Abstract
About 20% of the cancer incidences worldwide have been estimated to be associated with infections. However, the molecular mechanisms of exactly how they contribute to host tumorigenesis are still unknown. To evade host defense, pathogens hijack host proteins at different levels: sequence, structure, motif, and binding surface, i.e., interface. Interface similarity allows pathogen proteins to compete with host counterparts to bind to a target protein, rewire physiological signaling, and result in persistent infections, as well as cancer. Identification of host-pathogen interactions (HPIs)—along with their structural details at atomic resolution—may provide mechanistic insight into pathogen-driven cancers and innovate therapeutic intervention. HPI data including structural details is scarce and large-scale experimental detection is challenging. Therefore, there is an urgent and mounting need for efficient and robust computational approaches to predict HPIs and their complex (bound) structures. In this chapter, we review the first and currently only interface-based computational approach to identify novel HPIs. The concept of interface mimicry promises to identify more HPIs than complete sequence or structural similarity. We illustrate this concept with a case study on Kaposi’s sarcoma herpesvirus (KSHV) to elucidate how it subverts host immunity and helps contribute to malignant transformation of the host cells.
Keywords: Host-pathogen interaction prediction, Protein–protein interaction, Structural network, Superorganism network, Molecular mimicry, Interface mimicry
1. Introduction
1.1. Molecular Mimicry
Signaling pathways shape and convey the cell’s responses to stimuli from its environment; however, pathogens can circumvent this response by “repurposing” host signaling. Pathogens can interact with the host through proteins, metabolites, small molecules, and nucleic acids [1]. Direct protein-protein interactions are the most common interaction type (see Note 1). By interfering with key pathways pathogens can reshape physiological signaling, subverting the immune system, altering the cytoskeletal organization [2, 3], modifying membrane and vesicular trafficking [2, 4, 5], boosting pathogen entry into the host cell, changing the cell cycle regulation [6, 7], and modulating apoptosis [8]. All host-pathogen interactions (HPIs) aim to ensure pathogen survival within the host.
Pathogens evolved several strategies to cross-talk with their hosts. One powerful way is molecular mimicry, which has been extensively reviewed in our recent study [9]. There are four different levels of molecular (protein) mimicry: hijacking (1) both sequence and structure of a protein or a domain, (2) only structure without sequence homology, (3) sequence of a short motif—motif mimicry, and (4) structure of a binding surface without sequence similarity—interface mimicry. Global sequence and structural similarity is much rarer than interface similarity both within and across species. Thus, utilizing interface mimicry allows pathogens to target more host proteins. The concept of interface mimicry, proposed over two decades ago, suggested that proteins with different global structures can interact in similar ways, via similar binding surfaces [10-12]. Interfaces are frequently “reused” by distinct proteins [13], suggesting that these recurring architectures are favorable scaffolds [12].
Interface mimicry is often observed within (intraspecies/endogenous) [13-15] and across species (interspecies/host-pathogen/exogenous) [16, 17]. Similarity in endogenous and exogenous protein-protein interfaces permits pathogenic proteins to compete with their host counterparts [17], rewire host signaling, and cause infections, as well as cancer. Identification of the HPIs and the rewired host-pathogen superorganism protein interaction network, together with structural details, should provide critical insights into pathogenic virulence strategies underlying infections and pathogen-driven cancers, and hence help innovative therapeutics [18].
To date, the HPI networks show that different pathogens often target the same host pathway, and certain host pathways are attacked at several nodes to guarantee alteration of host cell signaling [19]. Although there are several available host-pathogen metaorganism interaction networks [19-29], there have been few attempts to integrate these HPI networks with the human 3D structural protein-protein interactions (PPIs) [17]. Traditional node-and-edge representation of the PPI networks simplifies the “big picture.” They depict which proteins interact, but not how. Structural networks allow a higher resolution with mechanistic insights, showing which residues are involved in the interaction and thus which binary interactions can co-occur or are mutually exclusive [16, 30] (see Note 2). The power of structural networks in displaying the details of endogenous signaling pathways was demonstrated earlier [30-33]. They are also vital to comprehend the mechanisms exerted by pathogens to avert and subvert host cell signaling and circumvent immune response [18]. Structures exhibit which endogenous PPIs are ablated by the HPIs, whether the virulence factors in different strains of the same pathogenic species have distinct HPIs, and possible outcomes of mutations on either the host or the pathogenic proteins.
The challenging large-scale experimental characterization of HPIs [34, 35], coupled with the scarcity of experimentally confirmed HPI data, especially structural details, escalates the demand for efficient and robust computational approaches to predict HPIs along with their complex (bound) structures. In this chapter, we first review available computational approaches to predict HPIs and present the only interface-based computational approach available to identify novel HPIs and their complex structures. Then, we illustrate the usefulness of our approach with a case study on Kaposi’s sarcoma herpesvirus (KSHV).
1.2. Review of Available Computational Tools to Identify HPIs
Several HPI databases have been developed for experimentally identified HPIs, including PHISTO [36], HPIDB [37], Proteopathogen [38], PATRIC [39], PHI-base [40], PHIDIAS [41], HoPaCI-DB [42], VirHostNet [43], ViRBase [44], VirusMentha [45], and HCVpro [46]. These databases comprise only a limited number of pathogens. Given that at least hundreds of different species can infect the host, thousands of HPIs are still unknown. Enriching of the host-pathogen interactome and construction of comprehensive HPI networks will still mostly rely on computational models in the near future [47]. Numerous studies computationally identified large-scale HPIs and built HPI networks for viruses and bacteria [20, 24, 48-56].
Although prediction of human PPIs is a well-established area, modeling of interspecies interactions is comparably new. Still, several attempts focused on computational approaches to identify HPIs [34], most of which rely on sequence homology [49, 52, 54, 57-63]. Homology-based approaches are successful only if the sequence similarity is high, but not all virulence factors have homologs in human. For instance, a secreted protein of H. pylori, VacA, does not have sequence similarity with any other known viral, bacterial, or eukaryotic proteins [64], but it alters signaling through several host pathways [65]. Thus, sequence-based methods cannot detect VacA’s HPIs, highlighting the importance of considering the 3D structures of proteins in predicting HPIs. There are also sequence-based comparative methods that consider structure [48, 55, 56, 61, 62, 66-70]; interologs (interacting homologs/conserved interactions) [71, 72]; and transcriptome data [73]. Available structure-based techniques often depend on global structural similarity rather than interface mimicry [55, 69]. One method combines interface data with sequence homology and gene expression, but the predicted interacting host and pathogenic proteins should satisfy a minimum of 80% sequence identity over at least 50% of template host PPI complexes [66]. To the best of our knowledge, none of the current approaches utilizes solely interface structures to model HPIs, except our recently developed interface-based method [74].
It has been suggested that the existing interface structures in PDB are diverse enough to cover majority of the endogenous PPIs [75-78] and hence success of template-based approaches to model endogenous PPIs is high [15] and expected to increase even more with the increase in the number of resolved PPI 3D structures [79] and advances in computational biology. Exogenous interactions are underrepresented in the PDB: there are not many exogenous interfaces. Since exogenous interfaces hijack endogenous ones, available endogenous and exogenous interfaces may represent most the structural host-pathogen interface space (see Note 2).
2. Methods
2.1. Modeling HPIs
Here, we review the first and to date only computational approach that utilizes solely interface mimicry to predict putative HPIs and their 3D structures as complexes [74]. Local structural resemblance is sufficient; there is no need for sequence similarity. This approach reveals not only targets of pathogenic proteins and how they interact, but also the host endogenous PPIs which may be disrupted by these potential HPIs. Figure 1 displays our workflow. Generally, the interacting protein partners are known from docking studies and the main purpose is to discern how they interact structurally. Therefore, inputs of the docking algorithms are structures of the two monomeric target proteins to be docked to each other. However, when dealing with HPIs, the main aim is to identify the interacting partners, as well as how they interact. Normally, the pathogenic proteins (one of the targets in a docking study) are known but not their partners in the host (second target). Hence, before performing docking, we need to identify those potential host interactors.
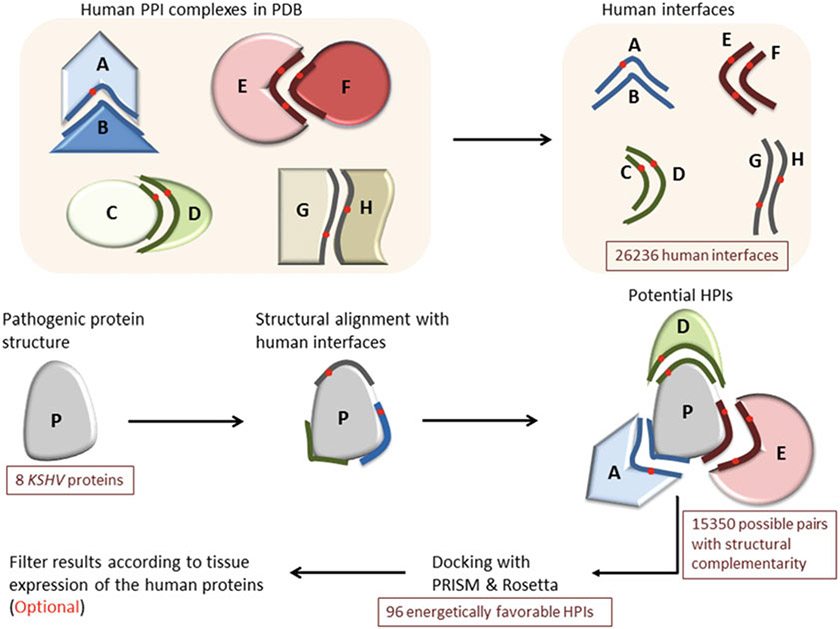
Fig. 1.
Workflow of our interface-based HPI modeling approach. In the first step, we extract human interfaces from the PDB. Then, we obtain the structures of the pathogenic proteins from the PDB. Before docking, we need to identify the potential HPI pairs since docking programs require two target proteins. To do that, we structurally align the pathogenic proteins with the human interfaces in our template set. If the pathogenic protein is aligned with the B-side of the interface, it can interact with the complementary A-side. After determining potential HPI pairs, we perform docking of these pairs with PRISM [81-84] and Rosetta (local refinement) [85-87] to select the energetically favorable ones. We further assess the likelihood that the HPI models take place in the cell based on the percent match of the interface residues with the template interface and probability of the template interface being a real biological interface. In the final optional step, we filter our energetically favorable HPI results according to tissue expression of the human proteins by checking whether the interactors of the pathogenic proteins are expressed in the same tissue where the pathogen resides
To accomplish this, we generate all known human interfaces— including endogenous and exogenous—in the PiFace interface database, as described in [14]. Each interface has two chains (partners/sides). There are 26,236 human interfaces in our template set. Then, we structurally align these interfaces with the pathogenic proteins by MultiProt [80]. The structural alignment thresholds for the number of matching interface residues and the hot spots follow the PRISM algorithm [81-84]. If the pathogenic protein is aligned with one side of the human interface, it may interact with the complementary side. Thus, the pathogenic protein can compete with the first side of the interface—with which it is structurally aligned—to bind to the second side, thereby abrogating the endogenous binary interaction in the template PPI (Fig. 1). Structural complementarity does not necessarily guarantee chemical complementarity and favorable interaction energy. For instance, 8 KSHV proteins aligned with 15,350 interfaces, but only 96 of them are energetically favorable. So, after detection of the potential partners in humans with structural complementarity, we check whether these potential HPI pairs have favorable interaction energy. To do that we perform docking with two programs: PRISM [81-84] and Rosetta (local refinement) [85-87]. We take HPIs as energetically favorable only if their Rosetta interface scores (I_sc) are below −5 and total energy scores are below zero. We also calculate Rosetta I_sc for the endogenous template PPIs and compare them with those of modeled HPIs to determine whether the pathogenic protein will outcompete the endogenous partner to bind to a target host protein with a higher affinity. For some template PPIs, Rosetta gives extremely low unrealistic I_sc, due to intermolecular disulfide bonds. To correct this, we calculate Rosetta I_sc with both including and disregarding the disulfide bonds. We consider the HPIs as favorable interactions if they have I_sc below −5 with both Rosetta scorings. Note that Rosetta I_sc does not have units nor reflects the real binding free energy. It only gives an idea whether an interaction is favorable or not.
To further evaluate the likelihoods of our HPI models, we calculate the “percent match” of the interfaces by taking the ratio of the number of interface residues that are aligned with the pathogenic protein to the number of interface residues in the endogenous template PPI. Each template interface is assigned with a weight based on the size of the endogenous template interface such that larger interfaces have higher weights. If the template interfaces have less than 30 residues (n < 30), the weight is 0.5; if 30 < n < 50, weight is 1; if 50 < n < 80 weight is 1.5; and if n > 80 (very large interface), the weight is 2. Score1 given in Table 1 is the product of the interface percent match and the corresponding interface weight.
Table 1.
HPIs for KSHV proteins
|
KSHV protein |
KSHV protein PDB |
Human protein |
Human protein PDB |
Template interface |
I_sc of HPI |
I_sc of PPI |
# of residues aligned |
# of residues in template interface |
% Match |
Weight | Sc1 | Probability of template interface being a biological interface |
Sc2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| K4 | 2fhtA | CCL4 | 2x6lB | 2x6lBD | −8.86 | −9.53 | 29 | 35 | 82.9 | 1 | 82.9 | 0.9 | 74.6 |
| K4 | 2fhtA | CXCR4 | 2k03D | 2k03CD | −6.76 | −11.26 | 25 | 55 | 45.5 | 1.5 | 68.2 | 0.48 | 32.7 |
| K6 | 1zxtA | CCL5 | 1u4lB | 1u4lAB | −8.90 | −11.30 | 26 | 34 | 76.5 | 1 | 76.5 | 0.77 | 58.9 |
| vCyclin | 1g3nC | CDK4 | 3g33A | 3g33AD | −6.07 | −8.24 | 39 | 51 | 76.5 | 1.5 | 114.7 | 0.98 | 112.4 |
| vCyclin | 1g3nC | CDK2 | 1w98A | 1w98AB | −5.41 | −13.69 | 48 | 94 | 51.1 | 2 | 102.1 | 1 | 102.1 |
| vIL6 | 1i1rB | IL12B | 3duhB | 3duhBD | −6.83 | −13.47 | 26 | 50 | 52.0 | 1.5 | 78.0 | 0.9 | 70.2 |
| vIL6 | 1i1rB | INAR1 | 3se4A | 3se4AB | −6.08 | −11.52 | 26 | 53 | 49.1 | 1.5 | 73.6 | 0.91 | 67.0 |
| vIRF1 | 4hlxA | UBP21 | 3i3tG | 3i3tGH | −5.88 | −16.50 | 18 | 51 | 35.3 | 1.5 | 52.9 | 0.97 | 51.4 |
| vFLIP | 3cl3A | TNR6 | 3ezqI | 3ezqIJ | −5.57 | −11.04 | 19 | 54 | 35.2 | 1.5 | 52.8 | 0.11 | 5.8 |
| vBCL2 | 1k3kA | ITA2B | 2vdkA | 2vdkAB | −5.19 | −13.52 | 20 | 63 | 31.7 | 1.5 | 47.6 | 1 | 47.6 |
I_sc refers to Rosetta interface score, where we ignored disulfide bonds. If the I_sc of modeled HPI is lower than I_sc of template PPI, it means that the pathogenic protein may have higher affinity to target protein than the endogenous partner of the target
We employ the EPPIC (Evolutionary Protein-Protein Interface Classifier) [88], to evaluate whether the template interfaces are real biological interfaces or crystal artifacts. The EPPIC server gives the probability of a particular interface to be biological. Score2 in Table 1 is the product of Score1 and the probability of being a biological interface. The higher the Score2, the more confidence we have that a particular HPI model would take place in the cell, as they are better mimics of real biological endogenous interfaces (see Note 3).
Finally, with an optional step, the results can be filtered according to tissue expression, checking whether the host partners of the pathogenic proteins are expressed in the same tissue where the pathogen resides. We take the tissue expression data from the Human Protein Atlas, which includes 19,709 human proteins, mapping to 7106 human PDBs [89, 90]. If the pathogen is a bacterial species, it resides in only certain tissues. For instance, Helicobacter pylori is mainly in the stomach and gastrointestinal tract, making it reasonable to focus on human proteins that are expressed in these tissues. However, if the pathogen is a virus, it can infect several different—if not all—tissues. Therefore, filtering according to tissue expression is an optional step depending on the pathogen type (see Note 4).
2.2. Constructing the Structural Superorganism Network
As we have the complex (bound) structures of the predicted HPIs, it is possible to construct the structural interspecies interaction network. Our template set serves as the human endogenous binary interactions. 26,236 interfaces map to 3366 distinct human PPIs. The predicted HPIs serve as exogenous interactions. So, all pairwise interactions in the structural network will have structures as complexes. The topological features of the resulting superorganism network can be calculated by the NetworkAnalyzer [91] application in Cytoscape [92]. Functional annotation of pathogenic targets in the host can be performed by DAVID [93, 94].
To compare the pathogen of interest with other bacteria/viruses, we can also build the structural interspecies network for all known HPIs in PDB. There are 299 HPIs in PDB between human and different bacterial, yeast, and viral species.
2.3. Case Study
Our interface-based HPI modeling method was successfully applied to H. pylori before and can be applied to any commensal or pathogenic microorganism. As a case study to illustrate the utility of the concept, here we applied it to KSHV, infection of which is associated with a blood/lymph vessel cancer—Kaposi’s sarcoma— and lymphoma [95]. We modeled its HPIs and constructed its structural superorganism network. We analyzed eight KSHV proteins, vCyclin, vFLIP, vBCL2, vIL6, vIRF1, vIRF2, and viral chemokines (K4 and K6). We found 96 putative HPIs. All our HPI models have 3D structures as complexes (see Note 5). Table 1 shows some examples from these 96 HPIs and Table 2 displays the human PPIs that are potentially disrupted by these HPIs.
Table 2.
Potentially disrupted endogenous host PPIs due to predicted KSHV HPIs
| KSHV protein | Human PPI disrupted by KSHV protein | PDB for the human PPI disrupted |
|---|---|---|
| K4 | CCL4-CCL4 | 2x6lBD |
| K4 | CXCR4-SDF1 | 2k03CD |
| K6 | CCL5-CCL5 | 1u4lAB |
| vCyclin | CDK4-CCND3 | 3g33AD |
| vCyclin | CDK2-CCNE1 | 1w98AB |
| vIL6 | IL12B-IL23A | 3duhBD |
| vIL6 | INAR1-IFNW1 | 3se4AB |
| vIRF1 | UBP21-RL40 | 3i3tGH |
| vFLIP | TNR6-FADD | 3ezqIJ |
| vBCL2 | ITA2B-ITB3 | 2vdkAB |
Our HPI candidates may elucidate the roles of KSHV in modulation of host signaling and contribution to malignant transformation. For instance, we found that KSHV chemokines and cytokines, like K4, K6, and vIL6, target many human chemokine and cytokine receptors (Fig. 2). Signaling through the cytokine and chemokine receptors is critical for T-cell recruitment to the infected host tissue to eradicate the pathogens and for regulation of their activation and differentiation [96]. Blockage of these pathways by the KSHV proteins may underlie the molecular mechanisms of evading the immune system and persistence of infection. We also found that vCyclin interferes with several CDKs (Fig. 2), thereby disrupting normal host cell cycle regulation, which may contribute to aberrant proliferation in malignant transformation.
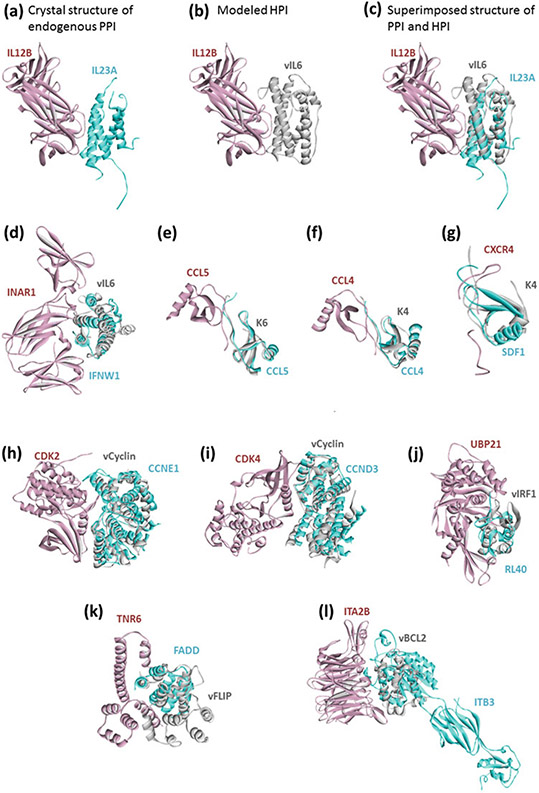
Fig. 2.
KSHV proteins mimic the human protein-protein interfaces, blocking human PPIs. (a) Endogenous human PPI between IL12B and IL23A. (b) Our HPI model between vIL6 and IL12B. (c) Superimposed view of PPI and HPI shows that vIL6 almost perfectly mimics the interface on IL23A to bind to IL12B. (d) through (l) also show the superimposed structures of endogenous human PPIs and modeled HPIs. Human proteins are shown in cyan and pink; and KSHV proteins are shown in gray. Gray proteins bind to pink proteins by hijacking the interface on cyan proteins (only the interface similarity is enough, no need for global structural similarity). Thus, they may block the pink-cyan protein interactions
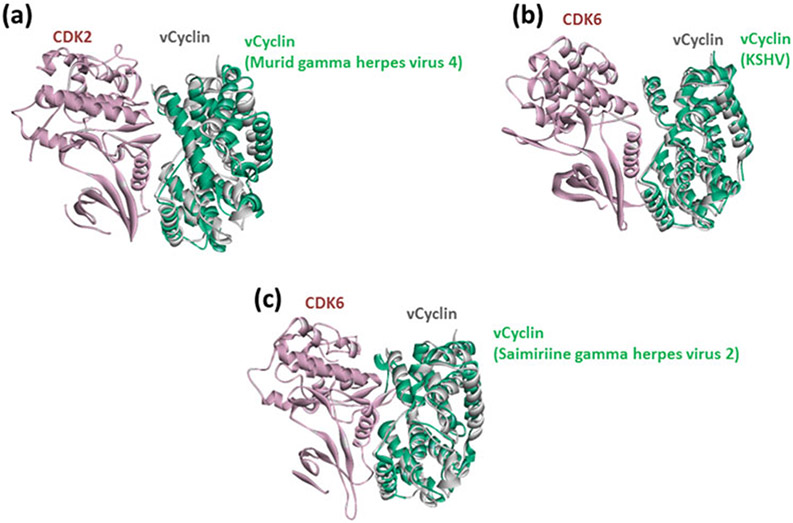
In addition to mimicked endogenous interfaces, hijacked exogenous interfaces can also be identified through our approach. A given pathogenic protein can mimic both human and pathogenic proteins from other species. For instance, we found that KSHV vCyclin mimics other viral vCyclin proteins to target human CDKs (Fig. 3).
Fig. 3.
KSHV proteins mimic not only host interactions, but also other HPIs from other species (a), (b), and (c). Figures show the superimposed structures of our HPI models for KSHV with the known exogenous interactions with proteins from other species. Pink proteins are from human, greens are proteins from other pathogens, and gray proteins are KSHV proteins. Gray proteins bind to pink proteins by hijacking the interfaces on green proteins
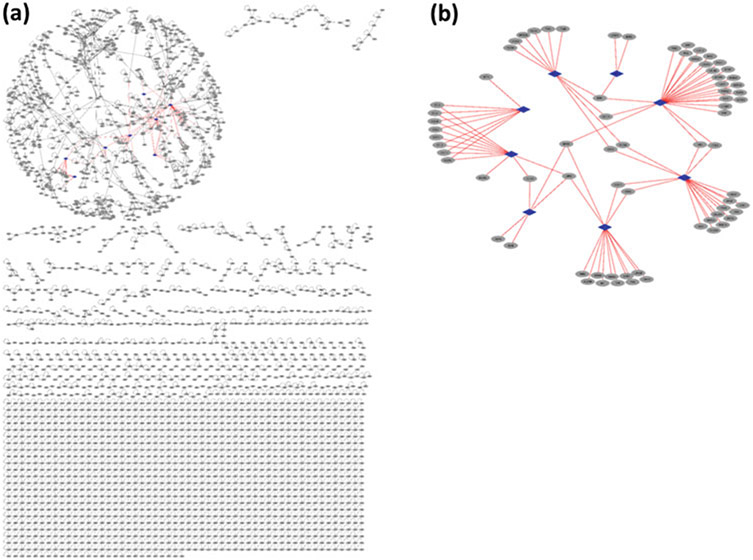
We also constructed the structural superorganism network between human and KSHV (Fig. 4). The endogenous human PPIs are template PPIs and the exogenous virus-human interactions are HPI models. There are 3366 human PPIs and 96 HPIs in this network. Our results indicate that KSHV proteins can potentially target the highly connected part of the network and hub proteins, like CDK2 in the human PPI network. Hub proteins are critical to many cellular functions, establishing pathway cross talk. It is an ingenious pathogen strategy, since by attacking only a single protein they can interfere with several pathways. Functional annotation of the KSHV-targeted human proteins is enriched in 17 KEGG pathways (Table 3). Among the highly enriched, there are cytokine and chemokine signaling, and viral carcinogenesis pathways.
Fig. 4.
Structural superorganism network for KSHV and human, where all binary interactions have structures as complexes. Endogenous human interactions (black edges) are obtained from crystal structures in PDB (template interface set), where human proteins are shown as gray circular nodes. Exogenous interactions (red edges) are our HPI models for 8 KSHV proteins, where viral proteins are shown as blue diamond nodes. (a) KSHV proteins target the highly connected part of the human PPI network. (b) Structural HPI network without the endogenous template interactions. Most targets of individual KSHV proteins are distinct, but some are shared across different KSHVproteins
Table 3.
| KEGG pathways | Number of genes enriched |
% | P value | KSHV-targeted human proteins |
|---|---|---|---|---|
| Cytokine-cytokine receptor interaction | 10 | 13.9 | 7.20E–05 | CCL3, CCL2, CCL13, TNR6, CCL4, ACVR1, CCL5, CXCR4, INAR1, IL12B |
| Chemokine signaling pathway | 9 | 12.5 | 9.80E–05 | RHOA, CCL3, CCL2, CCL13, CCL4, CCL5, JAK2, CCL14, CXCR4 |
| Herpes simplex infection | 8 | 11.1 | 5.60E–04 | CCL2, C1QBP, TNR6, CDK2, CCL5, JAK2, INAR1, IL12B |
| Measles | 7 | 9.7 | 6.10E–04 | TNR6, CCND3, CDK4, CDK2, JAK2, INAR1, IL12B |
| p53 signaling pathway | 5 | 6.9 | 1.90E–03 | TNR6, CCND3, CASP9, CDK4, CDK2 |
| Influenza A | 7 | 9.7 | 2.40E–03 | CCL2, TNR6, CASP9, CCL5, JAK2, INAR1, IL12B |
| Pathways in cancer | 10 | 13.9 | 3.50E–03 | RHOA, ITA2B, FGFR2, TNR6, CASP9, CDK4, CDK2, CXCR4, ARHGB, BMP2 |
| Hepatitis B | 6 | 8.3 | 5.70E–03 | CCNA2, TNR6, CASP9, CDK4, CDK2, INAR1 |
| Chagas disease (American trypanosomiasis) | 5 | 6.9 | 9.20E–03 | CCL3, CCL2, TNR6, CCL5, IL12B |
| Toll-like receptor signaling pathway | 5 | 6.9 | 9.80E–03 | CCL3, CCL4, CCL5, INAR1, IL12B |
| PI3K-Akt signaling pathway | 8 | 11.1 | 1.90E–02 | ITA2B, FGFR2, CCND3, CASP9, CDK4, CDK2, JAK2, INAR1 |
| African trypanosomiasis | 3 | 4.2 | 2.80E–02 | TNR6, HBA, IL12B |
| Small-cell lung cancer | 4 | 5.6 | 3.00E–02 | ITA2B, CASP9, CDK4, CDK2 |
| Glutathione metabolism | 3 | 4.2 | 6.20E–02 | GSTA4, GSTP1, GSTM2 |
| Cell cycle | 4 | 5.6 | 7.60E–02 | CCNA2, CCND3, CDK4, CDK2 |
| Viral carcinogenesis | 5 | 6.9 | 8.00E–02 | CCNA2, RHOA, CCND3, CDK4, CDK2 |
| Signaling pathways regulating pluripotency of stem cells | 4 | 5.6 | 1.00E–01 | FGFR2, ACVR1, JAK2, BMP2 |
3. Concluding Remarks
Insight into mechanisms of infectious diseases and pathogen-driven cancers at the molecular level is limited. Identification of novel HPIs and their atomistic details may illuminate how virulence factors modulate host signaling, and stimulate innovative therapeutics. Large-scale detection of HPIs will rely on computational techniques in the near future due to current limitations of experimental methodologies. Most computational approaches rely on sequence homology which constrains the application of these tools to pathogenic proteins that have no sequence homologs in human. Interface architectures are conserved within and across species regardless of the entire sequence and the structure of the proteins. Here we reviewed the first and only available interface-based method to uncover novel HPIs and their complex 3D structures. This approach predicts not only the HPIs, but also the potentially disrupted endogenous human PPIs. It can be applied to any microbial organisms, including commensals and pathogens.
Acknowledgments
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract number HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. This research was supported (in part) by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. This study utilized the high-performance computational capabilities of the Biowulf PC/Linux cluster at the National Institutes of Health (NIH), Bethesda, MD (http://biowulf.nih.gov).
Footnotes
Our approach is based on the reasonable assumption that pathogenic proteins may alter host signaling. However, interactions through metabolites and small molecules also have roles in modulation of the host responses. Moreover, interaction of a particular pathogen with other microbial species in the microbiota and different combination of bacterial species also affects the overall response.
Some of the limitations are as follows: coverage of endogenous human PPIs is low; available endogenous protein structures are biased toward permanent, not transient, interactions; disordered proteins are underrepresented in the PDB; and most pathogenic proteins lack crystal structures.
Both experimental and computational methods have false positives with varying rates. Although HPIs predicted here may have false positives, we cannot calculate the exact false-positive rate due to limited experimental HPI data. We tried to minimize the error rates by calculating the percent match of the HPI models with the corresponding template PPI and incorporating the probability of template interfaces being real biological interfaces. Predicted models should be tested by experiments. Computational screening of big data can provide possible leads to experiments guiding functional characterization while avoiding testing millions of possible binary combinations of host and pathogenic proteins.
In addition to filtering by tissue expression, HPI models can also be filtered by subcellular localization of the host proteins. For instance, if the pathogenic protein is found in the cytoplasm of the host cell, then it cannot interact with the host nuclear proteins. Since the large-scale subcellular localization data for all proteins are not available, it is a choice of the researchers to do so.
Proteins often assemble into multi-protein complexes. Modeling only pairwise interactions between host and pathogenic proteins may not be sufficient.
References
- 1.Durmus S, Cakir T, Ozgur A, Guthke R (2015) A review on computational systems biology of pathogen-host interactions. Front Microbiol 6:235. 10.3389/fmicb.2015.00235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stebbins CE, Galan JE (2001) Structural mimicry in bacterial virulence. Nature 412 (6848):701–705. 10.1038/35089000 [DOI] [PubMed] [Google Scholar]
- 3.Sal-Man N, Biemans-Oldehinkel E, Finlay BB (2009) Structural microengineers: pathogenic Escherichia coli redesigns the actin cytoskeleton in host cells. Structure 17(1):15–19. 10.1016/j.str.2008.12.001 [DOI] [PubMed] [Google Scholar]
- 4.Kahn RA, Fu H, Roy CR (2002) Cellular hijacking: a common strategy for microbial infection. Trends Biochem Sci 27 (6):308–314. 10.1016/S0968-0004(02)02108-4 [DOI] [PubMed] [Google Scholar]
- 5.Finlay BB, McFadden G (2006) Antiimmunology: evasion of the host immune system by bacterial and viral pathogens. Cell 124 (4):767–782. 10.1016/j.cell.2006.01.034 [DOI] [PubMed] [Google Scholar]
- 6.Moody CA, Laimins LA (2010) Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer 10(8):550–560. 10.1038/nrc2886 [DOI] [PubMed] [Google Scholar]
- 7.Filippova M, Song H, Connolly JL, Dermody TS, Duerksen-Hughes PJ (2002) The human papillomavirus 16 E6 protein binds to tumor necrosis factor (TNF) R1 and protects cells from TNF-induced apoptosis. J Biol Chem 277(24):21730–21739. 10.1074/jbc.M200113200 [DOI] [PubMed] [Google Scholar]
- 8.Shirin H, Sordillo EM, Kolevska TK, Hibshoosh H, Kawabata Y, Oh SH, Kuebler JF, Delohery T, Weghorst CM, Weinstein IB, Moss SF (2000) Chronic helicobacter pylori infection induces an apoptosis-resistant phenotype associated with decreased expression of p27(kip1). Infect Immun 68(9):5321–5328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guven-Maiorov E, Tsai CJ, Nussinov R (2016) Pathogen mimicry of host protein-protein interfaces modulates immunity. Semin Cell Dev Biol 58:136–145. 10.1016/j.semcdb.2016.06.004 [DOI] [PubMed] [Google Scholar]
- 10.Tsai CJ, Lin SL, Wolfson HJ, Nussinov R (1996) A dataset of protein-protein interfaces generated with a sequence-order-independent comparison technique. J Mol Biol 260 (4):604–620. 10.1006/jmbi.1996.0424 [DOI] [PubMed] [Google Scholar]
- 11.Tsai CJ, Lin SL, Wolfson HJ, Nussinov R (1996) Protein-protein interfaces: architectures and interactions in protein-protein interfaces and in protein cores. Their similarities and differences. Crit Rev Biochem Mol Biol 31 (2):127–152. 10.3109/10409239609106582 [DOI] [PubMed] [Google Scholar]
- 12.Keskin O, Nussinov R (2005) Favorable scaffolds: proteins with different sequence, structure and function may associate in similar ways. Protein Eng Des Sel 18(1):11–24. 10.1093/protein/gzh095 [DOI] [PubMed] [Google Scholar]
- 13.Keskin O, Nussinov R (2007) Similar binding sites and different partners: implications to shared proteins in cellular pathways. Structure 15(3):341–354. 10.1016/j.str.2007.01.007 [DOI] [PubMed] [Google Scholar]
- 14.Cukuroglu E, Gursoy A, Nussinov R, Keskin O (2014) Non-redundant unique interface structures as templates for modeling protein interactions. PLoS One 9(1):e86738. 10.1371/journal.pone.0086738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muratcioglu S, Guven-Maiorov E, Keskin O, Gursoy A (2015) Advances in template-based protein docking by utilizing interfaces towards completing structural interactome. Curr Opin Struct Biol 35:87–92. 10.1016/j.sbi.2015.10.001 [DOI] [PubMed] [Google Scholar]
- 16.Franzosa EA, Garamszegi S, Xia Y (2012) Toward a three-dimensional view of protein networks between species. Front Microbiol 3:428. 10.3389/fmicb.2012.00428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franzosa EA, Xia Y (2011) Structural principles within the human-virus protein-protein interaction network. Proc Natl Acad Sci U S A 108(26):10538–10543. 10.1073/pnas.1101440108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guven-Maiorov E, Tsai CJ, Nussinov R (2017) Structural host-microbiota interaction networks. PLoS Comput Biol 13(10):e1005579. 10.1371/journal.pcbi.1005579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhavsar AP, Guttman JA, Finlay BB (2007) Manipulation of host-cell pathways by bacterial pathogens. Nature 449(7164):827–834. 10.1038/nature06247 [DOI] [PubMed] [Google Scholar]
- 20.Uetz P, Dong YA, Zeretzke C, Atzler C, Baiker A, Berger B, Rajagopala SV, Roupelieva M, Rose D, Fossum E, Haas J (2006) Herpesviral protein networks and their interaction with the human proteome. Science 311(5758):239–242. 10.1126/science.1116804 [DOI] [PubMed] [Google Scholar]
- 21.von Schwedler UK, Stuchell M, Muller B, Ward DM, Chung HY, Morita E, Wang HE, Davis T, He GP, Cimbora DM, Scott A, Krausslich HG, Kaplan J, Morham SG, Sundquist WI (2003) The protein network of HIV budding. Cell 114(6):701–713 [DOI] [PubMed] [Google Scholar]
- 22.Calderwood MA, Venkatesan K, Xing L, Chase MR, Vazquez A, Holthaus AM, Ewence AE, Li N, Hirozane-Kishikawa T, Hill DE, Vidal M, Kieff E, Johannsen E (2007) Epstein-Barr virus and virus human protein interaction maps. Proc Natl Acad Sci U S A 104 (18):7606–7611. 10.1073/pnas.0702332104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Chassey B, Navratil V, Tafforeau L, Hiet MS, Aublin-Gex A, Agaugue S, Meiffren G, Pradezynski F, Faria BF, Chantier T, Le Breton M, Pellet J, Davoust N, Mangeot PE, Chaboud A, Penin F, Jacob Y, Vidalain PO, Vidal M, Andre P, Rabourdin-Combe C, Lotteau V (2008) Hepatitis C virus infection protein network. Mol Syst Biol 4:230. 10.1038/msb.2008.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shapira SD, Gat-Viks I, Shum BO, Dricot A, de Grace MM, Wu L, Gupta PB, Hao T, Silver SJ, Root DE, Hill DE, Regev A, Hacohen N (2009) A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell 139(7):1255–1267. 10.1016/j.cell.2009.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L, Villa NY, Rahman MM, Smallwood S, Shattuck D, Neff C, Dufford M, Lanchbury JS, Labaer J, McFadden G (2009) Analysis of vaccinia virus-host protein-protein interactions: validations of yeast two-hybrid screenings. J Proteome Res 8(9):4311–4318. 10.1021/pr900491n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khadka S, Vangeloff AD, Zhang C, Siddavatam P, Heaton NS, Wang L, Sengupta R, Sahasrabudhe S, Randall G, Gribskov M, Kuhn RJ, Perera R, LaCount DJ (2011) A physical interaction network of dengue virus and human proteins. Mol Cell Proteomics 10(12):M111.012187. 10.1074/mcp.M111.012187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jager S, Cimermancic P, Gulbahce N, Johnson JR, McGovern KE, Clarke SC, Shales M, Mercenne G, Pache L, Li K, Hernandez H, Jang GM, Roth SL, Akiva E, Marlett J, Stephens M, D’Orso I, Fernandes J, Fahey M, Mahon C, O’Donoghue AJ, Todorovic A, Morris JH, Maltby DA, Alber T, Cagney G, Bushman FD, Young JA, Chanda SK, Sundquist WI, Kortemme T, Hernandez RD, Craik CS, Burlingame A, Sali A, Frankel AD, Krogan NJ (2011) Global landscape of HIV-human protein complexes. Nature 481 (7381):365–370. 10.1038/nature10719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pichlmair A, Kandasamy K, Alvisi G, Mulhern O, Sacco R, Habjan M, Binder M, Stefanovic A, Eberle CA, Goncalves A, Burckstummer T, Muller AC, Fauster A, Holze C, Lindsten K, Goodbourn S, Kochs G, Weber F, Bartenschlager R, Bowie AG, Bennett KL, Colinge J, Superti-Furga G (2012) Viral immune modulators perturb the human molecular network by common and unique strategies. Nature 487 (7408):486–490. 10.1038/nature11289 [DOI] [PubMed] [Google Scholar]
- 29.Rozenblatt-Rosen O, Deo RC, Padi M, Adelmant G, Calderwood MA, Rolland T, Grace M, Dricot A, Askenazi M, Tavares M, Pevzner SJ, Abderazzaq F, Byrdsong D, Carvunis AR, Chen AA, Cheng J, Correll M, Duarte M, Fan C, Feltkamp MC, Ficarro SB, Franchi R, Garg BK, Gulbahce N, Hao T, Holthaus AM, James R, Korkhin A, Litovchick L, Mar JC, Pak TR, Rabello S, Rubio R, Shen Y, Singh S, Spangle JM, Tasan M, Wanamaker S, Webber JT, Roecklein-Canfield J, Johannsen E, Barabasi AL, Beroukhim R, Kieff E, Cusick ME, Hill DE, Munger K, Marto JA, Quackenbush J, Roth FP, DeCaprio JA, Vidal M (2012) Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nature 487(7408):491–495. 10.1038/nature11288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guven Maiorov E, Keskin O, Gursoy A, Nussinov R (2013) The structural network of inflammation and cancer: merits and challenges. Semin Cancer Biol 23(4):243–251. 10.1016/j.semcancer.2013.05.003 [DOI] [PubMed] [Google Scholar]
- 31.Guven-Maiorov E, Keskin O, Gursoy A, VanWaes C, Chen Z, Tsai CJ, Nussinov R (2015) The architecture of the TIR domain signalosome in the toll-like Receptor-4 signaling pathway. Sci Rep 5:13128. 10.1038/srep13128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guven-Maiorov E, Keskin O, Gursoy A, Nussinov R (2015) A structural view of negative regulation of the toll-like receptor-mediated inflammatory pathway. Biophys J 109 (6):1214–1226. 10.1016/j.bpj.2015.06.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Acuner-Ozbabacan ES, Engin BH, Guven-Maiorov E, Kuzu G, Muratcioglu S, Baspinar A, Chen Z, Van Waes C, Gursoy A, Keskin O, Nussinov R (2014) The structural network of Interleukin-10 and its implications in inflammation and cancer. BMC Genomics 15(Suppl 4):S2. 10.1186/1471-2164-15-S4-S2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nourani E, Khunjush F, Durmus S (2015) Computational approaches for prediction of pathogen-host protein-protein interactions. Front Microbiol 6:94. 10.3389/fmicb.2015.00094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brito AF, Pinney JW (2017) Protein-protein interactions in virus-host systems. Front Microbiol 8:1557. 10.3389/fmicb.2017.01557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Durmus Tekir S, Cakir T, Ardic E, Sayilirbas AS, Konuk G, Konuk M, Sariyer H, Ugurlu A, Karadeniz I, Ozgur A, Sevilgen FE, Ulgen KO (2013) PHISTO: pathogen-host interaction search tool. Bioinformatics 29 (10):1357–1358. 10.1093/bioinformatics/btt137 [DOI] [PubMed] [Google Scholar]
- 37.Kumar R, Nanduri B (2010) HPIDB--a unified resource for host-pathogen interactions. BMC Bioinformatics 11(Suppl 6):S16. 10.1186/1471-2105-11-S6-S16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vialas V, Nogales-Cadenas R, Nombela C, Pascual-Montano A, Gil C (2009) Proteopathogen, a protein database for studying Candida albicans--host interaction. Proteomics 9(20):4664–4668. 10.1002/pmic.200900023 [DOI] [PubMed] [Google Scholar]
- 39.Wattam AR, Abraham D, Dalay O, Disz TL, Driscoll T, Gabbard JL, Gillespie JJ, Gough R, Hix D, Kenyon R, Machi D, Mao C, Nordberg EK, Olson R, Overbeek R, Pusch GD, Shukla M, Schulman J, Stevens RL, Sullivan DE, Vonstein V, Warren A, Will R, Wilson MJ, Yoo HS, Zhang C, Zhang Y, Sobral BW (2014) PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res 42(Database issue):D581–D591. 10.1093/nar/gkt1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Urban M, Pant R, Raghunath A, Irvine AG, Pedro H, Hammond-Kosack KE (2015) The Pathogen-Host Interactions database (PHI-base): additions and future developments. Nucleic Acids Res 43(Database issue): D645–D655. 10.1093/nar/gku1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiang Z, Tian Y, He Y (2007) PHIDIAS: a pathogen-host interaction data integration and analysis system. Genome Biol 8(7):R150. 10.1186/gb-2007-8-7-r150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bleves S, Dunger I, Walter MC, Frangoulidis D, Kastenmuller G, Voulhoux R, Ruepp A (2014) HoPaCI-DB: host-Pseudomonas and Coxiella interaction database. Nucleic Acids Res 42(Database issue): D671–D676. 10.1093/nar/gkt925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guirimand T, Delmotte S, Navratil V (2015) VirHostNet 2.0: surfing on the web of virus/host molecular interactions data. Nucleic Acids Res 43(Database issue):D583–D587. 10.1093/nar/gku1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y, Wang C, Miao Z, Bi X, Wu D, Jin N, Wang L, Wu H, Qian K, Li C, Zhang T, Zhang C, Yi Y, Lai H, Hu Y, Cheng L, Leung KS, Li X, Zhang F, Li K, Li X, Wang D (2015) ViRBase: a resource for virus-host ncRNA-associated interactions. Nucleic Acids Res 43 (Database issue):D578–D582. 10.1093/nar/gku903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Calderone A, Licata L, Cesareni G (2015) VirusMentha: a new resource for virus-host protein interactions. Nucleic Acids Res 43 (Database issue):D588–D592. 10.1093/nar/gku830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kwofie SK, Schaefer U, Sundararajan VS, Bajic VB, Christoffels A (2011) HCVpro: hepatitis C virus protein interaction database. Infect Genet Evol 11(8):1971–1977. 10.1016/j.meegid.2011.09.001 [DOI] [PubMed] [Google Scholar]
- 47.Arnold R, Boonen K, Sun MG, Kim PM (2012) Computational analysis of interactomes: current and future perspectives for bioinformatics approaches to model the host-pathogen interaction space. Methods 57 (4):508–518. 10.1016/j.ymeth.2012.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Doolittle JM, Gomez SM (2011) Mapping protein interactions between dengue virus and its human and insect hosts. PLoS Negl Trop Dis 5(2):e954. 10.1371/journal.pntd.0000954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tyagi N, Krishnadev O, Srinivasan N (2009) Prediction of protein-protein interactions between Helicobacter pylori and a human host. Mol BioSyst 5(12):1630–1635. 10.1039/b906543c [DOI] [PubMed] [Google Scholar]
- 50.Xu Q, Xiang EW, Yang Q (2011) Transferring network topological knowledge for predicting protein-protein interactions. Proteomics 11 (19):3818–3825. 10.1002/pmic.201100146 [DOI] [PubMed] [Google Scholar]
- 51.Remmele CW, Luther CH, Balkenhol J, Dandekar T, Muller T, Dittrich MT (2015) Integrated inference and evaluation of host-fungi interaction networks. Front Microbiol 6:764. 10.3389/fmicb.2015.00764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Evans P, Dampier W, Ungar L, Tozeren A (2009) Prediction of HIV-1 virus-host protein interactions using virus and host sequence motifs. BMC Med Genet 2:27. 10.1186/1755-8794-2-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang M, Su S, Bhatnagar RK, Hassett DJ, Lu LJ (2012) Prediction and analysis of the protein interactome in Pseudomonas aeruginosa to enable network-based drug target selection. PLoS One 7(7):e41202. 10.1371/journal.pone.0041202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huo T, Liu W, Guo Y, Yang C, Lin J, Rao Z (2015) Prediction of host – pathogen protein interactions between Mycobacterium tuberculosis and Homo sapiens using sequence motifs. BMC Bioinformatics 16:100. 10.1186/s12859-015-0535-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Doolittle JM, Gomez SM (2010) Structural similarity-based predictions of protein interactions between HIV-1 and Homo sapiens. Virol J 7:82. 10.1186/1743-422X-7-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de Chassey B, Meyniel-Schicklin L, Aublin-Gex A, Navratil V, Chantier T, Andre P, Lotteau V (2013) Structure homology and interaction redundancy for discovering virus-host protein interactions. EMBO Rep 14 (10):938–944. 10.1038/embor.2013.130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Petrenko P, Doxey AC (2015) mimicMe: a web server for prediction and analysis of host-like proteins in microbial pathogens. Bioinformatics 31(4):590–592. 10.1093/bioinformatics/btu681 [DOI] [PubMed] [Google Scholar]
- 58.Krishnadev O, Srinivasan N (2011) Prediction of protein-protein interactions between human host and a pathogen and its application to three pathogenic bacteria. Int J Biol Macromol 48 (4):613–619. 10.1016/j.ijbiomac.2011.01.030 [DOI] [PubMed] [Google Scholar]
- 59.Dyer MD, Murali TM, Sobral BW (2007) Computational prediction of host-pathogen protein-protein interactions. Bioinformatics 23(13):i159–i166. 10.1093/bioinformatics/btm208 [DOI] [PubMed] [Google Scholar]
- 60.Doxey AC, McConkey BJ (2013) Prediction of molecular mimicry candidates in human pathogenic bacteria. Virulence 4(6):453–466. 10.4161/viru.25180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mahajan G, Mande SC (2017) Using structural knowledge in the protein data bank to inform the search for potential host-microbe protein interactions in sequence space: application to Mycobacterium tuberculosis. BMC Bioinformatics 18(1):201. 10.1186/s12859-017-1550-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mariano R, Wuchty S (2017) Structure-based prediction of host-pathogen protein interactions. Curr Opin Struct Biol 44:119–124. 10.1016/j.sbi.2017.02.007 [DOI] [PubMed] [Google Scholar]
- 63.Becerra A, Bucheli VA, Moreno PA (2017) Prediction of virus-host protein-protein interactions mediated by short linear motifs. BMC Bioinformatics 18(1):163. 10.1186/s12859-017-1570-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jones KR, Whitmire JM, Merrell DS (2010) A tale of two toxins: helicobacter pylori CagA and VacA modulate host pathways that impact disease. Front Microbiol 1:115. 10.3389/fmicb.2010.00115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Manente L, Perna A, Buommino E, Altucci L, Lucariello A, Citro G, Baldi A, Iaquinto G, Tufano MA, De Luca A (2008) The Helicobacter pylori’s protein VacA has direct effects on the regulation of cell cycle and apoptosis in gastric epithelial cells. J Cell Physiol 214 (3):582–587. 10.1002/jcp.21242 [DOI] [PubMed] [Google Scholar]
- 66.Davis FP, Barkan DT, Eswar N, McKerrow JH, Sali A (2007) Host pathogen protein interactions predicted by comparative modeling. Protein Sci 16(12):2585–2596. 10.1110/ps.073228407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Drayman N, Glick Y, Ben-nun-shaul O, Zer H, Zlotnick A, Gerber D, Schueler-Furman O, Oppenheim A (2013) Pathogens use structural mimicry of native host ligands as a mechanism for host receptor engagement. Cell Host Microbe 14(1):63–73. 10.1016/j.chom.2013.05.005 [DOI] [PubMed] [Google Scholar]
- 68.Aloy P, Bottcher B, Ceulemans H, Leutwein C, Mellwig C, Fischer S, Gavin AC, Bork P, Superti-Furga G, Serrano L, Russell RB (2004) Structure-based assembly of protein complexes in yeast. Science 303 (5666):2026–2029. 10.1126/science.1092645 [DOI] [PubMed] [Google Scholar]
- 69.Rajasekharan S, Rana J, Gulati S, Sharma SK, Gupta V, Gupta S (2013) Predicting the host protein interactors of Chandipura virus using a structural similarity-based approach. Pathog Dis 69(1):29–35. 10.1111/2049-632X.12064 [DOI] [PubMed] [Google Scholar]
- 70.Zhang A, He L, Wang Y (2017) Prediction of GCRV virus-host protein interactome based on structural motif-domain interactions. BMC Bioinformatics 18(1):145. 10.1186/s12859-017-1500-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee SA, Chan CH, Tsai CH, Lai JM, Wang FS, Kao CY, Huang CY (2008) Ortholog-based protein-protein interaction prediction and its application to inter-species interactions. BMC Bioinformatics 9(Suppl 12):S11. 10.1186/1471-2105-9-S12-S11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Krishnadev O, Srinivasan N (2008) A data integration approach to predict host-pathogen protein-protein interactions: application to recognize protein interactions between human and a malarial parasite. In Silico Biol 8 (3–4):235–250 [PubMed] [Google Scholar]
- 73.Schulze S, Henkel SG, Driesch D, Guthke R, Linde J (2015) Computational prediction of molecular pathogen-host interactions based on dual transcriptome data. Front Microbiol 6:65. 10.3389/fmicb.2015.00065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guven-Maiorov E, Tsai CJ, Ma B, Nussinov R (2017) Prediction of host-pathogen interactions for helicobacter pylori by interface mimicry and implications to gastric cancer. J Mol Biol 429(24):3925–3941. 10.1016/j.jmb.2017.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang QC, Petrey D, Deng L, Qiang L, Shi Y, Thu CA, Bisikirska B, Lefebvre C, Accili D, Hunter T, Maniatis T, Califano A, Honig B (2012) Structure-based prediction of protein-protein interactions on a genome-wide scale. Nature 490(7421):556–560. 10.1038/nature11503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang QC, Petrey D, Norel R, Honig BH (2010) Protein interface conservation across structure space. Proc Natl Acad Sci U S A 107 (24):10896–10901. 10.1073/pnas.1005894107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gao M, Skolnick J (2010) Structural space of protein-protein interfaces is degenerate, close to complete, and highly connected. Proc Natl Acad Sci U S A 107(52):22517–22522. 10.1073/pnas.1012820107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kundrotas PJ, Zhu Z, Janin J, Vakser IA (2012) Templates are available to model nearly all complexes of structurally characterized proteins. Proc Natl Acad Sci U S A 109 (24):9438–9441. 10.1073/pnas.1200678109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Franzosa EA, Xia Y (2012) Structural models for host-pathogen protein-protein interactions: assessing coverage and bias. Pac Symp Biocomput:287–298 [PubMed] [Google Scholar]
- 80.Shatsky M, Nussinov R, Wolfson HJ (2004) A method for simultaneous alignment of multiple protein structures. Proteins 56(1):143–156. 10.1002/prot.10628 [DOI] [PubMed] [Google Scholar]
- 81.Tuncbag N, Gursoy A, Nussinov R, Keskin O (2011) Predicting protein-protein interactions on a proteome scale by matching evolutionary and structural similarities at interfaces using PRISM. Nat Protoc 6(9):1341–1354. 10.1038/nprot.2011.367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Keskin O, Nussinov R, Gursoy A (2008) PRISM: protein-protein interaction prediction by structural matching. Methods Mol Biol 484:505–521. 10.1007/978-1-59745-398-1_30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Baspinar A, Cukuroglu E, Nussinov R, Keskin O, Gursoy A (2014) PRISM: a web server and repository for prediction of protein-protein interactions and modeling their 3D complexes. Nucleic Acids Res 42 (Web Server issue):W285–W289. 10.1093/nar/gku397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ogmen U, Keskin O, Aytuna AS, Nussinov R, Gursoy A (2005) PRISM: protein interactions by structural matching. Nucleic Acids Res 33 (Web Server):W331–W336. 10.1093/nar/gki585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gray JJ, Moughon S, Wang C, Schueler-Furman O, Kuhlman B, Rohl CA, Baker D (2003) Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J Mol Biol 331 (1):281–299 [DOI] [PubMed] [Google Scholar]
- 86.Wang C, Schueler-Furman O, Baker D (2005) Improved side-chain modeling for protein-protein docking. Protein Sci 14 (5):1328–1339. 10.1110/ps.041222905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang C, Bradley P, Baker D (2007) Protein-protein docking with backbone flexibility. J Mol Biol 373(2):503–519. 10.1016/j.jmb.2007.07.050 [DOI] [PubMed] [Google Scholar]
- 88.Duarte JM, Srebniak A, Scharer MA, Capitani G (2012) Protein interface classification by evolutionary analysis. BMC Bioinformatics 13:334. 10.1186/1471-2105-13-334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Uhlen M, Bjorling E, Agaton C, Szigyarto CA, Amini B, Andersen E, Andersson AC, Angelidou P, Asplund A, Asplund C, Berglund L, Bergstrom K, Brumer H, Cerjan D, Ekstrom M, Elobeid A, Eriksson C, Fagerberg L, Falk R, Fall J, Forsberg M, Bjorklund MG, Gumbel K, Halimi A, Hallin I, Hamsten C, Hansson M, Hedhammar M, Hercules G, Kampf C, Larsson K, Lindskog M, Lodewyckx W, Lund J, Lundeberg J, Magnusson K, Malm E, Nilsson P, Odling J, Oksvold P, Olsson I, Oster E, Ottosson J, Paavilainen L, Persson A, Rimini R, Rockberg J, Runeson M, Sivertsson A, Skollermo A, Steen J, Stenvall M, Sterky F, Stromberg S, Sundberg M, Tegel H, Tourle S, Wahlund E, Walden A, Wan J, Wernerus H, Westberg J, Wester K, Wrethagen U, Xu LL, Hober S, Ponten F (2005)A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics 4(12):1920–1932. 10.1074/mcp.M500279-MCP200 [DOI] [PubMed] [Google Scholar]
- 90.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Ponten F (2015) Proteomics. Tissue-based map of the human proteome. Science 347(6220):1260419. 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- 91.Yang H, Ke Y, Wang J, Tan Y, Myeni SK, Li D, Shi Q, Yan Y, Chen H, Guo Z, Yuan Y, Yang X, Yang R, Du Z (2011) Insight into bacterial virulence mechanisms against host immune response via the Yersinia pestis-human protein-protein interaction network. Infect Immun 79(11):4413–4424. 10.1128/IAI.05622-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11):2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Huang DW, Sherman BT, Lempicki RA (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37(1):1–13. 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Huang DW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4(1):44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 95.Dissinger NJ, Damania B (2016) Recent advances in understanding Kaposi’s sarcoma-associated herpesvirus. F1000Res 5:F1000. 10.12688/f1000research.7612.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Luther SA, Cyster JG (2001) Chemokines as regulators of T cell differentiation. Nat Immunol 2(2):102–107. 10.1038/84205 [DOI] [PubMed] [Google Scholar]