Abstract
Pentatricopeptide repeat (PPR) proteins are helical repeat-proteins that bind RNA in a modular fashion with a sequence-specificity that can be manipulated by the use of an amino acid code. As such, PPR repeats are promising scaffolds for the design of RNA binding proteins for synthetic biology applications. However, the in vivo functional capabilities of artificial PPR proteins built from consensus PPR motifs are just starting to be explored. Here, we report in vivo functions of an artificial PPR protein, dPPRrbcL, made of consensus PPR motifs that were designed to bind a sequence near the 5′ end of rbcL transcripts in Arabidopsis chloroplasts. We used a functional complementation assay to demonstrate that this protein bound its intended RNA target with specificity in vivo and that it substituted for a natural PPR protein by stabilizing processed rbcL mRNA. We targeted a second protein of analogous design to the petL 5′ UTR, where it substituted for the native stabilizing PPR protein PGR3, albeit inefficiently. These results showed that artificial PPR proteins can be engineered to functionally mimic the class of native PPR proteins that serve as physical barriers against exoribonucleases.
INTRODUCTION
The manipulation of gene expression represents a major challenge for both basic and applied biology. Progress in this field has been made possible by the discovery of natural products holding the potential to be tailored to powerful synthetic tools for genetic engineering. Posttranscriptional mechanisms play a prominent role in the control of gene expression and RNA binding proteins mediate these processes. Thus, the possibility to engineer RNA binding proteins with desired RNA binding specificity has attracted considerable attention (reviewed in 1,2). Pentatricopeptide repeat (PPR) proteins constitute one of the largest families of RNA binding proteins in eukaryotes comprising more than 400 members in higher plants (3). PPR proteins are nucleus-encoded proteins but they function almost exclusively in mitochondria or chloroplasts where they hold various biological activities: protein barriers to RNA degradation, translational activation, recruitment of effectors to specific RNA sites, regulation of important RNA cis-element and remodeling of local RNA structures (reviewed in 4). PPR proteins are characterized by a variable number of tandem repeats (from 2 to over 30) of a degenerate 35 amino acid motif that forms a helix-loop-helix structure (5,6). The consecutive repeats stack to form a right-handed super helix that binds RNA with sequence specificity in a one-repeat/one-nucleotide mechanism (7). The RNA base bound by a PPR repeat is primarily determined by the identity of amino acids at two positions, 5 and 35 (8–11). The modular architecture of PPR repeats and the existence of an amino acid code for RNA base recognition make them an attractive scaffold for the rational design of RNA binding proteins with desired sequence specificity and therefore, the control of RNA functions in vivo. In fact, the PPR code has been successfully used to reprogram natural PPR proteins in plants to bind new mRNA sequences in vivo that are different from their native ones. For example, the PPR code was used to reprogram the sequence specificity and in vivo function of the mitochondrial PPR protein RPF2 in Arabidopsis plants (12). RPF2 possesses 16 PPR repeats and targets two RNA sites sharing a strong sequence identity that are located within the 5′-UTRs of cox3 and nad9 genes to define the 5′ end processing of these transcripts by promoting a likely 5′-3′ endonucleolytic activity (13). Colas des Francs-Small et al. modified the amino acid composition of the RPF2 PPR tract to reprogram its in vivo specificity and bind a new RNA target within the mitochondrial nad6 ORF which induced its subsequent cleavage. Despite its relative success, the assay highlighted a major ab initio limitation for the engineering of natural PPR proteins: the nucleotide specificity of only two of the 16 PPR motifs in RPF2 could be manipulated, which greatly restricted the choice of the RNA target to a sequence sharing high identity with RPF2’s native targets in mitochondria. The difficulty to freely reprogram the binding specificity of natural PPR proteins was additionally illustrated by a recent study that exploited the maize RNA stabilizer and translation enhancer PPR10 and its cognate chloroplast atpH binding site to build an inducible switch for the expression of plastid transgenes in tobacco (14). In this study, a variant of PPR10 was successfully expressed from the tobacco nuclear genome to stimulate the expression of a chloroplast transgene whose mRNA stability and translation were under control of a modified version of the native PPR10 binding site. As for RPF2, however, the modification of PPR10 sequence specificity did not go further than 2 nucleotides.
Thus, in these two instances, the relative success of manipulating the specificity of natural PPR proteins in vivo is overshadowed by the inability to fully customize all of their PPR repeats to bind any chosen RNA sequence in vivo. The incapacity to recode the specificity of some PPR motifs in natural PPR proteins lies in their amino acid inconsistencies at positions that determine base specificity in relation to the PPR code and their unpredictable contribution to RNA binding (6,15). In order to circumvent the limitation of natural PPR proteins, several synthetic PPR scaffolds have been created (16–18). These artificial PPR proteins (called as well designer PPRs, dPPRs) derived from consensus PPR motifs that offer predictable and reliable sequence specificity in vitro (19). Several in vivo applications have been envisioned for dPPRs and each of these applications derived from two main functions that are naturally occupied by PPR proteins in organelles: the sequestration of RNA from interaction with other proteins or RNA, or the targeting of effectors to specific RNA sites (reviewed in 20). Therefore, artificial PPR proteins must fulfill these two activities in vivo in order to be implementable as tools for the manipulation of RNA functions in living organisms. Currently, there is only one example for the in vivo application of dPPRs (21). In this study, a dPPR protein was successfully engineered in transgenic Arabidopsis plants to capture a specific mRNA in chloroplasts, demonstrating that the artificial dPPR scaffold can selectively and reliably bind designated RNA in vivo. However, this study did not reveal whether dPPRs hold functional potentialities similar to that of natural PPRs in vivo.
Many natural PPR proteins are involved in the control of RNA stability in plant organelles where they specifically bind to intergenic regions of RNA precursors to stabilize and protect the adjacent RNA sequences from degradation by 5′→3′ or 3′→5′ exoribonucleases, thereby defining the 5′ or 3′ ends of the processed RNAs. To test whether dPPRs can fulfill these properties in vivo, we conducted functional complementation assays in Arabidopsis of natural PPR proteins that stabilize the 5′ end of mature chloroplast mRNAs by an RNase blockade mechanism. We showed that a dPPR made of 13 PPR repeats and programmed to bind the in vivo 5′ end of a particular mRNA efficiently substituted for the biological function of the endogenous PPR protein. Furthermore, we demonstrated that positioning the RNA binding site of the dPPR to a different genomic location than that of the natural PPR protein creates a new and functional 5′ end of the processed mRNA in vivo. In conclusion, these results showed that dPPRs hold functional capacities similar to those of natural PPRs by sequestering specific RNA sequences and preventing their access to exoribonucleases. This study provides an additional application of artificial dPPRs in the targeted control of RNA stability in vivo.
MATERIALS AND METHODS
Plant material
The mrl1 Arabidopsis T-DNA homozygote line (SALK_072806, Col-0 background) seeds were a generous donation of Olivier Vallon (22). Arabidopsis plants were grown on soil under controlled conditions: 12-h light (21°C): 12-h dark (18°C); 160–175 μmol photons m–2 s–1. Nicotiana benthamiana plants were grown on soil under standard greenhouse conditions: 16-h light (22°C): 8-h dark (18°C) cycles; 200–250 μmol photons m–2 s–1. Seeds for pgr3-4 homozygote line (FLAG_086B06, Was-0 background) were a gift of Toshiharu Shikanai (Kyoto University) and is the same allele previously analyzed by ribosome profiling (23). Seeds were germinated on Murashige and Skoog (MS) medium supplemented with 2% (w/v) sucrose and grown at 22°C s in 10-h light: 14-h dark (light intensities as stated in Figure legends) before being transferred to soil.
Complementation of mrl1 and pgr3 mutants with dPPR-encoding transgenes
The DNA sequences of Arabidopsis chloroplast transit peptide RecA and dPPRrbcL were codon-optimized for Arabidopsis and synthesized by GeneCust company. The two sequences were assembled using overlapping PCRs. RecA and dPPRrbcL were PCR-amplified using primers K450 Fw/K628 Rev and K627 Fw/K465 Rev, respectively, and assembled by overlapping PCR. Cloning was performed using Gateway technology (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer's protocols. The RecA-dPPRrbcL gene was first cloned into pDONR207 entry vector using BP clonase II and subsequently cloned into binary vectors pAUL1 (for C-terminal protein fusion with 3xHA tag) (24) or pMDC83 (for C-terminal protein fusion with GFP) (25) using LR clonase II. GV3101 Agrobacteria carrying the RecA-dPPRrbcL:pAUL1 construct were used for floral dip transformation of homozygous mrl1 plants and Agrobacteria carrying the RecA-dPPRrbcL:pMDC83 construct were used for tobacco leaf agroinfiltration and protein subcellular localization. Seeds obtained from T0 Arabidopsis plants were sown on soil and the transgenic seedlings were selected for resistance to BASTA® (Bayer AG, Leverkusen, Germany). 30 confirmed transgenic plants were screened for dPPRrbcL expression by immunoblot on leaf total protein extracts using HA antibodies (H9658 clone, Sigma-Aldrich) and three lines expressing gradual levels of dPPRrbcL protein were chosen for phenotypic analyses.
The dPPRpetL sequence was codon optimized for Arabidopsis, synthesized by Genewiz (South Plainfield, New Jersey) and cloned into a modified version of pCambia1300 that allows the C-terminal fusion of the protein with 3xFLAG, as described previously (21). This plasmid was used to transform Arabidopsis pgr3-4 plants by the floral dip method. Transgenic seedlings were selected for resistance to hygromycin on MS medium. Plants were grown for two weeks and pools of 6-to-8 seedlings were harvested for protein and RNA analysis.
Information about the primer sequences can be found in Supplementary Table S1. The DNA and protein sequences used in this study are provided in Supplementary Figure S1.
Immunoblot analysis
Total protein extraction was achieved by homogenizing leaf discs (9 mm diameter) in 150 μl of 2× Laemmli sample buffer (120 mM Tris–HCl, pH 6.8, 4% SDS, 20% glycerol, 2.5% ß-mercaptoethanol, 0.01% bromophenol blue). Samples were centrifuged at 18 000 g for 5 min at room temperature and 10 μl of the supernatant were analyzed by SDS-polyacrylamide gel electrophoresis (10% polyacrylamide). Proteins were transferred onto PVDF membrane by wet western transfer using the Mini Trans-Blot® Cell Assembly (Bio-Rad) in buffer containing 25 mM Tris, 192 mM glycine, 20% ethanol.
PsaD, AtpA, AtpB and PetD antibodies were described previously (26). The NdhL antibody was a gift of Toshiharu Shikanai (Kyoto University). PsbE and PsbA (D1) antibodies were purchased from Agrisera. Monoclonal anti-HA (H9658 clone), anti-FLAG (M2 clone) and anti-Myc (9E10 clone) antibodies were purchased from Sigma-Aldrich.
Subcellular protein localization
A. tumefaciens strain GV3101 carrying the RecA-dPPRrbcL:pMDC83 construct were cultured overnight, pelleted for 5 min at 3200 g and resuspended in 10 mM MES pH 5.6, 10 mM MgCl2, 150 μM acetosyringone to OD600 of 0.3. The bacterial suspension was incubated for 2 h at room temperature in the dark and used for infiltration of leaves of 3-week-old N. benthamina plants. Two days after infiltration, leaves were digested in protoplast extraction medium (0.01% Macerozyme R10, 0.1% Driselase, 0.2% Cellulase ‘Onozuka’, 4.3 g/l MS salt mix, 0.5 g/l MES pH 5.6, 20 g/l sucrose, 80 g/l mannitol) in the dark for 5 h at 30°C with gentle shaking (50 rpm). Protoplasts were examined under a Zeiss LSM 780 confocal microscope. GFP was excited with a 488 nm laser and emission was acquired between 493 and 556 nm. RFP and chlorophyll were excited with a 561 nm laser line and emissions were acquired between 588–641 and 671–754 nm, respectively.
Purification of recombinant dPPR proteins
DNA encoding dPPRrbcL without the transit peptide sequence was PCR-amplified (K461 Fw/K462 Rev) and cloned into the pMAL-TEV vector using BamHI/SalI restriction sites. DNA encoding dPPRpetL without the transit peptide sequence was PCR-amplified (K1245 Fw/K1227 Rev) and cloned in the destination vector pHMGWA (27) by a two-step Gateway® cloning following the manufacturer's instructions (Thermo Fisher). This destination vector expresses an N-terminal six-histidine in addition to the Maltose binding protein tag. Rosetta 2 E. coli cells containing the protein expression vector were grown to OD600 of 0.5 and protein expression was induced with 1 mM IPTG for 3.5 h at 20°C and 220 rpm shaking. Bacteria were pelleted and resuspended in lysis buffer (30 mM Tris pH 7.5, 450 mM NaCl, 5 mM ß-mercaptoethanol, cOmplete™ EDTA-free Protease Inhibitor Cocktail (Roche) with the addition of 5 mM of imidazole for rdPPRpetL) and lysed by sonication. Recombinant dPPRrbcL and dPPRpetL were purified by affinity chromatography to an amylose or Ni-IMAC resin.
Gel mobility shift assays
Synthetic RNA oligos were 5′ end-labeled with [γ-32P]-ATP. Binding was performed for 3 h at 25°C in 20 μl reactions containing increasing amounts of rdPPR, 20 pM of RNA, 100 mM NaCl, 40 mM Tris pH 7.5, 4 mM DTT, 0.5 mg/ml Heparin, 10% glycerol and 1 U RNasin Plus RNase Inhibitor (Promega). Reactions were separated on native 5% polyacrylamide gels in 0.5× TBE buffer at 4°C and gels were dried after electrophoresis and exposed to a phoshorimaging plate.
In vivo protein labeling
In vivo labeling was performed on leaf discs from 4-week-old plants as described (28). 35S-methionine pulse labeling was carried out for 15 min and total proteins were extracted as described previously. 100 000 cpm per sample were resolved on SDS-PAGE. After electrophoresis, the gel was stained in 50% methanol, 10% glacial acetic acid, 0.5g/l Coomassie brilliant blue R-250 and vacuum dried before being exposed to a phosphorimager plate.
RNA gel blot hybridization, cRT-PCR and primer extension assays
Total RNA was extracted from 2-week-old plants using Tri-Reagent (Molecular Research Center, Inc.) according to the TRIzol protocol (Thermo Fisher Scientific, Waltham, MA, USA). RNA was further purified by an additional phenol/chloroform extraction and ethanol precipitation. Five micrograms of total leaf RNA were resolved on denaturing formaldehyde gels as described (29). RNA was blotted onto Hybond-N+ membrane (GE Healthcare) by capillary transfer overnight using 20× SSC as blotting buffer. 60-mer DNA oligonucletoides were 5′ end-labeled using [γ-32P]-ATP and T4 Polynucleotide Kinase (PNK) (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer's instructions. Blots were hybridized overnight at 50°C in Church buffer (7% SDS, 0.5 M NaPhosphate pH 7.0, 1 mM EDTA) and washed twice with washing buffer (1× SSC, 0.1% SDS) for 5 min at the hybridization temperature. Results were visualized on an Amersham Typhoon imager. Analysis of petL RNA by RNA gel blot hybridization used 6 μg leaf RNA and a 5′-end labeled synthetic DNA probe complementary to the petL open reading frame. The blots were hybridized overnight at 48°C in Church buffer and washed five times for 10 min in 0.2% SDS and 5× SSC at 48°C.
cRT-PCR was carried out as described (30) except that SSIV RT (Thermo Fisher Scientific, Waltham, MA, USA) and GoTaq (Promega) enzymes were used. K801 Rev primer was used for cDNA synthesis and K801 Rev/K803 Fw primers for the subsequent PCR. To identify the primary transcript ends, RNA was treated with RNA 5′ Pyrophosphohydrolase (NEB) according to the manufacturer's instructions prior to RNA circularization. cRT-PCR products were cloned into pGEM®-T Easy vector (Promega) and more than 10 clones/cRT-PCR product were sequenced.
For primer extension assays of rbcL, 10 μg of total RNA were incubated in the presence of [γ-32P]-ATP 5′ end-labeled K626 Rev primer and 0.5 mM dNTPs at 75°C for 5 min. Temperature was then reduced to 50°C and SSIV RT enzyme mix (Thermo Fisher Scientific, Waltham, MA, USA) was added according to the manufacturer's protocol for First-Strand cDNA Synthesis Reaction. Reactions were incubated for 30 min at 50°C and stopped by adding one volume of RNA loading buffer (90% deionized formamide, 20 mM Tris pH 8.0, 20 mM EDTA, traces of bromophenol blue and xylene cyanole) and heating for 5 min at 95°C. Extension products were loaded onto an 8% polyacrylamide gel containing 8 M urea and run in 1× TBE buffer. The DynaMarker® Prestain Marker for Small RNA Plus (BioDynamics Laboratory Inc.) was run in parallel on the gel.
Primer extension analysis of the petL 5′ end was performed with leaf RNA (2 μg of the wild-type and 7 μg for the pgr3 mutant) and a 5′-end labeled synthetic DNA oligonucleotide as described previously (31). Products were resolved in an 8% denaturing polyacrylamide gel, and imaged with a Storm phosphorimager.
RIP-Seq and slot blot analysis
RIP-Seq analysis was performed as described previously (32). Briefly, stromal extracts (1 mg protein) were isolated from 2-week-old dPPRrbcL-1 or Col-0 plants and incubated with monoclonal anti-HA antibodies (Sigma). IgGs were captured with Protein A DynaBeads (Thermo Fisher Scientific, Waltham, MA, USA) and co-precipitated RNA was recovered by Trizol extraction followed by phenol/chloroform extraction and ethanol precipitation. The RNA was purified with the Monarch® RNA Cleanup Kit (NEB). 50 ng of RNA from each experiment were used for library generation with the NEBNext® Ultra™ II RNA Library Prep Kit according to the manufacturer's instructions. Deep sequencing (2 × 250 bp, v3 chemistry) was performed on a MiSeq sequencer (Illumina, San Diego, CA, USA) yielding 13.0 and 14.7 Mio (dPPRrbcL-1 replicate 1 and 2, respectively) as well as 15.5 and 13.1 Mio (Col-0 replicate 1 and 2, respectively) trimmed paired reads. To determine depth of coverage (reads/nucleotides), the primary reads were aligned to the Arabidopsis chloroplast genome (accession number NC_000932.1) using CLC Genomics Workbench 6.5.1 (Qiagen, Valencia, CA, USA) with the following parameters: mismatch cost = 2, insertion cost = 3, deletion cost = 3, length fraction = 0.5, similarity fraction = 0.8, global alignment = no, auto-detect paired distances = yes. Aligned reads were extracted as reads per nucleotide and the mean value of the two replicates was displayed across the entire chloroplast genome.
For slot blot analysis, the co-immunoprecipitated RNA was recovered as described above. One-fourth of the pellet RNA and 1/40th of the supernatants were heated at 70°C for 10 min in 2× SSPE buffer (300 mM NaCl, 20 mM NaH2PO4, and 2 mM EDTA at pH 7.4) and blotted onto Hybond-N+ membrane (GE Healthcare) using a Minifold I slot blot system (GE Healthcare). Probe labeling and hybridization were as described for RNA gel blot analysis. Some of the blots were stripped to be rehybridized with another gene probe. Results were visualized on an Amersham Typhoon imager and data quantification was performed with ImageQuant TL (GE Healthcare).
RESULTS
Customization of a dPPR targeting the 5′ end of mature rbcL mRNA
To test whether artificial PPR proteins built from consensus PPR motifs have similar functional capacities as natural P-type PPR proteins in vivo, we used an in vivo functional complementation assay of the Arabidopsis PPR protein MRL1. MRL1 is a PPR protein that is targeted to chloroplasts, where it stabilizes a processed isoform of RNA from the rbcL gene. rbcL encodes the large subunit of Ribulose-1,5-bisphosphate carboxylase-oxygenase (Rubisco) and is transcribed into a primary transcript whose 5′ end maps at position –177 from the start codon (Figure 1A) (22,33,34). This mRNA precursor is processed to a transcript possessing a shorter 5′-UTR mapping at position -69. Accumulation of the processed isoform requires MRL1, and processing is presumed to involve the canonical PPR barrier mechanism: 5′-to-3′ exonucleolytic degradation back to the MRL1 barrier bound immediately downstream of –69 (22). The RNA fragments bound by PPR proteins that stabilize RNA usually accumulate as small RNA footprints (sRNAs) whose termini coincide with the positions of the 5′ or 3′ ends of the mRNAs they stabilize in vivo (35–37). The MRL1 RNA footprint accumulates as a ∼20–30 nt sRNA whose 5′-end coincides with the position of the processed rbcL mRNA, due to protection by the bound protein (Figure 1A). Despite the loss of the processed rbcL mRNA, mrl1 mutant plants do not show a particular growth or physiological defect besides a slight reduction of RbcL protein content, suggesting that the primary rbcL transcript is translationally competent to produce sufficient RbcL (22). Therefore, mrl1 serves as an ideal surrogate plant to express an artificial PPR and test its ability to complement the in vivo function of a natural PPR in gene-specific RNA stabilization by monitoring the recovery of the mature rbcL mRNA.
Figure 1.
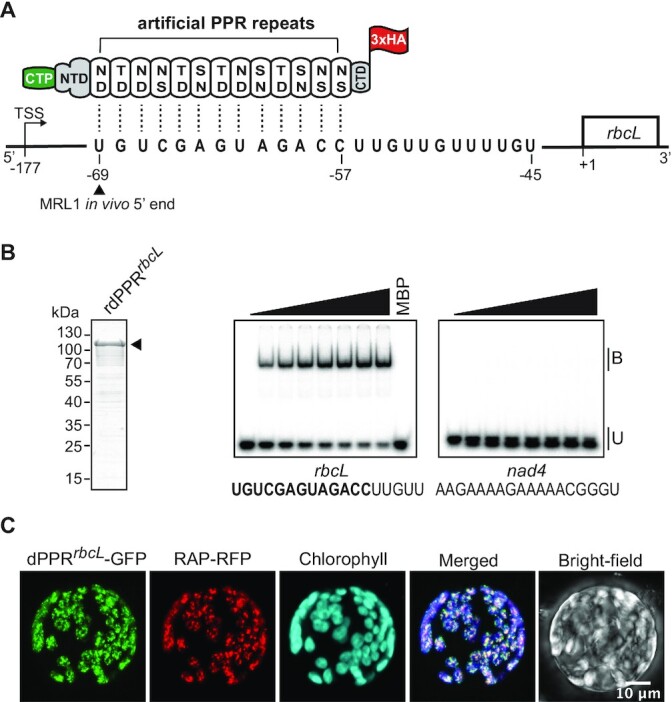
Engineering of an artificial PPR protein (dPPRrbcL) programmed to bind the 5′ end of mature rbcL mRNA in Arabidopsis chloroplasts. (A) Schematic of dPPRrbcL design. The engineered protein was made of 13 consensus PPR tandem repeats programmed to bind a 13 nucleotide sequence beginning 69 nt and finishing 57 nt upstream of rbcL start codon. The dPPR tract was flanked by the N- and C-terminal domains (NTD and CTD) of the maize PPR10 protein. The top and bottom amino-acids featured in each PPR repeat correspond to the amino acid positions 5 and 35 that determine base specificity. An N-terminal chloroplast transit peptide from RecA protein (CTP) and a C-terminal 3xHA epitope tag were added to the protein to allow the chloroplast import and immunodetection of the protein in vivo. The rbcL transcription start site maps at position -177 and the wild-type (WT) in vivo 5′ end of processed rbcL mRNA at position -69. The MRL1 footprint nucleotide sequence is shown. The genomic positions are given according to rbcL start codon. (B) Electrophoretic mobility shift assay (EMSA) showing preferential binding of recombinant dPPRrbcL (rdPPRrbcL) to rbcL 5′ end. (Left) An aliquot of the purified rdPPRrbcL used for binding assays was analyzed by SDS-PAGE and staining with Coomassie Blue. rdPPRrbcL runs on the gel at a predicted size of ∼118 kDa and is indicated by an arrowhead. (Right) rdPPRrbcL was used in EMSAs with radiolabeled RNA oligonucleotides (rbcL or nad4) whose sequences are shown below. The sequence of the designated dPPRrbcL binding site is highlighted in bold. Protein concentrations were 0, 22, 44, 66, 88, 110, 132 and 154 nM for rdPPRrbcL and 200 nM for the maltose binding protein (MBP). Bound (B) and unbound (U) RNAs are indicated. (C) Chloroplast localization of dPPRrbcL in N. benthamiana cells. The dPPRrbcL-GFP fusion protein was transiently co-expressed in tobacco protoplasts with the Arabidopsis chloroplast nucleoid-associated protein RAP fused to RFP. Fluorescence images of GFP, RFP, chlorophyll and a merged image are shown.
To this end, we conceived a protein, dPPRrbcL, targeting the in vivo 5′-end of processed rbcL mRNA according to the artificial PPR design described by Shen et al. (16). The dPPRrbcL was made of 13 consensus PPR repeats flanked by amino- and carboxy-terminal segments of the maize chloroplast-localized protein PPR10 (31) (Supplementary Figure S1), and the PPR tract was programmed to bind a 13 nucleotide sequence matching the processed 5′ end of the mature rbcL mRNA (Figure 1A). We chose a 13-motif design for dPPRrbcL due to evidence that longer artificial PPR tracts are prone to increased off-target binding (19). For the in vivo complementation assay, the dPPRrbcL additionally included the N-terminal chloroplast transit peptide of Arabidopsis RecA (first 68 amino acids) whose efficiency to target exogenous proteins to chloroplasts had already been demonstrated (38) and a C-terminal 3xHA tag for immunodetection (Figure 1A and Supplementary Figure S1). As a preliminary step to the functional complementation assay in vivo, we confirmed the RNA sequence binding specificity of dPPRrbcL by expressing and purifying the mature recombinant protein (lacking the cleaved transit peptide), rdPPRrbcL, in E. coli as an N-terminal protein fusion with the maltose binding protein and we used this protein in gel mobility shift assays (Figure 1B). The rdPPRrbcL bound a 18-nt RNA matching the in vivo 5′ end sequence of processed rbcL mRNA but did not bind an unrelated mitochondrial RNA sequence of the same size (nad4) demonstrating that, as expected, dPPRrbcL binds with specificity to its designated RNA target in vitro. Furthermore, rdPPRrbcL markedly bound its RNA ligand at protein concentrations in the nanomolar range showing that it binds RNA with high affinity.
Next, we tested the subcellular localization of the chimeric dPPRrbcL protein in plant cells. To this end, a dPPRrbcL-GFP fusion protein was transiently expressed in Nicotiana benthamiana leaves and leaf protoplasts were examined by confocal microscopy (Figure 1C). The GFP signal overlapped with the chlorophyll fluorescence demonstrating that dPPRrbcL-GFP was efficiently imported to chloroplasts. In addition, the protein was found in discrete foci within chloroplasts that colocalized with the fluorescence of the nucleoid marker RAP-RFP (39) as demonstrated by the degree of colocalization measured between the two fluorophores (Supplementary Figure 2). These results indicate that dPPRrbcL associates with the nucleoids in chloroplasts. This observation is in agreement with the reported association of MRL1 and several other PPRs controlling mRNA stability with the nucleoids of chloroplasts in maize (40). The results demonstrated that the customized dPPRrbcL was expressed in plant cells where it localized to the chloroplasts similarly to its natural PPR counterpart, MRL1.
dPPRrbcL can substitute for MRL1 to stabilize the 5′ end of rbcL mRNA in vivo
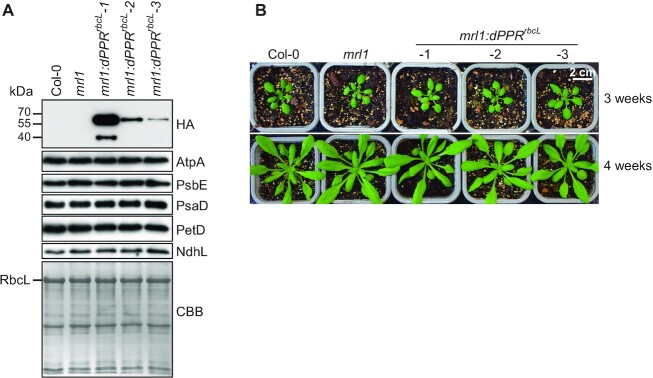
To test the in vivo activity of dPPRrbcL and its capacity to complement the biological function of MRL1, we generated transgenic Arabidopsis plants expressing dPPRrbcL in the mrl1 mutant background. Three independent transgenic lines expressing different levels of dPPRrbcL protein (Figure 2A) were selected for further phenotypic analyses. The expression of dPPRrbcL in the transgenic plants did not cause any growth phenotype as compared to mrl1 and wild-type (Col-0) plants (Figure 2B), and immunoblot analyses using antibodies against core subunits of the major thylakoid membrane complexes did not reveal any change in chloroplast protein accumulation (Figure 2A). These results suggest that expression of dPPRrbcL does not cause deleterious pleiotropic effects in plants.
Figure 2.
Phenotype of transgenic mrl1 Arabidopsis plants expressing dPPRrbcL. (A) Three mrl1:dPPRrbcL independent lines expressing gradual levels of dPPRrbcL were analyzed by immunoblots on total leaf protein extracts along with wild type (Col-0) and mrl1 mutant plants. The dPPRrbcL was detected with antibodies against the HA tag. The abundance of the chloroplast photosynthetic enzyme complexes in the different plant genotypes were analyzed with antibodies against core subunits: AtpA (ATP synthase), PsbE (Photosystem II), PsaD (Photosystem I), PetD (Cytochrome b6f) and NdhL (NDH complex). One of the membrane duplicates was stained with Coomassie brilliant blue (CBB) and serves as the protein loading control. The band corresponding to the large subunit of Rubisco (RbcL) is marked. (B) Visible phenotype of 3- (top) and 4-week-old (bottom) Col-0, mrl1 and transgenic mrl1:dPPRrbcL Arabidopsis plants.
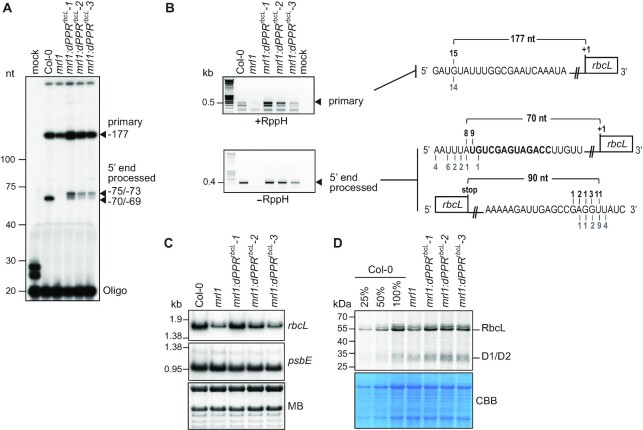
To determine whether dPPRrbcL stabilized the 5′ end of processed rbcL mRNAs in the mrl1 mutant, we performed a primer extension assay that allows the identification of both the primary and 5′-end processed rbcL transcripts (Figure 3A). As expected, both the primary and processed rbcL mRNAs were detected in wild-type plants whereas the processed RNA was absent in mrl1 mutant. However, we observed that mrl1 mutant also showed some reduction in the primary rbcL mRNA suggesting that the protein might have a secondary binding site upstream, or that its absence might expose an otherwise inaccessible endonuclease-sensitive site. In the three transgenic mrl1 lines expressing dPPRrbcL protein, a processed rbcL mRNA isoform accumulated, and the abundance of this isoform correlated with the abundance of the dPPRrbcL protein in each line. Altogether, these results demonstrated that the artificial dPPRrbcL had the capacity to control the stability of its RNA target in vivo and to complement the function of the natural PPR MRL1.
Figure 3.
Functional complementation assay of mrl1 Arabidopsis mutant by dPPRrbcL. (A) Primer extension analysis of rbcL mRNAs 5′ end accumulating in Arabidopsis chloroplasts. The extension products were separated on a 8% denaturing polyacrylamide gel. RNA samples from Col-0, mrl1 and the transgenic plant lines were analyzed. The arrowheads indicate the 5′ end of the primarily transcribed and processed rbcL mRNA. Their positions are given according to the rbcL start codon and their mapping by cRT-PCR. (B) cRT-PCR mapping of the termini of the primary (+RppH) and processed (-RppH) rbcL mRNAs in the Col-0 and complemented mrl1 plants. The arrowheads indicate the cRT-PCR bands that were excised from the agarose gel for sequencing. The diagrams on the right display the RNA sequences annotated with the 5′ or 3′ ends determined by cRT-PCR. The dPPRrbcL binding site is highlighted in bold and the number of clones mapping to each position is indicated above and underneath the RNA sequence for the Col-0 and mrl1:dPPRrbcL genotypes, respectively. RppH: RNA 5′ RNA 5′ Pyrophosphohydrolase. (C) RNA gel blot hybridizations of leaf RNAs from plants of the indicated genotypes using strand-specific oligonucleotide probes for rbcL and a chloroplast control psbE gene. A portion of one of the blots stained with methylene blue (MB) is shown to illustrate equal sample loading. (D) In vivo chloroplast protein synthesis analysis. Intact leaves from the indicated plant genotypes were 35S pulse labelled in presence of cycloheximide to block cytosolic translation. Leaf proteins were fractionated by SDS-PAGE. The autoradiograph and Coomassie blue stained gel (CBB) pictures are shown. The signal bands corresponding to radiolabeled RbcL and D1/D2 proteins are indicated.
The primer extension analysis revealed that the processed 5′ end of rbcL mRNA that accumulated in the transgenic plants was several nucleotides longer than that in the wild-type plants. We conducted cRT-PCR to map precisely the ends of the different rbcL mRNAs accumulating in the different genotypes (Figure 3B). An RNA treatment with the RNA 5′ pyrophosphohydrolase prior to cRT-PCR allowed to distinguish the 5′ ends of the primary rbcL mRNA from the processed ones. In agreement with the primer extension results, the cRT-PCR analysis revealed that the 5′ end of the primary transcript mapped 177 nt upstream of the rbcL start codon in all genotypes. In addition, the results confirmed that the major 5′ ends of the processed rbcL mRNA in the transgenic plants mapped at position –75/–73 from the rbcL start codon, 4 to 6 nucleotides upstream of the processed 5′ end in the wild-type. The position of the 5′-end that is stabilized by dPPRrbcL is as expected based on the fact that PPR10 (from which the N-terminal amino acid region of this dPPR is derived) protects several nucleotides upstream of the sequences bound by its canonical PPR motifs (41). Since we designated the first PPR repeat of dPPRrbcL to bind the immediate 5′ nucleotide of the MRL1-dependent processed rbcL mRNA, the engineered protein was expected to stabilize a processed rbcL mRNA with an end mapping several nucleotides 5′ to that in the wild-type.
Consistent with the loss of the mature rbcL mRNA in the mrl1 mutant, examination of the accumulation of rbcL transcripts by RNA gel blot hybridization showed that the level of rbcL mRNAs was reduced in the mutant compared to the wild-type (Figure 3C). In the complemented mrl1 lines, rbcL mRNA level was restored according to the level of dPPR expression in these plants with a full recovery of mRNA abundance in the strong expressor line that was comparable to the wild-type level.
Despite the reduction of rbcL mRNA abundance in mrl1, Coomassie staining of total leaf protein resolved by SDS-PAGE did not reveal a noticeable reduction of RbcL protein in the mutant compared to the wild type (Figure 2A).To assess RbcL synthesis rate in the different genotypes, we performed pulse labeling analysis using intact plant leaf tissues fed with the 35S-radiolabeled methionine (Figure 3D). The results showed that the loss of rbcL mRNA in mrl1 was accompanied by a small reduction in the rate of RbcL synthesis. This modest effect of MRL1 on Rubisco synthesis and accumulation is consistent with results reported in the original description of the mrl1 mutant (22). The level of rbcL mRNA correlated well with the abundance of neosynthesized RbcL protein in transgenic, wild-type and mrl1 mutant plants. These results indicate that the 5′ end processed rbcL mRNA contributes to the synthesis of RbcL protein in plants and confirmed that the 5′ end processed rbcL mRNA that was newly defined by dPPRrbcL in chloroplasts is functional in the transgenic plants.
Altogether, the molecular analyses demonstrated that the artificial dPPRrbcL protein has the capacity to complement the vivo function of the PPR protein, MRL1 and control RNA stability in chloroplasts.
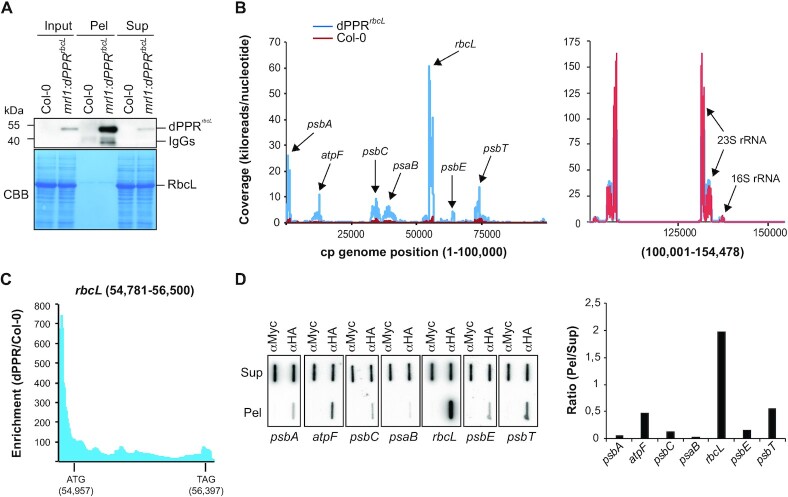
dPPRrbcL binds preferentially to the 5′ UTR of rbcL mRNA in vivo
To get a genome-wide view of dPPRrbcL RNA binding specificity in vivo, we performed RIP-seq analysis using chloroplast stroma isolated from the dPPRrbcL transgenic and wild-type plants in two replicate experiments (Figure 4A). The read values (reads/nucleotide) and read mapping of the two immunoprecipitation replicates for each genotype (wild type and mrl1:dPPRrbcL) were highly reproducible (Supplementary Figure S3). Therefore, the mean read values of the two replicates from the experimental and control immunoprecipitations were aligned to the Arabidopsis chloroplast genome (Figure 4B). The RNA immunoprecipitation using wild-type stroma yielded very low read coverage of the chloroplast genome except for the region covering the rRNA genes (Figure 4B, Supplementary Figure S3) whereas the experiment using the transgenic plants yielded several local peaks in the psbA, atpF, psbC, psaB, rbcL, psbE and psbT genes, with the rbcL peak being the most prominent (Figure 4B and Supplemental Table S2). Consistent with our experimental design, the genomic position of the prominent rbcL peak coincided with the rbcL 5′ UTR (Supplemental Table S2, Supplementary Figure S4 and Figure 4C). Calculation of the enrichment of RNAs in the experimental versus control immunoprecipitations showed that RNAs in the 5′ UTR of rbcL were enriched more than 700-fold whereas RNAs mapping within the rbcL ORF were more weakly enriched (less than 100-fold) demonstrating that dPPRrbcL primarily binds the 5′ UTR (Figure 4C). The RNAs from the other loci (psbA, atpF, psbC, psaB, psbE and psbT) were considered as potential off-targets and were examined in an independent RIP experiment. In this experiment, the experimental and control immunoprecipitations were both performed on stroma from the mrl1:dPPRrbcL transgenic plants but used different antibodies. The immunoprecipitated RNAs with HA antibodies were compared to those from an immunoprecipitation with Myc antibodies that do not recognize dPPRrbcL protein and the recovered RNAs from the pellets and supernatants were subsequently analyzed by slot blot hybridizations (Figure 4D). In agreement with the RIP-seq analysis, the slot blot data confirmed that rbcL, psbA, atpF, psbC, psaB, psbE, psbT RNAs were specifically recovered in the dPPRrbcL immunoprecipitation but with different degrees of enrichment. Quantification of the signals in the pellet versus supernatant showed that rbcL RNA was strongly enriched in the pellet whereas RNAs from psbA, atpF, psbC, psaB, psbE and psbT loci weakly coimmunoprecipitated with dPPRrbcL (Figure 4D). Altogether, the data argue that dPPRrbcL strongly associates to its designated rbcL RNA target in vivo while binding to a few off-targets with less affinity.
Figure 4.
Chloroplast genome-wide analysis of RNAs associated with dPPRrbcL in vivo. (A) Immunoblot analysis of an immunoprecipitation of dPPRrbcL performed with HA antibodies on stroma extracts from the complemented mrl1:dPPRrbcL or Col-0 Arabidopsis plants (negative control). Pel : immunoprecipitation pellet, Sup : supernatant; IgG : Immunoglobuline G. IgGs in the experimental pellet are detected by the secondary antibody used to probe the immunoblot. A portion of the blot stained with Coomassie blue is shown to display equal loading. (B) RIP-seq analysis of chloroplast RNAs that associates with dPPRrbcL in vivo. The mean coverage in kiloreads per nucleotide of two replicates for the experimental dPPRrbcL immunoprecipitation and negative Col-0 control are plotted along the chloroplast genome on the same graph. Since the read coverage was too high for the chloroplast rRNA loci, these were split and displayed in a separate graph to the right. The main peaks are labeled with the names of the locus they belong to. Data for replicate experiments are shown in Supplementary Figure S3 and the read counts are provided in Supplementary Table S2. (C) Local enrichment (Ratio dPPRrbcL/Col-0) of rbcL RNA. The highest enrichment (more than 700-fold) is found in the 5′ UTR region where the binding site of dPPRrbcL is located. The positions of the rbcL initiation (ATG) and stop (TAG) codons are indicated with their genomic position. (D) Slot blot hybridization analysis of RNAs that coimmunoprecipitate with dPPRrbcL in vivo. The experimental and control immunoprecipitations were both performed with mrl1:dPPRrbcL stroma but used different antibodies: α-HA that detects dPPRrbcL or α-Myc as a negative control. Four replicate blots were hybridized with strand and gene specific oligonucleotide probes. The same blot was used for hybridization with psbT and psbE probes, psbC and psaB probes and psbA and atpF probes after stripping. The hybridization signals in the pellet and supernatant of dPPRrbcL immunoprecipitation were quantified for each gene and their ratio is displayed to the right.
dPPRpetL can partially substitute for PGR3 to stabilize the 5′ end of petL mRNA in vivo.
To provide a second example, we addressed whether an artificial PPR protein can functionally substitute for the Arabidopsis PPR protein PGR3. PGR3 has three sites of action: (i) it binds the petL 5′-UTR, where it protects the downstream RNA from degradation and activates petL translation (42–44); (ii) it binds the ndhG 5′ UTR where it activates ndhG translation (45); and (iii) it binds the rpl14-rps8 intergenic region, where it stabilizes a processed 3′-end and mildly stimulates rps8 translation (23). The footprint of PGR3 on petL mRNA is represented in vivo by an abundant sRNA whose 5′-end matches that of the PGR3-dependent processed petL 5′-end (35–37) (Figure 5A).
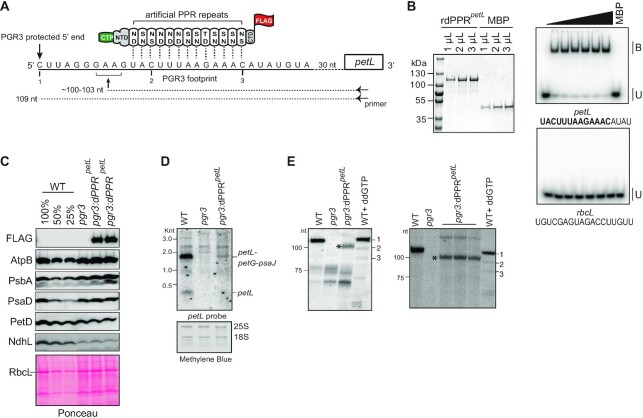
Figure 5.
Partial complementation of PGR3′s petL RNA stabilization function by dPPRpetL. (A) Schematic of dPPRpetL design. The protein consists of 13 consensus PPR motifs programmed to bind the indicated sequence in the petL 5′UTR. The amino acids at positions 5 (top) and 35 (bottom) in each dPPR motif are indicated. The dPPR tract was flanked by the N- and C-terminal regions (NTD and CTD) of maize PPR10, including PPR10’s chloroplast targeting peptide (CTP). A 3xFLAG tag was fused to the C-terminus. The 5′ end that is stabilized by PGR3 in vivo is marked with a vertical arrow, and the PGR3 footprint is underlined with a solid line. The horizontal arrow denotes the primer used for primer extension and the asterisk marks the 5′-end that is stabilized by dPPRpetL. The nucleotides marked 1, 2, and 3 are primer extension stops resulting from the use of the chain terminator ddG, which serve as size markers on the primer extension gels in panel E. (B) EMSA showing preferential binding of recombinant dPPRpetL (rdPPRpetL) to petL 5′ end. (Left) An aliquot of the purified rdPPRpetL and MBP used for binding assays was analyzed by SDS-PAGE and staining with Coomassie Blue. rdPPRpetL has a predicted size of ∼118 kDa. (Right) rdPPRpetL was used in EMSAs with radiolabeled RNA oligonucleotides (petL or rbcL) whose sequences are shown below. The sequence of the designated dPPRpetL binding site is highlighted in bold. Protein concentrations were 0, 5.6, 11.25, 22.5, 45, 90 and 118 nM for rdPPRpetLand 200 nM for the maltose binding protein (MBP). Bound (B) and unbound (U) RNAs are indicated. (C) Immunoblot analysis of pgr3 mutants expressing dPPRpetL. Plants used for this analysis were grown at low light intensity (60 μE). Results from plants grown at 120 μE are shown in Supplementary Figure S5B. Protein analyzed in each lane comes from pooled plants that were confirmed to have the indicated genotype by PCR. Replicate blots were probed with antibodies to the indicated proteins. The FLAG antibody detects dPPRpetL. The Ponceau S-stained blot is shown below to illustrate equal sample loading and the abundance of the large subunit of Rubisco (RbcL). (D) RNA gel blot hybridization demonstrating partial rescue of petL transcripts in pgr3 mutants expressing dPPRpetL. 2 μg of leaf RNA was loaded in each lane. The methylene blue stained blot is shown below to demonstrate equal sample loading. 25S and 18S are cytosolic rRNAs. (E) Primer extension assays demonstrating stabilization of the expected novel petL 5′ terminus by dPPRpetL. The two panels show analysis of four independent pools of pgr3 mutants expressing dPPRpetL. The positions of size markers are shown to the left. The last lane in each gel (WT + ddGTP) shows a primer extension reaction that used wild-type RNA and a trace amount of ddGTP to induce termination at C residues in the template (see panel A). The gel on the left used 4 μg of RNA from the WT (Ws-0) and pgr3 samples, and 14 μg from pgr3:dPPRpetL. The gel on the right used 2.5 μg of RNA from the WT (Ws-0) and pgr3 samples, and 7.5 μg from pgr3:dPPRpetL.
We designed an artificial PPR protein, dPPRpetL to bind a sequence within the PGR3 binding site in petL, but beginning several nucleotides downstream of the PGR3-dependent 5′ end (Figure 5A). The dPPRpetL design was not expected to complement PGR3 functions at ndhG and rpl14-rps16 because those binding sites involve contacts with PGR3’s C-terminal repeats (23,45), which are not represented by dPPR motifs in the synthetic protein (see Figure 6). The protein design is analogous to that of dPPRrbcL except for the use of a different chloroplast targeting sequence (PPR10 versus RecA) and C-terminal tag (FLAG versus HA) (Supplementary Figure S1). The native PPR10 chloroplast targeting peptide was previously shown to target a dPPR to Arabidopsis chloroplasts in vivo (21). As for dPPRrbcL, in vitro EMSA assays showed that recombinant dPPRpetL binds to its intended petL RNA target in vitro with specificity and high affinity (Figure 5B). We introduced the transgene expressing dPPRpetL into the genome of a null pgr3 mutant, and used immunoblot analysis to identify lines expressing dPPRpetL (Figure 5C). The pgr3 mutant has a barely discernable phenotype under the growth conditions we used, and the expression of dPPRpetL did not have an obvious effect on the phenotype (Supplementary Figure S5A). When grown at low light intensity, the pgr3 mutant exhibited a decrease in subunits of the cytochrome b6f complex (PetD) and NDH complex (NdhL) (Figure 5C), as expected based on results reported for different pgr3 alleles (42–44). When grown at a higher but still moderate light intensity, core subunits of other complexes were reduced as well (Supplemental Figure S5B). This is consistent with the mild defect in chloroplast translation reported for the same allele (23). Expression of dPPRpetL had little or no effect on the protein deficiencies. This is consistent with the fact that dPPRpetL was not expected to bind PGR3’s ndhG or rpl14-rps8 sites, together with its modest effects on petL mRNA as discussed below.
Figure 6.
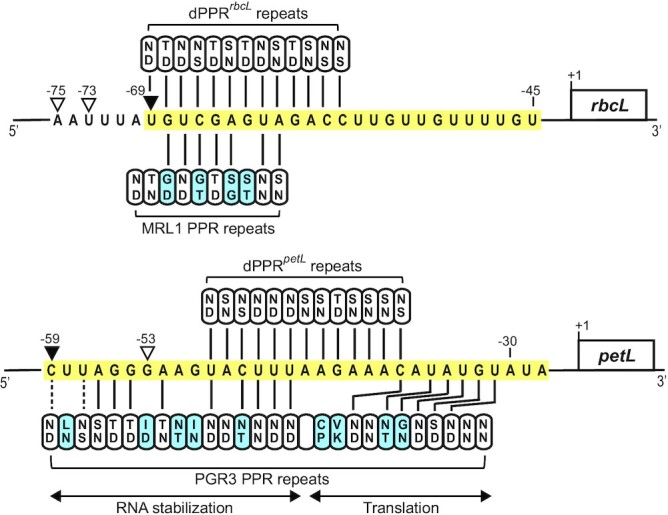
Alignments between dPPRrbcL, dPPRpetL and MRL1, PGR3 and their respective designated or predicted rbcL and petL binding sites. The in vivo footprints of MRL1 and PGR3 are highlighted in yellow. The MRL1- and PGR3- or dPPRrbcL- and dPPRpetL-dependent in vivo 5′ ends are indicated by black or white arrowheads, respectively. The specificity-determining amino acids in each PPR motif (positions 5 and 35) are indicated. The PPR motifs that exhibit canonical amino acid combinations according to the original PPR code (9) are colored in white while the others are in blue. PPR repeats whose amino acid combinations match the nucleotide binding prediction according to a recently extended PPR code (11) are indicated with black lines. Dotted lines indicate compatible PPR nucleotide binding but with low affinity. PGR3 PPR repeats promoting RNA stabilization or translation are underlined (44,45).
RNA gel blot hybridization showed the expected loss of petL transcripts from the pgr3 mutant (Figure 5D). Expression of dPPRpetL resulted in partial restoration of the major PGR3-dependent transcript isoform (a tricistronic petL-petG-psaJ RNA) in the transgenic plants. To probe further into the effects of dPPRpetL on petL RNA metabolism, we mapped petL transcript 5′ ends by primer extension (Figure 5E). Because petL transcripts are considerably less abundant in dPPR-expressing mutants than in the wild-type (Figure 5D), the amount of input RNA for the primer extension assays was adjusted with the intent of increasing signal strength for the dPPR-expressing samples (see Figure 5 legend). The wild-type RNA produced a single product of the expected length (109 nucleotides, transcript 1), and this was missing in the pgr3 mutant as expected. RNA extracted from four different pools of pgr3 mutants expressing dPPRpetL contained a novel shorter transcript whose 5′ end mapped ∼6 nucleotides downstream of the native end (marked with an asterisk). Because dPPRpetL was targeted to a sequence internal to the PGR3 binding site (Figure 5A), the position of the novel terminus matches that expected for the product of exonucleolytic degradation back to a dPPRpetL barrier (∼103 nt, see Figure 5A). These results show that an artificial PPR protein can substitute for PGR3’s petL RNA stabilization function, albeit weakly. Possible explanations for the weak complementation of pgr3 defects by dPPRpetL are discussed below.
DISCUSSION
The PPR motif has been proposed as a promising scaffold for the custom-design of RNA binding proteins with desired RNA sequence specificity for the control of RNA metabolism in vivo (20). The diversity of functions held by natural PPR proteins in vivo predicted many potential applications of PPR-based tools for the manipulation of RNA metabolism. However, first attempts to recode natural PPR proteins to bind new RNA sequences by targeted amino acid mutagenesis faced serious drawbacks due to irregularities found in the amino acid composition of some PPR repeats and the unpredictability of their binding specificity (6,9,15). In addition, many natural PPR proteins appeared to be insoluble when expressed in heterologous systems (7), which limits the ability to confirm sequence specificities of variants prior to in vivo applications. Synthetic consensus PPR repeats (dPPR) were established to overcome these issues (16–18) but their in vivo functionalities have just started to be investigated (21).
MRL1 and PGR3 are ‘pure’ (P-type) chloroplastic PPR proteins that lack any accessory domains. MRL1 and PGR3 consist of 10 and 27 PPR repeats, respectively (22,42). Whereas MRL1 specifically stabilizes processed rbcL mRNAs in Arabidopsis, PGR3 binds three RNA targets (petL 5′ UTR, ndhG 5′ UTR and rpl14-rps8 intergenic) with effects on RNA stability, translation, or both (23,42,45). However, we expected dPPRpetL to bind only the petL site because it did not include repeats that mimic the C-terminal region of PGR3, where specificity to the ndhG and rpl14-rps8 sites is encoded (23,45) (Figure 6). The footprints of MRL1 and PGR3 in the rbcL and petL 5′-UTR, respectively, are represented by abundant sRNA of ∼20–30 nucleotides that accumulate in vivo (Figure 6). However, like many other P-type PPR proteins, some PPR motifs in MRL1 and PGR3 lack canonical amino acids at the specificity-determining positions and the amino acid codes of some motifs do not match the presumed aligned nucleotides in the binding sites (Figure 6). Similarly to what has been reported for the well-characterized PPR protein PPR10 (15), some of these non-canonical interactions might still be critical for the high-affinity binding of these proteins to their RNA binding sites.
Our functional complementation assays of Arabidopsis mutants lacking MRL1 and PGR3 by dPPRs built from 13 synthetic PPR repeats add to the evidence that dPPRs bind their designated RNA targets with specificity in vivo, and additionally revealed that they can protect RNA downstream from their binding site against the action of exoribonucleases. Similarly to the action of native PPR proteins, this dPPR-dependent RNase blockade mechanism allowed the definition of novel 5′-end processed mRNAs in chloroplasts and control of RNA stability. Our RIP-seq experiment showed that dPPRrbcL binds weakly to several off target RNAs in vivo, but this had no apparent effects on plant growth, pigmentation, or the accumulation of photosynthetic complexes harboring plastid-encoded subunits. Weak off-target binding without phenotypic impact was also observed in the sole prior study of in vivo dPPR protein expression (21). Therefore, some degree of off-target binding can be tolerated and does not preclude the use of this protein design to achieve a variety of practical purposes.
Nevertheless, the dPPRrbcL and dPPRpetL data revealed interesting differences in their effects in comparison with their natural PPR counterparts. Whereas dPPRrbcL fully complemented the rbcL mRNA stabilization function of MRL1, dPPRpetL only weakly stabilized its cognate petL mRNA ligand as compared to PGR3. This difference might result simply from different expression levels. However, we suspect that the design of the dPPR tracts is a major factor underlying the different degree of complementation we observed. dPPRrbcL harbors more PPR repeats than its counterpart MRL1 (Figure 6). By contrast, dPPRpetL contains fewer than half of the PPR motifs as PGR3 (Figure 6). Thus, dPPRpetL provides many fewer protein-RNA contacts than does PGR3, which presumably results in much lower RNA binding affinity. Furthermore, roughly 15 of the nucleotides that are masked upon PGR3 binding at petL will be left exposed upon dPPRpetL binding; should these exposed sequences be susceptible to endonucleolytic cleavage, this would compromise the ability of dPPRpetL to stabilize thepetL 5′-end. PGR3 functions have been shown to partition between its N- and C-terminal PPR tracts: the petL RNA stabilization function requires only the N-terminal 16 PPR motifs, whereas the ndhG and petL translation activation functions also require its C-terminal ∼11 PPR motifs (44) (Figure 6). dPPRpetL is not expected to mimic this latter function because it was not designed to interact with the relevant nucleotides at petL (Figure 6).
In conclusion, our results demonstrate that synthetic PPR proteins can be used for the targeted stabilization of specific chloroplastic mRNAs and therefore, the manipulation of organellar gene expression. Finally, this study provides knowledge upon which to develop strategies for the regulation of RNA turn-over in plant organelles by the use of designer PPRs.
DATA AVAILABILITY
The NGS data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (46) and are accessible through GEO Series accession number GSE146249 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE146249).
Sequence data from this article can be found in the EMBL/GenBank libraries under the following accession numbers: MRL1 (At4g34830) and PGR3 (At4g31850) from A. thaliana, A. thaliana chloroplast genome (NC_000932.1).
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Olivier Vallon (CNRS UMR7141) for the gift of mrl1 seeds and Toshiharu Shikanai (Kyoto University) for the gift of pgr3 seeds and NdhL antibody.
Contributor Information
Nikolay Manavski, Institut de Biologie Moléculaire des Plantes, Centre National de la Recherche Scientifique (CNRS), Université de Strasbourg, 12 rue du Général Zimmer, 67084 Strasbourg, France.
Sébastien Mathieu, Institut de Biologie Moléculaire des Plantes, Centre National de la Recherche Scientifique (CNRS), Université de Strasbourg, 12 rue du Général Zimmer, 67084 Strasbourg, France.
Margarita Rojas, Institute of Molecular Biology, University of Oregon, Eugene, OR 97403 USA.
Louis-Valentin Méteignier, Institut de Biologie Moléculaire des Plantes, Centre National de la Recherche Scientifique (CNRS), Université de Strasbourg, 12 rue du Général Zimmer, 67084 Strasbourg, France.
Andreas Brachmann, Genetics, Faculty of Biology, Ludwig-Maximilians-University Munich, 82152 Planegg-Martinsried Germany.
Alice Barkan, Institute of Molecular Biology, University of Oregon, Eugene, OR 97403 USA.
Kamel Hammani, Institut de Biologie Moléculaire des Plantes, Centre National de la Recherche Scientifique (CNRS), Université de Strasbourg, 12 rue du Général Zimmer, 67084 Strasbourg, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Agence National de la Recherche [ANR-18-CE20-0013 to K.H.]; IdEx Unistra from the Investments for the future program of the French Government [to KH]; European Union's Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie action [794377 to N.M.]; U.S.-Israel Binational Agricultural Research and Development Grant [IS-5205-19 to A.B.]. Funding for open access charge: Centre National de la Recherche Scientifique.
Conflict of interest statement. None declared.
REFERENCES
- 1. Wei H., Wang Z.. Engineering RNA-binding proteins with diverse activities. Wiley Interdiscip Rev RNA. 2015; 6:597–613. [DOI] [PubMed] [Google Scholar]
- 2. Filipovska A., Rackham O.. Designer RNA-binding proteins: new tools for manipulating the transcriptome. RNA Biol. 2011; 8:978–983. [DOI] [PubMed] [Google Scholar]
- 3. O’Toole N., Hattori M., Andres C., Iida K., Lurin C., Schmitz-Linneweber C., Sugita M., Small I.. On the expansion of the pentatricopeptide repeat gene family in plants. Mol. Biol. Evol. 2008; 25:1120–1128. [DOI] [PubMed] [Google Scholar]
- 4. Barkan A., Small I.. Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 2014; 65:415–442. [DOI] [PubMed] [Google Scholar]
- 5. Small I.D., Peeters N.. The PPR motif - a TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 2000; 25:46–47. [DOI] [PubMed] [Google Scholar]
- 6. Cheng S., Gutmann B., Zhong X., Ye Y., Fisher M.F., Bai F., Castleden I., Song Y., Song B., Huang J.et al.. Redefining the structural motifs that determine RNA binding and RNA editing by pentatricopeptide repeat proteins in land plants. Plant J. 2016; 85:532–547. [DOI] [PubMed] [Google Scholar]
- 7. Yin P., Li Q., Yan C., Liu Y., Liu J., Yu F., Wang Z., Long J., He J., Wang H.W.et al.. Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature. 2013; 504:168–171. [DOI] [PubMed] [Google Scholar]
- 8. Takenaka M., Zehrmann A., Brennicke A., Graichen K.. Improved computational target site prediction for pentatricopeptide repeat RNA editing factors. PLoS One. 2013; 8:e65343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Barkan A., Rojas M., Fujii S., Yap A., Chong Y.S., Bond C.S., Small I.. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLos Genet. 2012; 8:e1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yagi Y., Hayashi S., Kobayashi K., Hirayama T., Nakamura T.. Elucidation of the RNA recognition code for pentatricopeptide repeat proteins involved in organelle RNA editing in plants. PLoS One. 2013; 8:e57286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yan J., Yao Y., Hong S., Yang Y., Shen C., Zhang Q., Zhang D., Zou T., Yin P.. Delineation of pentatricopeptide repeat codes for target RNA prediction. Nucleic Acids Res. 2019; 47:3728–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Colas des Francs-Small C., Vincis Pereira Sanglard L., Small I.. Targeted cleavage of nad6 mRNA induced by a modified pentatricopeptide repeat protein in plant mitochondria. Commun Biol. 2018; 1:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jonietz C., Forner J., Holzle A., Thuss S., Binder S.. RNA PROCESSING FACTOR2 is required for 5′ end processing of nad9 and cox3 mRNAs in mitochondria of Arabidopsis thaliana. Plant Cell. 2010; 22:443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rojas M., Yu Q., Williams-Carrier R., Maliga P., Barkan A.. Engineered PPR proteins as inducible switches to activate the expression of chloroplast transgenes. Nat Plants. 2019; 5:505–511. [DOI] [PubMed] [Google Scholar]
- 15. Miranda R.G., Rojas M., Montgomery M.P., Gribbin K.P., Barkan A.. RNA-binding specificity landscape of the pentatricopeptide repeat protein PPR10. RNA. 2017; 23:586–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shen C., Wang X., Liu Y., Li Q., Yang Z., Yan N., Zou T., Yin P.. Specific RNA recognition by designer pentatricopeptide repeat protein. Mol Plant. 2015; 8:667–670. [DOI] [PubMed] [Google Scholar]
- 17. Coquille S., Filipovska A., Chia T., Rajappa L., Lingford J.P., Razif M.F., Thore S., Rackham O.. An artificial PPR scaffold for programmable RNA recognition. Nat. Commun. 2014; 5:5729. [DOI] [PubMed] [Google Scholar]
- 18. Gully B.S., Shah K.R., Lee M., Shearston K., Smith N.M., Sadowska A., Blythe A.J., Bernath-Levin K., Stanley W.A., Small I.D.et al.. The design and structural characterization of a synthetic pentatricopeptide repeat protein. Acta Crystallogr. D. Biol. Crystallogr. 2015; 71:196–208. [DOI] [PubMed] [Google Scholar]
- 19. Miranda R.G., McDermott J.J., Barkan A.. RNA-binding specificity landscapes of designer pentatricopeptide repeat proteins elucidate principles of PPR-RNA interactions. Nucleic Acids Res. 2018; 46:2613–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yagi Y., Nakamura T., Small I.. The potential for manipulating RNA with pentatricopeptide repeat proteins. Plant J. 2014; 78:772–782. [DOI] [PubMed] [Google Scholar]
- 21. McDermott J.J., Watkins K.P., Williams-Carrier R., Barkan A.. Ribonucleoprotein capture by in vivo expression of a designer pentatricopeptide repeat protein in Arabidopsis. Plant Cell. 2019; 31:1723–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Johnson X., Wostrikoff K., Finazzi G., Kuras R., Schwarz C., Bujaldon S., Nickelsen J., Stern D.B., Wollman F.A., Vallon O.. MRL1, a conserved pentatricopeptide repeat protein, is required for stabilization of rbcL mRNA in Chlamydomonas and Arabidopsis. Plant Cell. 2010; 22:234–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rojas M., Ruwe H., Miranda R.G., Zoschke R., Hase N., Schmitz-Linneweber C., Barkan A.. Unexpected functional versatility of the pentatricopeptide repeat proteins PGR3, PPR5 and PPR10. Nucleic Acids Res. 2018; 46:10448–10459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lyska D., Engelmann K., Meierhoff K., Westhoff P.. pAUL: a gateway-based vector system for adaptive expression and flexible tagging of proteins in Arabidopsis. PLoS One. 2013; 8:e53787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Curtis M.D., Grossniklaus U.. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003; 133:462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McCormac D.J., Barkan A.. A nuclear gene in maize required for the translation of the chloroplast atpB/E mRNA. Plant Cell. 1999; 11:1709–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Busso D., Delagoutte-Busso B., Moras D. Construction of a set Gateway-based destination vectors for high-throughput cloning and expression screening in Escherichia coli. Anal. Biochem. 2005; 343:313–321. [DOI] [PubMed] [Google Scholar]
- 28. Romani I., Manavski N., Morosetti A., Tadini L., Maier S., Kuhn K., Ruwe H., Schmitz-Linneweber C., Wanner G., Leister D.et al.. A member of the Arabidopsis mitochondrial transcription termination factor family is required for maturation of chloroplast transfer RNAIle(GAU). Plant Physiol. 2015; 169:627–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Barkan A. Approaches to investigating nuclear genes that function in chloroplast biogenesis in land plants. Photosynthesis: Mol. Biol. Energy Capture. 1998; 297:38–57. [Google Scholar]
- 30. Manavski N., Guyon V., Meurer J., Wienand U., Brettschneider R.. An essential pentatricopeptide repeat protein facilitates 5′ maturation and translation initiation of rps3 mRNA in maize mitochondria. Plant Cell. 2012; 24:3087–3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pfalz J., Bayraktar O.A., Prikryl J., Barkan A.. Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J. 2009; 28:2042–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Meurer J., Schmid L.M., Stoppel R., Leister D., Brachmann A., Manavski N.. PALE CRESS binds to plastid RNAs and facilitates the biogenesis of the 50S ribosomal subunit. Plant J. 2017; 92:400–413. [DOI] [PubMed] [Google Scholar]
- 33. Luro S., Germain A., Sharwood R.E., Stern D.B.. RNase J participates in a pentatricopeptide repeat protein-mediated 5′ end maturation of chloroplast mRNAs. Nucleic Acids Res. 2013; 41:9141–9151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Castandet B., Germain A., Hotto A.M., Stern D.B.. Systematic sequencing of chloroplast transcript termini from Arabidopsis thaliana reveals >200 transcription initiation sites and the extensive imprints of RNA-binding proteins and secondary structures. Nucleic Acids Res. 2019; 47:11889–11905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ruwe H., Schmitz-Linneweber C.. Short non-coding RNA fragments accumulating in chloroplasts: footprints of RNA binding proteins. Nucleic Acids Res. 2012; 40:3106–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ruwe H., Wang G., Gusewski S., Schmitz-Linneweber C.. Systematic analysis of plant mitochondrial and chloroplast small RNAs suggests organelle-specific mRNA stabilization mechanisms. Nucleic Acids Res. 2016; 44:7406–7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhelyazkova P., Hammani K., Rojas M., Voelker R., Vargas-Suarez M., Borner T., Barkan A.. Protein-mediated protection as the predominant mechanism for defining processed mRNA termini in land plant chloroplasts. Nucleic Acids Res. 2012; 40:3092–3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Akashi K., Grandjean O., Small I.. Potential dual targeting of an Arabidopsis archaebacterial-like histidyl-tRNA synthetase to mitochondria and chloroplasts. FEBS Lett. 1998; 431:39–44. [DOI] [PubMed] [Google Scholar]
- 39. Kleinknecht L., Wang F., Stube R., Philippar K., Nickelsen J., Bohne A.V.. RAP, the sole octotricopeptide repeat protein in Arabidopsis, is required for chloroplast 16S rRNA maturation. Plant Cell. 2014; 26:777–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Majeran W., Friso G., Asakura Y., Qu X., Huang M., Ponnala L., Watkins K.P., Barkan A., van Wijk K.J.. Nucleoid-enriched proteomes in developing plastids and chloroplasts from maize leaves: a new conceptual framework for nucleoid functions. Plant Physiol. 2012; 158:156–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Prikryl J., Rojas M., Schuster G., Barkan A.. Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:415–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yamazaki H., Tasaka M., Shikanai T.. PPR motifs of the nucleus-encoded factor, PGR3, function in the selective and distinct steps of chloroplast gene expression in Arabidopsis. Plant J. 2004; 38:152–163. [DOI] [PubMed] [Google Scholar]
- 43. Cai W., Okuda K., Peng L., Shikanai T.. PROTON GRADIENT REGULATION 3 recognizes multiple targets with limited similarity and mediates translation and RNA stabilization in plastids. Plant J. 2011; 67:318–327. [DOI] [PubMed] [Google Scholar]
- 44. Fujii S., Sato N., Shikanai T.. Mutagenesis of individual pentatricopeptide repeat motifs affects RNA binding activity and reveals functional partitioning of Arabidopsis PROTON gradient regulation3. Plant Cell. 2013; 25:3079–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Higashi H., Kato Y., Fujita T., Iwasaki S., Nakamura M., Nishimura Y., Takenaka M., Shikanai T.. The pentatricopeptide repeat protein PGR3 is required for the translation of petL and ndhG by binding their 5′UTRs. Plant Cell Physiol. 2021; doi:10.1093/pcp/pcaa180. [DOI] [PubMed] [Google Scholar]
- 46. Edgar R., Domrachev M., Lash A.E.. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002; 30:207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The NGS data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (46) and are accessible through GEO Series accession number GSE146249 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE146249).
Sequence data from this article can be found in the EMBL/GenBank libraries under the following accession numbers: MRL1 (At4g34830) and PGR3 (At4g31850) from A. thaliana, A. thaliana chloroplast genome (NC_000932.1).