Abstract
The histone chaperone facilitates chromatin transactions (FACT) functions in various DNA transactions. How FACT performs these multiple functions remains largely unknown. Here, we found, for the first time, that the N-terminal domain of its Spt16 subunit interacts with the Set3 histone deacetylase complex (Set3C) and that FACT and Set3C function in the same pathway to regulate gene expression in some settings. We observed that Spt16-G132D mutant proteins show defects in binding to Set3C but not other reported FACT interactors. At the permissive temperature, induction of the GAL1 and GAL10 genes is reduced in both spt16-G132D and set3Δ cells, whereas transient upregulation of GAL10 noncoding RNA (ncRNA), which is transcribed from the 3′ end of the GAL10 gene, is elevated. Mutations that inhibit GAL10 ncRNA transcription reverse the GAL1 and GAL10 induction defects in spt16-G132D and set3Δ mutant cells. Mechanistically, set3Δ and FACT (spt16-G132D) mutants show reduced histone acetylation and increased nucleosome occupancy at the GAL1 promoter under inducing conditions and inhibition of GAL10 ncRNA transcription also partially reverses these chromatin changes. These results indicate that FACT interacts with Set3C, which in turn prevents uncontrolled GAL10 ncRNA expression and fine-tunes the expression of GAL genes upon a change in carbon source.
INTRODUCTION
In eukaryotic cells, histone post-translational modifications, including acetylation and methylation, play important roles in regulating gene transcription in response to both endogenous and environmental stimuli (1,2). For instance, in response to environmental cues, histone acetyltransferases (HATs) can be recruited to specific gene promoters to acetylate histone proteins, which in turn promotes gene transcription (3–6). On the other hand, histone deacetylases (HDACs), when recruited, reverse histone acetylation and in general result in gene repression (7–10). In addition to histone modifiers, histone chaperones, a group of proteins that can bind histones and regulate nucleosome assembly/disassembly but do not possess enzymatic activity, also regulate chromatin dynamics and gene transcription (11–13). However, it remains largely unexplored how histone chaperones function with histone-modifying enzymes in gene regulation.
F acilitates chromatin transactions (FACT) is a conserved histone chaperone that can both disassemble and reassemble nucleosomes (14–16). FACT consists of two essential subunits, Spt16 and Pob3, in budding yeast, which correspond to Spt16 and SSRP1 in higher eukaryotes. Human FACT was initially identified and named for its ability to facilitate in vitro transcription through a chromatin template (11). It has been proposed that FACT displaces H2A-H2B dimers from nucleosomes, which in turn facilitates the passage of RNA polymerase (Pol) II through the chromatin template (17–20). FACT can also change histone-DNA contact globally in vitro (19). In addition to participating in nucleosome disassembly, FACT is proposed to reassemble nucleosomes in the wake of RNA Pol II. Spt16 and Pob3 mutant alleles show the Spt– phenotype, indicating FACT’s roles in transcription initiation (21). Furthermore, Spt16 is enriched at actively transcribed genes (22). Depletion of yeast Spt16 using the temperature-sensitive allele spt16-G132D results in a global reduction in nucleosome occupancy and a dramatic increase in cryptic transcription, consistent with a role of FACT in nucleosome reassembly during transcription elongation (23–26). In addition to playing a role in gene transcription, FACT also plays important roles in DNA replication. Early studies revealed that FACT interacts with DNA polymerase α and can also be copurified with the replicative helicase MCM2-7 in both yeast and human cells (27,28). Using a partial separation-of-function allele, we have shown that FACT functions in the assembly of newly synthesized H3-H4 into nucleosomes following DNA replication (29). Recently, it has been shown that FACT is essential for replication via a chromatin template in an in vitro reconstituted DNA replication system (30). More recently, a structure of human FACT in complex with a nucleosome has been reported (31), providing novel insights into how FACT manipulates the nucleosome. Together, these findings suggest that FACT performs multiple functions during gene transcription and DNA replication, including nucleosome assembly and disassembly, likely by interacting with distinct proteins involved in these processes.
Both Spt16 and Pob3 contain multiple domains (32), and these domains are highly conserved from yeast to humans. For instance, Spt16 has an N-terminal peptidase domain, a dimerization domain, a middle domain (Spt16-M) and a C-terminal acidic domain. The N-terminal domain (NTD) of Spt16 is highly conserved among all known Spt16 homologs, although it is not essential for cell viability in yeast cells. It has been shown that deletion of the Spt16 NTD in yeast reduces the need for the SWI/SNF chromatin-remodeling complex for gene activation (33). It has been proposed that the Spt16 NTD is important for gene repression. The Spt16 NTD also interacts with Sas3, a subunit of the NuA3 HAT complex (34). Therefore, it is possible that this domain is also involved in gene activation through histone acetylation. Furthermore, yeast cells lacking the Spt16 NTD show sensitivity to high levels of hydroxyurea (HU) that interferes with DNA replication (35,36). Taken together, these findings indicate that the Spt16 NTD likely regulates both gene transcription and DNA replication.
Set3 is a component of the Set3 HDAC complex (Set3C). While Set3 contains a SET domain, it lacks histone methyltransferase activity; it also interacts with two proteins, Hos2 and Hst1, that have HDAC activity. Set3C contains seven subunits: Set3, Sif2, Snt1, Hos2, Hst1, Cpr1 and Hos4 (37). Sif2 and Snt1 interact with each other and form a 2:2 tetramer, which interacts with Set3 and other Set3C components. Hos4 interacts with both Hst1 and Hos2. Set3, Snt1, Hos2 and Hos4 are likely present only in the Set3 complex, whereas Sif2, Cpr1 and Hst1 can exist as free proteins. Moreover, deletion of HST1 does not disrupt Set3C integrity, suggesting that it is a peripheral component of Set3C (37). Hst1 also forms a complex with Sum1 to repress the transcription of genes involved in vegetative growth.
Set3C likely performs multiple cellular functions. It is reported to mediate the transgenerational inheritance of an activated chromatin state (38). Moreover, while histone deacetylation catalyzed by HDACs such as Rpd3 is generally involved in gene repression, Set3C has been found to be involved in both gene repression and activation (8,39). For instance, it has been shown that Set3C can repress the transcription of some meiotic genes and metabolic genes (37,40). However, Set3 and Hos2 are also required for maximal induction of the GAL1 and INO1 genes in response to changes in carbon sources (environmental stimuli) (8,41,42). While Set3C functions in gene repression most likely through histone deacetylation, it remains unclear how Set3C contributes to maximal expression of the GAL1 gene.
Multiple models have been proposed to explain how Set3C functions in gene activation. For instance, it has been suggested that Set3C represses noncoding transcription, which in turn facilitates activation of coding genes (40). Supporting this idea, most of the genes upregulated in cells lacking Set3 are overlapped with noncoding RNA (ncRNA) transcription (40). Moreover, Set3 contains a PHD (plant homeodomain) domain that binds H3K4me2, which recruits Set3C to the 5′ ends of transcribed units (41). In separate studies, a long ncRNA has been found to be transcribed from the 3′ end of the GAL10 gene under derepressed conditions (raffinose treatment), which prevents leakage expression of genes at the GAL locus under these conditions. Under induced conditions (galactose treatment), repression of GAL10 ncRNA is important for the maximal production of GAL1 and GAL10 (43–46). Therefore, it is possible that Set3C represses GAL10 noncoding transcription, which in turn promotes optimal production of GAL gene transcripts under inducing conditions.
We found that Spt16 interacts with Set3C through its NTD and functions together with Set3C in gene regulation in some but not all settings in which FACT participates. Moreover, we showed that the FACT–Set3C interaction likely contributes to recruitment of these molecules to the gene body of GAL1 under inducing conditions. Furthermore, FACT and Set3C repress the transcription of a ncRNA (GAL10 ncRNA), and this repression contributes to the maximal induction of the GAL1 gene. Finally, we revealed that FACT and Set3C regulate nucleosome occupancy and histone acetylation at the GAL1 promoter at least partly by repressing GAL10 ncRNA expression. Collectively, these results reveal a novel interaction between FACT and Set3C and a role of FACT–Set3C in repression of GAL10 ncRNA expression, which in turn fine-tunes GAL1 activation in response to environmental stimuli.
MATERIALS AND METHODS
Strains and media
All strains used in this paper are isogenic to W303-1: leu2-3, 112, ura3-1, his3-11,15, trp1-1, ade2-1, can1-100 (Supplementary Table S1). Yeast tagging and deletion mutant strains were generated by one step transformation and confirmed by western blot or PCR (47,48). Standard yeast media were used for yeast culture (49).
TAP purification and MS analysis
Tandem affinity purification was performed as described (49). Generally, 1-liter cells were grown in YPD media to an OD of 1.8–2.0. Cells were collected by centrifugation. The same volume of IP buffer (25 mM Tris pH 8.0, 100 mM NaCl, 1 mM EDTA, 10 mM MgCl2, 0.01% NP-40, 1 mM DTT) with protease inhibitors (1 mM PMSF, 1 mM benzamidine, 1 mM Pefabloc) and 15 KU/ml DNase I was added, and the cells were frozen drop by drop in liquid nitrogen. The frozen yeast cells were broken by Freezer Mill. Then, the cell powder was then thawed on ice, cleared by centrifugation at 20 000 g for 10 min. The supernatant was then added with 75 μg/ml ethidium bromide (EB) and placed on ice for 30 min to disrupt DNA-mediated interactions. The sample was again centrifuged at 20 000 g for 40 min. The resulting supernatant was incubated with 40 μl IgG beads for 2 h. The beads were extensively washed by IP buffer and bound protein was cut by TEV enzyme at 16°C for 2 h. The released proteins were bound to 30 μl calmodulin beads for 2 h. Then, the beads were washed. The bound proteins were eluted with SDS sample buffer and boiled for 3 min. The sample was separated on SDS-PAGE, and stained with silver staining for subsequent MS analysis, or for western blot.
Chromatin immunoprecipitation assay (ChIP)
ChIP assay was performed essentially as previously described (50). Briefly, cells were grown in SCM with 2% raffinose and 0.02% glucose (SC-raffinose) to an OD of about 1.0. The cells were then centrifuged and washed by cold ddH2O, and resuspended in synthetic complete media with 2% galactose (SC-galactose) for the indicated time. The cells were fixed with 1% formaldehyde for 20 min, quenched with 125 mM glycine for 5 min. After washing with cold TBS, the cells were resuspended in ChIP lysis buffer (50 mM HEPES/KOH pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate) with protease inhibitors (1 mM PMSF, 1 mM Pefabloc, 1 mM benzamidine, 1 mg/ml bacitracin) and equal amount of glass beads, and then broken by MP Biomedicals Fastprep-24. The cell pellet was resuspended in ChIP buffer and sonicated with bioruptor to an average DNA size of 500 bp. After centrifugation, the supernatant was incubated with IgG beads for 3 h at 4°C. The beads were then washed and boiled to release the crosslinked DNA. The eluted DNA was subsequently quantified using Q-PCR. For the H3K27Ac ChIP, the sonicated cell lysate was divided into two parts, 0.1 μl of anti-H3K27Ac antibody (homemade) or 0.5 μl of anti-H3 antibody (ab1791) was added separately and rotated at 4°C overnight. The next day, 20 μl of protein G beads were added to allow binding for 2 h. The protein G beads were then extensively washed, and the DNA was eluted for Q-PCR analysis. All ChIP experiments were performed at least three times with independent biological samples. The primers used are listed in Supplementary Table S2.
Micrococcal nuclease (MNase) protection assay
To determine the effect of set3Δ on nucleosome density of GAL1 promoter during galactose induction, we used Micrococcal nuclease (MNase) protection assay. Briefly, cells were induced in 2% galactose for 40 min, then fixed with 1% PFA for 20 min and quenched by 125 mM glycine for 5 min. The cells were then spheroplasted with zymolase, and the resulting spheroplasts were digested with MNase to the mononucleosome. Mononucleosomal DNA was gel purified and nucleosome density at GAL1 promoter region was determined by quantitative real-time PCR (QPCR) normalized to the PHO5 gene.
Drug sensitivity assay
MPA and 6-AU sensitivity assay was performed as described (41). Briefly, the cells were transformed with the empty vector pRS316 and grown on synthetic complete mix minus Uracil (SC-URA) media. The cells were diluted to an OD of 0.6, then 5-fold serially diluted. About 5 μl of each dilution was spotted on SCM-URA plate containing the indicated drugs. We took images after the cells grew at 30 or 25°C for 2–5 days.
RT-qPCR
Yeast total RNA was extracted by standard hot phenol method. GAL10 ncRNA level was determined by northern blot as described (44). GAL1 mRNA level was quantified by RT-qPCR. Briefly, 1 μg of total RNA was first treated with DNase I (Thermo Scientific) and then reverse-transcribed with oligo (dT), the cDNA was directly served as template for qPCR. Primers are listed in Supplementary Table S2.
RESULTS
Set3 and Sif2 are new binding partners of FACT
To investigate how the distinct functions of FACT are regulated, we used tandem-affinity purification to purify FACT-associated protein factors in wild-type (WT) Spt16 and three mutant forms of SPT16 (spt16-m, spt16-G132D and spt16-ΔN) that distinctly impact DNA replication and gene transcription (Supplementary Figure S1). Spt16-m has a defect in replication-coupled (RC) nucleosome assembly and has a minor effect on gene transcription (29). Spt16-G132D is a temperature-sensitive (ts) mutant that affects gene transcription with little impact on DNA replication at the permissive temperature (29,33). The impact of spt16-ΔN, a mutant without the conserved NTD (amino acids 2–484), on gene transcription and DNA replication is not clear (36). Based on SDS-PAGE analysis of purified proteins, we observed distinct proteins associated with WT FACT and different FACT mutants (Supplementary Figure S1). We then used mass spectrometry (MS) to identify these Spt16-associated proteins. In addition to Pob3, another subunit of FACT, we found that several proteins (e.g. Paf1, Ctr9, Chd1, Tbp7, Psh1, Sif2 and Set3; Supplementary Tables S3 and S4) were primarily associated with WT Spt16 and the Spt16-m mutant forms, but not with the Spt16-G132D or Spt16-ΔN mutant forms (Supplementary Tables S3 and S4). Paf1, Ctr9, Chd1, Tbp7 and Psh1 interact with FACT (51–53). The newly identified Spt16 binding partners Set3 and Sif2 are two core members of Set3C. Set3C consists of four core subunits: Set3, Sif2, Hos2 and Snt1 (37,41). Interestingly, Set3C functions in both gene repression and activation (8,40). We thus focused our current study on the FACT–Set3C interaction and the potential impact of this interaction on gene regulation.
The NTD of Spt16 is required for FACT–Set3C interaction
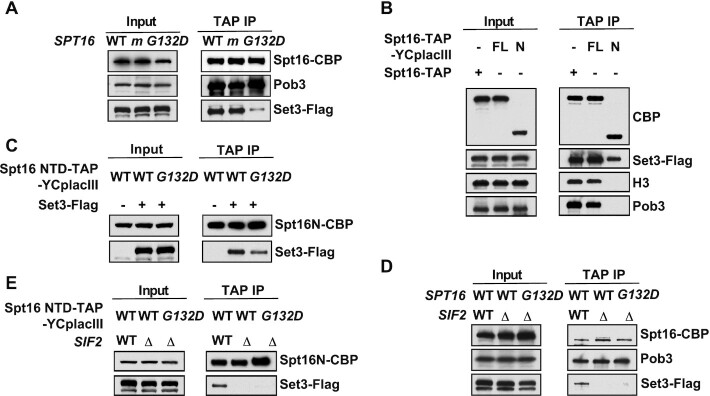
To validate the interaction between FACT and Set3C, we Flag-tagged SET3 and SIF2 and performed Spt16 immunoprecipitation (IP) experiments. Consistent with the MS data, WT Spt16 and the spt16-m mutant interacted with Set3. In contrast, the FACT–Set3 interaction was dramatically decreased in the spt16-G132D mutant cells at a permissive temperature (25°C) and barely detectable in the spt16-ΔN mutant (Figure 1A and Supplementary Figure S2A). Similar results were obtained when the FACT–Sif2 interaction was analyzed (Supplementary Figure S2B and S2C), suggesting that the N-terminus of Spt16 mediates the interaction between FACT and the Set3 complex. In addition, Spt16-m showed a defect in histone H3-H4 (29), but not Set3 and Sif2 binding (Supplementary Figure S2A and S2B). In contrast, the spt16-ΔN and spt16-G132D mutations did not appear to reduce the interaction between FACT and H3 (Supplementary Figure S2A and S2C). These results indicate that FACT interacts with Set3C and this interaction is unlikely to be mediated by histone H3-H4.
Figure 1.
FACT interacts with Set3C through the Spt16 NTD. (A) The N-terminal G132D mutation of SPT16 affects the Spt16–Set3 interaction, whereas mutation in the M-domain does not. Different forms of SPT16 (WT, m = spt16-K692A R693A and spt16-G132D) were TAP-tagged and subjected to tandem affinity purification. The purified protein complexes and the corresponding input samples were detected by western blot analysis using antibodies against calmodulin-binding peptide (CBP), Pob3 and Flag-tagged Set3. (B) The N-terminal domain (NTD) of Spt16 interacts with Set3. The full-length Spt16 and the NTD (amino acids 1–451) of Spt16 constructed in the YCp lac III plasmid with a TAP tag were expressed in a Set3-Flag-tagged strain and subjected to tandem affinity purification. The purified protein complexes and the corresponding input samples were detected by western blot analysis using antibodies against CBP, Flag-tagged Set3, H3 and Pob3. FL: full-length Spt16; N: NTD of Spt16. (C) G132D mutation affects the interaction of exogenously expressed Spt16-N with Set3. The Spt16-N proteins with or without the Spt16-G132D mutation constructed in the YCp lac III plasmid with a TAP tag were expressed in the no-tagged or Set3-Flag-tagged strain and subjected to tandem affinity purification. The purified protein complexes and the corresponding input samples were detected by western blot analysis using antibodies against CBP and Flag-tagged Set3. (D) The integrity of the Set3 complex is essential for the FACT–Set3C interaction. Spt16 protein complex was purified from WT or sif2Δ cells, and the purified protein complexes and the corresponding input samples were detected by western blot analysis using antibodies against CBP, Flag-tagged Set3, H3 and Pob3. (E) The integrity of the Set3 complex is also essential for the Spt16 NTD–Set3 interaction. The Spt16-N-TAP proteins with or without the Spt16-G132D mutation constructed in the YCp lac III plasmid were purified from WT or sif2Δ cells; the purified protein complexes and the corresponding input samples were detected by western blot analysis using antibodies against CBP and Flag-tagged Set3.
To test whether FACT interacts with Set3C directly, we first attempted to purify the Set3 complex. However, we were not able to purify a large amount of the Set3 complex from yeast cells for in vitro pull down assay despite repeated attempts. Next, we utilized similar approaches used by others to show direct interactions between two protein complexes (54,55). Briefly, we expressed the Spt16 NTD (amino acids 1–451) tagged with a TAP tag in Flag-tagged SET3 and SIF2 strains and analyzed the interaction of the Spt16 NTD with Set3C by IP. We observed that the Spt16 NTD protein interacted with both Set3 and Sif2 but to a lesser degree than full-length Spt16 (Figure 1B and Supplementary Figure S2D). Importantly, while H3 and Pob3 signals were readily detectable upon IP of full-length Spt16, they were not detectable upon IP of the Spt16 NTD, indicating that the FACT–Set3C interaction is also not bridged by proteins that interact with Pob3 as well as the large portion of Spt16 (amino acids 452–1035). Moreover, we observed that the Spt16 NTD containing the G132D mutation also displayed less binding with Set3C than the WT Spt16 NTD (Figure 1C and Supplementary Figure S2E). Collectively, these data demonstrate that the NTD of Spt16 is the primary domain that mediates the physical interaction between FACT and Set3C. However, we cannot eliminate the possibility that a protein interacts with Spt16 NTD and in turn bridges the FACT–Set3C interaction.
Previous data have revealed that several subunits, including Set3 and Sif2, are required for the integrity of Set3C (37). To further characterize the interaction between FACT and Set3C, we disrupted the Set3 complex by deleting either Set3 or Sif2, each of which is a core subunit of Set3C. We observed that the FACT–Set3 and FACT–Sif2 interactions were not detectable in the absence of SIF2 (Figure 1D) or SET3 (Supplementary Figure S2F), respectively. Similar results were obtained when Spt16-N proteins were used for immunoprecipitation (Figure 1E and Supplementary Figure S2G). We noted that deletion of SET3 did not affect the protein levels of Sif2 and vice versa. These results suggest that FACT interacts with multiple subunits of Set3C or that the integrity of Set3C is indispensable for the interaction between FACT and Set3C.
FACT also interacts with the catalytic subunit of the NuA3 HAT complex (Sas3) through the N-terminus of Spt16 (34). We confirmed that Sas3 coimmunoprecipitated with WT Spt16 and that this interaction was significantly reduced for the spt16-ΔN mutant (Supplementary Figure S2H). Interestingly, the spt16-G132D mutation did not affect Spt16–Sas3 interaction (Supplementary Figure S2I), indicating that Sas3 interacts with other regions in the Spt16 NTD. These results also indicate that the reduced interaction between Spt16-G132D and components of Set3C is unlikely due to the slight reductions in Spt16-G132D mutant protein levels in cells (Figure 1A and Supplementary Figure S2C). Therefore, we used the spt16-G132D mutant allele at the permissive temperature to dissect the function of FACT-Set3C interaction in gene regulation in subsequent experiments.
Spt16-G132D and Set3C mutations interact genetically
In the absence of SET3, yeast cells are sensitive to the transcription elongation inhibitor mycophenolic acid (MPA) (41,42). Spt16-G132D cells cannot grow at elevated temperature (37°C) (56,57). However, we observed that the FACT-Set3C interaction was weakened in spt16-G132D cells at the permissive temperature (25°C). Therefore, we monitored the phenotypes of set3Δ and spt16-G132D single or double mutants at this temperature. We observed that both the set3Δ and spt16-G132D mutant cells were sensitive to MPA, although spt16-G132D cells were more sensitive to MPA than set3Δ cells (Figure 2A). Interestingly, the MPA sensitivity of spt16-G132D set3Δ double-mutant cells was similar to that of set3Δ mutant cells, suggesting that SET3 depletion suppresses the MPA sensitivity of spt16-G132D cells. We noted that set3Δ did not suppress the temperature sensitivity of the spt16-G132D mutant (Figure 2A). A similar effect was observed when SIF2 was deleted from the spt16-G132D mutant cells (Supplementary Figure S3A). These results suggest that Set3C functions together with FACT in some but not all biological processes in which FACT participates.
Figure 2.
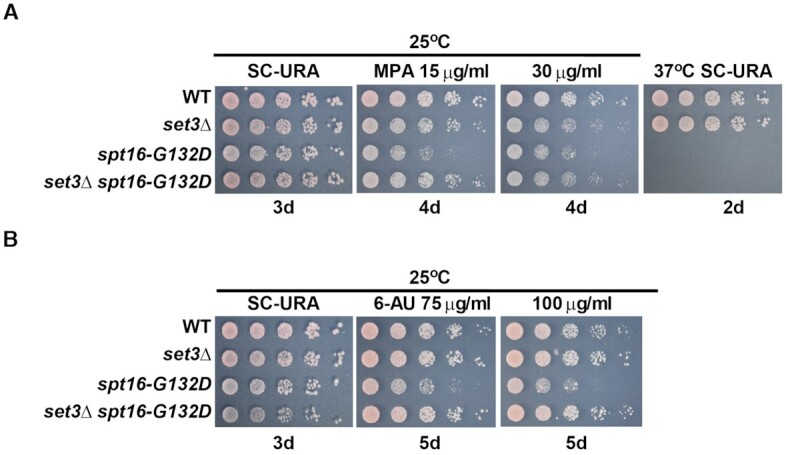
FACT genetically interacts with Set3C during gene transcription. set3Δ attenuates the MPA (A) and 6-AU (B) sensitivity of spt16-G132D cells. Five-fold serial dilutions of exponentially growing cells of the indicated genotypes were spotted onto SC-URA plates at 25 or 37°C, onto SC-URA plates containing 15 or 30 μg/ml MPA, and onto SCM-URA plates containing 75 or 100 μg/ml 6-AU at 25°C. Photographs of the SC-URA plates were taken on the indicated incubation days.
The spt16-G132D mutant cells were also sensitive to another inhibitor of transcription elongation, 6-azauracil (6-AU), at the permissive temperature (56,58), whereas the sif2Δ and set3Δ mutants were not (Figure 2B and Supplementary Figure S3A). Importantly, both spt16-G132D set3Δ and spt16-G132D sif2Δ were more resistant to 6-AU than the spt16-G132D single mutant (Figure 2B and Supplementary Figure S3A). Combined with the results of the MPA sensitivity analysis described above, these results show that the MPA and 6-AU sensitivity of spt16-G132D cells depends on the presence of intact Set3C.
Interestingly, we observed that spt16-ΔN cells did not show detectable sensitivity towards either MPA or 6-AU (Supplementary Figure S3B). One possible explanation for the differential drug sensitivity of spt16ΔN and spt16-G132D cells is that spt16ΔN mutation losses interactions with two histone modifying enzymes (Set3C and NuA3) with opposite effects on gene transcription. However, the spt16-ΔN mutation suppressed the MPA sensitivity of set3Δ cells (Supplementary Figure S3B). Taken together, these results indicate that FACT and Set3C function in the same pathway to regulate some aspects of gene transcription in which FACT is involved (see ‘Discussion’ section for more explanation) and strongly support the idea that FACT interacts with Set3C through the Spt16 NTD.
The associations of FACT and Set3C with actively transcribed GAL1 are mutually dependent
The regulation of GAL genes represents a classic transcriptional switch in response to changes in sugar in eukaryotic cells (59). The GAL gene cluster of Saccharomyces cerevisiae contains three genes (GAL1, GAL7 and GAL10) required for galactose metabolism (46). We noticed that Set3C is important for activation rather than repression of GAL genes (8,40). Moreover, FACT has been shown to play a role in GAL1 gene activation (19,24,60). However, the role of FACT and Set3C interaction during this process remains poorly understood. Therefore, we utilized this system to understand the function of the FACT–Set3C interaction in gene regulation.
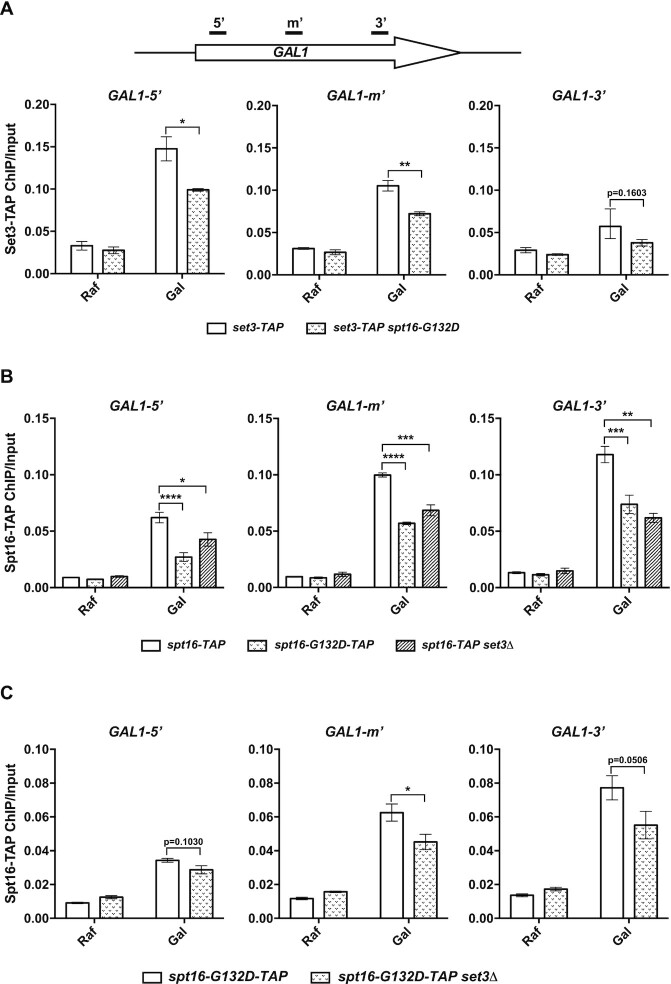
Both FACT and Set3C are recruited to transcribed regions (22,24,41,61). Therefore, we analyzed the effects of set3Δ and two spt16 mutations (spt16-G132D and spt16-ΔN) on the associations of Spt16 and Set3, respectively, with the GAL1 gene body. When cells are grown in SC-raffinose medium (Raf), the GAL1 gene is noninduced. However, the expression of GAL1 increases dramatically upon switching to galactose medium (Gal, inducing conditions). We observed that the levels of Set3 at the GAL1 coding region were higher in WT cells grown in Gal than in those grown in Raf, with a more robust increase at the 5′-GAL1 region than at the 3′-GAL1 region (Figure 3A), consistent with previous reports (8,62). Moreover, Set3 signals at the 5′ and middle regions of GAL1 were significantly lower in spt16-G132D mutant cells than in WT cells (Figure 3A). Similarly, deletion of the Spt16 NTD reduced the Set3 level at the GAL1 5′-coding region (Supplementary Figure S4A). Of note, neither the spt16-G132D allele nor the spt16-ΔN allele affected the protein level of Set3 (Supplementary Figure S4B and S4C).
Figure 3.
The FACT–Set3C interaction is important for the associations of these proteins with the gene body of actively transcribed GAL1. (A) spt16-G132D affects the loading of Set3C onto the gene body of actively transcribed GAL1. Cells were grown to the mid-logarithmic phase (OD600 = 1) in raffinose (Raf) at 25°C and shifted to galactose (Gal) for 40 min at 30°C. Set3-TAP ChIP analysis was performed using IgG Sepharose against the TAP tag. The precipitated DNA was analyzed by quantitative PCR (Q-PCR) using the primer sets indicated in the top panel. The average of three biological repeats is shown. The error bars indicate the ± 1 standard error values; Student’s t test was performed: *P< 0.05, **P< 0.01. (B) Spt16 mutation (spt16-G132D) and SET3 deletion affects the loading of FACT onto the gene body of actively transcribed GAL1. Spt16-TAP ChIP experiments were performed as described in (A). The average of three biological repeats is shown. The error bars indicate the ±1 standard error; Student’s t test was performed: *P< 0.05, **P< 0.01, ***P< 0.001, ****P< 0.0001. (C) SET3 deletion further affects the loading of FACT with G132D mutation onto the gene body of actively transcribed GAL1. Spt16-TAP ChIP experiments were performed as described in (A). The average of three biological repeats is shown. The error bars indicate the ± 1 standard error values; Student’s t test was performed: *P< 0.05.
We also observed that the levels of Spt16 across the GAL1 gene body were elevated in WT cells upon galactose addition, with slight enrichment at the 3′-GAL1 over 5′-GAL1 (Figure 3B and Supplementary Figure S4D) (62). Compared to WT Spt16, both spt16-G132D and spt16-ΔN mutants showed impaired Spt16 binding to GAL1 coding regions under inducing conditions (Figure 3B and Supplementary Figure S4D). The protein levels of Spt16-ΔN were similar to those of WT Spt16, whereas the protein levels of Spt16-G132D were slightly lower than those of WT Spt16 (Supplementary Figure S4E and S4F). Finally, the association of Spt16 with GAL1 coding regions was reduced in set3Δ mutant cells compared to WT cells (Figure 3B), and this reduction was unlikely due to the effect of set3Δ on Spt16 protein levels (Supplementary Figure S4E). Interestingly, the levels of Spt16 in the middle and at the 3′ end of GAL1 were reduced more in the spt16-G132D set3Δ double mutant than in the spt16-G132D single mutant (Figure 3C). Taken together, these results suggest that the associations of FACT and Set3C with active transcribed regions depend on each other or that the effects of Set3C and Spt16 mutations on the associations of their binding partner arise from defects in transcription in these mutant cells (see below and ‘Discussion’ section).
spt16-G132D and set3Δ have similar effects on GAL1 transcription
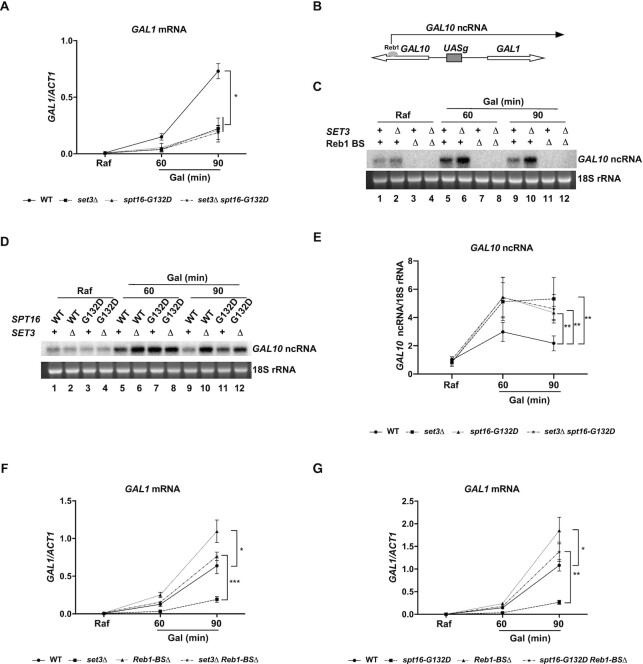
While HDACs are generally associated with gene repression, HDAC Set3C and Rpd3 display opposite effects on GAL1 gene transcription. Depletion of RPD3 results in increased transcription of GAL1, whereas deletion of SET3 delays the activation kinetics of GAL1 transcription compared to that in WT cells (8). We confirmed the differential impacts of set3Δ and rpd3Δ mutations on GAL1 transcription (Supplementary Figure S5A). Moreover, we observed similar effects on GAL10 transcription in the two corresponding mutant cell types (Supplementary Figure S5B). Next, we compared the effects of spt16-G132D and set3Δ mutations on transcription at the GAL gene cluster. We observed that the induction kinetics of GAL1 and GAL10 were impaired to similar extents in spt16-G132D mutant cells and set3Δ mutant cells. Importantly, the effect of set3Δ spt16-G132D double mutation on the transcription of the GAL1 and GAL10 genes was similar to the effects of spt16-G132D and set3Δ single mutation (Figure 4A and Supplementary Figure S5C). These results provide additional data to support the idea that FACT and Set3C interact and function in the same pathway to regulate gene transcription, at least in some settings.
Figure 4.
Set3C and FACT regulates the GAL1 expression by repressing the excessive expression of GAL10 ncRNA under inducing conditions. (A) The effects of spt16-G132D single and spt16-G132D set3Δ double mutations on the transcription of GAL1 mRNA under galactose inducing conditions. GAL1 levels were determined by RT-qPCR and normalized to ACT1 mRNA levels. The average of three biological repeats is shown. The error bars indicate the ±1 standard error; Student’s t test was performed: *P< 0.05; Raf: Raffinose; Gal: Galactose. (B) Schematic of the GAL1-10 locus showing the presence of GAL10 ncRNA. GAL10 ncRNA starts from the 3′ region of the GAL10 gene and runs across the GAL1 gene. GAL10 ncRNA transcription depends on the transcription factor Reb1. (C) SET3 deletion dramatically increases GAL10 ncRNA levels upon galactose induction. Cells were grown to the mid-logarithmic phase (OD600 = 1) in raffinose at 30°C and then transferred to galactose at 16°C for the indicated times (60 and 90 min). GAL10 ncRNA was detected by northern blot analysis, and 18S rRNA was used as a loading control. Three biological replicates were performed, and one representative result is shown; Raf: Raffinose; Gal: Galactose. (D) Effects of spt16-G132D single and spt16-G132D set3Δ double mutations on the transcription of GAL10 ncRNA under inducing conditions. Cells were grown to the mid-logarithmic phase (OD600 = 1) in Raf at 25°C and then transferred to Gal at 16°C for the indicated times (60 and 90 min). GAL10 ncRNA was detected by northern blot analysis, and 18S rRNA was used as a loading control. Three biological replicates were performed, and one representative result is shown. Raf: Raffinose; Gal: Galactose. (E) Quantification of the time course data from (D) after normalization to 18S rRNA. The average and SD of three biological repeats are shown for northern blot analysis. The error bars indicate the ±1 standard error; two-way ANOVA was used: **, P< 0.01. (F) Elimination of GAL10 ncRNA via deletion of the Reb1-binding site (Reb1-BSΔ) partially rescues the defects in the induction of the GAL1 gene in set3Δ mutant cells. GAL1 mRNA levels were determined by RT-qPCR and normalized to ACT1 mRNA levels. The average of three biological repeats is shown. The error bars indicate the ±1 standard error; Student’s t test was performed: *P< 0.05, ***P< 0.001. Raf: Raffinose; Gal: Galactose. (G) Elimination of GAL10 ncRNA via Reb1-BSΔ partially rescues the defects in the induction of the GAL1 gene in spt16-G132D mutant cells. GAL1 mRNA levels were determined by RT-qPCR and normalized to ACT1 mRNA levels. The average of three biological repeats is shown. The error bars indicate the ±1 standard error; Student’s t test was performed: *P< 0.05, **P< 0.01; Raf: Raffinose; Gal: Galactose.
The spt16-G132D and set3Δ mutations show similar effects on GAL10 ncRNA transcription
Most Set3C-affected genes have overlapping noncoding transcripts (40). Moreover, FACT functions to repress cryptic transcription initiation for a subset of yeast genes (62). A 4 kb GAL10 long ncRNA (GAL10 ncRNA) starting from the 3′ end of the GAL10 coding region is transcribed in the opposite direction of GAL10 under noninducing conditions (44) (Figure 4B). To understand how the HDAC complex Set3C and the histone chaperone FACT function in GAL1 activation, we first analyzed the effects of spt16-G132D and set3Δ mutation on GAL10 ncRNA transcription by northern blotting (Figure 4C). As the half-life of the GAL10 ncRNA transcript in WT cells is ∼8-17 min upon 2% galactose addition at 30°C (44,46), we tracked the expression of GAL10 ncRNA at different time points upon galactose addition at 16°C to capture the kinetics of GAL10 ncRNA expression. The transcription factor Reb1 binds to the promoter of GAL10 ncRNA (Figure 4B) and is essential for GAL10 ncRNA transcription (44). As controls, we also analyzed the effects of mutations in four putative Reb1-binding sites (Reb1-BSΔ mutant) on GAL10 ncRNA transcription. Mutations in Reb1-binding sites led to dramatic reductions in GAL10 ncRNA levels under all conditions, supporting the idea that Reb1 is essential for GAL10 ncRNA transcription (Figure 4C).
In WT cells grown in noninducing conditions (Raf), low levels of GAL10 ncRNA were detected. Under these conditions, deletion of SET3 had no apparent effect on the expression of GAL10 ncRNA, suggesting that Set3 has a minor role in the regulation of GAL10 ncRNA in Raf (Figure 4C). Under inducing conditions (Gal), we observed that the levels of GAL10 ncRNA in WT cells were apparently increased at 60 min and were reduced dramatically at 90 min upon galactose addition (Figure 4C, lanes 1, 5 and 9). Deletion of SET3 resulted in pronounced increases in GAL10 ncRNA at both 60 and 90 min upon galactose addition from the levels in WT cells (Figure 4C and Supplementary Figure S5D), suggesting that Set3 inhibits an unchecked increase in GAL10 ncRNA during galactose induction.
We next evaluated whether spt16-G132D also impinges on GAL10 ncRNA transcription in Gal (Figure 4D and E). Similar to the case in set3Δ cells, northern blot analysis indicated that GAL10 ncRNA in spt16-G132D mutant cells was increased at different time points after addition of galactose for induction. Importantly, GAL10 ncRNA levels were not further increased in spt16-G132D set3Δ double mutant cells compared to single-mutant cells of either cell line (Figure 4D and E). Together, these results indicate that FACT and Set3C function in the same pathway to repress GAL10 ncRNA transcription upon galactose induction, likely preventing unrestricted expression of GAL10 ncRNA, which may affect the induction of GAL gene expression.
The roles of FACT and Set3C in GAL1 activation are linked to their roles in GAL10 ncRNA transcription
Previous studies have demonstrated that GAL10 ncRNA can fine-tune the activation of GAL genes in response to metabolic changes (43,46). Therefore, we asked whether the effects of spt16-G132D and set3Δ mutations on GAL1 and GAL10 activation are due to their impacts on GAL10 ncRNA transcription. To do this, we first attenuated GAL10 ncRNA expression by mutating the Reb1-binding sites (Reb1-BSΔ) in WT and set3Δ mutant cells (44,45) and analyzed the impact of set3Δ and Reb1-BSΔ single mutation and set3Δ Reb1-BSΔ double mutation on GAL1 and GAL10 transcription. Consistent with previous results (44,45), rapid induction of GAL1 was observed in Reb1-BSΔ mutant cells compared to WT cells (Figure 4F and G), and this effect of Reb1-BSΔ on GAL1 and GAL10 transcription was opposite to that of set3Δ. Importantly, incorporation of the Reb1-BSΔ mutation in the set3Δ strain restored the induction profiles of GAL1 and GAL10 to levels similar to those in WT cells (Figure 4F and Supplementary Figure S5E). Similar results were also observed for the impact of depletion of the Reb1-binding sites in spt16-G132D cells on the induction of GAL1 and GAL10 transcription (Figure 4G and Supplementary Figure S5F). Of note, we site-specifically mutated the TATA box of the GAL10 promoter (Supplementary Figure S5H, top panel). While mutating the GAL10-TATA box led to a dramatic reduction of GAL10 gene expression, GAL10 ncRNA was not affected (Supplementary Figure S5G and S5H, left panel). No apparent changes were observed on GAL1 gene expression level in GAL10-TATA* mutation cells (Supplementary Figure S5H, right panel). Taken together, these results suggest that both Set3C and FACT promote the induction kinetics of the GAL genes at least partly through inhibition of excessive expression of GAL10 ncRNA upon galactose induction.
Spt16-dependent noncoding transcription at the GAL10 locus is detected in spt16-G132D cells under nonpermissive temperature
To understand how FACT and Set3C may function in GAL10 ncRNA transcription, we first investigated whether Spt16 and Set3 bound to the regions surrounding Reb1-binding sites. Unfortunately, this experiment was complicated by the fact that both FACT and Set3 are recruited to the GAL10 gene likely in a transcription-dependent manner. We previously found that depleting Spt16 by growing spt16-G132D cells at 37°C for 45 min leads to nucleosome fuzziness and position shifts in gene bodies and to an increase in the levels of Spt16-dependent noncoding transcripts (SNTs) (23). To understand how FACT may be involved in inhibition of GAL10 ncRNA induction, we reanalyzed micrococcal nuclease (MNase)-seq datasets for WT and spt16-G132D cells growing at the permissive temperature (T0) and a nonpermissive temperature (T45) for 45 min in glucose (repressive conditions). We observed that nucleosome occupancy and nucleosome positions at the GAL10 ncRNA region were barely changed when WT cells were shifted to 37°C. In contrast, nucleosome positions and occupancy were altered at this region in spt16-G132D cells growing at 37°C compared to 25°C (Supplementary Figure S6A). Moreover, high levels of noncoding transcription from the 3′ region of GAL10 were also detected at this region (Supplementary Figure S6B). These results indicate that FACT can be recruited to the GAL10 ncRNA locus to suppress noncoding transcription, at least under repressive conditions.
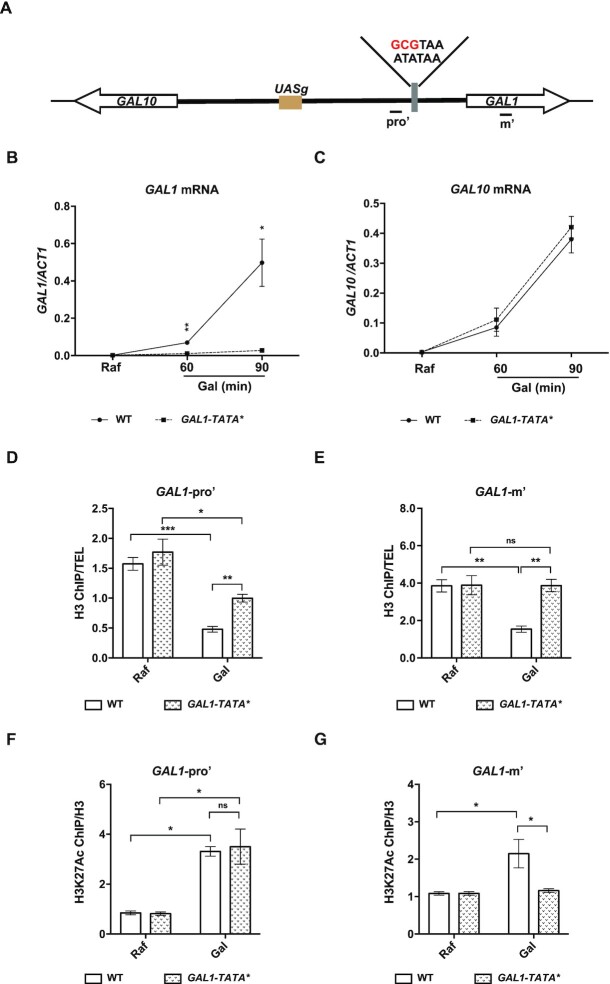
Mutation of the GAL1-TATA box has minor effects on changes in nucleosome dynamics at the GAL1 promoter
In general, nucleosome occupancy at promoters is inversely correlated with gene transcription (63–66). High nucleosome occupancy can in principle block the association of transcription factors with promoters, which in turn hinders gene transcription. To further dissect the role of Set3C and FACT in the GAL1 regulation, we analyzed the histone H3 levels at both the GAL1 promoter and gene body regions, which reflect the nucleosome occupancy at these regions, by H3 ChIP assays. When wild-type cells were shifted from raffinose condition to galactose condition, the H3 signals were apparently reduced at both the GAL1-promoter regions (GAL1-Pro’) and the GAL1-gene body regions (GAL1-m’) (Figure 5D and E), suggesting a nucleosome removal along with the GAL1 gene activation. To ask whether these nucleosomal changes are a consequence of ongoing gene transcription, we site-specifically mutated the TATA box of the GAL1 promoter (Figure 5A) (67). The transcription levels of GAL1 mRNA in GAL1-TATA box mutant cells (GAL1-TATA*) were dramatically reduced upon galactose induction (Figure 5B), consistent with the idea that the GAL1-TATA box is essential for GAL1 transcription. On the other hand, the GAL10 mRNA levels were not affected in the GAL1-TATA* mutant cells compared to the levels in WT cells under the same condition (Figure 5C). Under these conditions, nucleosome occupancy detected by H3 ChIP in the gene body region of GAL1 in GAL1-TATA* mutant cells did not change upon a change in carbon source from raffinose to galactose (Figure 5E), supporting that the on-going GAL1 transcription likely contributes to nucleosomal changes at the GAL1 gene boy in wild-type cells. Intriguingly, nucleosome occupancy detected by H3 ChIP at the GAL1 promoter region was also reduced in GAL1-TATA* mutant cells upon a change in carbon source from raffinose to galactose, while to a lesser degree than in WT cells (Figure 5D). These results suggest that changes in nucleosome occupancy at the gene body of GAL1 are due largely to gene transcription, whereas nucleosome changes at the GAL1 promoter might be independent of the on-going GAL1 transcription.
Figure 5.
Nucleosome removal at the GAL1 promoter upon induction impacts on the transcription of GAL genes. (A) A 3-bp substitution mutation within the GAL1 promoter TATA box region that can disrupt the binding of TATA-binding protein to the promoter is shown. Primers at the GAL1 promoter region (pro’) and the gene body region (m’) are shown. (B and C) GAL1 and GAL10 mRNA levels in cells with a GAL1-TATA mutation (GAL1-TATA*). Cells bearing the GAL1-TATA mutant site were grown in raffinose (Raf) at 30°C and then shifted to 16°C upon galactose (Gal) addition for the times indicated. GAL1 and GAL10 mRNA levels were determined by RT-qPCR and normalized to ACT1 mRNA levels. The average of three biological repeats is shown. The error bars indicate the ±1 standard error; Student’s t test was performed: *P< 0.05, **P< 0.01. (D) Nucleosome changes at the GAL1 promoter regions are compromised upon GAL1-TATA* mutation. WT and GAL1-TATA* mutant cells were grown in raffinose at 30°C and then shifted to medium containing 2% galactose for 40 min. H3 ChIP analysis was performed at the GAL locus in WT and GAL1-TATA* mutant cells. The H3-precipitated DNA was analyzed by quantitative PCR (Q-PCR) using the primer sets indicated in (A), and the levels were normalized to those obtained with a telomeric primer set (TEL). The error bars indicate the ±1 standard error; Student’s t test was performed: *P< 0.05, **P< 0.01; ns, not significant. (E) No apparent nucleosome changes at the GAL1 gene body region were detected upon GAL1-TATA* mutation. The experiment was performed as describe in (D). The error bars indicate the ±1 standard error; Student’s t test was performed: **P< 0.01, ns, not significant. (F) The levels of H3K27Ac at the promoter of GAL1 are increased upon GAL1-TATA* mutation. H3K27Ac ChIP analysis was performed in WT and GAL1-TATA* mutant cells as in (D). The H3K27Ac-precipitated DNA was analyzed by Q-PCR using the primer sets indicated in (A) and then normalized to the H3 occupancy. The average of three biological repeats is shown. The error bars indicate the ±1 standard error; Student’s t test was performed: *P< 0.05; ns, not significant. (G) No detectable changes of H3K27Ac levels at the gene body of GAL1 upon GAL1-TATA* mutation. H3K27Ac ChIP analysis was performed in WT and GAL1-TATA* mutant cells as in (F). The H3K27Ac-precipitated DNA was analyzed by Q-PCR using the primer sets indicated in the top panel (B) and then normalized to the H3 occupancy. The average of three biological repeats is shown. The error bars indicate the ±1 standard error; Student’s t test was performed: *P< 0.05, ns, not significant.
Set3C functions as an HDAC. We therefore examined the acetylation at lysine 27 of H3 (H3K27Ac) levels at the GAL1 regions during galactose induction. Since nucleosome occupancy was altered in WT cells, we normalized the H3K27Ac ChIP signals to the corresponding H3 ChIP signals. Under noninducing conditions, there were no apparent differences in H3K27Ac levels in GAL1-TATA* mutant cells compared to WT cells (Figure 5F and G). We found that H3K27Ac levels increased in WT cells at both promoter region and gene body of GAL1 upon GAL1 activation during the carbon source shift from raffinose to galactose (Figure 5F and G). In contrast, while H3K27Ac levels did not increase at the GAL1 gene body regions in GAL1-TATA* cells (Figure 5G), H3K27Ac levels at the GAL1 promoter regions also increased in GAL1-TATA* cells to a level similar to WT cells (Figure 5F). These results suggest that the increase in histone acetylation at the GAL1 gene body is also likely a consequence of increased transcription, but that the increase in histone acetylation levels at the GAL1 promoter region can occur in the absence of strong GAL1 induction. Collectively, these results strongly indicate that changes in nucleosome occupancy and H3K27Ac levels at the GAL1 promoter upon metabolic shifting can occur prior to strong induction of GAL1 expression.
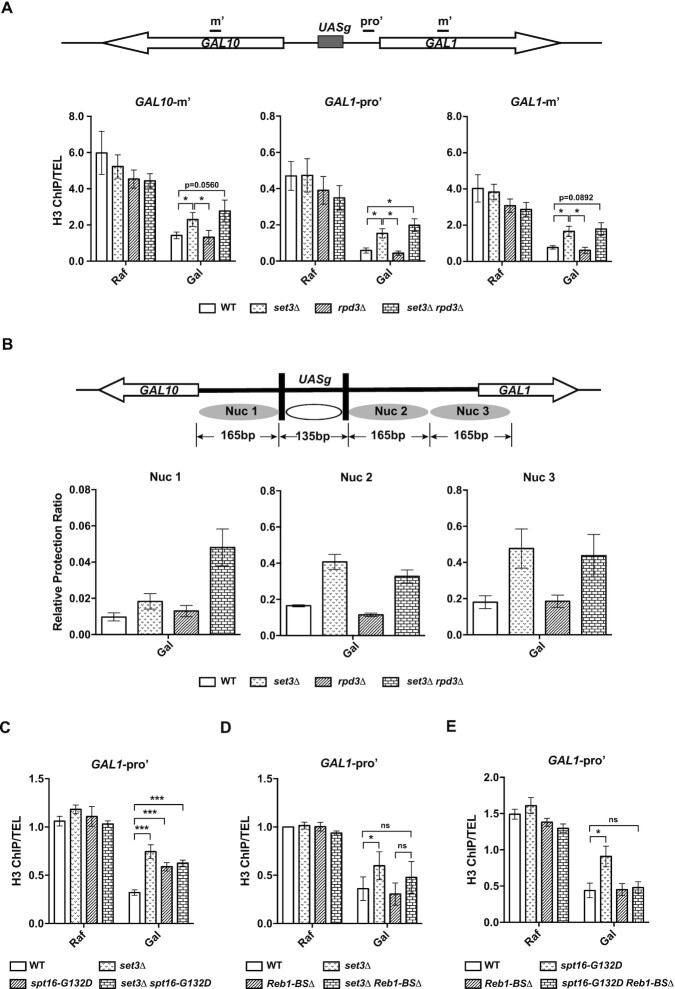
Effects of set3Δ and rpd3Δ mutations on nucleosome occupancy at the GAL1 promoter
It has been shown that an intergenic ncRNA (SRG1) causes repression of an adjacent coding gene (SER3) by affecting nucleosome occupancy of the promoter region of the SER3 gene (68). Moreover, a previous study has shown that the GAL10 ncRNA operates in cis to recruit the HDAC Rpd3 in order to repress GAL1 expression under repressive conditions (44). We therefore set out to evaluate whether GAL10 ncRNA could regulate nucleosome occupancy at the GAL1 promoter region, which is largely independent of GAL1 induction, to repress the induction of the GAL1 gene. To do this, we first analyzed nucleosome occupancy at the GAL1 promoter in set3Δ and rpd3Δ single and set3Δ rpd3Δ double mutant cells, as set3Δ and rpd3Δ mutations have opposite effects on GAL1 transcription (Supplementary Figure S5A). Under noninducing conditions, the H3 levels were comparable between WT and set3Δ, whereas the rpd3Δ single and set3Δrpd3Δ double mutants exhibited detectable but not significant decreases in H3 levels at the GAL1-GAL10 locus (Figure 6A, Raf). Upon galactose induction, we observed that the H3 levels were remarkably decreased at the promoter region (GAL1-Pro’) as well as the middle coding regions of GAL1 and GAL10 in WT cells (Figure 6A, compare Raf to Gal), suggesting a rapid removal of nucleosomes upon gene activation. Of note, the H3 level was significantly higher in set3Δ cells than in WT cells (Figure 6A), suggesting that nucleosome removal in set3Δ mutant cells is impaired than that in WT cells. These results are consistent with the observation that GAL1 transcription is lower in set3Δ mutant cells than in WT cells.
Figure 6.
Nucleosome occupancy at the GAL1 promoter in set3Δ and spt16-G132D mutant cells is higher than that in WT cells upon galactose induction. (A) The nucleosome levels across the GAL1/GAL10 locus in set3Δ cells are higher than those in WT cells or rpd3Δ cells in 2% galactose medium. H3 ChIP analysis was performed in the indicated strains after cells were shifted from Raf to Gal for 40 min at 30°C, and chromatin was precipitated with an antibody against H3. The precipitated DNA was analyzed by quantitative PCR (Q-PCR) using the primer sets indicated in the top panel, and the levels were normalized to those obtained with a telomeric primer set (TEL). The primer locations are shown schematically at the top. The average of three biological repeats is shown. The error bars indicate the ±1 standard error; Student’s t test was performed: *P< 0.05. (B) The nucleosomes at the GAL1 promoter regions in set3Δand set3Δrpd3Δcells exhibit enhanced resistance to MNase digestion. Cells with different genotypes were induced in Gal for 40 min at 30°C, and chromatin was subjected to MNase digestion. DNA was extracted, and Q-PCR was used to analyze the relative protection of nucleosomes within the GAL1 promoter. Representative results for two biological repeats are shown. The standard error is presented. (C) The effect of spt16-G132D on nucleosome occupancy at the GAL1 promoter during galactose induction is similar to that of set3Δ. H3 ChIP analysis was performed in the indicated strains after cells were shifted from Raf at 25°C to Gal for 40 min at 30°C, and chromatin was precipitated with an antibody against H3. The precipitated DNA was analyzed by Q-PCR using a GAL1-pro’ primer, and the levels were normalized to those obtained with a telomeric primer set (TEL). The average of three biological repeats is shown. The error bars indicate the ±1 standard error; Student’s t test was performed: ***P< 0.001. (D) Knockout of GAL10 ncRNA (Reb1-BSΔ) partially reverses the high nucleosome occupancy of set3Δ. H3 ChIP analysis was performed in the indicated strains after cells were shifted from Raf to Gal for 40 min at 30°C, and chromatin was precipitated with an antibody against H3. The precipitated DNA was analyzed by Q-PCR using the GAL1-pro primer, and the levels were normalized to those obtained with a telomeric primer set (TEL). The average of four biological repeats is shown. The error bars indicate the ±1 standard error; Student’s t test was performed: *P< 0.05, ns, not significant. (E) Knockout of GAL10 ncRNA (Reb1-BSΔ) fully restores the nucleosome occupancy of spt16-G132D to WT levels. H3 ChIP analysis was performed in the indicated strains after cells were shifted from Raf at 25°C to Gal for 40 min at 30°C, and chromatin was precipitated with an antibody against H3. The precipitated DNA was analyzed by Q-PCR using the GAL1-pro primer, and the levels were normalized to those obtained with a telomeric primer set (TEL). The average of three biological repeats is shown. The error bars indicate the ±1 standard error; Student’s t test was performed: *P< 0.05, ns, not significant.
H3 levels in rpd3Δ cells were also reduced to a similar degree to WT cells upon induction. In contrast, GAL1 expression in rpd3Δ mutant cells was higher than that in WT cells (Supplementary Figure S5A), consistent with published results (8). To rule out the possibility that our assay was unable to detect differences in nucleosome occupancy between rpd3Δ cells and WT cells, possibly due to the resolution of H3 chromatin IP (ChIP) assays based on sonication-sheared chromatin, we performed an MNase protection assay to further analyze the nucleosome occupancy at each of the three nucleosomes at the GAL1 promoter under galactose induction (69). In agreement with H3 ChIP results, nucleosome occupancy at all three nucleosomes localized at the promoter region was markedly higher in set3Δ mutant cells than in WT cells (Figure 6B and Supplementary Figure S7A). We again detected that the nucleosome occupancy in rpd3Δ single mutant cells was comparable to that in WT cells. These results suggest that the increase in gene transcription in rpd3Δ cells may not lead to an additional reduction in nucleosome occupancy at the GAL1 promoter region in rpd3Δ cells. These findings support the idea that both nucleosome occupancy and GAL1 transcription are altered in set3Δ mutant cells and that H3 ChIP assays can be used to detect nucleosome changes in these cells.
Interestingly, nucleosome occupancy at the GAL1 promoter was higher in set3Δ rpd3Δ double mutant cells than in rpd3Δ mutant cells and WT cells based on the H3 ChIP assay (Figure 6A) and MNase protection assays (Figure 6B and Supplementary Figure S7A). Moreover, we observed that GAL1 transcription in set3Δ rpd3Δ cells was lower than that in rpd3Δ mutant cells (Supplementary Figure S5A). Collectively, these results suggest that Set3C has more dominant roles in regulating gene transcription and nucleosome occupancy at GAL1 promoter regions under inducing conditions than the HDAC Rpd3.
The effects of spt16-G132D and set3Δ mutations on nucleosome occupancy at the GAL1 promoter depend on GAL10 ncRNA transcription
Next, we compared the effects of spt16-G132D and set3Δ mutations on nucleosome occupancy at the GAL1 promoter region upon galactose addition. Histone levels at the GAL1 promoter in spt16-G132D mutant cells were similar to those in set3Δ mutant cells but were higher than those in WT cells under galactose-mediated inducing conditions (Figure 6C). Moreover, the nucleosome occupancy at the GAL1 gene promoter in spt16-G132D set3Δ double mutant cells was similar to that in either spt16-G132D single mutant cells or set3Δ single mutant cells (Figure 6C). These results are consistent with the idea that FACT functions in the same pathway as Set3C to regulate nucleosome occupancy at the GAL1 promoter.
To test whether GAL10 ncRNA transcription has any role in the nucleosome occupancy observed in spt16-G132D and set3Δ mutant cells, we performed H3 ChIP experiments in Reb1-BSΔ mutant cells (Figure 6D). The histone H3 levels at the GAL1 promoter in Reb1-BSΔ mutant cells were similar to those in WT cells (Figure 6D). Moreover, the histone H3 levels of set3Δ Reb1-BSΔ double-mutant cells were similar to those of Reb1-BSΔ cells (Figure 6D). Similarly, the histone H3 levels of spt16-G132D Reb1-BSΔ cells were comparable to those of Reb1-BSΔ cells, which were lower than those of spt16-G132D mutant cells (Figure 6E). Collectively, these results suggest that the nucleosome occupancy changes observed in spt16-G132D and set3Δ cells are at least partly dependent on GAL10 ncRNA transcription.
Effects of FACT and Set3C on histone acetylation levels at the GAL1 promoter
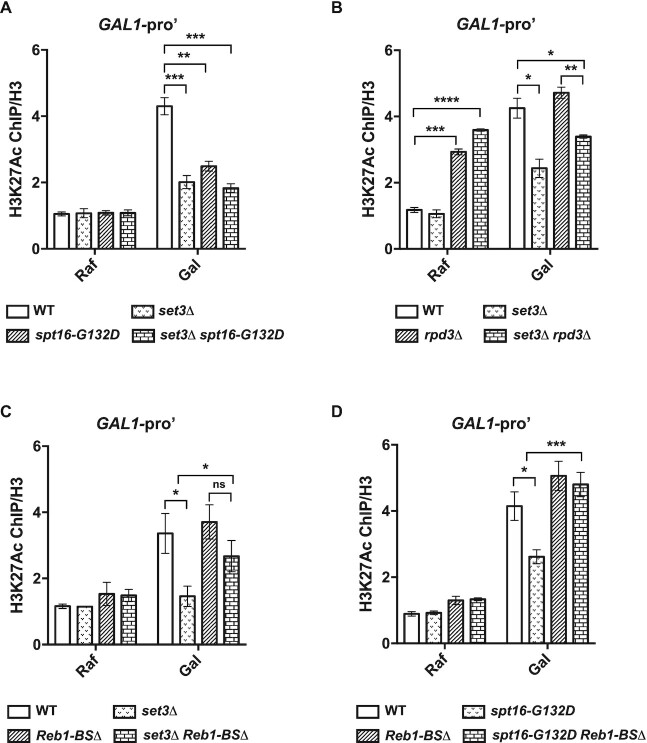
We next examined H3K27Ac levels at the GAL1 promoter during galactose induction. Since nucleosome occupancy was changed in set3Δ and spt16-G132D mutant cells, we normalized the H3K27Ac ChIP signals to the corresponding H3 ChIP signals. Under noninducing conditions, there were no apparent differences in H3K27Ac levels in set3Δ and spt16-G132D cells compared to WT cells (Figure 7A). Moreover, we observed markedly higher H3K27Ac levels at the GAL1 promoter in WT cells grown in Gal than in WT cells grown in Raf. This increase was impaired in both set3Δ and spt16-G132D single mutant cells. Moreover, the effect of the set3Δ G132D double mutation on H3K27Ac levels was similar to those of the set3Δ and spt16-G132D single mutations (Figure 7A). These results suggest that FACT and Set3C may functionally interact to affect the histone acetylation levels of the GAL1 promoter upon galactose addition.
Figure 7.
H3K27Ac levels at the GAL1 promoter are reduced in set3Δ and spt16-G132D mutant cells upon galactose induction. (A) The effect of spt16-G132D on H3K27Ac levels at the GAL1 promoter during galactose induction is similar to that of set3Δ. H3K27Ac ChIP analysis was performed in the indicated strains after cells were shifted from Raf at 25°C to Gal for 40 min at 30°C, and chromatin was precipitated with antibodies against H3K27Ac and H3. The H3K27Ac-precipitated DNA was analyzed by quantitative PCR (Q-PCR) using a GAL1-pro’ primer, and the levels were normalized to the H3 occupancy. The average of three biological repeats is shown. The error bars indicate the ±1 standard error; Student’s t test was performed: **P< 0.01, ***P< 0.001. (B) Set3C plays a dominant role in affecting H3K27Ac levels at the GAL1 promoter during galactose induction. H3K27Ac ChIP analysis was performed in the indicated strains after cells were shifted from Raf to Gal for 40 min at 30°C, and chromatin was precipitated with antibodies against H3K27Ac and H3. The H3K27Ac-precipitated DNA was analyzed by Q-PCR using the GAL1-pro primer, and the levels were normalized to the H3 occupancy. The average of three biological repeats is shown. The error bars indicate the ±1 standard error; Student’s t test was performed: *P< 0.05, **P< 0.01, ***P< 0.001, ****P< 0.0001. (C) Knockout of GAL10 ncRNA (Reb1-BSΔ) partially reverses the hypoacetylation at the GAL1 promoter in set3Δ cells. H3K27Ac ChIP analysis was performed in the indicated strains after cells were shifted from Raf to Gal for 40 min at 30°C, and chromatin was precipitated with antibodies against H3K27Ac and H3. The H3K27Ac-precipitated DNA was analyzed by Q-PCR using the GAL1-pro primer, and the levels were normalized to the H3 occupancy. The average of three biological repeats is shown. The error bars indicate the ±1 standard error; Student’s t test was performed: *P< 0.05. (D) Knockout of GAL10 ncRNA (Reb1-BSΔ) fully reverses hypoacetylation at the GAL1 promoter in spt16-G132D cells compared with WT cells. H3K27Ac ChIP analysis was performed in the indicated strains after cells were shifted from Raf at 25°C to Gal for 40 min at 30°C, and chromatin was precipitated with antibodies against H3K27Ac and H3. The H3K27Ac-precipitated DNA was analyzed by Q-PCR using the GAL1-pro primer, and the levels were normalized to the H3 occupancy. The average of three biological repeats is shown. The error bars indicate the ±1 standard error; Student’s t test was performed: *P< 0.05, ***P< 0.001.
It has been established that Rpd3 is a major HDAC regulating histone acetylation at the GAL gene locus under noninducing conditions (8). Consistent with this idea, we observed that deletion of RPD3 resulted in a dramatic increase in H3K27Ac under noninducing conditions at the GAL locus (Figure 7B and Supplementary Figure S7B). Interestingly, deletion of SET3 did not affect the levels of H3K27Ac under noninducing conditions (raffinose). Deletion of SET3 in rpd3Δ cells also did not affect the levels of H3K27Ac under noninducing conditions (raffinose) (Figure 7B and Supplementary Figure S7B). In contrast, H3K27Ac levels only at the GAL1 promoter were significantly lower in set3Δ rpd3Δ double mutant cells than in rpd3Δ mutant cells under inducing conditions (Figure 7B), and the levels of H3K27Ac were comparable at the GAL1 and GAL10 gene bodies in the two mutant cells (Supplementary Figure S7B). Moreover, both GAL1 and GAL10 mRNA levels showed dramatic decreases in set3Δ rpd3Δ mutant cells compared with rpd3Δ mutant cells upon galactose addition (Supplementary Figure S5A and S5B, (8)). We reasoned that this was due to delayed nucleosome removal in set3Δ cells upon galactose induction (Figure 6A and B) as well as reduction of H3K27Ac levels at the GAL1 promoter observed here, consistent with the idea that nucleosome occupancy changes at promoter regions are critical for gene transcription regulation.
As GAL10 ncRNA affects nucleosome occupancy, we next examined the effect of GAL10 ncRNA on histone acetylation at the GAL1 promoter. We observed that H3K27Ac levels increased slightly but not significantly in Reb1-BSΔ cells (Figure 7C). Interestingly, Reb1-BSΔ partially reversed the reductions in H3K27Ac levels at the GAL1 promoter in set3Δ cells (Figure 7C) and fully reversed those in spt16-G132D mutant cells (Figure 7D). These results support the idea that FACT and Set3C affect histone acetylation levels at the GAL1 promoter partially by repressing GAL10 ncRNA transcription under inducing conditions.
DISCUSSION
Eukaryotic gene transcription occurs in the context of chromatin and involves multiple layers of regulation to deal with the chromatin template in response to various signals. FACT is an essential histone chaperone functioning in multiple aspects of DNA replication and gene transcription. It has been proposed that FACT functions in these multiple processes by interacting with different proteins involved in DNA replication and gene transcription. Herein, we present multiple lines of evidence indicating that FACT interacts with Set3C to inhibit overproduction of GAL10 ncRNA, which in turn fine-tunes GAL1 expression in response to carbon source shifts. Thus, we reveal a new interaction and a mechanism by which FACT functions in gene regulation (Supplementary Figure S8).
The NTD of Spt16 interacts with distinct factors and regulates FACT function in different contexts
FACT, consisting of Spt16 and Pob3, is essential for cell proliferation. Both Spt16 and Pob3 contain multiple Pleckstrin homology (PH) domains, and these domains mediate interactions with histones or nucleosome substrates (32). Spt16 has an N-terminal peptidase domain, and this NTD is highly conserved among all known Spt16 homologs. Intriguingly, yeast cells expressing a Spt16 mutant lacking the NTD are viable, indicating that this domain may function in nonessential regulatory processes. It has been reported that the Spt16 NTD binds H3-H4 in vitro (70) as well as Sas3, the subunit of the NuA3 HAT complex, but not SAGA (34). Here, we show that FACT interacts with the HDAC Set3C and that this interaction is not bridged by histone H3-H4. Importantly, we found that the Spt16-G132D mutant shows defects in binding to Set3C but interacts with Sas3 similar to WT Spt16; these findings suggest that the Spt16 NTD interacts with distinct factors that in turn act with FACT to regulate different processes. Consistent with this idea, MS analysis suggested that the interactions between FACT and several proteins involved in DNA replication and/or DNA damage repair are also compromised in spt16-G132D and spt16-ΔN cells compared to cells with WT Spt16 (Supplementary Tables S3 and S4). Furthermore, yeast cells lacking the Spt16 NTD are sensitive to high levels of hydroxyurea (HU), which interferes with DNA replication (35,36). In the future, it would be interesting to dissect the interactions of FACT with these proteins involved in DNA replication and DNA damage repair.
FACT and Set3C participate in GAL1 activation likely through inhibition of GAL10 ncRNA
Supporting a direct interaction between FACT and Set3C, we observed that individual depletion of SET3 and SIF2, two components of Set3C, reduced the drug sensitivity of spt16-G132D cells towards transcription inhibitors but did not reduce the temperature sensitivity of these mutant cells. There are two nonmutually exclusive explanations for the suppression of MPA and 6-AU sensitivity of spt16-G132D cells by deletion of SET3 or SIF2. First, Set3C and FACT have opposite roles in gene transcription. This interpretation is similar to the proposed roles of Chd1 and FACT in gene regulation. It has been reported that Chd1 interacts with FACT physically (51,71). Deletion of CHD1 suppressed several defects of a spt16 mutant allele, and it was proposed that Chd1 and FACT have opposite roles in gene regulation (72). Second, in spt16-G132D mutant cells, Set3C is mis-localized to other regions, which in turn alters gene expression and renders spt16-G132D cells sensitive to MPA and 6-AU. Supporting this idea, we found that the chromatin association of Set3 at the GAL1 gene is compromised in spt16-G132D cells. Moreover, FACT and Set3C physically interact with each other. Nonetheless, the suppression of MPA and 6-AU sensitivity, but not temperature sensitivity of spt16-G132D cells by set3Δ is consistent with the idea that the Set3C and FACT function in the same pathway in some, but not all settings in which FACT participates in gene regulation (73).
Because previous studies have shown that FACT and Set3C are required for activation of the GAL1 gene (19,24,40,41,60), a well-studied model for gene regulation, we focused our analysis on the effects of spt16-G132D and set3Δ at the GAL locus. We found that spt16-G132D and set3Δ mutants showed similar defects in the induction of GAL genes after cells were switched from derepressed (raffinose) to inducing (galactose) conditions. Importantly, spt16-G132D set3Δ double-mutant cells showed the same defects in GAL gene transcription as spt16-G132D and set3Δ single-mutant cells under these conditions. Together, these results indicate that FACT interacts with Set3C and that these molecules work together to fine-tune the activation of GAL genes under inducing conditions.
Set3C contains two proteins, Hos2 and Hst1, which have HDAC activity. Histone deacetylation by HDACs generally represses genes. Therefore, the finding that Set3C is required for efficient activation of GAL genes, in contrast to the HDAC Rpd3, was surprising. It has been proposed that gene upregulation mediated by Set3C could be due to the effect of Set3C in repressing the expression of ncRNAs (40). Indeed, we also found that FACT and Set3C function in the same pathway to inhibit overproduction of GAL10 ncRNA transcripts, which are transcribed in the direction opposite to that of GAL10 transcription under inducing conditions. Finally, we show that inhibition of GAL10 ncRNA via mutation of the binding sites of the transcription factor Reb1, which is essential for GAL10 ncRNA transcription, reverses the effects of the spt16-G132D and set3Δ mutations on GAL1 transcription. These results indicate that both FACT and Set3C are required to repress GAL10 ncRNA upon galactose addition and that this repression is at least partially responsible for the role of Set3C in GAL1 gene activation, providing a mechanism for how an HDAC functions in gene activation.
FACT is best known for its role in gene activation. How, then, is FACT involved in repression of GAL10 ncRNA? It has been reported that noncoding transcription increases dramatically when FACT is depleted (23,74). Interestingly, we observed that noncoding transcription at the 3′ end of GAL10 was dramatically increased in spt16-G132D cells growing at a nonpermissive temperature, which led to depletion of Spt16-G132D mutant proteins. The increase in noncoding transcription likely arose from a loss of nucleosomes at this locus. In Schizosaccharomyces pombe, FACT is important for heterochromatin silencing (75). Recently, it has been shown that FACT acts with Swi6 to prevent histone turnover, which in turn contributes to the maintenance and inheritance of heterochromatin (76). Therefore, it is possible that FACT and Set3C repress histone turnover and thereby repress GAL10 ncRNA transcription.
FACT and Set3C together repress unrestricted transient induction of GAL10 ncRNA
Under noninducing conditions (raffinose), GAL10 ncRNA prevents transcriptional leakage of GAL1 (45). We and others observed that GAL10 ncRNA is transiently induced after Raf is switched to Gal. We speculate that this transient induction of GAL10 ncRNA may serve as a checkpoint for detecting the strength of external galactose signaling. GAL10 ncRNA is constitutively expressed at low levels prior to galactose addition to repress the leaky expression of GAL1. If yeast cells are exposed to a weak or short-term galactose stimulus, the transient induction of GAL10 ncRNA prevents GAL1 gene activation. Only strong and durable galactose induction signals can overcome repression by GAL10 ncRNA, which is important for optimal activation of the GAL1 gene. We propose that the transient induction of ncRNA ensures that yeast cells respond to continuous and possibly strong galactose stimulation but not to brief switching of medium from raffinose to galactose. This mechanism may be important for cells to adapt to carbon source changes economically.
After GAL1 is fully activated, GAL10 ncRNA expression is repressed, likely by FACT- and Set3C-mediated mechanisms. We and others have observed that both FACT and Set3C are enriched in the GAL10 gene body under inducing conditions, and this increased association of FACT and Set3C is likely due to increased GAL10 transcription. Once recruited, FACT and Set3C can suppress noncoding transcription, such as GAL10 ncRNA transcription. In this model, GAL10 ncRNA suppression arises after activation of the GAL1/GAL10 locus, which in turn prevents the interference of GAL10 ncRNA transcription with GAL1 and GAL10 transcription. In opposition to this idea, we observed that mutation of the GAL10-TATA box, while dramatically reducing GAL10 expression, had no apparent effects on GAL10 ncRNA production under noninducing and inducing conditions (Supplementary Figure S5G and S5H). In addition, it has been shown that H3K4me2 can recruit Set3C to 5′ transcribed units, which in turn repress noncoding transcription. Therefore, it is possible that FACT and Set3 are recruited to the 5′ end of GAL10 ncRNA by H3K4me2 and thereby suppress GAL10 ncRNA transcription. Future studies are needed to test these ideas.
Roles of FACT, Set3C and GAL10 ncRNA in nucleosomal changes at the GAL1 promoter regions
It has been proposed that both prior to and following induction, specific DNA-binding proteins, including the activator Gal4 and the chromatin-remodeling complex (RSC), are the predominant determinants of chromatin architecture at the regulatory locus of GAL1/10 genes (77). Consistent with this idea, we found that nucleosome occupancy and H3K27Ac levels are altered similarly in cells containing mutations in the GAL1-TATA box compared to WT cells under inducing conditions, whereas GAL1 induction is reduced dramatically in this mutant compared to WT cells under the same conditions. In contrast, nucleosome occupancy and H3K27Ac levels at the gene body of GAL1 are compromised in GAL1-TATA box mutant cells compared to WT cells, which is reflective of the reduction in GAL1 transcription. Moreover, we found that the nucleosome occupancy at the GAL1 promoter region was higher, whereas the histone acetylation levels were lower, in set3Δ and spt16-G132D mutant cells than in WT cells during galactose induction. Reb1-BSΔ partially rescued the nucleosome occupancy and histone acetylation levels of set3Δ mutant cells. These results suggest that GAL10 ncRNA transcription contributes to the changes in chromatin architecture at the GAL1 promoter observed in set3Δand spt16-G132D mutant cells. It has been reported that GAL10 ncRNA transcription can recruit the HDAC Rpd3 to alter the chromatin structure at the GAL1/GAL10 locus in cis to repress GAL1/10 transcription under conditions of concomitant weak induction and repression (44). Moreover, ncRNAs can also promote nucleosome assembly at downstream gene promoter regions to repress gene transcription (68). Therefore, it is possible that GAL10 ncRNA functions with FACT and Set3C to alter chromatin architecture at the GAL1/GAL10 regulatory locus, consequently fine-tuning the induction kinetics of GAL genes. Recently, an RNA-mediated feedback control of condensate formation model during transcriptional regulation was proposed (78). It would be interesting to test if transcriptional regulation at the GAL1 gene cluster region would incorporate a GAL10 ncRNA feedback mechanism.
In summary, we propose that FACT and Set3C cooperatively repress the expression of GAL10 ncRNA, facilitating the proper activation of GAL1. In the absence of Set3 or in spt16-G132D mutants defective in Set3 binding, GAL10 ncRNA levels are dramatically increased, which contributes to increases in nucleosome numbers and reductions in histone acetylation at the GAL1/10 regulatory regions, thereby impairing GAL gene activation (Supplementary Figure S8). Overall, our study reveals a synergetic mechanism of repression and activation of transcription at one gene cluster that involves cooperation between the histone chaperone FACT and the Set3 HDAC complex in response to carbon source changes. We suggest that this mechanism is important for rapid and economical adaptation of cells to new conditions.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Maria Vogelauer for her advice on the construction of GAL10 ncRNA deletion strains. We thank Dr. Zhiguo Zhang and Dr. Tingting Li for their critical readings and helpful discussions on this manuscript. We thank the Flow Cytometry Core at the National Center for Protein Sciences, Peking University, particularly Hongxia Lv and Liying Du, for their technical supports.
Author contributions: H.L. and S.L. conducted most of the experiments. Y.L. constructed some of the yeast strains used in this study and performed several protein purification and ChIP assays. Y.T. performed several protein purifications. S.C. performed the MS analysis. S.G. and J.Z. helped with the model drawings, Q.L., and J.F. supervised the project. H.L., S.L., J.F. and Q.L. wrote the manuscript.
Contributor Information
He Leng, State Key Laboratory of Protein and Plant Gene Research, School of Life Sciences and Peking-Tsinghua Center for Life Sciences, Peking University, Beijing 100871, China.
Shaofeng Liu, State Key Laboratory of Protein and Plant Gene Research, School of Life Sciences and Peking-Tsinghua Center for Life Sciences, Peking University, Beijing 100871, China.
Yang Lei, State Key Laboratory of Protein and Plant Gene Research, School of Life Sciences and Peking-Tsinghua Center for Life Sciences, Peking University, Beijing 100871, China.
Yuantao Tang, State Key Laboratory of Protein and Plant Gene Research, School of Life Sciences and Peking-Tsinghua Center for Life Sciences, Peking University, Beijing 100871, China.
Shijia Gu, State Key Laboratory of Protein and Plant Gene Research, School of Life Sciences and Peking-Tsinghua Center for Life Sciences, Peking University, Beijing 100871, China.
Jiazhi Hu, The MOE Key Laboratory of Cell Proliferation and Differentiation, Genome Editing Research Center, School of Life Sciences and Peking-Tsinghua Center for Life Sciences, Peking University, Beijing 100871, China.
She Chen, National Institute of Biological Sciences, Beijing 102206, China.
Jianxun Feng, State Key Laboratory of Protein and Plant Gene Research, School of Life Sciences and Peking-Tsinghua Center for Life Sciences, Peking University, Beijing 100871, China.
Qing Li, State Key Laboratory of Protein and Plant Gene Research, School of Life Sciences and Peking-Tsinghua Center for Life Sciences, Peking University, Beijing 100871, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [NSFC 31725015, 31830048, 31861143041 to Q.L.; 31671332 to J.F.]; Beijing Outstanding Young Scientist Program [BJJWZYJH01201910001005 to Q.L.]; National Key Research and Development Project of China [2019YFA0508903 to Q.L.]. Funding for open access charge: Beijing Outstanding Young Scientist Program [BJJWZYJH01201910001005 to Q.L.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Kouzarides T. Chromatin modifications and their function. Cell. 2007; 128:693–705. [DOI] [PubMed] [Google Scholar]
- 2. Lee J.S., Shukla A., Schneider J., Swanson S.K., Washburn M.P., Florens L., Bhaumik S.R., Shilatifard A.. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell. 2007; 131:1084–1096. [DOI] [PubMed] [Google Scholar]
- 3. Grant P.A., Duggan L., Cote J., Roberts S.M., Brownell J.E., Candau R., Ohba R., Owen-Hughes T., Allis C.D., Winston F.et al.. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997; 11:1640–1650. [DOI] [PubMed] [Google Scholar]
- 4. Ikeda K., Steger D.J., Eberharter A., Workman J.L.. Activation domain-specific and general transcription stimulation by native histone acetyltransferase complexes. Mol. Cell. Biol. 1999; 19:855–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang L., Liu L., Berger S.L.. Critical residues for histone acetylation by Gcn5, functioning in Ada and SAGA complexes, are also required for transcriptional function in vivo. Genes Dev. 1998; 12:640–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hassan A.H., Neely K.E., Workman J.L.. Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell. 2001; 104:817–827. [DOI] [PubMed] [Google Scholar]
- 7. Kadosh D., Struhl K.. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell. 1997; 89:365–371. [DOI] [PubMed] [Google Scholar]
- 8. Wang A., Kurdistani S.K., Grunstein M.. Requirement of Hos2 histone deacetylase for gene activity in yeast. Science. 2002; 298:1412–1414. [DOI] [PubMed] [Google Scholar]
- 9. Wu J., Suka N., Carlson M., Grunstein M.. TUP1 utilizes histone H3/H2B-specific HDA1 deacetylase to repress gene activity in yeast. Mol. Cell. 2001; 7:117–126. [DOI] [PubMed] [Google Scholar]
- 10. Vogelauer M., Wu J., Suka N., Grunstein M.. Global histone acetylation and deacetylation in yeast. Nature. 2000; 408:495–498. [DOI] [PubMed] [Google Scholar]
- 11. Orphanides G., LeRoy G., Chang C.H., Luse D.S., Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998; 92:105–116. [DOI] [PubMed] [Google Scholar]
- 12. Bortvin A., Winston F.. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science. 1996; 272:1473–1476. [DOI] [PubMed] [Google Scholar]
- 13. Schwabish M.A., Struhl K.. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol. Cell. 2006; 22:415–422. [DOI] [PubMed] [Google Scholar]
- 14. Formosa T. The role of FACT in making and breaking nucleosomes. Biochim. Biophys. Acta. 2012; 1819:247–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Winkler D.D., Luger K.. The histone chaperone FACT: structural insights and mechanisms for nucleosome reorganization. J. Biol. Chem. 2011; 286:18369–18374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Formosa T., Winston F.. The role of FACT in managing chromatin: disruption, assembly, or repair. Nucleic Acids Res. 2020; 48:11929–11941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rhoades A.R., Ruone S., Formosa T.. Structural features of nucleosomes reorganized by yeast FACT and its HMG box component, Nhp6. Mol. Cell. Biol. 2004; 24:3907–3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ruone S., Rhoades A.R., Formosa T.. Multiple Nhp6 molecules are required to recruit Spt16-Pob3 to form yFACT complexes and to reorganize nucleosomes. J. Biol. Chem. 2003; 278:45288–45295. [DOI] [PubMed] [Google Scholar]
- 19. Xin H., Takahata S., Blanksma M., McCullough L., Stillman D.J., Formosa T.. yFACT induces global accessibility of nucleosomal DNA without H2A-H2B displacement. Mol. Cell. 2009; 35:365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen P., Dong L., Hu M., Wang Y.Z., Xiao X., Zhao Z., Yan J., Wang P.Y., Reinberg D., Li M.et al.. Functions of FACT in breaking the nucleosome and maintaining its integrity at the single-nucleosome level. Mol. Cell. 2018; 71:284–293. [DOI] [PubMed] [Google Scholar]
- 21. Malone E.A., Clark C.D., Chiang A., Winston F.. Mutations in SPT16/CDC68 suppress cis- and trans-acting mutations that affect promoter function in Saccharomyces cerevisiae. Mol. Cell. Biol. 1991; 11:5710–5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Saunders A., Werner J., Andrulis E.D., Nakayama T., Hirose S., Reinberg D., Lis J.T.. Tracking FACT and the RNA polymerase II elongation complex through chromatin in vivo. Science. 2003; 301:1094–1096. [DOI] [PubMed] [Google Scholar]
- 23. Feng J., Gan H., Eaton M.L., Zhou H., Li S., Belsky J.A., MacAlpine D.M., Zhang Z., Li Q.. Noncoding transcription is a driving force for nucleosome instability in spt16 mutant cells. Mol. Cell. Biol. 2016; 36:1856–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mason P.B., Struhl K.. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol. Cell. Biol. 2003; 23:8323–8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kaplan C.D., Laprade L., Winston F.. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003; 301:1096–1099. [DOI] [PubMed] [Google Scholar]
- 26. Cheung V., Chua G., Batada N.N., Landry C.R., Michnick S.W., Hughes T.R., Winston F.. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol. 2008; 6:e277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wittmeyer J., Formosa T.. The Saccharomyces cerevisiae DNA polymerase alpha catalytic subunit interacts with Cdc68/Spt16 and with Pob3, a protein similar to an HMG1-like protein. Mol. Cell. Biol. 1997; 17:4178–4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tan B.C., Chien C.T., Hirose S., Lee S.C.. Functional cooperation between FACT and MCM helicase facilitates initiation of chromatin DNA replication. EMBO J. 2006; 25:3975–3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang J., Zhang X., Feng J., Leng H., Li S., Xiao J., Liu S., Xu Z., Xu J., Li D.et al.. The histone chaperone FACT contributes to DNA replication-coupled nucleosome assembly. Cell Rep. 2016; 14:1128–1141. [DOI] [PubMed] [Google Scholar]
- 30. Kurat C.F., Yeeles J.T.P., Patel H., Early A., Diffley J.F.X. Chromatin controls DNA replication origin selection, lagging-strand synthesis, and replication fork rates. Mol. Cell. 2017; 65:117–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu Y., Zhou K., Zhang N., Wei H., Tan Y.Z., Zhang Z., Carragher B., Potter C.S., D’Arcy S., Luger K.. FACT caught in the act of manipulating the nucleosome. Nature. 2020; 577:426–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kemble D.J., McCullough L.L., Whitby F.G., Formosa T., Hill C.P.. FACT disrupts nucleosome structure by binding H2A-H2B with conserved peptide motifs. Mol. Cell. 2015; 60:294–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Evans D.R., Brewster N.K., Xu Q., Rowley A., Altheim B.A., Johnston G.C., Singer R.A.. The yeast protein complex containing cdc68 and pob3 mediates core-promoter repression through the cdc68 N-terminal domain. Genetics. 1998; 150:1393–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. John S., Howe L., Tafrov S.T., Grant P.A., Sternglanz R., Workman J.L.. The something about silencing protein, Sas3, is the catalytic subunit of NuA3, a yTAF(II)30-containing HAT complex that interacts with the Spt16 subunit of the yeast CP (Cdc68/Pob3)-FACT complex. Genes Dev. 2000; 14:1196–1208. [PMC free article] [PubMed] [Google Scholar]
- 35. VanDemark A.P., Xin H., McCullough L., Rawlins R., Bentley S., Heroux A., Stillman D.J., Hill C.P., Formosa T.. Structural and functional analysis of the Spt16p N-terminal domain reveals overlapping roles of yFACT subunits. J. Biol. Chem. 2008; 283:5058–5068. [DOI] [PubMed] [Google Scholar]
- 36. O’Donnell A.F., Brewster N.K., Kurniawan J., Minard L.V., Johnston G.C., Singer R.A.. Domain organization of the yeast histone chaperone FACT: the conserved N-terminal domain of FACT subunit Spt16 mediates recovery from replication stress. Nucleic Acids Res. 2004; 32:5894–5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pijnappel W.W., Schaft D., Roguev A., Shevchenko A., Tekotte H., Wilm M., Rigaut G., Seraphin B., Aasland R., Stewart A.F.. The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev. 2001; 15:2991–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Harvey Z.H., Chakravarty A.K., Futia R.A., Jarosz D.F.. A prion epigenetic switch establishes an active chromatin state. Cell. 2020; 180:928–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Suka N., Suka Y., Carmen A.A., Wu J., Grunstein M.. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol. Cell. 2001; 8:473–479. [DOI] [PubMed] [Google Scholar]
- 40. Kim T., Xu Z., Clauder-Munster S., Steinmetz L.M., Buratowski S.. Set3 HDAC mediates effects of overlapping noncoding transcription on gene induction kinetics. Cell. 2012; 150:1158–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kim T., Buratowski S.. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell. 2009; 137:259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hang M., Smith M.M.. Genetic analysis implicates the Set3/Hos2 histone deacetylase in the deposition and remodeling of nucleosomes containing H2A.Z. Genetics. 2011; 187:1053–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cloutier S.C., Wang S., Ma W.K., Petell C.J., Tran E.J.. Long noncoding RNAs promote transcriptional poising of inducible genes. PLoS Biol. 2013; 11:e1001715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Houseley J., Rubbi L., Grunstein M., Tollervey D., Vogelauer M.. A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol. Cell. 2008; 32:685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lenstra T.L., Coulon A., Chow C.C., Larson D.R.. Single-molecule imaging reveals a switch between spurious and functional ncRNA transcription. Mol. Cell. 2015; 60:597–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Geisler S., Lojek L., Khalil A.M., Baker K.E., Coller J.. Decapping of long noncoding RNAs regulates inducible genes. Mol. Cell. 2012; 45:279–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Meth. Enzymol. 1991; 194:281–301. [DOI] [PubMed] [Google Scholar]
- 48. Lafontaine D., Tollervey D. One-step PCR mediated strategy for the construction of conditionally expressed and epitope tagged yeast proteins. Nucleic Acids Res. 1996; 24:3469–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li Q., Zhou H., Wurtele H., Davies B., Horazdovsky B., Verreault A., Zhang Z.. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell. 2008; 134:244–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Huang S., Zhou H., Tarara J., Zhang Z.. A novel role for histone chaperones CAF-1 and Rtt106p in heterochromatin silencing. EMBO J. 2007; 26:2274–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Krogan N.J., Kim M., Ahn S.H., Zhong G., Kobor M.S., Cagney G., Emili A., Shilatifard A., Buratowski S., Greenblatt J.F.. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol. Cell. Biol. 2002; 22:6979–6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gradolatto A., Rogers R.S., Lavender H., Taverna S.D., Allis C.D., Aitchison J.D., Tackett A.J.. Saccharomyces cerevisiae Yta7 regulates histone gene expression. Genetics. 2008; 179:291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Deyter G.M., Biggins S.. The FACT complex interacts with the E3 ubiquitin ligase Psh1 to prevent ectopic localization of CENP-A. Genes Dev. 2014; 28:1815–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Foltman M., Evrin C., De Piccoli G., Jones Richard C., Edmondson Rick D., Katou Y., Nakato R., Shirahige K., Labib K.. Eukaryotic replisome components cooperate to process histones during chromosome replication. Cell Rep. 2013; 3:892–904. [DOI] [PubMed] [Google Scholar]
- 55. Sengupta S., van Deursen F., de Piccoli G., Labib K.. Dpb2 integrates the leading-strand DNA polymerase into the eukaryotic replisome. Curr. Biol. 2013; 23:543–552. [DOI] [PubMed] [Google Scholar]
- 56. Formosa T., Eriksson P., Wittmeyer J., Ginn J., Yu Y., Stillman D.J.. Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J. 2001; 20:3506–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Schlesinger M.B., Formosa T.. POB3 is required for both transcription and replication in the yeast Saccharomyces cerevisiae. Genetics. 2000; 155:1593–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Orphanides G., Wu W.H., Lane W.S., Hampsey M., Reinberg D. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature. 1999; 400:284–288. [DOI] [PubMed] [Google Scholar]
- 59. Lohr D., Venkov P., Zlatanova J.. Transcriptional regulation in the yeast GAL gene family: a complex genetic network. FASEB J. 1995; 9:777–787. [DOI] [PubMed] [Google Scholar]
- 60. Biswas D., Dutta-Biswas R., Mitra D., Shibata Y., Strahl B.D., Formosa T., Stillman D.J.. Opposing roles for Set2 and yFACT in regulating TBP binding at promoters. EMBO J. 2006; 25:4479–4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Govind C.K., Qiu H., Ginsburg D.S., Ruan C., Hofmeyer K., Hu C., Swaminathan V., Workman J.L., Li B., Hinnebusch A.G.. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol. Cell. 2010; 39:234–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fleming A.B., Kao C.F., Hillyer C., Pikaart M., Osley M.A.. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol. Cell. 2008; 31:57–66. [DOI] [PubMed] [Google Scholar]
- 63. Bryant G.O., Prabhu V., Floer M., Wang X., Spagna D., Schreiber D., Ptashne M.. Activator control of nucleosome occupancy in activation and repression of transcription. PLoS Biol. 2008; 6:2928–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Almer A., Rudolph H., Hinnen A., Horz W.. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 1986; 5:2689–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tirosh I., Barkai N.. Two strategies for gene regulation by promoter nucleosomes. Genome Res. 2008; 18:1084–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yuan G.C., Liu Y.J., Dion M.F., Slack M.D., Wu L.F., Altschuler S.J., Rando O.J.. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005; 309:626–630. [DOI] [PubMed] [Google Scholar]
- 67. Kellis M., Patterson N., Endrizzi M., Birren B., Lander E.S.. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003; 423:241–254. [DOI] [PubMed] [Google Scholar]
- 68. Hainer S.J., Pruneski J.A., Mitchell R.D., Monteverde R.M., Martens J.A.. Intergenic transcription causes repression by directing nucleosome assembly. Genes Dev. 2011; 25:29–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Infante J.J., Law G.L., Young E.T.. Analysis of nucleosome positioning using a nucleosome-scanning assay. Methods Mol. Biol. 2012; 833:63–87. [DOI] [PubMed] [Google Scholar]
- 70. Stuwe T., Hothorn M., Lejeune E., Rybin V., Bortfeld M., Scheffzek K., Ladurner A.G.. The FACT Spt16 “peptidase” domain is a histone H3-H4 binding module. Proc. Natl. Acad. Sci. USA. 2008; 105:8884–8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kelley D.E., Stokes D.G., Perry R.P.. CHD1 interacts with SSRP1 and depends on both its chromodomain and its ATPase/helicase-like domain for proper association with chromatin. Chromosoma. 1999; 108:10–25. [DOI] [PubMed] [Google Scholar]
- 72. Biswas D., Dutta-Biswas R., Stillman D.J.. Chd1 and yFACT act in opposition in regulating transcription. Mol. Cell. Biol. 2007; 27:6279–6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Collins S.R., Miller K.M., Maas N.L., Roguev A., Fillingham J., Chu C.S., Schuldiner M., Gebbia M., Recht J., Shales M.et al.. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007; 446:806–810. [DOI] [PubMed] [Google Scholar]
- 74. Murawska M., Schauer T., Matsuda A., Wilson M.D., Pysik T., Wojcik F., Muir T.W., Hiraoka Y., Straub T., Ladurner A.G.. The chaperone FACT and histone H2B ubiquitination maintain S. pombe genome architecture through genic and subtelomeric functions. Mol. Cell. 2020; 77:501–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lejeune E., Bortfeld M., White S.A., Pidoux A.L., Ekwall K., Allshire R.C., Ladurner A.G.. The chromatin-remodeling factor FACT contributes to centromeric heterochromatin independently of RNAi. Curr. Biol. 2007; 17:1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Holla S., Dhakshnamoorthy J., Folco H.D., Balachandran V., Xiao H., Sun L.L., Wheeler D., Zofall M., Grewal S.I.S. Positioning heterochromatin at the nuclear periphery suppresses histone turnover to promote epigenetic inheritance. Cell. 2020; 180:150–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Floer M., Wang X., Prabhu V., Berrozpe G., Narayan S., Spagna D., Alvarez D., Kendall J., Krasnitz A., Stepansky A.et al.. A RSC/nucleosome complex determines chromatin architecture and facilitates activator binding. Cell. 2010; 141:407–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Henninger J.E., Oksuz O., Shrinivas K., Sagi I., LeRoy G., Zheng M.M., Andrews J.O., Zamudio A.V., Lazaris C., Hannett N.M.et al.. RNA-mediated feedback control of transcriptional condensates. Cell. 2021; 184:207–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.