Abstract
Telomere repeat-containing RNA (TERRA) has been identified in multiple organisms including Trypanosoma brucei, a protozoan parasite that causes human African trypanosomiasis. T. brucei regularly switches its major surface antigen, VSG, to evade the host immune response. VSG is expressed exclusively from subtelomeric expression sites, and we have shown that telomere proteins play important roles in the regulation of VSG silencing and switching. In this study, we identify several unique features of TERRA and telomere biology in T. brucei. First, the number of TERRA foci is cell cycle-regulated and influenced by TbTRF, the duplex telomere DNA binding factor in T. brucei. Second, TERRA is transcribed by RNA polymerase I mainly from a single telomere downstream of the active VSG. Third, TbTRF binds TERRA through its C-terminal Myb domain, which also has the duplex DNA binding activity, in a sequence-specific manner and suppresses the TERRA level without affecting its half-life. Finally, levels of the telomeric R-loop and telomere DNA damage were increased upon TbTRF depletion. Overexpression of an ectopic allele of RNase H1 that resolves the R-loop structure in TbTRF RNAi cells can partially suppress these phenotypes, revealing an underlying mechanism of how TbTRF helps maintain telomere integrity.
INTRODUCTION
Telomeres are dynamic nucleoprotein complexes at chromosome ends that help maintain genome integrity and chromosome stability (1). Earlier studies showed that telomeres are usually assembled into a heterochromatic structure and are typically associated with the silent chromatin markers (2). Yet, recent studies have identified telomeric repeat-containing long, non-coding RNA (lncRNA), TERRA, in many organisms (3). In addition, TERRA has been shown to be the product of telomeric repeat transcription in human cells (4), mouse ES cells (5), Saccharomyces cerevisiae (6), Schizosaccharomyces pombe (7), Arabidopsis thaliana (8), and Trypanosoma brucei (9,10). TERRA can be transcribed from multiple telomeres (4,11–16), but frequently not all telomeres are transcribed (9,17,18), and transcription from intrachromosomal telomeric sequences can be abundant (8). TERRA has been shown to play important roles in telomere protection (19–21), regulation of telomere length (22–24), and telomere recombination (25) in mammalian and yeast cells. TERRA may also play a role in gene expression regulation in mouse ES cells (21).
RNA-FISH analyses have shown that TERRA is associated with many telomeres in human and mouse cells (12,16,26). FISH analyses also revealed 1–3 TERRA foci in the S. pombe nucleus (7). In budding yeast, the MS2-tagged TERRA is seen to colocalize with its telomere of origin during the mid-late S phase (27). In contrast, most TERRA binding sites are not at the telomere but in distal intergenic and intronic regions in mouse cells (21). Whether the number of nuclear TERRA foci changes throughout the cell cycle has not been reported. On the other hand, the TERRA level has been reported to change in a cell cycle-dependent manner. In yeast cells, it dips in the S phase but peaks in the G2/M phase (28). In Hela cells, the TERRA level is high in the G1/S and decreases at the late S/G2 phase (29,30).
TERRA was first identified in T. brucei (31), a small fraction of which contains the poly(A) tail (10,31). In addition, the transcribed TERRA species and its 5′ phosphorylation status are dependent on T. brucei life cycle stages (3,10). However, TERRA subnuclear localization, its function and regulation in T. brucei are still not well known. T. brucei is the causative agent of human African trypanosomiasis, which is frequently fatal without treatment. While proliferating in extracellular spaces of its mammalian host, T. brucei sequentially expresses immunologically distinct VSGs, its major surface antigen proteins (32). This antigenic variation allows T. brucei to effectively evade the host immune response. At this life cycle stage, VSGs are exclusively transcribed by RNA Polymerase I (RNAP I) (33) from subtelomeric bloodstream form (BF) VSG expression sites (ESs), which are polycistronic transcription units (PTUs), with the VSG gene within 2 kb from the telomeric repeats and the promoter 40–60 kb upstream (Supplementary Figure S1A, top) (34,35). T. brucei has a large VSG gene pool (36), but only one VSG is fully expressed at any moment (37). Individual VSGs are also found in metacyclic ESs, which are subtelomeric monocistronic transcription units (with its promotor ∼ 5 kb upstream of the telomere) and can be expressed when the parasite resides in the salivary gland of its insect vector (Supplementary Figure S1A, bottom). VSG switching is frequently mediated by DNA recombination and sometimes through a transcriptional switch (38). DNA double-strand breaks (DSBs) have been shown to be a potent trigger for VSG switching (39,40). We have also shown that depletion of telomere proteins, such as TbTRF, the duplex telomere DNA binding factor (41), and its interacting factors TbRAP1 (42) and TbTIF2 (43), leads to telomere/subtelomere instability and higher VSG switching frequencies (9,43–45).
The UG-rich TERRA is transcribed in T. brucei when it is proliferating in a mammalian host (31). Interestingly, the TERRA level is not sensitive to α-amanitin (31), suggesting that TERRA is transcribed by RNAP I. We and others recently demonstrated that the telomere downstream of the active ES is indeed transcribed into TERRA (9,10), further suggesting that TERRA is transcribed by RNAP I as a read-through product in T. brucei. An excessive amount of TERRA can lead to telomere and subtelomere instability (46), as TERRA has a propensity to form R-loops with the telomeric repeat-containing dsDNA (20,47), and R-loops are known to induce DSBs and cause genome instability (19,48). Indeed, we have shown that depletion of TbRAP1 results in a cell growth arrest, higher TERRA and telomeric R-loop levels, more subtelomeric and telomeric DSBs, and an elevated VSG switching frequency (9). Therefore, it is important to regulate TERRA at a proper level. However, except for TbRAP1, no other factors have been known to regulate the TERRA level in T. brucei.
In this study, we show that TERRA has a very short half-life. Treating cells with the RNAP I inhibitor BMH-21 depletes >90% of TERRA, confirming that the majority of TERRA is transcribed by RNAP I in T. brucei. Using RNA FISH, we find that most T. brucei cells have only 1 - 3 TERRA foci. Surprisingly, the number of TERRA foci increases as cells progress through the cell cycle, which has not been reported in other TERRA-expressing organisms. In addition, dramatically bigger and fewer TERRA foci are frequently observed in cells depleted of TbTRF. We further demonstrate that TbTRF suppresses the TERRA and telomeric R-loop levels, and that loss of TbTRF causes more DSBs in the telomere repeats, revealing a mechanism of how TbTRF maintains the telomere integrity. TbTRF also exhibits a TERRA-binding activity that resides in its C-terminal Myb domain, which was previously found to be responsible for binding the duplex telomeric DNA (41,44). In addition, TbTRF binds the duplex telomeric DNA with a higher affinity than TERRA, and the two nucleic acid interaction interfaces may overlap.
MATERIALS AND METHODS
T. brucei strains
All T. brucei strains used in this study were derived from BF VSG2-expressing Lister 427 cells that express the T7 polymerase and the Tet repressor (Single Marker, aka SM) (49). SM/GFP-TbRAP1 and SM/GFP-TbTRF have one of the endogenous TbRAP1 or TbTRF alleles N-terminally tagged with GFP. SM/GFP-TbKU80 was established by transfecting the pLew82-GFP-TbKU80 plasmid (50) into SM cells. SM/TbTERT-GFP was established by transfecting the pCO57-TbTERT plasmid (51) into SM cells. The TbTRF RNAi strain was established by transfecting SM cells with the pZJMβ-TbTRF-Mid1 plasmid (41). The TbTRF+/– RNAi strain was described in (41), which was established by transfecting TbTRF+/– (TbTRF single-allele knockout) cells with the pZJMβ-TbTRF-Mid1 plasmid. The parent switching strain (S) was derived from SM for VSG switching analysis (52). The S/TRFi and S/TRFi+F2H-TbTRF strains were described in (44). All BF T. brucei cells were cultured in the HMI-9 medium supplemented with 10% FBS and appropriate antibiotics. pLew100-v5-2HA-RNase H1 was targeted to an rDNA spacer region in the TbTRF+/– RNAi cells to generate the TbTRF+/– RNAi + 2HA-RNase H1 strain.
TERRA northern hybridization
Total RNA was purified from 80 to 100 million cells using RNA STAT-60 (Tel. Test Inc.) twice and treated with 10 units of DNase I (ThermoFisher), followed by another round of purification with RNA STAT-60. The resulting RNA sample was treated with or without 20 units of RNase One (Promega) and 20 μg of RNase A (Sigma). For northern blotting, 10 μg of RNA was loaded in each lane. After electrophoresis, RNA was transferred to a Nylon membrane and hybridized with a radiolabeled (CCCTAA)4 oligo probe. The hybridization intensity (whole lane) was quantified for each sample using ImageQuant (GE) and the relative amount of TERRA level was calculated. The hybridization signals (TERRA species bigger than the largest rRNA precursor) were counted to estimate the average size of TERRA in each sample according to (53).
TERRA slot blot hybridizations
RNA isolation and DNase I digestion were performed the same way as described above. RNA samples were denatured at 65°C for 10 min in the presence of formamide and formaldehyde. 2 μg of RNA was spotted on the Nylon membrane. To prepare the (CCCTAA)n- [or (TTAGGG)n-] specific probe, the Klenow primer extension reaction was performed using a duplex TTAGGG repeats as the template in the presence of dA, dT and radioactive dC (or radioactive dG). Alternatively, TELC4 [(CCCTAA)4] and TELG4 [(TTAGGG)4] oligos were end-labeled by T4 Polynucleotide Kinase and used as probes.
RT-PCR analysis to determine the origin of TERRA
RNA isolation and DNase I digestion were performed the same way as described above. Reverse transcription was performed using TELC20, TELG20, or a random hexamer as the primer (9) and the SuperScript IV Reverse Transcriptase (ThermoFisher) according to the manufacturer's protocol. The RT products were PCR amplified using Taq polymerase (ThermoFisher) with primers specific to VSG2, VSG3, VSG9, tubulin and rDNA genes, 70 bp repeat, and a sequence at Chromosome 11 subtelomere (Chr 11sub).
TERRA FISH and TbTRF IF
In TERRA FISH, cells were fixed for 10 min at room temperature with 2% formaldehyde in 1 mM KCl/16 mM NaCl/0.2 mM MgSO4/0.4 mM Na2HPO4. Five million cells were spotted on each silanized coverslip. Cells were dehydrated by 70%, 85%, 95% and 100% ethanol (3 min each) at room temperature before hybridization with the TelC-Alexa488 or TelG-Cy3 probe (PNA Bio) in the hybridization buffer (62.2% formamide, 62.5 mg/ml Dextran Sulfate-500K, 2.5× SSC, 3.75 mg/ml BSA, 3.75 mg/ml Ficoll, 3.75 mg/ml Polyvinylpyrollidone) overnight at room temperature. As a control, some cells were treated with 200 μg/ml RNase A in 2× SSC for 1 hr at 37°C before hybridization. After hybridization, cells were washed with 2× SSC/50% formamide three times at 39°C for 5 min each, then with 2× SSC three times at 39°C for 5 min each, followed by washes with 2× SSC and 4× SSC for 5 min each at room temperature. DNA was stained with 0.5 μg/ml DAPI in 4× SSC followed by rinsing with 4× SSC for 5 min. Images were taken by a DeltaVision Elite deconvolution microscope (Applied precision/Olympus) and deconvolved using measured point spread functions. Images were processed in softWoRx.
For combined immunofluorescence (IF) and TERRA FISH, cells were fixed the same way as in TERRA FISH and permeabilized with 0.2% NP-40 for 5 min at room temperature. Cells were blocked with PBG (0.2% cold fish water gelatin/0.5% BSA/1× PBS) before they were incubated with a TbTRF rabbit antibody (41) followed by incubation with an Alexa-594 conjugated donkey anti-rabbit antibody (Jackson Immunology). Subsequently, TERRA FISH was performed the same way as described above.
RNA Immunoprecipitation (RNA IP)
50 μl of Dynabeads protein G (ThermoFisher) were washed and resuspended in 300 μl of antibody binding buffer (1× PBS/0.02% Tween 20). Antibodies were coupled to the beads at 4°C on a rotor. 200 million T. brucei cells resuspended in 1 ml NET-5 buffer (40 mM Tris–HCl, pH 7.5, 420 mM NaCl, 0.5% NP-40, 2 mg/ml aprotinin A, 1 mg/ml leupeptin, 1 mg/ml pepstatin A and 10 units of RNAsin) were lysed by freezing/thawing (at −80°C) three times followed by centrifugation at 13 krpm for 10 min at 4°C. Supernatant (lysate) was collected and equal volume of NET0.5 buffer (50 mM NaCl, 40 mM Tris–HCl, pH 7.5, 0.5% NP-40) was added. The diluted lysate was incubated with antibody and Dynabeads protein G at 4°C for 3 hrs. After washing three times with 1× PBS/0.02% Tween 20, IP samples were eluted (50 mM Tris–HCl, pH8.0, 10 mM EDTA, 1% SDS). RNA was extracted from all samples by phenol/chloroform followed by precipitation with ammonium acetate, ethanol, and glycogen. All samples were treated with DNase I before performing the RNA slot blot hybridization.
γH2A ChIP
200 million cells were cross-linked by 1% formaldehyde for 25 min at room temperature with constant mixing, and the cross-linking was stopped by 0.1 M glycine. Chromatin was sonicated by a BioRuptor (Diagenode) for four cycles (30-sec on/30-sec off) at the high output level. 10% of the lysate was saved as the input fraction, and the rest was equally divided into two fractions, each incubating with a γH2A antibody (9) or IgG conjugated with Dynabeads Protein G (ThermoFisher) for 3 hrs at 4°C. After extensive washing, immunoprecipitated products were eluted from the beads, and DNA was extracted by phenol chloroform and precipitated by ethanol followed by Southern slot blot hybridization with a telomere probe.
Electrophoretic mobility shift assay (EMSA)
The ds(TTAGGG)12 repeat DNA probe was prepared as described in (41).
Single-stranded (UUAGGG)12 and (CCCUAA)12 RNA probes were in vitro transcribed from HpaI/KpnI and NdeI/HindIII digested pTH12 plasmid (54), respectively, using the Maxiscript T7/SP6 kit (ThermoFisher) according to the manufacturer's protocol.
Recombinant proteins, GST-tagged TbTRF2-382, TbTRF-sMyb297-357, TbTRF-lMyb280-382, TbTRF-lMyb280-382R298E, TbTRF2-161, and TbTRF162-297, were partially purified from E. coli BL21-DE3 cells according to (41) and (55).
In EMSA, a radiolabeled DNA probe (1 ng) or RNA probe (2 ng) was incubated with recombinant GST-tagged TbTRF fragments in EMSA buffer (15 mM Tris–HCl, pH 7.5, 25 ng/μl E. coli DNA, 2.5 ng/μl beta-Casein, 4% Glycerol, and 4 U RNAseIn plus) for 30 min at room temperature. The mixture was separated on a 0.6% agarose gel in 0.1× TBE before the gel was dried and exposed to a phosphorimager.
Telomeric R-loop Assay
R-loop IP was performed as described in (56) with a few modifications. Genomic DNA was sonicated using a BioRuptor (Diagenode) at medium output for eight cycles with 30 sec pulse each. 10 μg of sonicated DNA was incubated with or without 50 U RNase H (ThermoFisher) for 3 hrs at 37°C followed by heat inactivation. Both RNase H-treated and untreated samples were divided equally for IP using IgG and S9.6 (Kerafast) antibodies. DNA samples were incubated with antibody-coupled Dynabeads protein G (ThermoFisher) for 2 hrs at 4°C. After extensive washing, IP products were eluted in elution buffer (1% SDS, 0.1 M NaHCO3) for 10 min at 65°C followed by Proteinase K treatment at 42°C for 1 hr in a Thermomixer (Eppendorf). The DNA was then extracted by phenol chloroform and precipitated by ethanol followed by Southern slot blot hybridization.
Estimation of the TERRA half-life
WT and TbTRF RNAi cells in log-phase growth were incubated with or without 100 ng/ml doxycycline for 24 hrs. Cells were then treated with 10 μg/ml Actinomycin D (Sigma) for various lengths of time. RNA was isolated from 50 million cells at each time point according to (9) followed by northern slot blot hybridization.
Examination of the TERRA level in BMH-21 treated cells
TbTRF RNAi cells in log-phase growth were incubated with or without 100 ng/ml doxycycline for 21 or 24 hrs. Both induced and uninduced cells were split equally to two fractions, which were treated with or without 3 μM of BMH-21 (Sigma) for 3 hrs (for cells incubated with doxycycline for 21 hrs) or 15 min (for cells incubated with doxycycline for 24 hrs). Subsequently, RNA was isolated from 50 million cells according to (9) followed by northern slot blot hybridization or RNA FISH analysis.
Quantitative RT-PCR to estimate the level of VSG2-TERRA-containing polycistronic transcript
TbTRF RNAi cells in log-phase growth were incubated with or without 100 ng/ml doxycycline for 24 and 30 hrs. RNA was isolated from 90 million cells according to (9). Reverse transcription was performed using TELC20 or a random hexamer as the primer and the SuperScript IV Reverse Transcriptase (ThermoFisher) according to the manufacturer's protocol. Quantitative PCR was performed using iTaq SYBR Green Supermix with ROX (Bio-Rad) according to the manufacturer's instructions. VSG2 specific primers were used for qPCR.
RESULTS
The number of nuclear TERRA foci is cell cycle-dependent in T. brucei
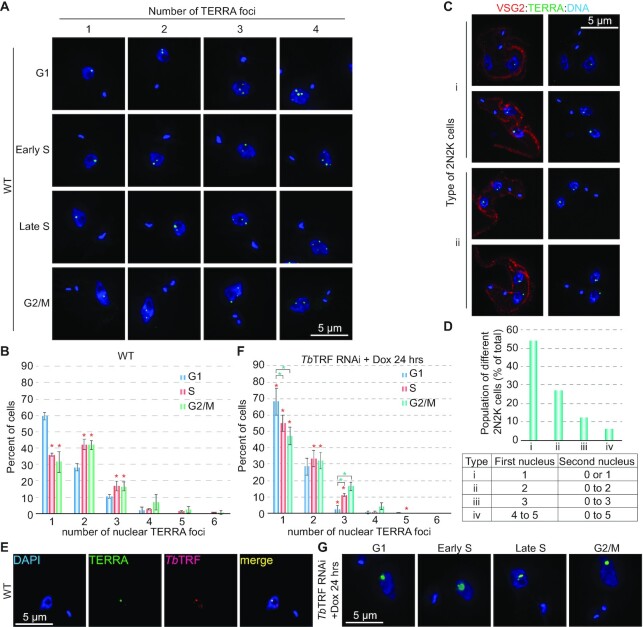
To better understand the functions of TERRA, we first determined the subnuclear localization of TERRA by RNA FISH using a (CCCTAA)n-containing TelC-Alexa488 PNA probe (PNA Bio). Only a few TERRA foci were observed in each T. brucei nucleus (Figure 1A), which were sensitive to RNase A treatment (Supplementary Figure S1B), indicating that the observed FISH signal was due to the telomeric RNA. As a control, RNA FISH using a (TTAGGG)n-containing TelG-Cy3 PNA probe (PNA Bio) was also performed, which did not show any signal (Supplementary Figure S1C), indicating that TERRA contains the G-rich telomere sequence.
Figure 1.
The number of nuclear TERRA foci increases as cells progress through the cell cycle. (A) Examples of WT cells with various numbers of TERRA foci at different cell cycle stages. TERRA was stained by a TelC-Alexa488 PNA probe (PNA Bio). DNA was stained by DAPI. All images are of the same scale, and a size bar is shown in one of the images in each panel. (B) Quantification of percent of WT cells with different numbers of nuclear TERRA foci at various cell cycle stages. A total of 310 G1, 403 S, and 253 G2/M nuclei were counted. Red asterisks mark significant differences when S or G2/M cells are compared to G1 cells. In this and other figures, error bars represent standard deviation. Unpaired t-tests were performed, and a significance threshold of p < 0.05 was set for the null hypothesis. (C) Examples of WT post-mitotic cells (2N2K) with 1–2 TERRA foci per nucleus. Examples of 2N2K cells with more nuclear TERRA foci can be found in Supplementary Figure S1D. VSG2 was stained to outline the cell surface and ensure the cells are at the post mitotic phase (each cell has 2N2K). Images of the same cells with (left) or without (right) the VSG2 staining are shown. (D) Quantification of percent of different types of WT post-mitotic cells. Different types of 2N2K cells are listed in the table at the bottom. A total of 240 cells were counted. (E) The brightest TERRA focus colocalizes with TbTRF. TbTRF is stained with a rabbit antibody (41). (F) Quantification of percent of TbTRF RNAi cells with different numbers of nuclear TERRA foci at various cell cycle stages after 24 hrs of RNAi induction. A total of 130–230 cells were counted at each cell cycle stage. Red asterisks mark significant differences between WT and TbTRF RNAi cells at each corresponding cell cycle stage. Green asterisks mark significant differences when S or G2/M cells are compared to G1 cells. (G) Examples of TERRA foci in TbTRF-depleted cells at various cell cycle stages.
We noticed that the number of TERRA foci varied in different cells. To determine the cell cycle stages in T. brucei, we followed the shape and number of kinetoplasts and the number of nuclei by DAPI staining (57,58). Cells in the G1 phase have one nucleus and one spherical kinetoplast (1N1K). Cells in the early and late S phase have one nucleus and one elongated or dumbbell-shaped kinetoplast (1N1eK) (58). Cells in the G2/M phase have one nucleus and 2 kinetoplasts (1N2K), and post-mitotic cells have two nuclei and two kinetoplasts (2N2K) (57). RNA FISH showed that of G1 cells, on average 60% had one bright nuclear TERRA focus, 28% had two, 11% had three and 2% had four TERRA foci (Figure 1B). Of cells in the S phase (both early and late), on average 36% had one nuclear TERRA focus, 42% had two, 18% had three and <5% had four or more foci (Figure 1B). G2/M cells had a very similar TERRA staining pattern as S cells (Figure 1B). Of these cells, on average 32% had one, 42% had two, 16% had three, and <10% had four or more foci (Figure 1B). However, compared to G1 cells, a significantly smaller portion of S and G2/M cells had one TERRA focus, but a significantly larger portion of S and G2/M cells had two or three nuclear TERRA foci (Figure 1B, red asterisks). Therefore, as T. brucei cells progress into the S phase and beyond, there are more TERRA nuclear foci per cell.
We subsequently examined how many TERRA foci per nucleus there are in post mitotic cells. To ensure that a cell is at the post-mitotic stage (2N2K per cell), we used a VSG2-specific rabbit antibody to mark the cell surface (VSG2 is expressed in these cells). In combined VSG2 immunofluorescence (IF) and TERRA FISH, we found that each of the divided nuclei in a post-mitotic cell had 1–5 TERRA foci, and most had only 1 or 2 foci (Figure 1C; Supplementary Figure S1D). For an easier reference, we defined the first nucleus as the one that always had equal or more foci than the second one and categorized four different types of post-mitotic cells (Figure 1D). In the first nucleus, >50% had a single TERRA focus (type i); ∼25% had two foci (type ii); ∼12% had three foci (type iii), and only ∼5% had four or more TERRA foci (type iv). Therefore, after mitosis, divided nuclei have roughly half the number of TERRA foci as those before mitosis and each nucleus has a similar TERRA staining pattern as the G1 phase cell (Figure 1B, D). Such cell cycle-dependent change in the number of nuclear TERRA foci has never been described in other TERRA-expressing organisms. On the other hand, the level of TERRA is cell cycle-regulated in budding yeast (28) and in human Hela cells (29, 30). However, since synchronizing T. brucei cells is not feasible without significant side effects (such as HU treatment), we currently cannot measure precisely whether the TERRA level changes throughout the cell cycle in T. brucei cells.
We noticed that in most cells, one (or occasionally two) TERRA foci were very bright, while the others were discernibly fainter (Figure 1A). Although T. brucei has nearly 250 telomeres, the telomeres are clustered together, and telomere FISH typically shows less than 10 foci per nucleus (41,59). In addition, we have observed that TbTRF, the duplex telomere DNA binding factor, is almost always colocalized with the telomere (41). Therefore, we stained TbTRF with a rabbit antibody (41) as a marker of the telomere. Combined TbTRF IF and TERRA FISH showed that the brightest TERRA focus (occasionally a couple of brightest TERRA foci) was colocalized with TbTRF. This colocalization was seen in cells at various cell cycle stages (Figure 1E; Supplementary Figure S1E), which is different from the scenario in budding yeast where TERRA is seen to colocalize with its telomere of origin in late S phase (27).
TbTRF depletion results in fewer TERRA foci
A recent study showed that TERRA can be recruited to telomeres in trans in human Hela cells, and depletion of TRF1 significantly increased TERRA co-localization at both short and long telomeres while depletion of TRF2 had a milder effect (60). To test whether TbTRF is involved in TERRA subnuclear localization, we performed TERRA FISH in TbTRF-depleted cells. To our surprise, we observed fewer TERRA foci when TbTRF was depleted (Figure 1F, G; Supplementary Figure S1F). Specifically, significantly larger fractions of TbTRF-depleted cells than WT cells (in G1, S and G2/M phases) had only one TERRA focus (Figure 1F, red asterisks), and significantly smaller fractions of TbTRF-depleted cells than WT cells (in S and G2/M phases) had two TERRA foci (Figure 1F, red asterisks). In addition, significantly smaller fractions of TbTRF-depleted cells than WT cells (in G1 and S phases) had three TERRA foci (Figure 1F, red asterisks). On the other hand, after depletion of TbTRF, there were still significantly more G1 cells than S or G2/M cells that had only one nuclear focus (Figure 1F, green asterisks), and there were significantly fewer G1 cells than S or G2/M cells that had three nuclear foci (Figure 1F, green asterisks), indicating that the cell-cycle regulated TERRA foci number pattern was still maintained in TbTRF-depleted cells. Therefore, depletion of TbTRF leads to more cells having a fewer number of TERRA foci, which is different from the scenario in Hela cells (60). Furthermore, most of the single TERRA foci in TbTRF-depleted cells appear much brighter and bigger than those in WT cells (Figure 1G; Supplementary Figure S1F). Some nuclei had a very large TERRA focus only slightly smaller than the nucleolus (Figure 1G; Supplementary Figure S1F), which was never found in WT cells (Figure 1A; Supplementary Figure S1E).
TbTRF suppresses the TERRA level but does not change the transcription origin of TERRA from the active telomere
We frequently observed very large TERRA foci in TbTRF-depleted cells, suggesting that TbTRF may also regulate the TERRA level. We first did northern blotting to examine the TERRA level in TbTRF RNAi cells. The telomerase RNA (TbTR) (61) level was detected as a loading control. Uninduced TbTRF RNAi cells had a comparable amount of TERRA as that in WT cells (Figure 2A, left), while after the induction for 24 and 30 hrs, the TERRA level was 3.8- and 4.4-fold of that in uninduced cells, respectively (Figure 2A, left). We further estimated the average TERRA size using the telomere length quantification method published in (53). In WT cells and TbTRF RNAi cells after 0, 24, and 30 hrs of induction, the average TERRA size is 3.19, 3.22, 3.44 and 3.35 thousand nucleotides, respectively, indicating that depletion of TbTRF causes a very subtle change in the TERRA size, if any. As a control, all RNA signals were abolished when the samples were treated with RNase A and RNAse One (Figure 2A, left). We also examined the TERRA level in TbTRF RNAi + F2H-TbTRF cells (Figure 2A, right), where an ectopic allele of FLAG-HA-HA (F2H)-tagged WT TbTRF was targeted to the rDNA spacer region. In these cells, adding doxycycline induced the expression of both TbTRF RNAi and the ectopic F2H-TbTRF (Supplementary Figure S2A). The TERRA level in TbTRF RNAi + F2H-TbTRF cells did not increase upon adding doxycycline (Figure 2A, right), indicating that the TbTRF RNAi knockdown phenotype was complemented by the ectopic TbTRF expression. For a more precise measurement of the TERRA level, we performed slot blot northern hybridization, which showed that the TERRA level increased 5- to 8-fold upon depletion of TbTRF in several independent TbTRF RNAi strains (Figure 2B; Supplementary Figure S2B, C).
Figure 2.
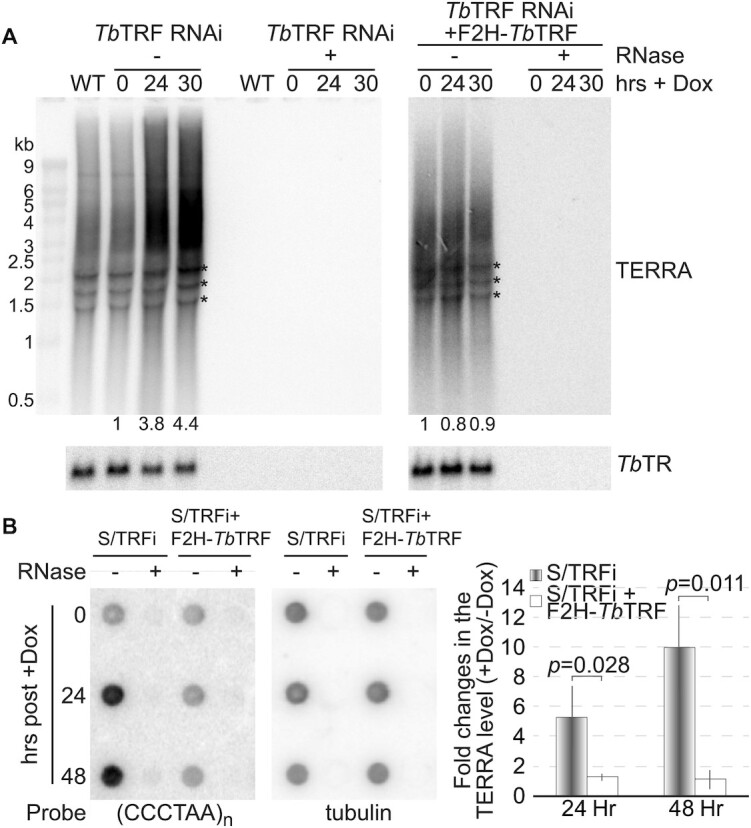
Depletion of TbTRF results in a higher TERRA level. (A) Northern analysis of TERRA in TbTRF RNAi cells with or without a complementary F2H-TbTRF allele. Total RNA was isolated from cells after the induction for 0, 24, and 30 hrs. Equal amounts of RNA samples were treated with or without RNase A and RNase One. TbTR was detected as a loading control. The relative TERRA levels (after normalization against the TbTR level) were quantified and indicated at the bottom of the blot. Asterisks represent rRNA precursors as non-specific hybridization signals. (B) A representative TERRA slot blot of samples from S/TRFi cells (TbTRF RNAi in the strain used for VSG switching analysis (44)) with or without a complementary F2H-TbTRF allele. Total RNA was isolated from cells after the induction for 0, 24 and 48 hrs. Equal amounts of RNA samples were treated with or without RNase A and RNase One. Tubulin was detected as a loading control. Average signal intensities were calculated from three slot blot analyses and are shown on the right. P-values of unpaired t-tests are shown.
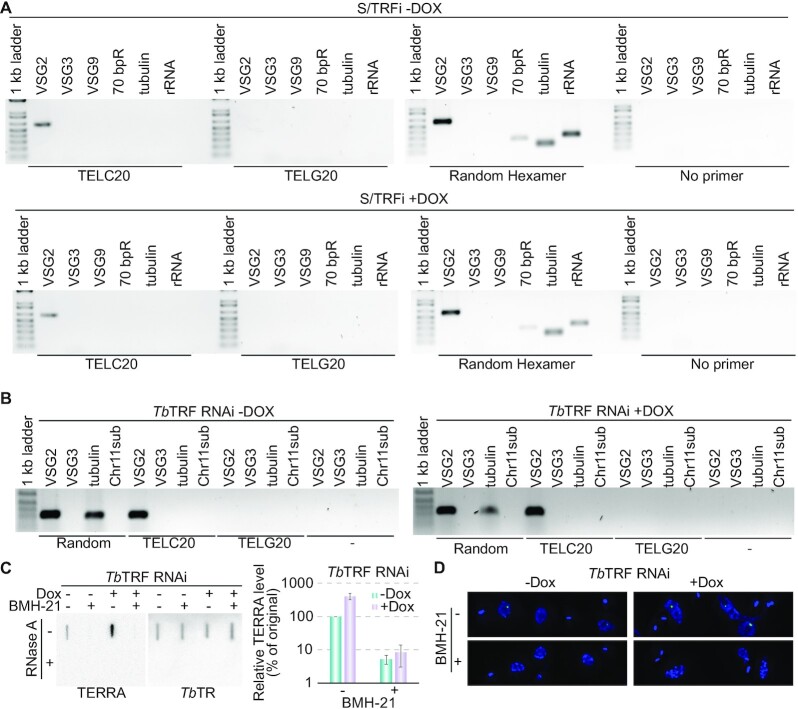
TERRA is expressed from the active VSG-adjacent telomere but not from those adjacent silent VSGs in WT cells (9). To determine the TERRA origin in TbTRF RNAi cells, we performed the RT-PCR experiment (Supplementary Figure S3A) as described previously (9). After using the CCCTAA repeat-containing TELC20 oligo (9) as a primer in reverse transcription, we used primers specific for the active VSG2 or silent VSG3 and VSG9 genes for PCR analysis. All these VSGs are located within 2 kb from the telomeric repeats in different ESs (34). In uninduced TbTRF RNAi cells, only VSG2-specific primers yielded clear PCR products (Figure 3A, top), as observed in WT cells (9). This indicated that TERRA was transcribed from the active VSG2-adjacent telomere, as a result of transcription read-through into the telomere region. In contrast, when using a TTAGGG repeat-containing TELG20 primer (9) for reverse transcription, no PCR products were detected for any genes tested. As a positive control, primers specific for all actively transcribed genes yielded PCR products when a random hexamer was used for reverse transcription (Figure 3A). It is interesting that we detected transcripts containing the 70 bp repeats (Figure 3A; Supplementary Figure S1A), although previous RNAseq analysis suggests that WT cells have a very low steady-state level of 70 bp repeat transcripts (62). Importantly, after depletion of TbTRF, we observed the same result as that in uninduced cells (Figure 3A, bottom). No PCR products were obtained using primers specific to the silent VSG3 or VSG9 genes, indicating that TERRA was still transcribed from the active ES- but not silent ES-adjacent telomeres. As a control, PCR using primers specific for VSG3 and VSG9 amplified their respective genomic sequences in TbTRF RNAi cells both before and after the RNAi induction (Supplementary Figure S3B, left).
Figure 3.
TERRA is transcribed from the active VSG-adjacent telomere in WT and TbTRF-depleted cells. (A) Total RNA was purified from the VSG2-expressing S/TRFi cells before (top) and after (bottom) induction for 24 hrs and reverse-transcribed using TELC20, TELG20, a random hexamer (as a positive control), or ddH2O (as a negative control) as the RT primer (labeled beneath each gel). The RT products were PCR amplified using primers specific to VSG2 (active), VSG3 (silent), VSG9 (silent), 70 bp repeats, tubulin, and rRNA (marked on top of each lane), and the PCR products were separated on agarose gels. (B) Total RNA was purified from the VSG2-expressing TbTRF RNAi cells before (left) and after (right) induction for 24 hrs and reverse-transcribed using TELC20, TELG20, and a random hexamer (as a positive control) as the RT primer (labeled beneath each gel). The RT products were PCR amplified using primers specific to VSG2 (active), VSG3 (silent), tubulin, and a subtelomeric locus on chromosome 11 (Chr11sub) (marked on top of each lane), and the PCR products were separated on agarose gels. (C) TERRA is sensitive to BMH-21, an RNAP I inhibitor. Left, a representative TERRA slot blot of samples from TbTRF RNAi cells before and 24 hrs after the induction of TbTRF RNAi with and without the BMH-21 treatment. TbTR was detected as a control. Right, quantification of relative TERRA levels in TbTRF RNAi cells treated with and without 3 μM BMH-21 for 3 hrs. Average was calculated from four slot blots. (D) representative images of nuclear TERRA foci in TbTRF RNAi cells treated with and without BMH-21 for 3 hrs.
The genome sequence of the Lister 427 strain T. brucei has recently been updated (35). Most subtelomeres of the megabase chromosomes harbor VSG gene arrays or VSG ESs. Among these, the VSG arrays and VSGs in silent ESs are not transcribed in WT cells (37), and only the active VSG ES and its downstream telomere are transcribed (9). However, we did not know whether VSG-free telomeres are transcribed or not. According to the available genome data, one of the chromosome 11 subtelomeres is free of any VSG (35). We were able to identify a 200 bp region within 2 kb from the telomere repeats on chromosome 11 to have a unique sequence. Using primers specific to this chromosome 11 subtelomere, we were able to amplify the expected PCR product using genomic DNA as the template (Supplementary Figure S3B, right). Subsequently, we performed the same RT-PCR experiment as described above. Both before and after depletion of TbTRF, the chromosome 11 subtelomere primers did not amplify any cDNA product after using TELC20 as the reverse transcription primer (Figure 3B), indicating that this VSG-free telomere is not transcribed in WT cells and that depletion of TbTRF does not lead to its transcription, either.
We previously showed that TERRA is transcribed from the active VSG-adjacent telomere, presumably as a result of RNAP I read-through activity (9). To further confirm that TERRA is an RNAP I transcription product, we treated the cells with the RNAP I inhibitor BMH-21 for 3 hrs according to (63) and performed TERRA northern slot blot hybridization (Figure 3C, left). TbTR was detected as a loading control. Treating cells with BMH-21 for 3 hrs did not affect cell growth (Supplementary Figure S3C) and was able to deplete the RNA of the VSG pseudogene in ES1 (ψES1) without affecting the TbTR level (Supplementary Figure S3D), indicating that BMH-21 specifically blocks RNAP I transcription as previously reported (63). In TbTRF RNAi cells, both before and after depletion of TbTRF, we found that the TERRA level decreased dramatically after the cells were treated with BMH-21 (Figure 3C). Using the TERRA signal level in uninduced TbTRF RNAi cells without the BMH-21 treatment as a reference (set as 100%), there was 5% of TERRA left after cells were treated with BMH-21 (Figure 3C, right). The TERRA level increased to >400% in induced TbTRF RNAi cells before the BMH-21 treatment and decreased to ∼8% after the BMH-21 treatment (Figure 3C, right). We further performed TERRA FISH in TbTRF RNAi cells that were treated with or without BMH-21 for 3 hrs. In both induced and uninduced TbTRF RNAi cells, the TERRA signal was positive in most cells not treated with BMH-21 but rarely observed in cells treated with BMH-21 (Figure 3D). Therefore, the TERRA level is sensitive to the RNAP I inhibitor BMH-21 in both WT and TbTRF-depleted cells, confirming that the majority of TERRA is transcribed by RNAP I from the active VSG-adjacent telomere.
TbTRF mildly suppresses the TERRA transcription but does not affect the TERRA half-life
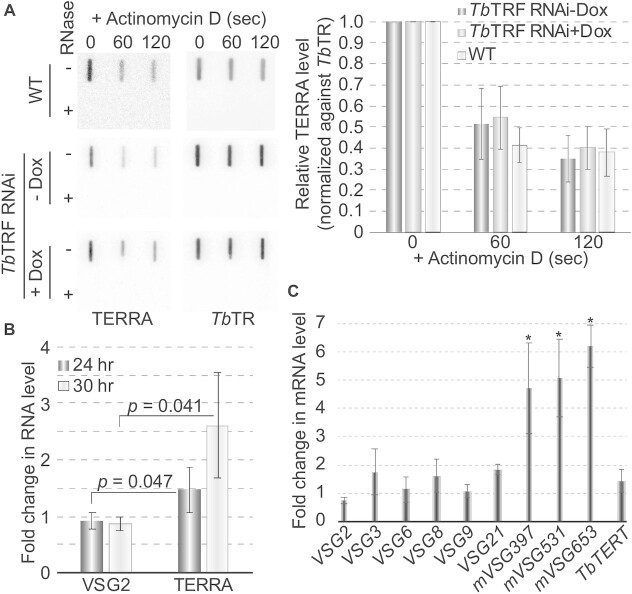
To explore how TbTRF affects the TERRA level, we measured the half-life of TERRA. Total RNA was isolated from WT cells treated with Actinomycin D for various lengths of time, and TERRA slot blot hybridization was performed. We found that after treating cells with Actinomycin D for 10 min, >90% of TERRA was degraded. To measure TERRA half-life more accurately, we treated T. brucei cells for a much shorter time. TERRA decayed very fast (Figure 4A), and its half-life is estimated to be ∼60 secs (Figure 4A, right). We repeated the same experiments in TbTRF RNAi cells before and after the induction of TbTRF RNAi for 24 hrs. The TERRA half-lives in these cells were approximately the same as that in WT cells (Figure 4A), indicating that TbTRF does not regulate TERRA stability. Subsequently, we treated TbTRF RNAi cells (both with and without a 24-hr doxycycline induction) with BMH-21 for only 15 min and examined TERRA level with northern slot blot hybridization (Supplementary Figure S3E). Using the TERRA signal level in uninduced and untreated TbTRF RNAi cells as a reference (100%), there was 7% of TERRA left after the uninduced cells were treated with BMH-21 (Supplementary Figure S3F). The TERRA level increased to more than 400% in induced TbTRF RNAi cells before the BMH-21 treatment and decreased to ∼11% after the BMH-21 treatment (Supplementary Figure S3F). Therefore, even with a 15-min BMH-21 treatment, the TERRA level was significantly decreased (Supplementary Figure S3E, F), further confirming that TERRA’s half-life is very short.
Figure 4.
TbTRF suppresses TERRA transcription mildly but does not affect its half-life. (A) The level of TERRA was estimated by northern slot blot after cells were treated with Actinomycin D for various lengths of time (left). TERRA hybridization signals were quantified by ImageQuant (right). Average values were calculated from four independent experiments. TbTR was detected as a loading control. In TbTRF RNAi cells, the TERRA level was estimated before (-Dox) and 24 hrs after induction of TbTRF RNAi (+Dox). (B) The level of the VSG2-TERRA-containing RNA increased mildly in TbTRF-depleted cells. Reverse transcription was performed using TELC20 (TERRA) and a hexamer containing a random sequence (VSG2) followed by qPCR using the VSG2-specific primers. The average fold changes in the RNA level were calculated from three to five independent experiments. (C) Metacyclic ES-linked VSGs were mildly derepressed in TbTRF-depleted cells. qRT-PCR was performed to estimate the relative RNA levels of various BF ES- and metacyclic ES-linked VSGs. TbTERT mRNA level was estimated as a control. Changes in mVSG397, mVSG531, and mVSG653 RNA levels are significantly higher (asterisks) than that in the TbTERT RNA level.
We also did quantitative RT-PCR (qRT-PCR) to estimate the level of the VSG2-TERRA-containing polycistronic transcript, in which TELC20 was used as the reverse transcription primer, and VSG2-specific primers were used in the qPCR (Supplementary Figure S3A). Depletion of TbTRF led to a mild but significant increase in the level of the VSG2-TERRA-containing transcript (Figure 4B), suggesting that TbTRF suppresses TERRA transcription. In contrast, the steady-state level of the VSG2 RNA was not increased (Figure 4B). We previously showed that TbTRF did not affect BF ES-linked VSG mRNA levels (42). Using qRT-PCR, we verified that the RNA levels of the silent VSGs in BF ESs (which are PTUs with promoters located 40–60 kb upstream, Supplementary Figure S1A, top) in TbTRF-depleted cells were only ∼1-2 fold of that in uninduced cells (Fig 4C). However, the RNA levels of VSGs in metacyclic ESs (which are monocistronic transcription units with promoters located ∼5 kb upstream, Supplementary Figure S1A, bottom) were mildly but significantly increased (5- to 6-fold) when TbTRF was depleted (Figure 4C), suggesting that TbTRF is also important for telomeric silencing but its effect does not spread beyond a few kbs upstream of the telomere.
TbTRF suppresses the telomeric R-loop level and helps maintain telomere integrity
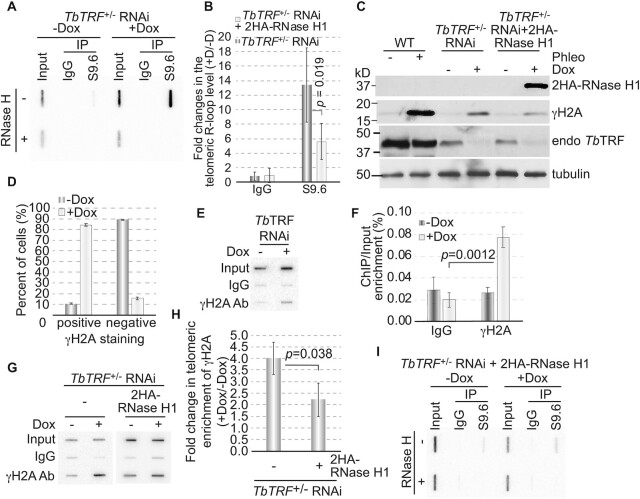
We previously showed that more telomeric R-loops were formed in TbRAP1-depleted cells that expressed a higher level of TERRA (9). Since it is unknown whether a higher level of TERRA always results in a higher amount of telomeric R-loops, we examined the telomeric R-loop level in TbTRF-depleted cells. We used S9.6, a monoclonal antibody specifically recognizing the RNA:DNA hybrid (64), to pull down genomic R-loops and did Southern hybridization using a telomere probe (Figure 5A). Upon induction of TbTRF RNAi for 24 hrs, we detected an increased amount of telomeric R-loops compared to that in uninduced cells (Figure 5A). Quantification of the slot blot hybridization signals showed that depletion of TbTRF increased the telomeric R-loop level ∼13-fold (Figure 5B). Therefore, TbTRF suppresses both telomeric R-loop and TERRA levels.
Figure 5.
Depletion of TbTRF leads to more telomeric R-loops and an increased amount of DNA damage at the telomere. (A, I) A representative Southern slot blot of input, IgG and S9.6 immunoprecipitated DNA samples in TbTRF+/– RNAi (A) and TbTRF+/– RNAi+2HA-RNase H1 (I) cells before (-Dox) and after (+Dox) adding doxycycline for 24 hrs. Samples were treated with or without RNase H (ThermoFisher) before IP. A (TTAGGG)n probe was used in the hybridization. (B) Quantification of the slot blot hybridization signals in both TbTRF+/– RNAi and TbTRF+/– RNAi+2HA-RNase H1 cells. Enrichment of the telomeric R-loop (over input) was calculated before and after adding doxycycline and the fold changes in this enrichment were plotted for both cells. Average values were calculated from four to five independent experiments. (C) Western analyses using the HA antibody (HA probe, Santa Cruz Biotechnologies), a TbTRF rabbit antibody (41), a γH2A rabbit antibody (9), and the tubulin antibody TAT-1 (86) in WT, TbTRF+/– RNAi and TbTRF+/– RNAi+2HA-RNase H1 cells. (D) Quantification of γH2A IF results. A majority of cells were positive for the γH2A staining after TbTRF was depleted. (E) ChIP using the γH2A antibody and IgG (as a negative control) were performed in TbTRF RNAi cells before and 24 hrs after the induction of RNAi. ChIP products were analyzed by slot blot hybridization with a telomere probe. (F) Quantification of three independent γH2A ChIP results in TbTRF RNAi cells. Averages of telomeric DNA enrichment in the ChIP experiments were calculated. (G) ChIP using the γH2A antibody and IgG were performed in TbTRF+/– RNAi and TbTRF+/– RNAi+2HA-RNase H1 cells before and 24 hrs after adding doxycycline. ChIP products were detected by Southern hybridization using a telomere probe. (H) Quantification of three independent γH2A ChIP results in TbTRF+/– RNAi and TbTRF+/– RNAi+2HA-RNase H1 cells. Enrichment of γH2A at the telomere (over input) was calculated before and after adding doxycycline and the fold changes in this enrichment were plotted for both cells. In (B), (F) and (H), p-values of unpaired t-test are shown.
R-loops frequently induce genome instability and cause DSBs (65). We next examined whether depletion of TbTRF increased the amount of DSBs at the telomere. γH2A (where T130 of histone H2A is phosphorylated) is deposited at DNA damage sites (66), so we used a γH2A antibody (9) to estimate the level of DNA damage in TbTRF-depleted cells. Western analysis showed that the γH2A level increased upon depletion of TbTRF (Figure 5C). As a positive control, the γH2A level also increased in phleomycin-treated WT cells (Figure 5C). In addition, in uninduced TbTRF RNAi cells, only a small fraction (∼10%) of cells showed the γH2A signal, while after TbTRF depletion, nearly 90% of cells are positive for the γH2A signal (Figure 5D). We also performed a ChIP analysis using the γH2A antibody and hybridized the ChIP product with a telomeric probe. More telomeric DNA was seen to associate with γH2A upon TbTRF depletion (Figure 5E, F), indicating that the TbTRF depletion indeed resulted in an increased amount of DNA damage at the telomere. Therefore, TbTRF has a role in maintaining telomere integrity. This can also explain why a transient depletion of TbTRF leads to an increase in the VSG switching frequency (44), as DSBs at the active VSG vicinity are known to be a potent inducer for VSG switching (39,40).
RNase H1 is known to resolve telomeric R-loops in T. brucei (9,67). Therefore, we introduced an inducible ectopic HA-HA (2HA) tagged RNase H1 in TbTRF+/– RNAi cells. Adding doxycycline to these cells induces both TbTRF depletion and 2HA-RNase H1 expression (Figure 5C; Supplementary Figure S4A). Importantly, the telomeric R-loop level only increased ∼5 fold in these cells after adding doxycycline (Figure 5B, I), indicating that overexpression of 2HA-RNase H1 partially suppressed the increase in the telomeric R-loop level caused by TbTRF depletion. In addition, induction of TbTRF RNAi resulted in a 4-fold increase in telomere-associated γH2A in TbTRF+/– RNAi cells but only a 2-fold increase in TbTRF+/– RNAi+2HA-RNase H1 cells (Figure 5G, H), suggesting that after 2HA-RNase H1 overexpression, a smaller increase of the telomeric R-loop level resulted in a smaller increase of the telomeric DNA damage amount. Therefore, TbTRF suppresses the telomeric R-loop level, which contributes to telomere integrity maintenance.
TbTRF binds (UUAGGG)n-containing RNA in vitro and in vivo
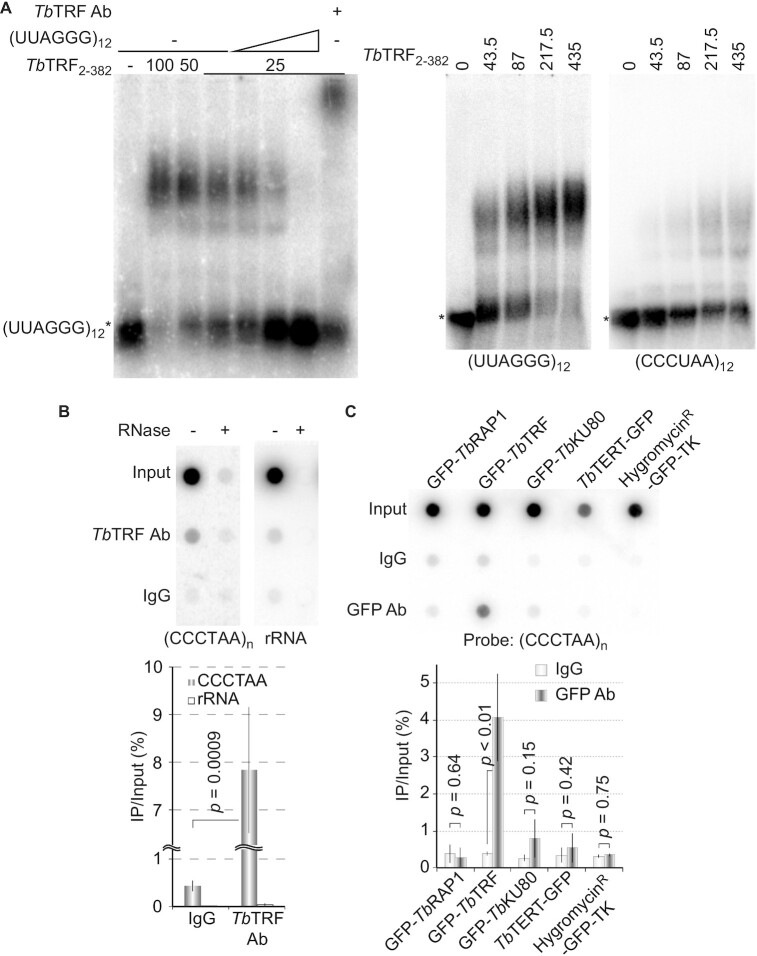
To explore how TbTRF regulates the TERRA level, we performed Electrophoretic Mobility Shift Assays (EMSAs) to determine whether TbTRF binds TERRA. Recombinant GST-tagged full-length TbTRF (Supplementary Figure S4B) (41) bound an RNA that contains (UUAGGG)12 (Figure 6A). It also bound a (CCCUAA)12-containing RNA substrate but much more weakly (Figure 6A, right). The TbTRF-(UUAGGG)12 complex was competed away by an unlabeled (UUAGGG)12-containing RNA (Figure 6A, left). In addition, this complex was super-shifted by the TbTRF antibody (Figure 6A, left) (41), confirming that the protein-RNA complex contained the TbTRF protein.
Figure 6.
(A) TbTRF binds to single stranded UUAGGG repeats directly. EMSA was performed using the recombinant GST-tagged TbTRF2-382 (numbers indicate aa positions) expressed from E.coli (41) and ss(UUAGGG)12 or ss(CCCUAA)12 as the probe. Non-radiolabeled (UUAGGG)12 was added as a competitor. The amount of TbTRF (ng) used is indicated on top of each lane. (B, C) TbTRF associates with TERRA in vivo. Cell lysates were incubated with TbTRF antibody (41) (B) or a rabbit anti-GFP antibody (ThermoFisher) (C). RNA was isolated from the IP products and analyzed by slot blot hybridization with a (CCCTAA)n-containing probe. A representative slot blot is shown at the top. Hybridization with an rRNA probe is used as a negative control in (B). The average enrichment of the RNA-IP product was calculated from three slot blot hybridizations and is shown at the bottom. P values of unpaired t-tests are shown in (B) and (C).
We subsequently examined whether TbTRF bound TERRA in vivo by RNA IP. A significant amount of TERRA (but not rRNA) was detected in the product of TbTRF IP using a TbTRF rabbit antibody (Figure 6B) (41). We also examined whether TbRAP1 (42), TbKU80 (a DNA end binding factor) (50,68), and TbTERT (the protein component of telomerase) (51) had any in vivo TERRA binding activity. For a better comparison, cells expressing GFP-tagged TbRAP1, TbTRF, TbKU80 and TbTERT were used (Supplementary Figure S4C). An additional strain that expressed a HygromycinR-GFP-TK fusion protein (69) was used as a negative control (Supplementary Figure S4C). We detected a significant amount of TERRA in the IP product using a rabbit GFP antibody (ThermoFisher) in GFP-TbTRF expressing cells (Figure 6C). As expected, the HygromycinR-GFP-TK fusion protein did not interact with TERRA, nor did TbRAP1 or TbTERT bind to TERRA under this condition. However, TbKU80 might interact with TERRA at a very low level (Figure 6C).
TbTRF’s TERRA binding activity resides in its Myb domain
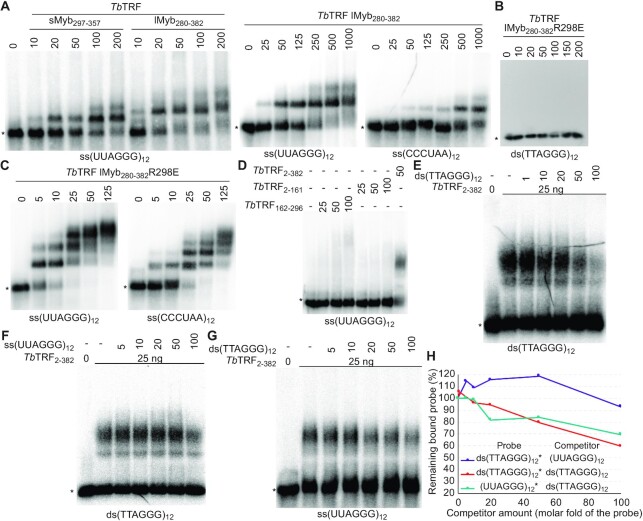
To determine which functional domain of TbTRF has the TERRA binding activity, we did EMSAs using various TbTRF recombinant fragments. We found that two GST-tagged TbTRF Myb fragments (containing aa 297–357 and aa 280–382, respectively, Supplementary Figure S4B) bound to the (UUAGGG)12 RNA (Figure 7A, left). Interestingly, the TbTRF-lMyb280-382 fragment was able to bind both UUAGGG and CCCUAA repeats (Figure 7A, right), although its affinity for UUAGGG repeats is much higher (Kd = 268 nM) than that for CCCUAA repeats (Kd = 17.171 μM). Interestingly, the RNA binding activity of TbTRF-lMyb280-382 was increased when its DNA binding activity was abolished. The TbTRF R298E mutant loses the duplex telomere DNA binding activity (Figure 7B) (44). Yet, this mutant was still able to bind both the UUAGGG and CCCUAA repeats and with a higher affinity (Kd = 5 and 36 nM, respectively), although it still preferred UUAGGG repeats (Figure 7C). In addition, when using radiolabeled UUAGGG repeats as the substrate, unlabeled UUAGGG repeats compete better than CCCUAA repeats (Supplementary Figure S4D). In contrast, TbTRF2-161 and TbTRF162-296 (Supplementary Figure S4B), both of which do not contain the Myb domain, did not show any robust RNA binding activity (Figure 7D), indicating that the TERRA-binding activity of TbTRF resides in its Myb domain.
Figure 7.
The TbTRF Myb domain has a TERRA binding activity. EMSA was performed using the recombinant GST-tagged TbTRF fragments containing aa 297–357 (A), aa 280–382 (A), aa 280–382 with the R298E mutation (B, C), aa 2–161 (D), aa 163–296 (D), and aa 2–382 (D–G) expressed from E. coli and ss(UUAGGG)12, ss(CCCUAA)12, or ds(TTAGGG)12 as the probe. The amount (ng) of recombinant proteins used in EMSA are indicated on top of each lane. Non-radiolabeled (UUAGGG)12 and ds(TTAGGG)12 were used as competitors in (E–G). Amounts of competitors are shown as molar folds of the radiolabeled probe. Asterisks indicate the positions of free probes. (H) Quantification of EMSA results shown in (E), (F) and (G). The remaining protein-probe complex in the presence of various amounts of the competitor is calculated as the percent of the complex amount in the absence of any competitor.
The fact that TbTRF-lMyb280-382R298E binds RNA substrates with a higher affinity than its WT counterpart suggests that the TbTRF RNA and DNA binding interfaces may overlap and the binding activities may compete with each other. Therefore, we further examined whether TbTRF bound the double-stranded telomere DNA or the single-stranded TERRA more strongly. Using radiolabeled ds(TTAGGG)12 or (UUAGGG)12 as the substrate, we found that competitions from the unlabeled ds(TTAGGG)12 or (UUAGGG)12 showed subtle differences (Figure 7E–G). Quantification of the amount of the TbTRF2-382-probe complex in the presence of various amounts of the competitors indicates that ds(TTAGGG)12 competes better for TbTRF2-382 binding than (UUAGGG)12 (Figure 7H). Therefore, the binding affinity of TbTRF to duplex telomere DNA is stronger than its affinity to TERRA, and both nucleic acid interaction interfaces may overlap with each other.
DISCUSSION
The TERRA transcription site in T. brucei
TERRA has been identified in multiple organisms (3). In yeast and mammalian cells, TERRA is transcribed by RNA Pol II from multiple telomeres (4,11–16). However, our observations reveal two unique features of TERRA in BF T. brucei: it is transcribed by RNAP I, and the majority of it is transcribed from the active VSG-adjacent telomere. First, the TERRA level is not sensitive to 1 mg/ml α-amanitin (31), and its level decreases ∼95% when the RNAP I-mediated transcription is inhibited by BMH-21. Therefore, TERRA is nearly all transcribed by RNAP I in BF T. brucei. Second, we previously found that TERRA is transcribed from the telomere downstream of the active ES but not from those downstream of silent ESs (9), which is confirmed in the current study. Third, one of the minichromosome subtelomeres has been reported to have an RNAP I promoter (70), although this has not been verified in the strain used in this study. Hence, it is possible that this minichromosome telomere is transcribed. However, ∼60% of cells in the G1 phase have only one nuclear TERRA focus, suggesting that transcription of the minichromosome subtelomere, if any, is at a very low level. Fourth, one of the chromosome 11 subtelomeres does not harbor any VSG gene array or VSG ES, and our RT-PCR analysis showed that TERRA is not transcribed from this telomere. Hence, RNA Pol II-mediated read-through of subtelomeric PTUs is unlikely a contributing factor to TERRA transcription in BF T. brucei. In summary, the majority of TERRA appears to be transcribed from the active VSG-adjacent telomere by RNAP I in T. brucei when it proliferates in its mammalian host, reflecting a significant evolution of TERRA transcription from protozoan to metazoan eukaryotes.
The number of nuclear TERRA foci is cell-cycle dependent
We demonstrate that the number of TERRA foci is cell cycle regulated in T. brucei, which has not been reported in other TERRA-expressing organisms. Using RNA FISH, we show that a majority of G1 cells have one or two nuclear TERRA foci, and the number of nuclear TERRA foci increases as the cell enters the S and G2/M phases. One possibility is that after DNA replication, the duplicated active ESs, now both transcribing TERRA, present two TERRA foci. Indeed, the active ES is replicated early in the S phase (71). However, most replicated active ESs appear to be tightly associated with each other until mitosis, as only one ES transcription site is observed from the S to G2 phases in most cells (72). Replicated sister chromatids are often positioned within a short distance from each other, but TERRA foci are usually dispersed in the nucleus. Therefore, it is unlikely that the increased number of TERRA foci simply results from replicated chromatids. We speculate that TERRA can be recruited to loci away from its transcription site (see below).
In yeast cells, the TERRA level dips in the S phase and peaks in the G2/M phase (28), while in Hela cells, the TERRA level is high in G1/S and decreases at the late S/G2 phase (29,30). Whether the T. brucei TERRA level also changes throughout the cell cycle is currently unknown. Although T. brucei cells can be arrested at the S phase by HU treatment (73), they are poorly synchronized after being released from HU, possibly due to its atypical cell cycle control (74). Therefore, it is unclear whether the TERRA level is cell-cycle regulated in T. brucei.
Does TERRA function in trans?
T. brucei has a large number of chromosomes (11 pairs of megabase chromosomes, 4–5 intermediate chromosomes, and ∼100 minichromosomes) and nearly 250 telomeres (75,76). However, telomeres are heavily clustered in T. brucei, and typically fewer than 10 telomere foci are seen per nucleus in telomere FISH studies (41,59,77). We frequently observed that among the few TERRA foci, one (or occasionally two) is noticeably brighter than the rest and is colocalized with TbTRF, indicating that TERRA is associated with the telomere chromatin. We also noticed that in nuclei with more than two TERRA foci, the fainter foci are frequently not colocalized with TbTRF. Since TbTRF is almost always colocalized with the telomere in combined IF and FISH analyses (41), this observation indicates that a subset of TERRA molecules are not associated with the telomere chromatin, which is similar to what has been described in mouse cells (21).
A recent study showed that in human cells TERRA can be recruited to telomeres in trans (60). It is likely that in T. brucei, TERRA is also recruited to non-telomeric loci after its transcription. Interestingly, TbTRF appears to facilitate the trans-localization of TERRA, as fewer number of nuclear TERRA foci are seen in cells depleted of TbTRF. This is different from the scenario in Hela cells, where both TRF1 and TRF2 suppress trans localization of TERRA (60). T. brucei has chromosome internal TTAGGG repeats (35), and it is possible that a small amount of TbTRF binds these chromosome internal TTAGGG repeat sequences even though they are not detected in IF/FISH analysis. Since TbTRF binds TERRA directly both in vivo and in vitro, one possible role for this protein-RNA interaction is that TbTRF helps to recruit TERRA to loci other than its transcription site, although TERRA may be recruited by other means as many TbTRF-depleted cells still have two or three nuclear TERRA foci.
TbTRF has a TERRA binding activity that resides in its Myb domain
TbTRF is a duplex telomere DNA binding factor that does not bind single-stranded DNA (41). It is surprising that TbTRF also has a strong affinity for the UUAGGG repeat-containing RNA. This TERRA binding activity is different from that of mammalian TRF2, which is mediated by its N-terminal basic GAR domain (78), as TbTRF does not have an N-terminal basic domain (41). In addition, we found that the Myb domain of TbTRF, which is responsible for recognizing the duplex telomere DNA (41), is also responsible for binding the TERRA RNA. Importantly, our data indicate that the Myb-mediated RNA binding is sequence-specific, as TbTRF strongly prefers UUAGGG to CCCUAA repeats, which is unusual for Myb domains. Most interestingly, a point mutation that disrupts TbTRF’s DNA binding activity (44) showed stronger RNA binding activity, suggesting that the DNA and TERRA binding activities of TbTRF may have overlapping (or partially overlapping) nucleic acid interaction interfaces. In addition, direct EMSA competition assays showed that TbTRF binds duplex telomere DNA stronger than TERRA. However, further investigation is necessary to determine the exact RNA binding interface in the TbTRF Myb domain.
The duplex telomere DNA and TERRA binding activities of TbTRF also make TbTRF a good candidate to recruit/retain TERRA to the TTAGGG repeats, either at the telomere or at chromosome internal regions. Although we have not observed any ternary complex of TbTRF with both the telomere DNA and the TERRA substrates in EMSA, TbTRF can interact with itself through its TRFH domain (41). It is therefore possible that the telomere DNA and TERRA are brought together by different TbTRF molecules that interact with each other.
TbTRF suppresses the TERRA level
TERRA has a very short half-life (∼60 secs), which is likely due to the fact that most TERRA molecules do not have a poly(A) tail (31). It is worth noting that VSG RNA accounts for ∼10% of total RNA (79), and RNAP I-mediated VSG transcription is at a very high level. Therefore, although TERRA has a very short half-life, there is still a significant amount of TERRA in the cell at any moment. TbTRF does not affect the TERRA half-life. However, qRT-PCR results showed that TbTRF is important for silencing subtelomeric VSGs located in metacyclic ESs where the promoter is ∼5 kb upstream of the telomere, although TbTRF does not affect BF ES silencing (42). In addition, removal of TbTRF from the telomere may allow a higher level of RNAP I read-through into the telomere repeat region. These likely explain why the level of the VSG2-TERRA-containing polycistronic transcript is mildly increased upon TbTRF depletion. Northern slot blot detected a higher fold increase in the TERRA level than the qRT-PCR analysis. This is likely because qRT-PCR only detects the VSG2-TERRA-containing transcript that has not been trans-spliced. On the other hand, VSG mRNAs have much longer half-lives (1-2 hrs) (80) and the active VSG is highly transcribed, which can explain why the steady state level of VSG2 RNA was not affected by TbTRF depletion.
TbTRF suppresses the telomeric R-loop level to help maintain telomere integrity
We showed that TbTRF suppresses the level of the telomeric R-loop. Interestingly, human TRF2 has been shown to facilitate telomeric R-loop formation while human TRF1 inhibits this function of TRF2 (81). Therefore, TbTRF has a similar effect as TRF1 but a different effect than TRF2 on telomeric R-loop formation. We hypothesize that TbTRF suppresses the telomeric R-loop level through two activities. First, the TbTRF-mediated telomeric silencing [although a more local effect than that mediated by TbRAP1 (42)] helps suppress the TERRA level, which in turn helps reduce the chance of telomeric R-loop formation. Second, it is possible that the TERRA-binding activity of TbTRF helps to disperse TERRA from its transcription site, preventing the accumulation of an excessive amount of TERRA at a single telomere and subsequent telomeric R-loop formation. Indeed, in TbTRF-depleted cells, we frequently observed a single large TERRA focus, and the number of nuclear TERRA foci was reduced compared to that in WT cells. Significantly, suppression of the telomeric R-loop level by TbTRF is important for maintaining telomere integrity, as overexpression of RNase H1 partially suppresses the increased telomeric R-loop level and the increased telomeric DNA damage amount observed in TbTRF-depleted cells. Previously we also found that TbRAP1 has a similar function and suppresses the telomeric R-loop level, which contributes to telomere and subtelomere integrity maintenance (9). In contrast, mammalian TRF2 prevents non-homologous end-joining (NHEJ)-mediated chromosome end-to-end fusions (82,83), suppresses telomere homologous recombination (84), but promotes the telomeric R-loop formation (81).
We speculate that telomeres in T. brucei and higher eukaryotes do not face the same genotoxic threats. First, the NHEJ machinery is absent in T. brucei (85), which is expected to greatly reduce the chance of telomere end fusions when compared to the situation in yeast and vertebrates. Second, at least one T. brucei telomere is transcribed at a very high level by RNAP I, while TERRA is transcribed by RNA Pol II in yeast and mammals. Presumably, more R-loops are likely to form at the T. brucei telomere, which are prone to cause DNA damage and need to be tightly controlled. Therefore, although TbTRF has the same role as its homologs in protecting natural chromosome ends, the underlying mechanism of TbTRF’s function is different from that of TRF homologs in yeasts and vertebrates.
It is intriguing that T. brucei telomeres have exactly the same sequences as those in vertebrates, yet the telomere biology has many different features in this parasite and higher eukaryotic cells. Identification of the conserved and unique aspects of T. brucei telomere biology not only helps us better understand the evolution of telomere proteins but also helps the development of anti-parasite agents in the future.
DATA AVAILABILITY
All data have been included in the manuscript, figures, and supplemental information.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Keith Gull for the TAT-1 antibody. We thank the Li lab members for their comments on the manuscript.
Notes
Present address: Unnati M. Pandya, Department of Obstetrics and Gynecology, Massachusetts General Hospital, 55 Fruit Street, Boston, MA 02114, USA.
Present address: Ranjodh Sandhu, Department of Microbiology and Molecular Genetics, University of California, One Shields Avenue, Davis, CA 95616, USA.
Present address: Vishal Nanavaty, Neuberg Center for Genomic Medicine, Neuberg Supratech Reference Laboratory, Ellisbridge Ahmedabad 380006, Gujarat, India.
Contributor Information
Arpita Saha, Center for Gene Regulation in Health and Disease, Department of Biological, Geological, and Environmental Sciences, College of Sciences and Health Professions, Cleveland State University, 2121 Euclid Avenue, Cleveland, OH 44115, USA.
Amit Kumar Gaurav, Center for Gene Regulation in Health and Disease, Department of Biological, Geological, and Environmental Sciences, College of Sciences and Health Professions, Cleveland State University, 2121 Euclid Avenue, Cleveland, OH 44115, USA.
Unnati M Pandya, Center for Gene Regulation in Health and Disease, Department of Biological, Geological, and Environmental Sciences, College of Sciences and Health Professions, Cleveland State University, 2121 Euclid Avenue, Cleveland, OH 44115, USA.
Marjia Afrin, Center for Gene Regulation in Health and Disease, Department of Biological, Geological, and Environmental Sciences, College of Sciences and Health Professions, Cleveland State University, 2121 Euclid Avenue, Cleveland, OH 44115, USA.
Ranjodh Sandhu, Center for Gene Regulation in Health and Disease, Department of Biological, Geological, and Environmental Sciences, College of Sciences and Health Professions, Cleveland State University, 2121 Euclid Avenue, Cleveland, OH 44115, USA.
Vishal Nanavaty, Center for Gene Regulation in Health and Disease, Department of Biological, Geological, and Environmental Sciences, College of Sciences and Health Professions, Cleveland State University, 2121 Euclid Avenue, Cleveland, OH 44115, USA.
Brittny Schnur, Center for Gene Regulation in Health and Disease, Department of Biological, Geological, and Environmental Sciences, College of Sciences and Health Professions, Cleveland State University, 2121 Euclid Avenue, Cleveland, OH 44115, USA.
Bibo Li, Center for Gene Regulation in Health and Disease, Department of Biological, Geological, and Environmental Sciences, College of Sciences and Health Professions, Cleveland State University, 2121 Euclid Avenue, Cleveland, OH 44115, USA; Case Comprehensive Cancer Center, Case Western Reserve University, 10900 Euclid Avenue, Cleveland, OH 44106, USA; Department of Inflammation and Immunity, Lerner Research Institute, Cleveland Clinic, 9500 Euclid Avenue, Cleveland, OH 44195, USA; Center for RNA Science and Therapeutics, Case Western Reserve University, 10900 Euclid Avenue, Cleveland, OH 44106, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NIH [R01 grant AI066095 to PI, Li]; NIH [S10 grant S10OD025252 to PI, Li]; GRHD at CSU. Funding for open access charge: NIH funding and partly supported by GRHD center at CSU.
Conflict of interest statement. None declared.
REFERENCES
- 1. de Lange T. Shelterin-mediated telomere protection. Annu. Rev. Genet. 2018; 52:223–247. [DOI] [PubMed] [Google Scholar]
- 2. Ottaviani A., Gilson E., Magdinier F.. Telomeric position effect: from the yeast paradigm to human pathologies. Biochimie. 2008; 90:93–107. [DOI] [PubMed] [Google Scholar]
- 3. Saha A., Nanavaty V.P., Li B.. Telomere and subtelomere R-loops and antigenic variation in trypanosomes. J. Mol. Biol. 2019; 432:4167–4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nergadze S.G., Farnung B.O., Wischnewski H., Khoriauli L., Vitelli V., Chawla R., Giulotto E., Azzalin C.M.. CpG-island promoters drive transcription of human telomeres. RNA. 2009; 15:2186–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chu H.P., Froberg J.E., Kesner B., Oh H.J., Ji F., Sadreyev R., Pinter S.F., Lee J.T.. PAR-TERRA directs homologous sex chromosome pairing. Nat. Struct. Mol. Biol. 2017; 24:620–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Luke B., Panza A., Redon S., Iglesias N., Li Z., Lingner J.. The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol. Cell. 2008; 32:465–477. [DOI] [PubMed] [Google Scholar]
- 7. Bah A., Wischnewski H., Shchepachev V., Azzalin C.M.. The telomeric transcriptome of Schizosaccharomyces pombe. Nucleic Acids Res. 2012; 40:2995–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vrbsky J., Akimcheva S., Watson J.M., Turner T.L., Daxinger L., Vyskot B., Aufsatz W., Riha K.. siRNA-mediated methylation of Arabidopsis telomeres. PLos Genet. 2010; 6:e1000986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nanavaty V., Sandhu R., Jehi S.E., Pandya U.M., Li B.. Trypanosoma brucei RAP1 maintains telomere and subtelomere integrity by suppressing TERRA and telomeric RNA:DNA hybrids. Nucleic Acids Res. 2017; 45:5785–5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Damasceno J.D., Silva G., Tschudi C., Tosi L.R.. Evidence for regulated expression of Telomeric Repeat-containing RNAs (TERRA) in parasitic trypanosomatids. Mem. Inst. Oswaldo Cruz. 2017; 112:572–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Iglesias N., Redon S., Pfeiffer V., Dees M., Lingner J., Luke B.. Subtelomeric repetitive elements determine TERRA regulation by Rap1/Rif and Rap1/Sir complexes in yeast. EMBO Rep. 2011; 12:587–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Azzalin C.M., Reichenbach P., Khoriauli L., Giulotto E., Lingner J.. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007; 318:798–801. [DOI] [PubMed] [Google Scholar]
- 13. Porro A., Feuerhahn S., Delafontaine J., Riethman H., Rougemont J., Lingner J.. Functional characterization of the TERRA transcriptome at damaged telomeres. Nat. Commun. 2014; 5:5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Feretzaki M., Renck Nunes P., Lingner J.. Expression and differential regulation of human TERRA at several chromosome ends. RNA. 2019; 25:1470–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Deng Z., Wang Z., Stong N., Plasschaert R., Moczan A., Chen H.S., Hu S., Wikramasinghe P., Davuluri R.V., Bartolomei M.S., Riethman H., Lieberman P.M.. A role for CTCF and cohesin in subtelomere chromatin organization, TERRA transcription, and telomere end protection. EMBO J. 2012; 31:4165–4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Feretzaki M., Lingner J.. A practical qPCR approach to detect TERRA, the elusive telomeric repeat-containing RNA. Methods. 2017; 114:39–45. [DOI] [PubMed] [Google Scholar]
- 17. Viceconte N., Loriot A., Abreu P.L., Scheibe M., Sola A.F., Butter F., De Smet C., Azzalin C.M., Arnoult N., Decottignies A.. PAR-TERRA is the main contributor to telomeric repeat-containing RNA transcripts in normal and cancer mouse cells. RNA. 2021; 27:106–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lopez de Silanes I., Grana O., De Bonis M.L., Dominguez O., Pisano D.G., Blasco M.A.. Identification of TERRA locus unveils a telomere protection role through association to nearly all chromosomes. Nat. Commun. 2014; 5:4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bettin N., Oss Pegorar C., Cusanelli E.. The emerging roles of TERRA in telomere maintenance and genome stability. Cells. 2019; 8:246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cusanelli E., Chartrand P.. Telomeric repeat-containing RNA TERRA: a noncoding RNA connecting telomere biology to genome integrity. Front Genet. 2015; 6:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chu H.P., Cifuentes-Rojas C., Kesner B., Aeby E., Lee H.G., Wei C., Oh H.J., Boukhali M., Haas W., Lee J.T.. TERRA RNA antagonizes ATRX and protects telomeres. Cell. 2017; 170:86–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang C., Zhao L., Lu S.. Role of TERRA in the regulation of telomere length. Int. J. Biol. Sci. 2015; 11:316–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Arora R., Azzalin C.M.. Telomere elongation chooses TERRA ALTernatives. RNA Biol. 2015; 12:938–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moravec M., Wischnewski H., Bah A., Hu Y., Liu N., Lafranchi L., King M.C., Azzalin C.M.. TERRA promotes telomerase-mediated telomere elongation in Schizosaccharomyces pombe. EMBO Rep. 2016; 17:999–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hu Y., Bennett H.W., Liu N., Moravec M., Williams J.F., Azzalin C.M., King M.C.. RNA-DNA hybrids support recombination-based telomere maintenance in fission yeast. Genetics. 2019; 213:431–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schoeftner S., Blasco M.A.. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 2008; 10:228–236. [DOI] [PubMed] [Google Scholar]
- 27. Cusanelli E., Romero C.A., Chartrand P.. Telomeric noncoding RNA TERRA is induced by telomere shortening to nucleate telomerase molecules at short telomeres. Mol. Cell. 2013; 51:780–791. [DOI] [PubMed] [Google Scholar]
- 28. Graf M., Bonetti D., Lockhart A., Serhal K., Kellner V., Maicher A., Jolivet P., Teixeira M.T., Luke B.. Telomere length determines TERRA and R-Loop regulation through the cell cycle. Cell. 2017; 170:72–85. [DOI] [PubMed] [Google Scholar]
- 29. Arnoult N., Van Beneden A., Decottignies A.. Telomere length regulates TERRA levels through increased trimethylation of telomeric H3K9 and HP1α. Nat. Struct. Mol. Biol. 2012; 19:948. [DOI] [PubMed] [Google Scholar]
- 30. Porro A., Feuerhahn S., Reichenbach P., Lingner J.. Molecular dissection of telomeric repeat-containing RNA biogenesis unveils the presence of distinct and multiple regulatory pathways. Mol. Cell. Biol. 2010; 30:4808–4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rudenko G., Van der Ploeg L.H.. Transcription of telomere repeats in protozoa. EMBO J. 1989; 8:2633–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Barry J.D., McCulloch R.. Antigenic variation in trypanosomes: enhanced phenotypic variation in a eukaryotic parasite. Adv. Parasitol. 2001; 49:1–70. [DOI] [PubMed] [Google Scholar]
- 33. Gunzl A., Bruderer T., Laufer G., Schimanski B., Tu L.C., Chung H.M., Lee P.T., Lee M.G.. RNA polymerase I transcribes procyclin genes and variant surface glycoprotein gene expression sites in Trypanosoma brucei. Eukaryot Cell. 2003; 2:542–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hertz-Fowler C., Figueiredo L.M., Quail M.A., Becker M., Jackson A., Bason N., Brooks K., Churcher C., Fahkro S., Goodhead I.et al.. Telomeric expression sites are highly conserved in Trypanosoma brucei. PLoS One. 2008; 3:e3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Müller L.S.M., Cosentino R.O., Förstner K.U., Guizetti J., Wedel C., Kaplan N., Janzen C.J., Arampatzi P., Vogel J., Steinbiss S.et al.. Genome organization and DNA accessibility control antigenic variation in trypanosomes. Nature. 2018; 563:121–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cross G.A.M., Kim H.S., Wickstead B. Capturing the variant surface glycoprotein repertoire (the VSGnome) of Trypanosoma brucei Lister 427. Mol. Biochem. Parasitol. 2014; 195:59–73. [DOI] [PubMed] [Google Scholar]
- 37. Cross G.A.M Identification, purification and properties of clone-specific glycoprotein antigens constituting the surface coat of Trypanosoma brucei. Parasitology. 1975; 71:393–417. [DOI] [PubMed] [Google Scholar]
- 38. Myler P.J., Allison J., Agabian N., Stuart K.. Antigenic variation in African trypanosomes by gene replacement or activation of alternative telomeres. Cell. 1984; 39:203–211. [DOI] [PubMed] [Google Scholar]
- 39. Boothroyd C.E., Dreesen O., Leonova T., Ly K.I., Figueiredo L.M., Cross G.A.M., Papavasiliou F.N. A yeast-endonuclease-generated DNA break induces antigenic switching in Trypanosoma brucei. Nature. 2009; 459:278–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Glover L., Alsford S., Horn D. DNA break site at fragile subtelomeres determines probability and mechanism of antigenic variation in African trypanosomes. PLoS Pathog. 2013; 9:e1003260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li B., Espinal A., Cross G.A.M. Trypanosome telomeres are protected by a homologue of mammalian TRF2. Mol. Cell. Biol. 2005; 25:5011–5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang X., Figueiredo L.M., Espinal A., Okubo E., Li B.. RAP1 is essential for silencing telomeric variant surface glycoprotein genes in Trypanosoma brucei. Cell. 2009; 137:99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jehi S.E., Wu F., Li B.. Trypanosoma brucei TIF2 suppresses VSG switching by maintaining subtelomere integrity. Cell Res. 2014; 24:870–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jehi S.E., Li X., Sandhu R., Ye F., Benmerzouga I., Zhang M., Zhao Y., Li B.. Suppression of subtelomeric VSG switching by Trypanosoma brucei TRF requires its TTAGGG repeat-binding activity. Nucleic Acids Res. 2014; 42:12899–12911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jehi S.E., Nanavaty V., Li B.. Trypanosoma brucei TIF2 and TRF suppress VSG switching using overlapping and independent mechanisms. PLoS One. 2016; 11:e0156746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yu T.Y., Kao Y.W., Lin J.J.. Telomeric transcripts stimulate telomere recombination to suppress senescence in cells lacking telomerase. Proc. Natl. Acad. Sci. USA. 2014; 111:3377–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Diman A., Decottignies A.. Genomic origin and nuclear localization of TERRA telomeric repeat-containing RNA: from Darkness to Dawn. FEBS J. 2018; 285:1389–1398. [DOI] [PubMed] [Google Scholar]
- 48. Toubiana S., Selig S.. DNA:RNA hybrids at telomeres - when it is better to be out of the (R) loop. FEBS J. 2018; 285:2552–2566. [DOI] [PubMed] [Google Scholar]
- 49. Wirtz E., Leal S., Ochatt C., Cross G.A.M. A tightly regulated inducible expression system for dominant negative approaches in Trypanosoma brucei. Mol. Biochem. Parasitol. 1999; 99:89–101. [DOI] [PubMed] [Google Scholar]
- 50. Janzen C.J., Lander F., Dreesen O., Cross G.A.M. Telomere length regulation and transcriptional silencing in KU80-deficient Trypanosoma brucei. Nucleic Acids Res. 2004; 32:6575–6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dreesen O., Li B., Cross G.A.M. Telomere structure and shortening in telomerase-deficient Trypanosoma brucei. Nuc Acids Res. 2005; 33:4536–4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kim H.S., Cross G.A.M. TOPO3alpha influences antigenic variation by monitoring expression-site-associated VSG switching in Trypanosoma brucei. PLoS Pathog. 2010; 6:e1000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li B., Oestreich S., de Lange T.. Identification of human Rap1: implications for telomere evolution. Cell. 2000; 101:471–483. [DOI] [PubMed] [Google Scholar]
- 54. Zhong Z., Shiue L., Kaplan S., de Lange T.. A mammalian factor that binds telomeric TTAGGG repeats in vitro. Mol. Cell. Biol. 1992; 12:4834–4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Afrin M., Gaurav A.K., Yang X., Pan X., Zhao Y., Li B.. TbRAP1 has an unusual duplex DNA binding activity required for its telomere localization and VSG silencing. Sci. Adv. 2020; 6:eabc4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wahba L., Costantino L., Tan F.J., Zimmer A., Koshland D. S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation. Genes Dev. 2016; 30:1327–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Woodward R., Gull K.. Timing of nuclear and kinetoplast DNA replication and early morphological events in the cell cycle of Trypanosoma brucei. J. Cell Sci. 1990; 95:49–57. [DOI] [PubMed] [Google Scholar]
- 58. Siegel T.N., Hekstra D.R., Cross G.A.M. Analysis of the Trypanosoma brucei cell cycle by quantitative DAPI imaging. Mol. Biochem. Parasitol. 2008; 160:171–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Perez-Morga D., Amiguet-Vercher A., Vermijlen D., Pays E.. Organization of telomeres during the cell and life cycles of Trypanosoma brucei. J. Eukaryot. Microbiol. 2001; 48:221–226. [DOI] [PubMed] [Google Scholar]
- 60. Feretzaki M., Pospisilova M., Valador Fernandes R., Lunardi T., Krejci L., Lingner J.. RAD51-dependent recruitment of TERRA lncRNA to telomeres through R-loops. Nature. 2020; 587:303–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sandhu R., Sanford S., Basu S., Park M., Pandya U.M., Li B., Chakrabarti K.. A trans-spliced telomerase RNA dictates telomere synthesis in Trypanosoma brucei. Cell Res. 2013; 23:537–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cestari I., McLeland-Wieser H., Stuart K.. Nuclear phosphatidylinositol 5-phosphatase is essential for allelic exclusion of variant surface glycoprotein genes in trypanosomes. Mol. Cell. Biol. 2019; 39: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kerry L.E., Pegg E.E., Cameron D.P., Budzak J., Poortinga G., Hannan K.M., Hannan R.D., Rudenko G.. Selective inhibition of RNA polymerase I transcription as a potential approach to treat African trypanosomiasis. PLoS Negl Trop Dis. 2017; 11:e0005432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Boguslawski S.J., Smith D.E., Michalak M.A., Mickelson K.E., Yehle C.O., Patterson W.L., Carrico R.J.. Characterization of monoclonal antibody to DNA.RNA and its application to immunodetection of hybrids. J. Immunol. Methods. 1986; 89:123–130. [DOI] [PubMed] [Google Scholar]
- 65. Li X., Manley J.L.. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005; 122:365–378. [DOI] [PubMed] [Google Scholar]
- 66. Glover L., Horn D. Trypanosomal histone gammaH2A and the DNA damage response. Mol. Biochem. Parasitol. 2012; 183:78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Briggs E., Crouch K., Lemgruber L., Lapsley C., McCulloch R.. Ribonuclease H1-targeted R-loops in surface antigen gene expression sites can direct trypanosome immune evasion. PLoS Genet. 2018; 14:e1007729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Conway C., McCulloch R., Ginger M.L., Robinson N.P., Browitt A., Barry J.D.. Ku is important for telomere maintenance, but not for differential expression of telomeric VSG genes, in African trypanosomes. J. Biol. Chem. 2002; 277:21269–21277. [DOI] [PubMed] [Google Scholar]
- 69. Afrin M., Kishmiri H., Sandhu R., Rabbani M.A.G., Li B. Trypanosoma brucei RAP1 has essential functional domains that are required for different protein interactions. mSphere. 2020; 5:e00027-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zomerdijk J.C.B.M., Kieft R., Borst P. A ribosomal RNA gene promoter at the telomere of a mini-chromosome in Trypanosoma brucei. Nucleic Acids Res. 1992; 20:2725–2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Devlin R., Marques C.A., Paape D., Prorocic M., Zurita-Leal A.C., Campbell S.J., Lapsley C., Dickens N., McCulloch R.. Mapping replication dynamics in Trypanosoma brucei reveals a link with telomere transcription and antigenic variation. Elife. 2016; 5:e12765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Landeira D., Bart J.M., Van Tyne D., Navarro M.. Cohesin regulates VSG monoallelic expression in trypanosomes. J. Cell Biol. 2009; 186:243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kabani S., Waterfall M., Matthews K.R.. Cell-cycle synchronisation of bloodstream forms of Trypanosoma brucei using Vybrant DyeCycle violet-based sorting. Mol. Biochem. Parasitol. 2010; 169:59–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. McKean P.G. Coordination of cell cycle and cytokinesis in Trypanosoma brucei. Curr. Opin. Microbiol. 2003; 6:600–607. [DOI] [PubMed] [Google Scholar]
- 75. Alsford S., Wickstead B., Ersfeld K., Gull K.. Diversity and dynamics of the minichromosomal karyotype in Trypanosoma brucei. Mol. Biochem. Parasitol. 2001; 113:79–88. [DOI] [PubMed] [Google Scholar]
- 76. Melville S.E., Leech V., Navarro M., Cross G.A.M. The molecular karyotype of the megabase chromosomes of Trypanosoma brucei stock 427. Mol. Biochem. Parasitol. 2000; 111:261–273. [DOI] [PubMed] [Google Scholar]
- 77. Chung H.M., Shea C., Fields S., Taub R.N., Van der Ploeg L.H., Tse D.B.. Architectural organization in the interphase nucleus of the protozoan Trypanosoma brucei: location of telomeres and mini-chromosomes. EMBO J. 1990; 9:2611–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Deng Z., Norseen J., Wiedmer A., Riethman H., Lieberman P.M.. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell. 2009; 35:403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Maudlin I.E., Kelly S., Schwede A., Carrington M.. VSG mRNA levels are regulated by the production of functional VSG protein. Mol. Biochem. Parasitol. 2020; 241:111348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ridewood S., Ooi C.P., Hall B., Trenaman A., Wand N.V., Sioutas G., Scherwitzl I., Rudenko G.. The role of genomic location and flanking 3′UTR in the generation of functional levels of variant surface glycoprotein in Trypanosoma brucei. Mol. Microbiol. 2017; 106:614–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lee Y.W., Arora R., Wischnewski H., Azzalin C.M.. TRF1 participates in chromosome end protection by averting TRF2-dependent telomeric R loops. Nat. Struct. Mol. Biol. 2018; 25:147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Smogorzewska A., Karlseder J., Holtgreve-Grez H., Jauch A., de Lange T.. DNA ligase IV-dependent NHEJ of deprotected mammalian telomeres in G1 and G2. Curr. Biol. 2002; 12:1635. [DOI] [PubMed] [Google Scholar]
- 83. van Steensel B., Smogorzewska A., de Lange T.. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998; 92:401–413. [DOI] [PubMed] [Google Scholar]
- 84. Wang R.C., Smogorzewska A., de Lange T.. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell. 2004; 119:355–368. [DOI] [PubMed] [Google Scholar]
- 85. Burton P., McBride D.J., Wilkes J.M., Barry J.D., McCulloch R.. Ku heterodimer-independent end joining in Trypanosoma brucei cell extracts relies upon sequence microhomology. Eukaryot Cell. 2007; 6:1773–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Woods A., Sherwin T., Sasse R., MacRae T.H., Baines A.J., Gull K.. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J. Cell Sci. 1989; 93:491–500. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data have been included in the manuscript, figures, and supplemental information.