Abstract
Profiling molecular features associated with the morphological landscape of tissue is crucial for investigating the structural and spatial patterns that underlie the biological function of tissues. In this study, we present a new method, spatial gene expression patterns by deep learning of tissue images (SPADE), to identify important genes associated with morphological contexts by combining spatial transcriptomic data with coregistered images. SPADE incorporates deep learning-derived image patterns with spatially resolved gene expression data to extract morphological context markers. Morphological features that correspond to spatial maps of the transcriptome were extracted by image patches surrounding each spot and were subsequently represented by image latent features. The molecular profiles correlated with the image latent features were identified. The extracted genes could be further analyzed to discover functional terms and exploited to extract clusters maintaining morphological contexts. We apply our approach to spatial transcriptomic data from different tissues, platforms and types of images to demonstrate an unbiased method that is capable of obtaining image-integrated gene expression trends.
INTRODUCTION
Until recently, numerous technologies have been developed to analyze spatial gene expression patterns that provide molecular profiling with spatial information in tissues (1). In particular, recent progress in spatial gene expression technologies that apply next-generation sequencing with spatial barcodes, fluorescence in situ hybridization (FISH), or in situ sequencing (ISS) has innovated experimental approaches to decipher the spatial heterogeneity of biological processes (2–7). A spatial context at a single-cell level of resolution has enabled the analysis of the location of heterogeneous cells and their spatial interactions in tumor tissues, as well as the brain, human heart, and inflammatory tissues (5–14).
Although spatial gene expression analyses have been actively developed and applied to various tissues and diseases, analytic methods that integrate transcriptome and imaging data are lacking. Despite the feasibility of analysis that combines gene expression, spatial interaction between different spots of spatial barcodes, and image patterns, most methods have regarded gene expression from spots as independent samples and interpreted similarly to single-cell RNA-sequencing (scRNA-seq) data (5,8–11,14). In particular, one of the advantages of spatial gene expression data is the additional information of coregistered images, which contain both morphological and functional patterns. In this regard, important genes related to the image features can be extracted and further utilized to interrogate molecular profiles underlying structural and morphological architectures.
In this study, we introduce a method for identifying spatial gene expression patterns by deep learning of tissue images (SPADE). SPADE extracts gene expression markers by incorporating morphological patterns of an image patch surrounding each spot that contains transcriptomic data. A convolutional neural network (CNN) was employed to define image latent features associated with gene expression. We present molecular markers of various tissues associated with the morphological landscape to discover not only a spatial trend of gene expression in tissues but also biological processes related to histological architecture.
MATERIALS AND METHODS
Data
The slide images and count data of gene expression for the spots from human breast cancer and adult mouse brain tissues were obtained from a publicly available dataset provided by 10× Genomics (https://www.10xgenomics.com/resources/datasets/). The distance between the center of neighboring spots and the diameter of each spot was 100 and 55 micrometers, respectively in both datasets. For human breast cancer, ‘Block A section 1’ data, which contains a total of 3813 spots and 33538 genes on a Hematoxylin and Eosin (H&E) stained slide, were used for the analysis. For mouse brain, ‘Adult Mouse Brain Section 2 (Coronal)’ data, which contains a total of 2807 spots and 32285 genes on an immunofluorescence stained slide with three channels (DAPI, Anti-NeuN, and Anti-GFAP stains) was utilized. For both tissues, high-resolution tissue images, scale factors, and coordinates of spots were included in the study.
The H&E stained slide images and count data of gene expression for the spots from mouse olfactory bulb and human prostate cancer tissues were obtained from a publicly available dataset provided by SciLife laboratory (https://www.spatialresearch.org/resources-published-datasets/). The distance between the center of neighboring spots and the diameter of each spot were 200 and 100 μm, respectively in both datasets. Among the 12 section slides of the olfactory bulb, ‘MOB Replicate 1’, which contains 267 spots and 16383 genes, was utilized for further analysis. Among the 12 section slides of the prostate, ’P3.3’, which contains 502 spots and 17355 genes and ‘P2.4’, which contains 448 spots and 15697 genes were utilized for downstream analysis. Tissue slide images, spot coordinates, and transformation matrix were downloaded. For prostate cancer data, a pathologic annotation for cancer tissue was additionally included in the analysis (8).
Image feature extraction from tissue images
A high-resolution H&E-stained slide was cropped into multiple square patches. The patch size depends on the size of the entire tissue image. The sizes of the images in breast cancer, olfactory bulb, prostate cancer (P3.3), prostate cancer (P2.4), and brain tissues were 2000 × 2000, 9931 × 9272, 2867 × 3276, 2918 × 3277 and 2000 × 2000 pixels, respectively, and the patch sizes were 48 × 48, 600 × 600, 200 × 200, 190 × 190 and 48 × 48 pixels (approximately 218 × 218, 476 × 476, 562 × 562, 559 × 559 and 218 × 218 μm), respectively. The center of the image patch was determined by sampling spot coordinates from the spatial transcriptomic data. Each image patch was provided as an input for a pretrained CNN (Figure 1A). As a pretrained CNN model, we used VGG-16, which was trained by the classification task of natural images of ImageNet data (15,16). The VGG-16 model was used as a feature extractor. Thus, the last layer, which consists of 1000 nodes for classification labels for ImageNet challenge, was removed. In addition, to apply a patch-based approach that can have variable patch size according to the size of the entire tissue image, convolutional-only layers were included. The last convolutional layer produced 2D images instead of vectors, thus a global-average pooling layer was added, considering the size-adaptive feature extractor (17). This feature extractor produced 512D vectors.
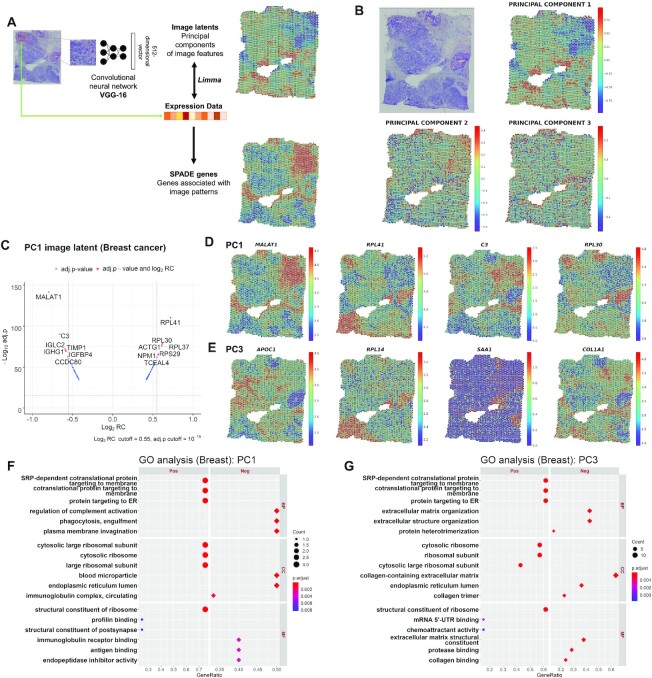
Figure 1.
Discovery of image-integrated spatially variable genes and functional terms in breast cancer data. (A) Multiple patches were extracted from a tissue slide image based on the coordinates of sampling spots. Each image patch was provided as an input to the pretrained convolutional neural network (CNN) model, VGG-16. A total of 512 image features extracted from the CNN were further processed with principal component analysis (PCA) to reduce the dimensions. SPADE genes were constructed by a linear model to identify gene expression correlated with the PC image latent of each spot. (B) Spatial mapping of the PC1, PC2 and PC3 image latent from breast cancer tissue. The PC values of each spot are visualized using colormaps. The maximum and minimum values of the colormap represent two standard deviations above and below the mean value, respectively. (C) Volcano plots for highly associated genes with PC1 image latent features. The cutoff for the log2 regression coefficient (RC) and adjusted P-value (Benjamini–Hochberg correction) is 0.55 and 10−15, respectively. Spatial expression of the top four genes representing the greatest contrast in the (D) PC1 and (E) PC3 image latent space. The top genes are presented in descending order of |log2RC| (FDR < 0.05). The normalized gene expression level of each spot is visualized with colormaps. The maximum and minimum values of the colormap represent two standard deviations above and below the mean expression, respectively. Gene ontology (GO) analysis for (F) PC1 and (G) PC3 SPADE genes showing positive or negative association with PC image latent in breast cancer data. The top 3 positive or negative GO terms for each subcategory, biological process (BP), cellular component (CC) and molecular function (MF), are exhibited in the left and right panel, respectively. The number of overlapping genes is expressed as the size of the dot, and the Benjamini-Hochberg adjusted P-value is exhibited with a colormap.
Dimension reduction for image features
To visualize the image features of all patches or gene features corresponding to spots, t-distributed stochastic neighbor embedding (t-SNE) was employed (18). t-SNE is a nonlinear method of reducing the dimensions of data and visualizing high-dimensional data in low-dimensional space. The perplexity was set at 30, and the initialization of embedding was based on principal component analysis (PCA).
PCA was performed to reduce the dimensionality of 512 features obtained from the VGG-16 model and extract principal components (PCs) (Figure 1A). These PCs were also mapped to the tissue image according to the location of patches to visualize spatial distribution patterns of image features. The whole process was implemented in Python version 3.7.0 with scikit-learn (ver. 0.21.3) and matplotlib (ver. 3.3.1).
SPADE genes
The spots with the number of unique genes less than 200 were excluded from the downstream analysis. A function SCTransform in the R package Seurat (version 3.1.5) was applied to normalize feature counts in each spot (19). To discover SPADE genes, a linear model was generated to fit scaled gene expression of all genes in each spatial transcriptomic dataset to PCs of image latent features. The empirical Bayes algorithm in the R package limma (version 3.44.3) was applied (20), and associated genes based on linear regression analysis for the value of PCs were ranked according to regression coefficient (RC) or corrected P-value with the Benjamini–Hochberg method (Figure 1A). The results for linear regression analysis for each PC were visualized by the R package EnhancedVolcano (version 1.6.0) (https://github.com/kevinblighe/EnhancedVolcano).
List of the genes presenting a false discovery rate (FDR) less than 0.05 in PCs which explain >2% of the variance in 512D image features were gathered to select SPADE genes. The number of utilized PCs were 5, 5, 9, 9 and 3 in the breast, olfactory, prostate (P3.3), prostate (P2.4) and brain tissues, respectively. SPADE genes were filtered by log2RC over 0.58, 0.02, 0.009 and 0.26 for breast, olfactory, prostate (P3.3), and brain tissue, respectively. The threshold was determined according to the results of limma, limiting the pooled number of SPADE genes from all selected PCs between 900 and 1100, except for the prostate tissue (P2.4) which had a total of 313 significant genes and the log2RC threshold was not applied. For spot clustering, SPADE genes derived from each PC were pooled and utilized for downstream analysis. The whole process was performed in R version 4.0.2.
Gene ontology analysis
Gene ontology (GO) analysis was implemented and visualized with the R package clusterProfiler (version 3.16.1) using the enrichGO function (21–23). The top enriched GO terms in subcategories including biological process (BP), cellular component (CC) and molecular function (MF) were extracted based on SPADE genes from each PC image feature. The GO analysis was performed separately in SPADE genes which show a positive or negative association with PC image latent. For a given list of genes, a gene count represents the number of overlapping genes with each GO term, a gene ratio the ratio of the gene count to the number of genes in the list, and background ratio (BgRatio) the ratio of the number of genes in each GO term to all of the genes in annotation database. P-value was calculated based on the hypergeometric model and multiple comparison correction was performed with the Benjamini–Hochberg false discovery rate with the cutoff of 0.05.
Clusters based on expression data of selected genes
The sampling spots were conventionally clustered according to the scRNA-seq analysis workflow in the R package Seurat (24). The spatial information of each spot was not included in this clustering process. The function SCTransform was performed to identify the top 1000 highly variable genes (HVGs), which show the variability of expression across spots. Dimension reduction with PCA followed by shared nearest neighbor (SNN) graph-based spot clustering using the Louvain algorithm was done (25,26). DEG analysis for each highly variable gene (HVG)-based cluster was performed by FindAllMarkers with both thresholds for log fold change and a minimum fraction of detected spots in each cluster as 0.25. The selected marker genes were visualized by a heatmap. As another clustering approach, SPADE genes were used to obtain an SNN graph of spots instead of HVG. Other methods were the same as the HVG-based clustering approach. Dimension reduction with PCA on SPADE genes followed by SNN graph-based spot clustering using the Louvain algorithm was performed as HVGs. The SPADE and HVG-based cluster numbers were rearranged such that the nth SPADE-based cluster shared most of the spots with the nth HVG-based cluster and the shared number of spots is in descending order. Marker genes for each SPADE-based cluster were also extracted by FindAllMarkers with the thresholds identical to HVGs and visualized by a heatmap.
The expression of SPADE genes and clusters was visualized using ComplexHeatmap (version 2.4.3) (27). The clusters generated from SPADE genes and HVGs were compared. For prostate cancer data, the pathologic annotation for each spot was compared with SPADE and HVG-based clusters. The agreement between the pathologic annotation and the two spot clusters was evaluated by adjusted rand indices. The computation was performed with R package mclust (version 5.4.6) (28). In addition, SPADE and HVG-based clusters were spatially mapped to tissue images. Mismatched spot clusters with different SPADE and HVG-based cluster numbers were mapped to the tissue images.
RESULTS
Markers and functional molecular features associated with morphological landscape of breast cancer tissue
We discovered important gene markers correlated to image features extracted by a CNN (Figure 1A). Five publicly available datasets were analyzed to identify gene expression markers associated with the morphological landscape, as defined as SPADE genes. Image latent features represented by 512D vectors were extracted by a pretrained CNN model, VGG-16 (15), from image patches surrounding spots that correspond to transcriptome data. To define highly variable image latent features, PCA was applied to the output of the VGG-16 for all patches corresponding to spots. SPADE genes were identified by linear regression analysis with PCs of image features.
First, SPADE was applied to human breast cancer tissue containing 3813 sampling spots. The dimensions of 512 image features were reduced by PCA, and PC1, PC2 and PC3 values of each spot were mapped to an H&E slide (Figure 1B). PC1 to PC3 explained 68.83%, 13.81% and 5.49% of the data variance in 512D image latent, respectively. Genes associated with PC1, PC2 and PC3 were identified (Figure 1C, Supplementary Figure S1, S2 and Table S1). The top 30 genes with FDRs below 0.05 were selected and then represented as a heatmap according to the increase in PCs (Supplementary Figure S3). The top 4 genes, MALAT1, RPL41, C3 and RPL30, from PC1 image latent, were mapped to the tissue (Figure 1D). The top 4 genes from PC2 and PC3 image latent also showed spatially variable expression according to morphological patterns (Supplementary Figure S4 and Figure 1E).
GO analysis (21,22) was performed with SPADE genes derived from PCs. PC1 SPADE genes presenting positive association with image feature were enriched with endoplasmic reticulum and ribosomal GO terms and negative association with complement system and humoral immunity (Figure 1F and Supplementary Table S2). Meanwhile, PC2 SPADE genes overrepresented GO terms regarding the metabolic process in a positive group (Supplementary Figure S5). PC3 overrepresented similar terms to PC1 in a positive group and extracellular matrix terms in a negative group (Figure 1G).
Markers and functional terms related to morphological patterns of olfactory bulb and prostate cancer tissues
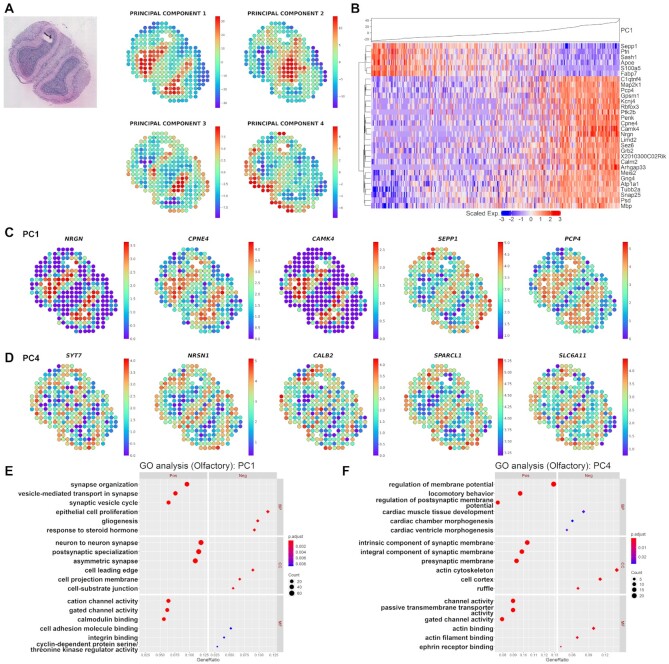
SPADE was applied to olfactory bulb and prostate cancer datasets which have sparser distances between the neighboring spots compared to the breast cancer data. The olfactory bulb included 267 spots with spatial gene expression data. PC1 to PC4 image latents which explain 64.23%, 10.23%, 5.36% and 3.39% of data variance were spatially mapped to the tissue (Figure 2A). The top associated genes with each PC value were presented as volcano plots, scatter plots and heatmaps (Figure 2B, Supplementary Figure S6–S8 and Table S1). Since none of the genes were significantly associated with PC2 image latent, it was excluded from further analysis (FDR < 0.05). The expression of the top genes gradually changed according to the increase of the PC value. When the top 5 genes were mapped to the tissue, it showed distinct gene expression patterns in different layers of the olfactory bulb (Figure 2C, D and Supplementary Figure S9). Among the top 5 marker genes from PC1, NRGN and CAMK4 are known markers for the granule cell layer (29).
Figure 2.
Investigation of morphological marker genes and functions in olfactory bulb data. (A) Spatial mapping of the PC1, PC2, PC3 and PC4 image latents. The PC values of each spot are visualized using colormaps. The maximum and minimum values of the colormap represent two standard deviations above and below the mean value, respectively. (B) Heatmap for the top 30 highly associated genes for log2RC in the PC1 image latent space from olfactory bulb tissue. Hierarchical clustering was performed for the top 30 genes, and the PC1 value in each of the spots is shown at the top. Spatial expression of the top 5 genes representing the greatest contrast in the (C) PC1 and (D) PC4 image latent space from olfactory bulb tissue. The top genes are presented in descending order of |log2RC| (FDR < 0.05). The normalized gene expression level of each spot is visualized with colormaps. The maximum and minimum values of the colormap represent two standard deviations above and below the mean expression, respectively. Gene ontology (GO) analysis for (E) PC1 and (F) PC4 SPADE genes showing positive or negative association with PC image latent in olfactory bulb data. The top 3 positive or negative GO terms for each subcategory, biological process (BP), cellular component (CC) and molecular function (MF), are exhibited in the left and right panel, respectively. The number of overlapping genes is expressed as the size of the dot, and the Benjamini-Hochberg adjusted P-value is exhibited with a colormap.
Functional enrichment analysis was performed and revealed the GO terms related to morphological patterns of the tissue. SPADE genes from PC1 image latent were enriched with ‘synapse organization’, ‘neuron to neuron synapse’ and ‘cation channel activity’ in a positive group and ‘epithelial cell proliferation’, ‘cell leading edge’ and ‘cell adhesion molecule binding’ in a negative group (Figure 2E). On the other hand, PC4 positive SPADE genes overrepresented terms related to membrane potential while negative SPADE genes regarding actin cytoskeleton (Figure 2F and Supplementary Table S2).
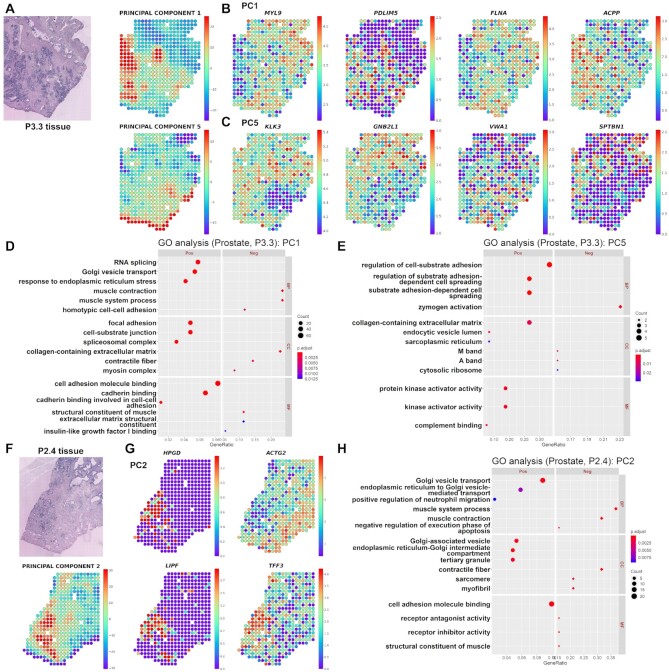
The utility of SPADE was further validated in prostate cancer tissue (P3.3) which was analyzed in a previous spatial transcriptomics paper (8). PC1, PC3 and PC5 values which explained 16.70%, 7.69% and 4.45% of data variance were spatially mapped to the tissue (Figure 3A and Supplementary Figure S10). There were no significantly associated genes with PC2 and only one associated gene (PXDN) in PC4 image latents, thus results from the two PCs were not visualized. The top associated genes with each PC were identified and exhibited by heatmaps for gene expression (Supplementary Figure S11–S13 and Table S1). The top genes were differentially expressed in cancer and non-cancer tissues as presented in Figure 3B, C and Supplementary Figure S14.
Figure 3.
Identification of morphologic markers and functional terms in prostate cancer data (P3.3 and P2.4). (A) Spatial mapping of the PC1 and PC5 image latents in P3.3 tissue. The PC values in each spot are visualized using colormaps. The maximum and minimum values of the colormap represent two standard deviations above and below the mean value, respectively. Spatial mapping of the top 4 genes showing the greatest contrast in (B) PC1 and (C) PC5 image latent space. The top genes are presented in descending order of |log2RC| (FDR < 0.05). The normalized gene expression level in each spot is visualized using colormaps. The maximum and minimum values of the colormap represent two standard deviations above and below the mean expression, respectively. Gene ontology (GO) analysis was performed in P3.3 tissue for (D) PC1 and (E) PC5 SPADE genes showing positive or negative association with PC image latent. The top 3 positive or negative GO terms for each subcategory, biological process (BP), cellular component (CC), and molecular function (MF), are exhibited in the left and right panel, respectively. The number of overlapping genes is expressed as the size of the dot, and the Benjamini–Hochberg adjusted P-value is exhibited with a colormap. (F) Spatial mapping of the PC2 image latent in P2.4 tissue. The PC values in each spot are visualized using a colormap. The maximum and minimum values of the colormap represent two standard deviations above and below the mean value, respectively. (G) Spatial mapping of the top 4 genes showing the greatest contrast in PC2 image latent space. The top genes are presented in descending order of |log2RC| (FDR < 0.05) in the top and bottom rows. (H) The GO analysis was implemented in P2.4 tissue for PC2 SPADE genes presenting a positive or negative association with PC2 image latent. The top 3 positive or negative GO terms for each subcategory, BP, CC and MF, are exhibited in the left and right panel, respectively. The number of overlapping genes is expressed as the size of the dot, and the Benjamini-Hochberg adjusted P-value is exhibited with a colormap.
The GO analysis was implemented for PC1, PC3 and PC5 SPADE genes. The PC1 overrepresented functional terms such as ‘RNA splicing’, ‘Golgi vesicle transport’, ‘focal adhesion’ and ‘cell adhesion molecule binding’ in a positive group while ‘muscle contraction’ and ‘collagen-containing extracellular matrix’ in a negative group (Figure 3D). On the other hand, PC3 genes were enriched with GO terms regarding ‘negative T-cell selection’ in a positive group and PC5 genes were enriched with ‘regulation of cell-substrate adhesion’ in a positive group (Figure 3E, Supplementary Figure S15 and Table S2).
Meanwhile, to assess the reproducibility of the SPADE in the heterogeneous cancer tissues, the analysis was repeated in another prostate cancer dataset (P2.4). PC1 to PC4 image latent which explained 16.38%, 14.63%, 8.61% and 5.96% of data variance were spatially mapped to the tissue (Figure 3F and Supplementary Figure S16A). Also, the top associated genes and GO terms in each PC were investigated (Figure 3G, H, Supplementary Figure S16B–D, S17 and Tables S1, S2). Pairwise cosine distances between 512D eigenvectors of PC image latent from P3.3 (data 1) and P2.4 (data 2) tissues were computed. It revealed that among all PC pairs, PC1 image feature from data 1 and PC2 or PC4 from data 2 extracted top 2 similar morphological patterns (Supplementary Figure S18A). The GO terms for PC1 SPADE genes from data 1 and PC2 genes from data 2 were compared (Figure 3D, H). Among the 229 positive and negative GO terms in data 1 and the 119 GO terms in data 2, 56 terms were overlapped. The top 10 shared GO terms with the highest gene ratio were related to muscle contraction, extracellular matrix and cell adhesion (Supplementary Figure S18B). Likewise, the 229 PC1 GO terms from data 1 and 382 PC4 GO terms from data 2 were compared and showed 47 overlapping genes (Figure 3D and Supplementary Figure S17B). The top shared terms were similar to the PC1–PC2 pairs (Supplementary Figure S18C).
Clustering based on SPADE genes in H&E stained tissue
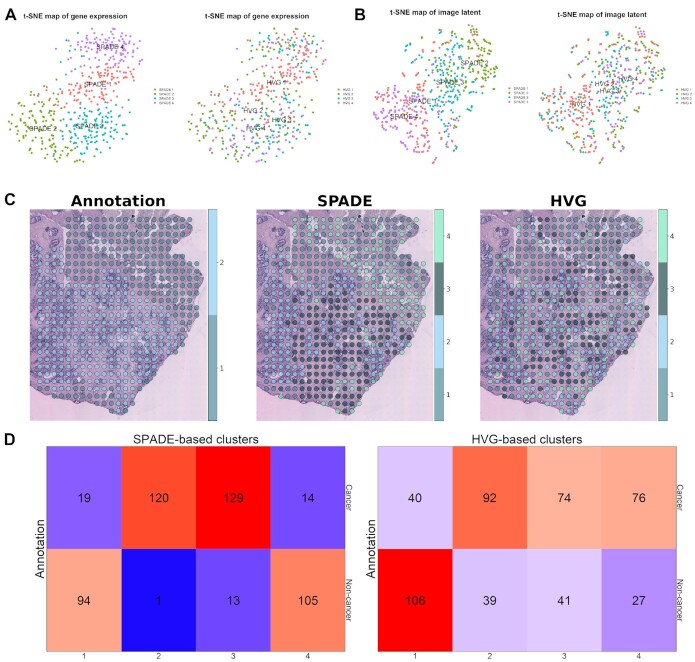
For the next step, spots from prostate cancer tissue were clustered with SPADE genes. In addition, the SPADE-based cluster was compared with the HVG-based cluster to evaluate performance to distinguish cancer from non-cancer tissue. A total of 910 SPADE genes were selected and spots were clustered with SNN graphs (25,26). The markers of clusters were identified (Supplementary Figure S19A and Table S3). As conventional methods use HVGs instead of image-related genes, the patterns of clusters are similar but different. The differentially expressed genes between HVG-based clusters were extracted (Supplementary Figure S19B and Table S3). A heatmap for the SPADE genes revealed distinct gene expression patterns in each SPADE-based cluster (Supplementary Figure S19C). The number of HVGs, SPADE genes and marker genes of clusters derived by HVG and SPADE are represented as a Venn diagram (Supplementary Figure S19D). The clusters based on SPADE genes and HVGs were visualized by two-dimensional t-SNE (18) of transcriptomic data (Figure 4A) and image latents (Figure 4B). Notably, both t-SNE maps of transcriptomic data (Figure 4A) were based on SPADE genes and exhibited overlapping, but different clustering results. In terms of histologic image features, SPADE-based clusters showed relatively separated patterns compared with HVG-based clusters (Figure 4B). The spatial mapping of the SPADE and HVG-based clusters are presented and compared with pathologic annotation for prostate cancer tissue (Figure 4C). Besides, the number of spots shared between the pathologic annotation and SPADE or HVG-based cluster was exhibited as heatmaps (Figure 4D). The adjusted rand index was 3.24 × 10−1 between pathologic annotation and SPADE-based cluster while 7.36 × 10−2 between the annotation and HVG-based cluster. Also, when considering spots in clusters 2 and 3 as cancer and 1 and 4 as non-cancer tissue, diagnostic accuracy was 90.51% in SPADE while in 60.40% HVG-based clusters.
Figure 4.
Spot clustering based on SPADE genes in prostate cancer (P3.3) data. (A) t-SNE plots of transcriptomic data from prostate cancer tissue. Both of the plots were generated based on transcriptomic profiles of SPADE genes. SPADE and HVG-based cluster identity is visualized in the left and right panel, respectively. (B) t-SNE plots for deep learning-derived image features from prostate cancer data. SPADE and HVG-based cluster identity is visualized in the left and right panel, respectively. (C) Spatial distribution of pathologic annotation and clusters based on SPADE genes or HVGs are mapped to the tissue slide. For the pathologic annotation, spot 1 stands for cancer and spot 2 for non-cancer tissues. (D) Cross tables exhibiting the number of overlapping spots between SPADE or HVG-based cluster and pathologic annotation are presented in the left and right panel, respectively. The top row in the cross table shows the spot numbers corresponding to cancer tissue and the bottom row for non-cancer tissue.
The spot clustering was also performed in breast cancer and olfactory bulb tissues. A total of 1073 and 968 SPADE genes were chosen from breast cancer and olfactory bulb data, respectively. For both tissues, the number of shared spots between SPADE and HVG-based clusters was visualized with heatmaps and both clusters were mapped to tissue slides (Supplementary Figure S20, 21). For breast cancer tissue, the highest number of mismatched spots was observed in SPADE 6-HVG 10 cluster (n = 249). The greater ratio of adjacent SPADE 6-HVG 10 mismatched spots to matched spot number was observed in SPADE 6-HVG 6 than SPADE 10-HVG 10 clusters (Supplementary Figure S22A). Meanwhile, in olfactory bulb tissue, the greatest number of mismatched spots was found between SPADE 2-HVG 6 cluster (n = 32). SPADE-based cluster considered spots inside the granule cell layer as one cluster (cluster 2) while the HVG-based cluster divided the layer into two clusters (clusters 2 and 6). The next greater number of mismatched spots was observed in the SPADE 3-HVG 5 cluster (n = 6). All of the spots were located at the boundary of the external plexiform layer (mainly cluster 3) and the mitral cell layer (mainly cluster 5) (Supplementary Figure S22B).
SPADE using immunofluorescence image
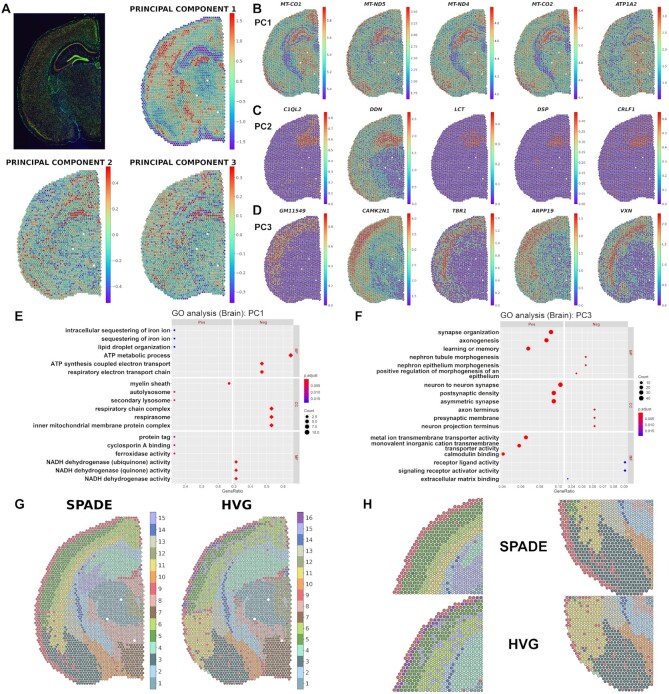
The performance of SPADE was evaluated in a tissue slide other than the H&E stain. The mouse brain tissue with three-channel immunofluorescence staining containing 2807 spots was selected for the analysis. PC1, PC2 and PC3 values which explain 79.64%, 7.73% and 3.57% of the variance in each spot were mapped to the tissue (Figure 5A). The results for linear regression analysis are presented as volcano plots (Supplementary Figure S23). Also, genes showing the greatest regression coefficient in PCs were identified (Supplementary Figure S24). The top 30 genes for PC1, PC2 and PC3 image latents exhibited contrasts in gene expression as the PC values changed (Supplementary Figure S25 and Table S1). The top 5 genes for PC1, PC2 and PC3 revealed distinguishable patterns of gene expression in different cortical layers and the hippocampus (Figure 5B–D).
Figure 5.
Application of SPADE in immunofluorescence stain of mouse brain. (A) Spatial mapping of the PC1, PC2 and PC3 image latents. The PC values in each spot are visualized using colormaps. The maximum and minimum values of the colormap represent two standard deviations above and below the mean value, respectively. Spatial mapping of the top 5 genes showing the greatest contrast in (B) PC1, (C) PC2 and (D) PC3 image latent space. The top genes are presented in descending order of |log2RC| (FDR < 0.05). The normalized gene expression level in each spot is visualized using a colormap. The maximum and minimum values of the colormap represent two standard deviations above and below the mean expression, respectively. Gene ontology (GO) analysis for (E) PC1 and (F) PC3 SPADE genes showing positive or negative association with PC image latent in mouse brain data. The top 3 positive or negative GO terms for each subcategory, biological process (BP), cellular component (CC) and molecular function (MF), are exhibited in the left and right panel, respectively. The number of overlapping genes is expressed as the size of the dot, and the Benjamini-Hochberg adjusted P-value is exhibited with a colormap. (G) Spatial distribution of SPADE and HVG-based spot clusters mapped to the mouse brain tissue. The background immunofluorescence image was removed such that the colors of each cluster are more clearly visualized. The cluster numbers for the SPADE or HVG-based cluster are exhibited in the right panel. (H) Enlarged images showing the spatial distribution of SPADE and HVG-based clusters in cortical layers (left panel) and amygdala (right panel). The images for the SPADE and HVG-based clusters are exhibited in the top and bottom panel, respectively.
Functional gene enrichment analysis showed that the top overrepresented GO terms in PC1 are ‘myelin sheath’ in a positive group and ‘ATP metabolic process’ in a negative group (Figure 5E). For PC2 and PC3, ‘neuron to neuron synapse and ‘postsynaptic density’ were discovered in a positive group and ‘myelin sheath’ and ‘axon terminus’ in a negative group (Figure 5F, Supplementary Figure S26 and Table S2).
Spot clustering was performed based on 1026 SPADE genes from PC1 to PC3 image latents, and marker genes of each SPADE-based cluster were extracted (Supplementary Figure S27A and Table S3). Also, conventional spot clustering with HVGs was done and marker discovery was performed. (Supplementary Figure S27B and Table S3). Heatmaps for SPADE genes showed distinct gene expression patterns across different SPADE-based and HVG-based clusters (Supplementary Figure S27C). Clusters defined by the two different genes were mapped to the tissue slide and spatial distribution of the clusters was visually assessed (Figure 5G). SPADE-based cluster showed a relatively homogeneous distribution of spots within the same cortical layer and amygdala while the HVG-based cluster presented heterogeneous distribution (Figure 5H). The top 2 mismatched clusters having the greatest number of spots were spatially mapped to the tissue and the difference between the two clustering methods was visually assessed (Supplementary Figure S28). SPADE 11-HVG 6 and SPADE 15-HVG 4 mismatched spot clusters were located at the subcortical layer and hippocampus, respectively (Supplementary Figure 28B, C). In summary, SPADE could better delineate the margins of both cortical and subcortical structures compared with HVG-based clustering.
DISCUSSION
SPADE, which integrates histological image patterns with spatial gene expression data, identified genes associated with the morphological landscape. The analysis of five different spatial transcriptomic datasets showed a flexible and scalable application of SPADE to various platforms, as well as tissue types and stain methods. The mapping of SPADE genes on tissue images showed spatial patterns that distinguish the morphological architectures. Moreover, SPADE was able to analyze biological processes related to the morphological heterogeneity of tissues by using conventional analytic methods, such as gene ontology. Since SPADE genes were related to heterogeneous image patterns, it was suggested that clustering based on these genes can effectively preserve morphological contexts.
SPADE can be applied as an investigative tool to unveil key genes or enriched biological processes related to the spatial and morphological heterogeneity of tissue. In breast cancer tissue, the MALAT1 gene showed the greatest variation in expression associated with PC1 image latent features (Figure 1D). MALAT1 is differentially expressed between cancer and stromal tissues and works as a tumor suppressor gene in breast cancer patients (30). As MALAT1 explained the spatial and cellular heterogeneity of tumor tissue, it can be assumed to be a key molecular feature underlying the morphological heterogeneity of the tumor microenvironment. Furthermore, the enriched GO terms for SPADE genes of breast cancer tissue included immune response and extracellular matrix (Figure 1F, G), which are important components of the tumor microenvironment, exhibited under H&E staining (31,32). The biological implications of SPADE genes were also found in olfactory bulb and prostate cancer tissues. NRGN, one of the spatial marker genes in the olfactory bulb (Figure 2C) and a gene when translated to a protein, binds to calmodulin and modifies the downstream signaling pathway in neurons, is localized in the granule cell layer of the olfactory bulb (33). MYL9, a component of the myosin light chain, is depleted in prostate cancer compared to non-cancer tissue (34). Besides, PDLIM5, a gene related to actin cytoskeleton organization, aids in the proliferation and migration of cancer cells (35). MYL9 and PDLIM5 were selected as PC1 or PC2 spatial markers in prostate cancer tissues (Figure 3B, Supplementary Figure S16B and Table S1) and among the GO terms, the muscle contraction and cell adhesion were highly enriched in SPADE genes of prostate cancer tissue (Figure 3D, E, H and Supplementary Figure S17). Meanwhile, in mouse brain immunofluorescence data, PC1 image features were associated with mitochondrial genes and corresponding negative GO terms (Figure 5B, E and Supplementary Figure S23A). There is little evidence that mitochondrial genes are differentially expressed across brain cortical and subcortical structures. However, the density of DAPI nuclear stain increases along with PC1 value and total unique molecular identifier (UMI) counts are significantly correlated with PC1 value (Supplementary Figure S29), thus PC1 image latent reflects the density of the nucleus in the patches. The expression of mitochondrial genes dominates in nucleus depleted regions of the tissue where PC1 value is low, probably resulting in the extraction of mitochondrial genes in top PC1 negative genes.
The molecular markers and functions identified by SPADE showed variable patterns according to different image latent features. In the case of P3.3 prostate cancer tissue, PC1 and PC5 image latent features represented the different morphologic patterns of cancer tissues (Figure 3A). The SPADE genes and molecular functions were selected accordingly, thus GO terms for PC1 and PC5 were similar but different. PC1 and PC5 SPADE genes overrepresented functions such as ‘Golgi vesicle transport’ and ‘regulation of cell-substrate adhesion’ which are related to the pathogenesis of prostate cancer (Figure 3D, E) (35,36). Also, in the breast cancer tissue, PC1 extracted image features in the cancer tissue while PC3 extracted in both extracellular matrix and cancer tissues (Figure 1B). GO terms associated with PC1 and PC3 were similar but different. PC1 negatively related genes included and ‘regulation of complement activation’, ‘phagocytosis’ and ‘immunoglobulin receptor binding’ which are associated with immune functions (Figure 1F). On the other hand, PC3 negatively related genes were mainly composed of extracellular matrix terms (Figure 1G). As different molecular functions were discovered according to the image latent features, SPADE could lead to the analysis of close interactions of molecular function and morphological patterns. Moreover, SPADE genes were extracted similarly in the morphologically close tissues. The PC1 image latent from P3.3 tissue extracted comparable histological features with PC2 or PC4 image latent from P2.4 and the GO terms in both PCs presented considerable overlap (Supplementary Figure S18). The shared GO terms were related to muscle system, extracellular matrix, and cell adhesion which are well represented morphologically under H&E staining. Therefore, SPADE is capable of extracting similar histological patterns and associated genes and functions across the replicate of heterogeneous cancer tissue.
One of the feasible applications of SPADE is the clustering of spots by preserving the morphological landscape. Since SPADE genes are representative genes responsible for morphological features, clustering of spots based on these genes could reflect the variability of image-level patterns. Accordingly, SPADE-based clusters better matched with pathologic annotation than HVG-based clusters (Figure 4C, D). It implies that SPADE-based clusters closely reflect visual patterns identified by pathologists to delineate the margin of cancer considering morphological features. Furthermore, when applied to brain tissue, the SPADE-based cluster could divide the cortical layers clearly and well-delineated the margin of the amygdala (Figure 5G). The spatial mapping of the top mismatched cluster SPADE 15-HVG 4 revealed that SPADE could divide the hippocampus into subregions while HVG-based clustering could not (Supplementary Figure S28C). Therefore, the SPADE-based clustering method can be used to cluster spots, more reflecting tissue structural architectures, such as human brain cortical layers, in an unsupervised manner because this type of architecture was conventionally defined by morphological features (37).
Besides, in accordance with the validation result for immunofluorescence tissue, SPADE can be combined with different types of images, such as non-invasive imaging and immunohistochemistry, which provide expression patterns of specific proteins if spatially coregistered images are available. In other words, by using different types of images, transcriptomes associated with spatial patterns of a specific protein or function can be analyzed to obtain key markers and investigate functional interactions.
There are several factors that may have influenced the SPADE analysis. Since image patches corresponding to spots are provided as inputs, the density of the spots and size of image patches may significantly affect the result. The distance between the center of spots in the human breast and mouse brain data from 10x genomics was approximately 100 micrometers, while in the olfactory bulb and prostate cancer dataset was 200 micrometers (5,8). Detailed morphological information could have been lost due to the sparse distribution of spots in the olfactory bulb and prostate cancer tissue, and the value of SPADE in exploring marker genes may have been underestimated. In spite of the concerns, SPADE could be applied to datasets with different spatial resolutions. Another inherent problem in SPADE is in the relatively large size of the spot compared with cell or nucleus. The morphological information obtained from the patches could not directly reflect single-cell level heterogeneity. It may lead to difficulty in interpreting the data, specifically in cancer tissues where a mixture of heterogeneous cell types present in a small region of the tissue. By integrating single-cell RNA-seq data and developing spatial transcriptomic data with higher resolution, the suggested limitation may be overcome in the future.
Our approach can help to elucidate the close relationship between molecular function and structure by identifying important genes responsible for the morphological landscape. Several methods have been proposed to find spatially variable genes employing location information of spots and focusing on the representative patterns of spatial gene expression (38–40). However, these methods did not employ image features, thus these methods identify spatially variable markers instead of markers associated with morphological features. Another analysis tool, SpaCell, utilized spatial transcriptomic data along with tissue image features derived from a deep neural network to identify cell type and classify the stage of disease (41). While SpaCell applied the deep learning model to classify cell type and disease stage labels in a supervised manner, feature extraction with SPADE provides unbiased information about morphologically important genes associated with histological features. Recently, a study extracted deep learning-based image features to predict spatial gene expression patterns (42). Compared with this prediction model, SPADE more concentrated on the relationship between morphology and gene features. As transcriptomic data could be combined with different types of data in addition to the deep-learning application, the integrative analysis may provide an opportunity to understand the function and structure of tissues.
In conclusion, SPADE is flexibly used to interrogate molecular profiles responsible for the tissue morphological landscape by listing important genes and biological processes. The integration of different types of data, images, and spatially resolved transcriptomes may help to elucidate the close relationship between structure and molecular functions, which may eventually lead to a comprehensive explanation of the pathophysiology of various diseases.
DATA AVAILABILITY
Five publicly available datasets were utilized in this study. Human breast cancer and mouse brain tissue datasets were downloaded from the 10x genomics homepage (https://www.10xgenomics.com/resources/datasets/) while mouse olfactory bulb and prostate cancer datasets were from SciLife laboratory homepage (https://www.spatialresearch.org/resources-published-datasets/). Python and R source code for SPADE is uploaded on https://github.com/mexchy1000/spade
Supplementary Material
ACKNOWLEDGEMENTS
Author's contributions: H.C. and D.S.L. designed the study. H.C. developed the model. S.B. collected and analyzed data. S.B. and H.C. modified the model. All authors contributed to the interpretation of the data and wrote the paper.
Contributor Information
Sungwoo Bae, Department of Molecular Medicine and Biopharmaceutical Sciences, Graduate School of Convergence Science and Technology, Seoul National University, Seoul, Republic of Korea; Department of Nuclear Medicine, Seoul National University Hospital, Seoul, Republic of Korea.
Hongyoon Choi, Department of Nuclear Medicine, Seoul National University Hospital, Seoul, Republic of Korea; Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea.
Dong Soo Lee, Department of Molecular Medicine and Biopharmaceutical Sciences, Graduate School of Convergence Science and Technology, Seoul National University, Seoul, Republic of Korea; Department of Nuclear Medicine, Seoul National University Hospital, Seoul, Republic of Korea; Department of Nuclear Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Research Foundation of Korea [NRF-2019R1F1A1061412, NRF-2019K1A3A1A14065446, NRF-2020M3A9B6038086]; Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea [HI19C0339]. Funding for open access charge: National Research Foundation of Korea [NRF-2019R1F1A1061412, NRF-2019K1A3A1A14065446, NRF-2020M3A9B6038086]; Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea [HI19C0339].
Conflict of interest statement. None declared.
REFERENCES
- 1. Strell C., Hilscher M.M., Laxman N., Svedlund J., Wu C., Yokota C., Nilsson M.. Placing RNA in context and space - methods for spatially resolved transcriptomics. FEBS J. 2019; 286:1468–1481. [DOI] [PubMed] [Google Scholar]
- 2. Ke R., Mignardi M., Pacureanu A., Svedlund J., Botling J., Wahlby C., Nilsson M.. In situ sequencing for RNA analysis in preserved tissue and cells. Nat. Methods. 2013; 10:857–860. [DOI] [PubMed] [Google Scholar]
- 3. Lubeck E., Coskun A.F., Zhiyentayev T., Ahmad M., Cai L.. Single-cell in situ RNA profiling by sequential hybridization. Nat. Methods. 2014; 11:360–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen K.H., Boettiger A.N., Moffitt J.R., Wang S., Zhuang X.. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015; 348:aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stahl P.L., Salmen F., Vickovic S., Lundmark A., Navarro J.F., Magnusson J., Giacomello S., Asp M., Westholm J.O., Huss M.et al.. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016; 353:78–82. [DOI] [PubMed] [Google Scholar]
- 6. Wang X., Allen W.E., Wright M.A., Sylwestrak E.L., Samusik N., Vesuna S., Evans K., Liu C., Ramakrishnan C., Liu J.et al.. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science. 2018; 361:eaat5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rodriques S.G., Stickels R.R., Goeva A., Martin C.A., Murray E., Vanderburg C.R., Welch J., Chen L.M., Chen F., Macosko E.Z.. Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution. Science. 2019; 363:1463–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Berglund E., Maaskola J., Schultz N., Friedrich S., Marklund M., Bergenstrahle J., Tarish F., Tanoglidi A., Vickovic S., Larsson L.et al.. Spatial maps of prostate cancer transcriptomes reveal an unexplored landscape of heterogeneity. Nat. Commun. 2018; 9:2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Moffitt J.R., Bambah-Mukku D., Eichhorn S.W., Vaughn E., Shekhar K., Perez J.D., Rubinstein N.D., Hao J., Regev A., Dulac C.et al.. Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science. 2018; 362:eaau5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Thrane K., Eriksson H., Maaskola J., Hansson J., Lundeberg J.. Spatially resolved transcriptomics enables dissection of genetic heterogeneity in stage III cutaneous malignant melanoma. Cancer Res. 2018; 78:5970–5979. [DOI] [PubMed] [Google Scholar]
- 11. Asp M., Giacomello S., Larsson L., Wu C., Furth D., Qian X., Wardell E., Custodio J., Reimegard J., Salmen F.et al.. A spatiotemporal Organ-Wide gene expression and cell atlas of the developing human heart. Cell. 2019; 179:1647–1660. [DOI] [PubMed] [Google Scholar]
- 12. Carlberg K., Korotkova M., Larsson L., Catrina A.I., Stahl P.L., Malmstrom V.. Exploring inflammatory signatures in arthritic joint biopsies with Spatial Transcriptomics. Sci. Rep. 2019; 9:18975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wen S., Ma D., Zhao M., Xie L., Wu Q., Gou L., Zhu C., Fan Y., Wang H., Yan J.. Spatiotemporal single-cell analysis of gene expression in the mouse suprachiasmatic nucleus. Nat. Neurosci. 2020; 23:456–467. [DOI] [PubMed] [Google Scholar]
- 14. Chen W.T., Lu A., Craessaerts K., Pavie B., Sala Frigerio C., Corthout N., Qian X., Lalakova J., Kuhnemund M., Voytyuk I.et al.. Spatial Transcriptomics and In Situ Sequencing to Study Alzheimer's Disease. Cell. 2020; 182:976–991. [DOI] [PubMed] [Google Scholar]
- 15. Simonyan K., Zisserman A.. Very deep convolutional networks for large-scale image recognition. 2014; arXiv doi:04 September 2014, preprint: not peer reviewedhttps://arxiv.org/abs/1409.1556v5.
- 16. Deng J., Dong W., Socher R., Li L., Li K., Fei-Fei L.. Imagenet: a large-scale hierarchical image database. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2009; 248–255. [Google Scholar]
- 17. Zhou B., Khosla A., Lapedriza A., Oliva A., Torralba A.. Learning deep features for discriminative localization. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016; 2921–2929. [Google Scholar]
- 18. Van Der Maaten L. Accelerating t-SNE using tree-based algorithms. J. Mach. Learn. Res. 2014; 15:3221–3245. [Google Scholar]
- 19. Hafemeister C., Satija R.. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019; 20:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K.. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015; 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T.et al.. Gene ontology: tool for the unification of biology. Nat. Genet. 2000; 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. The Gene Ontology Consortium. The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 2019; 47:D330–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yu G., Wang L.G., Han Y., He Q.Y.. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012; 16:284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M. III, Hao Y., Stoeckius M., Smibert P., Satija R.. Comprehensive integration of single-cell data. Cell. 2019; 177:1888–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Levine J.H., Simonds E.F., Bendall S.C., Davis K.L., Amir el A.D., Tadmor M.D., Litvin O., Fienberg H.G., Jager A., Zunder E.R.et al.. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell. 2015; 162:184–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xu C., Su Z.. Identification of cell types from single-cell transcriptomes using a novel clustering method. Bioinformatics. 2015; 31:1974–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gu Z., Eils R., Schlesner M.. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016; 32:2847–2849. [DOI] [PubMed] [Google Scholar]
- 28. Scrucca L., Fop M., Murphy T.B., Raftery A.E.. mclust 5: clustering, classification and density estimation using gaussian finite mixture models. R J. 2016; 8:289–317. [PMC free article] [PubMed] [Google Scholar]
- 29. Lein E.S., Hawrylycz M.J., Ao N., Ayres M., Bensinger A., Bernard A., Boe A.F., Boguski M.S., Brockway K.S., Byrnes E.J.et al.. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007; 445:168–176. [DOI] [PubMed] [Google Scholar]
- 30. Kwok Z.H., Roche V., Chew X.H., Fadieieva A., Tay Y.. A non-canonical tumor suppressive role for the long non-coding RNA MALAT1 in colon and breast cancers. Int. J. Cancer. 2018; 143:668–678. [DOI] [PubMed] [Google Scholar]
- 31. Garaud S., Buisseret L., Solinas C., Gu-Trantien C., de Wind A., Van den Eynden G., Naveaux C., Lodewyckx J.N., Boisson A., Duvillier H.et al.. Tumor infiltrating B-cells signal functional humoral immune responses in breast cancer. JCI Insight. 2019; 5:e129641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Henke E., Nandigama R., Ergun S.. Extracellular matrix in the tumor microenvironment and its impact on cancer therapy. Front. Mol. Biosci. 2019; 6:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gribaudo S., Bovetti S., Garzotto D., Fasolo A., De Marchis S.. Expression and localization of the calmodulin-binding protein neurogranin in the adult mouse olfactory bulb. J. Comp. Neurol. 2009; 517:683–694. [DOI] [PubMed] [Google Scholar]
- 34. Huang Y.Q., Han Z.D., Liang Y.X., Lin Z.Y., Ling X.H., Fu X., Cai C., Bi X.C., Dai Q.S., Chen J.H.et al.. Decreased expression of myosin light chain MYL9 in stroma predicts malignant progression and poor biochemical recurrence-free survival in prostate cancer. Med. Oncol. 2014; 31:820. [DOI] [PubMed] [Google Scholar]
- 35. Liu X., Chen L., Huang H., Lv J.M., Chen M., Qu F.J., Pan X.W., Li L., Yin L., Cui X.G.et al.. High expression of PDLIM5 facilitates cell tumorigenesis and migration by maintaining AMPK activation in prostate cancer. Oncotarget. 2017; 8:98117–98134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Migita T., Inoue S.. Implications of the Golgi apparatus in prostate cancer. Int. J. Biochem. Cell Biol. 2012; 44:1872–1876. [DOI] [PubMed] [Google Scholar]
- 37. Tasic B., Menon V., Nguyen T.N., Kim T.K., Jarsky T., Yao Z., Levi B., Gray L.T., Sorensen S.A., Dolbeare T.et al.. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat. Neurosci. 2016; 19:335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Edsgard D., Johnsson P., Sandberg R.. Identification of spatial expression trends in single-cell gene expression data. Nat. Methods. 2018; 15:339–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Svensson V., Teichmann S.A., Stegle O.. SpatialDE: identification of spatially variable genes. Nat. Methods. 2018; 15:343–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sun S., Zhu J., Zhou X.. Statistical analysis of spatial expression patterns for spatially resolved transcriptomic studies. Nat. Methods. 2020; 17:193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tan X., Su A., Tran M., Nguyen Q.. SpaCell: integrating tissue morphology and spatial gene expression to predict disease cells. Bioinformatics. 2020; 36:2293–2294. [DOI] [PubMed] [Google Scholar]
- 42. He B., Bergenstråhle L., Stenbeck L., Abid A., Andersson A., Borg Å., Maaskola J., Lundeberg J., Zou J.. Integrating spatial gene expression and breast tumour morphology via deep learning. Nat. Biomed. Eng. 2020; 4:827–834. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Five publicly available datasets were utilized in this study. Human breast cancer and mouse brain tissue datasets were downloaded from the 10x genomics homepage (https://www.10xgenomics.com/resources/datasets/) while mouse olfactory bulb and prostate cancer datasets were from SciLife laboratory homepage (https://www.spatialresearch.org/resources-published-datasets/). Python and R source code for SPADE is uploaded on https://github.com/mexchy1000/spade