Abstract
An intricate regulatory network controls the expression of Salmonella virulence genes. The transcriptional regulator HilD plays a central role in this network by controlling the expression of tens of genes mainly required for intestinal colonization. Accordingly, the expression/activity of HilD is highly regulated by multiple factors, such as the SirA/BarA two-component system and the Hcp-like protein HilE. SirA/BarA positively regulates translation of hilD mRNA through a regulatory cascade involving the small RNAs CsrB and CsrC, and the RNA-binding protein CsrA, whereas HilE inhibits HilD activity by protein-protein interaction. In this study, we show that SirA/BarA also positively regulates translation of hilE mRNA through the same mentioned regulatory cascade. Thus, our results reveal a paradoxical regulation exerted by SirA/BarA-Csr on HilD, which involves simultaneous opposite effects, direct positive control and indirect negative control through HilE. This kind of regulation is called an incoherent type-1 feedforward loop (I1-FFL), which is a motif present in certain regulatory networks and represents a complex biological problem to decipher. Interestingly, our results, together with those from a previous study, indicate that HilE, the repressor component of the I1-FFL reported here (I1-FFLSirA/BarA-HilE-HilD), is required to reduce the growth cost imposed by the expression of the genes regulated by HilD. Moreover, we and others found that HilE is necessary for successful intestinal colonization by Salmonella. Thus, these findings support that I1-FFLSirA/BarA-HilE-HilD cooperates to control the precise amount and activity of HilD, for an appropriate balance between the growth cost and the virulence benefit generated by the expression of the genes induced by this regulator. I1-FFLSirA/BarA-HilE-HilD represents a complex regulatory I1-FFL that involves multiple regulators acting at distinct levels of gene expression, as well as showing different connections to the rest of the regulatory network governing Salmonella virulence.
Author summary
To infect the intestine of a broad range of hosts, including humans, Salmonella is required to express a large number of genes encoding different cellular functions, which imposes a growth penalty. Thus, Salmonella has developed complex regulatory mechanisms that control the expression of virulence genes. Here we identified a novel and sophisticated regulatory mechanism that is involved in the fine-tuned control of the expression level and activity of the transcriptional regulator HilD, for the appropriate balance between the growth cost and the virulence benefit generated by the expression of tens of Salmonella genes. This mechanism forms an incoherent type-1 feedforward loop (I1-FFL), which involves paradoxical regulation; that is, a regulatory factor exerting simultaneous opposite control (positive and negative) on another factor. I1-FFLs are present in regulatory networks of diverse organisms, from bacteria to humans, and represent a complex biological problem to decipher. Interestingly, the I1-FFL reported here is integrated by ancestral regulators and by regulators that Salmonella has acquired during evolution. Thus, our findings reveal a novel I1-FFL of bacteria, which is involved in virulence. Moreover, our results illustrate the integration of ancestral and acquired factors into a regulatory motif, which can lead to the expansion of regulatory networks.
Introduction
Pathogenic bacteria have developed diverse regulatory mechanisms to control the appropriated spatiotemporal expression of virulence genes. Both activation and repression regulatory mechanisms are essential for bacteria to colonize different niches of hosts.
Salmonella enterica serovar Typhimurium (S. Typhimurium) contains a large number of virulence genes and a very complex regulatory network that control their expression. Two groups of genes mainly govern Salmonella virulence; those located in Salmonella Pathogenicity Island 1 (SPI-1) and those located in SPI-2. Both SPI-1 and SPI-2 encode a Type III Secretion System (T3SS), several effector proteins and chaperones, as well as transcriptional regulators [1,2]. The T3SS is a syringe-like multiprotein complex through which bacteria inject effector proteins directly into the cytoplasm of host cells. Effector proteins have distinct biological activities that alter different signal transduction pathways of eukaryotic host cells [3]. The SPI-1 genes mediate Salmonella invasion of cells at the intestinal epithelium, which generates enteritis, whereas the SPI-2 genes primarily mediate Salmonella replication within host cells [1,2]. Replication/survival inside macrophages within a membrane-bound compartment called the Salmonella-containing vacuole (SCV), leads to a systemic infection like typhoid fever [1,2]. Many other Salmonella virulence genes are located in distinct genomic regions, with their biological function being related to that of SPI-1 or SPI-2 [2,4,5].
Consistent with their role in intestinal infection, expression of the SPI-1 genes is induced when Salmonella resides in the intestinal lumen and in the cytosol of epithelial cells [6,7]. In addition, expression of the SPI-1 genes is regulated by different environmental cues commonly found in the intestine of hosts, such as short- and long-chain fatty acids, bile, oxygen level, osmolarity and pH [8–13]. In vitro, these genes are expressed in nutrient-rich media, such as lysogeny broth (LB), during the late exponential and early stationary growth phases [14–16]. Interestingly, different studies have reported that the SPI-1 genes show a bistable expression both in vitro [17–21] and in the gut lumen of mice [22], where only 10–50% of cells from clonal Salmonella populations express SPI-1. Moreover, it was shown that the two subpopulations of cells generated by this bistable expression (SPI-1ON and SPI-1OFF) cooperate for the successful invasion of host cells and intestinal colonization by Salmonella [20,21].
Expression of the SPI-1 and related genes is controlled by a very complex regulatory network involving many positive and negative regulators that act at the transcriptional, translational or post-translational level [2,23,24]. The AraC-like regulator HilD, encoded in SPI-1, is the apex of different regulatory cascades through which HilD controls the expression of the SPI-1 genes and many other genes located outside of SPI-1. HilD induces expression of: 1) the SPI-1 genes and other genes located outside of SPI-1, through the HilA, InvF or SprB regulators, encoded in SPI-1 [2,23–26]; 2) the SPI-2 genes and other genes located outside SPI-2, through the SsrA/SsrB two-component system, encoded in SPI-2 [15,27,28]; 3) the flagellar and chemotaxis genes, through the FlhD4C2 transcriptional complex [29,30]; and 4) several other genes located in distinct genomic regions, by direct interaction [26,31,32]. Furthermore, HilD forms a positive feedforward regulatory loop with the AraC-like regulators HilC and RtsA, which recognize the same DNA motif as HilD and form heterodimers with HilD; however, HilD has a dominant function over HilC and RtsA [33–35].
Consistently with its role as a master regulator for Salmonella virulence, the expression, concentration and activity of HilD are tightly controlled, representing the central point for regulating SPI-1 genes and many other related genes [2,23,24]. For instance, the Hcp-like protein HilE, encoded in a genomic island other than the SPIs, inhibits HilD activity by protein-protein interaction, thereby affecting homodimerization and DNA binding of HilD [36–38]. In addition, the SirA/BarA two-component system induces expression of hilD at the translational level [39]. In this two-component system, SirA and BarA are the response regulator and the sensor kinase, respectively [40–42].
Orthologs of SirA/BarA are present in many other bacteria. In Escherichia coli (UvrY/BarA), Pseudomonas spp. (GacA/GacS), and Vibrio cholerae (VarA/VarS), this system controls expression of numerous genes encoding different cellular activities, including virulence, motility, biofilm formation and metabolism [43,44]. This system has been better characterized in E. coli where UvrY/BarA was shown to form a regulatory cascade with the non-translated small RNAs (sRNAs) CsrB and CsrC, and the RNA binding protein CsrA [44,45]. CsrA represses numerous genes at the translational level by interacting with sequences overlapping the ribosome binding sites on target mRNAs [44,45]. In response to its phosphorylation by BarA, UvrY directly activates transcription of CsrB and CsrC, which contain several binding sites for CsrA [44,45]. Thus, CsrB/CsrC sequester CsrA and antagonize its ability to regulate expression of target transcripts [44,45]. Several studies have shown that the counterparts of the UvrY/BarA-CsrB/CsrC-CsrA global regulatory network function similarly in other bacteria [43,44]. For instance, the SirA/BarA system induces expression of the SPI-1 genes through CsrB and CsrC, which antagonize the CsrA-mediated translational repression of the hilD transcript [39,46,47].
Interestingly, expression of SPI-1 genes causes growth retardation of S. Typhimurium in laboratory conditions. This effect becomes even more pronounced when the SPI-1 genes are overexpressed, such as in the absence of HilE [19]. However, it remains poorly understood how S. Typhimurium fine-tunes the expression level of the SPI-1 genes to maintain an appropriate balance between the penalty on growth and the benefit for virulence.
In this study, we report that the SirA/BarA-CsrB/CsrC regulatory cascade induces the expression of hilE by counteracting CsrA-mediated translational repression on the hilE mRNA, which reveals that SirA/BarA, HilD and HilE form an incoherent type-1 feedforward loop. Additionally, we demonstrate that HilE is necessary to reduce the growth cost imposed by the expression of genes regulated by HilD, in laboratory conditions and in the intestine of mice. Thus, our results support that the control of HilD expression by the feedforward loop formed by SirA/BarA, HilD and HilE, plays a role in the fitness of S. Typhimurium during intestinal infection of hosts.
Results
CsrA directly represses the expression of hilE
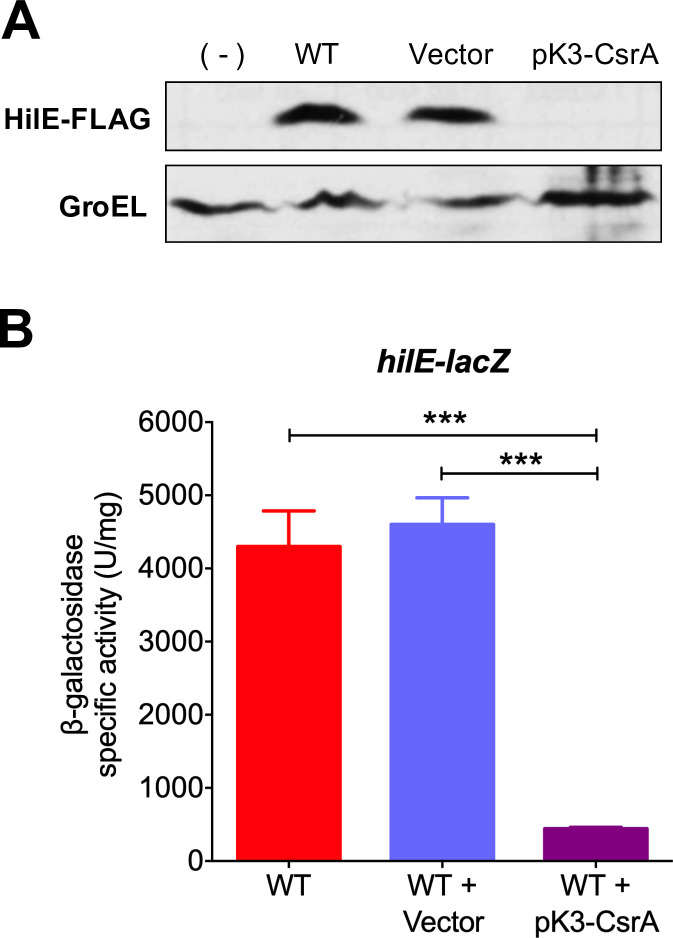
In a current project to analyze the global effect of CsrA on Salmonella we obtained circumstantial evidence that CsrA could negatively regulate the expression of hilE. To further investigate this phenomenon, we analyzed the chromosomal expression of FLAG-tagged HilE (HilE-FLAG) in the wild-type (WT) S. Typhimurium SL1344 strain carrying the pK3-CsrA plasmid expressing CsrA, when grown in conditions that favor the expression of SPI-1 genes (LB medium, at 37°C, with shaking; SPI-1-inducing conditions). Expression of CsrA from pK3-CsrA completely inhibited the amount of HilE-FLAG (Fig 1A). Similar results were obtained by assessing the expression of a hilE-lacZ translational fusion (Fig 1B). We were unable to evaluate the expression of HilE-FLAG and hilE-lacZ in the absence of CsrA because csrA mutants of S. Typhimurium exhibit severe growth defects [39,48]. These results indicate that CsrA negatively controls the expression of hilE.
Fig 1. CsrA represses expression of hilE.
(A) HilE-FLAG levels in the WT S. Typhimurium strain carrying a chromosomal FLAG-tagged hilE gene in the absence or presence of pMPM-K3 vector or the pK3-CsrA plasmid, which expresses CsrA from a constitutive promoter, were analyzed by Western blotting using monoclonal anti-FLAG antibodies. As a control for protein loading, the expression of GroEL was also determined using polyclonal anti-GroEL antibodies. A WT S. Typhimurium strain without the FLAG-tagged hilE gene was used as negative control (-). (B) β-galactosidase activity of the translational hilE-lacZ fusion contained in the philE-lacZ plasmid was determined in the WT S. Typhimurium strain in the absence or presence of pMPM-K3 vector or the pK3-CsrA plasmid. Data represent the average of three independent experiments done in duplicate. Error bars denote the standard deviations. Statistically different values are indicated (***, p-value < 0.0001). Western blot and β-galactosidase assays were performed with samples taken from bacterial cultures grown in LB at 37°C.
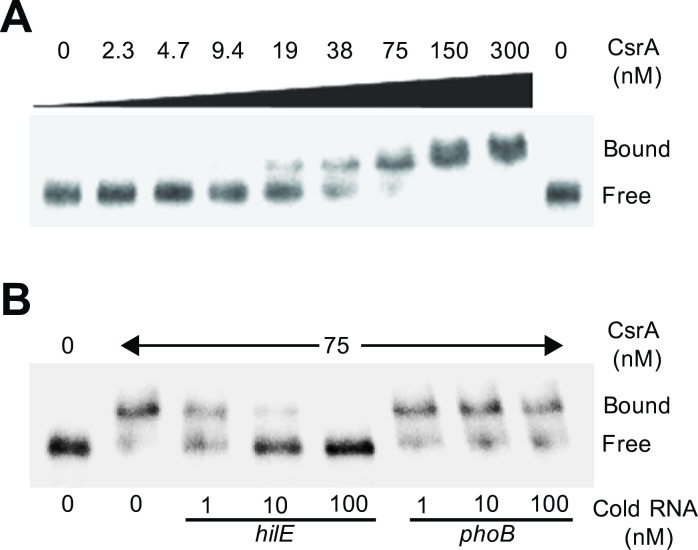
To determine whether CsrA regulates hilE directly, quantitative electrophoretic mobility shift assays (EMSAs) were performed using purified CsrA and the 5’-end-labelled leader RNA of hilE. A band with lower mobility was detected with concentrations of CsrA between 19 and 75 nM, indicating that CsrA formed a complex with the hilE transcript; at 150 and 300 nM CsrA a second complex with even lower mobility was also observed (Fig 2A). These data support that CsrA binds two sites on the hilE transcript. Nonlinear least-squares analysis of these EMSAs data yielded an apparent Kd value of 37 ± 13 nM CsrA for hilE mRNA. The specificity of the CsrA-hilE RNA interaction was evaluated by performing competition experiments with specific (hilE) and non-specific (phoB) unlabeled RNA competitors. Whereas unlabeled hilE RNA was an effective competitor, phoB RNA was not (Fig 2B). In agreement with these results, data from a previous global analysis by CLIP-seq showed that CsrA binds in vivo to a sequence located near the translation start codon of the hilE mRNA [49]. Thus, we conclude that CsrA binds specifically to the hilE leader transcript. Together, our results indicate that CsrA directly represses the expression of hilE.
Fig 2. CsrA specifically interacts with the leader region of the hilE transcript.
(A) CsrA binding to the hilE RNA was analyzed using EMSAs by incubating labeled hilE RNA (0.2 nM) with increasing concentrations of purified CsrA (0, 2.3, 4.7, 9.4, 19, 38, 75, 150 and 300 nM). Positions of bound and free RNA are marked. These experiments were performed three times and a representative gel is shown. (B) RNA competition experiment where labeled hilE RNA (0.2 nM) was combined with 1, 10 and 100 nM of unlabeled specific (hilE) or non-specific (phoB) competitor RNA and incubated with 0 and 75 nM of purified CsrA. Positions of bound and free RNA are marked. A representative gel from two independent assays is shown.
SirA/BarA induces the expression of hilE through CsrB/C
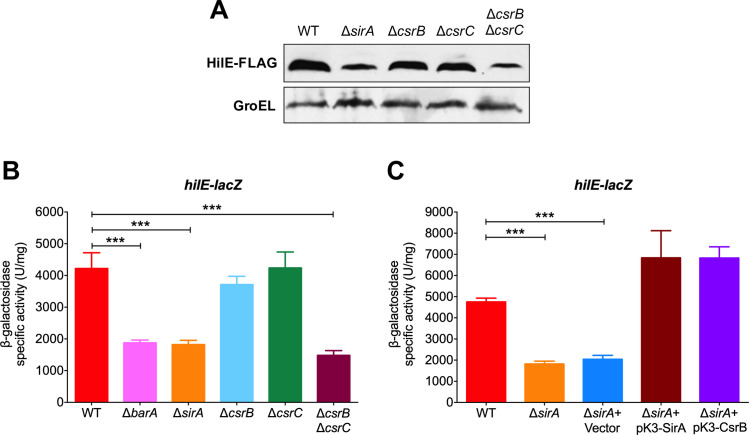
CsrA-mediated repression of target genes is counteracted by the two-component system SirA (UvrY)/BarA through the sRNAs CsrB and CsrC, which bind and sequester CsrA [44,45]. To define the complete regulatory cascade involving CsrA that controls the expression of hilE, the expression of HilE-FLAG and that of the lacZ-hilE fusion was monitored in the WT S. Typhimurium strain and its ΔsirA, ΔcsrB, ΔcsrC and ΔcsrB ΔcsrC derivative mutants, grown in SPI-1-inducing conditions. Both the amount of HilE-FLAG and the expression of hilE-lacZ were reduced in the ΔsirA mutant, as well as in the ΔcsrB ΔcsrC double mutant, compared with the WT strain (Fig 3A and 3B). Additionally, as shown in Fig 3B, activity of the hilE-lacZ fusion was reduced in the ΔbarA mutant lacking BarA, the cognate sensor kinase of the SirA response regulator [41,42]. In contrast, the ΔcsrB and ΔcsrC single mutants showed WT levels of HilE-FLAG and hilE-lacZ expression (Fig 3A and 3B), which is consistent with previous reports indicating that in S. Typhimurium, the absence of both CsrB and CsrC is required for observable effects on the expression of other target genes of the SirA/BarA-Csr cascade [39,50]. As expected, complementation of the ΔsirA mutant with the pK3-SirA plasmid expressing SirA, restored the activity of the hilE-lacZ fusion to levels even slightly higher than those of the WT strain (Fig 3C). Moreover, complementation of the ΔsirA mutant with the pK3-CsrB plasmid expressing CsrB, also restored the activity of the hilE-lacZ fusion (Fig 3C). Collectively, these results indicate that the SirA/BarA two-component system induces the expression of hilE through the sRNAs CsrB and CsrC, which counteract repression of this gene by CsrA.
Fig 3. SirA/BarA and CsrB/CsrC positively regulate the expression of hilE.
(A) Western blot analysis of HilE-FLAG expression in the WT S. Typhimurium strain and its derivative ΔsirA, ΔcsrB, ΔcsrC and ΔcsrB ΔcsrC mutants carrying a chromosomal FLAG-tagged hilE gene, using monoclonal anti-FLAG antibodies. As a protein loading control, the expression of GroEL was also determined using polyclonal anti-GroEL antibodies. (B) β-galactosidase activity of the translational hilE-lacZ fusion contained in the philE-lacZ plasmid was determined in the WT S. Typhimurium strain and its derivative ΔbarA, ΔsirA, ΔcsrB, ΔcsrC and ΔcsrB ΔcsrC mutants. (C) β-galactosidase activity of the translational hilE-lacZ fusion contained in the philE-lacZ plasmid was determined in the WT S. Typhimurium strain and its derivative ΔsirA mutant in the absence or presence of pMPM-K3 vector, pK3-SirA or pK3-CsrB plasmids, which express SirA and CsrB, respectively, from a constitutive promoter. Data represent the average of three independent experiments done in duplicate. Error bars symbolize the standard deviations. Statistically different values are indicated (***, p-value < 0.0001). Western blot and β-galactosidase assays were performed with samples taken from bacterial cultures grown in LB at 37°C.
HilE represses HilD-mediated expression of SPI-1, SPI-2 and SPI-5 genes
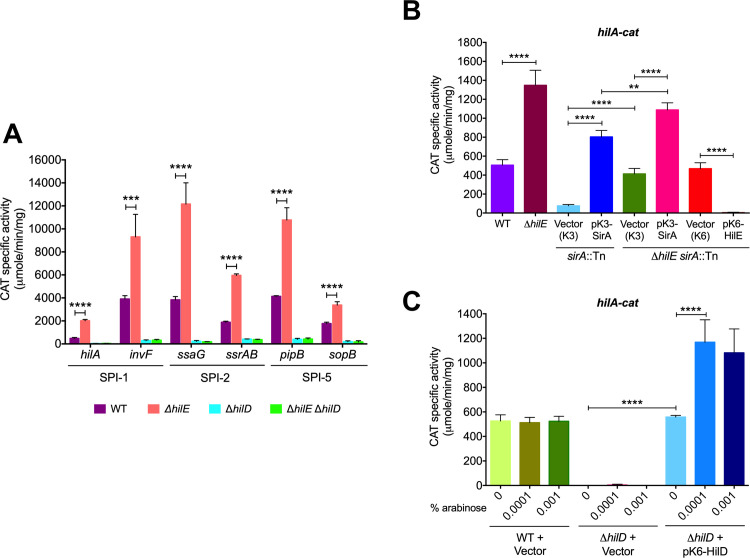
HilE negatively affects the activity of the transcriptional regulator HilD [36–38]. Consequently, HilE would decrease the expression of the large number of genes controlled by HilD. To further investigate this repressor role of HilE, we quantified the expression of two genes each from SPI-1 (hilA and invF), SPI-2 (ssrAB and ssaG) and SPI-5 (sopB and pipB), as cat transcriptional fusions. HilD positively regulates the expression of all of these genes directly or indirectly [15,23,27,33,51–53]. Experiments were performed in the WT S. Typhimurium strain and its ΔhilE derivative mutant grown in SPI-1-inducing conditions. As shown in Fig 4A, the expression of every gene increased significantly in the ΔhilE mutant compared with the WT strain. As expected, due to the absence of HilD, only low expression level was observed for these genes in the ΔhilD and ΔhilE ΔhilD mutants (Fig 4A). Consistent with our results, previous studies also indicate that the expression of hilA increases in the absence of HilE [36,54]. Additionally, we examined and compared the expression level of the hilA-cat fusion in the WT S. Typhimurium strain and its ΔhilE, sirA::Tn10d (transposon insertion in sirA) and ΔhilE sirA::Tn10d derivative mutants in the absence or presence of the pK3-SirA plasmid expressing SirA, the pMPM-K3/K6 vector, or the pK6-HilE plasmid expressing HilE. In agreement with previous studies indicating that SirA positively regulates SPI-1 genes [39,40,42,55], the hilA-cat fusion showed reduced expression levels in the sirA::Tn10d mutant containing the pMPM-K3 vector, with respect to the WT strain (Fig 4B). Surprisingly, the hilA-cat fusion was expressed in the ΔhilE sirA::Tn10d double mutant containing the pMPM-K3 vector at a level only slightly lower than in the WT strain (Fig 4B), revealing than in the absence of HilE, hilA is expressed independently of SirA. It is important to consider that the HilE expression is positively controlled by SirA (this study) but also by other regulators [56–59] as discussed later. In fact, our results show that a partial amount of HilE is present in the absence of SirA (Fig 3). Thus, our results suggest that in the absence of SirA the expression of hilA is repressed by two mechanisms: 1) CsrA inhibits translation of the hilD mRNA and 2) the diminished amount of HilE is able to inactivate background levels of HilD, which would avoid the positive autoregulation of HilD. It is reasonable to suggest that when SirA counteracts the CsrA-mediated repression of hilD, higher levels of HilE would be required to negatively control the activity of HilD, which can be reached with the positive regulation of the hilE expression by SirA.
Fig 4. HilE represses the expression of genes regulated by HilD.
(A) CAT activity of the hilA-cat, invF-cat, ssaG-cat, ssrAB-cat, pipB-cat and sopB-cat transcriptional fusions, contained in plasmids philA-cat, pinvF-cat, pssaG-cat, pssrAB-cat, ppipB-cat and psopB-cat, respectively, was determined in the WT S. Typhimurium strain and its derivative ΔhilE, ΔhilD and ΔhilE ΔhilD mutants. (B) CAT activity of the hilA-cat transcriptional fusion, contained in the philA-cat plasmid, was determined in the WT S. Typhimurium strain and its derivative ΔhilE, sirA::Tn10d and ΔhilE sirA::Tn10d mutants in the absence or presence of pMPM-K3 or pMPM-K6 vectors, the pK3-SirA plasmid expressing SirA from a constitutive promoter or the pK6-HilE plasmid expressing HilE from an arabinose-inducible promoter. Expression of HilE from pK6-HilE was induced by adding 0.001% L-arabinose to the medium. (C) CAT activity of the hilA-cat transcriptional fusion, contained in the philA-cat plasmid, was determined in the WT S. Typhimurium strain and its derivative ΔhilD mutant carrying the pMPM-K6 vector or the pK6-HilD plasmid, which expresses HilD from an arabinose-inducible promoter. Expression of HilD from pK6-HilD was induced with 0%, 0.0001% or 0.001% L-arabinose. CAT-specific activity was obtained from samples collected from bacterial cultures grown in LB at 37°C. Data represent the average of three independent experiments done in duplicate. Error bars indicate the standard deviations. Statistically different values are indicated (**, p-value < 0.01; ***, p-value < 0.001; ****, p-value < 0.0001).
The presence of the pK3-SirA plasmid further increased the expression of hilA-cat in the ΔhilE sirA::Tn10d mutant and, as could be expected, the presence of the pK6-HilE plasmid completely inhibited the expression of hilA-cat in the ΔhilE sirA::Tn10d mutant (Fig 4B). To note, the hilA-cat fusion showed similar expression levels in the ΔhilE and ΔhilE sirA::Tn10d + pK3-SirA mutants, as well as in the ΔhilD mutant complemented with the pK6-HilD plasmid overexpressing HilD (Fig 4B and 4C), which could suggest that in the absence of HilE (or overexpression of HilD) maximal expression levels of hilA are reached.
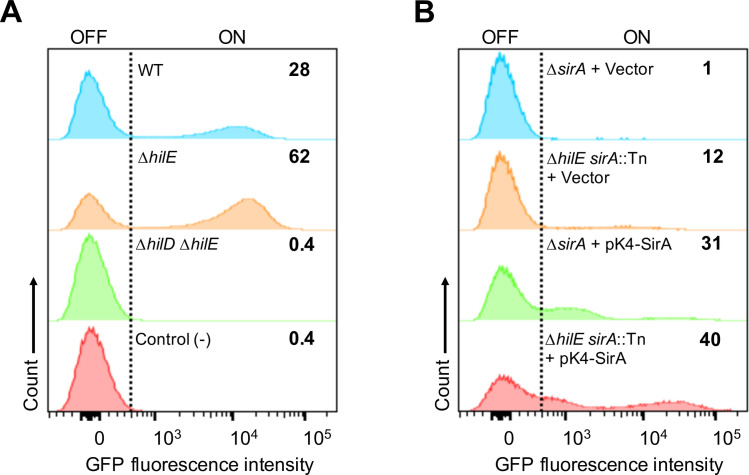
We next aimed to analyze the effect of HilE on the fraction of cells expressing SPI-1 genes in the Salmonella population. To do this, GFP fluorescence expression from an invF-gfp transcriptional fusion was quantified by flow cytometry in cultures of the WT S. Typhimurium strain and its ΔhilE and ΔhilE ΔhilD derivative mutants, as well as in cultures of the sirA::Tn10d and ΔhilE sirA::Tn10d mutants carrying the pK4-SirA plasmid expressing SirA or the pMPM-K4Ω vector, grown in SPI-1-inducing conditions. Similar to other reports [17–21], only 28% of the cells from cultures of the WT strain expressed invF-gfp, whereas 62% of the cells expressed this fusion in cultures of the ΔhilE mutant (Fig 5A). As expected, expression of invF-gfp in the ΔhilE ΔhilD mutant or the WT strain carrying the gfp reporter gene without a promoter showed nearly undetectable expression of GFP fluorescence (Fig 5A). In addition, the fraction of cells expressing the invF-gfp fusion was higher in the ΔhilE sirA::Tn10d double mutant than in the sirA::Tn10d single mutant, both in the presence of the vector pMPM-K4Ω or the pK4-SirA plasmid (Fig 5B). Intriguingly, complementation of the sirA::Tn10d and ΔhilE sirA::Tn10d mutants yielded a subpopulation of cells with an invF-gfp expression pattern somewhat different to that of cells from the WT strain and the ΔhilE mutant (Fig 5A and 5B), which seems to be an effect of the expression of SirA from a multicopy plasmid. Nevertheless, these results clearly show that HilE reduces the fraction of cells expressing SPI-1 genes in the Salmonella population.
Fig 5. HilE negatively controls the bistable expression of SPI-1.
Flow cytometry analysis of GFP fluorescence expression of the invF-gfp transcriptional fusion contained in the low copy pMPMA3ΔPlac PinvF-gfp[LVA]/R plasmid in different S. Typhimurium strains. (A) GFP expression in cultures of the WT strain and its derivative ΔhilE and ΔhilE ΔhilD mutants carrying the invF-gfp fusion, as well as in cultures of the WT strain carrying the gfp reporter gene without a promoter (negative control). (B) GFP expression in cultures of the sirA::Tn10d and ΔhilE sirA::Tn10d mutants carrying the invF-gfp fusion and the pMPM-K4Ω vector or the pK4-SirA plasmid expressing SirA. The percentage of the GFP+ population of each strain is indicated on the graphs. Data shown are representative of two independent experiments performed with ten independent bacterial colonies.
Collectively, these results reinforce that HilE plays a major role in regulating the SPI-1 genes by acting as a repressor of HilD. It is important to note that the presence of HilE only decreases, but does not eliminate, the HilD-mediated activation of target genes, or the fraction of cells from the Salmonella population that expresses these genes, in SPI-1-inducing conditions.
HilE reduces the deleterious effect on growth caused by expression of genes controlled by HilD
Interestingly, SirA/BarA induces the expression of both HilD [39] and HilE (this study), which would seem counterintuitive. What would be the role of the regulation of hilE by SirA/BarA?
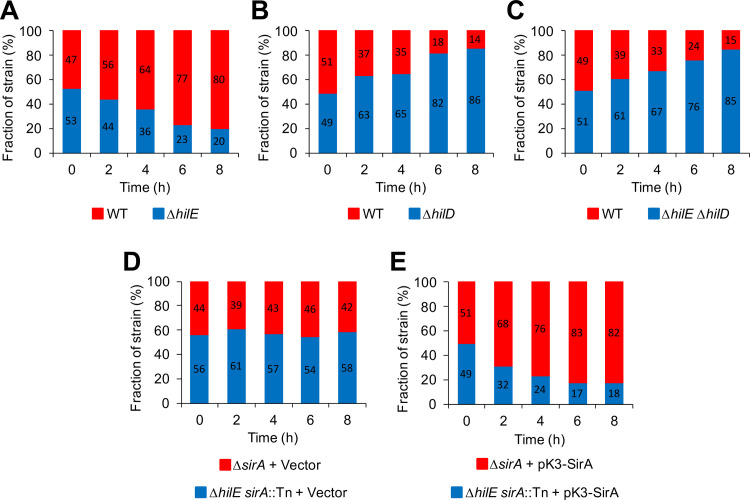
Overexpression of HilD or the absence of HilE retard growth of S. Typhimurium in competitive assays performed in LB, which was partially attributable to the overexpression of some genes regulated by HilD [19]. To further investigate this phenomenon, we monitored the growth of the WT S. Typhimurium strain and the ΔhilE, ΔhilD and ΔhilE ΔhilD mutants, in mixed cultures grown in SPI-1-inducing conditions. In the WT/ΔhilE mixed cultures, the WT strain outcompeted the ΔhilE mutant, representing 80% of the viable cells at the end of these assays (Fig 6A). In contrast, in the WT/ΔhilD mixed cultures, the WT strain was outcompeted by the ΔhilD mutant; only 14% of the viable cells at the end of these assays were WT (Fig 6B). These results are in agreement with those obtained previously, showing that the absence of HilE and HilD has negative and positive effects, respectively, on the growth of S. Typhimurium cultures [19]. Furthermore, we observed that the WT strain was also outcompeted by the ΔhilE ΔhilD mutant to a similar extent as the ΔhilD single mutant (Fig 6B and 6C), suggesting that the negative effect caused by the absence of HilE on the S. Typhimurium growth requires the presence of HilD.
Fig 6. Detrimental effect on bacterial growth by the expression of genes controlled by HilD.
In vitro competitive growth (A) between WT S. Typhimurium strain and isogenic ΔhilE mutant, (B) between WT S. Typhimurium strain and isogenic ΔhilD mutant, (C) between WT S. Typhimurium strain and isogenic ΔhilE ΔhilD mutant, (D) between the ΔsirA and ΔhilE sirA::Tn10d mutants carrying the pMPM-K3 vector, and (E) between the ΔsirA and ΔhilE sirA::Tn10d mutants carrying the pK3-SirA plasmid expressing SirA, was determined by counting CFUs for each strain from samples taken at 0, 2, 4, 6 and 8 h of bacterial mixed cultures grown in LB at 37°C with shaking. CFUs are represented as fraction (percentage) of each strain in the respective mixed culture. Data represent the average of three independent experiments.
Additionally, we analyzed the competitive growth between the ΔsirA and ΔhilE sirA::Tn10d mutants carrying the pMPM-K3 vector or the pK3-SirA plasmid expressing SirA. Interestingly, in the presence of the pMPM-K3 vector, the ΔsirA and ΔhilE sirA::Tn10d mutants grew similarly through the time assessed (Fig 6D). In contrast, in the presence of the pK3-SirA plasmid, the ΔhilE sirA::Tn10d mutant was outcompeted by the ΔsirA mutant; only 18% of the viable cells at the end of these assays were from the ΔhilE sirA::Tn10d mutant strain (Fig 6E). These results reveal that SirA is required for HilE to play a role in the fitness of S. Typhimurium.
Together, these results show that HilE reduces the deleterious effect on the S. Typhimurium growth caused by the expression of genes regulated by HilD in laboratory conditions.
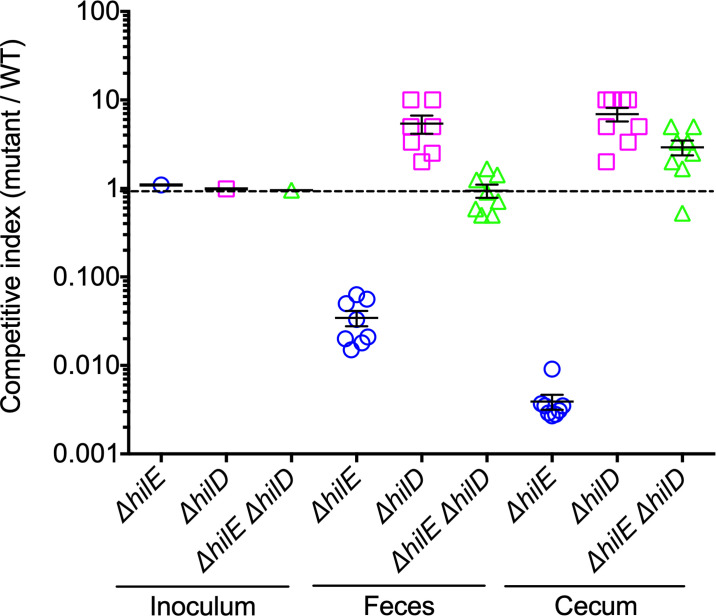
SPI-1-inducing conditions are considered to somehow mimic the intestinal environment found by Salmonella in hosts. Therefore, on the basis of our results described above, we investigated whether HilE is important for successful intestinal colonization by S. Typhimurium. For this analysis, we determined and compared the survival of the WT S. Typhimurium strain to that of the ΔhilE, ΔhilD and ΔhilE ΔhilD mutants, in the intestine of streptomycin-pretreated mice, a model used to study the intestinal colonization by S. Typhimurium [60]. Groups of eight mice were orally infected with a mix of an equal number of cells of the WT strain and each mutant strain. Three days post-infection, bacteria from feces and cecum of mice were counted, and a competitive index was obtained to determine the proportion between each mutant and the WT strain. The ΔhilE mutant showed a ~30-fold and ~250-fold reduction in survival with respect to the WT strain, in feces and cecum, respectively. In contrast, the survival of the ΔhilD and ΔhilE ΔhilD mutants was similar or slightly higher than that of the WT strain in both feces and cecum (Fig 7). Thus, the absence of HilE attenuates the intestinal colonization by S. Typhimurium in a HilD-dependent way. These results are in agreement with those from a previous study showing that HilE is important for the intestinal disease caused by long term infections of S. Typhimurium in mice (i.e. at 10 days post-infection), including colonization and induction of enteritis [20]. Furthermore, our results are also consistent with reports indicating that SPI-1 or HilD function is required to invade cells from the intestinal epithelium, and thus to induce enteritis, but not to colonize the cecum lumen [2,20,60–62]. Replication of Salmonella in the intestinal lumen has been shown to require genes for the use of specific nutrients present in the inflamed intestine, such as the eut, ttr and pdu genes [63–65], which, according to previous transcriptomic analyses, are not regulated by HilD [66]. These data reveal a crucial role for HilE during intestinal infection by S. Typhimurium.
Fig 7. HilE is required for the intestinal colonization of mice by S. Typhimurium.
Groups of eight streptomycin-pretreated mice were orally inoculated with a mixed bacterial suspension containing an equal amount of CFUs (0.5 x 106) of the WT S. Typhimurium strain and the respective mutant (WT + ΔhilE, WT + ΔhilD or WT + ΔhilE ΔhilD). At 3 days post-infection, CFUs from feces and cecum were counted for each strain and a Competitive Index (CI; CFUs mutant / CFUs WT) was obtained. A CI was also determined for the respective inoculum. Data for each mouse or inoculum were log-transformed and plotted. CIs are indicated as follows: WT/ΔhilE, blue circles; WT/ΔhilD, pink squares; and WT/ΔhilE ΔhilD, green triangles. Bars denote the standard error of the mean for each experimental group. CI of 1.0, represented by a horizontal dotted line, indicates that the two strains (WT and mutant) were present in equivalent numbers.
In all, together with previous reports, our data suggest that by inducing the expression of HilD and its negative regulator HilE, SirA/BarA exerts a fine-tuning regulatory control that cooperates to lessen the growth penalty produced by HilD-mediated expression of several tens of virulence factors, which is required for the successful intestinal colonization by S. Typhimiurium.
Discussion
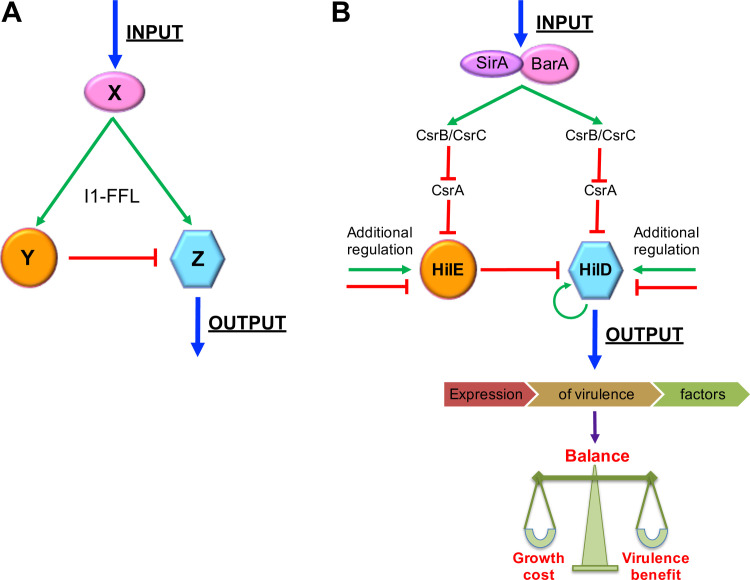
Regulatory networks are built up by recurring patterns of interactions between regulatory factors that are designed as network motifs, which are present in diverse organisms including bacteria, yeast, plants, animals, and humans [67,68]. One family of network motifs is the feedforward loop (FFL), where a factor X regulates factors Y and Z, and Y regulates Z. As the three regulatory interactions between X, Y, and Z can be either positive or negative, eight structural types of FFLs are possible [68,69]. One FFL commonly present in the best-studied transcriptional networks, those from E. coli and yeast, is the incoherent type-1 FFL (I1-FFL), where a factor X positively regulates factors Y and Z, while Y negatively regulates Z. Thus, X exerts an “incoherent” (or “paradoxical”) regulation on Z through two opposite effects [69,70] (Fig 8A), which represents a complex biological problem to decipher.
Fig 8. Structure and role of I1-FFLSirA/BarA-HilE-HilD in Salmonella virulence.
(A) Structure of the incoherent type-1 feedforward loop (I1-FFL) network motif, where the factor X activates factors Z and Y, and Y represses Z; thus, X exerts opposite controls on Z. (B) Structure and function of I1-FFLSirA/BarA-HilE-HilD. Molecules present in the mammalian intestines, such as short-chain fatty acids, have been shown to activate the SirA/BarA two-component system. SirA/BarA induces expression of both HilD and HilE, through the sRNAs CsrB and CsrC, which bind to CsrA and thus counteract CsrA-mediated repression on the hilD and hilE transcripts. HilE inhibits activity of HilD by protein-protein interaction. Therefore, SirA/BarA simultaneously exerts positive and negative control on HilD, which cooperates for the appropriate balance between the growth cost and the virulence benefit generated by the expression of tens of virulence genes regulated by HilD. Additional positive and negative regulation on HilD and HilE also would control the expression level of the genes regulated by HilD. See text for more details. Green arrows and red blunt-end lines indicate positive and negative control, respectively.
Our results from this study reveal a novel I1-FFL involved in gene expression in bacteria. We previously determined that in S. Typhimurium, the SirA/BarA two-component system induces the expression of the transcriptional regulator HilD, through the sRNAs CsrB and CsrC, which counteracts the CsrA-mediated repression of hilD translation [39]. HilD induces the expression of a large number of Salmonella virulence genes [2,15,26,29,31,32,66]. In this work, we show that the SirA/BarA/Csr system also regulates expression of HilE by a similar way. HilE negatively controls HilD activity by protein-protein interaction [36–38]. It is important to note that HilD is the only known target of HilE. Hence, the SirA/BarA-CsrB/C cascade exerts two opposite regulatory effects on HilD; the activation of the hilD mRNA translation, and the inhibition of HilD activity through HilE, thus constituting an I1-FFL (I1-FFLSirA/BarA-HilE-HilD) that involves regulation at the transcriptional, translational and post-translational levels (Fig 8B).
I1-FFLs can influence fold-change detection [71], adaptive tuning [72], response time [69,70] and response amplitude of gene expression [73]. Our results and those from a previous study [19] show that in the absence of HilE, the repressor factor of I1-FFLSirA/BarA-HilE-HilD (Fig 8B), the growth of S. Typhimurium is slowed down in laboratory conditions, which can be attributed to the overexpression of genes regulated by HilD that has a deleterious effect by a still unknown mechanism. We found that the absence of HilE negatively affects the S. Typhimurium growth only in the presence of SirA and HilD, the two other components of I1-FFLSirA/BarA-HilE-HilD. Additionally, our results and a prior report [20] indicate that the SPI-1 genes are expressed in vitro in a bistable fashion that involves a control by HilE. In the presence of HilE only about one third fraction of the S. Typhimurium population expresses SPI-1, while in the absence of HilE the fraction of cells expressing SPI-1 increases more than twofold. Moreover, our results and those reported previously [20] indicate that the absence of HilE attenuates the intestinal colonization by S. Typhimurium, an effect also mediated through HilD. Interestingly, in the intestine of mice, the SPI-1 genes are also expressed in a bistable fashion, and in the absence of HilE, the proportion of S. Typhimurium cells expressing SPI-1 increases, imposing a growth pressure that leads to the selection of avirulent hilD mutants [20]. Furthermore, the lack of HilE increases the invasion of S. Typhimurium into epithelial culture cells by 2-5-fold [36,54]. Altogether, these findings suggest that I1-FFLSirA/BarA-HilE-HilD is involved, together with other regulatory pathways, in the control of the precise amount and activity of HilD, resulting in an appropriate balance between the growth cost and the virulence benefit generated by the expression of the genes induced by this regulator (Fig 8B).
The SirA/BarA system seems to be directly activated by short-chain fatty acids such as acetate and formate, which are known to induce the expression of SPI-1 genes [8,74–76]. Therefore, these molecules commonly present in the mammalian intestines, probably in conjunction with other environmental cues, are expected to turn on I1-FFLSirA/BarA-HilE-HilD. However, several other positive and negative regulatory mechanisms acting on HilD or HilE provide additional inputs to I1-FFLSirA/BarA-HilE-HilD for fine tuning the expression level of the genes regulated by HilD (Fig 8B). Expression of HilD is controlled positively by transcriptional autoregulation, directly or through the feedforward loop that HilD forms with HilC and RtsA [32–34,77,78]. HilD is also controlled by Hfq, ArcA, LoiA, Fis and Hu at the transcriptional initiation level [79–82], by Gre factors acting during transcriptional elongation [83], as well as by Fur and FliZ acting at the transcriptional/post-translational and post-translational levels, respectively [84,85]. Whereas the expression or amount of HilD is controlled negatively by H-NS, IscR, SsrB/SsrA and PhoP/PhoQ at the transcriptional level [79,86–89], by StdE/StdF at the post-transcriptional level [90], by FnrS and ArcZ at the translational level [13], by CRP-cAMP at the post-translational level [91], as well as by the Lon protease and CpxR/CpxA that decrease the stability of HilD [78,92]. Additionally, controversial reports indicate that the acetyltransferase enzyme Pat decreases or increases the stability and controls the DNA-binding activity of HilD [93–95]. On another hand, expression of HilE is controlled positively by FimYZ, PhoP/PhoQ, PhoB/PhoR and LeuO at the transcriptional level [56–59], whereas it is controlled negatively by MIc and IsrM at the transcriptional and translational levels, respectively [96]. Thus, the three main components of I1-FFLSirA/BarA-HilE-HilD could connect in a dynamic way to this regulatory motif with the rest of the regulatory network controlling physiology and virulence in Salmonella.
Only a few I1-FFLs that control gene expression have been well-defined experimentally in bacteria. One of the best characterized is that involved in galactose metabolism in E. coli, where CRP-cAMP controls the expression of the gal and mgl genes by two opposite pathways, via direct positive regulation and negative regulation through GalS [70,73]. Additionally, in Pseudomonas aeruginosa, a quorum sensing system controlling virulence-related phenotypes is constituted by an I1-FFL, where LasR activates the expression of LasI, as well as that of RsaL, which in turn represses expression of LasI [97]. Furthermore, also in P. aeruginosa, the alternative sigma factor σ22 activates the expression of the virulence-associated enzyme AlgC, as well as that of the sRNA ErsA that represses translation of algC [98]. In addition, in Rhodobacter sphaeroides, two I1-FFLs are involved in the expression of photosynthesis-related genes. In one case, MppG exerts opposite transcriptional regulation on these genes, direct activation and indirect repression through AppA and PpsR, respectively [99], whereas in the other case, PrrA directly activates transcription of these genes but also represses their translation through the sRNA PcrZ [100,101]. Finally, in S. Typhimurium, the PhoP/PhoQ two-component system activates the expression of mgtC, encoding a virulence-associated protein, and that of the sRNA AmgR, while AmgR destabilizes mgtC mRNA [102], which also constitutes an I1-FFL.
Our study reveals a novel function for an I1-FFL, that is, fine tuning regulation to reduce the growth cost imposed by simultaneous expression of a high number of virulence genes. Our results and those from a previous study [20] provide evidence indicating that I1-FFLSirA/BarA-HilE-HilD is necessary for intestinal colonization by Salmonella. Moreover, our findings illustrate the integration of ancestral (e.g. SirA/BarA) and acquired regulators (e.g. HilD and HilE) into a specific regulatory motif, which can lead to the expansion of regulatory networks during evolution.
Materials and methods
Ethics statement
Animal experiments were conducted according to the standard operating protocols approved by the International Committee for Animal Care and Use from CICUAL-UNAM and by the Official Mexican Norm NOM-062-Z00-1999.
Bacterial strains, media and culture conditions
Bacterial strains used in this work are listed in Table 1. Bacterial cultures for β-galactosidase, chloramphenicol acetyltransferase and Western blot assays were grown in 250-ml flasks containing 50 ml of lysogeny broth (LB)-Miller, which were incubated at 37°C with shaking up to reach an O.D.600nm of 1.4 (5–8 h). Bacterial cultures for flow cytometry assays were grown in 16 x 150 mm glass tubes containing 5 ml of LB-Miller, which were incubated at 37°C with shaking up to reach an O.D.600nm of 1.2 (12–18 h). When necessary, culture medium was supplemented with ampicillin (200 μg/ml), streptomycin (100 μg/ml) or kanamycin (30 μg/ml).
Table 1. Bacterial strains and plasmids used in this study.
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| Strain | ||
| Salmonella Typhimurium | ||
| SL1344 | Wild type; xyl, hisG, rpsL; SmR | [114] |
| 14028s | Wild type | ATCC |
| JPTM5 | ΔhilD::kan | [15] |
| JPTM23 | ΔsirA::kan | [39] |
| JPTM27 | ΔsirA | [39] |
| JPTM39 | ΔbarA::kan | [39] |
| JPTM40 | ΔcsrB::kan | [39] |
| JPTM41 | ΔcsrC::kan | [39] |
| JPTM42 | ΔcsrB | [39] |
| JPTM43 | ΔcsrB ΔcsrC::kan | [39] |
| DTM56 | 14028s ΔhilE::kan | [78] |
| DTM133 | ΔcsrC | This study |
| DTM134 | ΔcsrB ΔcsrC | This study |
| DTM135 | ΔhilE::kan | This study |
| DTM136 | ΔhilE | This study |
| DTM137 | ΔhilE ΔhilD::kan | This study |
| DTM138 | 14028s hilE::3XFLAG-kan | This study |
| DTM139 | 14028s hilE::3XFLAG | This study |
| CJ035 | sirA::Tn10d | [40] |
| DTM140 | sirA::Tn10d | This study |
| DTM141 | ΔhilE sirA::Tn10d | This study |
| Escherichia coli | ||
| DH10β | Laboratory strain | (Invitrogen) |
| Plasmids | ||
| pKK232-8 | pBR322 ori vector containing a promoterless chloramphenicol acetyltransferase (cat) gene, ApR | [115] |
| philA-cat | pKK232-8 derivative containing a hilA-cat transcriptional fusion from nucleotides -410 to +446 | [15] |
| pinvF-cat | pKK232-8 derivative containing an invF-cat transcriptional fusion from nucleotides -306 to +213 | [15] |
| ppipB-cat | pKK232-8 derivative containing a pipB-cat transcriptional fusion from nucleotides -737 to +70 | [15] |
| psopB-cat | pKK232-8 derivative containing a sopB-cat transcriptional fusion from nucleotides -400 to +128 | [15] |
| pssaG-cat | pKK232-8 derivative containing a ssaG-cat transcriptional fusion from nucleotides -232 to +361 | [15] |
| pssrAB-cat | pKK232-8 derivative containing a ssrAB-cat transcriptional fusion from nucleotides -303 to +3054 | [15] |
| pMPMA3ΔPlac null-gfp[LVA]/R | P15A ori vector containing a promoterless gfp reporter gene encoding a destabilized variant of green fluorescent protein, GFP [LVA], ApR | [116] |
| pMPMA3ΔPlac PinvF-gfp[LVA]/R | pMPMA3ΔPlac null-gfp[LVA]/R derivative containing an invF-gfp transcriptional fusion | [116] |
| pRS414 | pBR322 ori vector for the construction of translational fusions to the lacZ reporter gene, ApR | [103] |
| philE-lacZ | pRS414 derivative containing a hilE-lacZ translational fusion from nucleotides -424 to +49 | This study |
| pMPM-K3 | P15A ori cloning vector, lac promoter, KanR | [104] |
| pK3-CsrA | pMPM-K3 derivative expressing CsrA from the lac promoter | [39] |
| pK3-CsrB | pMPM-K3 derivative expressing CsrB from the lac promoter | [39] |
| pK3-SirA | pMPM-K3 derivative expressing SirA from the lac promoter | [39] |
| pMPM-K4Ω | ColE1 ori vector, KanR | [104] |
| pK4-SirA | pMPM-K4Ω derivative expressing sirA from its own promoter | This study |
| pMPM-K6Ω | p15A ori vector, ara promoter, KanR | [104] |
| pK6-HilD | pMPM-K6Ω derivative expressing HilD from the ara promoter, KanR | [31] |
| pK6-HilE | pMPM-K6Ω derivative expressing HilE from the ara promoter, KanR | [38] |
| pCP20 | Plasmid expressing FLP recombinase from a temperature-inducible promoter, ApR | [106] |
| pSUB11 | pGP704 derivative template plasmid for FLAG epitope tagging | [105] |
The coordinates of the nucleotides for the cat and lacZ fusions are relative to the transcriptional start site reported for each gene. ApR, ampicillin resistance; KanR, kanamycin resistance; SmR, streptomycin resistance.
Plasmids
Table 1 shows the plasmids used in this study. To construct the philE-lacZ plasmid, a DNA fragment spanning the full-length intergenic region upstream hilE and the first ten codons of this gene, was amplified by PCR with the primers HilE-EcoRI-Fw1 (5’-TTAAATGAATTCACATAGTAAATATGTTCTATTG-3’) and HilE-BamHI-Rev1 (5’-GTTTTCAAGGATCCTGATCCGGCTTTCGCCTTC-3’), containing sites for EcoRI and BamHI restriction enzymes, respectively (underlined in the sequences). The PCR product was digested with EcoRI and BamHI and ligated into the pRS414 vector [103] digested with the same enzymes. To construct the pK4-SirA plasmid, a DNA fragment spanning sirA and its upstream intergenic region was amplified by PCR with the primers SirAF (5’-GCCGGATCCATCGCCTGCAGCATCAGC-3’) and SirARgfp (5’-ACCAAGCTTGTCATACATACGATAGACACCG-3’), containing sites for BamHI and HindIII restriction enzymes, respectively (underlined in the sequences). The PCR product was digested with BamHI and HindIII and ligated into the pMPM-K4Ω vector [104] digested with the same enzymes. The constructed recombinant plasmids were characterized by PCR amplification and sequencing.
Construction of mutants and strains expressing FLAG-tagged proteins
P22 transduction was used to transfer the ΔhilE::kan allele from DTM56 into the WT S. Typhimurium SL1344 strain, generating DTM135, to transfer the ΔhilD::kan allele from JPTM5 into DTM136, generating DTM137, and to transfer the sirA::Tn10d allele from CJ035 into the WT S. Typhimurium SL1344 and DTM136 strains, generating DTM140 and DTM141, respectively. The chromosomal hilE gene was FLAG-tagged in the WT S. Typhimurium 14028s strain by a previously reported method based on the λRed recombinase system [105], using the primers HilE-FLAG-5’ (5’-AGCAAAACACGGCGGCCTCTTCACCGACAGGCGCTGTGGCGAGACTACAAAGACCATGACGG-3’) and HilE-FLAG-3’ (5’-ATACAGCATCGCCCACTGCGAGTCCGCAAGCTTGTTTTGTCCCATATGAATATCCTCCTTAG-3’), generating DTM138; the sequences corresponding to the pSUB11 template plasmid are underlined. The kanamycin resistance cassette was excised from the JPTM41, JPTM43, DTM135 and DTM138 strains, by using helper plasmid pCP20 expressing the FLP recombinase, as described previously [106], generating strains DTM133, DTM134, DTM136 and DTM139, respectively. All modified strains were verified by PCR amplification and sequencing.
Enzymatic assays
β-galactosidase and chloramphenicol acetyltransferase (CAT) assays, as well as protein quantification to calculate the respective enzymatic specific activity, were performed as described previously [107,108].
Flow cytometry analysis
Samples containing ~107 cells were taken from bacterial cultures grown as described above. After washing with 0.22-μm-pore-size filtered 1X phosphate-buffered saline (PBS), cells were fixed for 20 min at room temperature in 400 μl of 2% (wt/vol) paraformaldehyde (Sigma) in 1X PBS. Fixed cells were centrifuged and resuspended in one ml of 1X PBS. GFP fluorescence was assessed on a FACSCanto II cytometer (BD Biosciences) equipped with the FACSDiva software (BD Biosciences). Data from 100 000 events were analyzed with the FlowJo v10 software (Tree Star Inc). Cells carrying the gfp reporter gene without a promoter were used to define the background level of fluorescence. Data shown represent the frequency of GFP positive cells.
Western blotting
Preparation and visualization of immunoblots were performed as described previously [88]. Monoclonal anti-FLAG M2 (Sigma) or polyclonal anti-GroEL (StressGen) antibodies were used at 1:3,000 and 1:100,000 dilutions, respectively. Horseradish peroxidase-conjugated anti-mouse or anti-rabbit antibodies (Pierce) diluted at 1:10,000 were used as secondary antibodies.
Electrophoretic mobility shift assays
The electrophoretic mobility shift assays (EMSAs) followed published procedures [109,110]. His-tagged CsrA (CsrA-H6) from E. coli was purified as described previously [111]. Note that CsrA from E. coli and S. Typhimurium are identical. RNA was synthesized in vitro using the RNA Maxx Transcription Kit (Agilent Technologies). PCR fragments used as templates in transcription reactions contained a T7 promoter and hilE sequence extending from +153 to +280 relative to the start of transcription. Gel-purified RNA was 5′-end labeled with [γ-32P]-ATP (7,000 Ci/mmol). RNA suspended in Tris-EDTA (TE) buffer was heated to 85°C for 3 min followed by slow cooling at room temperature for 10 min. Binding reactions (10 μl) contained 10 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 100 mM KCl, 200 ng/μl yeast RNA, 0.2 mg/ml bovine serum albumin (BSA), 7.5% glycerol, 20 mM dithiothreitol (DTT), 0.2 nM RNA, CsrA-H6 (various concentrations), and 0.1 mg/ml xylene cyanol. Competition assay mixtures also contained unlabeled competitor RNA. Reaction mixtures were incubated for 30 min at 37°C to allow CsrA-RNA complex formation. Samples were then fractionated through native 10% polyacrylamide gels using 0.5X Tris-borate-EDTA (TBE) as the gel running buffer. Radioactive bands were visualized with a Typhoon 9410 phosphorimager (GE Healthcare) and quantified using ImageQuant 5.2 software. Apparent equilibrium binding constants (Kd) of CsrA-hilE RNA interaction were calculated as described previously [112].
Competitive growth experiment
The WT S. Typhimurium SL1344 strain and its isogenic mutants were grown in 5 ml of LB at 37°C with shaking, until the cultures reached an OD600nm of 0.6. Next, mixed cultures were started by inoculating an equal amount of the initial culture of the respective strains (500 μl + 500 μl) in 250-ml flasks containing 50 ml of fresh LB without antibiotics, which were incubated at 37°C with shaking for 8 h. At 0, 2, 4, 6 and 8 h, culture samples were taken to determine colony forming units (CFUs) of each strain, by plating serial dilutions in 1X PBS on LB agar supplemented with the indicated antibiotics. For the WT+ΔhilE::kan, WT+ΔhilD::kan and WT+ΔhilE ΔhilD::kan mixed cultures, CFUs of the ΔhilE::kan, ΔhilD::kan or ΔhilE ΔhilD::kan mutants (kanamycin and streptomycin resistant) were obtained directly from the count on LB agar + kanamycin/streptomycin plates, whereas CFUs of the WT strain (streptomycin resistant, kanamycin susceptible) were determined by subtracting the number of CFUs on LB agar + kanamycin/streptomycin plates from that of CFUs on LB agar + streptomycin plates. For the ΔsirA/pMPM-K3+ΔhilE sirA::Tn10d/pMPM-K3 and ΔsirA/pK3-SirA+ΔhilE sirA::Tn10d/pK3-SirA mixed cultures, CFUs of the ΔhilE sirA::Tn10d/pMPM-K3 and ΔhilE sirA::Tn10d/pK3-SirA strains (kanamycin, tetracycline and streptomycin resistant) were obtained directly from the count on LB agar + kanamycin/tetracycline/streptomycin plates, whereas CFUs of the ΔsirA/pMPM-K3 and ΔsirA/pK3-SirA strains (kanamycin and streptomycin resistant, tetracycline susceptible) were determined by subtracting the number of CFUs on LB agar + kanamycin/tetracycline/streptomycin plates from that of CFUs on LB agar + kanamycin/streptomycin plates. To compare the growth of the two strains in the respective mixed culture (competitive growth), the number of CFUs for each strain is indicated as a fraction (percentage).
Competitive index assay
Pathogen-free BALB/c female mice (6- to 7-week-old) were obtained from the Experimental Medicine Research Unit, School of Medicine, UNAM, México. Maintenance, streptomycin treatment (50 mg) and euthanasia of the mice were performed as described previously [113]. Overnight cultures of the WT S. Typhimurium SL1344 strain and its isogenic ΔhilE, ΔhilD and ΔhilE ΔhilD mutants were diluted 1:100 in 5 ml of fresh LB and incubated at 37°C with shaking for about 3 h. Then, bacterial suspensions containing 0.5 x 107 CFUs/ml of each the WT strain and the respective mutant (WT + ΔhilE, WT + ΔhilD or WT + ΔhilE ΔhilD) were prepared in 1X PBS. Next, groups of eight streptomycin-pretreated mice were infected by orogastric route with 100 μl of the corresponding bacterial suspension. At 3 days post-infection, cecum and feces were harvested aseptically and homogenized in 1 ml of sterile and cold 1X PBS, solubilized in 2% Triton X-100, serially diluted and differentially plated on LB agar plates containing streptomycin or kanamycin to determine CFUs/mg. The number of CFUs for each strain was obtained as described in Competitive growth experiment. The competitive index was calculated by dividing the total CFUs of mutant strain by the total CFUs of the WT strain.
Statistical analysis
Data analyses were performed using GraphPad Prism version 6.0c for Mac OS X, GraphPad Software, La Jolla, California USA, using One-way ANOVA followed by Tukey’s multiple comparisons posttest. P-values < 0.05 were considered statistically significant.
Acknowledgments
We thank A. Téllez-Galván for helping with competitive growth experiments; C. Paredes-Amaya for constructing the philE-lacZ plasmid; L. C. Martínez for helping during early stages of this study; F. J. Santana, M. Fernández-Mora and A. F. Flores-Alcantar for technical assistance; E. Calva for providing resources and facilities for some CAT assays; O. Steele-Mortimer and J.A. Ibarra for providing plasmids carrying gfp, and I. Martínez-Flores for critical reading of the manuscript.
Data Availability
All relevant data are within the manuscript.
Funding Statement
This work was supported by grants from Consejo Nacional de Ciencia y Tecnología (CONACYT) / México (254531) and Dirección General de Asuntos del Personal Académico de la UNAM / México (IN202418) to V.H.B., from Consejo Nacional de Ciencia y Tecnología (CONACYT) / México (256263) to M.A.D.C., and from National Institutes of Health (GM059969) to P.B. J.N.-G. was supported by a master fellowship from CONACYT (429934). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Haraga A, Ohlson MB, Miller SI. Salmonellae interplay with host cells. Nat Rev Microbiol. 2008;6(1):53–66. doi: 10.1038/nrmicro1788 . [DOI] [PubMed] [Google Scholar]
- 2.Fàbrega A, Vila J. Salmonella enterica serovar Typhimurium skills to succeed in the host: virulence and regulation. Clin Microbiol Rev. 2013;26(2):308–41. Epub 2013/04/05. doi: 10.1128/CMR.00066-12 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deng W, Marshall NC, Rowland JL, McCoy JM, Worrall LJ, Santos AS, et al. Assembly, structure, function and regulation of type III secretion systems. Nat Rev Microbiol. 2017;15(6):323–37. Epub 2017/04/11. doi: 10.1038/nrmicro.2017.20 . [DOI] [PubMed] [Google Scholar]
- 4.Ilyas B, Tsai CN, Coombes BK. Evolution of Salmonella-host cell interactions through a dynamic bacterial genome. Front Cell Infect Microbiol. 2017;7:428. Epub 2017/10/17. doi: 10.3389/fcimb.2017.00428 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dos Santos AMP, Ferrari RG, Conte-Junior CA. Virulence factors in Salmonella Typhimurium: the sagacity of a bacterium. Curr Microbiol. 2019;76(6):762–73. Epub 2018/05/23. doi: 10.1007/s00284-018-1510-4 . [DOI] [PubMed] [Google Scholar]
- 6.Knodler LA, Vallance BA, Celli J, Winfree S, Hansen B, Montero M, et al. Dissemination of invasive Salmonella via bacterial-induced extrusion of mucosal epithelia. Proc Natl Acad Sci USA. 2010;107(41):17733–8. Epub 2010/09/30. doi: 10.1073/pnas.1006098107 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laughlin RC, Knodler LA, Barhoumi R, Payne HR, Wu J, Gomez G, et al. Spatial segregation of virulence gene expression during acute enteric infection with Salmonella enterica serovar Typhimurium. MBio. 2014;5(1):e00946–13. Epub 2014/02/06. doi: 10.1128/mBio.00946-13 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lawhon SD, Maurer R, Suyemoto M, Altier C. Intestinal short-chain fatty acids alter Salmonella Typhimurium invasion gene expression and virulence through BarA/SirA. Mol Microbiol. 2002;46(5):1451–64. doi: 10.1046/j.1365-2958.2002.03268.x . [DOI] [PubMed] [Google Scholar]
- 9.Altier C. Genetic and environmental control of Salmonella invasion. J Microbiol. 2005;43 Spec No:85–92. . [PubMed] [Google Scholar]
- 10.Hung CC, Garner CD, Slauch JM, Dwyer ZW, Lawhon SD, Frye JG, et al. The intestinal fatty acid propionate inhibits Salmonella invasion through the post-translational control of HilD. Mol Microbiol. 2013;87(5):1045–60. Epub 2013/01/08. doi: 10.1111/mmi.12149 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Golubeva YA, Ellermeier JR, Cott Chubiz JE, Slauch JM. Intestinal long-chain fatty acids act as a direct signal to modulate expression of the Salmonella pathogenicity island 1 type III secretion system. MBio. 2016;7(1):e02170–15. doi: 10.1128/mBio.02170-15 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eade CR, Hung CC, Bullard B, Gonzlez-Escobedo G, Gunn JS, Altier C. Bile acids function synergistically to repress invasion gene expression in Salmonella by destabilizing the invasion regulator HilD. Infect Immun. 2016. doi: 10.1128/IAI.00177-16 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim K, Golubeva YA, Vanderpool CK, Slauch JM. Oxygen-dependent regulation of SPI1 type three secretion system by small RNAs in Salmonella enterica serovar Typhimurium. Mol Microbiol. 2019;111(3):570–87. Epub 2018/11/30. doi: 10.1111/mmi.14174 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lundberg U, Vinatzer U, Berdnik D, von Gabain A, Baccarini M. Growth phase-regulated induction of Salmonella-induced macrophage apoptosis correlates with transient expression of SPI-1 genes. J Bacteriol. 1999;181(11):3433–7. doi: 10.1128/JB.181.11.3433-3437.1999 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bustamante VH, Martínez LC, Santana FJ, Knodler LA, Steele-Mortimer O, Puente JL. HilD-mediated transcriptional cross-talk between SPI-1 and SPI-2. Proc Natl Acad Sci USA. 2008;105(38):14591–6. doi: 10.1073/pnas.0801205105 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sridhar S, Steele-Mortimer O. Inherent variability of growth media impacts the ability of Salmonella Typhimurium to interact with host cells. PLoS One. 2016;11(6):e0157043. Epub 2016/06/10. doi: 10.1371/journal.pone.0157043 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hautefort I, Proenca MJ, Hinton JC. Single-copy green fluorescent protein gene fusions allow accurate measurement of Salmonella gene expression in vitro and during infection of mammalian cells. Appl Environ Microbiol. 2003;69(12):7480–91. Epub 2003/12/09. doi: 10.1128/AEM.69.12.7480-7491.2003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schlumberger MC, Muller AJ, Ehrbar K, Winnen B, Duss I, Stecher B, et al. Real-time imaging of type III secretion: Salmonella SipA injection into host cells. Proc Natl Acad Sci USA. 2005;102(35):12548–53. Epub 2005/08/19. doi: 10.1073/pnas.0503407102 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sturm A, Heinemann M, Arnoldini M, Benecke A, Ackermann M, Benz M, et al. The cost of virulence: retarded growth of Salmonella Typhimurium cells expressing type III secretion system 1. PLoS Pathog. 2011;7(7):e1002143. Epub 2011/08/11. doi: 10.1371/journal.ppat.1002143 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diard M, Garcia V, Maier L, Remus-Emsermann MN, Regoes RR, Ackermann M, et al. Stabilization of cooperative virulence by the expression of an avirulent phenotype. Nature. 2013;494(7437):353–6. Epub 2013/02/22. doi: 10.1038/nature11913 . [DOI] [PubMed] [Google Scholar]
- 21.Sánchez-Romero MA, Casadesús J. Contribution of SPI-1 bistability to Salmonella enterica cooperative virulence: insights from single cell analysis. Sci Rep. 2018;8(1):14875. Epub 2018/10/07. doi: 10.1038/s41598-018-33137-z . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ackermann M, Stecher B, Freed NE, Songhet P, Hardt WD, Doebeli M. Self-destructive cooperation mediated by phenotypic noise. Nature. 2008;454(7207):987–90. Epub 2008/08/23. doi: 10.1038/nature07067 . [DOI] [PubMed] [Google Scholar]
- 23.Golubeva YA, Sadik AY, Ellermeier JR, Slauch JM. Integrating global regulatory input into the Salmonella pathogenicity island 1 type III secretion system. Genetics. 2012;190(1):79–90. Epub 2011/10/25. doi: 10.1534/genetics.111.132779 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lou L, Zhang P, Piao R, Wang Y. Salmonella pathogenicity island 1 (SPI-1) and its complex regulatory network. Front Cell Infect Microbiol. 2019;9:270. Epub 2019/08/21. doi: 10.3389/fcimb.2019.00270 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Banda MM, Manzo R, Bustamante VH. HilD induces expression of a novel Salmonella Typhimurium invasion factor, YobH, through a regulatory cascade involving SprB. Sci Rep. 2019;9(1):12725. Epub 2019/09/06. doi: 10.1038/s41598-019-49192-z . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith C, Stringer AM, Mao C, Palumbo MJ, Wade JT. Mapping the regulatory network for Salmonella enterica serovar Typhimurium invasion. mBio. 2016;7(5). Epub 2016/09/08. doi: 10.1128/mBio.01024-16 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martínez LC, Banda MM, Fernández-Mora M, Santana FJ, Bustamante VH. HilD induces expression of Salmonella pathogenicity island 2 genes by displacing the global negative regulator H-NS from ssrAB. J Bacteriol. 2014;196(21):3746–55. Epub 2014/08/20. doi: 10.1128/JB.01799-14 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Banda MM, Zavala-Alvarado C, Pérez-Morales D, Bustamante VH. SlyA and HilD counteract H-NS-mediated repression on the ssrAB virulence operon of Salmonella enterica serovar Typhimurium and thus promote its activation by OmpR. J Bacteriol. 2019;201(8). Epub 2019/02/06. doi: 10.1128/JB.00530-18 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singer HM, Kuhne C, Deditius JA, Hughes KT, Erhardt M. The Salmonella Spi1 virulence regulatory protein HilD directly activates transcription of the flagellar master operon flhDC. J Bacteriol. 2014;196(7):1448–57. Epub 2014/02/04. doi: 10.1128/JB.01438-13 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mouslim C, Hughes KT. The effect of cell growth phase on the regulatory cross-talk between flagellar and Spi1 virulence gene expression. PLoS Pathog. 2014;10(3):e1003987. Epub 2014/03/08. doi: 10.1371/journal.ppat.1003987 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martínez-Flores I, Pérez-Morales D, Sánchez-Pérez M, Paredes CC, Collado-Vides J, Salgado H, et al. In silico clustering of Salmonella global gene expression data reveals novel genes co-regulated with the SPI-1 virulence genes through HilD. Sci Rep. 2016;6:37858. doi: 10.1038/srep37858 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petrone BL, Stringer AM, Wade JT. Identification of HilD-regulated genes in Salmonella enterica serovar Typhimurium. J Bacteriol. 2014;196(5):1094–101. Epub 2014/01/01. doi: 10.1128/JB.01449-13 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ellermeier CD, Ellermeier JR, Slauch JM. HilD, HilC and RtsA constitute a feed forward loop that controls expression of the SPI1 type three secretion system regulator hilA in Salmonella enterica serovar Typhimurium. Mol Microbiol. 2005;57(3):691–705. doi: 10.1111/j.1365-2958.2005.04737.x . [DOI] [PubMed] [Google Scholar]
- 34.Saini S, Ellermeier JR, Slauch JM, Rao CV. The role of coupled positive feedback in the expression of the SPI1 type three secretion system in Salmonella. PLoS Pathog. 2010;6(7):e1001025. Epub 2010/08/06. doi: 10.1371/journal.ppat.1001025 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Narm KE, Kalafatis M, Slauch JM. HilD, HilC, and RtsA form homodimers and heterodimers to regulate expression of the Salmonella pathogenicity island I type III secretion system. J Bacteriol. 2020;202(9). Epub 2020/02/12. doi: 10.1128/JB.00012-20 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baxter MA, Fahlen TF, Wilson RL, Jones BD. HilE interacts with HilD and negatively regulates hilA transcription and expression of the Salmonella enterica serovar Typhimurium invasive phenotype. Infect Immun. 2003;71(3):1295–305. doi: 10.1128/IAI.71.3.1295-1305.2003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grenz JR, Cott Chubiz JE, Thaprawat P, Slauch JM. HilE regulates HilD by blocking DNA binding in Salmonella enterica serovar Typhimurium. J Bacteriol. 2018;200(8). Epub 2018/01/31. doi: 10.1128/JB.00750-17 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paredes-Amaya CC, Valdes-García G, Juárez-González VR, Rudino-Pinera E, Bustamante VH. The Hcp-like protein HilE inhibits homodimerization and DNA binding of the virulence-associated transcriptional regulator HilD in Salmonella. J Biol Chem. 2018;293(17):6578–92. Epub 2018/03/15. doi: 10.1074/jbc.RA117.001421 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martínez LC, Yakhnin H, Camacho MI, Georgellis D, Babitzke P, Puente JL, et al. Integration of a complex regulatory cascade involving the SirA/BarA and Csr global regulatory systems that controls expression of the Salmonella SPI-1 and SPI-2 virulence regulons through HilD. Mol Microbiol. 2011;80(6):1637–56. doi: 10.1111/j.1365-2958.2011.07674.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnston C, Pegues DA, Hueck CJ, Lee A, Miller SI. Transcriptional activation of Salmonella Typhimurium invasion genes by a member of the phosphorylated response-regulator superfamily. Mol Microbiol. 1996;22(4):715–27. doi: 10.1046/j.1365-2958.1996.d01-1719.x . [DOI] [PubMed] [Google Scholar]
- 41.Altier C, Suyemoto M, Ruiz AI, Burnham KD, Maurer R. Characterization of two novel regulatory genes affecting Salmonella invasion gene expression. Mol Microbiol. 2000;35(3):635–46. Epub 2000/02/15. doi: 10.1046/j.1365-2958.2000.01734.x . [DOI] [PubMed] [Google Scholar]
- 42.Teplitski M, Goodier RI, Ahmer BM. Pathways leading from BarA/SirA to motility and virulence gene expression in Salmonella. J Bacteriol. 2003;185(24):7257–65. doi: 10.1128/JB.185.24.7257-7265.2003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lapouge K, Schubert M, Allain FH, Haas D. Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol Microbiol. 2008;67(2):241–53. Epub 2007/12/01. doi: 10.1111/j.1365-2958.2007.06042.x . [DOI] [PubMed] [Google Scholar]
- 44.Vakulskas CA, Potts AH, Babitzke P, Ahmer BM, Romeo T. Regulation of bacterial virulence by Csr (Rsm) systems. Microbiol Mol Biol Rev. 2015;79(2):193–224. doi: 10.1128/MMBR.00052-14 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Romeo T, Babitzke P. Global regulation by CsrA and its RNA antagonists. Microbiol Spectr. 2018;6(2). Epub 2018/03/25. doi: 10.1128/microbiolspec.RWR-0009-2017 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martínez LC, Martínez-Flores I, Salgado H, Fernández-Mora M, Medina-Rivera A, Puente JL, et al. In silico identification and experimental characterization of regulatory elements controlling the expression of the Salmonella csrB and csrC genes. J Bacteriol. 2014;196(2):325–36. Epub 2013/11/05. doi: 10.1128/JB.00806-13 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hung CC, Eade CR, Betteken MI, Pavinski Bitar PD, Handley EM, Nugent SL, et al. Salmonella invasion is controlled through the secondary structure of the hilD transcript. PLoS Pathog. 2019;15(4):e1007700. Epub 2019/04/25. doi: 10.1371/journal.ppat.1007700 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Altier C, Suyemoto M, Lawhon SD. Regulation of Salmonella enterica serovar Typhimurium invasion genes by csrA. Infect Immun. 2000;68(12):6790–7. doi: 10.1128/IAI.68.12.6790-6797.2000 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holmqvist E, Wright PR, Li L, Bischler T, Barquist L, Reinhardt R, et al. Global RNA recognition patterns of post-transcriptional regulators Hfq and CsrA revealed by UV crosslinking in vivo. EMBO J. 2016;35(9):991–1011. Epub 2016/04/06. doi: 10.15252/embj.201593360 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fortune DR, Suyemoto M, Altier C. Identification of CsrC and characterization of its role in epithelial cell invasion in Salmonella enterica serovar Typhimurium. Infect Immun. 2006;74(1):331–9. doi: 10.1128/IAI.74.1.331-339.2006 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schechter LM, Damrauer SM, Lee CA. Two AraC/XylS family members can independently counteract the effect of repressing sequences upstream of the hilA promoter. Mol Microbiol. 1999;32(3):629–42. doi: 10.1046/j.1365-2958.1999.01381.x . [DOI] [PubMed] [Google Scholar]
- 52.Schechter LM, Lee CA. AraC/XylS family members, HilC and HilD, directly bind and derepress the Salmonella Typhimurium hilA promoter. Mol Microbiol. 2001;40(6):1289–99. doi: 10.1046/j.1365-2958.2001.02462.x . [DOI] [PubMed] [Google Scholar]
- 53.Akbar S, Schechter LM, Lostroh CP, Lee CA. AraC/XylS family members, HilD and HilC, directly activate virulence gene expression independently of HilA in Salmonella Typhimurium. Mol Microbiol. 2003;47(3):715–28. doi: 10.1046/j.1365-2958.2003.03322.x . [DOI] [PubMed] [Google Scholar]
- 54.Fahlen TF, Mathur N, Jones BD. Identification and characterization of mutants with increased expression of hilA, the invasion gene transcriptional activator of Salmonella Typhimurium. FEMS Immunol Med Microbiol. 2000;28(1):25–35. doi: 10.1111/j.1574-695X.2000.tb01453.x . [DOI] [PubMed] [Google Scholar]
- 55.Ahmer BM, van Reeuwijk J, Watson PR, Wallis TS, Heffron F. Salmonella SirA is a global regulator of genes mediating enteropathogenesis. Mol Microbiol. 1999;31(3):971–82. Epub 1999/02/27. doi: 10.1046/j.1365-2958.1999.01244.x . [DOI] [PubMed] [Google Scholar]
- 56.Baxter MA, Jones BD. The fimYZ genes regulate Salmonella enterica serovar Typhimurium invasion in addition to type 1 fimbrial expression and bacterial motility. Infect Immun. 2005;73(3):1377–85. Epub 2005/02/26. doi: 10.1128/IAI.73.3.1377-1385.2005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saini S, Pearl JA, Rao CV. Role of FimW, FimY, and FimZ in regulating the expression of type 1 fimbriae in Salmonella enterica serovar Typhimurium. J Bacteriol. 2009;191(9):3003–10. Epub 2009/02/17. doi: 10.1128/JB.01694-08 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Espinosa E, Casadesús J. Regulation of Salmonella enterica pathogenicity island 1 (SPI-1) by the LysR-type regulator LeuO. Mol Microbiol. 2014;91(6):1057–69. Epub 2013/12/21. doi: 10.1111/mmi.12500 . [DOI] [PubMed] [Google Scholar]
- 59.Baxter MA, Jones BD. Two-component regulators control hilA expression by controlling fimZ and hilE expression within Salmonella enterica serovar Typhimurium. Infect Immun. 2015;83(3):978–85. doi: 10.1128/IAI.02506-14 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barthel M, Hapfelmeier S, Quintanilla-Martínez L, Kremer M, Rohde M, Hogardt M, et al. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun. 2003;71(5):2839–58. doi: 10.1128/IAI.71.5.2839-2858.2003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hapfelmeier S, Stecher B, Barthel M, Kremer M, Muller AJ, Heikenwalder M, et al. The Salmonella pathogenicity island (SPI)-2 and SPI-1 type III secretion systems allow Salmonella serovar Typhimurium to trigger colitis via MyD88-dependent and MyD88-independent mechanisms. J Immunol. 2005;174(3):1675–85. doi: 10.4049/jimmunol.174.3.1675 . [DOI] [PubMed] [Google Scholar]
- 62.Sekirov I, Gill N, Jogova M, Tam N, Robertson M, de Llanos R, et al. Salmonella SPI-1-mediated neutrophil recruitment during enteric colitis is associated with reduction and alteration in intestinal microbiota. Gut Microbes. 2010;1(1):30–41. doi: 10.4161/gmic.1.1.10950 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467(7314):426–9. Epub 2010/09/25. doi: 10.1038/nature09415 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, et al. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci USA. 2011;108(42):17480–5. doi: 10.1073/pnas.1107857108 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Faber F, Thiennimitr P, Spiga L, Byndloss MX, Litvak Y, Lawhon S, et al. Respiration of microbiota-derived 1,2-propanediol drives Salmonella expansion during colitis. PLoS Pathog. 2017;13(1):e1006129. Epub 2017/01/06. doi: 10.1371/journal.ppat.1006129 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Colgan AM, Kroger C, Diard M, Hardt WD, Puente JL, Sivasankaran SK, et al. The impact of 18 ancestral and horizontally-acquired regulatory proteins upon the transcriptome and sRNA landscape of Salmonella enterica serovar Typhimurium. PLoS Genet. 2016;12(8):e1006258. doi: 10.1371/journal.pgen.1006258 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U. Network motifs: simple building blocks of complex networks. Science. 2002;298(5594):824–7. Epub 2002/10/26. doi: 10.1126/science.298.5594.824 . [DOI] [PubMed] [Google Scholar]
- 68.Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8(6):450–61. Epub 2007/05/19. doi: 10.1038/nrg2102 . [DOI] [PubMed] [Google Scholar]
- 69.Mangan S, Alon U. Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci USA. 2003;100(21):11980–5. Epub 2003/10/08. doi: 10.1073/pnas.2133841100 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mangan S, Itzkovitz S, Zaslaver A, Alon U. The incoherent feed-forward loop accelerates the response-time of the gal system of Escherichia coli. J Mol Biol. 2006;356(5):1073–81. Epub 2006/01/13. doi: 10.1016/j.jmb.2005.12.003 . [DOI] [PubMed] [Google Scholar]
- 71.Goentoro L, Shoval O, Kirschner MW, Alon U. The incoherent feedforward loop can provide fold-change detection in gene regulation. Mol Cell. 2009;36(5):894–9. Epub 2009/12/17. doi: 10.1016/j.molcel.2009.11.018 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hong J, Brandt N, Abdul-Rahman F, Yang A, Hughes T, Gresham D. An incoherent feedforward loop facilitates adaptive tuning of gene expression. Elife. 2018;7. Epub 2018/04/06. doi: 10.7554/eLife.32323 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kaplan S, Bren A, Dekel E, Alon U. The incoherent feed-forward loop can generate non-monotonic input functions for genes. Mol Syst Biol. 2008;4:203. Epub 2008/07/17. doi: 10.1038/msb.2008.43 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Durant JA, Corrier DE, Ricke SC. Short-chain volatile fatty acids modulate the expression of the hilA and invF genes of Salmonella Typhimurium. J Food Prot. 2000;63(5):573–8. Epub 2000/05/29. doi: 10.4315/0362-028x-63.5.573 . [DOI] [PubMed] [Google Scholar]
- 75.Huang Y, Suyemoto M, Garner CD, Cicconi KM, Altier C. Formate acts as a diffusible signal to induce Salmonella invasion. J Bacteriol. 2008;190(12):4233–41. Epub 2008/04/22. doi: 10.1128/JB.00205-08 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chavez RG, Alvarez AF, Romeo T, Georgellis D. The physiological stimulus for the BarA sensor kinase. J Bacteriol. 2010;192(7):2009–12. doi: 10.1128/JB.01685-09 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Olekhnovich IN, Kadner RJ. DNA-binding activities of the HilC and HilD virulence regulatory proteins of Salmonella enterica serovar Typhimurium. J Bacteriol. 2002;184(15):4148–60. doi: 10.1128/JB.184.15.4148-4160.2002 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.De la Cruz MA, Pérez-Morales D, Palacios IJ, Fernández-Mora M, Calva E, Bustamante VH. The two-component system CpxR/A represses the expression of Salmonella virulence genes by affecting the stability of the transcriptional regulator HilD. Front Microbiol. 2015;6:807. doi: 10.3389/fmicb.2015.00807 . [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 79.Schechter LM, Jain S, Akbar S, Lee CA. The small nucleoid-binding proteins H-NS, HU, and Fis affect hilA expression in Salmonella enterica serovar Typhimurium. Infect Immun. 2003;71(9):5432–5. doi: 10.1128/IAI.71.9.5432-5435.2003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, et al. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 2008;4(8):e1000163. Epub 2008/08/30. doi: 10.1371/journal.pgen.1000163 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lim S, Yoon H, Kim M, Han A, Choi J, Choi J, et al. Hfq and ArcA are involved in the stationary phase-dependent activation of Salmonella pathogenicity island 1 (SPI1) under shaking culture conditions. J Microbiol Biotechnol. 2013;23(12):1664–72. Epub 2013/09/11. doi: 10.4014/jmb.1305.05022 . [DOI] [PubMed] [Google Scholar]
- 82.Jiang L, Feng L, Yang B, Zhang W, Wang P, Jiang X, et al. Signal transduction pathway mediated by the novel regulator LoiA for low oxygen tension induced Salmonella Typhimurium invasion. PLoS Pathog. 2017;13(6):e1006429. Epub 2017/06/03. doi: 10.1371/journal.ppat.1006429 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gaviria-Cantin T, El Mouali Y, Le Guyon S, Romling U, Balsalobre C. Gre factors-mediated control of hilD transcription is essential for the invasion of epithelial cells by Salmonella enterica serovar Typhimurium. PLoS Pathog. 2017;13(4):e1006312. Epub 2017/04/21. doi: 10.1371/journal.ppat.1006312 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ellermeier JR, Slauch JM. Fur regulates expression of the Salmonella pathogenicity island 1 type III secretion system through HilD. J Bacteriol. 2008;190(2):476–86. Epub 2007/11/13. doi: 10.1128/JB.00926-07 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chubiz JE, Golubeva YA, Lin D, Miller LD, Slauch JM. FliZ regulates expression of the Salmonella pathogenicity island 1 invasion locus by controlling HilD protein activity in Salmonella enterica serovar Typhimurium. J Bacteriol. 2010;192(23):6261–70. Epub 2010/10/05. doi: 10.1128/JB.00635-10 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Olekhnovich IN, Kadner RJ. Role of nucleoid-associated proteins Hha and H-NS in expression of Salmonella enterica activators HilD, HilC, and RtsA required for cell invasion. J Bacteriol. 2007;189(19):6882–90. doi: 10.1128/JB.00905-07 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vergnes A, Viala JP, Ouadah-Tsabet R, Pocachard B, Loiseau L, Meresse S, et al. The iron-sulfur cluster sensor IscR is a negative regulator of Spi1 type III secretion system in Salmonella enterica. Cell Microbiol. 2017;19(4). Epub 2016/10/30. doi: 10.1111/cmi.12680 . [DOI] [PubMed] [Google Scholar]
- 88.Pérez-Morales D, Banda MM, Chau NYE, Salgado H, Martínez-Flores I, Ibarra JA, et al. The transcriptional regulator SsrB is involved in a molecular switch controlling virulence lifestyles of Salmonella. PLoS Pathog. 2017;13(7):e1006497. Epub 2017/07/14. doi: 10.1371/journal.ppat.1006497 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Palmer AD, Kim K, Slauch JM. PhoP-mediated repression of the SPI1 type 3 secretion system in Salmonella enterica serovar Typhimurium. J Bacteriol. 2019;201(16). Epub 2019/06/12. doi: 10.1128/JB.00264-19 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.López-Garrido J, Casadesús J. Crosstalk between virulence loci: regulation of Salmonella enterica pathogenicity island 1 (SPI-1) by products of the std fimbrial operon. PLoS One. 2012;7(1):e30499. Epub 2012/02/01. doi: 10.1371/journal.pone.0030499 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.El Mouali Y, Gaviria-Cantin T, Sánchez-Romero MA, Gibert M, Westermann AJ, Vogel J, et al. CRP-cAMP mediates silencing of Salmonella virulence at the post-transcriptional level. PLoS Genet. 2018;14(6):e1007401. Epub 2018/06/08. doi: 10.1371/journal.pgen.1007401 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Takaya A, Kubota Y, Isogai E, Yamamoto T. Degradation of the HilC and HilD regulator proteins by ATP-dependent Lon protease leads to downregulation of Salmonella pathogenicity island 1 gene expression. Mol Microbiol. 2005;55(3):839–52. doi: 10.1111/j.1365-2958.2004.04425.x . [DOI] [PubMed] [Google Scholar]
- 93.Hung CC, Eade CR, Altier C. The protein acyltransferase Pat post-transcriptionally controls HilD to repress Salmonella invasion. Mol Microbiol. 2016;102(1):121–36. Epub 2016/06/25. doi: 10.1111/mmi.13451 . [DOI] [PubMed] [Google Scholar]
- 94.Sang Y, Ren J, Ni J, Tao J, Lu J, Yao YF. Protein acetylation is involved in Salmonella enterica serovar Typhimurium virulence. J Infect Dis. 2016;213(11):1836–45. Epub 2016/01/27. doi: 10.1093/infdis/jiw028 . [DOI] [PubMed] [Google Scholar]
- 95.Sang Y, Ren J, Qin R, Liu S, Cui Z, Cheng S, et al. Acetylation regulating protein stability and DNA-binding ability of HilD, thus modulating Salmonella Typhimurium virulence. J Infect Dis. 2017;216(8):1018–26. Epub 2017/03/23. doi: 10.1093/infdis/jix102 . [DOI] [PubMed] [Google Scholar]
- 96.Gong H, Vu GP, Bai Y, Chan E, Wu R, Yang E, et al. A Salmonella small non-coding RNA facilitates bacterial invasion and intracellular replication by modulating the expression of virulence factors. PLoS Pathog. 2011;7(9):e1002120. Epub 2011/09/29. doi: 10.1371/journal.ppat.1002120 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bondi R, Longo F, Messina M, D’Angelo F, Visca P, Leoni L, et al. The multi-output incoherent feedforward loop constituted by the transcriptional regulators LasR and RsaL confers robustness to a subset of quorum sensing genes in Pseudomonas aeruginosa. Mol Biosyst. 2017;13(6):1080–9. Epub 2017/05/04. doi: 10.1039/c7mb00040e . [DOI] [PubMed] [Google Scholar]
- 98.Ferrara S, Carloni S, Fulco R, Falcone M, Macchi R, Bertoni G. Post-transcriptional regulation of the virulence-associated enzyme AlgC by the σ22-dependent small RNA ErsA of Pseudomonas aeruginosa. Environ Microbiol. 2015;17(1):199–214. Epub 2014/09/05. doi: 10.1111/1462-2920.12590 . [DOI] [PubMed] [Google Scholar]
- 99.Imam S, Noguera DR, Donohue TJ. Global analysis of photosynthesis transcriptional regulatory networks. PLoS Genet. 2014;10(12):e1004837. Epub 2014/12/17. doi: 10.1371/journal.pgen.1004837 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mank NN, Berghoff BA, Hermanns YN, Klug G. Regulation of bacterial photosynthesis genes by the small noncoding RNA PcrZ. Proc Natl Acad Sci USA. 2012;109(40):16306–11. Epub 2012/09/19. doi: 10.1073/pnas.1207067109 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mank NN, Berghoff BA, Klug G. A mixed incoherent feed-forward loop contributes to the regulation of bacterial photosynthesis genes. RNA Biol. 2013;10(3):347–52. Epub 2013/02/09. doi: 10.4161/rna.23769 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee EJ, Groisman EA. An antisense RNA that governs the expression kinetics of a multifunctional virulence gene. Mol Microbiol. 2010;76(4):1020–33. Epub 2010/04/20. doi: 10.1111/j.1365-2958.2010.07161.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Simons RW, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53(1):85–96. Epub 1987/01/01. doi: 10.1016/0378-1119(87)90095-3 . [DOI] [PubMed] [Google Scholar]
- 104.Mayer MP. A new set of useful cloning and expression vectors derived from pBlueScript. Gene. 1995;163(1):41–6. doi: 10.1016/0378-1119(95)00389-n . [DOI] [PubMed] [Google Scholar]
- 105.Uzzau S, Figueroa-Bossi N, Rubino S, Bossi L. Epitope tagging of chromosomal genes in Salmonella. Proc Natl Acad Sci USA. 2001;98(26):15264–9. doi: 10.1073/pnas.261348198 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97(12):6640–5. doi: 10.1073/pnas.120163297 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Oropeza R, Sampieri CL, Puente JL, Calva E. Negative and positive regulation of the non-osmoregulated ompS1 porin gene in Salmonella typhi: a novel regulatory mechanism that involves OmpR. Mol Microbiol. 1999;32(2):243–52. doi: 10.1046/j.1365-2958.1999.01329.x . [DOI] [PubMed] [Google Scholar]
- 108.Puente JL, Bieber D, Ramer SW, Murray W, Schoolnik GK. The bundle-forming pili of enteropathogenic Escherichia coli: transcriptional regulation by environmental signals. Mol Microbiol. 1996;20(1):87–100. doi: 10.1111/j.1365-2958.1996.tb02491.x . [DOI] [PubMed] [Google Scholar]
- 109.Yakhnin H, Yakhnin AV, Baker CS, Sineva E, Berezin I, Romeo T, et al. Complex regulation of the global regulatory gene csrA: CsrA-mediated translational repression, transcription from five promoters by σ70 and σS, and indirect transcriptional activation by CsrA. Mol Microbiol. 2011;81(3):689–704. Epub 2011/06/24. doi: 10.1111/j.1365-2958.2011.07723.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yakhnin AV, Yakhnin H, Babitzke P. Gel mobility shift assays to detect protein-RNA interactions. Methods Mol Biol. 2012;905:201–11. Epub 2012/06/28. doi: 10.1007/978-1-61779-949-5_12 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mercante J, Suzuki K, Cheng X, Babitzke P, Romeo T. Comprehensive alanine-scanning mutagenesis of Escherichia coli CsrA defines two subdomains of critical functional importance. J Biol Chem. 2006;281(42):31832–42. Epub 2006/08/23. doi: 10.1074/jbc.M606057200 . [DOI] [PubMed] [Google Scholar]
- 112.Yakhnin AV, Trimble JJ, Chiaro CR, Babitzke P. Effects of mutations in the L-tryptophan binding pocket of the Trp RNA-binding attenuation protein of Bacillus subtilis. J Biol Chem. 2000;275(6):4519–24. Epub 2000/02/08. doi: 10.1074/jbc.275.6.4519 . [DOI] [PubMed] [Google Scholar]
- 113.Banda MM, López C, Manzo R, Rico-Pérez G, García P, Rosales-Reyes R, et al. HilD and PhoP independently regulate the expression of grhD1, a novel gene required for Salmonella Typhimurium invasion of host cells. Sci Rep. 2018;8(1):4841. Epub 2018/03/21. doi: 10.1038/s41598-018-23068-0 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hoiseth SK, Stocker BA. Aromatic-dependent Salmonella Typhimurium are non-virulent and effective as live vaccines. Nature. 1981;291(5812):238–9. doi: 10.1038/291238a0 . [DOI] [PubMed] [Google Scholar]
- 115.Brosius J. Plasmid vectors for the selection of promoters. Gene. 1984;27(2):151–60. doi: 10.1016/0378-1119(84)90136-7 . [DOI] [PubMed] [Google Scholar]
- 116.Ibarra JA, Knodler LA, Sturdevant DE, Virtaneva K, Carmody AB, Fischer ER, et al. Induction of Salmonella pathogenicity island 1 under different growth conditions can affect Salmonella-host cell interactions in vitro. Microbiology. 2010;156(Pt 4):1120–33. Epub 2009/12/26. doi: 10.1099/mic.0.032896-0 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the manuscript.