Abstract
Splicing is catalyzed by the spliceosome, a compositionally dynamic complex assembled stepwise on pre-mRNA. We reveal links between splicing machinery components and the intrinsically disordered ciliopathy protein SANS. Pathogenic mutations in SANS/USH1G lead to Usher syndrome—the most common cause of deaf-blindness. Previously, SANS was shown to function only in the cytosol and primary cilia. Here, we have uncovered molecular links between SANS and pre-mRNA splicing catalyzed by the spliceosome in the nucleus. We show that SANS is found in Cajal bodies and nuclear speckles, where it interacts with components of spliceosomal sub-complexes such as SF3B1 and the large splicing cofactor SON but also with PRPFs and snRNAs related to the tri-snRNP complex. SANS is required for the transfer of tri-snRNPs between Cajal bodies and nuclear speckles for spliceosome assembly and may also participate in snRNP recycling back to Cajal bodies. SANS depletion alters the kinetics of spliceosome assembly, leading to accumulation of complex A. SANS deficiency and USH1G pathogenic mutations affects splicing of genes related to cell proliferation and human Usher syndrome. Thus, we provide the first evidence that splicing dysregulation may participate in the pathophysiology of Usher syndrome.
Graphical Abstract
Graphical Abstract.
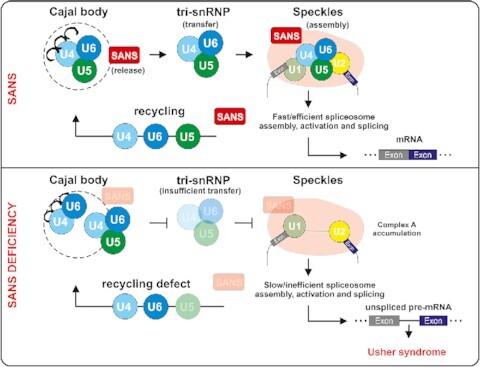
SANS acts as a factor required for the release of tri-snRNPs from Cajal bodies following by recruitment of the tri-snRNP complex to nuclear speckles for spliceosome assembly. SANS may also participate in the snRNP recycling back to the Cajal bodies.
INTRODUCTION
Pre-mRNA splicing is a fundamental process in eukaryotic cells that almost exclusively occurs in the nucleus. Splicing results in the removal of introns and the ligation of exons in a nascent precursor messenger RNA (pre-mRNA) to form mature RNA (mRNA) (1). Splicing allows the inclusion/exclusion of exons/introns in the mRNA that can give rise to the synthesis of multiple alternative protein isoforms from a single genomic locus. This process is catalyzed by the spliceosome, a highly dynamic macromolecular complex composed of five small nuclear ribonucleoproteins (U1, U2, U4/U6, U5 snRNPs) and numerous other polypeptides (1,2). During each splicing cycle, the spliceosome assembles de novo on the pre-mRNA by stepwise recruitment and assembly of the pre-formed snRNPs and non-snRNP proteins (3). The process starts with the accumulation of heterogeneous nuclear ribonucleoproteins (hnRNPs) for the splice site selection (complex H), then, U1 binds to the 5′ splice site (complex E) and U2 snRNP binds to the branch point site of the pre-mRNA, to form complex A. The U4/U6.U5 tri-snRNP is then recruited to complex A to form the pre-catalytic spliceosome (complex B). Complex B undergoes conformational and compositional rearrangements yielding the catalytically active complex B*, which catalyzes the first transesterification reaction. After additional rearrangements, complex C is formed catalyzing the second step of splicing which leads to excision of the intron lariat and ligation of the neighboring exons (2,4,5).
The biogenesis of snRNPs is an intricate process that occurs in cytosolic and nuclear compartments (6). Newly synthesized U snRNAs in the nucleus are exported to the cytoplasm for additional maturation steps, except U6 snRNA, which remains in the nucleus. Subsequently, they are reimported to the nucleus for additional maturation in Cajal bodies, where the final assembly of U2, U4/U6 and U4/U6.U5 snRNP complexes occurs (6–9). Next, mature snRNP complexes are released from Cajal bodies and are transferred to nuclear speckles, where splicing factors are stored until being supplied to active transcription/pre-mRNA splicing sites associated with nuclear speckles (10,11). During spliceosome activation and splicing, snRNPs are sequentially released from the spliceosome and recycled back to the Cajal bodies, where they are available for the formation of new snRNP complexes which facilitate another splicing cycle (2). However, little is known about the molecular transfer between these organelles per se, especially about the release and transfer of the mature U4/U6.U5 tri-snRNPs from Cajal bodies to nuclear speckles and their subsequent recruitment to pre-spliceosomes and the recycling pathways of snRNPs.
Changes in alternative splicing patterns are important for cell development and differentiation under normal conditions (12,13). Aberrant splicing patterns and defects in spliceosomal proteins have been implicated in human diseases (13–15). Notably, pathologic mutations in several genes coding for pre-mRNA-processing factors (PRPF) cause Retinitis pigmentosa (RP) (16). Here, we demonstrate that defects in a protein related to a syndromic retinal ciliopathy, the human Usher syndrome (USH) affects splicing patterns of genes associated with cell proliferation and USH.
USH is the most frequent cause of inherited combined deaf-blindness, a clinically and genetically complex disease (17,18); 10 USH genes have been assigned to three clinical types (USH1-3). USH1 is the most severe form characterized by profound hearing impairment, vestibular dysfunction and RP. Myosin VIIa (USH1B) and scaffold proteins such as harmonin (USH1C), whirlin (USH2D) and SANS (USH1G) organize a common USH protein interactome (19). The association of USH proteins with primary cilia, and defects in ciliogenesis and ciliary maintenance caused by their deficiencies characterize the USH disease as a ciliopathy (20–24). Ciliopathies are mainly considered as molecular defects in ciliary modules, such as ciliary transport or signaling (24); however, ciliopathy genes were also found to be related to the machineries associated with translation and DNA damage-repair processes (25–28). In the present study, we provide the first evidence for molecular and functional links between a ciliary protein, namely the USH1G protein SANS and the pre-mRNA splicing machinery.
The USH1G protein SANS is a scaffold protein comprised of several protein-binding domains able to form homodimers (Figure 1A) (29,30). Like other USH proteins, SANS is essential for auditory hair-cell development and is part of the signal transduction complex at the tips of stereocilia where it forms high-density protein condensates via liquid-liquid phase separation (31–33). In the eye, the role of SANS and the ophthalmic pathogenesis pathway for USH1G are less clear. SANS is expressed in the retinal photoreceptor cells and glia cells, where it is thought to organize networks of USH proteins that may promote mechanical stabilization of the photosensitive ciliary outer segment (24,34) and participate in intracellular transport processes (17,21,29,35). Recently, we reported a role for SANS in endocytosis and primary ciliogenesis (21,35,36). Here, we demonstrate the interaction of SANS with key components of the splicing machinery including splicing factor 3B subunit 1 (SF3B1) and SON, a large scaffolding protein known as a splicing cofactor. We show that SANS is necessary for the release and transfer of mature tri-snRNPs from Cajal bodies to nuclear speckles required for the assembly of catalytic spliceosomes. In addition, we provide evidence for a participation of SANS in the recycling of snRNPs back to the Cajal bodies. By these mechanisms SANS controls splicing of target genes including USH genes. Our results suggest that defective alternative splicing of USH genes may lead to the sensorineural disorders caused by pathogenic variations in the SANS/USH1G gene.
Figure 1.
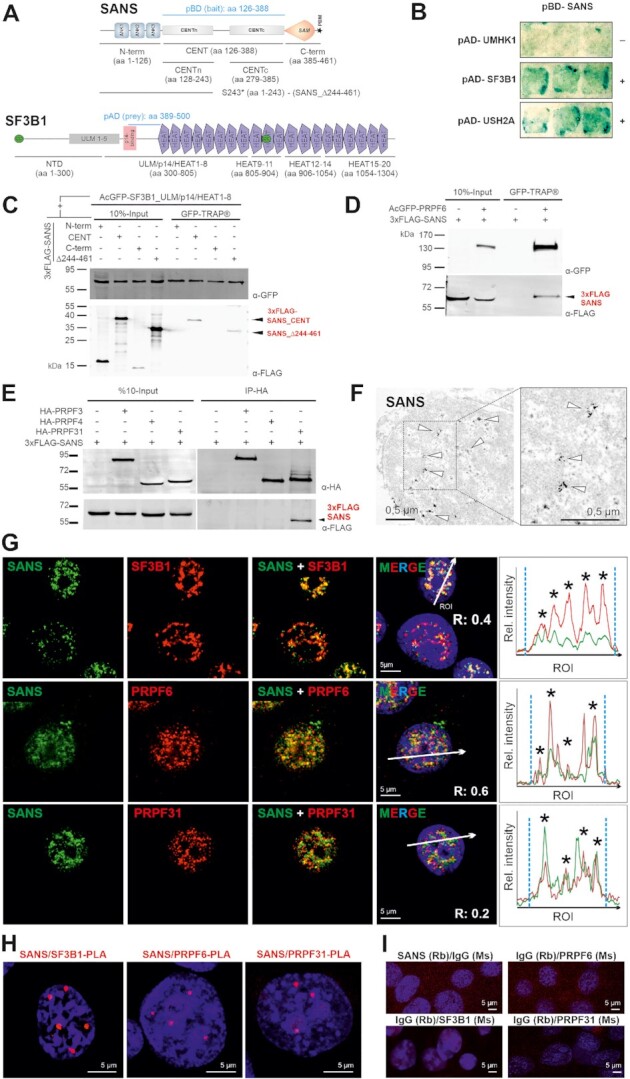
SANS interacts and co-localizes with SF3B1, PRPF6, and PRPF31 in the nucleus. (A) Domain structures of SANS and SF3B1. SANS is composed of three ankyrin (ANK) repeats, the central domain (CENT), divided into the N-terminal CENTn and the C-terminal CENTc, the sterile alpha motif (SAM) and PDZ binding motif (PBM, asterisk). SF3B1 contains the N-terminal domain (NTD) including UHM (U2AF-homology motif) ligand motifs (ULM1-5), p14-binding site, 20 HEAT (Huntingtin, elongation factor 3/EF3, protein phosphatase 2A/PP2A, and yeast kinase TOR1) repeats and two coiled-coil domains (CC, green). Bait and prey (light blue) used for the Y2H screen and deletion constructs are indicated. (B) 1:1 Y2H ß-galactosidase reporter assay shows binding of pBD-SANS_CENT bait to pAD-SF3B1 prey (aa 389–500) containing parts of the p14 and HEAT1 domain (middle panel) and to pAD-USH2A prey (bottom panel) used as positive control but no binding to U2AF homology motif kinase 1 (UMHK1) (negative control). (C) Western blot of anti-FLAG IPs from lysates of cells expressing different 3xFLAG-SANS and deletion constructs of acGFP-SF3B1. SF3B1 binds to SANS_CENT and the SANS truncation variant p.S243* (SANS_Δ244–461) reveal CENTn as binding site. (D, E) Western blot of GFP-TRAP® or anti-HA IPs from HEK293T cells co-expressing 3xFLAG-SANS and GFP- or HA-tagged PRPF31, respectively, demonstrates interaction of SANS with PRPF6 and PRPF31. (F) Immunoelectron microscopy (anti-SANS RpAb) showing SANS localization (arrowheads) in the nucleoplasm of a neuron from the inner nuclear layer of a human retina. (G) Double immunofluorescence of SANS (MmAb) with SF3B1, and SANS (RpAb), PRPF6, and PRPF31 in nuclei of HEK293T cells counterstained with DAPI. Intensity plots for regions of interest (white arrows) indicate co-localization of SANS with SC35 in nuclear speckles (asterisks). Blue vertical dashed lines indicate the nucleus extent. (H) PLAs for SANS in combination with SF3B1, PRPF6 or PRPF31 reveal their interaction in the nucleoplasm of HEK293T cells. (I) Negative controls for PLAs in H.
MATERIALS AND METHODS
Antibodies
We used the following primary antibodies: rabbit polyclonal antibodies against SANS (RpAb) (29,37), anti-SANS (mouse, monoclonal antibody; MmAb) (sc-514418, Santa Cruz), anti-FLAG (F1804, Sigma Aldrich), anti-PRPF6 (sc-48786, Santa Cruz), anti-HA (Roche), anti-GFP (ab6556, Abcam), anti-PRPF31 (PAB7154, Abnova), anti-hSNU114 (38), anti-Coilin (sc-55594, Santa Cruz), anti-SC35 (sc-53518, Santa Cruz), anti-SON (ATLAS, HPA023535), anti-SF3B1 (MmAb) (sc-514655, Santa Cruz), anti-SF3B1 (RmAb) (Will et al. 2001), anti-U5 52K (39), anti-SART1 (PA556663, Thermo Fisher) and anti-PRPF38 (40). Secondary antibodies were conjugated to the following fluorophores: Alexa488 (donkey-anti-rabbit, A21206, Molecular Probes,), CF 488 (donkey-anti-guinea pig, 20169-1, Biotrend,), Alexa488 (donkey-anti-mouse, A21202, Molecular Probes), Alexa 555 (donkey-anti-mouse, A31570, Molecular Probes), Alexa 568 (donkey-anti-rabbit, A10043, Invitrogen,) CF640 (donkey-anti-goat, 20179-1, Biotrend), Alexa 680 (donkey-anti-goat, A21084, Molecular Probes), Alexa 680 (goat-anti-rat, A21096, Molecular Probes) Alexa 680 (donkey-anti-rabbit, A10043), IRDye 800 (donkey-anti-mouse, 610–732-124, Rockland), Abberior STAR Orange (goat-anti-mouse, Abberior,), and Abberior STAR Red (goat-anti-Rabbit). Nuclear DNA was stained with DAPI (4’,6-diamidino-2-phenylindole) (1 mg/ml) (Sigma-Aldrich).
DNA constructs
3xFLAG-tagged SANS full length and deletion constructs were designed and cloned in house as previously described (29). HA-PRPF31 constructs were kind gifts from Dr. Utz Fischer (41). Deletion constructs of PRPF31 (N-term (aa1–165), C-term (aa 166–499)) were produced by PCR from the HA-PRPF31 plasmid and cloned into the pcDNA3.1 N-HA vector (Thermo Fisher). Deletion constructs of PRPF6 (NTD (aa 1–126), HAT1-6 (aa 293–592), HAT7-13 (aa 593–941)) were cloned into AcGFP-C1 vector (Addgene: 64607). RON (MST1R) minigene was a kind gift from Dr. Julian Koenig (IMB-Mainz), and USH1C minigene was amplified from the genomic DNA of HEK293T cells and cloned into the pcDNA5/FRT vector (Thermo Fisher). The pcDNA5/FRT-PRPF4 plasmid was constructed by cloning the PRPF4 gene fused to an N-terminal FLAG/HA tag into the pcDNA5/FRT plasmid (Thermo Fisher).
Yeast two-Hybrid (Y2H)
Yeast two-hybrid (Y2H) screens were performed using the HybriZAP two-hybrid cDNA synthesis kit (Stratagene, La Jolla, USA) as previously described (35). The DNA binding domain (pBD) was fused to the CENT domain of human SANS (amino acids 126–368) (NCBI: NM_173477) and used as bait in a bovine oligo-dT primed retinal cDNA library. The interactions were analyzed by evaluating the activation of the β-galactosidase reporter gene. The interaction between a protein pair was detected by staining the yeast clones in X-ß-gal colorimetric filter lift assay (LacZ reporter gene). For reciprocal 1:1 Y2H assay, bovine SF3B1 (aa389–500) prey fused to the DNA-activation domain (pAD) of the GAL4 transcription factor and human SANS_CENT domain fused to the DNA-binding domain (pBD) of GAL4 transcription factor were used as bait and as prey, respectively. Subsequently, bait and prey were transformed into yeast strains PJ694a and PJ694A, respectively, which were mated and yeast colonies were selected using HIS1, ADE3, LacZ, and MEL1 reporter genes to identify positive interactions confirmed by X-ß-gal colorimetric filter lift assay.
Immunoprecipitations/pull-down assays
GFP-TRAP® (Chromotek) was performed according to the manufacturer's protocol. Briefly, GFP-tagged proteins co-expressed with an indicated 3xFLAG-tagged protein in HEK293T cells. Sixteen hours post-transfection, cells were lysed with Triton X-100 lysis buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, and 0.5% Triton X-100) containing protease inhibitor cocktail (PI mix; Roche). Lysates were incubated with the beads for 2 h at 4°C followed by several washing steps with 10 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA. Bound proteins were eluted with 2x Laemmli buffer, separated by SDS-PAGE followed by western blotting. FLAG and HA immunoprecipitations (IP) were carried out by using the ANTI-FLAG® (Sigma) and Anti-HA Affinity Gel (Biotool) beads.
Mass spectrometry analysis
Proteins were separated on 4–12% Bis–Tris–HCl (pH 7.0) NuPAGE polyacrylamide gels (Invitrogen) and stained with G-colloidal Coomassie Brilliant Blue. Lanes of the Coomassie-stained gel were cut into 23 slices and processed for mass spectrometry as previously described (42). A comparison of the number of peptides identified for each protein in two samples was used as a reliable indication of the relative number of proteins when they are analyzed using identical chromatographic and mass-spectrometric conditions and the same mass spectrometer.
Cell culture and cell lines
Dulbecco's modified Eagle's medium (DMEM) and DMEM-F12 containing 10% heat-inactivated fetal calf serum (FCS) (Thermo Fisher) were used to culture human HEK293T (ATCC), HeLa cells (ATCC), Flp-In-293 (Thermo Fisher), and murine IMCD3 (ATCC) cells, respectively. Plasmids were delivered to the cells by using Plus Reagent and Lipofectamine LTX (Invitrogen) following the manufacturer's protocol. HEK293T cells stably expressing FLAG/HA-tagged human PRPF4 was generated using the pcDNA5/FRT-PRPF4 plasmid and Flp-In-293 cells according to the manufacturer's protocol (Thermo Fisher). Briefly, Flp-In-293 cells were transfected with a 9:1 ratio of pOG44:pcDNA5/FRT-PRPF4 plasmids using Lipofectamine 2000 (Invitrogen) and 48 h post-transfection, cells were selected in medium containing hygromycin. Stable clones resistant to hygromycin were expanded and tested for stable expression of FLAG-PRPF4 by Western blotting.
Knockdown and quantitative RT-PCR
siRNAs against human SANS (IDT), PRPF6 (Sigma), PRPF31 (IDT), SRSF1 (Sigma) SF3B1 (Sigma) and SON (Dharmacon) were purchased as indicated in Supplementary Table S3. Non-targeting control siRNA (siCtrl) was purchased from IDT. For knockdowns, HEK293T and HeLa cells were transfected with a final concentration of 20 nM siRNAs using Lipofectamine RNAiMAX (Invitrogen) following the manufacturer's protocol. Subsequently, total RNA was isolated from transfected cells using a Qiagen RNA isolation kit and converted to cDNA with SuperScript™ III First-Strand Synthesis SuperMix (ThermoFisher) according to manufacturer's protocols by using Oligo (dT) primers. For the validation of knockdowns, quantitative PCR (qPCR) was performed using gene specific primers (Supplementary Table S6) and iTaq Universal SYBR Green Supermix (Bio-Rad). Cq values were detected with a CFX Connect Real-Time PCR Detection System (Bio-Rad). Gene expression profiles were calculated with ΔΔCq methods based on normalization to GAPDH (for primers, see Supplementary Table S6). SANS siRNA knockdown efficiency was also validated by Western blotting and immunofluorescence analysis in HEK293T and HeLa cells (Supplementary Figure S3).
Immunocytochemistry and light microscopy
Cells were fixed with methanol for 10 min at –20°C or with 4% Paraformaldehyde (PFA) for 10 min at room temperature (RT) and washed with PBS. Permeabilization was performed with 0.1% Triton-X100 (Roth) for 5 min at RT, followed by washes with PBS and blocking for 45 min with 0.5% cold-water fish gelatin, 0.1% ovalbumin in PBS. Primary antibodies were incubated overnight at 4°C, followed by washing with PBS and incubation with secondary antibodies for 1 h at RT. After three washes, specimens were mounted with Mowiol 4.88 (Hoechst) and imaged using a Leica DM6000B microscope (Leica). For STED microscopy, we applied secondary antibodies from Abberior and imaged specimens on an Abberior 3D-STED Facility Line microscope (Abberior).
Image processing and quantification
Image processing was performed using a Leica DM6000B microscope (Leica), Leica imaging and ImageJ/Fiji software (43,44). Deconvolutions were carried out with the Leica program by using 5–12 Z-stacks with one iteration step and BlindDeblur Algorithm settings. Mean fluorescence intensities of the accumulations in Cajal bodies and nuclear speckles were quantified by the Cell Profiler program as previously applied (4). The ratio of the fluorescence intensities of Cajal bodies and nucleus or speckles and nuclei were compared between control and knockdowns. Mean intensities and standard deviation values were obtained from three independent experiments. The threshold settings were adjusted as follows: green channel: manual threshold: 0.1; red channel: manual threshold 0.05. The primary object sizes (Cajal bodies and nuclei) were set from 2 to 40 and 40 to 100 diameters of objects in pixel units, respectively. To ascertain the degree of association between various proteins based on their proximity in immunofluorescent images, we used the Coloc2 plugin of the ImageJ/Fiji program to calculate the Pearson correlation coefficient of this co-distribution.
Electron microscopy
For immunoelectron microscopy we applied a previously established pre-embedding labeling technique (45,46). Specimen were analyzed and documented in a Tecnai 12 BioTwin transmission electron microscope (FEI) as previously described. Images were processed using Adobe Photoshop CS (Adobe Systems).
Proximity ligation assay (PLA)
For in situ proximity ligation assay Duolink PLA probes anti-rabbitPLUS, anti-mouseMINUS, and Detection Reagent Red were purchased from Sigma. PLA was performed on HEK293T cells as previously described (29). Briefly, cells fixed in buffered 2% paraformaldehyde were incubated with the primary antibodies overnight at 4°C, followed by incubation with oligonucleotide-labelled secondary antibodies (‘PLA probes’) for 4 h at 37°C. After several washing steps hybridizing connector oligonucleotides were added and ligation was performed for 30 min at 37°C, to form a closed circle template. This was followed by rolling circle amplification for 100 min, addition of fluorescent-labelled oligonucleotides and analysis by fluorescence microscopy. For the negative controls, samples probed with only one protein-specific antibody and paired with either the rabbit or mouse IgG specific oligonucleotide-labelled antibody. Cells were mounted in Mowiol 4.88 (Hoechst) and analyzed with a Leica DM6000B microscope.
RNA-fluorescence in situ hybridization (RNA-FISH)
For RNA-fluorescence in situ hybridization, we used previously described snRNA probes 5′-end-labeled with Alexa-488 fluorescent dye (Invitrogen) in HEK293T cells (47,48). For RNA-FISH, we fixed and permeabilized cells as described for immunocytochemistry. Cells were incubated with RNA-FISH probes for 2 h at 40°C by using ViewRNA™ ISH Cell Assay Kit system (Thermo Fischer). Subsequently, cells were immune-stained and mounted in Mowiol 4.88 (Hoechst) and analyzed with a Leica DM6000B microscope.
Cell proliferation assay
The proliferation levels of the cell were tested WST-1 cell proliferation assay (Roche) kit according to the manufacturer's protocol. In brief, after the seeding 5×103 cells/well in 96 well plates, siRNA-mediated knockdowns were performed 24 h later as described above. Seventy-two hours after knockdown, cells were treated with 10 μl cell Proliferation Reagent WST-1 and incubated for 4 h at 37°C and 5% CO2. The metabolic activity of the cells was detected with Varioskan Flash (Thermo Fisher) at 460 nm wavelength to compare the difference between control and siRNA knockdowns.
Analysis of alternative splicing in vitro
Total RNAs were isolated and reverse transcribed using the Superscript III enzyme (Invitrogen) following the manufacturer's protocol. For the analysis of alternative splicing events of the minigenes and genes, forward and reverse primers against alternatively spliced exons were used to amplify each splice variant with BioTherm Taq polymerase (GeneCraft) (primer list, Supplementary Table S6). In brief, 48 h after siRNA transfection, minigenes were delivered to the siRNA-depleted cells using Lipofectamine 2000, and 24 h later, cells were harvested. Levels of splice variants were analyzed by TAPE 2200 capillary electrophoresis (Agilent) using high throughput D1000 ScreenTapes (Agilent). Data values were obtained from the 2200 Tape station controller program. Quantifications of the isoform ratios were carried out as previously described (1). Briefly, the median percent spliced in (PSI) indexes were obtained from biological triplicates of siRNA knockdowns. These values summarized as robust Z scores according to the effects of each siRNA treatment with the statistical tests explained in (1). Quantile-quantile plots (Q–Q plots) and Pearson′s correlation coefficient values were calculated in excel by using scaled Z-scores of each individual group (siPRPF6, siPRPF31 and siSRSF1) versus the siSANS group. Graphs were prepared by plotting siSANS groups against the siPRPF6, siPRPF31 and siSRSF1 groups, individually.
In vitro splicing and spliceosome assembly assays
In vitro splicing assays were performed as previously described (49). Briefly, nuclear extracts were prepared from HEK293T cells treated with control and SANS siRNAs. Radioactive [32P]-labelled MINX pre-mRNA was incubated with 40% (v/v) nuclear extracts in the splicing buffer containing 20 mM HEPES–KOH pH 7.9, 3 mM MgCl2, 65 mM KCl, 2 mM ATP and 20 mM creatine phosphate at 30°C for the indicated time points. The isolated RNAs either were separated on a denaturing 15% polyacrylamide gel containing 7 M urea and the signals were detected by autoradiography or analyzed by RT-qPCR. The cq values were normalized to controls to obtain relative fold change of snRNA bindings.
For the analysis of the spliceosomal complex formation, the splicing reactions (20 μl) were stopped at the indicated time points by the addition of 2 μl heparin (5 mg/ml) and were analyzed by 2% native-agarose gel electrophoresis followed by autoradiography. SANS depleted nuclear extracts are prepared from siSANS-treated HEK293T cells.
Purification of the tri-snRNP complex and rescue experiments
Purification of the tri-snRNP complex from HeLa nuclear extract was performed as previously described (50) with minor modifications. Briefly, the nuclear extract in the Roeder C buffer containing 20 mM HEPES pH 7.9, 1.5 mM MgCl2, 450 mM KCl, 0.2 mM EDTA and 25% sucrose, was clarified twice by centrifugation for 30 min. at 20 000 rpm. Next, 3 ml of the cleared supernatant was loaded onto a 20–50% sucrose gradient in 20 mM HEPES pH 7.9, 150 mM KCl, 5 mM MgCl2 and 0.1 mM EDTA in a Surespin rotor and centrifuged at 30 000 rpm for 40 h at 4°C. After gradient fractionation, fractions were analyzed by 4–12% NuPAGE (Invitrogen) and the U4, U5 and U6 snRNAs were detected by SYBR gold (Invitrogen) staining. tri-snRNP containing fractionations were pooled and centrifuged overnight in an S58A rotor at 25 000 rpm at 4°C. Pelleted tri-snRNPs were dissolved in G250 buffer containing 20 mM HEPES pH 7.9, 250 mM KCl, 5 mM MgCl2, 0.1 mM EDTA. To further purify the tri-snRNPs, the sample was loaded onto a 5–20% Sucrose gradient in gradient buffer (G150: 20 mM HEPES pH 7.9, 150 mM NaCl, 1.5 mM MgCl2 and 0.5 mM DTT) in a Surespin rotor and centrifuged for 16 h at 25 000 rpm at 4°C. After analyzing fractions on a 4–12% NuPAGE, tri-snRNP containing fractions were detected by SYBR gold staining. Purified tri-snRNP complex was directly used in the chase experiment at the zero time point.
Glycerol gradient ultracentrifugation
To analyze the levels of snRNPs by gradient fractionation, 200 μg of the siRNA-mediated SNAS depleted HEK293T nuclear extract or the control nuclear extract were diluted with an equal volume of G150 buffer and sedimented on linear 4 ml 10–30% (v/v) glycerol gradients in the G150 buffer. The ultracentrifugation was performed in a Sorvall TH-660 rotor for 14 h at 29,000 rpm (114 000 × g) and the gradients were separated into 24 fractions. The levels of snRNPs in each fraction were analyzed by Northern blotting using 5′-end radiolabeled DNA probes against U1, U2, U4, U6 and U5 snRNAs, as previously described (51).
RESULTS
SANS interacts with splicing factors SF3B1, PRPF6, and PRPF31
To identify putative interactive partners for SANS, we performed yeast two-hybrid (Y2H) screens of a retinal cDNA library with the central domain of the USH1G gene SANS (SANS_CENT, aa 126–388) (Figure 1A). Our screen revealed the splicing factor 3B subunit 1 (SF3B1) as a putative SANS binding partner. SF3B1, also known as SF3b155 or SAP155, is a component of the U2 snRNP complex of the spliceosome (52,53). 1:1 Y2H assays confirmed the interaction of SANS_CENT with the central region of SF3B1 (aa 389–500) (Figure 1B). Reciprocal co-immunoprecipitations (co-IPs) of recombinant GFP- and FLAG-tagged proteins co-expressed in HEK293T cells confirmed the interaction of both proteins and delineated the SF3B1_ULM/p14/HEAT1-8 domain and the SANS_CENT domain as binding sites (Figure 1C, Supplementary Figure S1A). Co-IPs in Supplementary Figure S1B demonstrated that SANS does not bind to the GFP control.
Because defects in several components of the spliceosomal U4/U6.U5 tri-snRNP lead to RP, the ocular phenotype in USH patients (16), we examined whether SANS also interacts with other components of the spliceosome. To this end, we focused on the U4/U6-specific protein PRPF31, the U5 protein PRPF6 as well as PRPF3 and PRPF4, which are all components of the U4/U6.U5 tri-snRNP complex and all linked to RP. Western blot analysis of co-IPs from HEK293T cells co-expressing FLAG-SANS and GFP-PRPF6 or HA-PRPF31, PRPF3 and PRPF4 revealed that both PRPF6 and PRPF31 also interact with SANS through its CENTn domain (Figure 1D, E and Supplementary Figure S1C-D′). In contrast, PRPF3 and PRPF4 did not co-IP with FLAG-SANS and likely do not interact in vivo (Figure 1E).
In silico structural analysis of SANS using the online tools FoldIndex© (54) and DisEMBL™ (55) predicted the SANS CENTn domain (aa 120–250) as an intrinsically disordered region (Supplementary Figure S1E). Such regions are characteristics of intrinsically disordered proteins (IDPs) which can bind to diverse proteins in a context-specific manner and their weak multivalent interactions are the basis for phase separation in cells (56).
SANS interacts with SF3B1 and PRPFs within the nucleus
Next, we addressed where in the cell SANS interacts with the spliceosomal proteins. Immunofluorescence and immunoelectron microscopy demonstrated the localization of SANS in nucleoplasma of the nucleus (Figure 1F). Its localization in the nucleus is consistent with in silico analysis using NLStradamus (57), which predicted a nuclear localization sequence (NLS) in the C-terminus of human SANS (aa436-RKKILGAVRRRR-aa447) (Figure 1A). This was confirmed by NucPred, which rated SANS with a high score (0.76) as a putative NLS-containing protein (58). Immunofluorescence analysis also revealed the apparent co-localization of SANS with SF3B1, PRPF6, and PRPF31 in the nucleoplasm (Figure 1G). The degree of co-distribution was confirmed by calculating Pearson coefficients above zero (R = 0.4, 0.6, and 0.2, respectively, for SF3B1, PRPF6, and PRPF31) and by fluorescence intensity plots (Figure 1G).
The nuclear co-localization of SANS and SF3B1, PRPF6 and PRPF31 was also confirmed by proximity ligation assays (PLAs). The PLAs using SANS-SF3B1, SANS-PRPF6 and SANS-PRPF31 pairs revealed PLA signals in the nucleus for all three combinations (Figure 1H), which were absent in the negative controls (Figure 1I). These data demonstrated a close spatial proximity of the probed proteins (∼30–40 nm) in the nucleus and suggested the presence of SANS in complexes with SF3B1, PRPF6 and PRPF31. Taken together, our results strongly support the molecular interaction of SANS with the splicing factors SF3B1, PRPF6 and PRPF31 in the nucleus.
SANS binds to spliceosomal snRNP complexes and pre-mRNA
To test whether SANS interacts with spliceosomal snRNP complexes, we performed co-IPs with nuclear extracts from HEK293T cells expressing FLAG-tagged SANS or PRPF4, a core component of the U4/U6.U5 tri-snRNP, in absence or in the presence of an in vitro synthetized MINX pre-mRNA to induce spliceosome formation (Supplementary Figure S2). Initial Northern blot analysis of PRPF4 IPs revealed strong hybridization signals for U4, U5 and U6 snRNAs indicating a strong pull-down of the U4/U6.U5 tri-snRNP by PRPF4 in the absence or in the presence of pre-mRNA (Supplementary Figure S2A, B). In contrast, SANS did not significantly pull down any of the U4, U5 and U6 snRNAs in the absence of MINX pre-mRNA while it pulled down these snRNAs after inducing spliceosome formation by the addition of MINX pre-mRNA under splicing conditions (Supplementary Figure S2A, B). To validate these data, we additionally analyzed the snRNAs in pull-downs of SANS and PRPF4 by RT-qPCR (Supplementary Figure S2C-D). Consistently, we observed results similar to the initial Northern blots (Supplementary Figure S2C). Quantification of three independent experiments revealed that snRNA binding to SANS was drastically increased under splicing conditions. Analysis of RT-qPCRs for snRNAs also demonstrated the presence of significant amounts of MINX pre-mRNA in both SANS and PRPF4 pull-downs when compared with the mock-transfected controls indicating the interaction of SANS with pre-mRNA (Supplementary Figure S2D). Taken together, our in vitro data suggests that SANS is not stably associated with snRNPs, but interacts with the spliceosomal complexes assembled on MINX pre-mRNA under splicing conditions.
SANS is present in nuclear speckles and its depletion alters the speckles morphology
Since nuclear speckles are important sites for splicing factor storage, modification and splicing (11), we tested whether SANS is present in these nuclear bodies. We co-stained SANS and the nuclear speckle marker SC35, also known as SRSF2. Deconvolution microscopy revealed the overlap of the fluorescent signals for SANS and SC35 in nuclei (Figure 2A), affirmed by merging fluorescence intensity plots and by a positive Pearson correlation coefficient value (R = 0.5) (Figure 2A–A′). 3D-STED super-resolution microscopy confirm the close association of both molecules at the nuclear speckles (Figure 2B) as previously found for other speckle-resident proteins by super-resolution microscopy (59).
Figure 2.
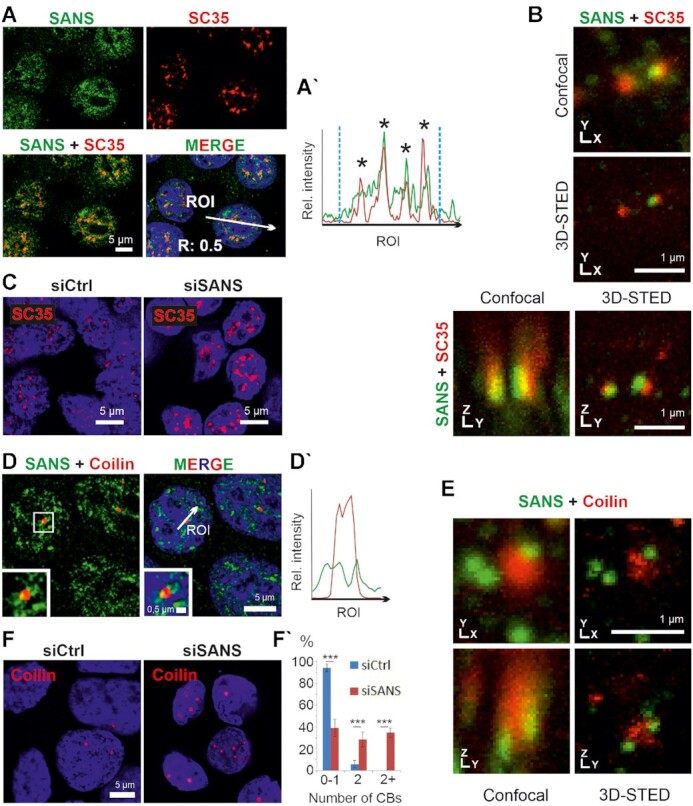
Localization of SANS in nuclear speckles and Cajal bodies. (A) Immunofluorescence staining of SANS (green) and the nuclear speckle marker/spliceosomal protein SC35 (red) in nuclei of HEK293T cells counterstained with DAPI (blue). (A′) Intensity plots for the regions of interest in A (white arrows) indicate co-localization of SANS and SC35 in nuclear speckles (asterisks). Blue vertical dashed lines indicate nuclear extensions. Positive Pearson′s correlation coefficient (R) supports co-localizations of both proteins. (B) 3D-STED microscopy images of SANS and SC35 co-stained speckles. XY and YZ axes demonstrate close association of SANS with SC35 in nuclear speckles. (C) Immunofluorescence of SC35 (red) in siRNA-treated HEK293T cells counterstained with DAPI (blue). Nuclear speckles size increases in SANS-depleted cells (siSANS) comparted with control siRNA-treated cells (siCtrl). (D) Double immunofluorescence of SANS (RpAb, green) and Coilin (red), a molecular marker for Cajal bodies in HEK293T cells counterstained with DAPI (blue). (D′) Fluorescence intensity plots for region of interest indicated by arrow in D reveals SANS localization at the periphery of Cajal bodies in nuclei. (E) 3D-STED microscopy images of SANS (RpAb, green) and Coilin (red). XY and YZ axes demonstrate localization of SANS at the periphery of Cajal bodies. (F) Immunofluorescence of Cajal bodies by anti-Coilin (red) in SANS-depleted HEK293T cells. (F′) Quantification of the number of Cajal bodies (CBs) in SANS-depleted cells; graph shows the percentage of cells with 0–1, 2 or more CBs. SANS depletion increases the number of Cajal bodies in nuclei (In 4 independent experiments 200 and 160 cells were counted in siCtrl and siSANS treated cells, respectively.). ***P values < 0.001.
To test whether SANS plays a functional role upon association with nuclear speckles, we next explored the effect of SANS depletion. To this end, we depleted SANS in HEK293T cells using a validated SANS siRNA (siSANS) and immunostained cells for SC35 (Figure 2C and Supplementary Figure S3A–D; Supplementary Table S1). Confocal microscopy revealed significantly increased and diffused SC35 positive speckles when compared with controls. To exclude off-target effects and cell specificity, we performed the same experiment with an additional siRNA (siSANS #2 recognizing a different region of SANS mRNA) and in HeLa cells as a second cell line. These showed similar diffused speckle formation (Supplementary Figures S3D and S4A). These data demonstrated that SANS has a functional role in nuclear speckles where large amounts of the spliceosome components are stored until they are supplied to active transcription/splicing sites or for subsequent post-transcriptional splicing (60,61).
SANS is located at the periphery of Cajal bodies, and its depletion increases the abundance of Cajal bodies
PRPF6 and PRPF31 participate in both the U4/U6.U5 tri-snRNP assembly and its integration into complex A to form pre-catalytic spliceosomes (62). The assembly and maturation of the tri-snRNP complex occur in separate membrane-less sub-nuclear compartments, called Cajal bodies (4). After maturation, tri-snRNP complexes are released from Cajal bodies and stored in nuclear speckles until being recruited to complex A formed at the active transcription/splicing sites (4,47,63). To investigate a possible role of SANS for the assembly of the tri-snRNP complex in Cajal bodies, we first determined whether SANS is localized to Cajal bodies by double immunofluorescence staining of SANS and Cajal body marker coilin, in HEK293T (Figure 2D–D′). Image analyses revealed that SANS is localized at the periphery of Cajal bodies. 3D-STED super-resolution microscopy corroborated the localization of SANS at the rim of Cajal bodies (Figure 2E).
Next, we investigated the role of SANS for the integrity and organization of Cajal bodies by analyzing the effect of SANS knockdown on them. Interestingly, siRNA-mediated SANS depletion led to increased abundance of Cajal bodies both in HEK293T and in HeLa cells (Figure 2F-F′and Supplementary Figure S4B–B′). Consistent with this, knockdown of SANS with a second siRNA (siSANS #2) increased the number of Cajal bodies, despite the milder effect, likely due to the reduced knockdown efficiency of this siRNA (∼50%) compared with siSANS (Supplementary Figure S3A, E–E′ and Supplementary Table S1).
Affinity proteomics reveals interactions of SANS with SON in the nucleus
To obtain a comprehensive view of splicing factors associated with SANS in the nucleus, we carried out a proteomic screen by affinity capture of SANS-binding proteins from nuclear extracts. We immunoprecipitated SANS-associated complexes from nuclear extracts of HEK293T cells expressing 3xFLAG-SANS and analyzed the protein content of the immunoprecipitates by mass spectrometry (MS) (Figure 3A). The MS data identified 730 proteins from the 3xFLAG-SANS nuclear extract with more than a five-fold change in peptide numbers when compared with the mock-transfected control (Supplementary Table S2A). Gene Ontology enrichment analysis for significantly enriched proteins by DAVID 6.8 (64) revealed ‘mRNA processing’ and ‘RNA splicing’ among the enriched biological processes and ‘nuclear speckles’ as one of the most enriched cellular components (Supplementary Table S2B–C).
Figure 3.
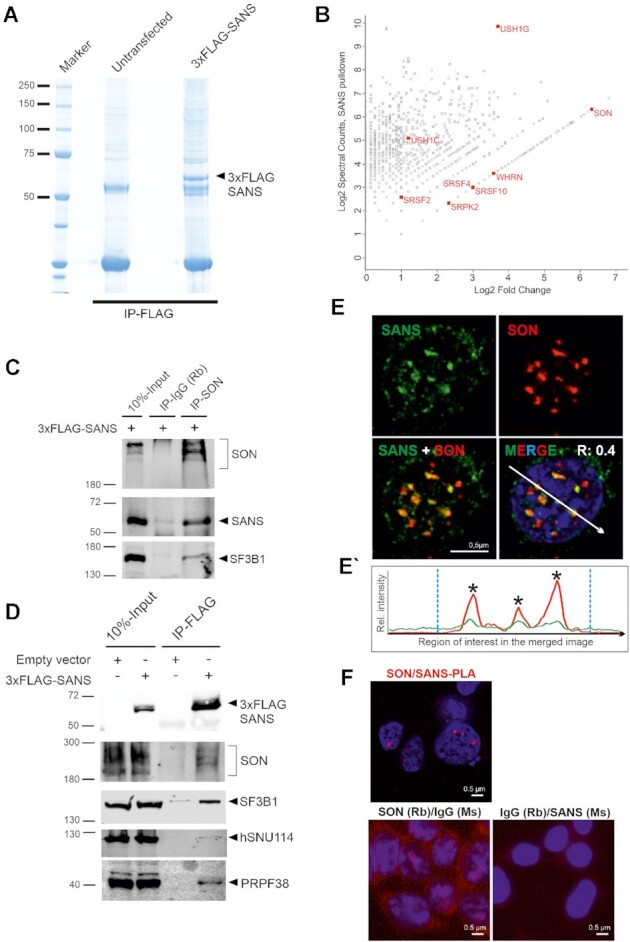
Affinity proteomics and validation of SANS interactions in the nucleus. (A) Coomassie blue staining of anti-FLAG immunoprecipitated proteins from nuclear extracts of control cells and cells expressing 3xFLAG-SANS, loaded onto a 4–12% NuPAGE. (B) Proteomic analysis of SANS interaction partners. Graph displays the relative abundance, in logarithmic scale, of proteins identified in 3xFLAG-SANS pull-down from nuclear extract compared with the control. Each dot represents a protein; SR-related splicing factor SON, SR splicing proteins and the SR protein kinase SRPK2 as well as the identified USH proteins are highlighted in red. (C) Validation of SON-SF3B1-SANS interactions by IP of intrinsic SON in cells expressing 3xFLAG-SANS. SON interacts with SANS and SF3B1. (D) Western blot analysis of anti-FLAG immunoprecipitations from control and 3xFLAG-SANS nuclear extracts revealed SANS interactions with SON, SF3B1, hSNU114, and PRPF38. (E) Double immunofluorescence staining of endogenous SANS (MmAb) and SON in HEK293T cells counterstained with DAPI for DNA. Positive Pearson′s correlation coefficient (R) supports co-localization of SANS and SON. (E′) Fluorescence intensity plots for the region of interest in D (white arrow) shows co-localization of SANS with SON in the nucleus. Blue vertical dashed lines indicate the boundaries of the nucleus. (F) PLAs demonstrate interaction of SANS with SON in cells. Red PLA signals are present in the nucleoplasm in SANS-SON PLAs but absent in the negative controls anti-SANS (MmAb) or rabbit anti-SON with mouse and rabbit IgG antibodies.
In addition to known interactors of SANS (e.g. harmonin/USH1C and whirlin/USH2D (30,65), the MS results identified SON, a large, SR protein-related splicing factor, as one of the most enriched proteins (Figure 3B, Supplementary Table S2A). SON serves as an important scaffold protein for the organization of splicing factors in nuclear speckles (66,67). Reciprocal co-IPs from nuclear extract of HEK293T cells expressing FLAG-SANS revealed efficient pull-down of SON with SANS as well as co-precipitation of SANS with anti-SON (Figure 3C–D). In agreement with the interaction of SANS-SON, immunocytochemistry and PLAs revealed co-localization and spatial proximity of SANS and SON in nuclear speckles in situ (Figure 3E–F). Furthermore, Western blotting of co-IPs for other spliceosomal core proteins demonstrated that SF3B1, hSNU114 and PRPF38 were also co-precipitated with SANS (Figure 3D). These data corroborate the interaction of SANS with spliceosomal complexes in nuclear speckles, and are in agreement with our results above (Figure 2C) showing that SANS is essential for the normal morphology of nuclear speckles.
The di- and mature tri-snRNP complexes accumulate in Cajal bodies of SANS-depleted cells
An increase in the number of Cajal bodies similar to what we observed after SANS knockdowns (Figure 2F) has previously been related to defects in the assembly or in the release of tri-snRNP complexes from Cajal bodies (4,8). To investigate the role of SANS in the formation of mature tri-snRNPs in Cajal bodies, we co-stained the U4/U6 di-snRNP protein PRPF31, the U5-specific-proteins CD2BP2/U5-52K and hSNU114 (2) or SART1, a protein specific for mature tri-snRNP complexes (68) together with the Cajal body scaffolding protein coilin in SANS-depleted cells. Confocal microscopy showed the accumulation of PRPF31 and SART1, but not CD2BP2/52K and hSNU114 in Cajal bodies of SANS-depleted cells when compared with controls (Figure 4A–D). Notably, a similar accumulation of PRPF31 in Cajal bodies was previously observed due to tri-snRNP assembly defects after depletion of the U5 protein PRPF6 (4) (Figure 4A). In addition, RNA-FISH followed by immunostaining for coilin consistently revealed the accumulation of U4 and U6 snRNAs, but not U5 snRNA, at Cajal bodies in SANS-depleted cells (Figure 4E–H). Thus, SANS deficiency leads to significant accumulation of di-and tri-snRNP complexes within Cajal bodies.
Figure 4.
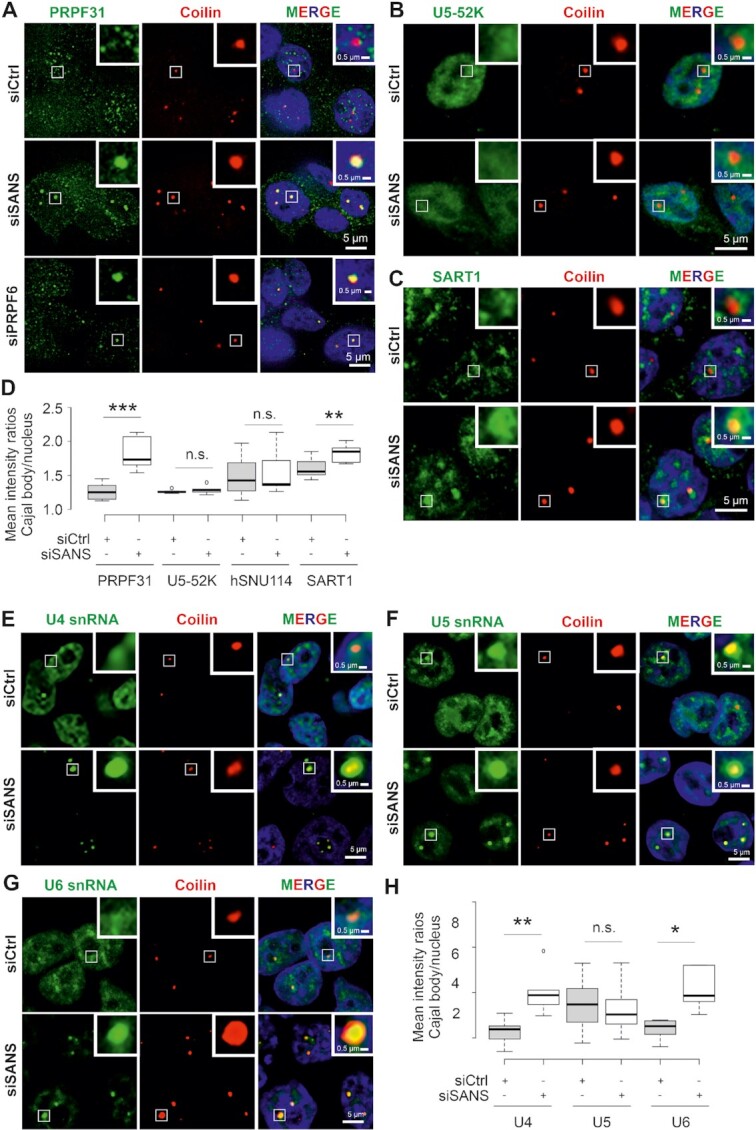
Effects of SANS depletion on Cajal bodies in HEK293T cells. (A–C) Double immunofluorescence staining for coilin (red) and PRPF31 in siCtrl- and siSANS- or siPRPF6-treated cells (A) and U5-52K (B) SART1 (C) in siCtrl- and siSANS-treated cells (for hSNU114 staining see Supplementary Figure S4D). (D) Quantification of mean intensity ratios of PRPF31, U5-52K and SART1 signals observed in Cajal bodies reveals the accumulation of PRPF31 and SART1 but not U5-52K in Cajal bodies in SANS-depleted cells. (E–G) Analysis of RNA-FISH labelling of U4 (E), U5 (F), and U6 (G), snRNAs (green) in Cajal bodies (anti-coilin, red) in siCtrl- and siSANS-treated cells. (H) Quantification of the mean intensity ratios of accumulation of snRNAs in Cajal bodies. SANS depletion leads to the significant accumulation of U4 and U6 snRNAs but not U5 snRNA in CBs. Blue, DAPI counterstaining for nuclear DNA. ***P values < 0.001, **P values < 0.01, *P values < 0.05.
Next, we addressed the question whether SANS deficiency also affects the integrity of tri-snRNPs in the nucleus. We depleted SANS in HEK293T cells stably expressing the FLAG-tagged U4/U6 snRNP protein PRPF4 and pulled down the tri-snRNP complex from nuclear extract by immunoprecipitating FLAG-PRPF4. MS revealed no significant changes neither in the peptide number of components of the tri-snRNP complex nor in its composition, when compared with the control (Figure 5A, Supplementary Table S3). This was confirmed by glycerol gradient fractionation of nuclear extracts (Supplementary Figure S5A). For this, we isolated RNA from the gradient fractions and probed for snRNAs by Northern blotting to quantify the amount of snRNPs. We detected only a slight increase in the amount of tri-snRNPs (fractions 17 and 19, Supplementary Figure S5A) in SANS-depleted extracts, but we did not observe significant defects in tri-snRNP formation between nuclear extracts of SANS-depleted cells and controls.
Figure 5.
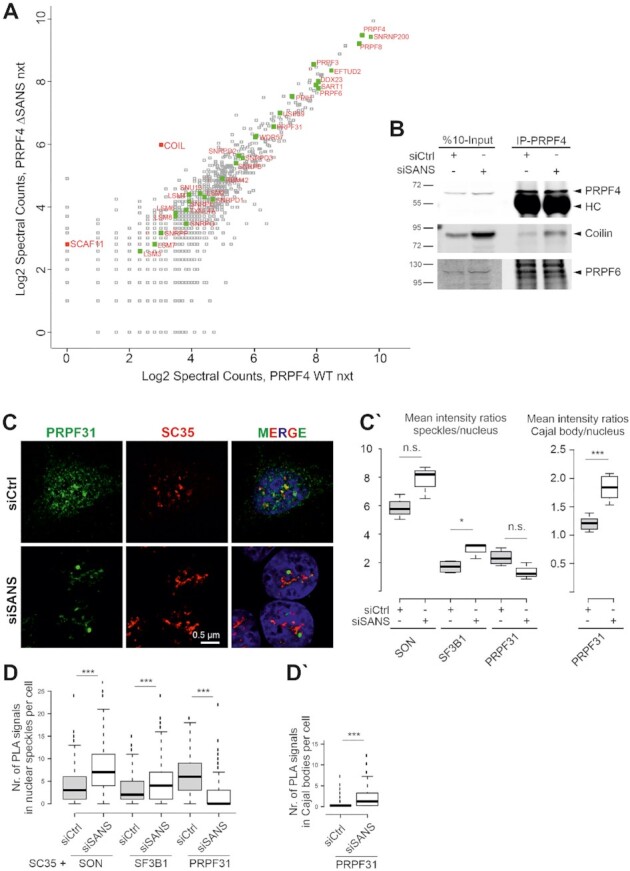
SANS depletion blocks release of tri-snRNP complexes from Cajal bodies and recruitment of mature tri-snRNP complexes to nuclear speckles. (A) Proteomic analysis of precipitated tri-snRNP complexes via FLAG-PRPF4 from nuclear extracts of control cells and SANS-depleted cells. Graph displays peptide spectral counts for each protein in logarithmic scale. Green: tri-snRNPs; red: coilin (COIL) and SC35-interacting protein (SCAF11). There was no significant change in the composition of the tri-snRNP in SANS-depleted nuclear extract, but the tri-snRNP was largely bound to the Cajal body scaffold protein coilin. (B) Western blot analysis of immunoprecipitation of intrinsic PRPF4 from lysates of HEK293T cells demonstrates stronger interactions of the tri-snRNP protein PRPF4 with coilin in SANS-depleted cells. SANS depletion does not affect tri-snRNP complex formation visualized by PRPF4 and PRPF6 interaction. (C) Double immunofluorescence staining reveals reduced PRPF31 staining in SC35 positive nuclear speckles in SANS-depleted cells indicating a defective recruitment of the tri-snRNP complex to nuclear speckles. (C′) Quantification of mean immunofluorescence intensity ratios for PRPF31, SON and SF3B1 observed in nuclear speckles and for PRPF31 in Cajal bodies (CB) versus nucleus. (D) Quantifications of PLA signals for SON-, SF3B1- and PRPF31-SC35 show an increase of SON and SF3B1, but a decrease of PRPF31 in nuclear speckles of SANS-depleted cells. (D′) Quantification of PLA signals for PRPF31-Coilin reveals the accumulation of PRPF31 in Cajal bodies of SANS-depleted cells. ***P values < 0.001, *P values < 0.05.
Interestingly, coilin was identified by MS as significantly enriched protein in the tri-snRNP fraction precipitated from the siRNA-mediated SANS-depleted nuclear extract (Supplementary Table S3, Figure 5A). We validated this result by immunoprecipitations of tri-snRNP complexes using antibodies against coilin, PRPF4 and PRPF6 (Figure 5B). While the levels of PRPF4 and PRPF6 were similar between the tri-snRNPs precipitated from control and that of the SANS-depleted cells, we observed an increased binding of coilin to tri-snRNPs precipitated from the SANS-depleted cells (Figure 5B).
In conclusion, our data indicated that SANS depletion leads to accumulation of di-snRNP and mature tri-snRNP complexes in Cajal bodies without affecting the tri-snRNP integrity.
SANS depletion decreases the recruitment of tri-snRNPs to nuclear speckles
To investigate the role of SANS in the transfer of tri-snRNPs from their assembly site in Cajal bodies to nuclear speckles, we first co-stained SC35 with the tri-snRNP protein PRPF31, the U2-specific protein SF3B1 or the splicing cofactor SON in SANS-depleted cells. Notably, the PRPF31 staining in nuclear speckles was decreased in SANS-depleted cells (Figure 5C) and PRPF31 was mainly localized in Cajal bodies (Figure 4A). In contrast, SF3B1 and SON staining slightly increased in nuclear speckles of SANS-depleted cells (Figure 5C′ and Supplementary Figure S5B-C). To examine the formation of spliceosomal complexes in the speckles, we performed in situ PLAs to visualize the interaction of SF3B1, SON and PRPF31 with SC35. Interestingly, we found a significant increase in the number of PLA signals for SF3B1-SC35 and SON-SC35 in nuclear speckles of SANS-depleted cells compared with controls (Figure 5D and Supplementary Figure S5D). In contrast, PRPF31-SC35 PLA signals were significantly decreased in nuclear speckles of SANS-depleted cells (Figure 5D and Supplementary Figure S5D), consistent with our previous immunostaining results: the Cajal body-specific PLA signals for PRPF31-coilin interaction were significantly increased in these cells (Supplementary Figure S4C and Figure 5C′–D′).
Taken together, these results demonstrated that SANS is important for the delivery of the mature tri-snRNP complex to nuclear speckles where tri-snRNPs are stored until being recruited to pre-spliceosomes sites for pre-mRNA splicing. In the absence of SANS, mature tri-snRNPs are retained in Cajal bodies leading to the drastic reduction of tri-snRNPs in nuclear speckles.
SANS depletion reduces kinetics of spliceosome assembly resulting in accumulation of complex A, a delay of complex B formation and inhibition of splicing
Since tri-snRNPs are recruited to complex A to form fully assembled spliceosomes (complex B), failure in the transfer of tri-snRNP complexes to nuclear speckles should result in the accumulation of complex A, and thus should have an impact on splicing per se (68). Thus, we examined the effects of SANS knockdown on the spliceosome assembly pathway in vitro. We assayed spliceosome complex formation on radiolabeled MINX pre-mRNAs using nuclear extracts prepared from control cells (treated with non-targeting control siRNA) and cells depleted of SANS using siSANS. Splicing reactions were stopped at different time points and spliceosomal complexes were separated by native agarose gel electrophoresis. Figure 6A shows a representative autoradiography from three independent experiments. It indicated substantial changes in the kinetics of the formation of spliceosomal complexes A and B under SANS-depleted conditions: we observed the accumulation of complex A along with the delayed formation of complex B in SANS-depleted nuclear extracts compared with the control (Figure 6A–A′, lane 9–14). Quantification of the autoradiograph of the native gel showed that in the control nuclear extract ∼45% of the MINX pre-mRNA was present in complex B already after 10 min, and this increased to ∼55% after 30 min (Figure 6A–A′, lanes 2–4). In contrast, in SANS-depleted nuclear extracts, complex A was accumulated, and the formation of complex B was significantly delayed, reaching only ∼35% after 30 min (Figure 6A–A′, lanes 9–11).
Figure 6.
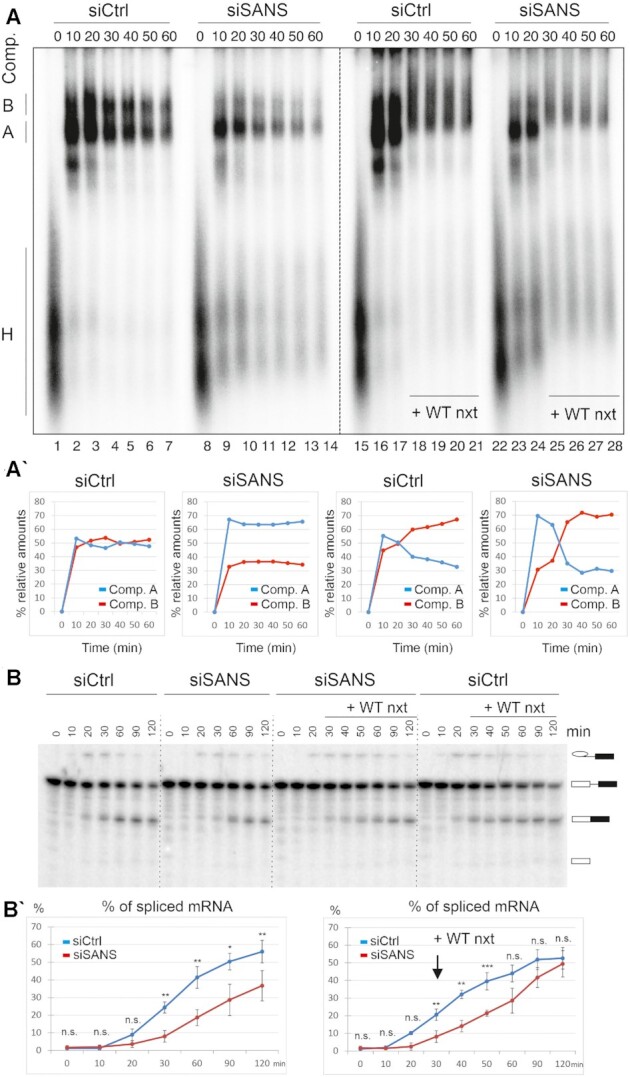
Analysis of the effects of SANS depletion on spliceosome complex formation and splicing. (A) Autoradiography of the time course of assembly of spliceosome complexes on 32P-radiolabelled MINX pre-mRNA analyzed by native agarose gel electrophoresis. Splicing reactions were incubated at 30°C for the indicated time points (0–60 min) and stopped by the addition of heparin. Wild type nuclear extract (WT nxt) was added after 20 min to chase the reactions (lanes: 18–21, 25–28). Positions of each spliceosomal complex are indicated, namely complex H, A and B on the left side of the gel. (A′) Graphical representation of relative amounts of complex A and B. Each time point in siCtrl and siSANS plots represents an average of three independent experiments with three nuclear extracts separately prepared from cells treated with the indicated siRNAs. The rescue experiments were performed in two independent repeats and were averaged. Depletion of SANS leads to the accumulation of complex A. (B) In vitro splicing of 32P radio-labelled MINX pre-mRNA with the control and SANS-depleted nuclear extracts. Splicing of pre-mRNA was analyzed by denaturing PAGE followed by autoradiography. (B′) Quantification of spliced mRNA. Each data point shows an average percentage of spliced mRNA (three independent experiments) for each time point. SANS depletion affects the kinetics of splicing leading to a decreased splicing efficiency (left panel). Addition of WT nxt restores the splicing efficiency (right panel). ***P values < 0.001, **P values < 0.01, *P values < 0.05.
Next, we tested whether the accumulated complex A upon SANS depletion is a functional intermediate or a dead-end complex. For this, we supplemented SANS-depleted nuclear extract with the wild-type nuclear extract (Figure 6A–A′, lanes 15–28). Autoradiography analysis revealed an effective rescue of the accumulated complex A shifted to complex B (Figure 6A–A′, lanes 18–21 and 25–28).
Owing to the altered kinetics of spliceosome assembly, we next tested the in vitro splicing of pre-mRNA with control and SANS-depleted nuclear extracts. Aliquots were taken at various time points of the MINX pre-mRNA splicing reaction followed by RNA isolation and separation of splicing products by denaturing PAGE (Figure 6B). Quantification of the autoradiographs from three independent experiments revealed that at 120 min only 37% of the pre-mRNA was spliced in siRNA-mediated SANS-depleted extracts, while 56% of the pre-mRNA was already spliced in the control reaction at this time point (Figure 6B′). The impaired splicing was fully rescued by the addition of wild-type nuclear extract to the SANS-depleted extract after 30 min, yielding ∼50% of spliced mRNA (Figure 6B′). We conclude that the reduced kinetics of spliceosome assembly upon SANS depletion leads to impaired pre-mRNA splicing in vitro.
Purified tri-snRNP complex converts the complex A to complex B in SANS-depleted nuclear extract
Next, we tested whether the tri-snRNP complex is the spliceosomal subunit affected in the siRNA-mediated SANS-depleted extract and thus required for converting complex A to the pre-catalytic spliceosome (complex B). Therefore, we supplemented SANS-depleted nuclear extracts with purified native tri-snRNPs (Supplementary Figure S6). Indeed, the in vitro spliceosomal complex formation experiment using radiolabeled MINX pre-mRNA demonstrated that while complex A accumulates in SANS-depleted extracts, the addition of the purified tri-snRNPs efficiently rescued the conversion of complex A to complex B. It is worth to mention that the addition of the purified tri-snRNPs did not change the kinetics of the control reaction. Taken together, our results indicated that SANS is important for the supply of the tri-snRNP complexes to the sites of active splicing, required for the assembly of the spliceosome complex B.
SANS regulates splicing in liaison with pre-mRNA splicing factors in cells
So far, we have shown that SANS is required for the spliceosome assembly. We next investigated whether SANS affects splicing of target genes in liaison with core splicing factors in cells. Initially, we analyzed the effects of SANS and PRPF6 depletion on the splicing of RON (MST1R) and USH1C minigenes expressed in HEK293T cells (Figure 7A, A′). Analysis of the relative percentage of spliced minigene variants indicated that SANS depletion leads to an accumulation of non-spliced RON and USH1C pre-mRNAs, by 61% and 29%, respectively, as compared with the control siRNA treated cells. In addition, we observed similar results in PRPF6-depleted cells namely 61% and 20%, respectively.
Figure 7.
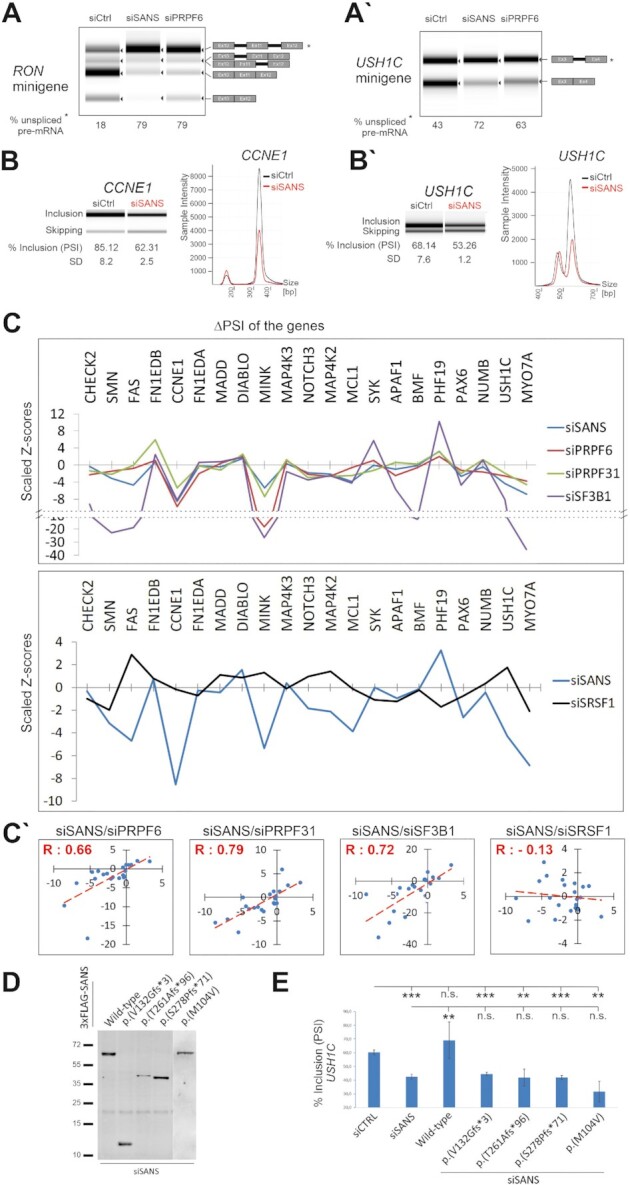
SANS regulates in vitro splicing of minigene reporters as well as splicing of genes related to cell proliferation and the human Usher syndrome. (A–A′) Splicing of RON and USH1C minigenes in SANS- and PRPF6-depleted cells. Percentage of unspliced mRNA is indicated below lanes. (B–B′) High throughput capillary electrophoresis analyses of alternatively spliced variants of the CCNE1 gene (exon 9) and USH1C (exon 11) gene after siRNA-mediated depletion of SANS. Relative intensities of the exon included and skipped isoforms obtained from the representative gel images are shown. PSI (% inclusion) and SD indicate, respectively, the percentage spliced-in median and the standard deviation values from three independent experiments. (C) Graphs showing the splicing perturbation profiles of selected endogenous genes upon knockdown of PRPF6, PRPF31, SF3B1 (upper panel), SRSF1 (lower panel) or SANS. Scaled Z-scores represent changes toward inclusion (>0) or skipping (<0) for each alternative splicing event after siRNA mediated knockdowns. SANS depletion exhibits very similar perturbation profiles to PRPF6, PRPF31, and SF3B1 knockdowns (upper panel), but to SRSF1 (lower panel). (C′) Quantile-quantile (Q-Q) plots are showing positive correlation between SANS, PRPF6, PRPF31 and SF3B1 (Pearson′s correlation coefficient values indicated with positive R values) but not with SRSF1 (negative R value). (D) Western blot analysis showing the expression of SANS pathogenic variants in the SANS-depleted cells. (E) Capillary electrophoresis analyses of alternatively spliced variants of USH1C (exon 11) gene after siRNA-mediated depletion of SANS following by overexpression of the wild-type and pathogenic USH1G variants. Only wild-type SANS, but none of the pathogenic USH1G variants restored the siSANS-induced splicing deficiency in USH1C. Relative intensities of the exon included and skipped isoforms obtained from the representative gel images are shown. Quantification of PSI (% inclusion) and SD values shows the percentage spliced-in median and the standard deviation values from three independent experiments. ***P values < 0.001, *P values < 0.05.
Next, we examined whether SANS also affects the alternative splicing of intrinsic genes in HEK293T cells. For this, we assessed 19 previously described genes that are alternatively spliced and translated to functional protein isoforms (Figure 7C) (1). In addition, we included two USH genes USH1C and MYO7A (USH1B) known to be alternatively spliced (69,70). After siRNA-mediated knockdowns, we isolated mRNA from cells followed by RT-PCR using splice variant-specific primers to detect alternatively spliced products. As an example, in Figure 7B, we show analyses of the two genes CCNE1 and USH1C, demonstrating a highly significant change in skipping exon 9 or exon 11, respectively (P values < 0.005).
To depict the splicing perturbation after knockdown of SANS and splicing factors (PRPF6, PRPF31 and SF3B1 as well as SRSF1), we calculated the percent-spliced-in (PSI) for each gene in three independent experiments (Supplementary Tables S4 and S5). The scaled Z-scores for the PSIs are plotted in Figure 7C and show the directional change and magnitude of each splicing event. Our results demonstrated that SANS knockdown resulted in a perturbation profile that correlated well with PRPF6, PRPF31 and SF3B1 siRNA-mediated knockdowns (Figure 7C, upper graph, Supplementary Figure S3B). Analyses of Z-scores for SANS, PRPF6, PRPF31 and SF3B1 obtained in three independent experiments revealed positive Pearson′s correlation coefficients between these molecules (R: 0.66, R: 0.79 and R: 0.72, respectively). In addition, quantile-quantile (Q–Q) plots of the Z-scores revealed a positive inclination line (Figure 7C′) demonstrating similarities between the function of these molecules. As a negative control, we knocked down the serine/arginine-rich splicing factor 1 (SRSF1), which has been previously identified as peripheral component of the spliceosome (1). siSRSF1-mediated depletions gave rise to a different perturbation profile when compared with knockdowns of SANS or the three core splicing factors (Figure 7C, lower graph, Supplementary Figure S3B). Furthermore, no correlation was observed between siSANS and siSRSF1 (negative R: –0.13) (Figure 7C′). Altogether, our results demonstrated that SANS participates in splicing and that its absence leads to similar splicing defects, as seen in knockdowns of the essential spliceosomal proteins SF3B1, PRPF6 and PRPF31.
Depletion of SANS and its spliceosomal interaction partners decreases cell proliferation
The data obtained for splicing of the 19 test genes above indicated that that SANS is important for alternative splicing of genes related to cell proliferation (Figure 7C, Supplementary Table S5B). Moreover, functional annotation of SANS interaction partners highlighted the cell cycle as one of the enriched pathways (Supplementary Table S2). To investigate a possible role for SANS in the cell cycle, we quantified cell proliferation by WST-1 colorimetric assay after siRNA-mediated knockdowns of SANS, PRPF31, SON, and SRSF1, respectively (validation of siRNAs, Supplementary Figure S3B). While SRSF1 depletion reduced cell proliferation only to ∼25% of the level of controls, the depletion of SANS, PRPF31, and SON significantly decreased cell proliferation to >80% when compared with controls (Supplementary Figure S7, Supplementary Table S5). These data further confirmed that SANS and its spliceosomal interaction partners are important for cell proliferation through regulation of alternative splicing of cell proliferation-related genes.
Pathogenic variants of SANS/USH1G affect splicing of the USH1C gene
In humans, pathogenic variants of SANS lead to Usher Syndrome type 1G (USH1G), characterized by combined hearing disorder and RP. Since our previous results showed that SANS deficiency significantly changes the alternative splicing of the USH1C gene, we examined whether pathogenic variants of SANS/USH1G can also affect the splicing of USH1C. We expressed wild-type or four different pathogenic variants of SANS, harboring frame-shift or missense mutations in cells depleted of endogenous SANS, and analyzed the splicing of USH1C (Figure 7D–E). Our data revealed that the expression of wild-type SANS restored USH1C splicing defects and indicated that aberrant splicing caused by SANS depletion can be reversed. In contrast, none of the pathogenic variants of SANS was able to restore the splicing defect of USH1C in SANS-depleted cells (Figure 7D–E). This demonstrated that pathogenic variants in SANS leading to USH1G disrupt the regulatory function of SANS in splicing of target genes.
DISCUSSION
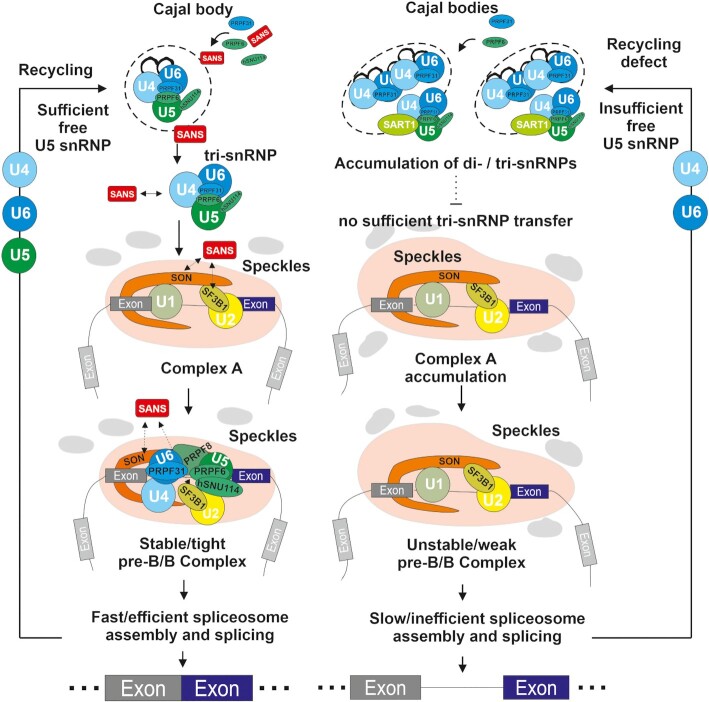
We found previously unidentified and unexpected links between the ciliary USH1G protein SANS and the pre-mRNA splicing machinery. Splicing is catalyzed by the spliceosome, a highly dynamic macromolecular complex comprised of more than 150–200 proteins contributing to the sequential assembly and regulation of the spliceosome (1,2). Although previous studies have indicated the participation of SANS in several cellular functions it has not hitherto been related to the splicing machinery. Here, we show that SANS interacts with proteins related to spliceosomal complexes and that it mediates the intra-nuclear transfer of tri-snRNPs between sub-nuclear membrane-less organelles during spliceosome assembly and snRNP recycling; thereby SANS plays a significant role in pre-mRNA splicing (Figure 8).
Figure 8.
Graphical representation of proposed role of SANS in the splicing process. In the nucleus, the USH1G protein SANS controls the tri-snRNP turnover. SANS depletion leads to the accumulation of tri-snRNP intermediates in Cajal bodies and stalls spliceosome assembly at the complex A stage. This can occur due to an imbalance in the recycling of snRNPs components and/or defective release of tri-snRNP complexes from Cajal bodies and their subsequent transfer to nuclear speckles for formation of pre-spliceosomes (complex B). Consistently, cells attempt to compensate low levels of mature tri-snRNP complexes in nuclear speckles by increasing the number of Cajal bodies.
An intrinsically disordered region of SANS interacts with proteins of the pre-mRNA splicing machinery in membrane-less organelles of the nucleus
SANS is a multivalent scaffold protein that participates in membrane adhesion complexes (29,35), is associated with microtubule-based intracellular transport (17,30,31,35,71), regulates endocytosis, and is involved in primary ciliogenesis (21). For these functions, SANS interacts with a variety of proteins including several proteins related to the human USH (17,21,29–32,34,35,71) and other ciliopathies (72). While, PDZ-domain-containing scaffold proteins bind to the type I PDZ-binding motif and the SAM domain at the C-terminus of SANS (21,29,35,73), the majority of interacting proteins bind to SANS via the CENT domain (17,29,30).
In the present study, we demonstrate that core proteins of the pre-mRNA splicing machinery namely SF3B1, PRPF6 and PRPF31 interact with SANS in cells through the N-terminal region of the SANS CENT domain (CENTn). In silico structural analysis predicted SANS_CENTn as an intrinsically disordered region (IDR) (Supplementary Figure S1E). IDRs can adopt their three-dimensional structures for binding to diverse target proteins in a cell-context-specific manner. This explains the promiscuous interaction of SANS_CENTn with numerous proteins in diverse cellular processes carried out in different cell compartments. Intrinsically disordered proteins (IDPs) are found mainly in macromolecular assemblies and regulate dynamic processes by often transient interactions with target proteins (56,74). While the binary direct interaction of SANS with SF3B1 was confirmed by both reciprocal co-IPs and Y2H assays, we cannot rule out that the interaction of SANS with PRPF31 and PRPF6 shown by reciprocal co-IPs is indirect via other splicing factors or even RNA. By exploiting these properties, SANS may only transiently and/or weakly interact with spliceosome molecules, which would explain why it has so far not been found in the proteome of purified spliceosomes.
IDPs drive the formation of the membrane-less organelles Cajal bodies and nuclear speckles by liquid-liquid phase separation (LLPS) (9,75,76). Recently, it has been suggested that SANS generates protein condensates via LLPS in the tip-link complex of the stereocilia of inner ear hair cells (33). It is thought that SANS changes the material properties in the tip-link complex and in its absence, the complex loses its integrity (32). This feature of SANS is in line with the significant changes in the nuclear speckle morphology observed upon SANS depletion (Figure 2C, Supplementary Figures S3D and S4A). We propose that the multivalent properties of SANS also promote changes in the properties of nuclear speckles (77,78), which remains to be further investigated.
SANS facilitates the release of tri-snRNPs from Cajal bodies and participates in the recycling of U5 snRNPs back to Cajal bodies
Pre-mRNA splicing is a highly dynamic process, characterized by stepwise assembly and release of the spliceosomal U1, U2, U4, U6 and U5 snRNP complexes (2,79). Northern blot analysis of SANS pull-downs from nuclear extract did not show enrichment of snRNAs. We concluded that SANS does not directly act as a core splicing factor in the catalytic processes of the splicing. However, under splicing conditions and in the presence of the MINX pre-mRNA, SANS could pull down U1, U2, U4, U6 and U5 snRNP complexes suggesting a possible role for SANS in the regulation of splicing activity of at least a subset of genes.
The U4/U6.U5 tri-snRNP complex is a protein-rich particle consisting of more than two dozen polypeptides (2), which assembles and fully matures in Cajal bodies within the nucleus. Failure of tri-snRNP complex maturation, for example in the absence PRPF6, leads to the accumulation of incomplete/immature tri-snRNP intermediates in Cajal bodies (for confirmation see Figure 4A) and the formation of new Cajal bodies (4,8), which is thought to be a mechanism that compensates this maturation defect and enhances the kinetics of snRNP complex assembly (4,8). Notably, we observed a similar phenotype in SANS depleted cells: the number of Cajal bodies was drastically increased and SANS deficiency led to the accumulation of the U4/U6-specific protein PRPF31 and tri-snRNP complex-specific protein SART1 and U4 and U6 snRNAs in Cajal bodies. However, SANS deficiency did not change the abundance of U5 snRNP-specific proteins U5-52K and hSNU114 and U5 snRNAs in Cajal bodies. These results may support a role for SANS in the regulation of the tri-snRNP complex assembly which should be evident from the accumulation of the U4/U6 di-snRNP intermediates (47,49) or from the composition of the isolated tri-snRNP complex. However, the abundance of fully maturated tri-snRNP complexes indicated by the presence SART1 and their accumulation in SANS-depleted cells strongly argues against a prominent role of SANS in tri-snRNP complex assembly. In addition, the analysis of snRNPs by glycerol gradient fractionation as well as the MS analysis in SANS-depleted nuclear extracts showed no significant deficit in the composition of the tri-snRNP complex when compared with those isolated from a wild-type extract (Figure 5A, B, Supplementary Figure S5A and Supplementary Table S3). Furthermore, while snRNPs are co-localized with the Cajal body scaffold protein coilin and thus are located in the core of Cajal bodies, where their assembly is driven (80), microscopy analysis consistently demonstrated the localization of SANS at the periphery of Cajal bodies. Taken together, these findings are not consistent with a key function for SANS in the assembly of the tri-snRNP complex in Cajal bodies. Instead, they suggest a role for SANS in the liberation of assembled tri-snRNPs from Cajal bodies.
Strikingly, in contrast to U4/U6 components, U5 snRNP-specific proteins and snRNA did not accummulate in Cajal bodies in SANS deficient cells (Figure 4). This phenomenon was also observed in a previous study after inhibition of spliceosome disassembly, which was associated with insufficient recycling and delivery of U5 snRNPs back to Cajal bodies (81). Therefore, it is likely that SANS also plays a role in the recycling pathway of splicing molecules, which is also consistent with its interaction with core spliceosome components.
SANS participates in the transfer and recruitment of tri-snRNPs to nuclear speckles and promotes the stable assembly of pre-catalytic spliceosomes
Our microscopy analyses showed that SANS is present in nuclear speckles—the nuclear compartment where snRNP and non-snRNP splicing factors are stored to be supplied to transcription/splicing sites (11,61). Our present data demonstrated that the depletion of SANS leads to an increase in size and diffluence of the nuclear speckles, the morphological alterations previously found in splicing-inhibited cells (79). This phenotype was thought to be due to the release of incomplete spliceosomal sub-complexes from the speckles (79) and is in line with the role of SANS as a multi-valent IDP in driving phase separation of nuclear speckles discussed above.
Our in vitro interaction studies demonstrated that SANS interacts with several components of the spliceosome indicating that SANS might be involved in the regulation of spliceosome activity and snRNP recycling. Strikingly our data also revealed the interaction of SANS with SON in nuclear speckles (Figure 3). SON acts as an important coactivator in splicing and facilitates the formation and stabilization of spliceosomes on weak splice sites by recruiting SR proteins, serine/arginine-rich‐splicing factors known as potent regulators of splicing and RNA shuttling (82). Through this function, SON contributes to efficient splicing of genes related to cell cycle progression (67,83). Interestingly, our data showed that SANS also promotes splicing of genes related to cell proliferation and its depletion inhibits cell proliferation (Figure 7C, Supplementary Figure S7). Moreover, we found several SR proteins such as SC35/SRSF2 in the SANS affinity proteomics data (Figure 3B) suggesting that SANS and SON may cooperate as a complex in these activities.
A crucial step before catalytic activation of the spliceosome is the recruitment of the U4/U6.U5 tri-snRNP to complex A, to form the pre-catalytic spliceosomal complex B (63,84). Our in vitro biochemical assays using SANS-depleted nuclear extract and a model pre-mRNA showed the accumulation of complex A corroborating a role for SANS in the formation of stable pre-catalytic spliceosomes (complex B; Figure 6). Analysis of the recruitment of SON, SF3B1 and PRPF31 to nuclear speckles by double-immunofluorescence staining and PLAs for the interaction with SC35 revealed that SANS is required to recruit the tri-snRNP component PRPF31, but not SON and the U2 snRNP component SF3B1, to nuclear speckles (Figure 4 and Supplementary Figure S5). Conversely, SANS deficiency led to slight increases in the staining of SON and SF3B1 in nuclear speckles and to significant increases of PLA signals for SON/SC35 and SF3B1/SC35 complexes, which reflect the accumulation of spliceosomal complexes in nuclear speckles.
Prior to the transfer of the tri-snRNP to nuclear speckles, the assembled complex is released from the Cajal bodies. The mechanisms underlying the intra-nuclear transfer processes of snRNPs between nuclear organelles such as Cajal bodies and speckles are unknown. Here, we provide the first evidence that SANS plays an important role in the release of mature tri-snRNPs from Cajal bodies, and their recruitment to nuclear speckles where assembled spliceosomal complexes are stored for pre-mRNA splicing. Based on our findings, we suggest the following mechanism (Figure 8): the tri-snRNP complexes are assembled in the core of Cajal bodies, diffuse to the periphery where SANS coordinates the release of tri-snRNPs from Cajal bodies for the translocation to nuclear speckles. In addition, SANS may participate in the recycling pathway of U5 snRNP components during the splicing process from speckles back to Cajal bodies. In the absence of SANS, di-snRNP complexes accumulate due to insufficient amounts of U5 snRNPs (see also Figure 4). This leads to deprivation of tri-snRNPs in nuclear speckles required for the formation of the pre-catalytic spliceosomal complex B. Consequently, complex A accumulates leading to significant alterations in splicing kinetics (Figures 5 and 6). The accumulated complex A in the SANS-depleted nuclear extract is not a dead-end complex. It can be rescued to complex B by the addition of wild-type nuclear extract or isolated tri-snRNP complexes. Furthermore, overexpression of wild-type SANS can rescue the effect of SANS knockdown and increases splicing activity (Figure 7E).
SANS is required for correct constitutive and alternative pre-mRNA splicing
The in vitro splicing assays with MINX pre-mRNA and splicing experiments in cells including minigene reporter splicing assays as well as the analysis of alternative splicing of intrinsic intron-containing genes showed that SANS has a significant impact on constitutive and alternative pre-mRNA splicing. Strikingly, SANS depletion strongly inhibited constitutive and alternative splicing of the RON minigene and the constitutive splicing of the USH1C minigene (Figure 7A–A′). In addition, we observed very similar splicing perturbation profiles with positive correlations after the siRNA knockdown of SANS and that of the core components of the spliceosome, namely SF3B1, PRPF6, and PRPF31 (Figure 7C–C′). In contrast, depletion of SRFS1, a non-snRNP splicing factor known to be involved in splicing regulation (1), gave rise to a different perturbation profile. These findings suggest that SANS interacts not only physically, but also functionally, with core components of the pre-mRNA splicing machinery. Thus, the splicing data obtained in cells are in agreement with the in vitro biochemical results, in that both underline the importance of SANS for the delivery of tri-snRNP complexes to nuclear speckles, and thereby for the kinetics of spliceosome assembly.
SANS/USH1G mutations lead to USH due to splicing defects in other USH1 genes
Pathogenic variants in USH1G cause human USH1, a ciliopathy characterized by sensory neuronal degenerations leading to profound hearing loss, vestibular dysfunction and vision loss in the form of Retinitis pigmentosa (RP) (24). While in the inner ear the essential roles of SANS in hair cell differentiation and the mechano-electrical signal transduction complex can well explain the pathophysiology in USH1G (31–33), the ophthalmic pathogenesis in the retina is still puzzling (85). Links between the USH1G protein SANS—or any other USH1 proteins—and the splicing machinery of cells has not been reported yet. Here, we provide the first evidence that perturbation of splicing of target genes can participate in the pathogenesis of the human USH1 phenotype. Our knockdown experiments demonstrate that SANS is essential for pre-mRNA splicing in the nucleus. However, not only depletion of SANS but also USH causing mutations in SANS/USH1G lead to splicing defects (Figure 7E). It is feasible that these mutations alter the physical interaction of expressed SANS variants with components of the splicing machinery, since interaction domains are affected. Alternatively, pathogenic mutations in SANS/USH1G can alter the cellular and nuclear distribution of SANS (29) and thereby SANS might not be any longer recruitable for splicing. In either case, the disruption of splicing resulting from SANS defects cause splicing errors in other USH1 genes, such as USH1C and MYO7A/USH1B known to be spliced in the retina (69,70). It is therefore conceivable that the USH1 phenotype in the retina is not directly caused by SANS, but by mis-splicing of other USH1 molecules. Direct testing of this hypothesis must be deferred for future studies.
Evidence for a common pathomechanism of SANS and PRPFs deficiencies leading to Retinitis pigmentosa
Interestingly, mutations in several pre-mRNA-processing factors of the U4/U6.U5 tri-snRNP—such as PRPF31 (RP11), PRPF6 (RP60), PRPF8 (RP13), PRPF3 (RP18), SNRNP200 (RP33) and PRPF4 (RP70)—also result in retina-specific phenotypes (16,51,86). Our present study shows that deficiencies in PRPFs and SANS cause nearly identical effects on splicing which is in line with significant congruencies between the phenotypes caused by defects in SANS/USH1G and by those in PRPF genes suggesting similar pathomechanisms leading to RP. In this regard, it is noteworthy that both the reduction in expression level due to haploinsufficiency of PRPF31 in RP11 (∼60/∼80% in mRNA, 40–60% reduction in protein level) (51) and the knockdown of SANS/USH1G (∼60/∼80% in mRNA, 40% reduction in protein level) (present study) significantly affected the splicing of target genes. Mono-allelic mutations in the PRPF31 gene disrupt retina-specific splicing programs and affect ciliogenesis (51,87). Equally, we have previously shown that SANS depletion alters ciliogenesis (21,29) and here we demonstrate that SANS depletion affects the splicing programs of target genes, particularly that of USH1C, known to be specifically spliced in the retina (69).
Since splicing is a fundamental process that occurs in likely all cell types, mutations in tri-snRNP-related genes cause a retinal phenotype only (in the case of RP) or defects in both the retina and the inner ear (in the case of USH). Interestingly, murine animal models for Prpf genes and Sans/Ush1g do not recapitulate the human ocular phenotype (19,23,88,89). Pre-mRNA splicing factor deficiency may dysregulate specific splicing programs that are known to be specifically implemented in the human retina (90,91). Recently, it was shown that the truncated pathogenic form of PRPF31 protein is only expressed in ocular cells derived from RP11 patients, while it was absent in other RP11 patient-derived cells, and that in these non-ocular cells the levels of wild-type protein were nearly equal between normal and patient cells (51). It is thought that the expression of the pathogenic variant protein exacerbates splicing defects, and thus ocular cells are most strongly affected. Nevertheless, it should not be concealed that the PRPF pathogenic variants lead to autosomal dominant RP and USH1G is a recessive disease. However, recent studies show that there are recessive pathogenic variants of splicing factors such as CWC27, SNRNP200 also causing RP (23,92). Taken together, there is evidence that the pathomechanisms and pathways leading to RP in patients with PRPF or USH1G pathogenic variants converge and thereby provide common targets for future therapeutic strategies.
CONCLUSION
This study has deciphered the nuclear function of the USH1 protein SANS as a pre-mRNA splicing regulator (Figure 8). In the nucleus, SANS acts as a factor required for the release of tri-snRNPs from Cajal bodies and their transfer to nuclear speckles. Moreover, SANS may participate in the recycling of snRNPs during splicing. Thereby, SANS contributes to the assembly of the pre-catalytic spliceosome on target pre-mRNAs. SANS depletion stalls the splicing process at complex A and reduces the kinetics of spliceosome assembly. Consequently, SANS deficiency alters constitutive and alternative splicing of genes related to cell proliferation and to the human USH. Pathogenic variants of USH1G/SANS lead to aberrant splice variants of other USH genes which may underlie the pathophysiology of the retinal degenerations associated with USH1G. In addition, our results further support findings of earlier studies, demonstrating that the RP phenotype in the eye can arise from changes in splicing efficiency.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD023942.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs Utz Fischer, Carlo Rivolta and Julian Koenig for providing several plasmids used in the study; Drs Kerstin Nagel-Wolfrum, Karl Fath, Cyrill Girard, Dimitry Agofanov and Julian Koenig for valuable discussions and proofreading; IMB Genomics core facility for the support applying capillary electrophoresis; Mrs Elisabeth Sehn for her technical expertise with TEM and her valuable help. We also thank to Abberior Company for their valuable support on super-resolution microscopy.
Author contributions: A.Y. set up and carried out most of the experiments. S.M.J., S.L. and A.Y. set up and carried out the in vitro splicing assays and generated stable cell lines. A.K.W. and J.S. helped with several western blot and immunofluorescence analyses. H.U. and S.M.J carried out the mass-spectrometric analyses. A.Y., S.M.J. and U.W. designed the studies and A.Y. wrote the manuscript under supervision by S.M.J., R.L. and U.W.
Contributor Information
Adem Yildirim, Molecular Cell Biology, Institute of Molecular Physiology, Johannes Gutenberg-University of Mainz, Germany.
Sina Mozaffari-Jovin, Department of Cellular Biochemistry, Max-Planck-Institute for Biophysical Chemistry, Goettingen, Germany; Medical Genetics Research Center, Mashhad University of Medical Sciences, Mashhad, Iran; Department of Medical Genetics, Faculty of Medicine, Mashhad University of Medical Sciences, Mashhad, Iran; Bioanalytical Mass Spectrometry, Max-Planck-Institute for Biophysical Chemistry, Goettingen, Germany.
Ann-Kathrin Wallisch, Molecular Cell Biology, Institute of Molecular Physiology, Johannes Gutenberg-University of Mainz, Germany.
Jessica Schäfer, Molecular Cell Biology, Institute of Molecular Physiology, Johannes Gutenberg-University of Mainz, Germany.
Sebastian E J Ludwig, Department of Cellular Biochemistry, Max-Planck-Institute for Biophysical Chemistry, Goettingen, Germany.
Henning Urlaub, Bioanalytical Mass Spectrometry, Max-Planck-Institute for Biophysical Chemistry, Goettingen, Germany; Bioanalytics, Department of Clinical Chemistry, University Medical Center Goettingen, Germany.
Reinhard Lührmann, Department of Cellular Biochemistry, Max-Planck-Institute for Biophysical Chemistry, Goettingen, Germany.
Uwe Wolfrum, Molecular Cell Biology, Institute of Molecular Physiology, Johannes Gutenberg-University of Mainz, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
GeneRED, IPP Programme Mainz; FAUN-Stiftung, Nuremberg; Foundation Fighting Blindness (FFB) [PPA-0717-0719-RAD]; RP Fighting Blindness (GR595); NIMAD (989278). Funding for open access charge: Foundation Fighting Blindness (FFB) grants and other third-party resources of our lab.
Conflict of interest statement. None declared.
REFERENCES
- 1. Papasaikas P., Tejedor J.R., Vigevani L., Valcarcel J.. Functional splicing network reveals extensive regulatory potential of the core spliceosomal machinery. Mol. Cell. 2015; 57:7–22. [DOI] [PubMed] [Google Scholar]
- 2. Will C.L., Luhrmann R.. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011; 3:a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wahl M.C., Will C.L., Luhrmann R.. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009; 136:701–718. [DOI] [PubMed] [Google Scholar]
- 4. Novotny I., Malinova A., Stejskalova E., Mateju D., Klimesova K., Roithova A., Sveda M., Knejzlik Z., Stanek D.. SART3-dependent accumulation of incomplete spliceosomal snRNPs in Cajal bodies. Cell Rep. 2015; 10:429–440. [DOI] [PubMed] [Google Scholar]
- 5. Michaud S., Reed R.. An ATP-independent complex commits pre-mRNA to the mammalian spliceosome assembly pathway. Genes Dev. 1991; 5:2534–2546. [DOI] [PubMed] [Google Scholar]
- 6. Fischer U., Englbrecht C., Chari A.. Biogenesis of spliceosomal small nuclear ribonucleoproteins. Wiley Interdiscip. Rev. RNA. 2011; 2:718–731. [DOI] [PubMed] [Google Scholar]
- 7. Roithova A., Klimesova K., Panek J., Will C.L., Luhrmann R., Stanek D., Girard C.. The Sm-core mediates the retention of partially-assembled spliceosomal snRNPs in Cajal bodies until their full maturation. Nucleic Acids Res. 2018; 46:3774–3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stanek D. Cajal bodies and snRNPs - friends with benefits. RNA Biol. 2017; 14:671–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Neugebauer K.M. Special focus on the Cajal Body. RNA Biol. 2017; 14:669–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mao Y.S., Zhang B., Spector D.L.. Biogenesis and function of nuclear bodies. Trends Genet. 2011; 27:295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Galganski L., Urbanek M.O., Krzyzosiak W.J.. Nuclear speckles: molecular organization, biological function and role in disease. Nucleic Acids Res. 2017; 45:10350–10368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Calarco J.A., Saltzman A.L., Ip J.Y., Blencowe B.J.. Technologies for the global discovery and analysis of alternative splicing. Adv. Exp. Med. Biol. 2007; 623:64–84. [DOI] [PubMed] [Google Scholar]
- 13. Daguenet E., Dujardin G., Valcarcel J.. The pathogenicity of splicing defects: mechanistic insights into pre-mRNA processing inform novel therapeutic approaches. EMBO Rep. 2015; 16:1640–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Harbour J.W. Genomic, prognostic, and cell-signaling advances in uveal melanoma. Am. Soc. Clin. Oncol. Educ Book. 2013; 33:388–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Quesada V., Ramsay A.J., Lopez-Otin C.. Chronic lymphocytic leukemia with SF3B1 mutation. N. Engl. J. Med. 2012; 366:2530. [DOI] [PubMed] [Google Scholar]
- 16. Tanackovic G., Ransijn A., Ayuso C., Harper S., Berson E.L., Rivolta C.. A missense mutation in PRPF6 causes impairment of pre-mRNA splicing and autosomal-dominant retinitis pigmentosa. Am. J. Hum. Genet. 2011; 88:643–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Overlack N., Kilic D., Bauss K., Marker T., Kremer H., van Wijk E., Wolfrum U.. Direct interaction of the Usher syndrome 1G protein SANS and myomegalin in the retina. Biochim. Biophys. Acta. 2011; 1813:1883–1892. [DOI] [PubMed] [Google Scholar]
- 18. Mathur P., Yang J.. Usher syndrome: hearing loss, retinal degeneration and associated abnormalities. Biochim. Biophys. Acta. 2015; 1852:406–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wolfrum U. Ahuja S. Usher Syndrome. 2011; Nova Science Publishers, Inc; 51–73. [Google Scholar]
- 20. Liu X., Vansant G., Udovichenko I.P., Wolfrum U., Williams D.S.. Myosin VIIa, the product of the Usher 1B syndrome gene, is concentrated in the connecting cilia of photoreceptor cells. Cell Motil. Cytoskeleton. 1997; 37:240–252. [DOI] [PubMed] [Google Scholar]
- 21. Bauss K., Knapp B., Jores P., Roepman R., Kremer H., Wijk E.V., Marker T., Wolfrum U.. Phosphorylation of the Usher syndrome 1G protein SANS controls Magi2-mediated endocytosis. Hum. Mol. Genet. 2014; 23:3923–3942. [DOI] [PubMed] [Google Scholar]
- 22. Jansen F., Kalbe B., Scholz P., Mikosz M., Wunderlich K.A., Kurtenbach S., Nagel-Wolfrum K., Wolfrum U., Hatt H., Osterloh S.. Impact of the Usher syndrome on olfaction. Hum. Mol. Genet. 2016; 25:524–533. [DOI] [PubMed] [Google Scholar]
- 23. Bujakowska K.M., Liu Q., Pierce E.A.. Photoreceptor cilia and retinal ciliopathies. Cold Spring Harb. Perspect. Biol. 2017; 9:a028274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. May-Simera H., Nagel-Wolfrum K., Wolfrum U.. Cilia - the sensory antennae in the eye. Prog. Retin. Eye Res. 2017; 60:144–180. [DOI] [PubMed] [Google Scholar]
- 25. Chaki M., Airik R., Ghosh A.K., Giles R.H., Chen R., Slaats G.G., Wang H., Hurd T.W., Zhou W., Cluckey A.et al.. Exome capture reveals ZNF423 and CEP164 mutations, linking renal ciliopathies to DNA damage response signaling. Cell. 2012; 150:533–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Choi H.J., Lin J.R., Vannier J.B., Slaats G.G., Kile A.C., Paulsen R.D., Manning D.K., Beier D.R., Giles R.H., Boulton S.J.et al.. NEK8 links the ATR-regulated replication stress response and S phase CDK activity to renal ciliopathies. Mol. Cell. 2013; 51:423–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Attanasio M. Ciliopathies and DNA damage: an emerging nexus. Curr. Opin. Nephrol. Hypertens. 2015; 24:366–370. [DOI] [PubMed] [Google Scholar]
- 28. Gascue C., Tan P.L., Cardenas-Rodriguez M., Libisch G., Fernandez-Calero T., Liu Y.P., Astrada S., Robello C., Naya H., Katsanis N.et al.. Direct role of Bardet-Biedl syndrome proteins in transcriptional regulation. J. Cell Sci. 2012; 125:362–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sorusch N., Bauss K., Plutniok J., Samanta A., Knapp B., Nagel-Wolfrum K., Wolfrum U.. Characterization of the ternary Usher syndrome SANS/ush2a/whirlin protein complex. Hum. Mol. Genet. 2017; 26:1157–1172. [DOI] [PubMed] [Google Scholar]
- 30. Adato A., Michel V., Kikkawa Y., Reiners J., Alagramam K.N., Weil D., Yonekawa H., Wolfrum U., El-Amraoui A., Petit C.. Interactions in the network of Usher syndrome type 1 proteins. Hum. Mol. Genet. 2005; 14:347–356. [DOI] [PubMed] [Google Scholar]
- 31. Lefevre G., Michel V., Weil D., Lepelletier L., Bizard E., Wolfrum U., Hardelin J.P., Petit C.. A core cochlear phenotype in USH1 mouse mutants implicates fibrous links of the hair bundle in its cohesion, orientation and differential growth. Development. 2008; 135:1427–1437. [DOI] [PubMed] [Google Scholar]
- 32. Caberlotto E., Michel V., Foucher I., Bahloul A., Goodyear R.J., Pepermans E., Michalski N., Perfettini I., Alegria-Prevot O., Chardenoux S.et al.. Usher type 1G protein sans is a critical component of the tip-link complex, a structure controlling actin polymerization in stereocilia. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:5825–5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. He Y., Li J., Zhang M.. Myosin VII, USH1C, and ANKS4B or USH1G together form condensed molecular assembly via liquid-liquid phase separation. Cell Rep. 2019; 29:974–986. [DOI] [PubMed] [Google Scholar]
- 34. Sahly I., Dufour E., Schietroma C., Michel V., Bahloul A., Perfettini I., Pepermans E., Estivalet A., Carette D., Aghaie A.et al.. Localization of Usher 1 proteins to the photoreceptor calyceal processes, which are absent from mice. J. Cell Biol. 2012; 199:381–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Maerker T., van Wijk E., Overlack N., Kersten F.F., McGee J., Goldmann T., Sehn E., Roepman R., Walsh E.J., Kremer H.et al.. A novel Usher protein network at the periciliary reloading point between molecular transport machineries in vertebrate photoreceptor cells. Hum. Mol. Genet. 2008; 17:71–86. [DOI] [PubMed] [Google Scholar]
- 36. Sorusch N., Yildirim A., Knapp B., Janson J., Fleck W., Scharf C., Wolfrum U.. SANS (USH1G) molecularly links the human Usher syndrome protein network to the intraflagellar transport module by direct binding to IFT-B proteins. Front. Cell Dev. Biol. 2019; 7:216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Overlack N., Maerker T., Latz M., Nagel-Wolfrum K., Wolfrum U.. SANS (USH1G) expression in developing and mature mammalian retina. Vision Res. 2008; 48:400–412. [DOI] [PubMed] [Google Scholar]
- 38. Fabrizio P., Laggerbauer B., Lauber J., Lane W.S., Luhrmann R.. An evolutionarily conserved U5 snRNP-specific protein is a GTP-binding factor closely related to the ribosomal translocase EF-2. EMBO J. 1997; 16:4092–4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Laggerbauer B., Liu S., Makarov E., Vornlocher H.P., Makarova O., Ingelfinger D., Achsel T., Luhrmann R.. The human U5 snRNP 52K protein (CD2BP2) interacts with U5-102K (hPrp6), a U4/U6.U5 tri-snRNP bridging protein, but dissociates upon tri-snRNP formation. RNA. 2005; 11:598–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Keiper S., Papasaikas P., Will C.L., Valcarcel J., Girard C., Luhrmann R.. Smu1 and RED are required for activation of spliceosomal B complexes assembled on short introns. Nat. Commun. 2019; 10:3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Linder B., Hirmer A., Gal A., Ruther K., Bolz H.J., Winkler C., Laggerbauer B., Fischer U.. Identification of a PRPF4 loss-of-function variant that abrogates U4/U6.U5 tri-snRNP integration and is associated with retinitis pigmentosa. PLoS One. 2014; 9:e111754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fabrizio P., Dannenberg J., Dube P., Kastner B., Stark H., Urlaub H., Luhrmann R.. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol. Cell. 2009; 36:593–608. [DOI] [PubMed] [Google Scholar]
- 43. Schindelin J., Rueden C.T., Hiner M.C., Eliceiri K.W.. The ImageJ ecosystem: an open platform for biomedical image analysis. Mol. Reprod. Dev. 2015; 82:518–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B.et al.. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012; 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sedmak T., Wolfrum U.. Intraflagellar transport molecules in ciliary and nonciliary cells of the retina. J. Cell Biol. 2010; 189:171–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sedmak T., Sehn E., Wolfrum U.. Immunoelectron microscopy of vesicle transport to the primary cilium of photoreceptor cells. Methods Cell Biol. 2009; 94:259–272. [DOI] [PubMed] [Google Scholar]
- 47. Schaffert N., Hossbach M., Heintzmann R., Achsel T., Luhrmann R.. RNAi knockdown of hPrp31 leads to an accumulation of U4/U6 di-snRNPs in Cajal bodies. EMBO J. 2004; 23:3000–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Taneja K.L., Lifshitz L.M., Fay F.S., Singer R.H.. Poly(A) RNA codistribution with microfilaments: evaluation by in situ hybridization and quantitative digital imaging microscopy. J. Cell Biol. 1992; 119:1245–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Makarova O.V., Makarov E.M., Liu S., Vornlocher H.P., Luhrmann R.. Protein 61K, encoded by a gene (PRPF31) linked to autosomal dominant retinitis pigmentosa, is required for U4/U6*U5 tri-snRNP formation and pre-mRNA splicing. EMBO J. 2002; 21:1148–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Agafonov D.E., Kastner B., Dybkov O., Hofele R.V., Liu W.T., Urlaub H., Luhrmann R., Stark H.. Molecular architecture of the human U4/U6.U5 tri-snRNP. Science. 2016; 351:1416–1420. [DOI] [PubMed] [Google Scholar]
- 51. Buskin A., Zhu L., Chichagova V., Basu B., Mozaffari-Jovin S., Dolan D., Droop A., Collin J., Bronstein R., Mehrotra S.et al.. Disrupted alternative splicing for genes implicated in splicing and ciliogenesis causes PRPF31 retinitis pigmentosa. Nat. Commun. 2018; 9:4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Golas M.M., Sander B., Will C.L., Luhrmann R., Stark H.. Molecular architecture of the multiprotein splicing factor SF3b. Science. 2003; 300:980–984. [DOI] [PubMed] [Google Scholar]
- 53. Cretu C., Schmitzova J., Ponce-Salvatierra A., Dybkov O., De Laurentiis E.I., Sharma K., Will C.L., Urlaub H., Luhrmann R., Pena V.. Molecular architecture of SF3b and structural consequences of its cancer-related mutations. Mol. Cell. 2016; 64:307–319. [DOI] [PubMed] [Google Scholar]
- 54. Prilusky J., Felder C.E., Zeev-Ben-Mordehai T., Rydberg E.H., Man O., Beckmann J.S., Silman I., Sussman J.L.. FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics. 2005; 21:3435–3438. [DOI] [PubMed] [Google Scholar]
- 55. Linding R., Jensen L.J., Diella F., Bork P., Gibson T.J., Russell R.B.. Protein disorder prediction: implications for structural proteomics. Structure. 2003; 11:1453–1459. [DOI] [PubMed] [Google Scholar]
- 56. Wright P.E., Dyson H.J.. Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 2015; 16:18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nguyen Ba A.N., Pogoutse A., Provart N., Moses A.M.. NLStradamus: a simple Hidden Markov Model for nuclear localization signal prediction. BMC Bioinformatics. 2009; 10:202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Brameier M., Krings A., MacCallum R.M.. NucPred–predicting nuclear localization of proteins. Bioinformatics. 2007; 23:1159–1160. [DOI] [PubMed] [Google Scholar]
- 59. Fei J., Jadaliha M., Harmon T.S., Li I.T.S., Hua B., Hao Q., Holehouse A.S., Reyer M., Sun Q., Freier S.M.et al.. Quantitative analysis of multilayer organization of proteins and RNA in nuclear speckles at super resolution. J. Cell Sci. 2017; 130:4180–4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cremer T., Cremer C.. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2001; 2:292–301. [DOI] [PubMed] [Google Scholar]
- 61. Spector D.L., Lamond A.I.. Nuclear speckles. Cold Spring Harb. Perspect. Biol. 2011; 3:a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schutze T., Ulrich A.K., Apelt L., Will C.L., Bartlick N., Seeger M., Weber G., Luhrmann R., Stelzl U., Wahl M.C.. Multiple protein-protein interactions converging on the Prp38 protein during activation of the human spliceosome. RNA. 2016; 22:265–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Liu S., Rauhut R., Vornlocher H.P., Luhrmann R.. The network of protein-protein interactions within the human U4/U6.U5 tri-snRNP. RNA. 2006; 12:1418–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Huang da W., Sherman B.T., Lempicki R.A.. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009; 4:44–57. [DOI] [PubMed] [Google Scholar]
- 65. Sorusch N., Bauss K., Plutniok J., Samanta A., Knapp B., Nagel-Wolfrum K., Wolfrum U.. Characterization of the ternary Usher syndrome SANS/ush2a/whirlin protein complex. Hum. Mol. Genet. 2017; 26:1157–1172. [DOI] [PubMed] [Google Scholar]
- 66. Sharma A., Takata H., Shibahara K., Bubulya A., Bubulya P.A.. Son is essential for nuclear speckle organization and cell cycle progression. Mol. Biol. Cell. 2010; 21:650–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lu X., Goke J., Sachs F., Jacques P.E., Liang H., Feng B., Bourque G., Bubulya P.A., Ng H.H.. SON connects the splicing-regulatory network with pluripotency in human embryonic stem cells. Nat. Cell Biol. 2013; 15:1141–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Makarova O.V., Makarov E.M., Luhrmann R.. The 65 and 110 kDa SR-related proteins of the U4/U6.U5 tri-snRNP are essential for the assembly of mature spliceosomes. EMBO J. 2001; 20:2553–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Reiners J., Reidel B., El-Amraoui A., Boeda B., Huber I., Petit C., Wolfrum U.. Differential distribution of harmonin isoforms and their possible role in Usher-1 protein complexes in mammalian photoreceptor cells. Invest. Ophthalmol. Vis. Sci. 2003; 44:5006–5015. [DOI] [PubMed] [Google Scholar]
- 70. Chen Z.Y., Hasson T., Kelley P.M., Schwender B.J., Schwartz M.F., Ramakrishnan M., Kimberling W.J., Mooseker M.S., Corey D.P.. Molecular cloning and domain structure of human myosin-VIIa, the gene product defective in Usher syndrome 1B. Genomics. 1996; 36:440–448. [DOI] [PubMed] [Google Scholar]
- 71. Papal S., Cortese M., Legendre K., Sorusch N., Dragavon J., Sahly I., Shorte S., Wolfrum U., Petit C., El-Amraoui A.. The giant spectrin betaV couples the molecular motors to phototransduction and Usher syndrome type I proteins along their trafficking route. Hum. Mol. Genet. 2013; 22:3773–3788. [DOI] [PubMed] [Google Scholar]
- 72. Sorusch N., Wunderlich K., Bauss K., Nagel-Wolfrum K., Wolfrum U.. Usher syndrome protein network functions in the retina and their relation to other retinal ciliopathies. Adv. Exp. Med. Biol. 2014; 801:527–533. [DOI] [PubMed] [Google Scholar]
- 73. van Wijk E., Kersten F.F., Kartono A., Mans D.A., Brandwijk K., Letteboer S.J., Peters T.A., Marker T., Yan X., Cremers C.W.et al.. Usher syndrome and Leber congenital amaurosis are molecularly linked via a novel isoform of the centrosomal ninein-like protein. Hum. Mol. Genet. 2009; 18:51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. De Biasio A., Ibanez de Opakua A., Cordeiro T.N., Villate M., Merino N., Sibille N., Lelli M., Diercks T., Bernado P., Blanco F.J.. p15PAF is an intrinsically disordered protein with nonrandom structural preferences at sites of interaction with other proteins. Biophys. J. 2014; 106:865–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Protter D.S.W., Rao B.S., Van Treeck B., Lin Y., Mizoue L., Rosen M.K., Parker R.. Intrinsically disordered regions can contribute promiscuous interactions to RNP granule assembly. Cell Rep. 2018; 22:1401–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Alberti S., Dormann D.. Liquid-liquid phase separation in disease. Annu. Rev. Genet. 2019; 53:171–194. [DOI] [PubMed] [Google Scholar]
- 77. Banani S.F., Lee H.O., Hyman A.A., Rosen M.K.. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017; 18:285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Banani S.F., Rice A.M., Peeples W.B., Lin Y., Jain S., Parker R., Rosen M.K.. Compositional control of phase-separated cellular bodies. 2016; 166:651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Girard C., Will C.L., Peng J., Makarov E.M., Kastner B., Lemm I., Urlaub H., Hartmuth K., Luhrmann R.. Post-transcriptional spliceosomes are retained in nuclear speckles until splicing completion. Nat. Commun. 2012; 3:994. [DOI] [PubMed] [Google Scholar]
- 80. Novotny I., Blazikova M., Stanek D., Herman P., Malinsky J.. In vivo kinetics of U4/U6.U5 tri-snRNP formation in Cajal bodies. Mol. Biol. Cell. 2011; 22:513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Stanek D., Pridalova-Hnilicova J., Novotny I., Huranova M., Blazikova M., Wen X., Sapra A.K., Neugebauer K.M.. Spliceosomal small nuclear ribonucleoprotein particles repeatedly cycle through Cajal bodies. Mol. Biol. Cell. 2008; 19:2534–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Howard J.M., Sanford J.R.. The RNAissance family: SR proteins as multifaceted regulators of gene expression. Wiley Interdiscip Rev RNA. 2015; 6:93–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ahn E.Y., DeKelver R.C., Lo M.C., Nguyen T.A., Matsuura S., Boyapati A., Pandit S., Fu X.D., Zhang D.E.. SON controls cell-cycle progression by coordinated regulation of RNA splicing. Mol. Cell. 2011; 42:185–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Boesler C., Rigo N., Anokhina M.M., Tauchert M.J., Agafonov D.E., Kastner B., Urlaub H., Ficner R., Will C.L., Luhrmann R.. A spliceosome intermediate with loosely associated tri-snRNP accumulates in the absence of Prp28 ATPase activity. Nat. Commun. 2016; 7:11997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Zou J., Chen Q., Almishaal A., Mathur P.D., Zheng T., Tian C., Zheng Q.Y., Yang J.. The roles of USH1 proteins and PDZ domain-containing USH proteins in USH2 complex integrity in cochlear hair cells. Hum. Mol. Genet. 2017; 26:624–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Matera A.G., Wang Z.. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 2014; 15:108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Wheway G., Schmidts M., Mans D.A., Szymanska K., Nguyen T.M., Racher H., Phelps I.G., Toedt G., Kennedy J., Wunderlich K.A.et al.. An siRNA-based functional genomics screen for the identification of regulators of ciliogenesis and ciliopathy genes. Nat. Cell Biol. 2015; 17:1074–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Williams D.S. Usher syndrome: animal models, retinal function of Usher proteins, and prospects for gene therapy. Vision Res. 2008; 48:433–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Graziotto J.J., Farkas M.H., Bujakowska K., Deramaudt B.M., Zhang Q., Nandrot E.F., Inglehearn C.F., Bhattacharya S.S., Pierce E.A.. Three gene-targeted mouse models of RNA splicing factor RP show late-onset RPE and retinal degeneration. Invest. Ophthalmol. Vis. Sci. 2011; 52:190–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Farkas M.H., Grant G.R., White J.A., Sousa M.E., Consugar M.B., Pierce E.A.. Transcriptome analyses of the human retina identify unprecedented transcript diversity and 3.5 Mb of novel transcribed sequence via significant alternative splicing and novel genes. BMC Genomics. 2013; 14:486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zambelli F., Pavesi G., Gissi C., Horner D.S., Pesole G.. Assessment of orthologous splicing isoforms in human and mouse orthologous genes. BMC Genomics. 2010; 11:534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Xu M., Xie Y.A., Abouzeid H., Gordon C.T., Fiorentino A., Sun Z., Lehman A., Osman I.S., Dharmat R., Riveiro-Alvarez R.et al.. Mutations in the spliceosome component CWC27 cause retinal degeneration with or without additional developmental anomalies. Am. J. Hum. Genet. 2017; 100:592–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD023942.