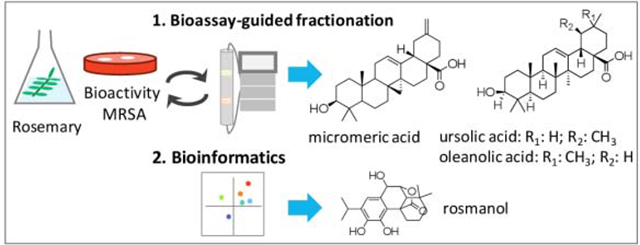
Abstract
Natural products have been a primary source of medicines throughout the history of human existence. It is estimated that close to 70% of small molecule pharmaceuticals on the market are derived from natural products. With increasing antibiotic resistance, natural products remain an important source for the discovery of novel antimicrobial compounds. The plant rosemary (Rosmarinus officinalis), has been widely and commonly used as a food preservative due to its antimicrobial potential. To evaluate the antimicrobial profile of this plant, we used bioassay-guided fractionation and bioinformatics approaches. Through bioassay-guided fractionation, we tested in vitro activities of a R. officinalis extract and fractions thereof, as well as pure compounds micromeric acid (1), oleanolic acid (2), and ursolic acid (3) against methicillin-resistant Staphylococcus aureus (MRSA). Compounds 1 and 3 showed complete inhibition of MRSA (with MIC values of 32 μg/mL and 8 μg/mL, respectively) while compound 2 displayed only partial inhibition (MIC > 64 μg/mL). In addition, we utilized orthogonal partial least square-discriminant analysis (OPLS-DA) and selectivity ratio (SR) analysis to correlate the isolated compounds 1–3 with the observed antimicrobial activity, as well as to identify antimicrobials present in trace quantities. For mass spectrometry (MS) data collected in the negative ionization mode, compound 1 was the most positively correlated with activity, while for MS data collected in the positive ion mode, compounds 2-3 had the highest positive correlation. Using the bioinformatics approaches, we highlighted additional antimicrobials associated with the antimicrobial activity of R. officinalis, including genkwanin (4), rosmadial (5a) and/or 16-hydroxyrosmadial (5b), rosmanol (6), and hesperetin (7). Compounds 1–3 resulting from the bioassay-guided fractionation were identified by MS-MS fragmentation patterns and 1H NMR spectra. Among the compounds highlighted by the biochemical analysis, compound 6 was identified by comparison with its commercial standard by employed ultra-performance liquid chromatography-high resolution mass spectrometry (UHPLC-HRMS), while 4, 5a-b and 7 were putatively identified based on MS data and in comparison, with the literature. This is the first reported antimicrobial activity of micromeric acid (1) against MRSA.
Keywords: Rosmarinus officinalis, rosemary, bioassay-guided fractionation, bioinformatics, biochemometrics, MRSA, natural products
Graphical Abstract
1. Introduction
The use of natural products, including medicinal plants, remains widely popular today with approximately 80% of the world’s population relying on herbal products and related supplements as part of their health care regimen [1]. Rosmarinus officinalis L., or rosemary, synonymously known as Salvia rosmarinus Spenn., is an aromatic shrubby herb that is a member of Lamiaceae, native to the Mediterranean region [2]. R. officinalis extract has been reported to demonstrate antimicrobial activity against the bacterial pathogen methicillin-resistant Staphylococcus aureus (MRSA), although the chemical compounds responsible for that activity were not identified [3]. According to the 2019 CDC report, MRSA is one of the most common antibiotic-resistant bacterial pathogens and is estimated to cause more than 323,000 cases and 10,600 deaths annually in the United States alone [4].
In this study, our goal was to use two different approaches, bioassay-guided fractionation and bioinformatics, to evaluate the antimicrobial profile of R. officinalis against MRSA USA300 LAC strain AH1263 [5], with the long-term objective being the utilization of this combination approach to efficiently improve the antimicrobial screening processes in natural products.
Bioassay or bioactivity-guided fractionation has been the gold standard in natural products, in which the extracts are subjected to chemical fractionations and bioassays to simplify the complexity and pinpoint certain compounds with significant biological effects [6]. It was reported in 2012 that bioassay-guided fraction was used in more than 1,500 published articles in ISI Web of Science, with hundreds more citations applying various forms of the same method [6]. While the bioassay-guided fractionation method can be employed to effectively isolate the most abundant antimicrobials, it also comes with several limitations in that the botanical materials contain thousands of individual compounds and the methodology tends to overlook some of the bioactive constituents that are present in low amounts [7].
Here we sought to demonstrate the use of bioinformatics approaches to correlate the chemical constituents with the bioactivity of R. officinalis and identify the additional antimicrobials that are not identified through bioassayguided fractionation. Both orthogonal partial least square-discriminant analysis (OPLS-DA) and selectivity ratio (SR) analysis were utilized toward this goal. OPLS-DA is an extension of partial least square-discriminant analysis (PLS-DA) and utilizes orthogonal signal correction filter to remove variabilities that are not appropriate to the dataset, which in turn allows the analysis to be targeted and robust against noise [7]. SR plots compare the correlation and covariance to the residual variance and provide a numerical scale to differentiate between constituents that correlate with biological activity and those that do not [8].
2. Experimental
2.1. General experimental procedures
LC-HRMS analysis was completed using a Thermo-Fisher Q-Exactive Plus Orbitrap mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) connected to an Acquity UPLC system (Waters, Milford, MA, USA) with a reverse phase UPLC column (BEH C18, 1.7 μm, 2.1 × 50 mm, Waters Corporation, Milford, MA, USA). All fractions were analyzed at 0.1 mg mL−1 in MeOH with 5 μL injections. The gradient was comprised of solvent A (H2O with 0.1% formic acid) and solvent B (acetonitrile with 0.1% formic acid). The gradient began with 90:10 (A:B) from 0–0.5 min, and increased linearly to 0:100 (A:B) over 19.5 min, then it was held at 100% B for 0.5 min, before returning to starting conditions over 0.5 min with an isocratic hold from 21.0–22.0 min. Analysis was performed in positive and negative switching mode over the m/z range of 150–1500 with: capillary voltage at −0.7 V, capillary temperature at 310°C, S-lens RF level at 80.00, spray voltage at 3.7 kV, sheath gas flow at 50.15, and auxiliary gas flow at 15.16. The three most intense ions were fragmented with a higher-energy collisional dissociation (HCD) of 65.0. Flash chromatography was conducted with a CombiFlash RF system (Teledyne-Isco) with a photo-diode array (PDA) detector. The HPLC separations were performed with a preparative Gemini (5 μm; 250 × 21.20 mm) column, at a flow rate of 21.2 mL/min, with a Varian Prostar HPLC system (Agilent, Santa Clara, CA, USA) equipped with a Prostar 210 pumps and a Prostar 335 photodiode array detector (PDA), with the collection and analysis of data using Galaxie Chromatography Workstation software. The NMR data was collected using a JOEL ECS-400 spectrometer, which was equipped with a JOEL normal geometry broadband Royal probe, and a 24-slot autosampler, and operated at 400 MHz for 1H (JOEL USA, Inc.) All analytes including oleanolic acid, ursolic acid, and rosmanol standards were acquired through Sigma-Aldrich (St. Louis, MO, USA) and were spectroscopic or microbiological grade. All solvents were purchased from Fisher Scientific (Waltham, MA, USA), in Optima or HPLC or ACS-grade dependent upon the experiment.
2.2. Plant material
Rosmarinus officinalis L. was collected on July 25, 2019 from the Dunleath Community Garden in Guilford County, North Carolina (N 36°4’53.616”, W 79°47’6.18”). The voucher specimen was deposited at the herbarium at the University of North Carolina at Chapel Hill (Accession number NCU670047, Catalog number NCU00433752). Stems and leaves were dried at room temperature until crisp before extraction. The plant name was verified on http://www.theplantlist.org/ on August 10th, 2020.
2.3. Extraction
R. officinalis stems and leaves (30.60 g of dried mass) were ground using a Wiley Mill Standard Model No. 3 (Arthur Thomas Company) and extracted in MeOH (160 g/L) at room temperature for 24 hours. The supernatant MeOH layer in the extract was decanted using a borosilicate glass side-arm flask and funnel. The supernatant was dried down using a Rotovap (Heidolph, Schwabach, Germany). The MeOH was recycled to repeat the process for a total of three times. The MeOH extract was dried under nitrogen, yielding 3.2 g.
2.4. Chromatographic separation
The fractionation scheme is provided in Figure S1. A portion of the methanol extract (900 mg) was dissolved in CHCl3, adsorbed onto Celite 545 (Acros Organics) and was separated with normal-phase flash chromatography (40 g silica gel column) with a 55.0-minute hexane/CHCl3/MeOH gradient at a 40 mL/min flow rate (Figure S4). This generated a total of 8 fractions. Fraction RO-5 (115 mg) went through a second stage of normal-phase flash chromatography (Figure S5), after dissolving in chloroform, absorbing onto Celite 545 (Acros Organics), with a 35.0-minute hexane/CHCl3/MeOH gradient and was separated using a 15.5 g silica gel column at a flow rate of 30 mL/min. This fractionation yielded 6 simplified fractions, in which the bioactive fraction RO-5–4 (approximately 10 mg) was purified further via preparative HPLC using a gradient system 80:20 to 90:10 of CH3CN:H2O with 0.1% formic acid over 30 minutes (Figure S6). This yielded 4 fractions: RO-5–4-1 (3.88 mg eluted at 4.0 min), RO-5–4-2 (2.27 mg eluted at 12.5 min), RO-5–4-3 (3.15 mg eluted at 15.9 min), and RO-5–4-4 (5.4 mg eluted at 0.5 min).
2.5. Antimicrobial assays
Antimicrobial activity was evaluated according to Clinical Laboratory Standards Institute (CLSI) guidelines [9] against a clinically relevant strain of methicillin resistant S. aureus (MRSA USA300 LAC strain AH1263) [5]. Cultures were grown from a single colony isolate in Müeller-Hinton broth (MHB) and diluted to 1.0 × 105 CFU/mL based on absorbance at 600 nm (OD600).
Samples were screened in triplicates at final concentrations of 10 and 100 μg/mL. Samples were dissolved in DMSO and diluted with MHB to prepare final concentrations containing 2% DMSO in each well of 96-well cell culture plate (Corning Incorporated, Corning, NY, USA). Minimal inhibitory concentrations (MICs) were identified by testing micromeric acid (1), oleanolic acid (2), and ursolic acid (3) in 96-well cell culture plate (Corning Incorporated, Corning, NY, USA) at concentration ranging from 0.00–64.0 μg/mL in MHB. MIC was defined as the lowest concentration at which no statistically significant difference between the wells that did not contain bacteria and the treated samples were found. In both screening and MIC assays, levofloxacin (98% purity, Sigma-Aldrich) was used as a positive control at 10 μg/mL while 2% DMSO served as a negative control. The samples were incubated while shaken at 37°C for approximately 18 – 24 hours and OD600 values were measured using a Synergy H1 microplate reader (Biotek, Winooski, VT, USA). Percent inhibition and MIC values were calculated based on the CLSI standard protocols [9].
2.6. Bioinformatics analysis
Raw data were converted to .mzML format with Proteowizard (http://proteowizard.sourceforge.net/). MZmine 2.53 software ((http://mzmine.github.io/) was employed for data processing. In both ionization modes, the exact mass detector was used for mass detection with a noise level set at 5×103. The ADAP chromatogram builder [10] was set to a minimum group size scan of 5, a minimum group intensity threshold of 5×103, a minimum highest intensity of 1×104 and a m/z tolerance of 0.003 Da. The wavelet algorithm ADAP was used for deconvolution: the intensity window was used as signal to noise (S/N) estimator, the S/N threshold was set at 10, the minimum feature height at 1×105, the coefficient area threshold at 110, a peak duration range of 0.0 to 1.0 min, and a retention time wavelet range of 0.0 to 0.1 min. The peak lists were de-isotoped with the isotope peaks grouper with a m/z tolerance of 0.015 Da, a retention time (RT) tolerance of 0.01 min, a maximum charge of 3 and the most intense isotope was the representative one. Each series of triplicates was aligned independently with the Join aligner method, with a RT tolerance of 0.1 min, a m/z range of 0.002, a weight for RT of 1 and for m/z of 2. The obtained aligned peak lists were filtered with the feature list row filter module to keep only rows that have features detected in each replicate. After filtering, these peak lists were all aligned together as well as with the blank analyses (n=7) with the same parameters. Rows with features detected in more than 4 blank analyses were removed. For further analysis, the peak areas, m/z values, and retention times for detected individual ions in triplicate injections were exported from the data matrix to Excel (Microsoft, Redmond, WA, USA). A peak area of 0 was assigned to the samples that did not contain a particular marker ion, to maintain a consistent number of variables throughout the dataset. The dataset is available on the GNPS server (MassIVE MSV000086110, https://doi.org/doi:10.25345/C5XB4V )(temporary private access during submission process, password “rosemary”). It includes raw and mzML files, as well as the resulting tables exported from MZmine.
The averages of triplicate injections were used after verifying that replicates clustered in the dendrogram analysis (Figure S7 and S8). The resulting data matrix from Excel (Microsoft, Redmond, WA, USA) was merged with bioactivity data set in the form of percent inhibition against MRSA (at 100 μg/mL concentration) to form a final analytical matrix. Biochemometric analysis was performed using Sirius version 11.5 (Pattern Recognition Systems AS, Bergen, Norway). An internally cross-validated PLS (partial least squares) model was constructed using 100 iterations, at a significance level of 0.010. Selectivity ratios from the final PLS model were calculated using algorithms internal to Sirius.
Features of interest were annotated by combining the determination of the molecular formula and data from the literature. In the module for predicting the molecular formulae of MZmine, the m/z tolerance was set at 0.002 Da, the isotopic score at a minimum of 60% and the element count heuristics were taken into account. The CAMERA identification module was used with the parameters described in https://mzmine.github.io/ADAP_user_manual.pdf. In addition, the identification module for adduct search was used with a RT tolerance set at 0.02 min, a m/z tolerance at 0.0015 Da and a maximum relative adduct peak height at 10,000%. To complete the list of adducts included in MZmine, common adducts described in https://fiehnlab.ucdavis.edu/staff/kind/metabolomics/ms-adduct-calculator/ were imported in the module.
3. Results and discussion
3.1. Bioassay-guided fractionation approach
Bioassay-guided fractionation was completed in three stages (Figure S1) to simplify the R. officinalis extract. With each stage, bioactive constituents were concentrated further, resulting in the elevated percent inhibition values (Figure 2), most significantly going from first stage to second stage fractions. The most bioactive second stage fraction (RO-5–4) was chromatographically separated and resulted in four subfractions, in which three (RO-5–4-1 through RO-5–4-3) of them demonstrated significant inhibitions ranging from 75–100% against MRSA strain (Figures 2, S1). MS-MS (Figure S2) and 1H NMR (Figure S3) analyses were acquired. RO-5–4-1 was confirmed to be compound 1 by 1H NMR in comparison to the literature (Figures S3A, S3B) [11]. RO-5–4-2 and RO-5–4-3 were found to contain compounds 2 and 3, respectively, in comparison to the fragmentation patterns (Figure S2) and 1H NMR spectra (Figure S3) of the standards [12,13]. In particular, RO-5–4-2 had two methyl singlets (δH 0.89 and δH 0.92), where RO-5–4-3 had the corresponding methyl doublets (δH 0.85, and δH 0.93). While RO-5–4 fraction had MIC value of 32 μg/mL, compounds 1 and 3 showed antimicrobial activities against MRSA with MIC values of 32 μg/mL (70 μM) and 8 μg/mL (18 μM) respectively. Compound 2 only demonstrated partial inhibition of MRSA (MIC > 64 μg/mL (140 μM)). This is the first reported antimicrobial activity of 1 against MRSA. Compounds 2 and 3 have previously been reported to have activity against S. aureus strain ATCC 25923, with MIC values of 3.75 mg/mL and 0.01 mg/mL, respectively [14]. Hence, the bioassay-guided fractionation enabled identification of two antimicrobial constituents (1 and 3) present in the most active second stage fraction, RO-5–4 (Figure S2), and also resulted in the isolation of compound 2 with very weak activity.
Figure 2.
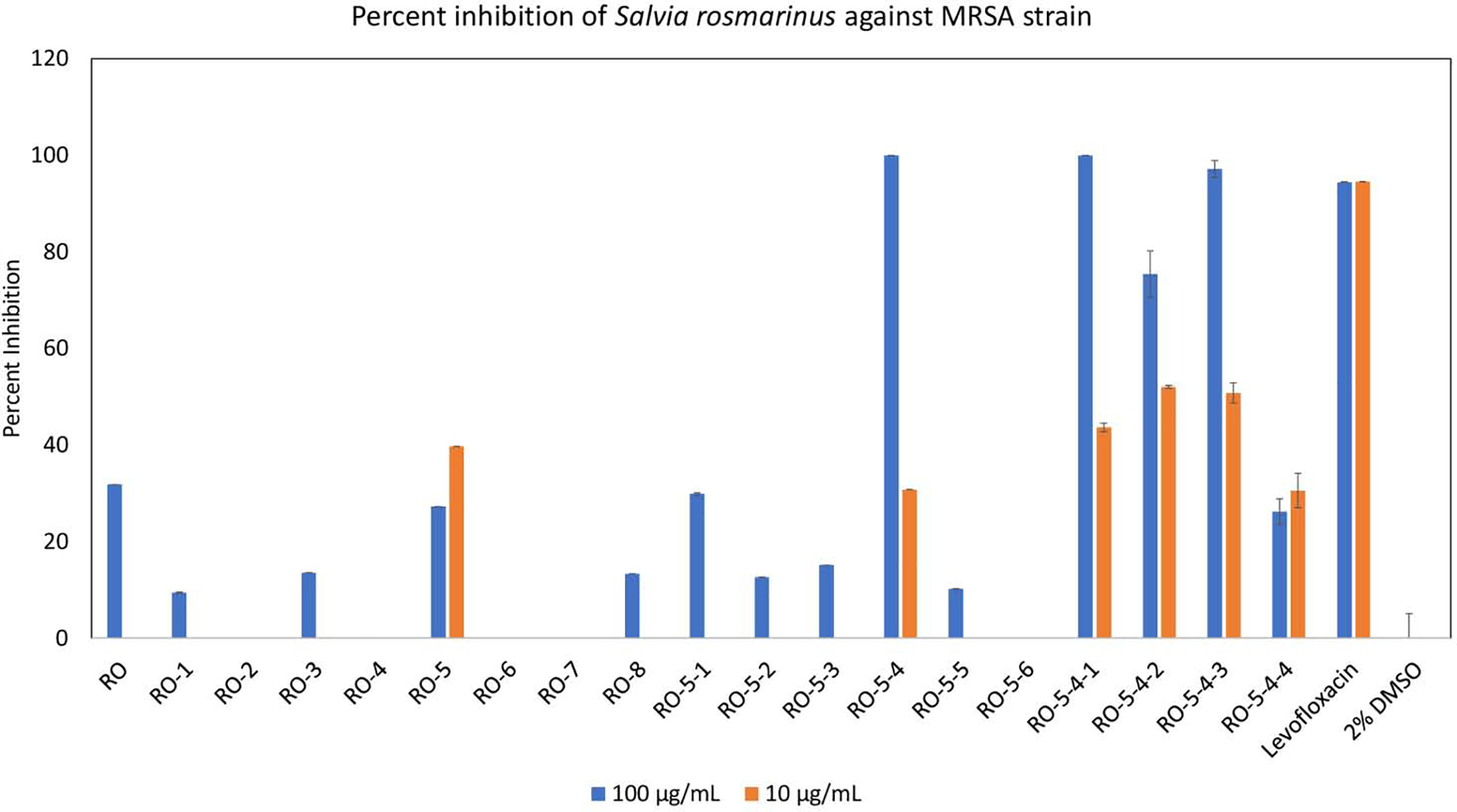
Percent inhibition of methicillin resistant Staphylococcus aureus (MRSA) strain AH1263 growth by Rosmarinus officinalis methanol extract (RO) and fractions therefrom. The fractionation scheme can be seen in Figure S1. The initial extract (RO) exhibited partial inhibition at high concentration (100 μg/mL) but no inhibition at low concentration (10 μg/mL). Fractionation of the initial extract (RO) yielded eight fractions (RO-1 through RO-8), of which one fraction (RO-5) possessed strong antimicrobial activity. Fraction RO-5 was subsequently fractionated into six sub-fractions (RO-5–1 through RO-5–6), of which RO-5–4 was active. Fraction RO-5–4 was separated into four sub-fractions, RO-5–4-1 through RO-5–4-4, all of which exhibited significantly high inhibitions of MRSA growth at 100 μg/mL. Mean percent inhibition was calculated from triplicate assays wells and error bars represent standard error of the triplicates. The known antimicrobial agent levofloxacin served as positive control, and vehicle (2% DMSO) served as negative control.
3.2. Bioinformatics approach
We UHPLC-HRMS to evaluate differences in chemical compositions of the first stage fraction with the most potent antimicrobial activity (RO-5) and its subfractions (RO-5–1 through RO-5–6) (Figure S1) at 100 μg/mL concentrations. Only these fractions of R. officinalis were included since the second stage fractions were previously found to be the most effective for the bioinformatics process [15].
To perform bioinformatics, the chemical information came from MS analysis in the form of mass over charge-retention time (m/z-RT) pairs and antimicrobial assays provided the required percent inhibition values (Figure 2). Both OPLS-DA (Figures 3, 5) and SR plot (Figures 4, 6) analyses were performed on the MS data sets collected in the positive ionization and negative ionization modes.
Figure 3.
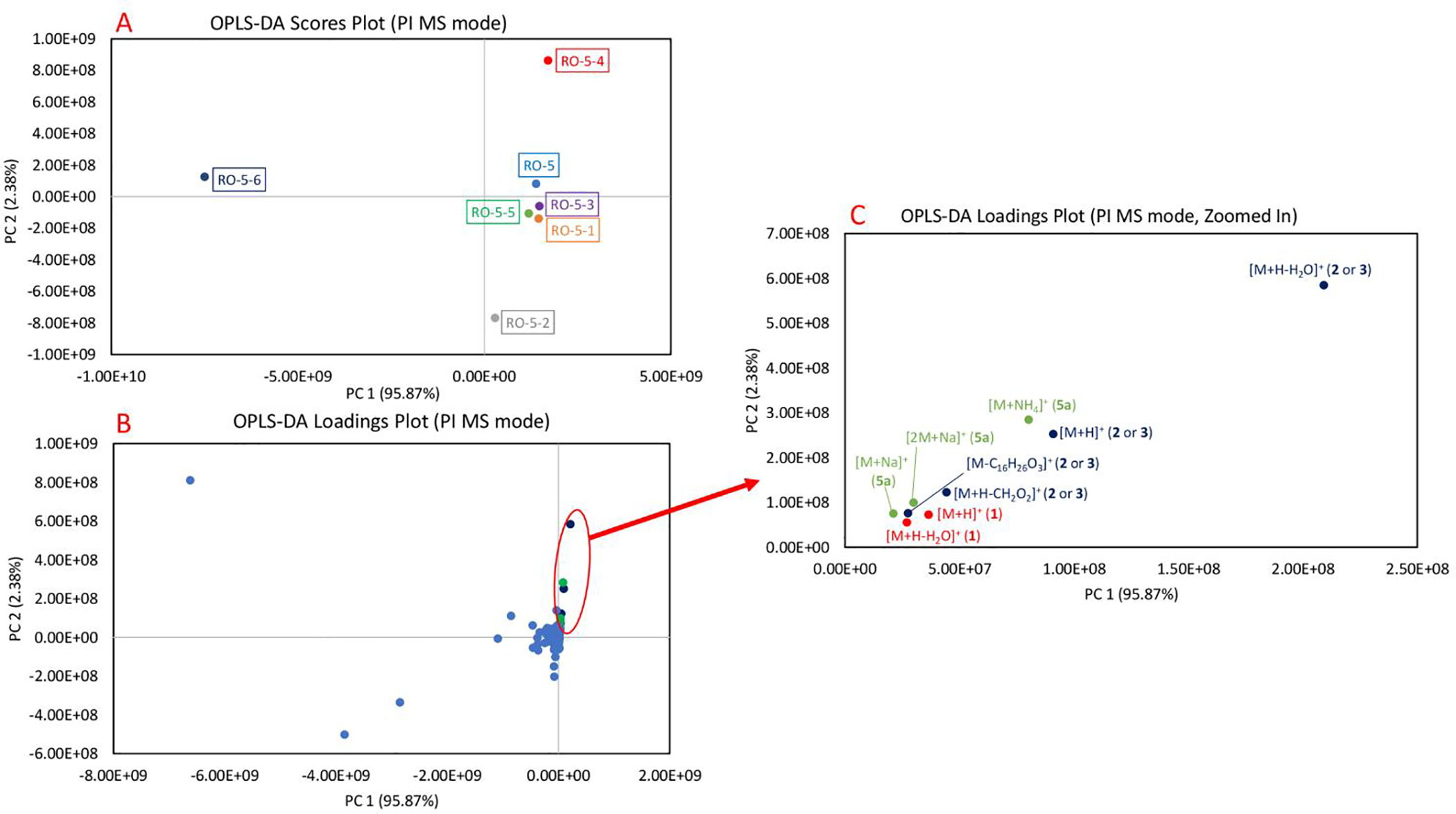
OPLS-DA plots of Rosmarinus officinalis fractions (first stage RO-5 fraction and its subfractions, RO-5–1 through RO-5–6) in positive (PI) MS mode. The separation pattern of the data points is supervised by the percent inhibition values against MRSA. 3A. OPLS-DA scores plot of the fractions. Each fraction is plotted as an individual data point and color-coded. RO-5–4 having the highest inhibition against MRSA is visualized in the quadrant I, while RO-5–6 with no inhibition against MRSA is in the quadrant II. Other fractions with moderate bioactivities cluster closely to one another near the center of the plot. 3B. OPLS-DA loadings plot of the fractions. The detected features, which exist in the same quadrant as RO-5–4, are clustered in the quadrant I and their presence is highly correlated to the bioactivity of the fraction. 3C. OPLS-DA loadings plot, zoomed in on the features correlating to the bioactivity. The features are color-coded based on the compounds. Features corresponding to micromeric acid (1) and oleanolic acid (2) or ursolic acid (3) were identified to be positively correlating to the bioactivity of RO-5–4. [M+H+H2O]+ of oleanolic acid (2) or ursolic acid (3) was identified to be the most positively correlated ion to the bioactivity of RO-5–4. Additional tentative annotations of the features can be seen in Table 1.
Figure 5.
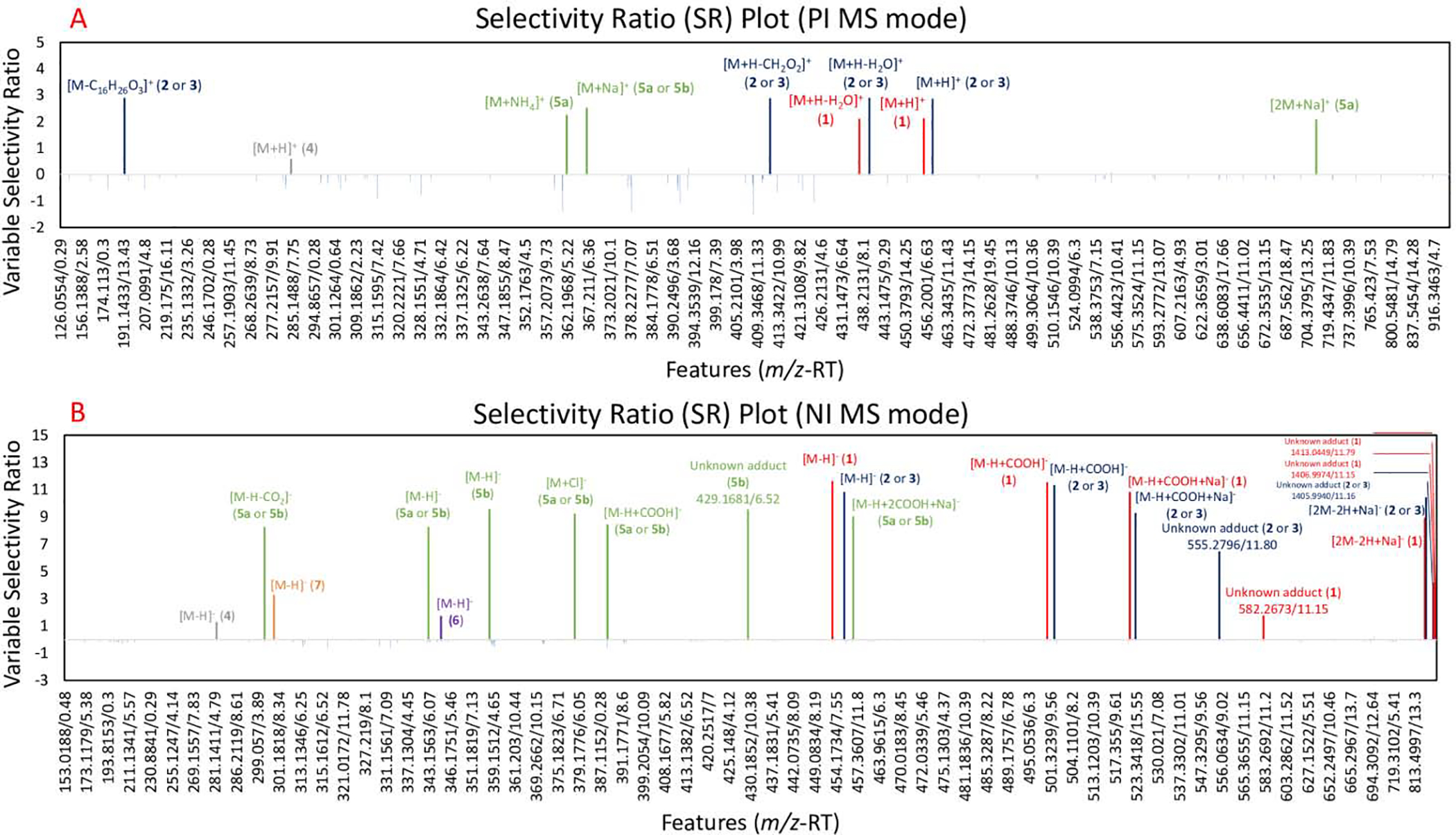
Selectivity ratio (SR) plot for multivariate statistical analysis for ions detected in first stage RO-5 fraction and its subfractions, RO-5–1 through RO-5–6. The data was collected using UPLC-HRMS. Percent inhibition measurements against MRSA after 18–24 hours were used as a dependent variable. The x-axis shows the features (identified by a unique m/z and retention time) detected in the metabolomics dataset. Features are shown in the order of increasing m/z value and color-coded based on compounds. Variable selectivity ratio is shown on the y-axis and higher ratio indicates stronger correlation with the percent inhibition. 5A. SR plot analysis in positive (PI) MS mode. 5B. SR plot analysis in negative (NI) MS mode. In both figures, micromeric acid in red (1), oleanolic acid (2) or ursolic acid (3) in blue, and their related features had high SR ratios, confirming their positive correlation to the bioactivity of RO-5–4 fraction. Unknown adducts are labelled along with their features (m/z-RT). Additional tentative annotations of the features can be seen in Table 1.
Figure 4.
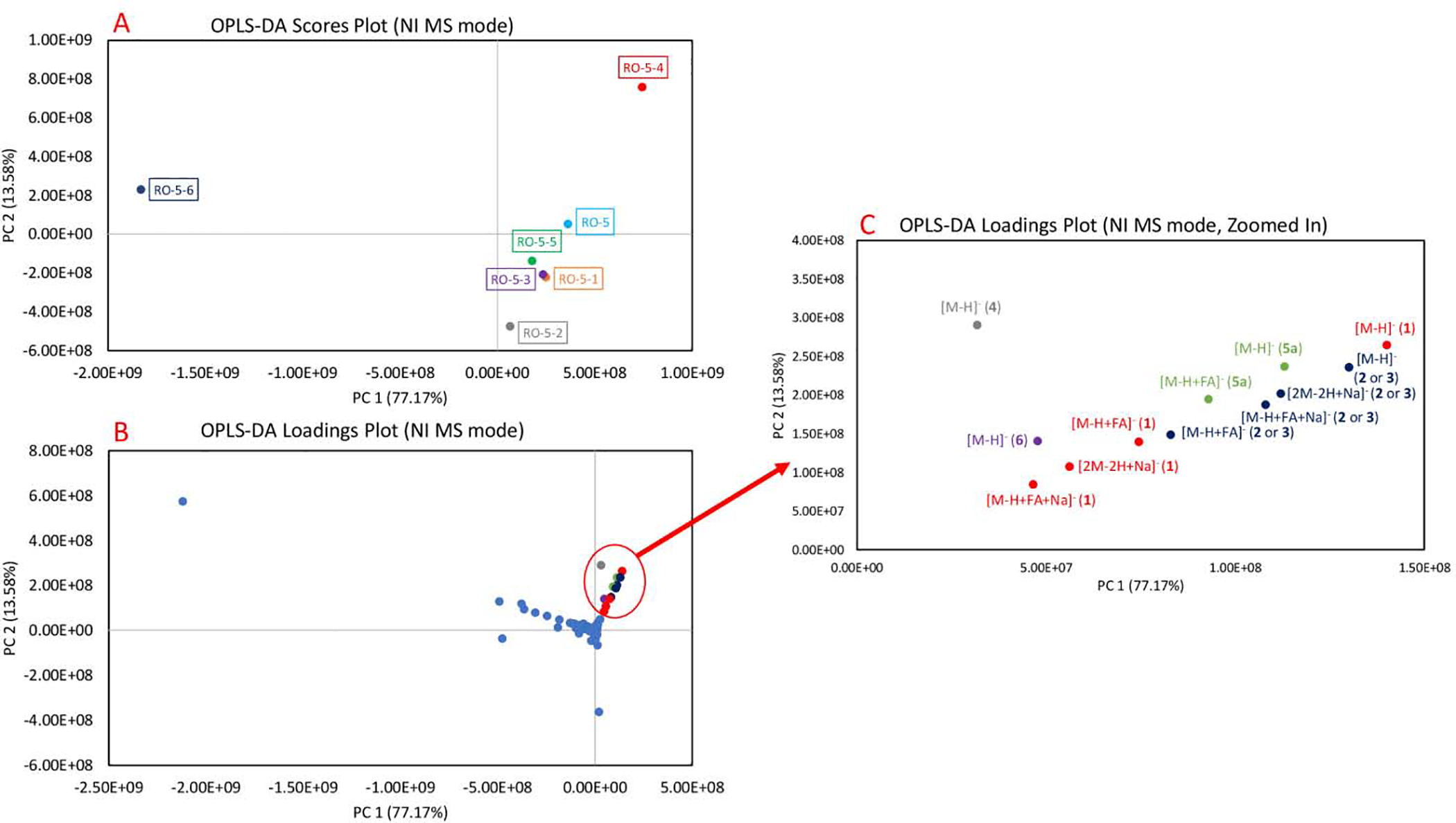
OPLS-DA plots of Rosmarinus officinalis fractions (first stage RO-5 fraction and its subfractions, RO-5–1 through RO-5–6) in negative (NI) MS mode. The separation pattern of the data points is supervised by the percent inhibition values against MRSA. 5A. OPLS-DA scores plot of the fractions. Each fraction is plotted as an individual data point and color-coded. RO-5–4 having the highest inhibition against MRSA is visualized in the quadrant I, while RO-5–6 with no inhibition against MRSA is in the quadrant II. Other fractions with moderate bioactivities cluster closely to one another near the center of the plot. 5B. OPLS-DA loadings plot of the fractions. The detected features, which exist in the same quadrant as RO-5–4, are clustered in the quadrant I and their presence is highly correlated to the bioactivity of the fraction. 5C. OPLS-DA loadings plot, zoomed in on the features correlating to the bioactivity. The features are color-coded based on the compounds. Similar to thepositive mode, micromeric acid (1), oleanolic acid (2) or ursolic acid (3), and their adducts were identified to be positively correlating to the bioactivity of RO-5–4. The precursor ion [M−H]− of micromeric acid (1) was identified as the most positively correlated ion to the bioactivity of RO-5–4. Additional tentative annotations of the features can be seen in Table 1.
OPLS-DA is a supervised methodology that was employed here to integrate mass spectrometry datasets, which provide information on the chemical constituents, and bioactivity assays, which give percent inhibition values against MRSA. This analysis can be described in two plots, “scores” (Figure 3A) in which each fraction represents a single data point and “loadings” (Figure 3B) in which each individual features (m/z-RT pairs) found in the analyzed fractions are plotted as single data points [7]. The plots involve mapping the dataset onto a series of latent variables in a two-dimensional space [7]. Although more than two principal components (PC) exist, it is typical to plot data using the two PCs (often PC1 and PC2) that introduce the most variability across the analyzed fractions [7]. The location of the detected chemical constituent on the loadings plot explains the locations of the fractions on the corresponding scores plot, which in turn assists in the identification of bioactive compounds which correlate to the observed antimicrobial activities [7].
In both positive and negative ionization modes, PC1 explains the differences between the fractions and detected ions more than PC2 due to the higher percentage: Figure 3 (PC1 = 95.87%, PC2 = 2.38%) and Figure 4 (PC1 = 77.17%, PC2 = 13.58%). Since OPLS-DA is guided by the MRSA growth inhibition values, the fraction with most potent antimicrobial activity, RO-5–4, clusters by itself in the quadrant I, the fraction with the least potent antimicrobial activity, RO-5–6 clusters by itself in the quadrant II, while other partially inhibitory fractions cluster together in the centers of the scores plots (Figures 3A, 4A). Similarly, in the loading plots (Figures 3B, 4B), we observed that the features correlating to the antimicrobial activity of RO-5–4 cluster in the quadrant I.
We identified nine features in positive ionization mode analysis (Figure 3C) and twelve features in negative ionization mode analysis (Figure 4C), which correlate to antimicrobial activity of RO-5–4. As compounds 2 and 3 are very similar structurally, they have been observed as co-eluting in our chromatographic conditions and they couldn’t be differentiated. Compounds 1 and 2–3, and their in-source features (adducts and neutral losses) were identified to be the features of importance. In the positive ionization mode, the [M+H-H2O]+ neutral loss of 2–3 was the most positively correlating feature in positive ionization mode (Figure 3C) and in the negative ionization mode, the deprotonated molecular ion [M−H]− of 1 was the highest correlating feature in negative ionization mode to the observed bioactivity of RO-5–4 (Figure 4C). This further validates the results from the bioassay-guided fractionation approach since compounds 1–3 were isolated from RO-5–4 (Figure S1) and exhibited antimicrobial activity when tested in isolation. More features corresponding to the compounds (4–6) were highlighted which will be discussed more in the SR analysis section (Figures 3C, 5C). Detailed annotations of the features can be seen in Table 1 for both positive and negative ionization modes.
Table 1.
Identifications and annotations of the features with positive selectivity ratios (SR) in UPLC-HRMS.
| Positive Ionization (PI) MS mode | ||||||||
| Identifications/Annotations | SR | Loadingsa | m/z | RT (min) | Ion descriptionb | Molecular Formula | Reference | MSIc |
| oleanolic acid (2) and ursolic acid (3) | 2.88 | Y | 439.3576 | 11.80 | [M+H-H2O]+ (m/z-18.010865) | C30H46O2 | ||
| oleanolic acid (2) and ursolic acid (3) | 2.88 | Y | 191.1797 | 11.80 | [M-C16H26O3]+ (m/z-266.1887) | C14H22 | ||
| oleanolic acid (2) and ursolic acid (3) | 2.87 | Y | 411.3627 | 11.80 | [M+H-CH2O2]+ (m/z-46.005480) | C29H46O | ||
| oleanolic acid (2) and ursolic acid (3) | 2.85 | Y | 457.3684 | 11.80 | [M+H]+ | C30H48O3 | [12] | 1 |
| rosmadial (5a) | 2.51 | Y | 367.1523 | 6.52 | [M+Na]+ (m/z+21.98194) | C20H24O5 | [21] | 3 |
| rosmadial (5a) | 2.24 | Y | 362.1968 | 6.52 | [M+NH4]+ (m/z+17.02655) | C20H24O5 | ||
| micromeric acid (1) | 2.11 | Y | 455.3525 | 11.19 | [M+H]+ | C30H46O3 | [11] | 1 |
| micromeric acid (1) | 2.09 | Y | 437.3422 | 11.20 | [M+H-H2O]+ (m/z-18.010865) | C30H44O2 | ||
| rosmadial (5a) | 2.07 | Y | 711.3148 | 6.52 | [2M+Na]+ | C40H48O10 | ||
| genkwanin (4) | 0.56 | N | 285.0762 | 6.30 | [M+H]+ | C16H12O5 | [22] | 2 |
| Negative Ionization (NI) MS mode | ||||||||
| Identity/ annotation | SR | Loadingsa | m/z | RT (min) | Ion description | Molecular Formula | Reference | MSIc |
| micromeric acid (1) | 11.6 | Y | 453.3389 | 11.15 | [M-H]− | C30H46O3 | [11] | 1 |
| micromeric acid (1) | 11.5 | Y | 499.3446 | 11.15 | [M+COOH]− (m/z+46.00547) | C30H46O3 | ||
| oleanolic acid (2) and ursolic acid (3) | 11.3 | Y | 501.3601 | 11.80 | [M+COOH]−(m/z+46.00547) | C30H48O3 | ||
| oleanolic acid (2) and ursolic acid (3) | 10.8 | Y | 455.3542 | 11.80 | [M-H]− | C30H48O3 | [12] | 1 |
| micromeric acid (1) | 10.8 | Y | 521.3262 | 11.15 | [M-H+NaCOOH]− (m/z+67.987425) | C30H46O3 | ||
| oleanolic acid (2) and ursolic acid (3) | 10.4 | Y | 933.6978 | 11.81 | [2M-2H+Na]− | C60H64O6 | ||
| 16-hydroxyrosmadial (5b) | 9.54 | N | 359.1511 | 6.52 | [M-H]− | C20H24O6 | [23] | 3 |
| unknown feature [16-hydroxyrosmadial (5b)] | 9.5 | N | 429.1681 | 6.52 | - | - | ||
| oleanolic acid (2) and ursolic acid (3) | 9.25 | Y | 523.3418 | 11.80 | [M-H+NaCOOH]− (m/z+67.987425) | C31H48O5 | ||
| rosmadial (5a) | 9.19 | N | 379.133 | 6.52 | [M+Cl]− (m/z+35.976678) | C22H26O9 | ||
| rosmadial (5a) | 9 | N | 457.1494 | 6.52 | [M-H+2HCOO+Na]− (m/z+113.992905) | C22H26O9 | ||
| micromeric acid (1) | 8.9 | Y | 929.6664 | 11.15 | [2M-2H+Na]− | C60H90O6 | ||
| rosmadial (5a) | 8.4 | Y | 389.1619 | 6.52 | [M+COOH]− (m/z+46.00547) | C20H24O5 | ||
| rosmadial (5a) | 8.23 | Y | 343.1561 | 6.52 | [M-H]− | C20H24O5 | [21] | 3 |
| rosmadial (5a) | 8.22 | N | 299.1662 | 6.52 | [M-H-CO2]− (m/z-43.989830) | C19H24O3 | ||
| unknown feature [oleanolic acid (2) and ursolic acid (3)] | 6.43 | N | 555.2796 | 11.80 | - | - | ||
| unknown feature [micromeric acid (1)] | 4.13 | N | 1405.994 | 11.16 | - | - | ||
| unknown feature [oleanolic acid (2) and ursolic acid (3)] | 4.13 | N | 1406.997 | 11.15 | - | - | ||
| unknown feature [oleanolic acid (2) and ursolic acid (3)] | 4.13 | N | 1413.045 | 11.79 | - | - | ||
| hesperetin (7) | 3.23 | N | 301.073 | 4.58 | [M-H]− | C16H14O6 | [24] | 2 |
| unknown feature [micromeric acid (1)] | 1.72 | N | 582.2673 | 11.15 | - | |||
| rosmanol (6) | 1.69 | Y | 3.5.1716 | 5.82 | [M-H]− | C20H26O5 | [25] | 1 |
| genkwanin (4) | 1.26 | Y | 283.062 | 6.30 | [M-H]− | C16H12O5 | [22] | 2 |
Features detected in the loading plots of OPLS-DA analysis. Y: detected in quadrant I, N: not detected in quadrant I.
Annotations were performed with a m/z tolerance of 2 mDa
MSI: Metabolomic Standard Initiative, level of identification proposed in Sumner et al. 2007 [26].
Bioinformatics analysis can also be conducted using selectivity ratio (SR) analysis, which quantitatively measures the level of correlation between the observed biological effect and each of the features (m/z-RT pairs) in the dataset [8]. For this dataset, the higher (more positive) SR values represent positive correlations with antimicrobial activity. In the SR analysis for the positive ionization MS data set, the positive SR ranged from 0.56 to 2.88 (Figure 5A). The two features with the highest SR (SR = 2.88), [M+H-H2O]+ and [M-C16H26O3]+, were annotated as features of compounds 2 or 3 (Figure 5A), further validating the OPLS-DA analysis (Figure 3) which indicated that these compounds are highly correlated to the antimicrobial activity. In the negative ionization mode SR analysis, the positive SR ranged from 1.26 to 11.60 (Figure 5B). The features with the highest SR values were annotated as features of compounds 1 and 2–3 (Figure 5B), again validating the OPLS-DA analysis (Figure 4).
In addition to the compounds already identified by the bioassay-guided fractionation approach, other compounds were highlighted by the biochemometric approach. In both ionization modes, the precursor ions, [M+H]+ and [M−H]−, of compound 4 were highlighted with positive SR values (Figure 5). Furthermore, the SR plots showed features that could be related to compound 5a; however, the presence of a feature at the same retention time in the negative ionization mode annotated as the [M−H]− deprotonated molecular ion of 5b could indicate that all related features to compound 5a came from compound 5b, which would lose one oxygen during the ionization process (Figures 5). In addition to these features, the negative ionization mode SR plot highlighted the [M−H]− deprotonated molecular ions of compounds 6 and 7. The annotation of compound 6 was validated by comparison with a commercial standard by UHPLC-HRMS. These data are strengthened with literature comparison, as compounds 6 has been previously shown to MICs ranging from 15.6–62.5 μg/mL against MRSA strains [16] and compound 7 has been shown to have MIC of 500 g/mL against MRSA [17] while compound 4 has previously been reported to have antimicrobial activity against Bacillus cereus and Escherichia coli strains although not against MRSA [18]. To our knowledge, no previous studies were conducted on the antimicrobial activities of compound 5a or 5b against MRSA. Detailed annotations including SR values can be seen in Table 1.
Previous reports have often attributed the antimicrobial activity of R. officinalis to volatile constituents that the plant contains [19]. While the analyses conducted here did not detect these constituents in the R. officinalis extract, it is possible that they could have been present below the limit of detection for UHPLC-HRMS and may have contributed to the observed antimicrobial activity of the extracts. Future experiments to explore this possibility, possibly employing gas chromatography-mass spectrometry as an analytical technique, would we warrant.
The bioinformatics approach enabled not only the identification of the highly positive correlations of 1–3 but also of compound 6, which was not detected by the bioassay-guided fractionation approach. Furthermore, it allowed the putative annotations of compounds 4, 5a or 5b, and 7 using their m/z and RT values and in comparison, with the literature. Therefore, the combination of bioassay-guided fractionation and bioinformatics can lead to the isolation of abundant compounds, correlations of these compounds with bioactivity, and the highlighting of potential antimicrobials present in low quantities. This captures a more complete assessment of the antimicrobial profile of a given natural product than using either technique alone. Further studies would be required to confirm the identity of the annotated compounds and to evaluate their bioactivities individually and in combination via synergistic antimicrobial assays [20].
Supplementary Material
Figure 1.
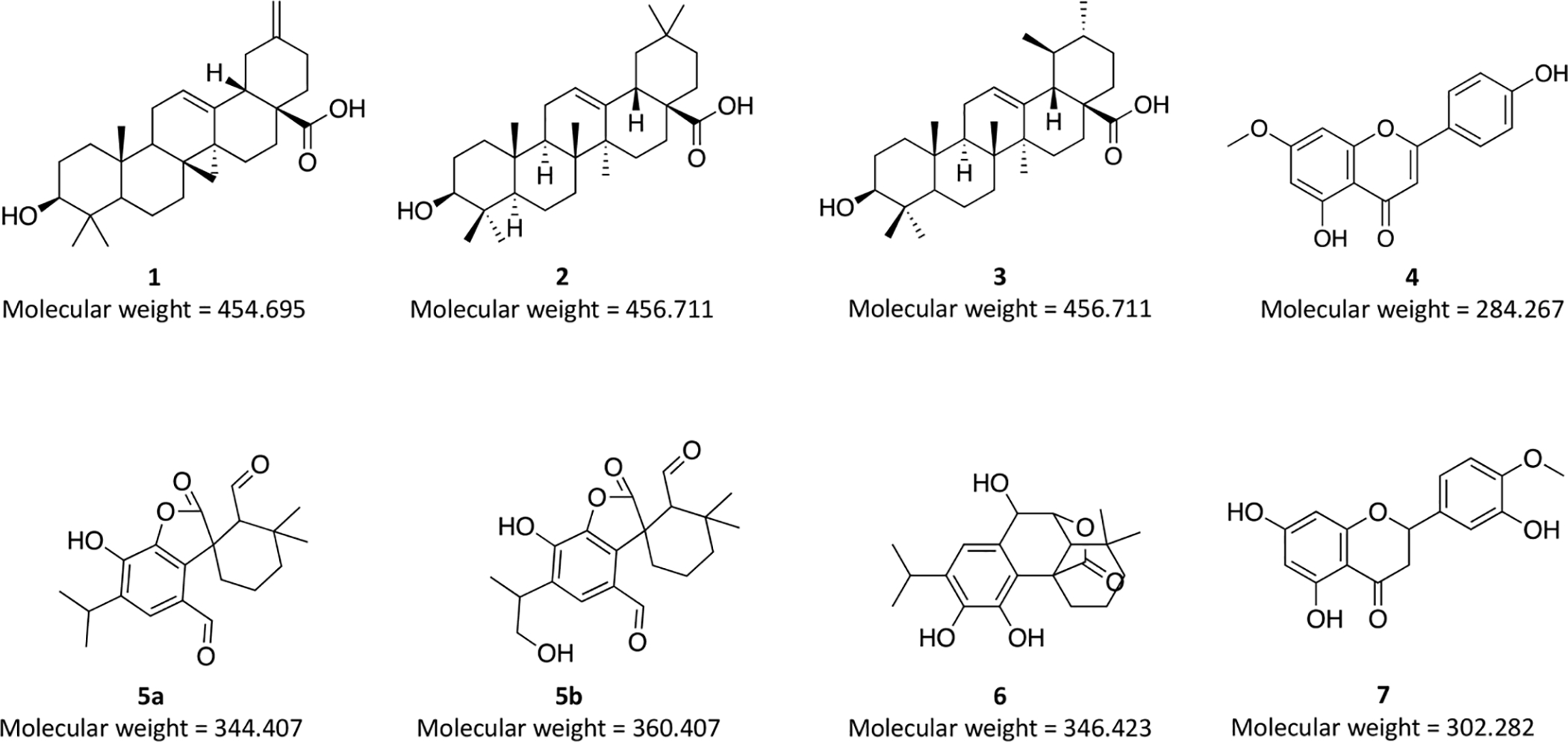
Structures of isolated compounds: micromeric acid (1), oleanolic acid (2) and ursolic acid (3) and structures of putatively identified compounds: genkwanin (4), rosmadial (5a), 16-hydroxyrosmadial (5b), rosmanol (6), and hesperetin (7) from Rosmarinus officinalis. Configuration of compounds 1–3 was assigned via 1H NMR. The annotation of compound 6 was validated by comparison with a commercial standard by UHPLC-HRMS, while others were tentatively identified through m/z and RT values and in comparison with the literature. Molecular weights of the compounds (in g/mol) are denoted below each structure.
Highlights.
First report of the antimicrobial activity of micromeric acid (1) against MRSA
Positive correlations of isolated compounds to the bioactivity
Putative identifications of additional compounds using bioinformatics
Advantage of using both bioassay-guided fractionation and bioinformatics
Acknowledgements
We thank Dr. Daniel A. Todd (Director of Triad Mass Spectrometry Facility, University of North Carolina) for the assistance in running UPLC-HRMS and Dr. Alexander R. Horswill (University of Colorado Anschutz Medical Campus) for providing the Staphylococcus aureus strains.
Funding
This research was supported in part by grant number R15 AT010191–01 from the National Center for Complementary and Integrative Health, a component of the National Institutes of Health. SLK was supported by the same agency via F31 AT010558.
Abbreviations:
- OPLS-DA
orthogonal partial least square-discriminant analysis
Footnotes
Conflict of Interest
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bodeker G, Ong CK, Grundy CK, Burford G, Shein K, WHO global atlas of traditional, complementary and alternative medicine. World Health Organization, Kobe, Japan, 2005, pp. 1–210. [Google Scholar]
- 2.Borrás-Linares I, Stojanović Z, Quirantes-Piné R, Arráez-Román D, Švarc-Gajić J, Fernández-Gutiérrez A, Segura-Carretero A, Rosmarinus officinalis leaves as a natural source of bioactive compounds, Int. J. Mol. Sci 15 (2014), 20585–20606. 10.3390/ijms151120585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manilal A, Sabu KR, Shewangizaw M, Aklilu A, Seid M, Merdekios B, Tsegaye B, In vitro antibacterial activity of medicinal plants against biofilm-forming methicillin-resistant Staphylococcus aureus: Efficacy of Moringa stenopetala and Rosmarinus officinalis extracts, Heliyon. 6 (2020), e03303. 10.1016/j.heliyon.2020.e03303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention (CDC)., 2019. Antibiotic resistance threats in the United States. Centers for Disease Control and Prevention. vii, 95. https://www.cdc.gov/drugresistance/biggest-threats.html (Accessed 31 October, 2020) [Google Scholar]
- 5.Boles BR, Thoendel M, Roth AJ, Horswill AR, Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation, PloS. One. 5 (2010), e10146. 10.1371/journal.pone.0010146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weller MG, A unifying review of bioassay-guided fractionation, effect-directed analysis and related techniques, Sensors. 12 (2012), 9181–9209. 10.3390/s120709181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sugimoto M, Kawakami M, Robert M, Soga T, Tomita M, Bioinformatics tools for mass spectroscopy-based metabolomic data processing and analysis, Curr. Bioinform 7 (2012), 96–108. 10.2174/157489312799304431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rajalahti T, Arneberg R, Berven FS, Myhr K, Ulvik RJ, Kvalheim OM, Biomarker discovery in mass spectral profiles by means of selectivity ratio plot, Chemometr. Intell. Lab. 95 (2009), 35–48. 10.1016/j.chemolab.2008.08.004 [DOI] [Google Scholar]
- 9.Clinical and Laboratory Standards Institute (CLSI), Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically (11th Ed.). National Committee for Clinical Laboratory Standards, Wayne, PA, USA, 2018, pp. 7–63. [Google Scholar]
- 10.Myers OD, Sumner SJ, Li S, Barnes S, Du X, One step forward for reducing false positive and false negative compound identifications from mass spectrometry metabolomics data: New algorithms for constructing extracted ion chromatograms and detecting chromatographic peaks, Anal. Chem 89 (2017), 8696–8703. 10.1021/acs.analchem.7b00947 [DOI] [PubMed] [Google Scholar]
- 11.Altinier G, Sosa S, Aquino RP, Mencherini T, Loggia RD, Tubaro A, Characterization of topical anti-inflammatory compounds in Rosmarinus officinalis L, J. Agric. Food. Chem 55 (2007), 1718–1723. 10.1021/jf062610 [DOI] [PubMed] [Google Scholar]
- 12.Razborsek MI, Voncina DB, Dolecek V, Voncina E, Determination of oleanolic, betulinic and ursolic acid in Lamiaceae and mass spectral fragmentation of their trimethylsilylated derivatives, Chromatographia. 57 (2008), 433–440. 10.1365/s10337-008-0533-6 [DOI] [Google Scholar]
- 13.Samarakoon SR, Ediriweera MK, Wijayabandara L, Fernando N, Tharmarajah L, Tennekoon KH, Piyathilaka P, Adhikari A, Isolation of cytotoxic triterpenes from the Mangrove plant, Scyphiphora Hydrophyllacea C.F.Gaertn (Rubiaceae), Trop. J. Pharm. Res 17 (2018), 475–481. 10.4314/tjpr.v17i3.13 [DOI] [Google Scholar]
- 14.Kamatou GPP, Van Vuuren SF, Van Heerden FR, Seaman T, Viljoen AM, Antibacterial and antimycobacterial activities of South African Salvia species and isolated compounds from S. Chamelaeagnea, S. Afr. J. Bot 73 (2007), 552–557. 10.1016/j.sajb.2007.05.001 [DOI] [Google Scholar]
- 15.Britton ER, Kellogg JJ, Kvalheim OM, Cech NB, Biochemometrics to identify synergists and additives from botanical medicines: a case study with Hydrastis canadensis (goldenseal), J. Nat. Prod 81 (2018), 484–493. 10.1021/acs.jnatprod.7b00654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chabán MF, Karagianni C, Joray MB, Toumpa D, Sola C, Crespo MI, Palacios SM, Athanassopoulos CM, Carpinella MC, Antibacterial effects of extracts obtained from plants of Argentina: Bioguided isolation of compounds from the anti-infectious medicinal plant Lepechinia Meyenii, J. Ethnopharmacol 239 (2019), 111930–111938. 10.1016/j.jep.2019.111930 [DOI] [PubMed] [Google Scholar]
- 17.Denny S, West PWJ, Mathew TC, Antagonistic interactions between the flavonoids hesperetin and naringenin and β-lactam antibiotics against Staphylococcus aureus, Br. J. Biomed. Sci 65 (2008), 145–147. 10.1080/09674845.2008.11732819 [DOI] [PubMed] [Google Scholar]
- 18.Kumarasamy Y, Cox PJ, Jaspars M, Nahar L, Sarker SD, Bioactivity of hirsutanolol, oregonin and genkwanin, isolated from the seeds of Alnus Glutinosa (Betulaceae), Nat. Prod. Comm 1 (2006), 641–644. 10.1177/1934578X0600100808 [DOI] [Google Scholar]
- 19.Abdallah FB, Lagha R, Gaber A Biofilm inhibition and eradication properties of medicinal plant essential oils against methicillin-resistant Staphylococcus aureus clinical isolates, Pharmaceut. 13 (2020), 369. 10.3390/ph13110369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caesar LK, Cech NB Synergy and antagonism in natural product extracts: when 1+1 does not equal 2. Nat. Prod. Rep. 36 (2019), 869–888. 10.1039/c9np00011a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perez-Fons L, Garzon MT, Micol V, Relationship between the antioxidant capacity and effect of rosemary (Rosmarinus officinalis L.) polyphenols on membrane phospholipid order, J. Agr. Food Chem 58 (2010), 161–171. 10.1021/jf9026487 [DOI] [PubMed] [Google Scholar]
- 22.Kontogianni VG, Tomic G, Nikolic I, Nerantzaki AA, Sayyad N, Stosic-Grujicic S, Stojanovic I, Gerothanassis IP, Tzakos AG, Phytochemical profile or Rosmarinus officinalis and Salvia officinalis extracts and correlation to their antioxidant and anti-proliferative activity, Food. Chem 136 (2013), 120–129. 10.1016/j.foodchem.2012.07.091 [DOI] [PubMed] [Google Scholar]
- 23.Luis JG, and Andres LS, C-16 hydroxylated abietane diterpenes from Salvia mellifera, Phytochem. 33 (1993), 635–638. 10.1016/0031-9422(93)85463-2 [DOI] [Google Scholar]
- 24.Valleverdu-Queralt A, Regueiro J, Martinez-Huelamo M, Rinaldi Alvarenga JF, Leal LN, Lamueala-Raventos RM, A comprehensive study on the phenolic profile of widely used culinary herbs and spices: rosemary, thyme, oregano, cinnamon, cumin and bay, Food. Chem. 154 (2014), 299–307. 10.1016/j.foodchem.2013.12.106 [DOI] [PubMed] [Google Scholar]
- 25.Nakatani N, and Inatani R, Two antioxidative diterpenes from rosemary (Rosmarinus officinalis L.) and a revised structure for rosmanol, Agric. Biol. Chem 48 (1984), 2081–2085. 10.1080/00021369.1984.10866436 [DOI] [Google Scholar]
- 26.Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, Fan TWM, Fiehn O, Goodacre R, Griffin J, Hankemeier T, Hardy N, Harnly J, Higashi R, Kopka J, Lane AN, Lindon JC, Marriott P, Nicholls AW, Relly MD, Thaden JJ, Viant MR, Proposed minimum reporting standards for chemical analysis. Chemical Analysis Working Group (CAWG) Metabolomic Standards Initiative (MSI), Metabolomics. 3 (2007), 211–221. 10.1007/s11306-007-0082-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


