Abstract
Background
Diagnosis of COVID-19 in symptomatic patients and screening of populations for SARS-CoV-2 infection require access to straightforward, low-cost and high-throughput testing. The recommended nasopharyngeal swab tests are limited by the need of trained professionals and specific consumables and this procedure is poorly accepted as a screening method In contrast, saliva sampling can be self-administered.
Methods
In order to compare saliva and nasopharyngeal/oropharyngeal samples for the detection of SARS-CoV-2, we designed a meta-analysis searching in PubMed up to December 29th, 2020 with the key words “(SARS-CoV-2 OR COVID-19 OR COVID19) AND (salivary OR saliva OR oral fluid)) NOT (review[Publication Type]) NOT (PrePrint[Publication Type])” applying the following criteria: records published in peer reviewed scientific journals, in English, with at least 15 nasopharyngeal/orapharyngeal swabs and saliva paired samples tested by RT-PCR, studies with available raw data including numbers of positive and negative tests with the two sampling methods. For all studies, concordance and sensitivity were calculated and then pooled in a random-effects model.
Findings
A total of 377 studies were retrieved, of which 50 were eligible, reporting on 16,473 pairs of nasopharyngeal/oropharyngeal and saliva samples. Meta-analysis showed high concordance, 92.5% (95%CI: 89.5–94.7), across studies and pooled sensitivities of 86.5% (95%CI: 83.4–89.1) and 92.0% (95%CI: 89.1–94.2) from saliva and nasopharyngeal/oropharyngeal swabs respectively. Heterogeneity across studies was 72.0% for saliva and 85.0% for nasopharyngeal/oropharyngeal swabs.
Interpretation
Our meta-analysis strongly suggests that saliva could be used for frequent testing of COVID-19 patients and “en masse” screening of populations.
Introduction
Propagation of infections by SARS-CoV-2, the coronavirus causing the COVID-19 pandemic, occurs from asymptomatic as well as symptomatic carriers [1]. To reduce the circulation of the virus in the population, SARS-CoV-2 carriers need to be identified rapidly and isolated as soon as possible, ideally before the onset of symptoms. When the virus has disseminated throughout a whole country, massive testing becomes of utmost urgent importance to combat the pandemic [2–4].
The recommended diagnosis of SARS-CoV-2 infection from the World Health Organisation is based on real time RT-qPCR detection of viral RNA in respiratory specimen such as nasopharyngeal swabs (NP), bronchial aspiration (BA), throat swab and sputum [5]. The American Centres For Disease Control and Prevention and the European Centre for Disease Prevention and Control now recommend viral testing from the respiratory system such as nasal or oral swabs or saliva [6, 7]. The French health regulatory authority (Haute Autorité de Santé) has recently included in its recommendations the use of saliva samples for the detection of SARS-CoV-2 in symptomatic individuals for whom nasopharyngeal sampling is difficult, and for mass testing within schools, universities and among health workers [8].
To make the diagnostic acceptable to the largest number of people, especially asymptomatic individuals, massive testing should be based on a sampling procedure that is inexpensive, easy to set up and well accepted by the population [9]. In contrast to nasopharyngeal swabbing, saliva sampling meets these criteria. Saliva sampling is fast, non-invasive, inexpensive and painless. It does not require trained professional with personal protective equipment nor other material than a simple plastic tube, and can be self-administered.
To evaluate saliva sampling for the detection of SARS-CoV-2, we conducted a meta-analysis on studies published in peer-reviewed journals until the 29th of December 2020 comparing the detection of SARS-CoV-2 using RT-PCR on paired nasopharyngeal/oropharyngeal and saliva samples in the same individuals sampled at the same time.
Methods
Search strategy and selection criteria
Literature search in PubMed (https://pubmed-ncbi-nlm-nih-gov) run the 29th of December 2020 with search word (SARS-CoV-2 OR COVID-19 OR COVID19) AND (salivary OR saliva OR oral fluid)) NOT (review[Publication Type]) NOT (PrePrint[Publication Type]) identified 377 articles. We included publications if they met the following eligibility criteria:
Records published in peer reviewed scientific journals;
Records published in English;
Data curation based on examination of the title and abstract, searching for research articles assessing viral RNA presence in saliva vs nasopharyngeal and/or oropharyngeal swabs;
Availability of nasopharyngeal (and/or oropharyngeal) swabs and saliva data on specimens sampled on the same individuals at the same time;
Detection of SARS-CoV-2 using the same RT-PCR method on both samples;
More than 15 individuals included in the study.
Data analysis
From each eligible article, we extracted: the number of individuals positive for SARS-CoV-2 in both nasopharyngeal/oropharyngeal swabs and saliva (a), those positive only in nasopharyngeal/oropharyngeal swab (b), those positive only in saliva (c) and (d) those negative in both nasopharyngeal swab and saliva (Table 1). From these data, we calculated the concordance of the same test (RTqPCR for 49 studies and RTdPCR for 1 study) on the two types of sample (a+d)/(a+b+c+d). We also computed the sensitivity of the test on each type of sample. The estimation of the sensitivity of a test requires a reference diagnosis. Since nasopharyngeal swab sampling has been shown to produce false negatives by RTqPCR [10], sensitivities for the saliva and the nasopharyngeal swab are defined here respectively as (a+c)/(a+b+c) and (a+b)/(a+b+c), considering as true positive any individual with a positive result on one or the other sample. This definition of a positive individual is also in agreement with the US-CDC and the ECDC directives on SARS-Cov-2 testing.
Table 1. Studies comparing SARS-CoV-2 detection in paired saliva and nasopharyngeal samples meeting inclusion criteria.
| Reference | Number of tested individuals | Concordance | Reference: S+ ou N+ | |||||
|---|---|---|---|---|---|---|---|---|
| Saliva + Nasoph. + | Saliva—Nasoph. + | Saliva + Nasoph. - | Saliva—Nasoph. - | Total | Sensitivity of S | Sensitivity of N | ||
| a | b | c | d | n = a+b+c+d | (a+d)/n | (a+c)/p | (a+b)/p | |
| Aita | 7 | 1 | 0 | 35 | 43 | 97.7% | 87.5% | 100.0% |
| Altawalah | 287 | 57 | 18 | 529 | 891 | 91.6% | 84.3% | 95.0% |
| Azzi | 22 | 4 | 33 | 54 | 113 | 67.3% | 93.2% | 44.1% |
| Babady | 16 | 1 | 1 | 69 | 87 | 97.7% | 94.4% | 94.4% |
| Barat | 30 | 7 | 1 | 421 | 459 | 98.3% | 81.6% | 97.4% |
| Berenger | 52 | 11 | 6 | 6 | 75 | 77.3% | 84.1% | 91.3% |
| Bhattacharya | 53 | 5 | 0 | 16 | 74 | 93.2% | 91.4% | 100.0% |
| Binder | 10 | 1 | 1 | 7 | 19 | 89.5% | 91.7% | 91.7% |
| Borghi | 79 | 28 | 7 | 187 | 301 | 88.4% | 75.4% | 93.9% |
| Braz-Silva | 37 | 15 | 18 | 131 | 201 | 83,6% | 78,6% | 74,3% |
| Byrne | 12 | 2 | 0 | 96 | 110 | 98.2% | 85.7% | 100.0% |
| Cassinarri | 8 | 5 | 1 | 17 | 31 | 80.6% | 64.3% | 92.9% |
| Caulley | 34 | 22 | 14 | 1869 | 1939 | 98.1% | 68.6% | 80.0% |
| Chau | 19 | 0 | 1 | 7 | 27 | 96.3% | 100.0% | 95.0% |
| Chen | 49 | 6 | 3 | 0 | 58 | 84.5% | 89.7% | 94.8% |
| Güçlü | 23 | 4 | 4 | 33 | 64 | 87.5% | 87.1% | 87.1% |
| Hanege | 29 | 9 | 0 | 0 | 38 | 76.3% | 76.3% | 100.0% |
| Hanson | 75 | 5 | 6 | 268 | 354 | 96.9% | 94.2% | 93.0% |
| Hasanoglu | 27 | 21 | 3 | 9 | 60 | 60.0% | 58.8% | 94.1% |
| Iwasaki | 8 | 1 | 1 | 66 | 76 | 97.4% | 90.0% | 90.0% |
| Jamal 1 | 44 | 20 | 8 | 19 | 91 | 69.2% | 72.2% | 88.9% |
| Jamal 2 | 42 | 14 | 9 | 10 | 75 | 69.3% | 78.5% | 86.2% |
| Kandel | 39 | 4 | 3 | 383 | 429 | 98.4% | 91.3% | 93.5% |
| Kojima | 20 | 3 | 6 | 16 | 45 | 80.0% | 89.7% | 79.3% |
| Landry | 28 | 5 | 2 | 89 | 124 | 94.4% | 85.7% | 94.3% |
| Leung | 38 | 7 | 13 | 37 | 95 | 78.9% | 87.9% | 77.6% |
| Matic | 15 | 6 | 1 | 52 | 74 | 90.5% | 72.7% | 95.5% |
| McCormick-Baw | 47 | 2 | 1 | 106 | 156 | 98.1% | 96.0% | 98.0% |
| Migueres | 34 | 7 | 3 | 79 | 123 | 91.9% | 84.1% | 93.2% |
| Moreno-Contreras | 19 | 9 | 6 | 37 | 71 | 78.9% | 73.5% | 82.4% |
| Nagura-Ikeda | 84 | 19 | 0 | 0 | 103 | 81.6% | 81.6% | 100.0% |
| Otto | 45 | 0 | 4 | 43 | 92 | 95.7% | 100.0% | 91.8% |
| Pasomsub | 16 | 3 | 2 | 179 | 200 | 97.5% | 85.7% | 90.5% |
| Procop | 38 | 0 | 1 | 177 | 216 | 99.5% | 100.0% | 97.4% |
| Rao | 73 | 11 | 76 | 57 | 217 | 59.9% | 93.1% | 52.5% |
| Sakanashi | 15 | 0 | 4 | 9 | 28 | 85.7% | 100.0% | 78.9% |
| Senok | 19 | 7 | 9 | 366 | 401 | 96.0% | 80.0% | 74.3% |
| Skolimowska | 15 | 3 | 1 | 112 | 131 | 96.9% | 84.2% | 94.7% |
| Sorelle | 32 | 7 | 0 | 44 | 83 | 91.6% | 82.1% | 100.0% |
| Sui | 14 | 0 | 2 | 0 | 16 | 87.5% | 100.0% | 87.5% |
| Torres | 46 | 54 | 8 | 835 | 943 | 93.4% | 50.0% | 92.6% |
| Uwamino | 32 | 15 | 11 | 138 | 196 | 86.7% | 74.1% | 81.0% |
| Vaz | 67 | 4 | 2 | 82 | 155 | 96.1% | 94.5% | 97.3% |
| Vogels | 49 | 5 | 4 | 3776 | 3834 | 99.8% | 91.4% | 93.1% |
| Williams | 33 | 6 | 1 | 49 | 89 | 92.1% | 85.0% | 97.5% |
| Wong | 104 | 18 | 37 | 70 | 229 | 76.0% | 88.7% | 76.7% |
| Wyllie | 34 | 9 | 13 | 13 | 69 | 68.1% | 83.9% | 76.8% |
| Yee | 69 | 18 | 10 | 203 | 300 | 90.7% | 81.4% | 89.7% |
| Yokota | 42 | 4 | 6 | 1872 | 1924 | 99.5% | 92.3% | 88.5% |
| Zhu | 382 | 60 | 15 | 487 | 944 | 92.1% | 86.9% | 96.7% |
| Total | 2412 | 525 | 376 | 13160 | 16473 | 94,5% | 84,2% | 88,7% |
The overall concordance and sensitivities have been estimated in a meta-analysis via Generalized Linear Mixed Models (GLMM), using a fixed-effect model, and also a random-effect model in case of over dispersion of the observations [11, 12]. Dispersion of effect sizes was evaluated using the Higgins I2 estimate of heterogeneity along with the Cochran’s Q in fixed-effect models [13] and using the tau2 estimate (between-study variance which is the variance of the distribution of true effect size) in random-effects models. The tau2 was calculated using the maximum likelihood estimator. As some studies have a sample size too small for the normality hypothesis, confidence intervals for each study were computed using the Clopper-Pearson method [14] also called “exact” binomial interval. Those for the overall estimates are based on normal approximation. Results are presented as forest plots.
All analyses were done with R version 4.0.3, using the package “meta” version 4.15–1 (2020-09-30) [15, 16].
Results
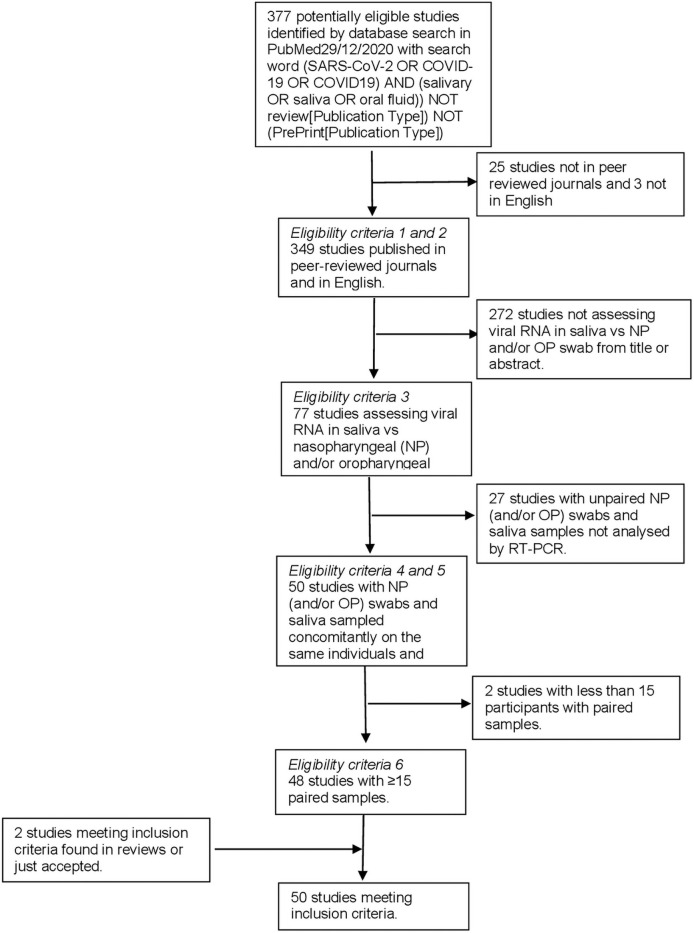
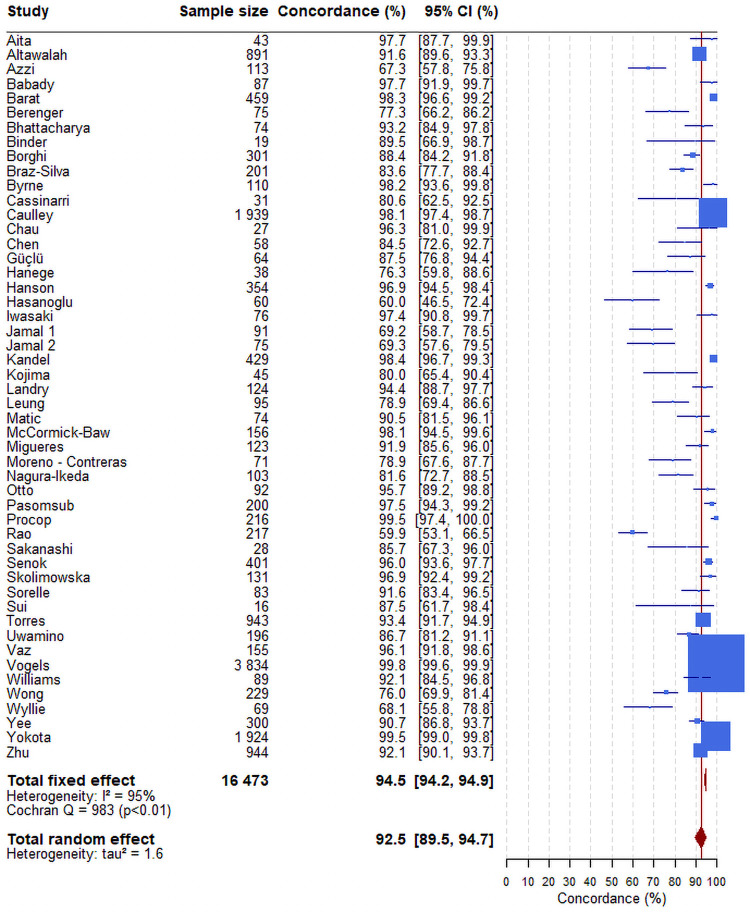
Forty-eight studies comparing SARS-CoV-2 loads in NP swabs and saliva samples collected concurrently in the same individuals using the same technique and providing positivity and negativity in both samples have been identified in PubMed [17–64]. We also included 2 articles not identified in the original Pubmed keywords search while fulfilling eligibility criteria (Table 1 and Fig 1) [65, 66]. In total 16,473 paired samples were analysed. The number of paired samples per study varied between 16 and 3834. Meta-analysis showed an overall concordance of 92.5% (95%CI: 89.5–94.7) across studies (Fig 2).
Fig 1. Evidence search and selection.
Fig 2. Forest plot of the concordance between the results of RTqPCR tests on nasopharyngeal and saliva samples.
The confidence intervals for each study are computed using the Clopper-Pearson method. Those for the overall estimates (fixed-effect or random-effect) are based on normal approximation. The blue box size is proportional to the number of positive tests. The red line corresponds to the value of the overall concordance of the random-effect model. This vertical line enables to locate the studies having an estimate concordance higher than 92.5%.
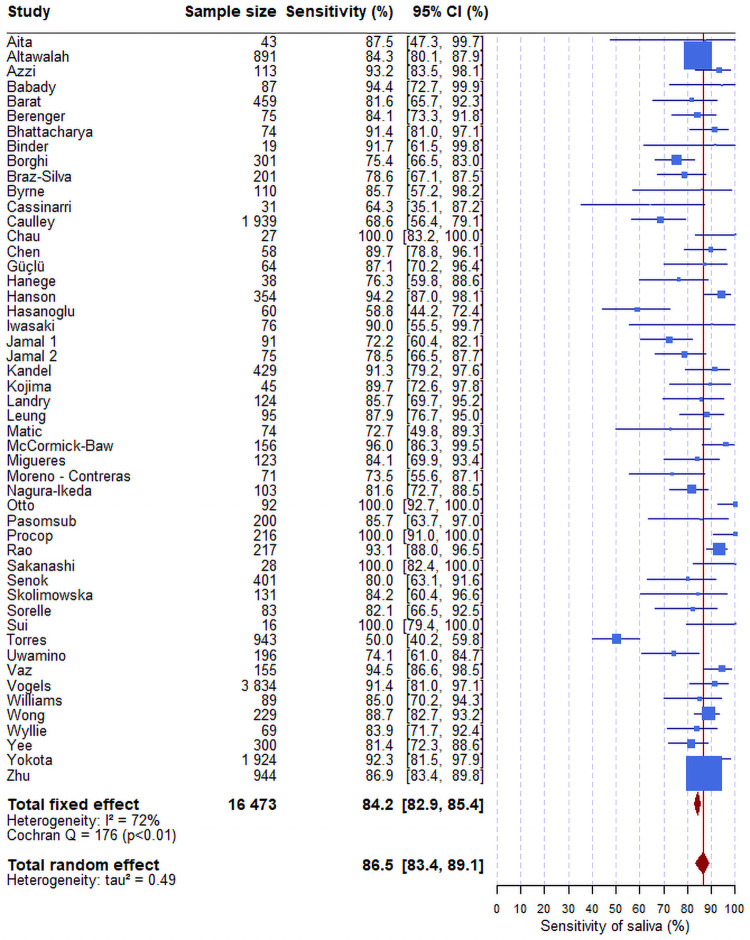
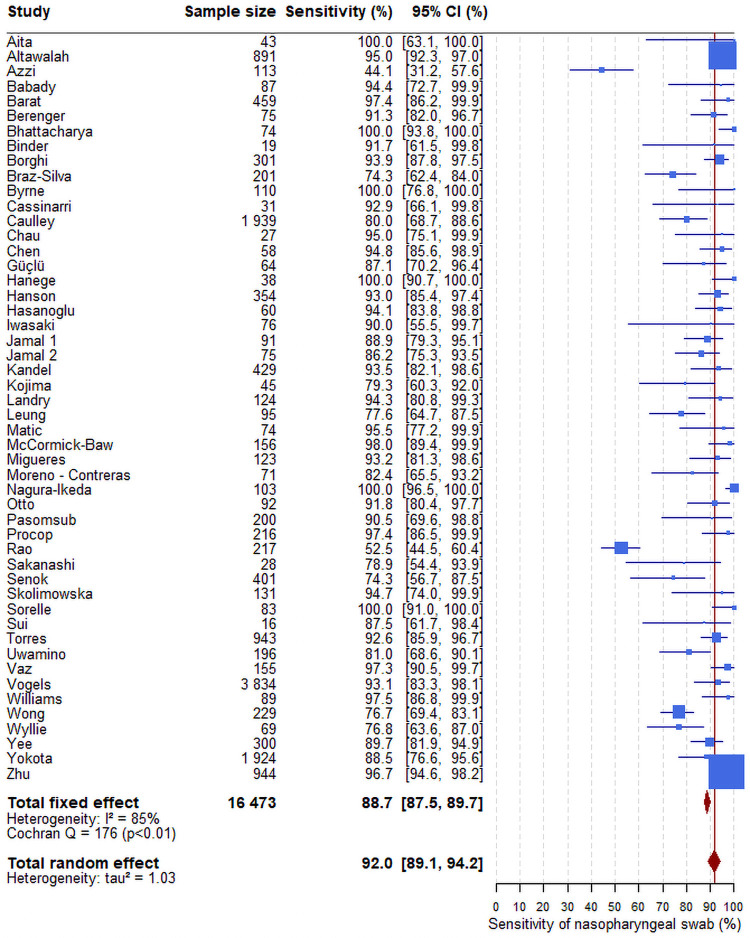
The overall sensitivity of the RT-PCR test from saliva samples was 86.5% (95%CI: 83.4–89.1) (Fig 3) versus 92.0% from nasopharyngeal swabs (95%CI: 89.1–94.2) (Fig 4). There was no association between the sensitivities of the saliva (S1 Fig) or of nasopharyngeal swab (S2 Fig) estimated in each study and the prevalence of the virus in the same study. If the sensitivity of the saliva was lower in populations of asymptomatic individuals than in population of individuals with symptoms, one would expect to observe a lower sensibility of the saliva in the studies with a low prevalence of infection.
Fig 3. Forest plot of the sensitivity of RTqPCR test on saliva.
The confidence intervals for each study are computed using the Clopper-Pearson method. Those for the overall estimates (fixed-effect or random-effect) are based on normal approximation. The blue box size is proportional to the number of positive tests. The difference between fixed-effect and random-effect overall sensitivity (respectively 84.2%, 86.5%) is low. The red line corresponds to the value of the overall sensitivity of the random-effect model. This vertical line enables to locate the studies having an estimate sensitivity higher than 86.5%. The heterogeneity estimator I2 is equal to 72%, which means a higher level of heterogeneity.
Fig 4. Forest plot of the sensitivity of RTqPCR test on nasopharyngeal sample.
The confidence intervals for each study are computed using the Clopper-Pearson method. Those for the overall estimates (fixed-effect or random-effect) are based on the normal approximation. The blue box size is proportional to the number of positive tests. The red line corresponds to the value of the overall sensitivity of the random-effect model. This vertical line enables to locate the studies having an estimate sensitivity higher than 92.0%. The heterogeneity estimator I2 is equal to 85%, which means a higher level of heterogeneity.
In our fixed-effect model meta-analysis, saliva gave I2 of 72% and nasopharyngeal gave I2 of 85%, both corresponding to high heterogeneity. Therefore, a random-effect model was performed to assess the overall sensitivities of the RTqPCR tests, taking into account the fact that the studies did not originate from one single population. The variance of saliva was 0.49 and that of nasopharyngeal was 1.03. As a sensitivity analysis, we also used various other methods to estimate the sensitivity, the confidence interval and the tau2 and all yielded very similar results (Figs 2–4).
Discussion
This meta-analysis reviewed 50 studies and concluded to a high concordance between nasopharyngeal and saliva samples for the detection of SARS-CoV-2 by RT-PCR. Although sensitivity was slightly lower on saliva samples than on nasopharyngeal samples, both values are above the 80% sensitivity cut-off recommended by health regulatory authorities such as the French Haute Autorité de Santé [67].
For computing concordance and sensitivities, we considered here the reference as SARS-CoV-2 positivity by RT-PCR either in saliva and/or nasopharyngeal samples since the presence of the virus in any sample is indicative of virus carriage.
In the context of mass screening, most participants are asymptomatic. Among the 50 studies analysed, only one included exclusively asymptomatic participants [63] and 8 studies included both symptomatic and asymptomatic participants but it was impossible to separate the data between the two populations [17, 20, 25, 29, 51, 56, 58, 65]. One study of contact cases included a larger number of asymptomatic subjects as compared to study of symptomatic subjects [49]. We did not observe any difference in concordance of the tests in these particular studies involving asymptomatic participants (Table 1). Formal comparison of nasopharyngeal and saliva samples from asymptomatic individuals is challenging: it would require to screen a large population for a small number of positive cases detected since the prevalence is usually low in this population. On the contrary, the symptomatic population expectedly contains higher percentage of positive subjects, as symptoms usually timely correlates with the highest viral load, which allow an easier comparison of both sampling procedures. Anyhow, viral load in saliva of presymptomatic subjects remains in the range of detection of the RT-PCR test for several days both in saliva [68] and nasopharyngeal samples [69]. In addition, both asymptomatic and symptomatic subjects appear to be contagious [1] with similarities in their viral load evolution [70–72]. Moreover, S3 Fig shows that in France the proportion of positive cases in the symptomatic tested population is consistently about 5 times larger than in the asymptomatic tested population, independently of viral prevalence over time. This and the fact that neither saliva nor nasopharyngeal sensitivities are affected in a screening-like context (the low prevalence being taken as a proxy, S1 and S2 Figs) strongly predict that viral detection is expected to exhibit similar performance in both populations.
Our meta-analysis showed large heterogeneity between studies. Sources of heterogeneity are both biological and technical. Biological heterogeneity may come from the fact that a given individual may carry the virus in only one of the saliva or nasopharyngeal specimens, or from the timing of sampling during the course of contamination. Technical heterogeneity comes from differences in the sampling and in RT-PCR methods. Among the 50 studies meeting the inclusion criteria for the meta-analysis, 29 studies report saliva collection in sterile containers (urine tubes or vials) without any additional solution. The other studies diluted the saliva in various viral transport media or phosphate buffer saline with or without bovine serum albumin. Other sources of variability come from differences in the amplified region of SARS-Cov-2 or to different positivity threshold between studies; most studies do not present viral load data but only cycle thresholds (Ct) for specific amplification of SARS-Cov-2 sequences and not even Ct differences (ΔCt) with a human reference gene.
Altogether, our meta-analysis of 50 studies including 16,473 paired samples shows high concordance (92.5%) between nasopharyngeal/oropharyngeal swabs and saliva, with a 5% higher sensitivity for the nasopharyngeal/oropharyngeal (92.0%) as compared to saliva (86.5%). While that might have been a liability in the context of individual diagnosis, it is not such a concern for mass screening, especially given the major advantage of the saliva sampling in terms of logistics. Previous meta-analyses of paired nasopharyngeal and saliva samples included less than 15 peer-reviewed studies or preprints and reported average sensitivity of 91%, 85% and 83.4% in 4, 16 and 5 studies respectively [73–75]. However, these studies used nasopharyngeal positivity as the reference, which did not seem relevant in our context.
To prevent a shortage of analytic reagents and to cut the costs necessarily associated to mass screening strategies, several recent publications have proposed mass testing methods based on saliva sampling either through extraction-free protocols [76–78] or through pre-extraction sample pooling [79–81]. In addition, sample pooling has gained a recognized interest for recurrent screening programs from the Centres of Disease Control recommendations [82], and surveillance protocols implemented in higher education institutions across the world, e.g. the State University of New York (United States) [83], Liège University (Belgium) [84], Heidelberg University (Germany) [85] as well as at Nottingham University (United Kingdom) [86].
In conclusion, this meta-analysis conclusively demonstrates that saliva is as valid as nasopharyngeal sampling for the detection of SARS-CoV-2 infections in symptomatic as well as asymptomatic carriers. In contrast to nasopharyngeal swabs, saliva sampling is simple, fast, non-invasive, inexpensive, painless and it thus uniquely applicable for surveillance, screening and diagnosis.
Supporting information
(TIF)
(TIF)
Week 28 corresponds to the results published the 13th of July 2020. Sources: Points épidémiologiques hebdomadaires.
(TIF)
(DOC)
Acknowledgments
We wish to thank members of the FranceTest group for useful discussions and support: Drs. Philippe Froguel, Eric Karsenti, Franck Molina, Jean Rossier and Claire Wyart. We also thank Yannick Marie and Prof. Marc Sanson for setting up the saliva tests at the ICM.
Data Availability
All relevant data are within the manuscript and its Supporting information files.
Funding Statement
The authors received no specific funding for this work but wish to thank ‘Investissements d’avenir’ Agence Nationale de la Recherche (ANR-10-IAIHU-06) for supporting the Paris Brain Institute ICM. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ferretti L, Wymant C, Kendall M, Zhao L, Nurtay A, Abeler-Dorner L, et al. Quantifying SARS-CoV-2 transmission suggests epidemic control with digital contact tracing. Science. 2020;368(6491). doi: 10.1126/science.abb6936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Larremore DB, Wilder B, Lester E, Shehata S, Burke JM, Hay JA, et al. Test sensitivity is secondary to frequency and turnaround time for COVID-19 screening. Sci Adv. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lavezzo E, Franchin E, Ciavarella C, Cuomo-Dannenburg G, Barzon L, Del Vecchio C, et al. Suppression of a SARS-CoV-2 outbreak in the Italian municipality of Vo’. Nature. 2020;584(7821):425–9. doi: 10.1038/s41586-020-2488-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xing Y, Wong GWK, Ni W, Hu X, Xing Q. Rapid Response to an Outbreak in Qingdao, China. N Engl J Med. 2020;383(23):e129. doi: 10.1056/NEJMc2032361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laboratory testing for 2019 novel coronavirus (2019-nCoV) in suspected human cases. https://www.who.int/publications-detail/laboratory-testing-for-2019-novel-coronavirus-in-suspected-human-cases-20200117 (accessed Jan. 3, 2021).
- 6.Overview of Testing for SARS-CoV-2 (COVID-19). https://www.cdc.gov/coronavirus/2019-ncov/hcp/testing-overview.html?CDC_AA_refVal=https%3A%2F%2Fwww.cdc.gov%2Fcoronavirus%2F2019-ncov%2Fhcp%2Fclinical-criteria.html (accessed Jan. 3, 2021).
- 7.Diagnostic testing and screening for SARS-CoV-2. https://www.ecdc.europa.eu/en/covid-19/latest-evidence/diagnostic-testing (accessed Jan. 4, 2021).
- 8.Detection of SASR-CoV-2 in saliva. https://www.has-sante.fr/upload/docs/application/pdf/2021-02/ac_2021_0007_rt-pcr_salivaire_covid-19.pdf accessed Apr. 21, 2021).
- 9.Siegler AJ, Hall E, Luisi N, Zlotorzynska M, Wilde G, Sanchez T, et al. Willingness to Seek Diagnostic Testing for SARS-CoV-2 With Home, Drive-through, and Clinic-Based Specimen Collection Locations. Open Forum Infect Dis. 2020;7(7):ofaa269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woloshin S, Patel N, Kesselheim AS. False Negative Tests for SARS-CoV-2 Infection—Challenges and Implications. N Engl J Med. 2020;383(6):e38. doi: 10.1056/NEJMp2015897 [DOI] [PubMed] [Google Scholar]
- 11.Schwarzer G, Chemaitelly H, Abu-Raddad LJ, Rucker G. Seriously misleading results using inverse of Freeman-Tukey double arcsine transformation in meta-analysis of single proportions. Res Synth Methods. 2019;10(3):476–83. doi: 10.1002/jrsm.1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stijnen T, Hamza TH, Ozdemir P. Random effects meta-analysis of event outcome in the framework of the generalized linear mixed model with applications in sparse data. Stat Med. 2010;29(29):3046–67. doi: 10.1002/sim.4040 [DOI] [PubMed] [Google Scholar]
- 13.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–60. doi: 10.1136/bmj.327.7414.557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agresti A, Coull BA. Approximate Is Better than "Exact" for Interval Estimation of Binomial Proportions. The American Statistician. 1998;52(2):119–26. [Google Scholar]
- 15.Team RC. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. 2020. [Google Scholar]
- 16.Balduzzi S, Rucker G, Schwarzer G. How to perform a meta-analysis with R: a practical tutorial. Evid Based Ment Health. 2019;22(4):153–60. doi: 10.1136/ebmental-2019-300117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aita A, Basso D, Cattelan AM, Fioretto P, Navaglia F, Barbaro F, et al. SARS-CoV-2 identification and IgA antibodies in saliva: One sample two tests approach for diagnosis. Clin Chim Acta. 2020;510:717–22. doi: 10.1016/j.cca.2020.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altawalah H, AlHuraish F, Alkandari WA, Ezzikouri S. Saliva specimens for detection of severe acute respiratory syndrome coronavirus 2 in Kuwait: A cross-sectional study. J Clin Virol. 2020;132:104652. doi: 10.1016/j.jcv.2020.104652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Azzi L, Carcano G, Gianfagna F, Grossi P, Gasperina DD, Genoni A, et al. Saliva is a reliable tool to detect SARS-CoV-2. J Infect. 2020;81(1):e45–e50. doi: 10.1016/j.jinf.2020.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Babady NE, McMillen T, Jani K, Viale A, Robilotti EV, Aslam A, et al. Performance of Severe Acute Respiratory Syndrome Coronavirus 2 Real-Time RT-PCR Tests on Oral Rinses and Saliva Samples. J Mol Diagn. 2020. doi: 10.1016/j.jmoldx.2020.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barat B, Das S, Giorgi V, Henderson DK, Kopka S, Lau AF, et al. Pooled Saliva Specimens for SARS-CoV-2 Testing. medRxiv. 2020. doi: 10.1101/2020.10.02.20204859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berenger BM, Conly JM, Fonseca K, Hu J, Louie T, Schneider AR, et al. Saliva collected in universal transport media is an effective, simple and high-volume amenable method to detect SARS-CoV-2. Clin Microbiol Infect. 2020. doi: 10.1016/j.cmi.2020.10.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhattacharya D, Parai D, Rout UK, Dash P, Nanda RR, Dash GC, et al. Saliva for diagnosis of SARS-CoV-2: first report from India. J Med Virol. 2020. doi: 10.1002/jmv.26719 [DOI] [PubMed] [Google Scholar]
- 24.Binder RA, Alarja NA, Robie ER, Kochek KE, Xiu L, Rocha-Melogno L, et al. Environmental and Aerosolized Severe Acute Respiratory Syndrome Coronavirus 2 Among Hospitalized Coronavirus Disease 2019 Patients. J Infect Dis. 2020;222(11):1798–806. doi: 10.1093/infdis/jiaa575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borghi E, Massa V, Carmagnola D, Dellavia C, Parodi C, Ottaviano E, et al. Saliva sampling for chasing SARS-CoV-2: A Game-changing strategy. Pharmacol Res. 2020:105380. doi: 10.1016/j.phrs.2020.105380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Braz-Silva PH, Mamana AC, Romano CM, Felix AC, de Paula AV, Fereira NE, et al. Performance of at-home self-collected saliva and nasal-oropharyngeal swabs in the surveillance of COVID-19. J Oral Microbiol. 2020;13(1):1858002. doi: 10.1080/20002297.2020.1858002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Byrne RL, Kay GA, Kontogianni K, Aljayyoussi G, Brown L, Collins AM, et al. Saliva Alternative to Upper Respiratory Swabs for SARS-CoV-2 Diagnosis. Emerg Infect Dis. 2020;26(11):2770–1. doi: 10.3201/eid2611.203283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cassinari K, Alessandri-Gradt E, Chambon P, Charbonnier F, Gracias S, Beaussire L, et al. Assessment of multiplex digital droplet RT-PCR as a diagnostic tool for SARS-CoV-2 detection in nasopharyngeal swabs and saliva samples. Clin Chem. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chau NVV, Thanh Lam V, Thanh Dung N, Yen LM, Minh NNQ, Hung LM, et al. The natural history and transmission potential of asymptomatic SARS-CoV-2 infection. Clin Infect Dis. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen L, Zhao J, Peng J, Li X, Deng X, Geng Z, et al. Detection of SARS-CoV-2 in saliva and characterization of oral symptoms in COVID-19 patients. Cell Prolif. 2020;53(12):e12923. doi: 10.1111/cpr.12923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guclu E, Koroglu M, Yurumez Y, Toptan H, Kose E, Guneysu F, et al. Comparison of saliva and oro-nasopharyngeal swab sample in the molecular diagnosis of COVID-19. Rev Assoc Med Bras (1992). 2020;66(8):1116–21. doi: 10.1590/1806-9282.66.8.1116 [DOI] [PubMed] [Google Scholar]
- 32.Hanege FM, Kocoglu E, Kalcioglu MT, Celik S, Cag Y, Esen F, et al. SARS-CoV-2 Presence in the Saliva, Tears, and Cerumen of COVID-19 Patients. Laryngoscope. 2020. [DOI] [PubMed] [Google Scholar]
- 33.Hanson KE, Barker AP, Hillyard DR, Gilmore N, Barrett JW, Orlandi RR, et al. Self-Collected Anterior Nasal and Saliva Specimens versus Health Care Worker-Collected Nasopharyngeal Swabs for the Molecular Detection of SARS-CoV-2. J Clin Microbiol. 2020;58(11). doi: 10.1128/JCM.01824-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hasanoglu I, Korukluoglu G, Asilturk D, Cosgun Y, Kalem AK, Altas AB, et al. Higher viral loads in asymptomatic COVID-19 patients might be the invisible part of the iceberg. Infection. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iwasaki S, Fujisawa S, Nakakubo S, Kamada K, Yamashita Y, Fukumoto T, et al. Comparison of SARS-CoV-2 detection in nasopharyngeal swab and saliva. J Infect. 2020;81(2):e145–e7. doi: 10.1016/j.jinf.2020.05.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jamal AJ, Mozafarihashjin M, Coomes E, Anceva-Sami S, Barati S, Crowl G, et al. Sensitivity of midturbinate versus nasopharyngeal swabs for the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Infect Control Hosp Epidemiol. 2020:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jamal AJ, Mozafarihashjin M, Coomes E, Powis J, Li AX, Paterson A, et al. Sensitivity of nasopharyngeal swabs and saliva for the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clin Infect Dis. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kandel C, Zheng J, McCready J, Serbanescu MA, Racher H, Desaulnier M, et al. Detection of SARS-CoV-2 from Saliva as Compared to Nasopharyngeal Swabs in Outpatients. Viruses. 2020;12(11). doi: 10.3390/v12111314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kojima N, Turner F, Slepnev V, Bacelar A, Deming L, Kodeboyina S, et al. Self-Collected Oral Fluid and Nasal Swab Specimens Demonstrate Comparable Sensitivity to Clinician-Collected Nasopharyngeal Swab Specimens for the Detection of SARS-CoV-2. Clin Infect Dis. 2020. doi: 10.1093/cid/ciaa1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Landry ML, Criscuolo J, Peaper DR. Challenges in use of saliva for detection of SARS CoV-2 RNA in symptomatic outpatients. J Clin Virol. 2020;130:104567. doi: 10.1016/j.jcv.2020.104567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leung EC, Chow VC, Lee MK, Lai RW. Deep throat saliva as an alternative diagnostic specimen type for the detection of SARS-CoV-2. J Med Virol. 2020. doi: 10.1002/jmv.26258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matic N, Stefanovic A, Leung V, Lawson T, Ritchie G, Li L, et al. Practical challenges to the clinical implementation of saliva for SARS-CoV-2 detection. Eur J Clin Microbiol Infect Dis. 2020. doi: 10.1007/s10096-020-04090-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCormick-Baw C, Morgan K, Gaffney D, Cazares Y, Jaworski K, Byrd A, et al. Saliva as an Alternate Specimen Source for Detection of SARS-CoV-2 in Symptomatic Patients Using Cepheid Xpert Xpress SARS-CoV-2. J Clin Microbiol. 2020;58(8). doi: 10.1128/JCM.01109-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Migueres M, Mengelle C, Dimeglio C, Didier A, Alvarez M, Delobel P, et al. Saliva sampling for diagnosing SARS-CoV-2 infections in symptomatic patients and asymptomatic carriers. J Clin Virol. 2020;130:104580. doi: 10.1016/j.jcv.2020.104580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moreno-Contreras J, Espinoza MA, Sandoval-Jaime C, Cantu-Cuevas MA, Baron-Olivares H, Ortiz-Orozco OD, et al. Saliva Sampling and Its Direct Lysis, an Excellent Option To Increase the Number of SARS-CoV-2 Diagnostic Tests in Settings with Supply Shortages. J Clin Microbiol. 2020;58(10). doi: 10.1128/JCM.01659-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nagura-Ikeda M, Imai K, Tabata S, Miyoshi K, Murahara N, Mizuno T, et al. Clinical Evaluation of Self-Collected Saliva by Quantitative Reverse Transcription-PCR (RT-qPCR), Direct RT-qPCR, Reverse Transcription-Loop-Mediated Isothermal Amplification, and a Rapid Antigen Test To Diagnose COVID-19. J Clin Microbiol. 2020;58(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pasomsub E, Watcharananan SP, Boonyawat K, Janchompoo P, Wongtabtim G, Suksuwan W, et al. Saliva sample as a non-invasive specimen for the diagnosis of coronavirus disease 2019: a cross-sectional study. Clin Microbiol Infect. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Procop GW, Shrestha NK, Vogel S, Van Sickle K, Harrington S, Rhoads DD, et al. A Direct Comparison of Enhanced Saliva to Nasopharyngeal Swab for the Detection of SARS-CoV-2 in Symptomatic Patients. Journal of Clinical Microbiology. 2020;58(11):e01946–20. doi: 10.1128/JCM.01946-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rao M, Rashid FA, Sabri F, Jamil NN, Zain R, Hashim R, et al. Comparing nasopharyngeal swab and early morning saliva for the identification of SARS-CoV-2. Clin Infect Dis. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sakanashi D, Asai N, Nakamura A, Miyazaki N, Kawamoto Y, Ohno T, et al. Comparative evaluation of nasopharyngeal swab and saliva specimens for the molecular detection of SARS-CoV-2 RNA in Japanese patients with COVID-19. J Infect Chemother. 2021;27(1):126–9. doi: 10.1016/j.jiac.2020.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Senok A, Alsuwaidi H, Atrah Y, Al Ayedi O, Al Zahid J, Han A, et al. Saliva as an Alternative Specimen for Molecular COVID-19 Testing in Community Settings and Population-Based Screening. Infect Drug Resist. 2020;13:3393–9. doi: 10.2147/IDR.S275152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Skolimowska K, Rayment M, Jones R, Madona P, Moore LSP, Randell P. Non-invasive saliva specimens for the diagnosis of COVID-19: caution in mild outpatient cohorts with low prevalence. Clin Microbiol Infect. 2020;26(12):1711–3. doi: 10.1016/j.cmi.2020.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.SoRelle JA, Mahimainathan L, McCormick-Baw C, Cavuoti D, Lee F, Thomas A, et al. Saliva for use with a point of care assay for the rapid diagnosis of COVID-19. Clin Chim Acta. 2020;510:685–6. doi: 10.1016/j.cca.2020.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sui Z, Zhang Y, Tu J, Xie J, Huang W, Peng T, et al. Evaluation of saliva as an alternative diagnostic specimen source for SARS-CoV-2 detection by RT-dPCR. J Infect. 2020. doi: 10.1016/j.jinf.2020.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Torres M, Collins K, Corbit M, Ramirez M, Winters CR, Katz L, et al. Comparison of saliva and nasopharyngeal swab SARS-CoV-2 RT-qPCR testing in a community setting. J Infect. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Uwamino Y, Nagata M, Aoki W, Fujimori Y, Nakagawa T, Yokota H, et al. Accuracy and stability of saliva as a sample for reverse transcription PCR detection of SARS-CoV-2. J Clin Pathol. 2021;74(1):67–8. doi: 10.1136/jclinpath-2020-206972 [DOI] [PubMed] [Google Scholar]
- 57.Vaz SN, Santana DS, Netto EM, Pedroso C, Wang WK, Santos FDA, et al. Saliva is a reliable, non-invasive specimen for SARS-CoV-2 detection. Braz J Infect Dis. 2020;24(5):422–7. doi: 10.1016/j.bjid.2020.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vogels CBF, Watkins AE, Harden CA, Brackney DE, Shafer J, Wang J, et al. SalivaDirect: A simplified and flexible platform to enhance SARS-CoV-2 testing capacity. Med. doi: 10.1016/j.medj.2020.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Williams E, Bond K, Zhang B, Putland M, Williamson DA. Saliva as a Noninvasive Specimen for Detection of SARS-CoV-2. J Clin Microbiol. 2020;58(8). doi: 10.1128/JCM.00776-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wong S, Tse H, Siu HK, Kwong TS, Chu MY, Yau FYS, et al. Posterior oropharyngeal saliva for the detection of SARS-CoV-2. Clin Infect Dis. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wyllie AL, Fournier J, Casanovas-Massana A, Campbell M, Tokuyama M, Vijayakumar P, et al. Saliva or Nasopharyngeal Swab Specimens for Detection of SARS-CoV-2. N Engl J Med. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yee R, Truong T, Pannaraj PS, Eubanks N, Gai E, Jumarang J, et al. Saliva is a Promising Alternative Specimen for the Detection of SARS-CoV-2 in Children and Adults. J Clin Microbiol. 2020. doi: 10.1101/2020.10.25.20219055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yokota I, Shane PY, Okada K, Unoki Y, Yang Y, Inao T, et al. Mass screening of asymptomatic persons for SARS-CoV-2 using saliva. Clin Infect Dis. 2020. doi: 10.1093/cid/ciaa1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhu J, Guo J, Xu Y, Chen X. Viral dynamics of SARS-CoV-2 in saliva from infected patients. J Infect. 2020;81(3):e48–e50. doi: 10.1016/j.jinf.2020.06.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Caulley L, Corsten M, Eapen L, Whelan J, Angel JB, Antonation K, et al. Salivary Detection of COVID-19. Ann Intern Med. 2020. doi: 10.7326/M20-4738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Otto MP, Darles C, Valero E, Benner P, Dutasta F, Janvier F. Posterior oropharyngeal salivafor the detection of SARS-CoV-2. Clin Infect Dis. 2020. doi: 10.1093/cid/ciaa1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.COVID-19: HAS positions antigenic tests in three situations Press release—Posted on 09 Oct 2020 https://www.has-sante.fr/jcms/p_3212125/fr/covid-19-la-has-positionne-les-tests-antigeniques-dans-trois-situations.
- 68.Winnett A, Cooper MM, Shelby N, Romano AE, Reyes JA, Ji J, et al. SARS-CoV-2 Viral Load in Saliva Rises Gradually and to Moderate Levels in Some Humans. medRxiv. 2020:2020.12.09.20239467. doi: 10.1101/2020.12.09.20239467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mina MJ, Parker R, Larremore DB. Rethinking Covid-19 Test Sensitivity—A Strategy for Containment. N Engl J Med. 2020;383(22):e120. doi: 10.1056/NEJMp2025631 [DOI] [PubMed] [Google Scholar]
- 70.Arons MM, Hatfield KM, Reddy SC, Kimball A, James A, Jacobs JR, et al. Presymptomatic SARS-CoV-2 Infections and Transmission in a Skilled Nursing Facility. N Engl J Med. 2020;382(22):2081–90. doi: 10.1056/NEJMoa2008457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.He X, Lau EHY, Wu P, Deng X, Wang J, Hao X, et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020;26(5):672–5. doi: 10.1038/s41591-020-0869-5 [DOI] [PubMed] [Google Scholar]
- 72.Widders A, Broom A, Broom J. SARS-CoV-2: The viral shedding vs infectivity dilemma. Infect Dis Health. 2020;25(3):210–5. doi: 10.1016/j.idh.2020.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Czumbel LM, Kiss S, Farkas N, Mandel I, Hegyi A, Nagy A, et al. Saliva as a Candidate for COVID-19 Diagnostic Testing: A Meta-Analysis. Front Med (Lausanne). 2020;7:465. doi: 10.3389/fmed.2020.00465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kivela JM, Jarva H, Lappalainen M, Kurkela S. Saliva-based testing for diagnosis of SARS-CoV-2 infection: A meta-analysis. J Med Virol. 2020. doi: 10.1002/jmv.26613 [DOI] [PubMed] [Google Scholar]
- 75.Ricco M, Ranzieri S, Peruzzi S, Valente M, Marchesi F, Balzarini F, et al. RT-qPCR assays based on saliva rather than on nasopharyngeal swabs are possible but should be interpreted with caution: results from a systematic review and meta-analysis. Acta Biomed. 2020;91(3):e2020025. doi: 10.23750/abm.v91i3.10020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ott IM, Strine MS, Watkins AE, Boot M, Kalinich CC, Harden CA, et al. Simply saliva: stability of SARS-CoV-2 detection negates the need for expensive collection devices. medRxiv. 2020. doi: 10.1101/2020.08.03.20165233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ranoa DRE, Holland RL, Alnaji FG, Green KJ, Wang L, Brooke CB, et al. Saliva-Based Molecular Testing for SARS-CoV-2 that Bypasses RNA Extraction. bioRxiv. 2020:2020.06.18.159434. [Google Scholar]
- 78.Vogels C.B.F. B DE, Wang J., Kalinich C.C., Ott I.M., Kudo E., Lu P., et al. SalivaDirect: simple and sensitive molecular diagnostic test for SARS-Cov-2 surveillance. medRxive. 2020. [Google Scholar]
- 79.Gavars D, Gavars M, Perminovs D, Stasulans J, Stana J, Metla Z, et al. Saliva as testing sample for SARS-CoV-2 detection by RT-PCR in low prevalence community setting. medRxiv. 2020:2020.10.20.20216127. [Google Scholar]
- 80.Sahajpal NS, Mondal AK, Ananth S, Njau A, Ahluwalia P, Chaubey A, et al. SalivaAll: Clinical validation of a sensitive test for saliva collected in healthcare and community settings with pooling utility for SARS-CoV-2 mass surveillance. medRxiv. 2020:2020.08.26.20182816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Watkins A.E. F EP, Weinberger Daniel M., Vogels Chantal B.F., Brackney Doug E., Casanovas-Massana Arnau, Campbell Melissa, et al. Pooling saliva to increase SARS-CoV-2 testing capacity. MedRxiv. 2020. doi: 10.1101/2020.09.02.20183830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Interim Guidance for Use of Pooling Procedures in SARS-CoV-2 Diagnostic, Screening, and Surveillance Testing. https://www.cdc.gov/coronavirus/2019-ncov/lab/pooling-procedures.html (accessed Jan. 4, 2021).
- 83.SUNY COVID-19 Case Tracker. https://www.suny.edu/COVID19-tracker/ (accessed Jan. 4, 2021).
- 84.Screening in practice. https://www.coronavirus.uliege.be/cms/c_12199182/en/screening-in-practice (accessed Jan. 4, 2021).
- 85.New Corona testing strategies for the general public. https://www.virusfinder.de/en/ (accessed Jan. 4, 2021).
- 86.Fogarty A, Joseph A, Shaw D. Pooled saliva samples for COVID-19 surveillance programme. Lancet Respir Med. 2020;8(11):1078–80. doi: 10.1016/S2213-2600(20)30444-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
Week 28 corresponds to the results published the 13th of July 2020. Sources: Points épidémiologiques hebdomadaires.
(TIF)
(DOC)
Data Availability Statement
All relevant data are within the manuscript and its Supporting information files.