Abstract
We report a novel and straightforward fluorescence recovery assay which enables the detection of protein–DNA interactions and simultaneously determines relative binding affinities of sequence-specific DNA-binding proteins for a variety of DNA sequences in a multiplexed format. The detection of protein–DNA binding is accomplished by monitoring fluorescence recovery during exonuclease digestion of DNA sequences that are modified with fluorophore–quencher pairs. Retardation of fluorescence recovery occurs with binding of the protein to the putative DNA binding element, which arrests exonuclease digestion. The assay detects protein–DNA binding in a homogeneous solution simply, quickly, and reliably without using radioisotopes. Multiplexing is possible by labeling different DNA sequences with spectrally distinct dyes, allowing simultaneous analysis of experimental and control binding reactions in the same mixture.
DNA-binding proteins are critically important in the regulation of a variety of essential cellular processes, such as genome replication, active gene transcription, cell division, and DNA repair.1-4 Transcription factors are one of the largest classes of DNA-binding proteins, regulating cell development, differentiation, growth, and physiology.5 Given the established significance of transcription factors in regulating a wide variety of biological processes, DNA-binding proteins are also of interest for their potential importance in disease diagnosis and drug development.6 For example, estrogen receptors (ERs) are a class of ligand-induced transcription factors. Upon activation, they bind to specific DNA sequences, known as estrogen response elements (EREs) mediating the developmental and physiological responses to the steroid hormone estrogen.7 As such, ERs have been studied intensely to elucidate the mechanisms by which they regulate target gene transcription and to understand their roles in the development of cancers in estrogen-sensitive tissues, such as the breast and endometrium.8
Current methods for the detection of specific DNA–protein binding include gel mobility shift assays9 and DNA footprinting assays.10 The electrophoretic mobility shift assays separate protein-bound and free DNA species in nondenaturing polyacrylamide or agarose gel. Electrophoretic mobility based methods are not only time-consuming and tedious but also usually require radioisotopes and laborious optimization procedures. Recently, several fluorescence-based approaches for measuring specific protein–DNA interactions have been developed, including “molecular beacon” based assays, fluorescence polarization assays, protein–DNA FRET assays, and a “no-shift” transcription factor assay.11-23 In comparison to conventional methods, fluorescence detection is convenient, sensitive, and by circumventing the use of radioisotopes, environmentally friendly.24 In addition, fluorescence-based techniques are often homogeneous systems that allow direct measurement of binding in solution. However, for methods developed to date, neither electrophoretic mobility based methods nor existing fluorescence-based techniques allow for detection of sequence-specific protein–DNA binding and appropriate control reactions in a multiplexed system.
Here we describe a novel and straightforward multiplexed fluorescence recovery assay, which enables the detection of protein–DNA interactions and simultaneously determines relative binding affinities of DNA-binding proteins for a variety of sequences in a multiplexed format (Scheme 1). The fluorescence recovery assay is a multiplexed system that relies on the exonuclease digestion of double-stranded (ds) DNA–substrates and recovery of fluorescence. This novel multiplexed fluorophore recovery method possesses advantages over the conventional electrophoretic mobility based methods that include detection of protein–DNA binding in homogeneous solution in a fast, straightforward, and reliable format without relying on radioisotopic labels. The multiplexed feature of the fluorescence recovery assay also provides an advantage over the existing fluorescence-based methods since it allows one to analyze a variety of DNA sequences in the same reaction mixture. The multiplexed measurements are possible by modifying two or more specific DNA sequences with different fluorophore–quencher pairs, as demonstrated in the proof-of-concept experiments described below.
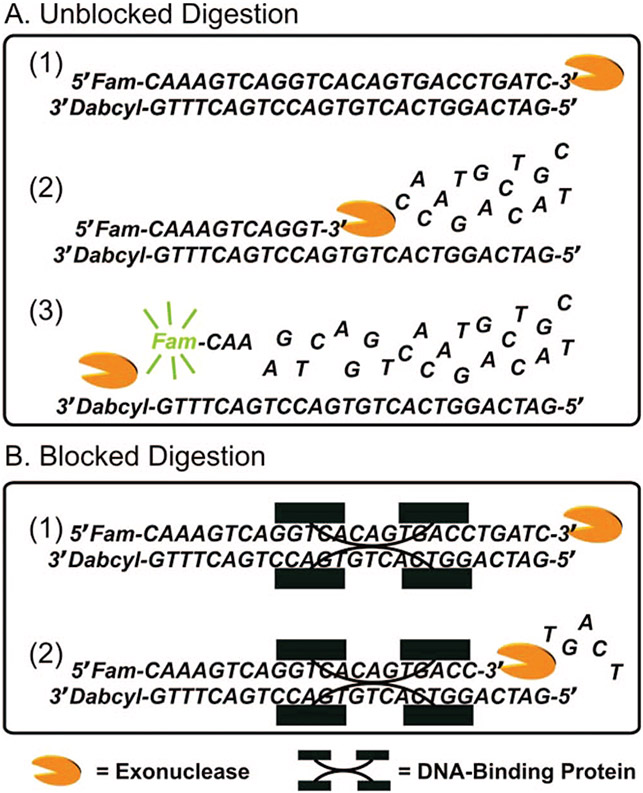
Scheme 1. Schematic Illustration of the Fluorescence Recovery Systema.
a The fluorophore and quencher are covalently linked to the 5′ of the sense and 3′ of the antisense ERE ODNs, respectively. Exonuclease III digests the single-strand sense DNA from the unmodified ends with specificity from the 3′ to 5′ termini, allowing the fluorophore and quencher labels to separate, causing fluorescence recovery (“Unblocked Digestion”). In the presence of the sequence-specific DNA-binding protein, Exo III digestion is inhibited through the formation of a DNA–protein complex. Therefore, the fluorophore is not be released from the ds DNA (“Blocked Digestion”). The sequence corresponding to the consensus ERα binding site is underlined.
EXPERIMENTAL SECTION
Chemicals and Materials.
Nanopure water (18 MΩ: Barnstead International) was used in all experiments and to prepare all buffers. ERα and vitamin D3 receptor proteins were obtained from Invitrogen (Carlsbad, CA). Exonuclease III (Exo III) was purchased from Promega (Madison, WI). All other reagents were purchased from Sigma-Aldrich (St. Louis, MO).
Methods.
In a typical experiment, the 5′ end of a sense strand was modified with a fluorophore (e.g., FAM, HEX, and Cy5). The 3′ end of the complementary strand was modified with a quencher (e.g., Dabcyl and BHQ2). The sense/antisense sequences and their various fluorophore–quencher modifications are shown in Table 1. To obtain DNA duplex, the sense strand and the antisense strand were mixed in a 1.5 mL microcentrifuge tube (Ambion Inc., RNase/DNase free, non-stick) at 1 μM concentration in the hybridization buffer (50 mM Tris–HCl, pH 8, 100 mM NaCl), heated for 5 min at 90 °C, and cooled to 25 °C overnight. Hybridization of the sense and antisense oligodeoxynucleotides (ODNs) brings the fluorophore and quencher in close proximity for efficient fluorescence quenching. The DNA duplex (with a final concentration 100 nM of each) were incubated for 20 min at room temperature in a buffer containing 10% glycerol, 50 mM KCl, 25 mM HEPES, pH 7.9, 1 mM MgCl2, 1 mM DTT, and 4 mg/mL BSA in a final volume of 100 μL with or without 130 nM of purified ERα. Exo III was added to the above solution. The hydrolysis of the ODNs and the release of the fluorophores were monitored by fluorescence spectroscopy (Flurorlog, HORIBA Group). For the competition experiment, a 100-fold excess of the unmodified Vit ERE ODN was added. All fluorescence measurements were carried out at room temperature in 100 μL quartz cuvettes.
Table 1.
Oligonucleotides Designed for the Fluorescence Recovery Assaya
| Vit EREs | 5′ FAM–CAAAGTCA GGTCACAGTGACC TGATC 3′ |
| Vit EREanti-s | 5′ GATCAGGTCACTGTGACCTGACTTTG–Dabcyl 3′ |
| Vit EREs mt | 5′ HEX–CAAAGTCAttTCACAGTGACC TGATC 3′ |
| Vit EREanti-s mt | 5′ GATCAGGTCACTGTGAaaTGACTTTG–Dabcyl 3′ |
| VEGFs | 5′ Cy5–ATCTGCAA GAGCACCCTGCCC TCTGG 3′ |
| VEGFanti-s | 5′ CCAGAGGGCAGGGTGCTCTTGCAGAT–BHQ2 3′ |
| VEGFs mt | 5′ Texas red–ATCTGCAAGAGCACCCTGCtt TCTGG 3′ |
| VEGFanti-s mt | 5′ CCAGAaaGCAGGGTGCTCTTGCAGAT–BHQ2 3′ |
The bold bases represent the core ERE sequences tested, and the mutated bases are illustrated in lower case letters.
RESULTS AND DISCUSSION
In a typical assay, a fluorophore was covalently linked to the 5′ terminus of a “sense” ODN strand whose fluorescence is quenched by a neighboring quencher group covalently linked to the 3′ terminus of a complementary “antisense” strand. These hybridized ds ODNs emit low fluorescence. Exo III has a double-strand-specific, nonprocessive 3′ to 5′ exodeoxyribonuclease activity.25,26 When Exo III was added to the solution containing free ds ODNs modified with a fluorophore–quencher pair, the ds ODNs were digested from the unmodified ends with specificity from the 3′ to 5′ termini. The digestion of the ODN duplex allows the fluorophore and quencher labels to separate, causing a fluorescence increase which can be easily quantified (Scheme 1). However, in the presence of the sequence-specific DNA-binding protein, the protein binding sterically inhibits Exo III digestion by forming a relatively stable DNA–protein complex. Consequently, only a partial fluorescence recovery, relative to the unbound ds ODNs, was seen after adding Exo III (Scheme 1). This fluorescence recovery ratio depends on both the DNA-binding protein concentration and its binding affinity. In principle, it also can provide semiquantitative information pertaining to the relative binding affinities of one or more proteins for a variety of DNA sequences by using various fluorophore–quencher modifications.
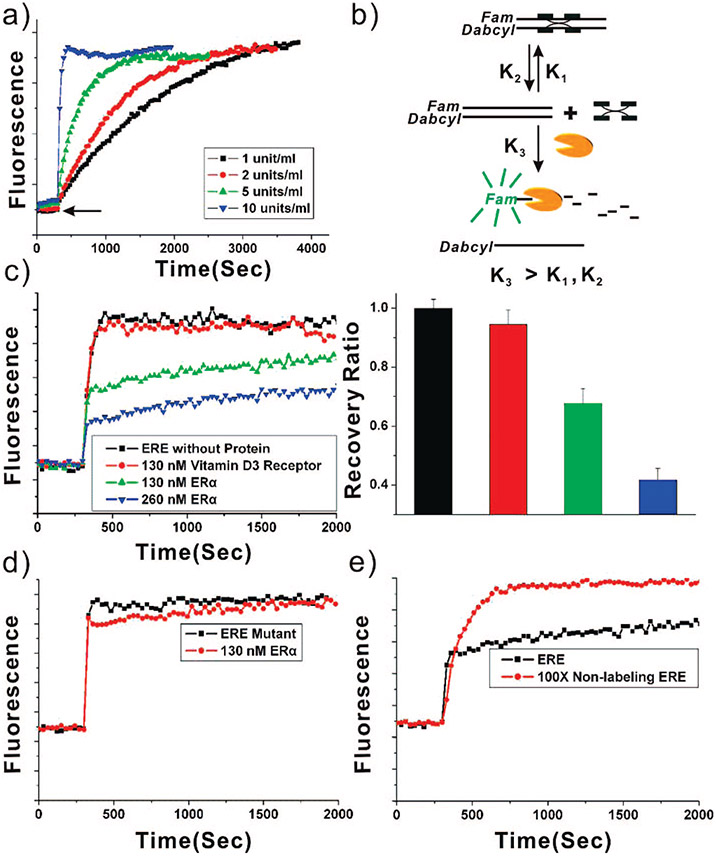
In order to evaluate the performance of the fluorescence recovery assay, we chose transcription factor ERα, which binds to specific DNA sequences known as EREs.27 A 26 bp classical Xenopus laevis vitellogenin A2 (Vit) ERE sequence (Table 1) was used as a basis for verifying the fluorescence recovery system. The 5′ end of the Vit ERE’s sense strand was modified with a 6-FAM fluorophore. The 3′ end of the complementary strand, Vit EREanti-s, was modified with a Dabcyl quencher (Table 1). Hybridization of the sense and antisense ODNs brings the fluorophore and quencher in close proximity for efficient fluorescence quenching. In solution, the quenched FAM exhibits very little fluorescence (Figure 1a). When Exo III is added to this solution (designated by the arrow in Figure 1a), the sense strand of the ds Vit ERE ODN substrate is digested, and the fluorophore becomes separated from the quencher, resulting in a substantial increase in fluorescence. The fluorescence reached a maximum when ds ODNs were digested to the point of dissociation. Note that the time for the full recovery of fluorescence is related to the concentration of Exo III (1, 2, 5, 10 U/mL) (Figure 1a). At 10 U/mL nuclease concentration, fluorescence is recovered in seconds after the addition of Exo III. Importantly, after incubation of Vit ERE with 130 nM ERα for 20 min, only ~68% of the fluorescence was recovered 1000 s after adding the Exo III. Note, in the presence of the protein, the fluorescence intensity increased slowly after an initial sharp rise because ERα bound to ds Vit ERE sterically inhibits Exo III digestion of the DNA. A higher concentration of ERα (260 nM) led to further attenuation of the fluorescence recovery (~42%). One possibility for this kinetic phenomenon is that the exonuclease digests the free duplex DNA, which slowly drives protein dissociation and releases more free duplex DNA for further digestion (Figure 1b). To test the specificity of this system, we used an unrelated DNA-binding protein (vitamin D3 receptor), a mutant Vit ERE (Vit EREmt, Table 1), and competing Vit ERE at a 100 times the concentration of the labeled sequence without fluorophore labeling as controls. Nearly full fluorescence recovery (~94%) was observed when vitamin D3 receptor (130 nM) was incubated with ds Vit ERE and challenged with Exo III (Figure 1c, Table 2). A nearly full fluorescence recovery (~94%) from ds Vit EREmt was also observed in the presence of ERα (Figure 1d, Table 2). Similarly, a nearly full fluorescence recovery from ds Vit ERE was detected in the competing control experiment (Figure 1e). These data, taken together with the negative control data, demonstrate that this fluorescence recovery system can be used to detect sequence-specific DNA–protein binding quickly and accurately in a homogeneous solution.
Figure 1.
Experimental optimization of the single-target fluorophore recovery system. (a) Kinetic FAM fluorescence emission curves (λex = 495 nm, λem = 515 nm) for the digestion reaction as a function of time for various concentrations of Exo III (1, 2, 5, and 10 U/mL). (b) Scheme showing exonuclease digest of the free duplex DNA, driving the DNA–protein complex dissociation. (c) Kinetic FAM fluorescence emission curves in the absence of protein (black squares), presence of 130 nM vitamin D3 receptor (red circles), 130 nM ERα (green up triangles), and 260 nM ERα (blue down triangles). The concentration of digestion enzyme Exo III is 10 U/mL. (d) Kinetic FAM fluorescence emission curves for Vit EREmt in the presence (red circles) and absence (black squares) of 130 nM ERα. The concentration of digestion enzyme Exo III is 10 U/mL. (e) Kinetic FAM fluorescence emission curves of Vit ERE in the presence (red circles) and absence (black squares) of 100× competing Vit ERE without fluorophore labeling, containing 130 nM ERα. The concentration of digestion enzyme Exo III is 20 U/mL. Inset: fluorescence recovery ratio at 1000 s after adding Exo III.
Table 2.
Fluorescence Recovery Ratio
| single-target fluorescence recovery experiment |
fluorescence recovery percentagea |
|---|---|
| ERE without ERα | 100 ± 3 |
| vitamin D3 Receptor (130 nM) | 94 ± 5 |
| ERα 130 nM | 68 ± 5 |
| ERα 260 nM | 42 ± 4 |
| EREmt | 94 ± 5 |
| multiplex fluorescence recovery experiment |
fluorescence recovery percentage comparisona |
| ERE Vs EREmt | 68 ± 4:98 ± 2 |
| VEGF Vs VEGFmt | 84 ± 3:102 ± 2 |
| ERE Vs VEGF | 65 ± 5:90 ± 3 |
Data are expressed as mean ± SD, n = 3.
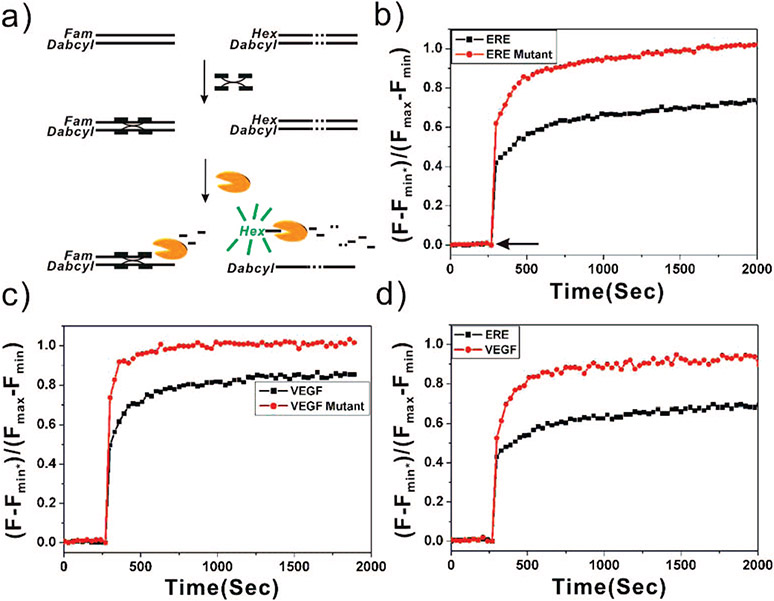
In order to determine the specificity of protein binding to a particular DNA sequence, several control experiments were conducted. In a gel mobility shift assay, the negative or positive control experiment must be set up in a different reaction mixture. One valuable feature of this fluorescence recovery system is multiplexed detection in one solution using multiple sequences with different dye modifications (Figure 2a). We first tested the capabilities of the system by adding both ds Vit ERE and ds Vit EREmt ODNs labeled with different fluorophore–quencher pairs: FAM with Dabcyl and HEX with Dabcyl, respectively. Multiplexed detection was accomplished by measuring both FAM and HEX signals at different wavelengths in the same reaction. In a typical experiment, equal concentrations of ds Vit ERE (FAM–Dabcyl) and ds Vit EREmt (HEX–Dabcyl) (100 nM of each) were incubated with ERα (130 nM) for 20 min at room temperature. By comparing the emission maxima of the fluorophores with and without DNA-binding protein, we could clearly detect a partially recovered fluorophore signal (Figure 2, parts a and b). After adding Exo III (20 U/mL, designated by an arrow in Figure 2b), the fluorescence was monitored in real time. At the time of 1000 s, ~68% of the FAM fluorophore signal had recovered at 515 nm (Figure 2b, black line; Table 2). After this initial sharp recovery, the fluorescence intensity then slowly increases. This phenomenon was also observed in the ds Vit ERE DNA (FAM–Dabcyl) single-component system (Figure 1c). In contrast, near total recovery of HEX fluorescence at 555 nm was observed (Figure 2b, red line). This result is very similar to that which was observed with ds Vit EREmut (HEX–Dabcyl) in the single-target system (Figure 1d). Those experiments show that the assay can easily differentiate a target sequence from a mutant with a 2 bp mismatch.
Figure 2.
Application of multiplexed fluorescence recovery detection. (a) Schematic representation of multiplexed fluorescence recovery detection. (b) Normalized kinetic fluorescence emission curves for the mixture of FAM (λex = 495 nm, λem = 515 nm, black line) released from Vit ERE and HEX (λex = 535 nm, λem = 555 nm, red line) released from Vit EREmt, respectively. (c) Normalized kinetic fluorescence emission curves for Cy5 (λex = 650 nm, λem = 670 nm, black line) released from VEGF and Texas red (λex = 585 nm, λem = 605 nm, red line) released from VEGFmt, respectively. (d) Normalized kinetic fluorescence emission curves of FAM (black line) released from Vit ERE and Cy5 (red line) released from VEGF.
Subsequent to the characterization of the first classical Vit ERE sequence,27 many variants of these sequences that possess various affinities for the activated ER have been identified.7,28 To test the validity and generality of this novel fluorescence recovery system for identifying other sequences capable of ER binding, we evaluated the system in the context of the vascular endothelial growth factor (VEGF) sequence (Table 1),29 which is homologous but not identical to the consensus ERE. This sequence has been demonstrated to exhibit specific ER binding by a gel mobility shift assay. In our system, fluorophore–quencher pairs Cy5–BHQ2 were linked to sense VEGFs and the complementary antisense VEGFanti-s ODNs, respectively. ds VEGF (Cy5–BHQ2) and ds VEGFmt (Texas red–BHQ2) were incubated together with ERα for the binding reaction. Upon adding Exo III, ~84% of Cy5 was recovered and ~102% of Texas red was recovered after 1000 s (Figure 2c, Table 2). Analysis of the data involved the comparison of fluorescence intensity change with the control experiment in the absence of ERα. Data are expressed as mean ± standard deviation (SD) unless otherwise indicated. Importantly, these data are consistent with the conclusion that Vit ERE has a larger binding affinity for ERα than VEGF (by comparing the degree of fluorescence recovery of Vit ERE ~68% to VEGF ~84%). Note that in this assay, the higher the binding affinity of the protein–DNA complex, the lower the degree of fluorescence recovery. We hypothesize that the stronger the binding affinity of DNA for protein, the more duplex DNA will bind to the specific protein, thus leaving less free duplex available in the solution for enzymatic digestion, resulting in less fluorophore released from the duplex DNA.
To further address the advantages of this novel multiplexed fluorescence recovery method over the existing fluorescence-based methods, we compared the relative binding affinity of ERα to a variety of sequences in a multiplexed format. ds Vit ERE (FAM–Dabcyl) and ds VEGF (Cy5–BHQ2) (100 nM of each) were incubated with ERα (130 nM) for 20 min at room temperature. After addition of Exo III (20 U/mL) to this solution, ~65% FAM and ~90% Cy5 (Figure 2d, Table 2) fluorescence were recovered after 1000 s, respectively. This indicates the fluorescence recovery system also can determine relative protein–DNA binding affinity in one-pot fashion. Proteins with higher binding affinities for DNA have a slower on–off rate, and in this assay they would have a smaller amount of fluorescence recovery at the same enzyme digestion time point. The order of binding affinity shown in this multiplexed fluorescence recovery system was Vit ERE (65% recovery) > VEGF (90% recovery), which is consistent with results obtained from gel mobility shift assays.29 Our results verified the relative binding affinities for different ERE variants in a multiplexed system. In the multiplexed system, the sequence-specific binding protein ERα was present with different ERE variants in the same binding reaction. The sequences with higher binding affinity to ERα competed with those with lower binding affinity. The binding affinities were reflected by the multicolor fluorescence recovery detection.
CONCLUSIONS
In conclusion, we have developed a novel method for detecting DNA–protein binding activity. Our multiplexed assay presents several advantages over currently available assays, such as the gel-shift assay, an existing molecular beacon assay, and the commercial no-shift assay. These include speed, low complexity, generality, multiplexing, and low cost. For example, when compared to the gel-shift assay, this assay obviates the need for radioisotopes. It is fast, allowing completion in 30 min, including positive and negative controls, and it allows one to determine relative protein binding affinities and do control experiments, all in a single reaction mixture. In comparison to the molecular beacon assay, the method is simpler because it requires only two oligonucleotides instead of four. In addition, it does not require fluorophore- and quencher-modified nucleotides near the binding site. This not only simplifies synthesis and lowers cost but also prevents these moieties from interfering with protein binding. Importantly, this assay is an off–on system with the capacity to show a full dynamic range of fluorescence intensity change, providing an advantage over the fluorescence polarization method which is limited to a dynamic range of fluorescence polarization changes and relatively short DNA sequences. Last, this new method is more general than the commercial “no-shift” assay, which requires antibodies for each protein of interest. In principle, this method can be used for the high-throughput screening of DNA–protein binding and inhibitors of DNA-binding proteins. As such, this novel method adds to the growing base of emerging assays for proteins, nucleic acids, and small molecules.30-43
ACKNOWLEDGMENT
Xiaoyang Xu and Zhen Zhao contributed equally to this work. This work was supported by NIH Grants R01 HD20677, NIH U54 HD041859, and the NCI through a CCNE and the NSF through an NSEC. C.A.M. is grateful for an NIH Director’s Pioneer Award. Z.Z. acknowledges the American Association of University Women for an International Fellowship.
References
- (1).Ren B; Robert F; Wyrick JJ; Aparicio O; Jennings EG; Zeitlinger IJ; Schreiber J; Hannett N; Kanin E; Volkert TL; Wilson CJ; Bell SP Science 2000, 290, 2306–2309. [DOI] [PubMed] [Google Scholar]
- (2).Sen R; Baltimore D Cell 1986, 47, 921–928. [DOI] [PubMed] [Google Scholar]
- (3).Helin K Curr. Opin. Genet. Dev 1998, 8, 28–35. [DOI] [PubMed] [Google Scholar]
- (4).Aboussekhra A; Biggerstaff M; Shivji MK; Vilpo JA; Moncollin V; Podust VN; Protic M; Hubscher U; Egly JM; Wood RD Cell 1995, 80, 859–868. [DOI] [PubMed] [Google Scholar]
- (5).Pabo CO; Sauer RT Annu. Rev. Biochem 1992, 61, 1053–1095. [DOI] [PubMed] [Google Scholar]
- (6).Herley LH Nat. Rev. Cancer 2002, 2, 188–200. [DOI] [PubMed] [Google Scholar]
- (7).Nilsson S; Mäkelä S; Treuter E; Tujague M; Thomsen J; Enmark GE; Pettersson K; Warner M; Gustafsson J Physiol. Rev 2001, 81, 1535–1565. [DOI] [PubMed] [Google Scholar]
- (8).Raegan O; Martin CF; Elinor KK; Ulla H Mol. Endocrinol 2004, 18, 1859–1875.15031323 [Google Scholar]
- (9).Garner MM; Revzin A Nucleic Acids Res. 1981, 9, 3047–3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Galas DJ; Schmitz A Nucleic Acids Res. 1978, 9, 3157–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Hill JJ; Royal CA Methods Enzymol. 1997, 278, 390–416. [DOI] [PubMed] [Google Scholar]
- (12).Reedstrom RJ; Brown MP; Grillo A; Roen D; Royer CA J. Mol. Biol 1997, 273, 572–585. [DOI] [PubMed] [Google Scholar]
- (13).Bjornson KP; Moore KJ; Lohman TM Biochemistry 1996, 35, 2268–2282. [DOI] [PubMed] [Google Scholar]
- (14).Hey T; Lipps G; Krauss G Biochemistry 2001, 40, 2901–2910. [DOI] [PubMed] [Google Scholar]
- (15).Bailey M; Hagmar P; Millar DP; Davidson BE; Tong G; Haralambidis J; Sawyer WH Biochemistry 1995, 34, 15802–15812. [DOI] [PubMed] [Google Scholar]
- (16).Parkhurst KM; Brenowitz M; Parkhurst LJ Biochemistry 1996, 35, 7459–7465. [DOI] [PubMed] [Google Scholar]
- (17).Wang K; Rodgers ME; Toptygin D; Munsen VA; Brand L Biochemistry 1998, 37, 41–50. [DOI] [PubMed] [Google Scholar]
- (18).Gourves AS; LeGac NT; Villani G; Boehmer PE; Johnson NP J. Biol. Chem 2000, 275, 10864–10869. [DOI] [PubMed] [Google Scholar]
- (19).Lima LM; Foguel D; Silva JL Proc. Natl. Acad. Sci. U.S.A 2000, 97, 14289–14294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Ozers MS; Hill JJ; Ervin K; Wood JR; Nardulli AM; Royer CA; Gorski J J. Biol. Chem 1997, 272, 30405–30411. [DOI] [PubMed] [Google Scholar]
- (21).Heyduk T; Lee JC Proc. Natl. Acad. Sci. U.S.A 1990, 87, 1744–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Bruggink F; Hayes S Nat. Methods 2004, 1, 177–179. [Google Scholar]
- (23).Wang JL; Li T; Guo X; Lu Z Nucleic Acids Res. 2005, 33, 2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Heyduk T; Heyduk E Nat. Biotechnol 2002, 20, 171–176. [DOI] [PubMed] [Google Scholar]
- (25).Henikoff S Gene 1984, 28, 315–319. [DOI] [PubMed] [Google Scholar]
- (26).Xu X; Han MS; Mirkin CA Angew. Chem., Int. Ed 2007, 46, 3468–3470. [DOI] [PubMed] [Google Scholar]
- (27).Klein-Hitpass L; Schorpp M; Wagner U; Ryffel GU Cell 1986, 46, 1053–1061. [DOI] [PubMed] [Google Scholar]
- (28).Sanchez R; Nguyen D; Rocha W; White JH; Mader S BioEssays 2002, 24, 244–254. [DOI] [PubMed] [Google Scholar]
- (29).Hyder SM; Nawaz Z; Chiappetta C; Stancel GM Cancer Res. 2000, 60, 3183–3190. [PubMed] [Google Scholar]
- (30).Mirkin CA; Letsinger RL; Mucic RC; Storhoff JJ Nature 1996, 382, 607–609. [DOI] [PubMed] [Google Scholar]
- (31).Elghanian R; Storhoff JJ; Mucic RC; Letsinger RL; Mirkin CA Science 1997, 277, 1078–1080. [DOI] [PubMed] [Google Scholar]
- (32).He L; Musick MD; Nicewarner SR; Salinas FG; Benkovic SJ; Natan MJ; Keating CD J. Am. Chem. Soc 2000, 122, 9071–9077. [Google Scholar]
- (33).Maxwell DJ; Taylor JR; Nie S J. Am. Chem. Soc 2002, 124, 9606–9612. [DOI] [PubMed] [Google Scholar]
- (34).Yang CJ; Martinez K; Lin H; Tan W J. Am. Chem. Soc 2006, 128, 9986–9987. [DOI] [PubMed] [Google Scholar]
- (35).Li D; Shlyahovsky B; Elbaz J; Willner I J. Am. Chem. Soc 2007, 129, 5804–5805. [DOI] [PubMed] [Google Scholar]
- (36).Liu J; Lu Y Angew. Chem., Int. Ed 2007, 46, 7587–7590. [DOI] [PubMed] [Google Scholar]
- (37).Han MS; Lytton-Jean AKR; Oh BK; Heo J; Mirkin CA Angew. Chem., Int. Ed 2006, 45, 1807–1810. [DOI] [PubMed] [Google Scholar]
- (38).Bailey RC; Kwong GA; Radu CG; Witt ON; Heath JR J. Am. Chem. Soc 2007, 129, 1959–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Cao YC; Jin R; Mirkin CA Science 2002, 197, 1536–1540. [DOI] [PubMed] [Google Scholar]
- (40).Niemeyer CM; Simon U Eur. J. Inorg. Chem 2005, 18, 3641–3655. [Google Scholar]
- (41).Georganopoulou DG; Chang L; Nam JM; Thaxton CS; Mufson EJ; Klein WL; Mirkin CA Proc. Natl. Acad. Sci. U.S.A 2005, 102, 2273–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Xu X; Georganopoulou DG; Hill HD; Mirkin CA Anal. Chem 2007, 79, 6650–6654. [DOI] [PubMed] [Google Scholar]
- (43).Rosi NL; Mirkin CA Chem. Rev 2005, 105, 1547–1562. [DOI] [PubMed] [Google Scholar]