Abstract
Objective
We sought to determine whether genetic risk modifies the effect of environmental risk factors for multiple sclerosis (MS). To test this hypothesis, we tested for statistical interaction between polygenic risk scores (PRS) capturing genetic susceptibility to MS and environmental risk factors for MS in UK Biobank.
Methods
People with MS were identified within UK Biobank using ICD-10–coded MS or self-report. Associations between environmental risk factors and MS risk were quantified with a case-control design using multivariable logistic regression. PRS were derived using the clumping-and-thresholding approach with external weights from the largest genome-wide association study of MS. Separate scores were created including major histocompatibility complex (MHC) (PRSMHC) and excluding (PRSnon-MHC) the MHC locus. The best-performing PRS were identified in 30% of the cohort and validated in the remaining 70%. Interaction between environmental and genetic risk factors was quantified using the attributable proportion due to interaction (AP) and multiplicative interaction.
Results
Data were available for 2,250 people with MS and 486,000 controls. Childhood obesity, earlier age at menarche, and smoking were associated with MS. The optimal PRS were strongly associated with MS in the validation cohort (PRSMHC: Nagelkerke's pseudo-R2 0.033, p = 3.92 × 10−111; PRSnon-MHC: Nagelkerke's pseudo-R2 0.013, p = 3.73 × 10−43). There was strong evidence of interaction between polygenic risk for MS and childhood obesity (PRSMHC: AP = 0.17, 95% CI 0.06–0.25, p = 0.004; PRSnon-MHC: AP = 0.17, 95% CI 0.06–0.27, p = 0.006).
Conclusions
This study provides novel evidence for an interaction between childhood obesity and a high burden of autosomal genetic risk. These findings may have significant implications for our understanding of MS biology and inform targeted prevention strategies.
Susceptibility to multiple sclerosis (MS) is multifactorial with genetic and environmental determinants.1-3 Environmental exposures associated with MS risk include smoking, solvent exposure, childhood obesity, vitamin D deficiency, increasing latitude, and infectious mononucleosis (IM).2,3 The largest genome-wide association study (GWAS) meta-analysis of MS risk performed by the International Multiple Sclerosis Genetics Consortium (IMSGC) revealed 233 independent signals that account for ∼48% of the estimated heritability of MS.1 Attempts to model MS risk using polygenic risk scores (PRS) have had some success,4-6 supporting the view that MS susceptibility is influenced by common variants across the genome, in addition to the contribution from the major histocompatibility complex (MHC).
A large proportion of MS risk remains unexplained despite the well-described genetic architecture.1 One potential explanation for this “missing risk” is the presence of gene-environment interactions, whereby the effect of certain genes or variants may depend on exposure to environmental risk factors.
Evidence from Scandinavian and North American cohorts suggests that environmental influences on MS risk can be modified by the HLA genotype. The deleterious effects of childhood obesity, smoking, IM, and solvent exposure on MS risk are potentiated among carriers of the HLA DRB1*15 allele and those lacking the protective HLA A*02 genotype.7-10 It is not currently known whether gene-environment interactions in MS extend beyond the HLA locus.11,12
In this work, we harnessed the power of UK Biobank to extend our understanding of how common genetic variation interacts with environmental factors associated with MS development. We achieved this by first performing a large case-control study to confirm the role of established risk factors in this cohort and by developing and validating PRS for MS, which both included and excluded the MHC. Finally, we used these data to look for potential interactions between polygenic risk and environmental factors associated with MS.
Methods
Data Sources
UK Biobank is a longitudinal cohort study described in detail elsewhere.13 In brief, participants between the ages of 40 and 69 years were recruited between 2006 and 2010 from across the United Kingdom. Participants underwent genotyping, donated body fluid samples, and answered a range of questions about lifestyle, environmental, and demographic factors. Health records were linked to participants using Hospital Episode Statistics (HES), primary care data, and the death register. Phenotype data are composed of survey data, linked health care records, anthropometric measurements, and a variety of other biochemical and imaging data (which were not used in this study).
Identification of Cases and Controls
Cases were defined by ICD-coded diagnoses (ICD-10-G35; ICD-9-3409), self-reported MS diagnosis, and a GP-coded diagnosis, or through death registration. Age at diagnosis was determined using the first recorded MS diagnostic code (see supplementary methods for further details, links.lww.com/NXI/A488). Controls were unmatched UK Biobank participants without a coded diagnosis of MS. Individuals diagnosed with MS before age 20 years were excluded because of difficulties establishing the timing of exposures relative to MS onset. Furthermore, the age of 20 years has been used in previous studies and excludes a minimal number of MS cases, and this is safely outside of the range of normal pubertal timing. Participant flow through the study is depicted in figure e-1, links.lww.com/NXI/A487; diagnostic codes used are provided in supplementary data (table e-1, links.lww.com/NXI/A488). To ensure that our results were robust to the definition of MS, we conducted a sensitivity analysis restricting the analysis to participants whose MS diagnosis was corroborated by at least 2 sources (out of self-report, HES code, GP report, and death register; see table e-2, links.lww.com/NXI/A488).
Genotype Data
Genotyping and quality control protocols are described in detail elsewhere.14 Imputed HLA alleles were provided by UK Biobank. HLA alleles were imputed to four-digit resolution using the HLA*IMP:02 software with a multipopulation reference panel (see biobank.ctsu.ox.ac.uk/crystal/crystal/docs/HLA_imputation.pdf). We extracted each participant's allelic dosage for the MS risk allele HLA-DRB1*15:01 and the protective allele HLA-A*02:01 by thresholding posterior allele probabilities at 0.7 as suggested by UK Biobank. These 2 HLA alleles were used because they have the largest effect sizes across multiple studies.2 Genetic principal components (PCs) were supplied by UK Biobank (field ID 22009).
Definition of Exposures
Exposures were selected if they pertained to early life/adolescence (to mitigate the risk of reverse causation) and were previously associated with MS in at least one other observational cohort. Selected exposures were captured from baseline data recorded in UKB, along with age, ethnicity, sex, birth latitude, and Townsend deprivation index at recruitment (table e-1, links.lww.com/NXI/A488).
We examined the following 10 early life/environmental exposures: month of birth, having been breastfed as a child, childhood body size at age 10 years (a proxy for childhood obesity15,16), exposure to maternal smoking around the time of birth, age at menarche (females), age at voice breaking (males), age at first sexual intercourse, smoking status before age 20 years, birth weight, and infectious mononucleosis before age 20 years. Where multiple data points were available for a participant, the first recorded reading was used.
Childhood body size was dichotomized, and participants were classified as “not overweight” if they answered “thinner” or “average” and “overweight” if they answered “plumper.” Smoking status was characterized as “ever” or “never” smoking. Age at menarche was treated as a continuous variable, and analyses regarding menarche were restricted to women. IM status before age 20 years was defined using the source of first report fields. Participants whose IM diagnosis was reported after age 20 years were coded as having not had IM. Vitamin D status was not included, as vitamin D levels are only available from the initial visit (i.e., at study recruitment), which in most cases was subsequent to diagnosis.
Case-Control Study
For each risk factor, we built a multivariable logistic regression model modeling MS status as the outcome, with age, sex, ethnicity, current deprivation status, and birth latitude as potential confounding covariates.17
The strength of evidence for association with MS was determining using the model likelihood ratio, comparing the full model with a null model comprising only the confounding covariates. Strong evidence for association was defined using a Bonferroni-adjusted p-value threshold to maintain an alpha of 0.05 (pthreshold = 0.05/10 = 0.005). Risk factors robustly associated with MS at alpha < 0.05 were then combined in a multivariable model including the most potent genetic risk factors, HLA DRB1*15:01 and HLA A*02:01, to assess whether their effects showed evidence of independent association with MS.
Development of PRS for MS
A variety of PRS were created using the clumping-and-thresholding approach with external weights derived from the IMSGC discovery stage meta-analysis (supplementary methods, links.lww.com/NXI/A488). We created scores both including the MHC region (PRSMHC) and excluding this region (PRSnon-MHC). To validate the PRS, the data set was divided randomly into a training set (30%, nMS = 589, ncontrol = 112,724) and a testing set (70%, nMS = 1,237, ncontrol = 263,159, figure e-1, links.lww.com/NXI/A487). To determine the optimal PRS, we constructed multivariable logistic regression models for each PRS with MS status as the outcome with age, sex, Townsend deprivation index, and the first 4 genetic PCs as confounding covariates. For the sensitivity analysis excluding MS cases with only one source of diagnostic code report, MS case numbers were 395 (training set) and 871 (testing set).
PRS performance was evaluated using Nagelkerke's pseudo-R2 metric, which is analogous to the R2 derived from linear regression models. Nagelkerke's pseudo-R2 was calculated comparing the full model including the PRS with a null model comprising the confounding covariates alone. This procedure was repeated for all 64 scores (table e-3, links.lww.com/NXI/A488). Altering the number of PCs adjusted for did not substantially alter the results (figures e-2 and e-3, links.lww.com/NXI/A487). Further validation is described in the supplementary methods, links.lww.com/NXI/A488.
PRS × Environment Interactions
The optimal PRSMHC and PRSnon-MHC were used to look for evidence of genome-wide gene-environment interactions using exposures identified as significantly associated with MS in the case-control study. All interaction analyses were conducted in the testing set to avoid PRS overfitting. Interaction was assessed on the additive and multiplicative scales (supplementary methods for full details, links.lww.com/NXI/A488). Multiplicative interaction was quantified using the interaction term beta from logistic models, and additive interaction was quantified using the attributable proportion due to interaction (AP).
HLA × PRS Interactions
To determine whether non-MHC genetic risk of MS modulates the effects of the most potent MHC risk allele, DRB1*15:01, we calculated additive and multiplicative interaction statistics using the methods described previously, considering both the DRB 1*15:01 genotype (dominant-coding) and the non-MHC PRS as independent covariates.
Association of PRS With Disease Measures
To determine whether the MS-PRS was associated with age at first report and claiming of disability benefits, we constructed regression models in the testing set. For age at first MS diagnostic code report, values were normalized using the inverse-rank normalization. Linear regression models were constructed, using age, sex, Townsend score, and the first 4 PCs as covariates. Claiming of disability benefits was assessed using the UKB field “Attendance/disability/mobility allowance” (field 6,146) and recoded this as a binary variable (i.e., participants were coded as “1” if they claimed any of the blue badge, attendance allowance, or disability living allowance and as “0” if not). Logistic regression models were then constructed using the same covariates as above (age, sex, Townsend score, and first 4 genetic PCs).
Ethical Approval
This work was performed using data from UK Biobank (REC approval 11/NW/0382). All participants gave informed consent on Biobank registration and are free to withdraw from the study at any point, at which point their data are censored and cannot be included in further analyses.
Computing
This research was supported by the High-Performance Cluster computing network hosted by Queen Mary University of London.18 Statistical analyses were performed in R version 3.6.1. Extraction of European individuals from the 1,000 genomes reference genome was conducted using vcftools. Construction of the PRS, application of the PRS to individuals, and quality control were performed in PLINK 1.9 and PLINK2.
Data Availability
UK Biobank data are available on request from biobank.ctsu.ox.ac.uk/crystal/. MS IMSGC GWAS data are available on request from imsgc.net/?page_id=31. All codes used in this study are available on GitHub (@benjacobs123456).
Results
Population Demographics
Phenotype and genotype data were available for 488,276 UK Biobank participants comprising 2,276 people with MS and 486,000 unmatched controls. The median age at first MS report was 43.5 years (IQR 16.1, figure e-4, links.lww.com/NXI/A487). Demographic characteristics are shown in table 1. Characteristics of individuals with MS were consistent with published observational data (72.7% female, 98.1% White British). One thousand six hundred fifty-five individuals were included in the sensitivity analysis (table e-2, links.lww.com/NXI/A488).
Table 1.
Demographic Characteristics of Included Participants and Results From the Case-Control Study
Exposures Associated With MS in UK Biobank
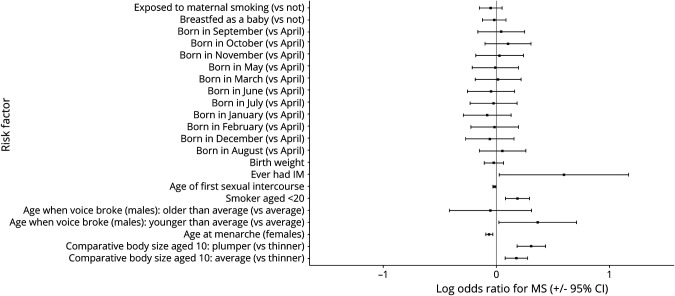
There was strong evidence for association between 3 of the 10 risk factors examined and MS (pBonf < 0.05): higher childhood body size at age 10 years (“plumper than average” vs “thinner than average”: OR 1.36, 95% CI 1.20–1.55), smoking before age 20 years (OR 1.21, 95% CI 1.08–1.34), and earlier menarche (OR 0.94, 95% CI 0.91–0.97, figure 1, table 1). The effects of these 3 risk factors remained similar in a combined model incorporating HLA DRB1*15:01 and HLA A*02:01 genotype (table e-4, links.lww.com/NXI/A488).
Figure 1. ORs and 95% CIs for the Association of Each Exposure With MS.
ORs and CIs are from the output of a multivariable logistic regression with the following covariates: age, sex, ethnicity, birth latitude, current deprivation status, and the exposure in question. For menarche (females only) and voice breaking (males only), sex was not included as a covariate.
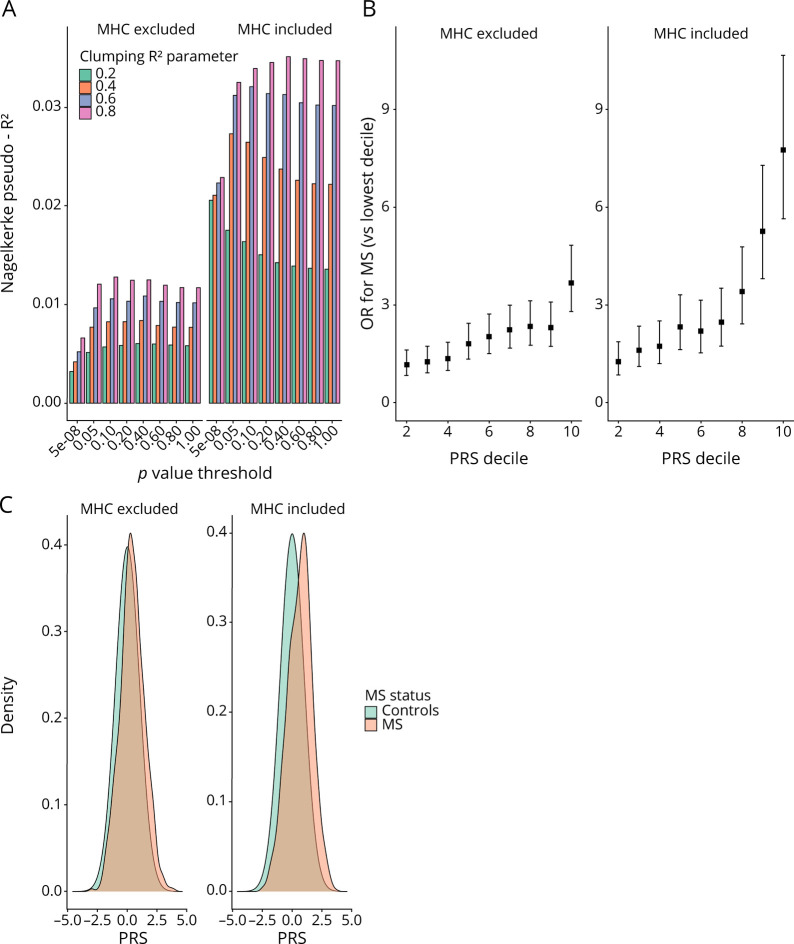
Development and Validation of PRS for MS
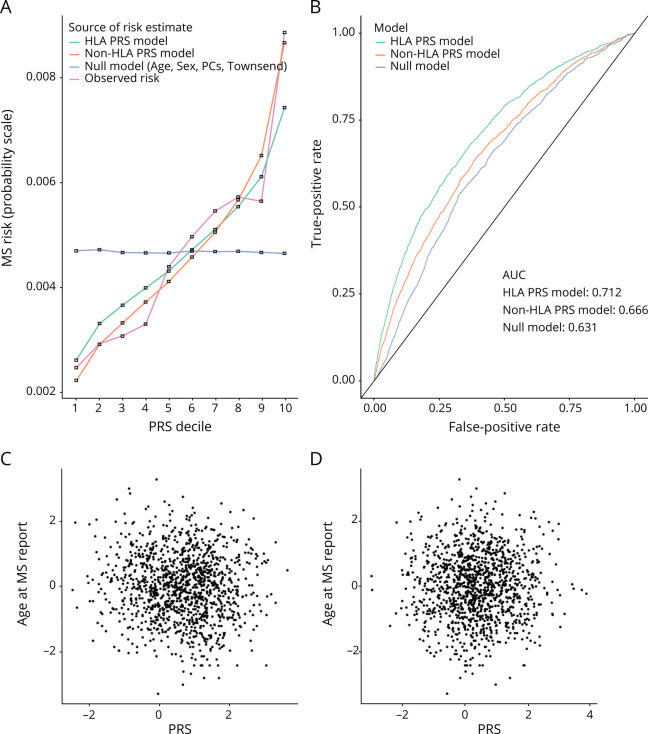
The optimal PRSMHC and PRSnon-MHC explained 3.5% and 1.3% of MS risk in the training set, respectively (figure 2a, table e-3, links.lww.com/NXI/A488, figures e-2 and e-3, links.lww.com/NXI/A487). Both scores were strongly associated with MS in the testing set (PRSMHC: Nagelkerke's pseudo-R2 0.033, p = 3.92 × 10−111; PRSnon-MHC: Nagelkerke's pseudo-R2 0.013, p = 3.73 × 10−43, figure 2, table e-5, links.lww.com/NXI/A488). Both scores were reasonably well calibrated (figure 3A) with good discriminative performance (AUCMHC 0.71, AUCnon-MHC 0.67, AUCnull 0.63; figure 3B). There was no evidence of association between the PRSMHC or PRSnon-MHC and either age at MS report (figure 3, C and D) or claiming of disability benefits (pMHC = 0.44, pnon-MHC = 0.96, figure e-5, links.lww.com/NXI/A487).
Figure 2. (A) Nagelkerke's Pseudo-R2 Metric for Each of the Individual PRS Used.
The R2 was calculated by comparing the model fit (age, sex, Townsend deprivation index, the first 4 genetic PCs, and PRS) vs the null model (age, sex, Townsend deprivation index, and the first 4 genetic PCs). A variety of p value thresholds and clumping parameters were used to create different PRS. Note that the clumping R2 refers to the linkage disequilibrium threshold within which variants were “clumped” and is a different quantity from the Nagelkerke pseudo-R2. PRS are shown both including and excluding the major histocompatibility complex region. (B) ORs and 95% CIs for MS for individuals in each PRS decile (reference: lowest decile). ORs were calculated from logistic regression models with the following covariates: age, sex, first 4 genetic PCs, and PRS. (C) Histogram showing PRS distributions among MS cases and controls. MHC = major histocompatibility complex; PC = principal component; PRS = polygenic risk score.
Figure 3. (A) Calibration Plot Showing Absolute MS Disease Probabilities Within Each PRS Decile (of the Non-MHC PRS).
Other lines represent the mean fitted disease probabilities for models incorporating the MHC PRS, the non-MHC PRS, and null covariates alone (age, sex, deprivation, and genetic PCs). (B) Receiver operating characteristic (ROC) curves demonstrating the discriminative performance (i.e., ability to distinguish MS cases from controls) of each PRS. The null model, MHC PRS, and non-MHC PRS are shown. (C) Scatter plots showing no relationship between MHC PRS and normalized age at MS report. (D) Scatter plots showing no relationship between non-MHC PRS and normalized age at MS report. HLA = human leukocyte antigen; MHC = major histocompatibility complex; PC = principal component; PRS = polygenic risk score.
PRS Interactions With Environmental Risk Factors and DRB1*15:01
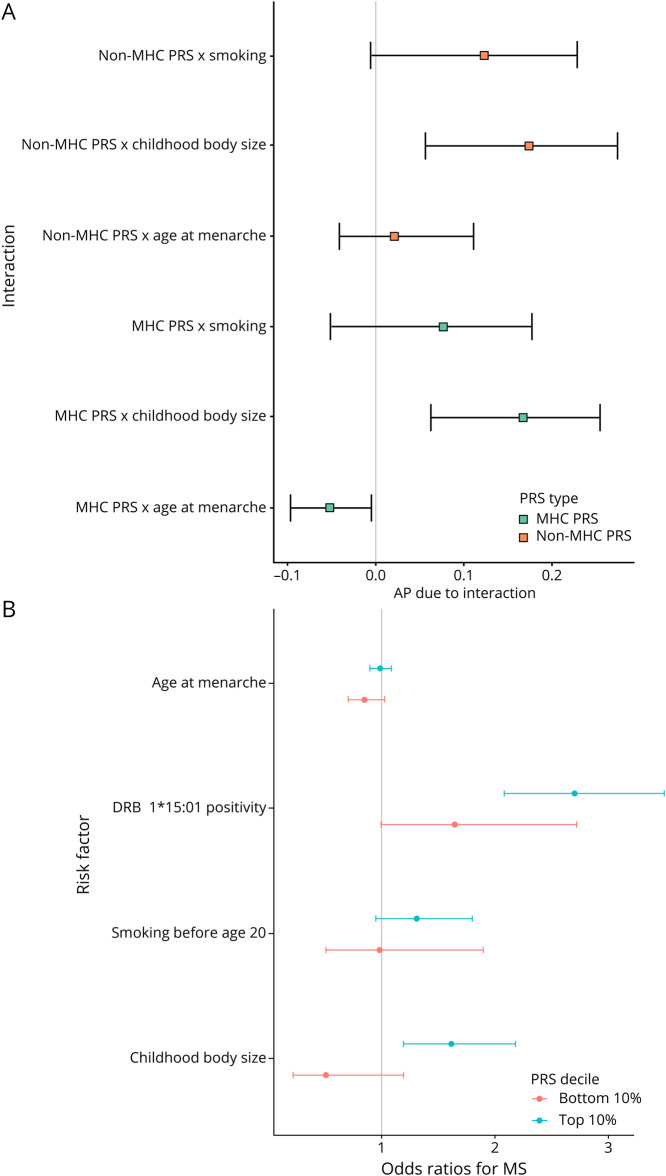
We found strong evidence of interaction on the additive scale between the PRSMHC and PRSnon-MHC and childhood body size (PRSMHC: AP = 0.17, 95% CI 0.06–0.25, p = 0.004; PRSnon-MHC: AP = 0.17, 95% CI 0.06–0.27, p = 0.006). We found weaker evidence for interaction on this scale between age at menarche and the PRSMHC (AP = −0.05, 95% CI −0.10 to 0.00, p = 0.033; figure 4A, table 2), consistent with a previous report,19 but this estimate did not surpass the multiple testing threshold (table 2). There was a lack of strong evidence for other pairwise additive interactions (figure 4) or for multiplicative interactions (figure e-6, links.lww.com/NXI/A487, table e-6, links.lww.com/NXI/A488). There was evidence of additive interaction between the PRSnon-MHC and HLA DRB1*15:01 carriage (AP 0.24, 95% CI 0.17–0.30, p = 0.0002, figure 4B) but no evidence of multiplicative interaction (beta 0.060, p = 0.30). We found similar results with a more stringent case definition (only counting individuals as having MS if their diagnosis was supported by more than 1 source of report; table e-7, links.lww.com/NXI/A488, figures e-7, e-8, e-9, links.lww.com/NXI/A487). All CIs for the estimates overlapped between the primary and sensitivity analysis.
Figure 4. (A) Forest Plot Demonstrating Attributable Proportion due to Interaction (AP) and 95% CIs for Interactions Between Environmental Exposures and Genetic Risk Factors for MS.
If there is no interaction, the AP is 0. AP > 1 indicates positive interaction (combined effects exceed the sum of the individual effects) and vice versa. CIs are derived from taking the 2.5th and 97.5th percentiles of 10,000 bootstrap replicates. (B) Forest plot demonstrating ORs and 95% CIs for participants in the top and bottom polygenic risk score deciles. The outcome in each case is MS status, and the exposures of interest are childhood body size, age at menarche, smoking before age 20 years, and carriage of the HLA DRB1*15:01 allele. ORs are from the output of the logistic regression model of the form MS risk ∼ age + sex + first 4 genetic PCs. Models were built separately for individuals with the highest 10% of genetic risk scores and the lowest 10% of genetic risk scores (“top” and “bottom” decile, respectively). MHC = major histocompatibility complex; PC = principal component; PRS = polygenic risk score.
Table 2.
AP due to Interaction, 95% CIs, and 2-Sided p Values for Each of the PRS × E Interactions Examined
Discussion
In this study, we harnessed the scale and breadth of UK Biobank to study >2000 MS cases and >480,000 controls, providing the first evidence that the effect of an established risk factor for MS (childhood obesity) may be potentiated by an individual's genome-wide genetic risk for MS. We show that this effect persists even when the MHC locus is excluded from the PRS. By using data from the largest GWAS of MS susceptibility to derive and validate PRS for MS, both incorporating and excluding the MHC region, we demonstrate supportive evidence for a gene-gene interaction. This work shows that the effect of DRB1*15:01 on MS susceptibility may be potentiated among individuals with a high background genetic risk for MS in this cohort. To our knowledge, our study is the first to demonstrate that the polygenic risk of an individual for MS may alter the effect of established environmental risk factors on their risk of MS.3 These findings are especially interesting in the context of evidence from mendelian randomization studies supporting a causal role for childhood obesity in the pathogenesis of MS.20,21
Previous studies of gene-environment interactions in MS have focused on interactions between HLA alleles and environmental risk factors. Specifically, evidence suggests that carriage of high-risk HLA haplotypes containing DRB1*15:01 and lacking A*02:01 enhances the deleterious association of childhood obesity, smoking, infectious mononucleosis, and solvent exposure with MS risk.2,7-9 The intuitive biological explanation for such interactions is that high-risk HLA alleles may promote presentation of epitopes, e.g., from cigarette smoke or within adipose tissue, in such a way that mimics myelin peptides and triggers CNS-directed autoimmunity. Beyond the MHC, there has been relatively limited study of how genetic variation modulates the effect of environmental risk factors for MS,11,12 probably in large part because of the relatively small number of data sets with sufficient power, deep phenotyping, and high-quality genetic data required for such analyses.
In this study, we created 64 individual PRS, both including and excluding the MHC locus on chromosome 6, which is the strongest single genetic determinant of MS risk and accounts for ∼20% of the SNP heritability of MS in Europeans.1 Both the non-MHC and MHC PRS were strongly associated with MS risk in both training and testing sets. The non-MHC PRS in this study captured a small proportion of overall MS liability but was robustly associated with MS. Previous efforts using the PRS from the IMSGC explained up to ∼3% of variance.5 The best-performing non-MHC PRS in this study explained ∼1% of MS variance. This discrepancy could be explained by several factors, including the relatively low number of cases in UK Biobank, the possibility of missed cases, the possibility of controls misclassified as cases, differences in population structure, restriction according to self-declared ethnicity with an additional genetic PC analysis, and some SNPs not being available and/or failing QC checks in Biobank. Nevertheless, despite low overall variance, the validity of the PRS is underscored by the monotonic relationship between the PRS and OR of MS, the robust model fit when using the PRS to model MS risk, reasonable discriminative capacity, and good calibration.
There are several important caveats to this work. Most importantly, although we are able to observe and measure statistical interaction—that is, deviation from a model whereby the effects of genetic and environmental risk factors are combined additively (in the case of the AP) or multiplicatively (in the case of multiplicative interaction—statistical interaction does not straightforwardly imply biological interaction, nor does it necessarily imply interaction that is meaningful in terms of real-life disease prediction or prevention). We were unable to demonstrate replication in a truly independent cohort (dividing the cohort into training and testing sets does not yield a genuinely independent cohort). Our findings have limited generalizability for non-European groups because UK Biobank participants are predominantly White. MS diagnosis in this cohort is derived from linked health care records or self-report and so do not carry the same degree of certainty as criteria-defined MS. Equally, it is conceivable that there are “missed” cases in the data set, that is, individuals with MS who do not have a coded diagnosis available through linked health care records. However, MS prevalence in UK Biobank approaches the expected UK prevalence,22 suggesting that the overwhelming majority of individuals with MS are correctly identified. The UKB cohort is highly selected, and is enriched for individuals living near assessment centers, from more affluent socioeconomic groups than the general population, for White British individuals, and (intentionally) for individuals older than 40 years (the minimum age at recruitment). These factors carry a risk of introducing various biases, for example, through collider bias, which may induce spurious associations and destroy true associations. We emphasize that these findings require replication in other independent cohorts. Our findings concerning gene-gene interactions could be replicated in “genetics-only” cohorts such as the IMSGC, and we would encourage others to attempt to replicate this finding in large GWAS cohorts (with many more cases than the ∼2000 in UKB), so we can ascertain whether it is robust.
Our failure to replicate the previously reported interactions between HLA genotypes, smoking, and childhood body size2,7-9 could be explained by methodologic differences between our study and the published literature: this cohort is likely to differ in key respects from the Kaiser Permanente and EIMS cohorts in that UK Biobank participants are predominantly White, from relatively affluent parts of the United Kingdom, are self-selecting, and are middle-aged (recruitment from 40 to 69 years); we control for different covariates in our interaction analyses (using PCs to account for ancestry), and we used imputed HLA alleles to four-digit resolution; UK Biobank survey data are also prone to recall bias as it is retrospective. We would interpret the lack of HLA-environment interactions in our study with caution as an absence of evidence rather than evidence of absence.
The key variables used in this study are retrospective or cross-sectional (e.g., MS diagnosis, self-reported body size in childhood, and self-reported smoking status). Not only are these subject to recall bias, but more importantly our results are not revealing about predicting an individual's risk of developing MS. To demonstrate predictive power, these results need to be replicated in a longitudinal cohort. In addition, the metric we focus on, “comparative body size at age 10 years,” is clearly not a perfect proxy for childhood obesity. Furthermore, childhood obesity itself is not equivalent with obesity during earlier life or sustained throughout adolescence. The extent and timing of obesity during childhood and adolescence determine the importance of BMI for MS risk, and clearly, a snapshot of body size at age 10 years does not reflect the complexity of BMI changes during the first 20 years of life.9,20,21 Other limitations to this study include the limited overall variance explained by optimal PRS, the relatively small absolute number of people with MS, and the imperfect nature of self-reported phenotypes. Furthermore, some exposures known to be strongly associated with MS were either unavailable (e.g., vitamin D status before diagnosis) or so underreported as to be unreliable (e.g., infectious mononucleosis).
Despite these limitations, our study also has some strengths. We use the UK Biobank data set, which provides a unique opportunity to study gene-environment interactions on a large scale. The vast number of controls in UKB adds substantial power. We tune and test the PRS in separate samples, which is important to prevent overfitting of the PRS to the data. We use an agnostic approach to develop the PRS, using a range of clumping-and-thresholding parameters to discover the optimal structure of the PRS, allowing us to discover a significant improvement in predictive power from using a large number of variants weakly associated with MS over using strictly “GWAS-significant” hits (p < 5e-8). These optimal parameters also reiterate the polygenic architecture of MS.
We evaluate interactions on both the multiplicative and additive scales, as has become standard practice to avoid missing biologically significant interactions.2 We additionally evaluate the relationship between the PRS and proxies for clinical characteristics of MS, including age at diagnosis and claiming of disability benefits. We evaluated whether the effect of DRB1*15:01 is modulated by polygenic risk, as has been demonstrated for high-effect variants in the LDL-R (causing familial hypercholesterolemia) and BRCA (causing breast cancer),23 and find evidence in support of this hypothesis. Clearly, this finding is easily replicated in the IMSGC cohort, and we would urge caution in overinterpreting the finding without confirmation in this far larger cohort of cases.
This study thus provides novel evidence that childhood body size interacts with non-HLA MS genetic risk. Demonstrating benefit for preventive measures in rare, complex diseases such as MS is a challenge because of the low population incidence and the small effects of individual interventions. Power can be enhanced by enriching for high-risk individuals and by selecting individuals who are likely to experience the greatest benefit from the intervention. As the effect of childhood body size on MS risk appears greater among individuals with a high genome-wide genetic risk, trials attempting to demonstrate the benefit of targeting childhood obesity may benefit from risk-stratifying individuals using this approach. Further efforts are required to localize the variants and genes that account for the observed interaction effects, which should help to shed further light on the biology of these risk factors and improve efforts to individualize MS risk prediction algorithms in the future.
Acknowledgment
The authors thank the relevant consortia for making their data available. MS GWAS data were taken from the MS Chip discovery summary statistics. IMSGC summary statistics are available through request on the website: nettskjema.no/answer/imsgc-data-access.html. The authors thank the Queen Mary University High Performance Computing team for their help with computing resources. The authors thank the participants and researchers involved in UK Biobank, who have created an exceptional resource. UK Biobank data are available on request through their website. Code used in this article is available on GitHub (@benjacobs123456).
Glossary
- AP
attributable proportion
- GWAS
genome-wide association study
- HES
Hospital Episode Statistics
- IM
infectious mononucleosis
- IMSGC
International Multiple Sclerosis Genetics Consortium
- MHC
major histocompatibility complex
- MS
multiple sclerosis
- PC
principal component
- PRS
polygenic risk score
Appendix. Authors

Contributor Information
Benjamin Meir Jacobs, Email: ben.meir.jacobs@gmail.com.
Alastair J. Noyce, Email: a.noyce@qmul.ac.uk.
Jonathan Bestwick, Email: j.p.bestwick@qmul.ac.uk.
Daniel Belete, Email: d.belete@nhs.net.
Gavin Giovannoni, Email: g.giovannoni@qmul.ac.uk.
Study Funding
Barts Charity (grant ref MGU0365).
Disclosure
The authors report no disclosures relevant to the manuscript. Go to Neurology.org/NN for full disclosures.
References
- 1.International Multiple Sclerosis Genetics Consortium. Multiple sclerosis genomic map implicates peripheral immune cells and microglia in susceptibility. Science. 2019;365(6460):eaav7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olsson T, Barcellos LF, Alfredsson L. Interactions between genetic, lifestyle and environmental risk factors for multiple sclerosis. Nat Rev Neurol. 2017;13(1):25-36. [DOI] [PubMed] [Google Scholar]
- 3.Alfredsson L, Olsson T. Lifestyle and environmental factors in multiple sclerosis. Cold Spring Harb Perspect Med. 2019;9(4):a028944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Disanto G, Dobson R, Pakpoor J, et al. The refinement of genetic predictors of multiple sclerosis. PLoS One. 2014;9(5):e96578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The International Multiple Sclerosis Genetics Consortium (IMSGC). Evidence for polygenic susceptibility to multiple sclerosis—the shape of things to come. Am J Hum Genet. 2010;86(4):621-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dobson R, Ramagopalan S, Topping J, et al. A risk score for predicting multiple sclerosis. PLoS One. 2016;11(11):e0164992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hedström AK, Bomfim IL, Barcellos LF, et al. Interaction between passive smoking and two HLA genes with regard to multiple sclerosis risk. Int J Epidemiol. 2014;43(6):1791-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hedström AK, Sundqvist E, Bäärnhielm M, et al. Smoking and two human leukocyte antigen genes interact to increase the risk for multiple sclerosis. Brain. 2011;134(pt 3):653-664. [DOI] [PubMed] [Google Scholar]
- 9.Hedström AK, Lima Bomfim I, Barcellos L, et al. Interaction between adolescent obesity and HLA risk genes in the etiology of multiple sclerosis. Neurology. 2014;82(10):865-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hedström AK, Hössjer O, Katsoulis M, Kockum I, Olsson T, Alfredsson L. Organic solvents and MS susceptibility: interaction with MS risk HLA genes. Neurology. 2018;91(15):e455-e462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Briggs FBS, Acuna B, Shen L, et al. Smoking and risk of multiple sclerosis: evidence of modification by NAT1 variants. Epidemiology. 2014;25(4):605-614. [DOI] [PubMed] [Google Scholar]
- 12.Briggs FB. Nicotinic acetylcholine receptors α7 and α9 modifies tobacco smoke risk for multiple sclerosis. Mult Scler. 2020. doi: 10.1177/1352458520958361. [DOI] [PubMed] [Google Scholar]
- 13.Sudlow C, Gallacher J, Allen N, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12(3):e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bycroft C, Freeman C, Petkova D, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simmonds M, Llewellyn A, Owen CG, Woolacott N. Predicting adult obesity from childhood obesity: a systematic review and meta-analysis. Obes Rev. 2016;17(2):95-107. [DOI] [PubMed] [Google Scholar]
- 16.He J, Cai Z, Fan X. Accuracy of using self-reported data to screen children and adolescents for overweight and obesity status: a diagnostic meta-analysis. Obes Res Clin Pract. 2017;11(3):257-267. [DOI] [PubMed] [Google Scholar]
- 17.Murray S, Bashir K, Penrice G, Womersley SJ. Epidemiology of multiple sclerosis in Glasgow. Scott Med J. 2004;49(3):100-104. [DOI] [PubMed] [Google Scholar]
- 18.King T, Butcher S, Zalewski L. Apocrita—High Performance Computing Cluster for Queen Mary University of London; 2017. Accessed February 5, 2021. zenodo.org/record/438045. [Google Scholar]
- 19.Bove R, Chua AS, Xia Z, Chibnik L, De Jager PL, Chitnis T. Complex relation of HLA-DRB1*1501, age at menarche, and age at multiple sclerosis onset. Neurol Genet. 2016;2(4):e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mokry LE, Ross S, Timpson NJ, Sawcer S, Davey Smith G, Richards JB. Obesity and multiple sclerosis: a mendelian randomization study. PLoS Med. 2016;13(6):e1002053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacobs BM, Noyce AJ, Giovannoni G, Dobson R. BMI and low vitamin D are causal factors for multiple sclerosis: a Mendelian Randomization study. Neurol Neuroimmunol Neuroinflamm. 2020;7(2):e662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Multiple sclerosis: prevalence, incidence and smoking status—data briefing [online]. Accessed February 5, gov.uk/government/publications/multiple-sclerosis-prevalence-incidence-and-smoking-status/multiple-sclerosis-prevalence-incidence-and-smoking-status-data-briefing.
- 23.Fahed AC, Wang M, Homburger JR, et al. Polygenic background modifies penetrance of monogenic variants for tier 1 genomic conditions. Nat Commun. 2020;11(1):3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
UK Biobank data are available on request from biobank.ctsu.ox.ac.uk/crystal/. MS IMSGC GWAS data are available on request from imsgc.net/?page_id=31. All codes used in this study are available on GitHub (@benjacobs123456).