Abstract
Background
Egypt was among the first 10 countries in Africa that experienced COVID-19 cases. The sudden surge in the number of cases is overwhelming the capacity of the national healthcare system, particularly in developing countries. Central to the containment of the ongoing pandemic is the availability of rapid and accurate diagnostic tests that could pinpoint patients at early disease stages. In the current study, we aimed to (1) Evaluate the diagnostic performance of the rapid antigen test (RAT) “Standard™ Q COVID-19 Ag” against reverse transcriptase quantitative real-time PCR (RT-qPCR) in eighty-three swabs collected from COVID-19 suspected individuals showing various demographic features, clinical and radiological findings. (2) Test whether measuring laboratory parameters in participant’s blood would enhance the predictive accuracy of RAT. (3) Identify the most important features that determine the results of both RAT and RT-qPCR.
Methods
Diagnostic measurements (e.g. sensitivity, specificity, etc.) and receiver operating characteristic curve were used to assess the clinical performance of “Standard™ Q COVID-19 Ag”. We used the support vector machine (SVM) model to investigate whether measuring laboratory indices would enhance the accuracy of RAT. Moreover, a random forest classification model was used to determine the most important determinants of the results of RAT and RT-qPCR for COVID-19 diagnosis.
Results
The sensitivity, specificity, and accuracy of RAT were 78.2, 64.2, and 75.9%, respectively. Samples with high viral load and those that were collected within one-week post-symptoms showed the highest sensitivity and accuracy. The SVM modeling showed that measuring laboratory indices did not enhance the predictive accuracy of RAT.
Conclusion
“Standard™ Q COVID-19 Ag” should not be used alone for COVID-19 diagnosis due to its low diagnostic performance relative to the RT-qPCR. RAT is best used at the early disease stage and in patients with high viral load.
Keywords: Accuracy, Diagnosis, SARS-CoV-2, Rapid antigen test
Introduction
Infection with coronavirus (CoV) diseases (COVID-19), which is caused by novel severe acute respiratory syndrome CoV-2 (SARS-CoV-2), was firstly reported in Wuhan city, Hubei province, China in December 2019 [1]. On 11 March 2020, the World Health Organization (WHO) declared COVID-19 as a pandemic of global concern [2]. There has been a tremendous increase in the number of confirmed cases as well as death records worldwide. As of 24 March 2021, ∼123 million COVID-19 cases were confirmed worldwide and more than two million deaths were reported [3]. Egypt was among the first 10 countries in Africa that experienced COVID-19 cases [4]. By this date, Egypt has reported 197 confirmed cases with 5.9% of them died of the pandemic [3]. The sudden and unprecedented surge in the number of reported cases is overwhelming the capacity of the national healthcare system, particularly in developing countries [5]. Central to the containment of the ongoing pandemic is the availability of rapid and accurate diagnostic tests that could pinpoint patients at early disease stages before further spread occurs.
Reverse transcriptase quantitative real-time PCR (RT-qPCR) assay has been the gold standard for diagnosing COVID-19 in many health sectors and laboratories [[6], [7], [8]]. However, this assay is often done in large centralized hospitals or laboratories away from the access of local inhabitants; it requires long time-to-results, skilled staff, and specialized instruments and is of high cost [9]. This is particularly the case in many developing countries, including Egypt [10]. In Egypt, the RT-qPCR diagnosis is mostly done in Cairo, the capital, or in capitals of governorates leading to an overall turn-around-time of ∼24 h at best between shipping the samples and obtaining the results [11]. Indeed, suspected individuals often go first to the local clinics for an emergency where RT-qPCR might not be available. The RT-qPCR may thus not be able to cope with the testing or screening needs in the low and middle-income countries due to limited infrastructure, low fund, and limited human resources [12]. To fill in this gap and to improve this situation, rapid antigen tests (RATs) are being developed and are in use as point-of-care diagnostic tools [13]. They offer the advantage of being quick and can be done simply without the need for special equipment [[14], [15], [16]]. Determining the diagnostic performance of commercialized RATs is crucial because this gives an indication of their reliability and clinical utility during the time of the pandemic.
With the availability of both RT-qPCR and RAT for diagnosis of COVID-19 suspected individuals, it is worth comparing their diagnostic performance to unravel advantages and disadvantages of using both tests, and thus determine the preference of using any of them over the other in various pandemic scenarios. This is done taken into account the fundamental differences between both assays in methodology and applications. There have been many RAT available for diagnosis of COVID-19 (reviewed in Refs. [9,12]), yet their clinical applicability is questionable because their accuracy has shown to be low as compared to RT-qPCR, their diagnostic performance is highly variable, even when the same assay was applied in two different population with different ethnicity background [15,17] and their accuracy is host- and virus-dependent [6,17]. Similar to other countries, several RATs have been under development in Egypt, but the available studies lack detailed characterization of RAT performance, especially the impact of patient’s criteria, clinical features, sampling time, and viral load on the test performance. Actually, only one RAT (BIOCREDIT COVID-19 antigen test) has been recently evaluated in Egypt [18] and details were given on the influence of patient’s criteria on test performance. In the current study, we aimed to (1) Evaluate the diagnostic performance of the rapid antigen test (RAT) “Standard™ Q COVID-19 Ag” against RT-qPCR in eighty-three swabs collected from COVID-19 suspected individuals showing various demographic features, clinical and radiological findings. (2) Investigate whether measuring laboratory parameters in participant’s blood would enhance the predictive accuracy of this RAT. (3) Identify the most important features that determine the results of both RAT and RT-qPCR.
Material and methods
Study setting, sample size, and participant’s data
This is a cross-sectional study conducted at the premises of Zagazig University Hospitals and the affiliated Scientific & Medical Research Centre, Faculty of Medicine, Zagazig University in the period from June 2020 through October 2020. The sample size was estimated using the online tool Openepi version 3.1 [19] considering the hypothesized frequency of outcome in the population (p) equal to 50% ± 5, confidence limits as % of 100 (absolute ± %)(d): 5%, design effect (for cluster surveys-DEFF): 1 and sample size at confidence level 95%: 152. Eighty-three individuals were enrolled in the study after being referred to the COVID-19 isolation unit in the previously mentioned settings. The inclusion criterion for participants was that he/she should be suspected of having COVID-19 infection. According to WHO case definition for COVID-19 [20], participant should have experienced acute onset of fever and cough in addition to either be working in areas with high risk of virus transmission, recently travelled to areas with community transmission or working in a health care setting. Participant’s metadata (age and gender), symptoms, radiological findings, and fourteen laboratory measurements (Hemoglobin; HB, urea, platelet, white blood cells; WBC, creatinine, alanine aminotransferase; ALT, aspartate aminotransferase; AST, lactate dehydrogenase; LDH, serum ferritin; S. ferritin, C. reactive protein; CRP, prothrombin time; PT, international normalized ratio; INR and polymorphonuclear leukocytes; PNL) were collected at the time of doing the test. The laboratory measurements were done at the Clinical Pathology Department, Faculty of Medicine, Zagazig University.
Sample collection and preparation
Eighty-three nasopharyngeal (NP) and oropharyngeal swabs (OP) were obtained by trained health staff at the isolation units. Based on the recommendation from the Center for Disease Control and Prevention (CDC) [21] and to maximize sensitivity and to limit the use of test resources, the two NP and OP swabs which were taken from one participant, were admixed in a 3-ml tube containing viral transport medium (VTM, Ismailia free zone, Egypt. Ref: 1/V T01.001.0001) and stored at −80 °C until further analyses.
RNA extraction and detection of SARS-CoV-2 RNA by standard RT-qPCR
RNA extraction was done under BSL-2 on 410 μl of the VTM of both swabs using the QIAamp® Viral RNA mini kit (cat. no. 52906, Qiagen) according to the manufacturer’s recommendation. During the extraction, RNase-free DNase set (cat no. 79254, Qiagen) was used to treat the RNA samples to eliminate the possibility of genomic DNA contamination. RNA quality and quantity were determined with the Nanodrop S1000 spectrophotometer (Thermo Fisher Scientific). A one-step RT-qPCR was done on extracted RNA using a real-time PCR kit (Primerdesign Ltd, Ref: Z-Path-COMD-19-CE, UK) in Stratagene Mx3000P qPCR System (Agilent). This assay targets the RNA-dependent RNA polymerase (RdRP) gene within SARS-CoV-2. The genesig® kit detects 0.58 copies/μl of SARS-CoV-2 viral RNA with a confidence ≥95%. This concentration therefore serves as the limit of detection of the kit. The 20 μl reaction mix formed of 10 μl 2X RT-qPCR Master Mix, 2 μl of COVID-19 Primer & Probe and 8 μl sample extract. A positive control template and negative amplification control with nuclease-free water were included in each run. In the one-step protocol, the reverse transcription (complementary DNA; cDNA formation) was done by heating the mix at 55 °C for 10 min. and the cDNA was heated at 95 °C for 2 min. (initial denaturation) followed by 45 cycles, each consists of denaturation at 95 °C for 10 s, annealing, and extension at 60 °C for 1 min. The cycle threshold (Ct) values were recorded for each sample. The analyzed samples were considered negative if they have a Ct value ≥40 or when no Ct values were reported. For positive samples, SARS-CoV-2 RNA content was categorized according to the Ct values into strong (Ct < 29), moderate (Ct = 29−36) and weak (Ct = 37−39).
Detection of SARS-CoV-2 antigen by rapid antigen test (RAT)
The RT-qPCR-characterized samples were tested with the RAT Standard™ Q COVID-19 Ag test (SD Biosensor, Inc., Republic of Korea). The Standard™ Q COVID-19 Ag test is an immunoassay that detects SARS-CoV-2 nucleocapsid protein by lateral flow technique. The test device consists of a membrane with control and test lines that are pre-coated with mouse monoclonal anti-SARS-CoV-2 antibody and anti-chicken IgG antibody, respectively. The anti-SARS-CoV-2 antibody is conjugated with color particles and is used as detectors for SARS-CoV-2 antigen. A colored test line would be visible, with various intensities, if SARS-CoV-2 nucleocapsid protein were present in the specimen. The test procedure was all done under BSL-2 following the manufacturer’s instruction. Briefly, the VTM, containing the NP and OP fluids, was first vortexed for 20 s and only 100 μl thereof was placed into the sample port of the cassette and incubated at room temperature for 15−30 min until reading the results in a blinded approach (i.e. without knowing the RT-qPCR results of the samples).
Data analyses and machine learning modeling
Continuous variables (e.g. age) were expressed as median ± SD and were compared using the Mann Whitney U test. Categorical variables were expressed as numbers and percentages and were compared using or Fisher’s exact test. Correlation and agreement between RAT and RT-qPCR results were calculated using Pearson’s correlation (r) and Cohn’s kappa (κ), respectively [22]. Measurements of diagnostic performance of RAT (sensitivity, specificity, positive predictive value, negative predictive value, accuracy, and likelihood ratio) for the whole subjects and subject’s subgroups were calculated on contingency tables containing the numbers of each outcome. The confidence intervals (CI) were calculated using the Wilson-Brown method [23]. Participant’s categories based on Ct values were defined following a previous report [15]. A receiver operating characteristic curve (ROC) was generated to provide another assessment for the diagnostic power of the RAT. These analyses were done using GraphPad Prism version 8.0.0 for Windows, GraphPad Software, San Diego, California USA, (www.graphpad.com). To investigate whether measuring blood parameters would by any means enhance the predictive accuracy of the RAT and thus raises its clinical utility, a support vector machine (SVM) model with Monte-Carlo cross-validation was applied as described previously [24] and the performance of top-ranked combination (best model) was evaluated for sensitivity, specificity, and accuracy using class probability analysis. This analysis was done on data from 68 subjects (the other 15 subjects had no data on any of the laboratory parameters). Random forest classification was utilized to reveal the demographic and clinical parameters that are most important in determining individuals with positive and negative results for both RAT and RT-qPCR. In both SVM and random forest models, the singular value decomposition method was used to impute the missing values [25]. These analyses were done using Metaboanalyst online server [26].
Results
Demographic, clinical, and laboratory characteristics of enrolled subjects
The demographic and baseline clinical characteristics of the participants are summarized in Table 1 and S1. The range of ages was 22–87 (Median = 55.5 ± 18.5). More than half (59%, 49/83) of the subjects were male and 41% (34/83) were females. Data on the sampling time post-symptoms were available in only 70 participants. Samples were collected between 0−7 days post-symptoms in 54.2% (38/70), between 8−16 days post-symptoms in 38.5% (27/70) and >16 day post-symptoms in 5.7% (4/70) of the participants. In only one participant (1.2%), symptoms appeared 5 days after sampling. Clinical data of 47 (56.6%) subjects were only available, the majority of whom (89.3%, 42/47) were symptomatic as they showed at least one of the following symptoms [fever (n = 40), pharyngitis (n = 38), chest pain and dyspnea (n = 37, each), cough (n = 36) and diarrhea (n = 8)] and 5 subjects (10.6%) were asymptomatic. Radiological data were available in 44 participants, 84% (37/44) of whom showed radiological findings of various grades and 7 participants (15.9%) had no radiological findings.
Table 1.
Frequency of demographic and clinical characteristics of study participants and its association with RAT results.
| Feature | Subgroup | No. (%) | P-value* |
|---|---|---|---|
| Age | Median: 55.5 ± 18.4 (SDc) | 0.8 | |
| Gender | Male | 49 (59) | 0.56 |
| Female | 34 (41) | ||
| Radiology | With findings | 37 (44.5) | 0.56 |
| No findings | 7 (8.4) | ||
| Not reported | 39 (46.9) | ||
| Days post-symptom onset | 0−7 | 38 (45.7) | 0.42 |
| 8−16 | 27 (32.5) | ||
| >16 | 4 (4.8) | ||
| Symptoms appeared after sampling | 1 (1.2) | ||
| Symptomatologyb | Symptomatic | 42 (50.6) | 0.14 |
| Asymptomatic | 5 (6.2) | ||
| Not reported | 36 (43.3) | ||
| Fevera | 40 (95.2) | 0.87 | |
| Pharyngitisa | 38 (90.4) | 0.73 | |
| Chest paina | 37 (88) | 0.44 | |
| Dyspneaa | 37 (88) | 0.44 | |
| Cougha | 36 (85.7) | 0.4 | |
| Diarrheaa | 8 (19) | 0.44 | |
The percentages were calculated by dividing the number of subjects showing particular symptom/number of symptomatic subjects.
The percentages were calculated by dividing the number of symptomatic, asymptomatic, and not reported subjects by the total number of enrolled subjects.
SD: standard deviation.
P-value refers to the significance of the difference between the results of RAT in the respective subgroups.
Reverse transcriptase quantitative real-time PCR (RT-qPCR) detection of COVID-19 patients
The average Ct values of COVID-19 subjects was 31.1 ± 7.4 (min = 15.7, max = 40.5). The majority of the subjects (83.1%, 69/83) were positive by RT-qPCR and 16.8% (14/83) were negative. As shown in Table 2 , according to the Ct values, 29 (42%), 29(42%) and 11 (15.9%) subjects have strong, moderate, and weak RT-qPCR positive results, respectively.
Table 2.
Diagnostic performance of Standard™ Q COVID-19 Ag test against the RT-qPCR at various Ct categories.
| Features | Subgroup | Standard™ Q COVID-19 Ag test | RT-qPCR test |
Sensitivity % (CI) | Specificity % (CI) | Accuracy % | Likelihood ratio | |
|---|---|---|---|---|---|---|---|---|
| Positive (n = 69) | Negative (n = 14) | |||||||
| All | Ct values | Positive (n = 59) | 54 | 5 | 78.2 (0.67−0.86) | 64.2 (0.38−0.83) | 75.9 | 2.1 |
| Negative (n = 24) | 15 | 9 | ||||||
| Subgroups of RT-qPCR positive patientsa | Strongly positive (<29) | Positive | 28 | 28 | 96.5 (0.83−0.99) | 44 (0.31−0.57) | 63.2 | 1.7 |
| Negative | 1 | 22 | ||||||
| Moderately positive (29−36) | Positive | 21 | 38 | 72.4 (0.54−0.85) | 29.6 (0.19−0.42) | 44.5 | 1 | |
| Negative | 8 | 16 | ||||||
| Weakly positive (37−39) | Positive | 5 | 54 | 45.4 (0.21−0.71) | 23.9 (0.15−0.35) | 26.8 | 0.59 | |
| Negative | 6 | 17 | ||||||
Test performance for RT-PCR subgroups was calculated considering the negative PCR results are the ones that lack the respective group feature.
Diagnostic performance of Standard™ Q COVID-19 Ag
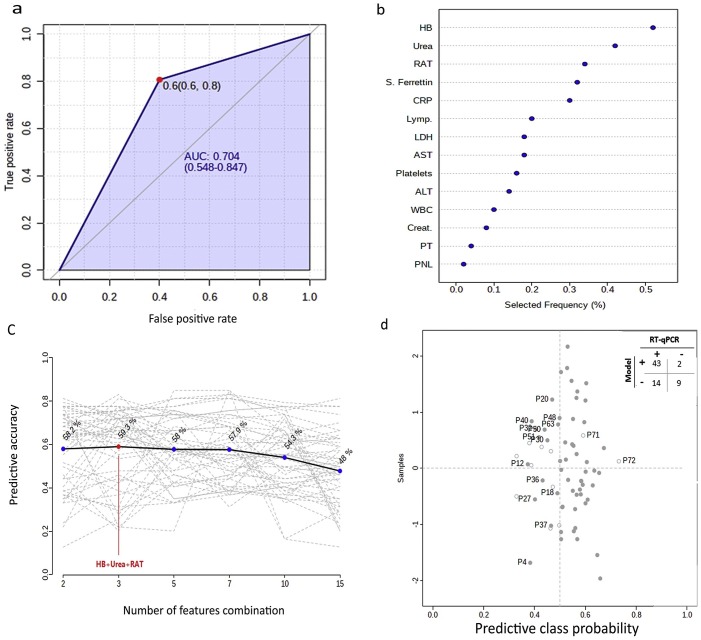
As shown in Table 2 and Fig. 1 , out of the 69 RT-qPCR positive subjects, 54 were positive by the RAT (sensitivity = 78.2%). Of the 14 RT-qPCR negative subjects, nine were negative by the RAT (specificity = 64.2%). The RAT showed that 15 and 5 subjects had false-negative and false-positive results, respectively. The accuracy (overall concordance rate) of the RAT tests was 75.9%. Overall, the results of RT-qPCR and the RAT correlated weakly positive (r = 0.3, P-value = 0.005) and agreed fairly [Cohen’s kappa (κ) = 0.3 (CI = 0.16−0.59), SD = 0.1, P-value = 0.0006)] in a significant manner. The Ct values were significantly different (P-value < 0.0001) between RAT positive and negative subjects. The RAT showed the highest sensitivity (96.5%), specificity (44%), and accuracy (63.2%) in COVID-19 patients that have the highest viral nucleic acid content (Ct values < 29) compared to the groups with the lower viral load (Ct values ≥ 29). Other measures for accuracy in subject’s subgroups based on Ct values are shown in Table S2. The ROC analysis yielded an AUC value of 0.7 ± 0.08 (CI = 0.0.5−0.8, P-value = 0.02) (Fig. 2 a).
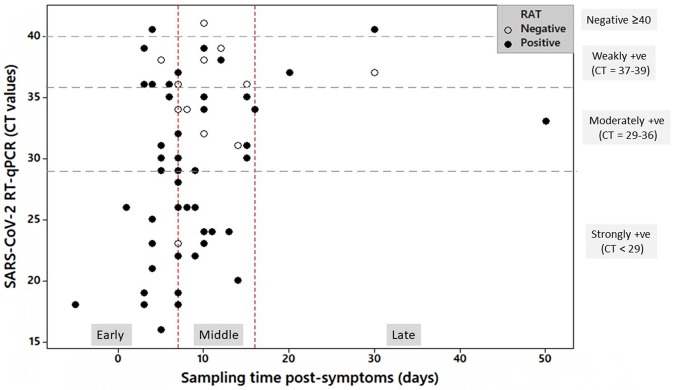
Fig. 1.
Distribution of participants with positive and negative results of RAT according to sampling time post-symptoms in days (x-axis) and Ct values as determined by RT-qPCR (y-axis). Sampling time post-symptom onset was classified into early (0–7 d), middle (8–16 d), and late (>16 d). RT-qPCR categories are indicated on the right side of the graph.
Fig. 2.
Diagnostic performance of RAT A. Receiver operating characteristic curve (ROC) analyses showing the diagnostic performance of the RAT with an area under the curve (AUC) of 0.7 B–C. Support vector machine model. B. Top-ranked features based on their frequency of being selected after the cross-validation is the model. C. Plot showing the accuracy of feature combination in predicting the COVID-19 positive subjects as determined by RT-qPCR. The most accurate classifier gave an accuracy of 59.3% for the top 3-feature as revealed in B. D. Predicted class probability analyses evaluating the performance of the 3-features model. Each dot refers to the average prediction of one subject after cross-validation. Dark and light-colored dots indicate positive and negative cases by RT-qPCR. The misclassified subjects by the 3-feature model are labeled. The classification boundary for COVID-19 positive subjects lies at the center of the x-axis (x = 0.5, vertical dotted line). Values >0.5 indicate a probability of COVID-19 positive and closer to 1 indicate high probability. The confusion matrix shows the summary of the model performance.
Effect of subjects’ demographic, clinical criteria, and sampling time on the results of the RAT
Table 3 and S2 show the diagnostic performance of the RAT in various subject’s subgroups. Although RAT results did not differ significantly between male and female subjects, it was slightly more sensitive and more accurate in female (sensitivity = 78.5% and accuracy = 79.4%) than in male (sensitivity = 76.1% and accuracy = 71.4%) participants and its specificity in female was almost twice (83.3%) than in male subjects (42.8%). High sensitivity and accuracy were evident when swabs were collected 0−7 days post-symptoms (n of subjects = 38) followed in order by the case when swabs were taken at 8−16 (n of subjects = 27) and >16 days post-symptoms (n of subjects = 4). There were no significant differences in the RAT results between symptomatic, radiology-positive subjects and asymptomatic, radiology-negative subjects, respectively. However, RAT was more sensitive and accurate in symptomatic subjects relative to the asymptomatic ones. RAT proved positive in 3 (60%) out of the 5 asymptomatic participants, one of these was considered asymptomatic COVID-19 carrier at the time of sampling as evidenced by the high RNA content (Ct = 17.6), showed strong positive RAT (i.e. strong line intensity) and showed symptoms 5-days after the sampling. RAT showed higher sensitivity and accuracy in subjects with no radiological findings than those with radiological findings. No radiological findings were evident in seven participants, 6 of whom were positive by both RAT and RT-qPCR.
Table 3.
Diagnostic performance of Standard™ Q COVID-19 Ag test in different subgroups of participants.
| Features | Subgroup | Standard™ Q COVID-19 Ag test | RT-qPCR test |
Sensitivity% (CIa) | Specificity% (CI) | Accuracy% | Likelihood ratio | |
|---|---|---|---|---|---|---|---|---|
| Positive | Negative | |||||||
| Gender | Male (n = 49) | Positive | 32 | 4 | 76.1 (0.61−0.86) | 42.8 (0.27−0.64) | 71.4 | 1.33 |
| Negative | 10 | 3 | ||||||
| Female (n = 34) | Positive | 22 | 1 | 78.5 (0.60−0.89) | 83.3 (0.15−0.74) | 79.4 | 4.71 | |
| Negative | 5 | 5 | ||||||
| Symptomatology (total n = 47) | Symptomatic (n = 42) | Positive | 32 | 0 | 76.1 (0.63−0.88) | NA | 76.1 | NA |
| Negative | 10 | 0 | ||||||
| Asymptomatic (n = 5) | Positive | 2 | 1 | 66.6 (0.61−0.98) | 50 (0−0.94) | 60 | 0.66 | |
| Negative | 1 | 1 | ||||||
| Radiology (total n = 44) | With findings (n = 37) | Positive | 28 | 1 | 75.6 (0.59−0.86) | 0 (0−0.94) | 73.6 | 0.75 |
| Negative | 9 | 0 | ||||||
| No findings (n = 7) | Positive | 6 | 1 | 85.7 (0.48−0.95) | 0 (0−1) | 75 | 0.75 | |
| Negative | 2 | 0 | ||||||
| Days post symptom onset (total n = 69)b | 0−7 (n = 38) | Positive | 31 | 1 | 83.7 (0.68−0.92) | 0 (0−0.94) | 81.5 | 0.83 |
| Negative | 6 | 0 | ||||||
| 8−16 (n = 27) | Positive | 19 | 0 | 70 (0.51−0.84) | NA | 70.3 | NA | |
| Negative | 8 | 0 | ||||||
| >16 (n = 4) | Positive | 2 | 1 | 66.6 (0.11−0.98) | 0 (0−0.94) | 50 | 0.66 | |
| Negative | 1 | 0 | ||||||
CI: confidence intervals.
This number includes one participant that showed symptoms 5 days after the sampling.
Importance of combining laboratory parameters and RAT in diagnosing COVID-19 patients
Considering the intermediate diagnostic power (AUC = 0.7) (Fig. 2a) and the low diagnostic performance of the RAT (Table 2, Table 3), we investigated whether combining laboratory indices measured in blood with the results of RAT would enhance the true identification of COVID-19 patients by RAT. First, the SVM model revealed that HB, urea, RAT, and S. ferritin were the top-4 parameters most frequently selected during the model building and cross-validation followed by the other features (Fig. 2b). The various combination between these parameters (i.e. those listed in ascending order in Fig. 2b) yielded various accuracies in predicting true COVID-19 cases. The highest prediction accuracy (59.3%) was obtained when combining RAT with both HB and urea (top 2-features in Fig. 2b). Coupling RAT with HB, urea, S. ferritin, and CRP (top 5- ranked features) yielded slightly lower prediction accuracy (58%) than the one produced by the 3-feature model. Combining all features together revealed low prediction accuracy of 48%. Subsequent evaluation of the “top 3-feature” model by predictive class probability analyses (Fig. 2d) revealed a sensitivity of 75.4%, where this model correctly identified 43 as positive subjects out of the 57 true positive ones (as determined by RT-qPCR). The model specificity was 81.8%, where the model correctly identified 9 negative subjects out of the 11 true negative ones. The misclassified subjects are labeled in Fig. 2d.
Important determinants for RT-qPCR and RAT results by the random forest classification model
As shown in Fig. S1, it was found that the sampling time post-symptom onset was the most significant parameter that determines the results in both assays. The order of importance of other parameters differed between the RT-qPCR and the RAT. Dyspnea and radiological findings were the top two parameters for the RT-qPCR results, whereas the subject's age and chest pain were the most important features of the RAT.
Discussion
In this study, we aimed to evaluate the diagnostic performance of Standard™ Q COVID-19 Ag test, a recently commercialized RAT in Egypt, and to investigate the patient’ features that could influence test performance. Determining the diagnostic performance of commercialized RAT is crucial because this gives an indication of their reliability and clinical utility during the time of the pandemic. Our data suggest that RAT has low performance as compared to the RT-qPCR. Combining laboratory parameters with RAT did not enhance RAT predictive accuracy. To the best of our knowledge, this study deems the first one in Egypt that provides a detailed evaluation of the diagnostic performance of a RAT against RT-qPCR on Egyptian subjects.
Indeed, RT-qPCR and RAT differ in terms of protocols, core idea, targets and application. As both of the test were in use during COVID-19 pandemic, comparing their ability to identify truly infected individuals have become an important and logical point of research [6,17].
Given that the ideal RAT should have a sensitivity >95% and a specificity of 100% [15], The Standard™ Q COVID-19 Ag studied here showed less than optimal performance. The observed 78.2% sensitivity means that RAT test has falsely considered 21.7% (15/69) of the COVID-19 true positive cases as non-infected. Similarly, a specificity of 64.2% means that RAT has falsely considered 35.7% (5/14) of the COVID-19 negative subjects as positive. The lack of sensitivity of the RAT could lead to disease dissemination among the population if the missed patients are infectious. Actually, an RT-qPCR-positive subject does not necessarily mean that he/she is infectious. Our data indicated the majority of the 15 false-negative patients by RAT had a low viral load, although being symptomatic (Table S1). Since we did not isolate live viruses from those patients, their infectiousness remains unknown and the presence of symptoms does not imply that the person is infectious as shown previously for COVID-19 patients with low viral load [27]. Symptoms in those “low or no viral load” groups could be attributed to virus-induced end-organ damage, as shown in their radiological findings, rather than the presence of replicating virus. On the other side, the lack of specificity could lead to extra cost due to the wrong decision of isolation or advising needless therapy. At the time of writing this paper, we are analyzing clinical data from a big Egyptian cohort, which might solidify some of these conclusions. The current RAT showed higher sensitivity and lower specificity when it was applied in 262 Ugandan subjects [15]. RAT had higher sensitivity (98.3%) and higher specificity (98.7%) than our results when applied on 454 subjects from Thailand [17]. This indicates that test results might be race/ethnicity- dependent. Our data add to the already known diversity in RATs results. The sensitivity of the current RAT was higher than that obtained by BIOCREDIT COVID-19 Ag test (43.1%) applied on nasal swabs in Egypt [18]. In two independent studies, Ag Respi-Strip (Coris Bioconcept, Gembloux, Belgium) exhibited specificity of 100% and sensitivity ranged from 30 to 50% [14,16]. Fluorescence RAT done on 239 participants in China showed low sensitivity of 68% and maximum specificity (100%) [28]. The fluorescence immunochromatographic assay produced 93.9% sensitivity and 100% specificity when used on 127 subjects from Chile [29]. The differences in test performance could be due to variabilities in the participant’s clinical features, sample type and processing, PCR protocol, and viral load in samples. When evaluating any RAT performance, it is worth noting that misdiagnosis of COVID-19 patients could be due to the presence of many virus variants and thus difference between the virus strain contained in the sample and the virus strain against which the antibodies coated in the RAT were raised. This is highlighted knowing that Standard™ Q COVID-19 Ag was designed to detect the original WUHAN-01 strain and that mutation rate is high in the antibody-target SARS-CoV-2 N protein [30]. It is therefore recommended to continuously evaluate and update the validity of this and other RAT when applied in different communities that might experience other SARS-CoV-2 strains especially with the beginning of the second wave.
Our data showed that the Standard™ Q COVID-19 Ag was more sensitive and more accurate in patients with a high viral load than those with low viral load. Similar results were shown for the same RAT in Uganda [15] and for other qualitative [14,29,31] and quantitative [32] RATs. In parallel, RAT showed the highest sensitivity and accuracy in the samples collected during the first-week post-symptoms, and sampling time was the top important feature that determines the results of both RT-qPCR and RAT as revealed by our random forest classification (Fig. S1). These findings support previous studies that reported a 14% decrease in sensitivity of fluorescence immunochromatographic assay when performed on samples collected between 8–12 days post-symptoms relative to earlier samples [29]. It is already known that SARS-CoV-2 load in upper respiratory tract samples often peaks few days after symptom onset [33,34]. This complements our results since 17 out of the 28 subjects with RAT positive and strong RT-qPCR were sampled between 0−7 days post-symptoms. Taken together, this suggests a triple relationship between the high diagnostic performance of RAT, high viral load in the sample, and the early time of sampling post-symptoms and highlights the clinical utility of RAT in severely affected patients with high viral load and at early stages of COVID-19 infection.
Many studies exist that analyzed the performance of RAT, yet limited studies correlate patient’s clinical and radiological features to the RAT performance. Our observation that Standard™ Q COVID-19 Ag test has higher sensitivity and accuracy in symptomatic than asymptomatic subjects is in line with the previous study done on 3410 Italian patients using the same assay, where the RAT’s sensitivity declined from 89.9% in symptomatic subjects to 50% in the asymptomatic ones. Our analyses indicated that the Standard™ Q COVID-19 Ag test was able to detect, albeit with a very faint line, RT-qPCR-negative subjects who were asymptomatic and had no radiological alteration. This highlights the importance of subjecting asymptomatic suspected individuals to the test and that RAT might be sensitive enough to truly detect asymptomatic carriers, who likely account for a significant portion of disease transmission events among humans [35]. Additional analyses are needed to better evaluate this RAT in asymptomatic subjects, since our study included low number of this category (n = 5). Our data indicate the low clinical value of radiological analyses in determining COVID-19 patients relative to RT-qPCR or even the RAT since all participants who had no radiological alteration proved positive by RT-qPCR (4 of them have high Ct value > 25) and five of them were also positive by RAT. Obviously, additional analyses are needed to generalize these observations.
From a diagnosis point of view, particularly in the absence of RT-qPCR in local sites, it might be useful to combine RAT results with laboratory measurements in patient’s blood in pursuit of enhancing RAT performance in detecting patients. The machine learning approach employed here enabled us to test this hypothesis. The best-obtained and validated model (formed of RAT plus HB and urea) gave a predictive accuracy of 59.3% and other models with more features, that are COVID-19 related, gave even lower accuracy than this one. This analysis scheme suggests that using laboratory parameters might not afford the desired improvement in the diagnostic performance of the RAT studied here, and possibly other RAT. Another point to consider for clinicians is the parameters that should be taken into consideration when performing the test given the differences in the results between RT-qPCR and RAT. The vast difference between determinants of both assays (as shown by the random forest classification model) suggests that the differences between the results of both assays have reflected on the parameters to be considered as determinants for the assay.
We acknowledge that this analysis is limited by some factors that should be taken into account in future studies: the small sample size and the unavailability of some participant’s data were due to logistic hurdles during the pandemic time. Obviously, additional samples are required for evaluating RAT. The accelerated pressure for obtaining results precluded us from evaluating the influence of sample processing procedures on the RAT accuracy, such an important factor that might alter test results. We do believe that the strength of this study lies in its performance in real-life settings. We were able to link viral load, sampling time, clinical symptoms, and laboratory parameters to the assay results and to test, by a machine learning approach, the effect of measuring blood parameters on enhancing RAT performance.
Conclusion
Based on the real-world data described here, Standard™ Q COVID-19 Ag has the disadvantage of low diagnostic performance relative to the RT-qPCR; its sensitivity varies with sampling time and with the amount of viral nucleic acid contained in the sample. This test is best used for subjects with high viral load and when done early after COVID-19 symptom onset. Pending its application on large scale, our data recommend against using this test alone for COVID-19 diagnosis. RAT, therefore, has no benefit in replacing or reducing the use of RT-qPCR assay for COVID-19 diagnosis at the time of the pandemic.
Availability of data and material
Data from this study are available as supplementary materials. Any other data are available from the corresponding authors on reasonable request.
Code availability
Not applicable.
Ethics approval
The study was approved by the ethical committee at the Faculty of Medicine, Zagazig University (IRB number: 6263, issued on 14.07.2020).
Consent to participate
Written informed consent was obtained from each enrolled participant.
Consent for publication
Participants have agreed to publish their data anonymously in this work.
Funding
This study did not receive any funding.
Conflicts of interest/competing interests
The authors declare no conflict of interest.
Acknowledgment
The authors thank staff members at the “Zagazig Scientific and Medical Research Center, Zagazig University” for their assistance during the laboratory work. This work did not receive any funding.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jiph.2021.06.002.
Appendix A. Supplementary data
The following are Supplementary data to this article:
Demographic and clinical features of the study participants.
Diagnostic criteria for RAT for participant’s subgroups.
Random forest classification model showing the ranked importance of subjects’ demographic and clinical features in predicting the results of RAT and RT-qPCR assays. The features are ranked in an ascending order according to the mean decrease in accuracy (x-axis) when the respective feature was permuted.
References
- 1.Akashi Y., Suzuki H., Ueda A., Hirose Y., Hayashi D., Imai H. Analytical and clinical evaluation of a point-of-care molecular diagnostic system and its influenza A/B assay for rapid molecular detection of the influenza virus. J Infect Chemother. 2019;25(8):578–583. doi: 10.1016/j.jiac.2019.02.022. [DOI] [PubMed] [Google Scholar]
- 2.WHO (11 March 2020) Coronavirus diseases (COVID-19) global situation report-51.
- 3.WHO (11 January 2020) COVID-19 global situation report.
- 4.Nkengasong J.N., Mankoula W. Looming threat of COVID-19 infection in Africa: act collectively, and fast. Lancet. 2020;395(10227):841–842. doi: 10.1016/S0140-6736(20)30464-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leung T.Y.M., Chan A.Y.L., Chan E.W., Chan V.K.Y., Chui C.S.L., Cowling B.J. Short- and potential long-term adverse health outcomes of COVID-19: a rapid review. Emerg Microbes Infect. 2020;9(1):2190–2199. doi: 10.1080/22221751.2020.1825914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang Y.W., Schmitz J.E., Persing D.H., Stratton C.W. Laboratory diagnosis of COVID-19: current issues and challenges. J Clin Microbiol. 2020;58(6) doi: 10.1128/JCM.00512-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen L., Cui S., Zhang D., Lin C., Chen L., Wang Q. Comparison of four commercial RT-PCR diagnostic kits for COVID-19 in China. J Clin Lab Anal. 2020 doi: 10.1002/jcla.23605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Kasteren P.B., van der Veer B., van den Brink S., Wijsman L., de Jonge J., van den Brandt A. Comparison of seven commercial RT-PCR diagnostic kits for COVID-19. J Clin Virol. 2020;128 doi: 10.1016/j.jcv.2020.104412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peeling R.W., Olliaro P.L., Boeras D.I., Fongwen N. Scaling up COVID-19 rapid antigen tests: promises and challenges. Lancet Infect Dis. 2021 doi: 10.1016/S1473-3099(21)00048-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anjum F.R., Anam S., Rahman S.U. Novel Coronavirus disease 2019 (COVID-19): new challenges and new responsibilities in developing countries. Hum Vaccin Immunother. 2020;16(10):2370–2372. doi: 10.1080/21645515.2020.1766939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sheridan C. Fast, portable tests come online to curb coronavirus pandemic. Nat Biotechnol. 2020;38(5):515–518. doi: 10.1038/d41587-020-00010-2. [DOI] [PubMed] [Google Scholar]
- 12.Olalekan A., Iwalokun B., Akinloye O.M., Popoola O., Samuel T.A., Akinloye O.M. COVID-19 rapid diagnostic test could contain transmission in low- and middle-income countries. Afr J Lab Med. 2020;9(1) doi: 10.4102/ajlm.v9i1.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wee L.E., Fua T.P., Chua Y.Y., Ho A.F.W., Sim X.Y.J., Conceicao E.P. Containing COVID-19 in the emergency department: the role of improved case detection and segregation of suspect cases. Acad Emerg Med. 2020;27(5):379–387. doi: 10.1111/acem.13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lambert-Niclot S., Cuffel A., Le Pape S., Vauloup-Fellous C., Morand-Joubert L., Roque-Afonso A.M. Evaluation of a rapid diagnostic assay for detection of SARS-CoV-2 antigen in nasopharyngeal swabs. J Clin Microbiol. 2020;58(8) doi: 10.1128/JCM.00977-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nalumansi A., Lutalo T., Kayiwa J., Watera C., Balinandi S., Kiconco J. Field evaluation of the performance of a SARS-CoV-2 antigen rapid diagnostic test in Uganda using nasopharyngeal samples. Int J Infect Dis. 2020 doi: 10.1016/j.ijid.2020.10.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scohy A., Anantharajah A., Bodeus M., Kabamba-Mukadi B., Verroken A., Rodriguez-Villalobos H. Low performance of rapid antigen detection test as frontline testing for COVID-19 diagnosis. J Clin Virol. 2020;129 doi: 10.1016/j.jcv.2020.104455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaimayo C., Kaewnaphan B., Tanlieng N., Athipanyasilp N., Sirijatuphat R., Chayakulkeeree M. Rapid SARS-CoV-2 antigen detection assay in comparison with real-time RT-PCR assay for laboratory diagnosis of COVID-19 in Thailand. Virol J. 2020;17(1):177. doi: 10.1186/s12985-020-01452-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abdelrazik A.M., Elshafie S.M., Abdelaziz H.M. Potential use of antigen-based rapid test for SARS-CoV-2 in respiratory specimens in Low-Resource settings in Egypt for symptomatic patients and high-risk contacts. Lab Med. 2020 doi: 10.1093/labmed/lmaa104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dean A.G.S.K., Soe M.M. 2013. Open source epidemiologic statistics for public health. Cited 2020/11/29 2020. [Google Scholar]
- 20.WHO (16 December 2020) WHO COVID-19: Case definitions: updated in public health surveillance for COVID-19, published 16 December 2020.
- 21.CDC . 2020. Interim guidelines for collecting, handling, and testing clinical specimens for COVID-19. [Google Scholar]
- 22.Watson P.F., Petrie A. Method agreement analysis: a review of correct methodology. Theriogenology. 2010;73(9):1167–1179. doi: 10.1016/j.theriogenology.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 23.Brown L.D., Cai T.T., DasGupta A. Interval estimation for a binomial proportion. Stat Sci. 2001;16(2):101–117. [Google Scholar]
- 24.de Araujo L.S., Ribeiro-Alves M., Leal-Calvo T., Leung J., Durán V., Samir M. Reprogramming of small noncoding RNA populations in peripheral blood reveals host biomarkers for latent and active Mycobacterium tuberculosis infection. mBio. 2019;10(6) doi: 10.1128/mBio.01037-19. e01037-01019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stacklies W., Redestig H., Scholz M., Walther D., Selbig J. pcaMethods—a bioconductor package providing PCA methods for incomplete data. Bioinformatics. 2007;23(9):1164–1167. doi: 10.1093/bioinformatics/btm069. [DOI] [PubMed] [Google Scholar]
- 26.Pang Z., Chong J., Li S., Xia J. MetaboAnalystR 3.0: toward an optimized workflow for global metabolomics. Metabolites. 2020;10(5) doi: 10.3390/metabo10050186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singanayagam A., Patel M., Charlett A., Lopez Bernal J., Saliba V., Ellis J. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro Surveill. 2020;25(32) doi: 10.2807/1560-7917.ES.2020.25.32.2001483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diao B., Wen K., Chen J., Liu Y.-P., Yuan Z., Han C. Diagnosis of acute respiratory syndrome coronavirus 2 infection by detection of nucleocapsid protein. medRxiv. 2020 doi: 10.1101/2020.03.07.20032524. [DOI] [Google Scholar]
- 29.Porte L., Legarraga P., Vollrath V., Aguilera X., Munita J.M., Araos R. Evaluation of a novel antigen-based rapid detection test for the diagnosis of SARS-CoV-2 in respiratory samples. Int J Infect Dis. 2020;99:328–333. doi: 10.1016/j.ijid.2020.05.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rahman M.S., Islam M.R., Alam A., Islam I., Hoque M.N., Akter S. Evolutionary dynamics of SARS-CoV-2 nucleocapsid protein and its consequences. J Med Virol. 2020 doi: 10.1002/jmv.26626. [DOI] [PubMed] [Google Scholar]
- 31.Abdelrazik A.M., Elshafie S.M., Abdelaziz H.M. Potential use of antigen-based rapid test for SARS-CoV-2 in respiratory specimens in low-resource settings in Egypt for symptomatic patients and high-risk contacts. Lab Med. 2020 doi: 10.1093/labmed/lmaa104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hansen G., Marino J., Wang Z.X., Beavis K.G. Clinical performance of the point-of-care cobas Liat for detection of SARS-CoV-2 in 20 minutes: a multicenter study. J Clin Microbiol. 2021;59(2) doi: 10.1128/jcm.02811-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382(12):1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 35.Cloutier L., Merindol N., Pépin G., Marcoux-Huard C., Vasil P.-A., Houle C. Asymptomatic carriers of COVID-19 in a confined adult community population in Quebec: a cross-sectional study. Am J Infect Control. 2021;49(1):120–122. doi: 10.1016/j.ajic.2020.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Demographic and clinical features of the study participants.
Diagnostic criteria for RAT for participant’s subgroups.
Random forest classification model showing the ranked importance of subjects’ demographic and clinical features in predicting the results of RAT and RT-qPCR assays. The features are ranked in an ascending order according to the mean decrease in accuracy (x-axis) when the respective feature was permuted.
Data Availability Statement
Data from this study are available as supplementary materials. Any other data are available from the corresponding authors on reasonable request.