacked heat map, plotted separately for each strain, IES left and right boundaries, and CBS, all with its own color scale. (E) RT-PCR revealing transcriptional activation of transposable elements (TEs) in the EZL1 complex mutants. RNA samples were collected from WT and the EZL1 complex mutants at late conjugation (10 h post-mixing). An hAT element and a Tc1 element were tested. ngoA, a gene expressed at high and constant levels during conjugation was used as the positive/loading control. TPB2, encoding a domesticated piggyBac transposase only expressed at late conjugation, was used to validate conjugation progress. Top: representative gel images. Bottom: quantification of RT-qPCR results (triplicate); expression levels in the EZL1 complex mutants were normalized against that of WT cells. (F) A PCR assay revealing increased excision of the Tc1 transposable element in the EZL1 complex mutants. Top: A schematic for excision of a Tc1 element, a functional element with recent duplications in the Tetrahymena MIC genome. The red arrows represent the nested PCR primers used in the transposition assay shown below. The boundaries of the IESs lie outside the PCR primers, so the somatically rearranged DNA will not complicate this assay. Bottom: Mobilization of the Tc1 element at low levels in WT cells but at dramatically increased levels in the mutants. To monitor excision at the Tc1 element, nested PCR was performed on total genomic DNA purified at the end of conjugation (24 h post-mixing) from the specified cells. PCR from a MDS region was included as the positive/loading control (Figure 2B, bottom panel).
To confirm the global role of PcG proteins in programmed genome rearrangement in Tetrahymena, we sequenced the purified developing MAC from ΔEZL1, ΔRNF1 and ΔPDD1 cells at the end of conjugation (Figure 2C, D). The WT developing MAC were used as the positive control, showing gaps of minimal sequencing coverage at IESs and CBSs in representative genomic regions (Figure 2C). Views of the same regions showed that ΔEZL1, ΔRNF1, and ΔPDD1 cells were deficient in processing TPB1/TPB6-dependent IES (i.e. exonic IES; Figure 2C, left panel), TPB2-dependent IES (i.e. regular IES; Figure 2C, middle panel), and CBS (Figure 2C, right panel). Furthermore, we performed composite analysis of Illumina sequencing coverage across the left and right boundaries of well-defined IES (Figure 2D, left panels). For WT cells, there was clear transition from high coverage to low coverage as one moves from MDS to IES across the boundaries, with the residue coverage of IESs (at levels <10% of flanking MDSs) attributable to contaminating MIC. For ΔEZL1 and ΔPDD1 cells, IES coverage was generally high and comparable with the flanking MDS (>80% of flanking MDSs). Similar analysis of ΔRNF1 cells showed IES coverage levels slightly lower than those in ΔEZL1 and ΔPDD1 cells (>75% of flanking MDSs; most likely due to increased parental MAC contamination), but still much higher than WT cells (Figure 2D, left panels). Composite analysis of CBSs corroborated that they also failed to be processed in ΔEZL1, ΔRNF1, and ΔPDD1 cells (Figure 2D, right panel). These results indicate that the EZL1 complex as well as PDD1 is required for programmed genome rearrangement in Tetrahymena.
Transposable elements (TEs) are controlled by RNAi-dependent Polycomb repression in Tetrahymena. To validate the role played by the EZL1 complex, we first tested RNA transcript levels from two (2) TEs in all six (6) null mutants of the EZL1 complex, with WT cells as the control (Figure 2E). Both are class I TEs—DNA transposons of hAT (Figure 2E, left panels) and Tc1/mariner superfamilies (Figure 2E, right panels)—which underwent recent transposition and may still be capable of mobilization (47). RT-PCR primed by oligo(dT) showed that RNA transcripts, most likely poly(A)-tailed mRNA, were expressed at much higher levels in the mutants than in WT cells; expression variance in the mutants may be attributed to transcriptional mis-regulation and epigenetic instability, as well as variations in sample preparation (Figure 2E). We also examined ncRNA transcripts and scnRNAs from the M element (likely derived from a degenerate TE), and found that compared with WT cells, they persisted much longer in the EZL1 complex KO strains as well as RNAi-deficient cells (Supplemental Figure S10). These results support widespread transcriptional de-repression in the absence of the EZL1 complex.
We next focused on the Tc1 element, which has been shown to mobilize in RNAi and Polycomb repression-deficient Tetrahymena cells (47). Tc1/mariner elements mobilize through a ‘cut-and-paste’ mechanism, leaving behind a ‘vacant’ locus (101,102), which was preferentially amplified by PCR due to its much smaller size relative to the original locus (Figure 2F). Only a small amount of the PCR product corresponding to the ‘vacant’ locus was detected from WT cells, reflecting the rarity of this transposition event, while the PCR product was much more abundant in the EZL1 complex KO strains, supporting its critical role in controlling TEs (Figure 2F). TE mobilization occurs predominantly, if not exclusively, in the new MIC (47). It may not be a coincidence that abnormally high levels of H3K27 methylation were often observed in the new MIC of the EZL1 complex KO strains (Figure 1F). H3K27 methylation in the new MIC, most likely catalyzed by EZL2 (103), may be a response to TE mobilization, as part of the conserved pathways affecting chromatin in DNA damage and repair (104).
The EZL1 complex interacts with the nuclear RNAi component EMA1
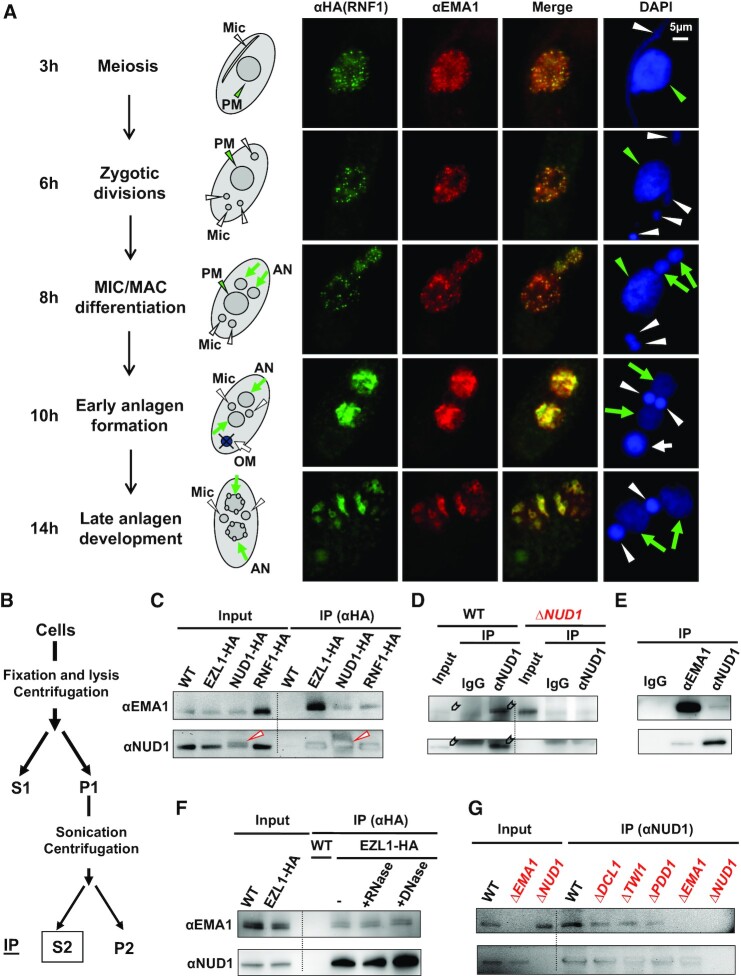
Next, we addressed the question concerning how the EZL1 complex is recruited to specific loci for heterochromatin formation. IF staining showed that the EZL1 complex localized in the parental MAC during early conjugation and in the developing MAC during late conjugation (Figure 3A and Supplemental Figure S3B), exhibiting a trafficking pattern reminiscent of TWI1 (42) and its associated proteins including EMA1, a putative RNA helicase facilitating base-pairing between scnRNA and nascent transcripts (39,105). The EZL1 complex localization largely overlapped with that of EMA1 (Figure 3A). The co-localization started in the parental MAC at early conjugation (Figure 3A: 3 and 6 h post-mixing), during which long ncRNA and scnRNA begin to accumulate (37,39,42), and EZL1-catalyzed H3K27 methylation is first detected (46). Initially, the EZL1 complex and EMA1 distributed diffusively or formed fine speckles in the parental MAC (Figure 3A: 3 h). As conjugation progressed, they gradually coalesced into fewer but more distinct foci (Figure 3A: 6 h). They diminished from the degrading parental MAC (referred to as the old MAC); they reappeared in the developing MAC, starting from early stages of MIC/MAC differentiation (Figure 3A: 8 h). The EZL1 complex and EMA1 initially distributed diffusively in the developing MAC (Figure 3A: 10 h). There they also coalesced over time, culminating in co-localization in DNA elimination bodies at late conjugation (Figure 3A: 14 h). As EMA1 is a key nuclear RNAi component required for EZL1-dependent heterochromatin formation (39), our result strongly suggests that it may play a direct role in recruiting the EZL1 complex.
Figure 3.
The EZL1 complex interacts with the nuclear RNAi component EMA1. (A) Co-localization of the EZL1 complex and EMA1. RNF1-HA cells were stained with anti-HA (green) and anti-EMA1 (red) antibodies, and counterstained with DAPI (blue). Representative images of several conjugation stages (at 3, 6, 8, 10 and 14 h post-mixing) are shown: meiosis, zygotic divisions, MIC/MAC differentiation, early anlagen (developing MAC) formation and late anlagen development. The schematic illustrates the typical nuclear morphology. Parental MAC (PM): green arrowhead; developing MAC (AN): green arrows; MIC (Mic): white arrowheads; old MAC (OM): white arrow. (B) A schematic of crosslinking IP. Cells were crosslinked with formaldehyde, and the solubilized nuclear fraction was used as IP input. (C) Co-IP of NUD1 and EMA1 in tagged cells. WT and various tagged cells were processed for crosslinking IP with the anti-HA antibody. The anti-EMA1 (top) and anti-NUD1 antibodies (bottom) were used for immunoblotting. Note the slight up-shift in the migration of NUD1-HA compared with endogenous NUD1 (arrowhead). (D) Co-IP of NUD1 and EMA1 in WT cells. The anti-NUD1 antibody was used for crosslinking IP; the anti-EMA1 (top) and anti-NUD1 antibodies (bottom) were used for immunoblotting (signal indicated by block arrows; the background band in the NUD1 panel corresponds to the IgG heavy chain); pre-bleed IgG (IgG) as negative control. (E) Reciprocal IP of NUD1 and EMA1. WT cells were processed for crosslinking IP with the pre-bleed IgG (IgG), anti-EMA1, and anti-NUD1 antibodies, respectively; the anti-EMA1 (top) and anti-NUD1 antibodies (bottom) were used for immunoblotting. (F) Nucleic acid-independent association of NUD1 and EMA1. After crosslinking IP, the anti-HA agarose was treated with RNase A or DNase I before wash and elution; no nuclease treatment (–) as the control. The anti-EMA1 (top) and anti-NUD1 antibodies (bottom) were used for immunoblotting. (G) Association of NUD1 and EMA1 in various mutants. WT and the designated mutants were processed for crosslinking IP with the anti-NUD1 antibody. The anti-EMA1 (top) and anti-NUD1 antibodies (bottom) were used for immunoblotting.
The strong co-localization pattern prompted us to examine whether there were physical interactions between the EZL1 complex and EMA1. As conventional immunoprecipitation (IP) could only detect the core complex components in association with EZL1, we adopted a procedure involving mild crosslinking, optimized for detecting weak or transient interactions (Figure 3B) (106). In this way, we identified EMA1 in association with the EZL1 complex by affinity purification from HA-tagged strains (Figure 3C and Supplemental Table S1). Furthermore, EMA1 was detected when pulling down the EZL1 complex using a NUD1 antibody from WT cells, but not from ΔNUD1 cells, nor with the pre-bleed IgG (Figure 3D). Reciprocally, NUD1 was detected in the EMA1 IP (Figure 3E). The co-IP of the EZL1 complex and EMA1 was not affected by RNase A or DNase I treatment (Figure 3F), indicating an interaction independent of nucleic acids. Furthermore, the co-IP was not affected in ΔDCL1, ΔTWI1 and ΔPDD1 cells (Figure 3G), suggesting a more direct interaction. Taken together, our results support physical interactions between the EZL1 complex and EMA1, establishing a critical link for nuclear RNAi-dependent recruitment of PcG proteins.
The EZL1 complex and EMA1 are co-transcriptionally tethered to chromatin in a nuclear RNAi-dependent manner
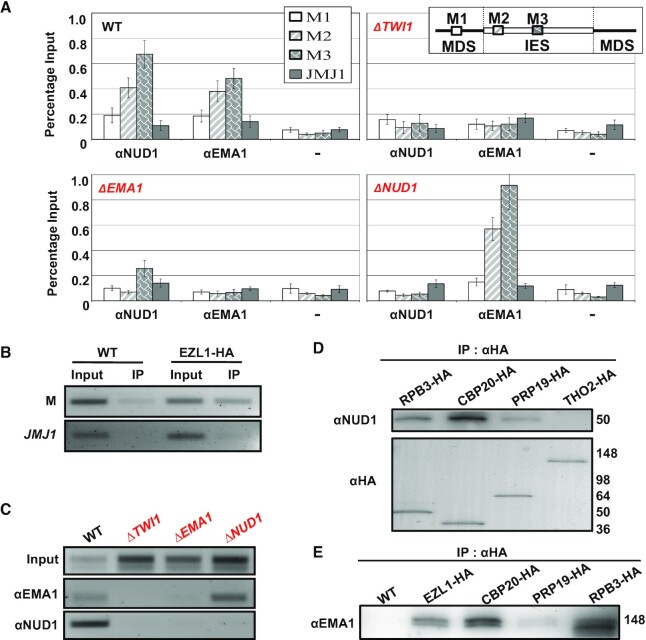
We have previously demonstrated by chromatin immunoprecipitation (ChIP) that H3K27 and H3K9 methylation, both dependent on EZL1, are enriched in IES before their eventual excision (46,48). Here we performed ChIP to map the distribution of the EZL1 complex and EMA1, focusing on the M element, an IES retained in the null mutants of the EZL1 complex (Supplemental Figure S9A–C) (46), the nuclear RNAi machinery (TWI1, DCL1 and EMA1) (39,40,42), and the chromodomain effector PDD1 (50). In WT cells, both EMA1 and NUD1 were enriched in the M element, but not in the flanking MDS or a gene-coding locus (Figure 4A). However, binding of the IES by EMA1 and NUD1 was abolished in ΔTWI1 cells (Figure 4A). Similarly, NUD1 binding was abolished in ΔEMA1 cells (Figure 4A). On the contrary, in ΔNUD1 cells, EMA1 binding was not affected (Figure 4A). Our result supports nuclear RNAi-dependent chromatin association of the EZL1 complex (Figures 3A and 5A).
Figure 4.
The EZL1 complex and EMA1 are co-transcriptionally tethered to the chromatin in a nuclear RNAi-dependent manner. (A) Association of the EZL1 complex and EMA1 with an IES, M element. WT, ΔTWI1, ΔEMA1, and ΔNUD1 cells (10 h post-mixing) were processed for ChIP with the anti-EMA1 and anti-NUD1 antibodies; pre-bleed IgG was used as the negative control (–). ChIP samples were analyzed by qPCR for levels of the associated IES (M2 and M3), as well as the flanking MDS (M1) and a gene coding region (JMJ1) as negative control. (B) Association of the EZL1 complex with the M element transcripts. IP samples (with the anti-HA antibody) from WT and EZL1-HA cells (10 h post-mixing) were reverse transcribed and analyzed by PCR for levels of associated transcripts from the M element, as well as JMJ1 as negative control. (C) Association of the EZL1 complex and EMA1 with the M element transcripts in various mutants. IP samples (with the anti-EMA1 and anti-NUD1 antibodies) from WT, ΔTWI1, ΔEMA1 and ΔNUD1 cells (10 h post-mixing) were reverse transcribed and analyzed by PCR for levels of the M element transcripts. (D) Differential interactions between the EZL1 complex and the transcriptional/co-transcriptional machineries. The designated strains were processed for crosslinking IP with the anti-HA antibody at late conjugation (10 h post-mixing), as previously reported (47). The anti-HA and anti-NUD1 antibodies were used for immunoblotting. Note that similar amounts of bait proteins were recovered, as shown by the anti-HA immunoblotting. (E) WT and various tagged strains were processed for crosslinking IP with the anti-HA antibody at late conjugation (10 h post-mixing). The anti-EMA1 antibody was used for immunoblotting.
Figure 5.
Nuclear RNAi and histone methylation-binding drive dispersion of Polycomb bodies. (A) RNAi-dependent localization of the EZL1 complex. WT and the designated RNAi mutants at early (top panels: 6 h post-mixing) and late conjugation (bottom panels: 10 h post-mixing) were stained with the anti-NUD1 antibody (green) and counterstained with DAPI (blue). Parental MAC (PM): green arrowhead; developing MAC (AN): green arrows; MIC (Mic): white arrowheads; old MAC (OM): white arrow. Granularity analysis of the EZL1 complex foci in the images was provided in Supplemental Figure S11C. (B) Co-localization of the EZL1 complex and EMA1 in ΔTWI1 cells. Cells at early (left panels: 6 h post-mixing) and late conjugation (right panels: 10 h post-mixing) were co-stained with the anti-NUD1 (green) and anti-EMA1 (red) antibodies; counterstained with DAPI (blue). Note their abnormal aggregation at the nuclear periphery or DAPI-poor regions. (C) Co-localization of the EZL1 complex, EMA1, and PDD1 in WT and EZL1 H721A cells (methyltransferase dead). EZL1-HA WT and EZL1-HA H721A cells were co-stained with the anti-HA (green) and anti-EMA1 antibodies (red; top panels), or the anti-HA (green) and anti-PDD1 antibodies (red; bottom panels); counterstained with DAPI (blue). Note abnormal aggregation in EZL1 H721A cells. (D) Co-localization of the EZL1 complex, EMA1 and PDD1 in WT and PDD1 Y49A cells (methyl-binding deficient). PDD1-HA WT and PDD1-HA Y49A cells were co-stained with the anti-HA (green) and anti-EMA1 antibodies (red; top panels), or the anti-HA (green) and anti-NUD1 antibodies (red; bottom panels); counterstained with DAPI (blue). Note abnormal aggregation in PDD1 Y49A cells.
Nascent RNA transcripts can function as an assembly platform for recruiting chromatin-modifying complexes to regulate transcription (20–22). Using RNA-immunoprecipitation (RIP), we examined the specific association of the EZL1 complex and EMA1 with transcripts from the M element, which accumulated in the mutants as well as WT cells (Supplemental Figure S10). These transcripts were enriched in the RIP sample from EZL1-tagged cells, but not the untagged control cells (Figure 4B). Additionally, no enrichment was observed for the highly abundant JMJ1 mRNA transcripts (Figure 4B). Despite accumulation of M element transcripts in ΔTWI1, ΔEMA1, as well as ΔNUD1 cells (Supplemental Figure S10), their association with NUD1 was abolished (Figure 4C), arguing against direct association of the EZL1 complex and ncRNA. Association between EMA1 and M element transcripts was abolished in ΔTWI1 cells, while it was not affected in ΔNUD1 cells (Figure 4C). Together, the ChIP and RIP results support a linear pathway: in a nuclear RNAi-dependent manner, EMA1 is first tethered to chromatin via nascent transcripts, presumably guided by TWI1 associated with homologous scnRNA (39); EMA1 in turn recruits the EZL1 complex, leading to transcriptional silencing and heterochromatinization of IES.
In Tetrahymena, RNA polymerase II (Pol II) synthesizes long ncRNA—precursors to scnRNA—in the meiotic MIC (38), and is most likely also responsible for nascent transcripts targeted by scnRNA in the parental MAC and the developing MAC, conforming to the paradigm established in other eukaryotes (22). Pol II transcription also involves a cascade of co-transcriptional events, including the addition and recognition of the 5′ cap structure, splicing, packaging and exporting (107). To understand how nuclear RNAi and Polycomb repression are coordinated with the transcriptional and co-transcriptional processes, we HA-tagged key components of corresponding molecular machineries: RPB3 for Pol II, CBP20 for the cap-binding complex, PRP19 for the splicing complex, and THO2 for the RNA packaging and exporting complex (Figure 4D). With crosslinking IP, we found that NUD1 interacted strongly with RPB3 and CBP20 (Figure 4D), supporting the co-transcriptional nature of the EZL1 complex recruitment. Only weak interactions were detected with PRP19, and no interaction with THO2 was detected (Figure 4D). This result strongly suggests that 5′ end processing play a more important role than the downstream co-transcriptional events in recruiting the EZL1 complex. Additionally, we were able to pull down substantial amount of EMA1 with RPB3, CBP20, as well as EZL1, but not much with PRP19 (Figure 4E), further supporting EMA1’s role in RNAi and Polycomb repression-dependent co-transcriptional gene silencing. We conclude that Pol II and the cap binding complex provide the co-transcriptional context for base-pairing between scnRNA and nascent transcripts, targeting chromatin modifying activities to the proximity of transcription sites.
Nuclear RNAi and histone methylation-binding drive dispersion of Polycomb bodies
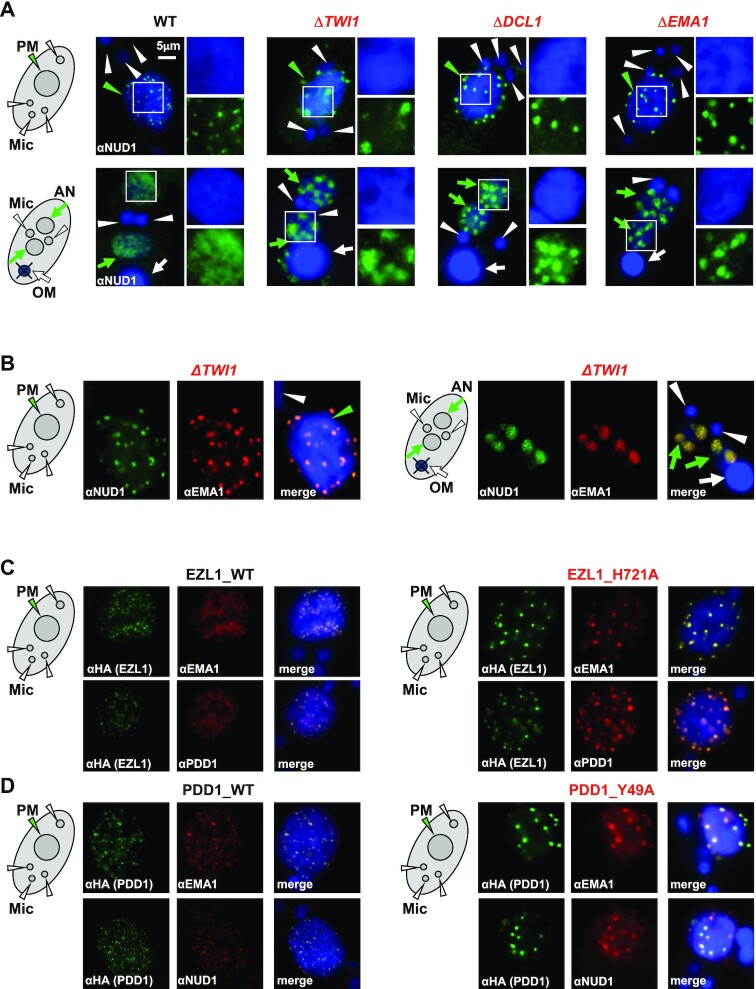
The dynamic distributions of the EZL1 complex, PDD1, and the nuclear RNAi component EMA1 are reminiscent of Polycomb bodies in metazoan cells (51). We’ll hereafter refer to these structures as Tetrahymena Polycomb bodies, based on conservation of their composition, function, as well as morphology. We next tracked how Tetrahymena Polycomb bodies were perturbed in various mutants. IF staining showed that the EZL1 complex aggregated into abnormally large foci in the parental MAC and the early developing MAC of RNAi deficient mutants, including ΔDCL1, ΔTWI1 and ΔEMA1 (Figure 5A). EMA1 also exhibited premature aggregation in the parental MAC and the developing MAC of ΔTWI1 cells, instead of gradually forming distinct foci as in WT cells (Figures 5B, 6C; Supplemental Figure S11). Even in the aggregated state, EMA1 still co-localized with the EZL1 complex (Figure 5B), consistent with co-IP of the EZL1 complex and EMA1 in ΔTWI1 cells (Figure 3G). These foci were often detected in regions weakly stained with DAPI or at the nuclear periphery, suggesting poor association with chromatin (Figure 6C). This is consistent with the ChIP result showing disrupted chromatin association of the EZL1 complex and EMA1 in ΔTWI1 cells (Figure 4A), which is most likely attributable to disrupted ncRNA binding of the EZL1 complex and EMA1 in ΔTWI1 cells (Figure 4C).
Foci formation was also affected by the histone methyltransferase activity of the EZL1 complex. Here, we focused on the parental MAC to avoid complications from zygotic expression in the developing MAC. As the parental MAC share with the developing MAC the same set of players (TWI1, EMA1, the EZL1 complex and PDD1), ncRNA transcription (39), and H3K27 methylation dependent on nuclear RNAi and the EZL1 complex ((46) and this study), the same underlying mechanism likely applies there. Abnormally large foci were observed in cells only expressing EZL1 H721A (Figure 5C), mutating the conserved residue in the SET domain critical for catalysis (90). Importantly, co-localization of the EZL complex, EMA1, and PDD1 was not affected in the methyltransferase-dead mutant (Figure 5C). Abnormally large foci containing the EZL1 complex, EMA1 and PDD1 were also observed in cells carrying PDD1 Y49A mutation (Figure 5D), targeting a conserved aromatic residue in PDD1’s canonical chromodomain required for binding tri-methylated lysine (108). Still, co-localization of the EZL1 complex, EMA1, and PDD1 was preserved in the methyl-binding deficient mutant (Figure 5D). The coordinated changes in the localization of the EZL1 complex, EMA1 and PDD1 in EZL1 H721A and PDD1 Y49A mutants strongly suggest that the histone modification writer/reader coupling forms a positive feedback loop to reinforce chromatin association of the EZL1 complex, complementing the scnRNA/ncRNA-mediated pathway. However, unlike the strong writer/reader coupling found in RNAi-dependent H3K9 methylation of Schizosaccharomyces pombe (20–22), neither PDD1 protein as a whole nor its methyl-binding capability was absolutely required for EZL1-dependent H3K27 and H3K9 methylation in conjugating Tetrahymena cells (Supplementary Figure S12; in apparent contrast to a previous report claiming that H3K9 methylation is missing in ΔPDD1 cells (48)). These results support an attenuated writer/reader coupling underlain by the weak interaction between the EZL1 complex and PDD1 (Supplementary Figure S5).
The EZL1 complex and PDD1 drive coalescence of Polycomb bodies
We next investigated what drove coalescence of Polycomb bodies in Tetrahymena. We noted that in RNAi-deficient mutants (ΔDCL1, ΔTWI1, and ΔEMA1), similar aggregation was observed for the EZL1 complex (Figure 5A) and PDD1 (Figure 6A), implicating both in driving coalescence. In ΔPDD1 cells, both the EZL1 complex and EMA1 could only form fine speckles in the parental MAC, and remained distributed diffusively in the developing MAC (Figure 6B). There was still strong overlap between the EZL1 complex and EMA1 in this altered distribution (Figure 6B), consistent with their co-IP in ΔPDD1 cells (Figure 3G). The lack of aggregation in ΔPDD1 cells was also observed for H3K27me3 and H3K9me3 staining (Supplemental Figure S12A). In contrast, PDD1 still formed large foci in ΔNUD1 cells (Figure 6A, D). These results therefore argue that PDD1 plays a major role in coalescing, and can coalesce independent of the EZL1 complex, possibly even by itself. We also noted that while the EZL1 complex and EMA1 were dispersed in ΔPDD1 cells (Figure 6B), they were aggregated in the PDD1 Y49A mutant (Figure 5D). This suggests that coalescence is not driven by PDD1’s canonical chromodomain; more likely, its chromoshadow domain and intrinsically disordered hinge regions are involved (66,71,109).
Intriguingly, when the EZL1 complex is abolished in ΔNUD1 cells, EMA1 and PDD1 localization is decoupled. EMA1 remained dispersed in DAPI-rich regions in the parental MAC and the developing MAC throughout conjugation in ΔNUD1 cells (Figure 6C). This abnormal localization pattern of EMA1 was similar to that of TWI1 in WT as well as in ΔNUD1 cells (Supplemental Figure S11), whose diffusive distribution can be attributed to its co-transcriptionally tethering to the chromatin (Figure 4A, C) (39). In striking contrast, PDD1 aggregated into large foci in DAPI-poor regions, no longer co-localized with EMA1 in ΔNUD1 cells (Figure 6D). This phenotype was distinct from that of the EZL1 H721A mutant, which showed coordinated aggregation of EMA1 and PDD1 (Figure 5C). These results demonstrate that independent of its methyltransferase activity, the EZL1 complex act as a nexus between the two opposing forces driving Polycomb body dispersion and coalescence, most likely by physically bridging the interaction between EMA1 and PDD1. These interactions underpin the coordinated distribution of nuclear RNAi components and PcG proteins in conjugating Tetrahymena cells.
DISCUSSION
A nuclear RNAi-dependent Polycomb repression pathway arose early in evolution
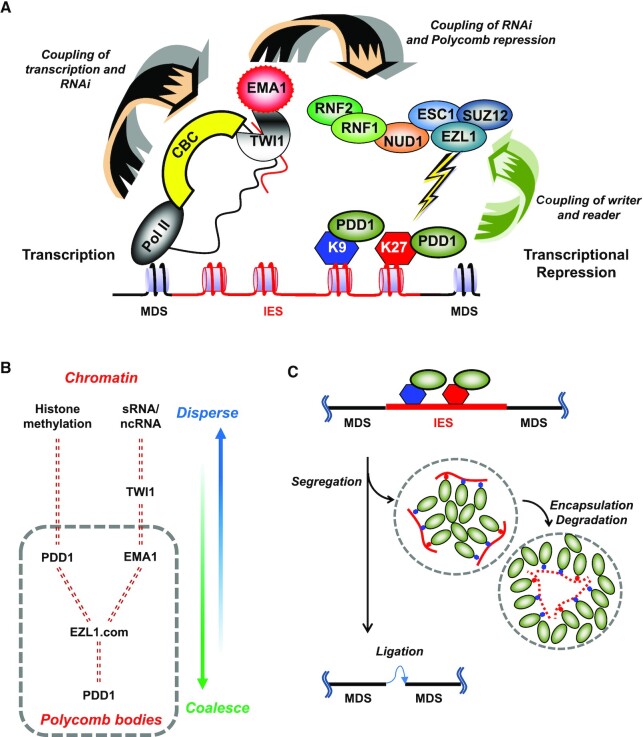
PcG proteins’ involvement in transcriptional repression in eukaryotes is deep-rooted and widespread (110). In the ciliate Tetrahymena, three E(z) homologues are necessary for heterochromatin formation in MAC and/or MIC at various development stages (46,103). Our results demonstrate that the EZL1 complex—a prototypical PRC—is recruited co-transcriptionally in a nuclear RNAi-dependent manner (Figure 7A). The recruitment platform is assembled around the complementary scnRNA and nascent transcripts, integrating the nuclear RNAi machinery, the transcription machinery, and certain co-transcriptional processing machineries. We posit that EMA1—a key component of the nuclear RNAi machinery (39)—plays a direct role in recruiting the EZL1 complex, based upon experimental evidence including co-immunoprecipitation, co-localization in the same cytological structure, and co-occupancy of the same genomic locus, in WT and various mutants. The EZL1 complex recruitment is further reinforced by interacting with PDD1, a chromodomain effector recognizing EZL1-catalyzed H3K27 and H3K9 methylation (46,48). This pathway, featuring dual tethering of the EZL1 complex to the chromatin, is reminiscent of the H3K9 methylation and heterochromatin formation pathway established in S. pombe, and reflects the general theme for co-transcriptional gene silencing in nuclear RNAi (20–22). It also echoes the plant PRC2 recruitment pathway depending on long ncRNA (111), suggesting evolution conservation.
Figure 7.
RNAi and PcG proteins coordinate to repress transcription and reorganize genome. (A) A model for RNAi-dependent recruitment of PcG proteins and their role in heterochromatin formation. Facilitated by the cap-binding complex (CBC), TWI1 and EMA1, scnRNA interacts with complementary nascent ncRNA transcripts generated by RNA polymerase II. The interaction recruits to the chromatin the EZL1 complex, which deposits the heterochromatic marks (H3K27 and H3K9 methylation) that are subsequently recognized by chromodomain effectors including PDD1. Heterochromatinization in turn leads to transcription repression and ultimately DNA elimination in Tetrahymena. (B) A model for the two opposing forces driving dispersion and coalescence of Polycomb bodies. Chromatin association drives dispersion, while PDD1-mediated phase separation drives coalescence. Dashed double lines represent interactions between key players. Note that PDD1 plays two distinct roles in this process: as a histone methylation binder driving dispersion, and as a scaffold protein of the nuclear condensate driving coalescence. See text for details. (C) Coalescence of Polycomb bodies drives programmed genome rearrangement in Tetrahymena. See text for details.
In fungal and metazoan PRC2, the writer/reader coupling is mainly mediated by EED binding of H3K27me3 (112–116). The chromatin association of PRC2 is reinforced by the N-terminal part of SUZ12, either in itself or through interaction with other factors (92). This is in strong contrast to the Tetrahymena EZL1 complex's dependence on PDD1. Nonetheless, a scenario similar to Tetrahymena is found in Arabidopsis, in which LHP1, another chromodomain-containing protein, interacts with PRC2 and is required for binding of H3K27me3 as well as its maintenance (117–119). This distinction may represent a divergence in the evolution of PRC2, as fungi and metazoa branched off protists and plants.
Phase separation is implicated in the dynamic behavior of Polycomb bodies
Nuclear bodies in conjugating Tetrahymena cells—containing PDD1 and many other key players in the RNAi-dependent Polycomb repression pathway—have long been known to exhibit dynamic behavior similar to metazoan Polycomb bodies. This is best illustrated as they coalesce into micron-sized membraneless structures at critical development stages (49,63). The spherical geometry and capacity to fuse support that they are nuclear condensates formed by liquid-liquid phase separation. Importantly, PDD1 not only serves as a reader protein specifically recognizing H3K9/H3K27 methylation, but also has hallmark features of a scaffold protein (Figure 7B)—defined as a component essential for forming a biomolecule condensate (120). Its absence abolishes Polycomb bodies, while its over-expression, by itself in asexually propagating Tetrahymena cells, leads to condensate formation (67). Furthermore, PDD1 interacts with RNA, mostly mediated by its intrinsically disordered hinge regions (66); this interaction is regulated by phosphorylation (highly enriched in the hinge regions), which directly affects condensate formation (66,71). A mutation in PDD1’s chromoshadow domain, required for its dimerization, also abolishes the condensate (66,109). Other components in the Tetrahymena Polycomb bodies, including the EZL1 complex and EMA1, are likely clients (Figure 7B): Their retention in the compartment is dependent on direct or indirect interactions with a scaffold protein, in this case PDD1. Current evidence therefore strongly suggests that Polycomb bodies in conjugating Tetrahymena cells are nuclear condensates formed by phase separation. A rigorous demonstration of their physical nature in future studies will help to shed more light on the repressed state and its maintenance.
Dynamics behavior of biomolecule condensates is also affected by interactions at their interface, countering phase separation driven by interactions between scaffolds and clients: increasing interactions at the interface favor dispersion, while decreasing interactions favor demixing and fusion (121). Building on this theoretical framework as well as our results, we propose that these two opposing forces drive the dynamic distribution of Polycomb bodies in conjugating Tetrahymena cells (Figure 7B). On the one hand, nuclear RNAi and histone methylation-binding lead to chromatin association and dispersion of Polycomb bodies; on the other hand, PDD1-mediated phase separation promotes coalescence of Polycomb bodies in the nucleoplasm. Chromatin exclusion from the mature Polycomb bodies is further supported by their poor DAPI-staining. By sequestering the uncommitted EZL1 complex, this process limits non-specific interactions and buffers fluctuations in protein concentrations (122). Similar dynamic behavior is also observed for Polycomb bodies during embryogenesis in Drosophila (123,124), which is also regulated by the RNAi machinery (29). This evolutionarily conserved network of interactions potentially allows Polycomb repression to be precisely tuned according to developmental needs.
Coalescence of Polycomb bodies drives programmed genome rearrangement
Coalescence of Polycomb bodies in conjugating Tetrahymena cells occurs in the same development window as programmed genome rearrangement. The most prominent event in programmed genome rearrangement is DNA elimination, which excises most of the germline-specific sequences as internally eliminated sequences (IES), while retaining MAC-destined sequences (MDS). Coalescence of Polycomb bodies and DNA elimination also affect each other. Polycomb bodies fail to coalesce in many mutants defective in DNA excision, including 1) loss-of-function mutants of TPB2—a domesticated piggyBac transposase that functions as the DNA excisase in Tetrahymena (64,65,125) and Paramecium (126), and 2) KO of LIA5—homologous to TPB2 and possibly an integral component of the excisase complex (127), as is the case in Paramecium (128). Reciprocally, in several PDD1 mutants defective in coalescence of Polycomb bodies but not histone methylation and its binding, DNA elimination is compromised (66,71). Based on these reports and our results, we propose that coalescence of Polycomb bodies, by mechanically pulling the IES-containing chromatin tethered at their interface, separates IES from MDS (Figure 7C). As the transposase-catalyzed DNA excision, a transesterification reaction, is fully reversible, physical segregation of IES and MDS may be essential to shift the equilibrium and drive DNA elimination to completion. Once separated, IES may be further sequestered in sub-compartments (Figure 7C), like the hollowed spherical structure of DNA elimination bodies formed by PDD1 in late conjugation (49). This kind of structure can also be generated by phase separation (129), and allow IES to be safely degraded without affecting MDS (Figure 7C). The mechanical force generated by coalescence of Polycomb bodies may also promote chromosome fragmentation, another key event in programmed genome rearrangement in Tetrahymena. The spatial reorganization role played by PcG proteins in Tetrahymena is reminiscent of contact between chromatin regions under Polycomb repression observed in Drosophila (130,131). Our model is also consistent with the general framework that nuclear condensates mechanically sense and restructure the genome (59).
DATA AVAILABILITY
The compiled annotations for Tetrahymena MAC and MIC genome sequence were obtained from Tetrahymena Genome Database (www.cilate.org). Illumina sequencing was performed at the DNA Sequencing Core of University of Michigan.
The GenBank accession numbers of EZL1 (TTHERM_00335780), ESC1 (TTHERM_00442420), SUZ12 (TTHERM_00149839), RNF1 (TTHERM_00637350), RNF2 (TTHERM_00522660) and NUD1 (TTHERM_00171760) are EF446990, EU814895, GU550521, EU814896, EU814897, and GU550522, respectively. Illumina sequencing data of genomic DNA samples from the developing MAC can be accessed at PRJNA362353: SRR5184454 (WT), PRJNA593846: SRR11769410 (ΔEZL1), PRJNA593846: SRR10593974 (ΔRNF1) and PRJNA593846: SRR11769409 (ΔPDD1).
Supplementary Material
ACKNOWLEDGEMENTS
WT Tetrahymena strains CU427 and CU428 were obtained from Tetrahymena Stock Center. ΔDCL1 and ΔPDD1 strains were provided by Douglas L. Chalker. ΔTWI1 and ΔEMA1 strains, and anti-TWI1 and anti-EMA1 antibody were provided by Mochizuki Kazufumi.
Notes
Present address: Yifan Liu, Department of Biochemistry & Molecular Medicine, University of Southern California Keck School of Medicine, Los Angeles, CA 90033, USA.
Contributor Information
Jing Xu, School of Life Science, Shanxi University, Taiyuan, Shanxi 030006, China.
Xiaolu Zhao, Department of Pathology, University of Michigan, Ann Arbor, MI 48109, USA; Institute of Evolution & Marine Biodiversity, Ocean University of China, Qingdao 266003, China.
Fengbiao Mao, Department of Pathology, University of Michigan, Ann Arbor, MI 48109, USA.
Venkatesha Basrur, Proteomics Resource Facility, Department of Pathology, University of Michigan, Ann Arbor, MI 48109, USA.
Beatrix Ueberheide, Laboratory of Mass Spectrometry and Gaseous Ion Chemistry, the Rockefeller University, New York, NY 10065, USA.
Brian T Chait, Laboratory of Mass Spectrometry and Gaseous Ion Chemistry, the Rockefeller University, New York, NY 10065, USA.
C David Allis, Laboratory of Chromatin Biology and Epigenetics, the Rockefeller University, New York, NY 10065, USA.
Sean D Taverna, Department of Pharmacology and Molecular Sciences and the Center for Epigenetics, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA.
Shan Gao, Institute of Evolution & Marine Biodiversity, Ocean University of China, Qingdao 266003, China.
Wei Wang, School of Life Science, Shanxi University, Taiyuan, Shanxi 030006, China.
Yifan Liu, Department of Pathology, University of Michigan, Ann Arbor, MI 48109, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
J.X. was supported by the National Natural Science Foundation of China (No. 31572253) and Natural Science Foundation of Shanxi Province (201901D111008). V.B. was supported by Department of Pathology at the University of Michigan. B.T.C. was supported by NIH (5P41 GM103314). C.D.A. was supported the Rockefeller University. C.F. was supported by NIH (R01 GM077582). R.S.C. was supported by NSF (1158346). S.D.T. was supported by NIH (R01 GM106024) and the Institute for Basic Biomedical Sciences at Johns Hopkins University School of Medicine. S.G. was supported by the Taishan Scholar Program of Shandong Province, the Marine S&T Fund of Shandong Province for Pilot National Laboratory for Marine Science and Technology (Qingdao) (2018SDKJ0406-2) and Fundamental Research Funds for the Central Universities (201841005). W.W. was supported by the National Natural Science Foundation of China (No. 31872224). Y.L. was supported by NIH (R01 GM087343), Department of Pathology at the University of Michigan, and Department of Biochemistry & Molecular Medicine at the University of Southern California Keck School of Medicine.
Conflict of interest statement. None declared.
REFERENCES
- 1. Di Croce L., Helin K.. Transcriptional regulation by Polycomb group proteins. Nat. Struct. Mol. Biol. 2013; 20:1147–1155. [DOI] [PubMed] [Google Scholar]
- 2. Schuettengruber B., Bourbon H.M., Di Croce L., Cavalli G.. Genome regulation by polycomb and Trithorax: 70 years and counting. Cell. 2017; 171:34–57. [DOI] [PubMed] [Google Scholar]
- 3. Yu J.R., Lee C.H., Oksuz O., Stafford J.M., Reinberg D.. PRC2 is high maintenance. Genes Dev. 2019; 33:903–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gil J., O’Loghlen A.. PRC1 complex diversity: where is it taking us. Trends Cell Biol. 2014; 24:632–641. [DOI] [PubMed] [Google Scholar]
- 5. Chittock E.C., Latwiel S., Miller T.C., Muller C.W.. Molecular architecture of polycomb repressive complexes. Biochem. Soc. Trans. 2017; 45:193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Simon J.A., Kingston R.E.. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell. 2013; 49:808–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Simon J., Chiang A., Bender W., Shimell M.J., O’Connor M.. Elements of the Drosophila bithorax complex that mediate repression by Polycomb group products. Dev. Biol. 1993; 158:131–144. [DOI] [PubMed] [Google Scholar]
- 8. Beisel C., Paro R.. Silencing chromatin: comparing modes and mechanisms. Nat. Rev. Genet. 2011; 12:123–135. [DOI] [PubMed] [Google Scholar]
- 9. Brockdorff N. Noncoding RNA and Polycomb recruitment. RNA. 2013; 19:429–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Davidovich C., Cech T.R.. The recruitment of chromatin modifiers by long noncoding RNAs: lessons from PRC2. RNA. 2015; 21:2007–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Khalil A.M., Guttman M., Huarte M., Garber M., Raj A., Rivea Morales D., Thomas K., Presser A., Bernstein B.E., van Oudenaarden A.et al.. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl Acad. Sci. U.S.A. 2009; 106:11667–11672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhao J., Ohsumi T.K., Kung J.T., Ogawa Y., Grau D.J., Sarma K., Song J.J., Kingston R.E., Borowsky M., Lee J.T.. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell. 2010; 40:939–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tsai M.C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F., Shi Y., Segal E., Chang H.Y.. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010; 329:689–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Davidovich C., Zheng L., Goodrich K.J., Cech T.R.. Promiscuous RNA binding by Polycomb repressive complex 2. Nat. Struct. Mol. Biol. 2013; 20:1250–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kaneko S., Son J., Shen S.S., Reinberg D., Bonasio R.. PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat. Struct. Mol. Biol. 2013; 20:1258–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kaneko S., Son J., Bonasio R., Shen S.S., Reinberg D.. Nascent RNA interaction keeps PRC2 activity poised and in check. Genes Dev. 2014; 28:1983–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cifuentes-Rojas C., Hernandez A.J., Sarma K., Lee J.T.. Regulatory interactions between RNA and polycomb repressive complex 2. Mol. Cell. 2014; 55:171–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. da Rocha S.T., Boeva V., Escamilla-Del-Arenal M., Ancelin K., Granier C., Matias N.R., Sanulli S., Chow J., Schulz E., Picard C.et al.. Jarid2 is implicated in the initial Xist-induced targeting of PRC2 to the inactive X chromosome. Mol. Cell. 2014; 53:301–316. [DOI] [PubMed] [Google Scholar]
- 19. Kaneko S., Bonasio R., Saldana-Meyer R., Yoshida T., Son J., Nishino K., Umezawa A., Reinberg D.. Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol. Cell. 2014; 53:290–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009; 457:413–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Grewal S.I., Elgin S.C.. Transcription and RNA interference in the formation of heterochromatin. Nature. 2007; 447:399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Castel S.E., Martienssen R.A.. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat. Rev. Genet. 2013; 14:100–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhao J., Sun B.K., Erwin J.A., Song J.J., Lee J.T.. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008; 322:750–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ogawa Y., Sun B.K., Lee J.T.. Intersection of the RNA interference and X-inactivation pathways. Science. 2008; 320:1336–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kanellopoulou C., Muljo S.A., Dimitrov S.D., Chen X., Colin C., Plath K., Livingston D.M.. X chromosome inactivation in the absence of Dicer. Proc. Natl Acad. Sci. U.S.A. 2009; 106:1122–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pal-Bhadra M., Bhadra U., Birchler J.A.. Cosuppression inDrosophila: gene silencing of Alcohol dehydrogenase by white-Adh transgenes is Polycomb dependent. Cell. 1997; 90:479–490. [DOI] [PubMed] [Google Scholar]
- 27. Pal-Bhadra M., Bhadra U., Birchler J.A.. RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol. Cell. 2002; 9:315–327. [DOI] [PubMed] [Google Scholar]
- 28. Pal-Bhadra M., Leibovitch B.A., Gandhi S.G., Rao M., Bhadra U., Birchler J.A., Elgin S.C.. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004; 303:669–672. [DOI] [PubMed] [Google Scholar]
- 29. Grimaud C., Bantignies F., Pal-Bhadra M., Ghana P., Bhadra U., Cavalli G.. RNAi components are required for nuclear clustering of Polycomb group response elements. Cell. 2006; 124:957–971. [DOI] [PubMed] [Google Scholar]
- 30. Portoso M., Cavalli G.. Morris KV RNA and the Regulation of Gene Expression: A Hidden Layer of Complexity. 2008; Caister Academic Press; 29–43. [Google Scholar]
- 31. Pirrotta V., Li H.B.. A view of nuclear Polycomb bodies. Curr. Opin. Genet. Dev. 2012; 22:101–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Laugesen A., Hojfeldt J.W., Helin K.. Molecular mechanisms directing PRC2 recruitment and H3K27 methylation. Mol. Cell. 2019; 74:8–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chalker D.L., Meyer E., Mochizuki K.. Epigenetics of ciliates. Cold Spring Harb. Perspect. Biol. 2013; 5:a017764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Noto T., Mochizuki K.. Whats, hows and whys of programmed DNA elimination in Tetrahymena. Open Biol. 2017; 7:170172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Karrer K.M. Nuclear dualism. Methods Cell Biol. 2012; 109:29–52. [DOI] [PubMed] [Google Scholar]
- 36. Duharcourt S., Lepere G., Meyer E.. Developmental genome rearrangements in ciliates: a natural genomic subtraction mediated by non-coding transcripts. Trends Genet. 2009; 25:344–350. [DOI] [PubMed] [Google Scholar]
- 37. Chalker D.L., Yao M.C.. Nongenic, bidirectional transcription precedes and may promote developmental DNA deletion inTetrahymena thermophila. Genes Dev. 2001; 15:1287–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mochizuki K., Gorovsky M.A.. RNA polymerase II localizes in Tetrahymena thermophila meiotic micronuclei when micronuclear transcription associated with genome rearrangement occurs. Eukaryot Cell. 2004; 3:1233–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Aronica L., Bednenko J., Noto T., DeSouza L.V., Siu K.W., Loidl J., Pearlman R.E., Gorovsky M.A., Mochizuki K.. Study of an RNA helicase implicates small RNA-noncoding RNA interactions in programmed DNA elimination in Tetrahymena. Genes Dev. 2008; 22:2228–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Malone C.D., Anderson A.M., Motl J.A., Rexer C.H., Chalker D.L.. Germ line transcripts are processed by a Dicer-like protein that is essential for developmentally programmed genome rearrangements of Tetrahymena thermophila. Mol. Cell. Biol. 2005; 25:9151–9164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mochizuki K., Gorovsky M.A.. A Dicer-like protein in Tetrahymena has distinct functions in genome rearrangement, chromosome segregation, and meiotic prophase. Genes Dev. 2005; 19:77–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mochizuki K., Fine N.A., Fujisawa T., Gorovsky M.A.. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in Tetrahymena. Cell. 2002; 110:689–699. [DOI] [PubMed] [Google Scholar]
- 43. Mochizuki K., Gorovsky M.A.. Conjugation-specific small RNAs in Tetrahymena have predicted properties of scan (scn) RNAs involved in genome rearrangement. Genes Dev. 2004; 18:2068–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Noto T., Kurth H.M., Kataoka K., Aronica L., DeSouza L.V., Siu K.W., Pearlman R.E., Gorovsky M.A., Mochizuki K.. The Tetrahymena argonaute-binding protein Giw1p directs a mature argonaute-siRNA complex to the nucleus. Cell. 2010; 140:692–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu Y., Mochizuki K., Gorovsky M.A.. Histone H3 lysine 9 methylation is required for DNA elimination in developing macronuclei in Tetrahymena. Proc. Natl Acad. Sci. U.S.A. 2004; 101:1679–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu Y., Taverna S.D., Muratore T.L., Shabanowitz J., Hunt D.F., Allis C.D.. RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena. Genes Dev. 2007; 21:1530–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhao X., Xiong J., Mao F., Sheng Y., Chen X., Feng L., Dui W., Yang W., Kapusta A., Feschotte C.et al.. RNAi-dependent Polycomb repression controls transposable elements in Tetrahymena. Genes Dev. 2019; 33:348–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Taverna S.D., Coyne R.S., Allis C.D.. Methylation of histone h3 at lysine 9 targets programmed DNA elimination in tetrahymena. Cell. 2002; 110:701–711. [DOI] [PubMed] [Google Scholar]
- 49. Madireddi M.T., Coyne R.S., Smothers J.F., Mickey K.M., Yao M.C., Allis C.D.. Pdd1p, a novel chromodomain-containing protein, links heterochromatin assembly and DNA elimination in Tetrahymena. Cell. 1996; 87:75–84. [DOI] [PubMed] [Google Scholar]
- 50. Coyne R.S., Nikiforov M.A., Smothers J.F., Allis C.D., Yao M.C.. Parental expression of the chromodomain protein Pdd1p is required for completion of programmed DNA elimination and nuclear differentiation. Mol. Cell. 1999; 4:865–872. [DOI] [PubMed] [Google Scholar]
- 51. Pirrotta V. Bazett-Jones D.P., Dellaire G.. The Functional Nucleus. 2016; NY: Springer Berlin Heidelberg; 157–173. [Google Scholar]
- 52. Mao Y.S., Zhang B., Spector D.L.. Biogenesis and function of nuclear bodies. Trends Genet. 2011; 27:295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Plys A.J., Davis C.P., Kim J., Rizki G., Keenen M.M., Marr S.K., Kingston R.E.. Phase separation of Polycomb-repressive complex 1 is governed by a charged disordered region of CBX2. Genes Dev. 2019; 33:799–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tatavosian R., Kent S., Brown K., Yao T., Duc H.N., Huynh T.N., Zhen C.Y., Ma B., Wang H., Ren X.. Nuclear condensates of the Polycomb protein chromobox 2 (CBX2) assemble through phase separation. J. Biol. Chem. 2019; 294:1451–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Banani S.F., Lee H.O., Hyman A.A., Rosen M.K.. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017; 18:285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shin Y., Brangwynne C.P.. Liquid phase condensation in cell physiology and disease. Science. 2017; 357:eaaf4382. [DOI] [PubMed] [Google Scholar]
- 57. Grau D.J., Chapman B.A., Garlick J.D., Borowsky M., Francis N.J., Kingston R.E.. Compaction of chromatin by diverse Polycomb group proteins requires localized regions of high charge. Genes Dev. 2011; 25:2210–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lau M.S., Schwartz M.G., Kundu S., Savol A.J., Wang P.I., Marr S.K., Grau D.J., Schorderet P., Sadreyev R.I., Tabin C.J.et al.. Mutation of a nucleosome compaction region disrupts Polycomb-mediated axial patterning. Science. 2017; 355:1081–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shin Y., Chang Y.C., Lee D.S.W., Berry J., Sanders D.W., Ronceray P., Wingreen N.S., Haataja M., Brangwynne C.P.. Liquid nuclear condensates mechanically sense and restructure the genome. Cell. 2018; 175:1481–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gibson B.A., Doolittle L.K., Schneider M.W.G., Jensen L.E., Gamarra N., Henry L., Gerlich D.W., Redding S., Rosen M.K.. Organization of chromatin by intrinsic and regulated phase separation. Cell. 2019; 179:470–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Larson A.G., Elnatan D., Keenen M.M., Trnka M.J., Johnston J.B., Burlingame A.L., Agard D.A., Redding S., Narlikar G.J.. Liquid droplet formation by HP1alpha suggests a role for phase separation in heterochromatin. Nature. 2017; 547:236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Strom A.R., Emelyanov A.V., Mir M., Fyodorov D.V., Darzacq X., Karpen G.H.. Phase separation drives heterochromatin domain formation. Nature. 2017; 547:241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Janetopoulos C., Cole E., Smothers J.F., Allis C.D., Aufderheide K.J.. The conjusome: a novel structure in Tetrahymena found only during sexual reorganization. J. Cell Sci. 1999; 112:1003–1011. [DOI] [PubMed] [Google Scholar]
- 64. Vogt A., Mochizuki K.. A domesticated PiggyBac transposase interacts with heterochromatin and catalyzes reproducible DNA elimination in Tetrahymena. PLoS Genet. 2013; 9:e1004032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cheng C.Y., Vogt A., Mochizuki K., Yao M.C.. A domesticated piggyBac transposase plays key roles in heterochromatin dynamics and DNA cleavage during programmed DNA deletion in Tetrahymena thermophila. Mol. Biol. Cell. 2010; 21:1753–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kataoka K., Mochizuki K.. Phosphorylation of an HP1-like protein regulates heterochromatin body assembly for DNA elimination. Dev. Cell. 2015; 35:775–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kataoka K., Mochizuki K.. Heterochromatin aggregation during DNA elimination in Tetrahymena is facilitated by a prion-like protein. J. Cell Sci. 2017; 130:480–489. [DOI] [PubMed] [Google Scholar]
- 68. Yao M.C., Yao C.H., Halasz L.M., Fuller P., Rexer C.H., Wang S.H., Jain R., Coyne R.S., Chalker D.L.. Identification of novel chromatin-associated proteins involved in programmed genome rearrangements in Tetrahymena. J. Cell Sci. 2007; 120:1978–1989. [DOI] [PubMed] [Google Scholar]
- 69. Nikiforov M.A., Smothers J.F., Gorovsky M.A., Allis C.D.. Excision of micronuclear-specific DNA requires parental expression of pdd2p and occurs independently from DNA replication in Tetrahymena thermophila. Genes Dev. 1999; 13:2852–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Nikiforov M.A., Gorovsky M.A., Allis C.D.. A novel chromodomain protein, pdd3p, associates with internal eliminated sequences during macronuclear development in Tetrahymena thermophila. Mol. Cell. Biol. 2000; 20:4128–4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kataoka K., Noto T., Mochizuki K.. Phosphorylation of an HP1-like protein is a prerequisite for heterochromatin body formation in Tetrahymena DNA elimination. Proc. Natl Acad. Sci. U.S.A. 2016; 113:9027–9032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gao S., Xiong J., Zhang C., Berquist B.R., Yang R., Zhao M., Molascon A.J., Kwiatkowski S.Y., Yuan D., Qin Z.et al.. Impaired replication elongation in Tetrahymena mutants deficient in histone H3 Lys 27 monomethylation. Genes Dev. 2013; 27:1662–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sweet M.T., Allis C.D.. Spector D.L., Goldman R.D., Leinwand L.A.. Cells: A Laboratory Manual. 1998; 1:Cold Spring Harbor Laboratory Press. [Google Scholar]
- 74. Cassidy-Hanley D.M. Tetrahymena in the laboratory: strain resources, methods for culture, maintenance, and storage. Methods Cell Biol. 2012; 109:237–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sweet M.T., Allis C.D.. Isolation and purification of Tetrahymena nuclei. Cold Spring Harb. Protoc. 2006; 2006:pdb.prot4500. [DOI] [PubMed] [Google Scholar]
- 76. Allis C.D., Dennison D.K.. Identification and purification of young macronuclear anlagen from conjugating cells ofTetrahymena thermophila. Dev. Biol. 1982; 93:519–533. [DOI] [PubMed] [Google Scholar]
- 77. Hamilton E.P., Kapusta A., Huvos P.E., Bidwell S.L., Zafar N., Tang H., Hadjithomas M., Krishnakumar V., Badger J.H., Caler E.V.et al.. Structure of the germline genome of Tetrahymena thermophila and relationship to the massively rearranged somatic genome. Elife. 2016; 5:e19090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.Proc, G.P.D. . The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ramirez F., Ryan D.P., Gruning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dundar F., Manke T.. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids. Res. 2016; 44:W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Margueron R., Reinberg D.. The Polycomb complex PRC2 and its mark in life. Nature. 2011; 469:343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Morey L., Helin K.. Polycomb group protein-mediated repression of transcription. Trends Biochem. Sci. 2010; 35:323–332. [DOI] [PubMed] [Google Scholar]
- 84. Levine S.S., Weiss A., Erdjument-Bromage H., Shao Z., Tempst P., Kingston R.E.. The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol. Cell. Biol. 2002; 22:6070–6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Saurin A.J., Shao Z., Erdjument-Bromage H., Tempst P., Kingston R.E.. A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature. 2001; 412:655–660. [DOI] [PubMed] [Google Scholar]
- 86. Shao Z., Raible F., Mollaaghababa R., Guyon J.R., Wu C.T., Bender W., Kingston R.E.. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell. 1999; 98:37–46. [DOI] [PubMed] [Google Scholar]
- 87. Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R.S., Zhang Y.. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002; 298:1039–1043. [DOI] [PubMed] [Google Scholar]
- 88. Czermin B., Melfi R., McCabe D., Seitz V., Imhof A., Pirrotta V.. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002; 111:185–196. [DOI] [PubMed] [Google Scholar]
- 89. Muller J., Hart C.M., Francis N.J., Vargas M.L., Sengupta A., Wild B., Miller E.L., O’Connor M.B., Kingston R.E., Simon J.A.. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002; 111:197–208. [DOI] [PubMed] [Google Scholar]
- 90. Kuzmichev A., Nishioka K., Erdjument-Bromage H., Tempst P., Reinberg D.. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002; 16:2893–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Cao R., Zhang Y.. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol. Cell. 2004; 15:57–67. [DOI] [PubMed] [Google Scholar]
- 92. Kasinath V., Faini M., Poepsel S., Reif D., Feng X.A., Stjepanovic G., Aebersold R., Nogales E.. Structures of human PRC2 with its cofactors AEBP2 and JARID2. Science. 2018; 359:940–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Frapporti A., Miro Pina C., Arnaiz O., Holoch D., Kawaguchi T., Humbert A., Eleftheriou E., Lombard B., Loew D., Sperling L.et al.. The Polycomb protein Ezl1 mediates H3K9 and H3K27 methylation to repress transposable elements in Paramecium. Nat. Commun. 2019; 10:2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Wang H., Wang L., Erdjument-Bromage H., Vidal M., Tempst P., Jones R.S., Zhang Y.. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004; 431:873–878. [DOI] [PubMed] [Google Scholar]
- 95. Illingworth R.S., Moffat M., Mann A.R., Read D., Hunter C.J., Pradeepa M.M., Adams I.R., Bickmore W.A.. The E3 ubiquitin ligase activity of RING1B is not essential for early mouse development. Genes Dev. 2015; 29:1897–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Pengelly A.R., Kalb R., Finkl K., Muller J.. Transcriptional repression by PRC1 in the absence of H2A monoubiquitylation. Genes Dev. 2015; 29:1487–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Francis N.J., Kingston R.E., Woodcock C.L.. Chromatin compaction by a polycomb group protein complex. Science. 2004; 306:1574–1577. [DOI] [PubMed] [Google Scholar]
- 98. Eskeland R., Leeb M., Grimes G.R., Kress C., Boyle S., Sproul D., Gilbert N., Fan Y., Skoultchi A.I., Wutz A.et al.. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol. Cell. 2010; 38:452–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Lin C.G., Lin I.T., Yao M.C.. Programmed minichromosome elimination as a mechanism for somatic genome reduction in Tetrahymena thermophila. PLos Genet. 2016; 12:e1006403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Feng L., Wang G., Hamilton E.P., Xiong J., Yan G., Chen K., Chen X., Dui W., Plemens A., Khadr L.et al.. A germline-limited piggyBac transposase gene is required for precise excision in Tetrahymena genome rearrangement. Nucleic Acids Res. 2017; 45:9481–9502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Plasterk R.H., Izsvak Z., Ivics Z.. Resident aliens: the Tc1/mariner superfamily of transposable elements. Trends Genet. 1999; 15:326–332. [DOI] [PubMed] [Google Scholar]
- 102. Tellier M., Bouuaert C.C., Chalmers R.. Mariner and the ITm superfamily of transposons. Microbiol Spectr. 2015; 3:MDNA3-0033-2014. [DOI] [PubMed] [Google Scholar]
- 103. Papazyan R., Voronina E., Chapman J.R., Luperchio T.R., Gilbert T.M., Meier E., Mackintosh S.G., Shabanowitz J., Tackett A.J., Reddy K.L.et al.. Methylation of histone H3K23 blocks DNA damage in pericentric heterochromatin during meiosis. Elife. 2014; 3:e02996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Hauer M.H., Gasser S.M.. Chromatin and nucleosome dynamics in DNA damage and repair. Genes Dev. 2017; 31:2204–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Bednenko J., Noto T., DeSouza L.V., Siu K.W., Pearlman R.E., Mochizuki K., Gorovsky M.A.. Two GW repeat proteins interact withTetrahymena thermophila argonaute and promote genome rearrangement. Mol. Cell. Biol. 2009; 29:5020–5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Smart S.K., Mackintosh S.G., Edmondson R.D., Taverna S.D., Tackett A.J.. Mapping the local protein interactome of the NuA3 histone acetyltransferase. Protein Sci. 2009; 18:1987–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Aguilera A. Cotranscriptional mRNP assembly: from the DNA to the nuclear pore. Curr. Opin. Cell Biol. 2005; 17:242–250. [DOI] [PubMed] [Google Scholar]
- 108. Jacobs S.A., Khorasanizadeh S.. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002; 295:2080–2083. [DOI] [PubMed] [Google Scholar]
- 109. Schwope R.M., Chalker D.L.. Mutations in Pdd1 reveal distinct requirements for its chromodomain and chromoshadow domain in directing histone methylation and heterochromatin elimination. Eukaryot Cell. 2014; 13:190–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Ringrose L., Paro R.. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu. Rev. Genet. 2004; 38:413–443. [DOI] [PubMed] [Google Scholar]
- 111. Tian Y., Zheng H., Zhang F., Wang S., Ji X., Xu C., He Y., Ding Y.. PRC2 recruitment and H3K27me3 deposition at FLC require FCA binding of COOLAIR. Sci Adv. 2019; 5:eaau7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Oksuz O., Narendra V., Lee C.H., Descostes N., LeRoy G., Raviram R., Blumenberg L., Karch K., Rocha P.P., Garcia B.A.et al.. Capturing the onset of PRC2-mediated repressive domain formation. Mol. Cell. 2018; 70:1149–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Jiao L., Liu X.. Response to comment on “Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2”. Science. 2016; 354:1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Jiao L., Liu X.. Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2. Science. 2015; 350:aac4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Justin N., Zhang Y., Tarricone C., Martin S.R., Chen S., Underwood E., De Marco V., Haire L.F., Walker P.A., Reinberg D.et al.. Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2. Nat. Commun. 2016; 7:11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Margueron R., Justin N., Ohno K., Sharpe M.L., Son J., Drury W.J. 3rd, Voigt P., Martin S.R., Taylor W.R., De Marco V.et al.. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009; 461:762–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Derkacheva M., Steinbach Y., Wildhaber T., Mozgova I., Mahrez W., Nanni P., Bischof S., Gruissem W., Hennig L.. Arabidopsis MSI1 connects LHP1 to PRC2 complexes. EMBO J. 2013; 32:2073–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Zhou Y., Tergemina E., Cui H., Forderer A., Hartwig B., Velikkakam James G., Schneeberger K., Turck F.. Ctf4-related protein recruits LHP1-PRC2 to maintain H3K27me3 levels in dividing cells inArabidopsis thaliana. Proc. Natl Acad. Sci. U.S.A. 2017; 114:4833–4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Exner V., Aichinger E., Shu H., Wildhaber T., Alfarano P., Caflisch A., Gruissem W., Kohler C., Hennig L.. The chromodomain of LIKE HETEROCHROMATIN PROTEIN 1 is essential for H3K27me3 binding and function during Arabidopsis development. PLoS One. 2009; 4:e5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Banani S.F., Rice A.M., Peeples W.B., Lin Y., Jain S., Parker R., Rosen M.K.. Compositional control of phase-separated cellular bodies. Cell. 2016; 166:651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Maharana S., Wang J., Papadopoulos D.K., Richter D., Pozniakovsky A., Poser I., Bickle M., Rizk S., Guillen-Boixet J., Franzmann T.M.et al.. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science. 2018; 360:918–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Klosin A., Oltsch F., Harmon T., Honigmann A., Julicher F., Hyman A.A., Zechner C.. Phase separation provides a mechanism to reduce noise in cells. Science. 2020; 367:464–468. [DOI] [PubMed] [Google Scholar]
- 123. Bantignies F., Roure V., Comet I., Leblanc B., Schuettengruber B., Bonnet J., Tixier V., Mas A., Cavalli G.. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell. 2011; 144:214–226. [DOI] [PubMed] [Google Scholar]
- 124. Cheutin T., Cavalli G.. Progressive polycomb assembly on H3K27me3 compartments generates polycomb bodies with developmentally regulated motion. PLos Genet. 2012; 8:e1002465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Mutazono M., Noto T., Mochizuki K.. Diversification of small RNA amplification mechanisms for targeting transposon-related sequences in ciliates. Proc. Natl Acad. Sci. U.S.A. 2019; 116:14639–14644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Baudry C., Malinsky S., Restituito M., Kapusta A., Rosa S., Meyer E., Betermier M.. PiggyMac, a domesticated piggyBac transposase involved in programmed genome rearrangements in the ciliate Paramecium tetraurelia. Genes Dev. 2009; 23:2478–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Shieh A.W., Chalker D.L.. LIA5 is required for nuclear reorganization and programmed DNA rearrangements occurring duringtetrahymena macronuclear differentiation. PLoS One. 2013; 8:e75337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Bischerour J., Bhullar S., Denby Wilkes C., Regnier V., Mathy N., Dubois E., Singh A., Swart E., Arnaiz O., Sperling L.et al.. Six domesticated PiggyBac transposases together carry out programmed DNA elimination in Paramecium. Elife. 2018; 7:e37927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Feric M., Vaidya N., Harmon T.S., Mitrea D.M., Zhu L., Richardson T.M., Kriwacki R.W., Pappu R.V., Brangwynne C.P.. Coexisting liquid phases underlie nucleolar subcompartments. Cell. 2016; 165:1686–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Sexton T., Yaffe E., Kenigsberg E., Bantignies F., Leblanc B., Hoichman M., Parrinello H., Tanay A., Cavalli G.. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012; 148:458–472. [DOI] [PubMed] [Google Scholar]
- 131. Rao S.S., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S.et al.. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014; 159:1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P.. Integrative genomics viewer. Nat. Biotechnol. 2011; 29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The compiled annotations for Tetrahymena MAC and MIC genome sequence were obtained from Tetrahymena Genome Database (www.cilate.org). Illumina sequencing was performed at the DNA Sequencing Core of University of Michigan.
The GenBank accession numbers of EZL1 (TTHERM_00335780), ESC1 (TTHERM_00442420), SUZ12 (TTHERM_00149839), RNF1 (TTHERM_00637350), RNF2 (TTHERM_00522660) and NUD1 (TTHERM_00171760) are EF446990, EU814895, GU550521, EU814896, EU814897, and GU550522, respectively. Illumina sequencing data of genomic DNA samples from the developing MAC can be accessed at PRJNA362353: SRR5184454 (WT), PRJNA593846: SRR11769410 (ΔEZL1), PRJNA593846: SRR10593974 (ΔRNF1) and PRJNA593846: SRR11769409 (ΔPDD1).