Introduction
Although the incidence of head and neck cancers across almost all major anatomic subsites is decreasing, oropharyngeal squamous cell carcinoma (OPSCC) is unique in that it has steadily increased in incidence over the last 40 years.1 Global estimates suggest that approximately 20 to 25% of all OPSCC is related to high risk human papillomavirus (HPV), particularly HPV type 16.2 In many countries, including the United States, HPV-related OPSCC is much more prevalent, with 70% or more of all OPSCC patients having HPV-related tumors.
HPV-related OPSCC tends to affect slightly younger, former or never smoking males, although the age gap between it and traditional OPSCC appears to be closing over time.3 In these patients, tumor HPV-status is a well-proven, independent prognostic factor for survival.4 Thus, accurate assignment of tumor HPV status is critical for appropriate patient counseling, staging, and consideration of de-escalation of care or enrollment in clinical trials. In the United States, overexpression of p16 protein by immunohistochemistry (IHC), with greater than 70% nuclear and cytoplasmic staining of at least moderate to strong intensity, is considered both a suitable prognostic marker and surrogate marker of transcriptionally active high-risk HPV. It is currently recommended by both the College of American Pathologists and the American Society of Clinical Oncology for largely practical reasons, despite its obvious shortcoming as only an indirect test for transcriptionally-active HPV.5,6
Recent studies have identified a subgroup of patients with discordant p16 and HPV-specific testing results, raising concern for the performance of p16 IHC or HPV specific testing alone. Discordant patients may be HPV positive and p16 negative, or more commonly, p16 positive and HPV negative (undetectable by HPV specific testing).7 Early studies have estimated that approximately 5 to 10% of all OPSCC demonstrate overexpression of p16 that is not associated with HPV infection. This is the so-called “false positive” problem surrounding p16 testing alone.8,9 Interestingly, the available data suggest that those with discordant testing (p16+/HPV mRNA- or p16-/HPV mRNA+) may experience varying clinical outcomes that fall somewhere between patients who are truly double positive (p16+/hrHPV mRNA+) and double negative (p16-/hrHPV mRNA-).9–11
Studies specifically examining this phenomenon and its significance for patients have emanated primarily from Europe where the HPV fraction is variable, but significantly lower overall than in the United States. More importantly, these studies have been performed with either DNA PCR or DNA in situ hybridization (ISH) for HPV detection rather than with mRNA-based HPV testing, which is considered the reference standard. To examine the phenomenon of discordant p16 and high-risk HPV mRNA status in our Western population, we performed a multi-institutional, retrospective study of well characterized OPSCC patient cohorts using p16 IHC and reverse transcriptase PCR (RTPCR) for high risk HPV mRNA.12–18 Our objectives were to show the rate of discordant testing and to characterize the demographics, clinicopathologic features, and prognosis for those patients. We also sought to develop an algorithm for predicting who might benefit from HPV specific testing in addition to p16 IHC that might be an alternative in having to perform p16 and HPV mRNA testing in all patients with OPSCC.
Materials and Methods
A multi-institutional, retrospective cohort of patients diagnosed with OPSCC at Vanderbilt University Medical Center (VUMC) and Washington University in St. Louis (WU) between 2003 and 2017, respectively, was gathered for study.
Vanderbilt University Medical Center
Incident, previously untreated cases of OPSCC were identified though the Vanderbilt Research Derivative (RD), an IRB-approved, identified, searchable database of more than 3.5 million electronic health records (EHR) from patients seen at VUMC.19 Patients diagnosed between June 1, 2000 and July 9, 2018 were identified using the following International Classification of Diseases for Oncology, 3rd Edition (ICD-O-3) codes: C01.9, C02.4, C05.1, C05.2, C05.8, C09.0, C09.1, C09.8, C09.9, C10.0, C10.2, C10.3, C10.8, C10.9, C14.0 and C14.2. Clinical data were abstracted through manual review of the EHR. Patients for whom an OPC diagnosis could not be confirmed, those with a prior history of cancer (other than non-melanoma skin cancer), patients who were immunocompromised, and those without an in-house tumor specimen collected prior to treatment were excluded. For patients who met inclusion criteria, all surgical pathology specimen slides and pathology files created as part of the original diagnosis were reviewed by trained head and neck pathologists (JSL and/or MM). Tumor specimens of insufficient size for RNA extraction, those containing less than 25% tumor, those with original histologic diagnoses inconsistent with the findings of the pathology re-review, or with tumor blocks missing and/or exhausted were excluded.
Washington University in St. Louis
A prior published study examining morphology and correlation with high-risk HPV mRNA status was utilized for the current study. In short, 70 consecutive patients from 2011 and 2012 who had OPSCC biopsy or resection specimens and had been tested for p16 IHC in routine clinical practice were tested for high-risk HPV mRNA by RTPCR, using the same methods as the VUMC patients.20 Medical records were manually reviewed to obtain baseline characteristics and outcome data. Patient specimens in both the VUMC and WU cohorts were both classified for morphology using a single established system into keratinizing, nonkeratinizing, and nonkeratinizing with maturation as previously described (Figure 1).21,22
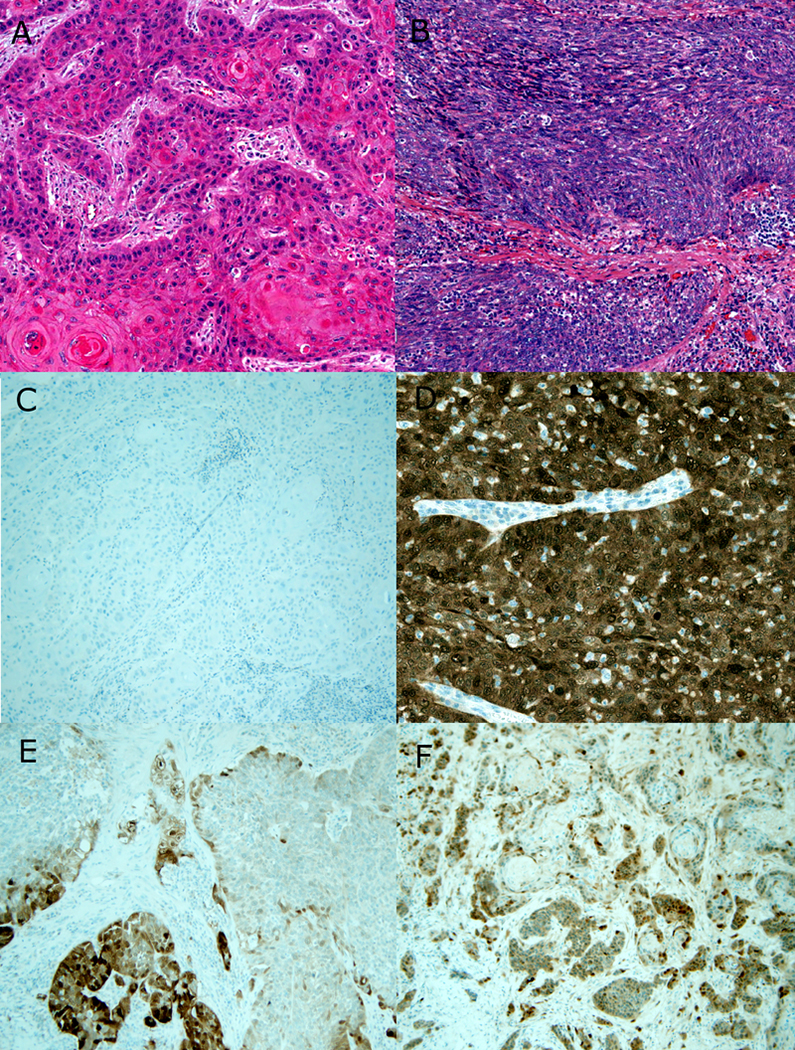
Figure 1 –
Morphology and p16 IHC in OPSCC. A) Typical keratinizing SCC with angulated nests of cells with dense, eosinophilic cytoplasm, scattered keratin material, and prominent stromal desmoplastic reaction (8X magnification; hematoxylin and eosin). B) Typical nonkeratinizing SCC with large nests of blue cells, defined borders with no stromal reaction, and tumor cells with round to spindled nuclei lacking prominent nucleoli and with brisk mitosis/apoptosis (8X magnification; hematoxylin and eosin). C) Negative p16 immunohistochemistry for the keratinizing type SCC (10X magnification). D) Strongly and diffusely positive p16 immunohistochemistry for the nonkeratinizing SCC (20X magnification). E) Example of a negative p16 immunohistochemical stain with approximately 30 to 40% of cells positive (10X magnification). F) Example of an equivocal p16 immunohistochemical stain with approximately 60% of cells positive (10X magnification).
Approval from the respective institutional review boards had been obtained prior to study (VUMC Human Research Protection Program Protocols #151366 and #171218 and WU Human Research Protection Office Protocol #201108311).
p16 Immunohistochemistry
p16 IHC was performed at two different laboratories and times. For VUMC, IHC was performed for p16 using the E6H4 antibody (Ventana Medical Systems, Inc.; monoclonal; 1:1 dilution) on a Leica Bond automated instrument (Leica Biosystems, Inc.) on 4 μm formalin-fixed, paraffin-embedded tissue sections with antigen retrieval consisting of 10 minutes in the ER1 proprietary antigen retrieval solution. Primary antibody solution was diluted using Leica’s BOND primary antibody diluent. The Bond Polymer Refine detection system was used for visualization. Slides were then dehydrated, cleared, and coverslipped. For WU, p16 IHC had been performed in routine clinical practice using the E6H4 antibody (Ventana Roche MTM Laboratories; monoclonal; 1:1 dilution) on 4 μm formalin-fixed, paraffin-embedded tissue sections using a Ventana Benchmark automated immunostainer. The VUMC p16 stains were reviewed by one of the study head and neck specialist pathologists (MM and/or JSL) while the WU p16 stains had been reviewed in routine clinical practice which included three head and neck pathologists, two of whom are on this current study (RC or JSL). Staining was interpreted for percentage of nuclear and cytoplasmic staining with a cutoff of 70% for positivity (Figure 1). Cases with staining between 50 and 70% were described and flagged as equivocal.
Expression Profiling of High-Risk HPV Using RT-PCR
Formalin fixed paraffin embedded (FFPE) tissue blocks were retrieved from surgical pathology archives and cut into 10 μm sections on glass slides. Tumor rich areas were circled on a representative H&E slide with a dotting pen by a head and neck pathologist (JSL, RDC, or MM) for macro-dissection. Then, total RNA was extracted from the identified tumor regions with the miRNeasy FFPE Kit (Qiagen Inc, Valencia, Calif) according to the manufacturer’s protocol. Total DNA was extracted from the same identified tumor regions using a QIAamp DNA FFPE Tissue Kit (Qiagen). In this way, we were able to focus on the profiling of the tumors with minimal contamination from adjacent normal tissues. HPV RT-qPCR assays were used to profile the expression of HPV E6 and E7 in OPSCC with total RNA extracted from the tumor blocks. All oligo primers in the assays were purchased from Sigma-Aldrich. Reverse transcription (RT) reaction was performed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Real-time PCR was then performed using Power SYBR Green PCR Master Mix (Applied Biosystems) and 500 nM HPV type-specific primers. HPV assays (profiling E6 and E7 transcripts separately from each of the 13 HPV types) were individually performed on a 384-well PCR plate. The PCR running protocol was 95°C for 10 min, followed by 36 cycles of amplification (95°C for 10 sec, 58°C for 15 sec and 60°C for 15 sec). HPV positivity was determined by the detection of both HPV E6 and E7 transcripts based on normalized expression levels with RT-qPCR.12
Statistical Analysis
Differences in the distribution of baseline covariates were assessed using one-way analyses of variance for continuous variables and Pearson contingency table chi-squared tests for categorical variables. Survival curves were plotted using the Kaplan-Meier method. The effect of baseline covariates on disease-free and overall survival were estimated by Cox proportional hazards regression. Patients determined to not have undergone definitive treatment (palliative chemotherapy or radiation, no treatment, or chemotherapy only) were excluded from survival analysis. Factors included in multivariable regression were determined a priori and included concordance (or lack thereof) between p16 IHC and HPV RTPCR, age, race, smoking status, tumor staging, p16 status, HPV mRNA status, and treatment regimen. Statistical analysis was performed with Stata (StataCorp, College Station, TX).
Cox proportional hazards ratio was used for multivariate regression analysis with Breslow method for ties, with particular attention to differences in those who were p16-positive/HPV mRNA positive, p16-negative/HPV mRNA negative, and either p16-negative/HPV mRNA positive or p16-positive/HPV mRNA negative (discordant). Patients determined to not have undergone definitive treatment (palliative chemotherapy or radiation, no treatment, or chemotherapy only) were excluded from survival analysis. Factors included in multivariate regression were determined a priori and included concordance (or lack thereof) between p16 IHC and HPV RTPCR, age, race, smoking status, tumor staging, p16 status, HPV mRNA status, and treatment regimen. Statistical analysis was performed with Stata (StataCorp, College Station, TX). Overall survival was defined as follow up time from the completion of treatment to time of death or to last follow up if surviving. Disease free survival was defined as follow up time from the completion of treatment to disease recurrence, death, or to last follow up if surviving and disease free.
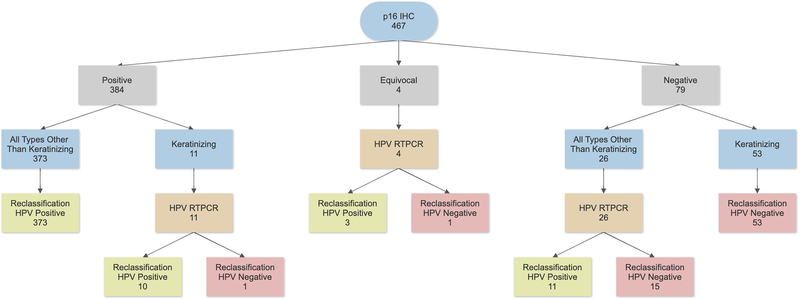
Because it has been suggested that 1) borderline p16 IHC (50–70% staining, by strict CAP guidelines, a negative result) is often still positive for transcriptionally-active high-risk HPV23 and because 2) p16 results not matching the other characteristics of the patients, particularly morphology, can be used to apply HPV-specific testing selectively as an “arbiter”, we applied a reclassification scheme to our patient results with the thought experiment as if starting only with p16 IHC and the other clinical and pathologic characteristics. This allowed us to compare whether a selective application of HPV mRNA testing after p16 testing would provide a reasonable compromise between the extremes of p16 IHC alone and p16 IHC plus high-risk HPV mRNA testing for all patients (Figure 2).
Figure 2 –
HPV reclassification scheme using p16 IHC and morphology to determine the need for, and application of, HPV mRNA testing in routine clinical practice.
Results
Participant Characteristics
A total of 467 OPSCC patients were identified and tested for p16 IHC and high-risk HPV mRNA. As expected for a large U.S. OPSCC cohort, patients were primarily Caucasian men (411, 88%) in their upper 50s (mean 57.1, SD 9.6), most of whom were current or former smokers (64%). Most patients had either T1 or T2 tumors (71%). Most were p16 positive (82%), and/or high-risk HPV mRNA positive (84%). In patients treated with curative intent, 250 (54%) underwent surgery with postoperative adjuvant therapy, 159 (34%) underwent definitive chemoradiation, and 44 (9%) underwent surgery alone.
Most of the tumors were nonkeratinizing (378 overall, or 81%). Of these the most common pattern was nonkeratinizing (264 or 57%) or nonkeratinizing with maturation (114 or 24%), and nonkeratinizing morphology was strongly associated with p16 and HPV positivity (93% and 95%, respectively) (Table 1). Comparing p16 and HPV specific testing, 81% of patients were double positive (p16-positive/HPV mRNA positive), 14% double negative (p16-negative/HPV mRNA negative), and 4.9% discordant, of whom 3.4% were p16-negative/HPV mRNA positive and just 1.5% p16-positive/HPV mRNA negative.
Table 1:
Patient Characteristics
| Double Positive | Discordant | Double Negative | Total | Significance | ||
|---|---|---|---|---|---|---|
| (n=377) | (n=23) | (n=67) | (n=467) | (P) | ||
| Demographics/Baseline Characteristics | ||||||
| Age, Mean years (SD) | 57 (9) | 57 (13) | 58 (11) | 57.1 (9.6) | 0.69 | |
| Sex, Male, N (%) | 342 (91) | 21 (91) | 48 (72) | 411 (88.0) | < 0.001 | |
| Race, Caucasian, N (%) | 360 (95) | 21 (91) | 45 (67) | 426 (91.2) | < 0.001 | |
| Smoking Status, N (%) | Never Smoker | 154 (41) | 7 (30) | 6 (9) | 167 (35.8) | < 0.001 |
| Former or Current Smoker | 221 (59) | 16 (70) | 61 (91) | 298 (63.8) | ||
| p16-Status, N (%) | Positive | 384 (82.2) | ||||
| Negative | 83 (17.8) | |||||
| HPV mRNA, N (%) | Positive | 393 (84.2) | ||||
| Negative | 74 (15.9) | |||||
| Initial Classification, N (%) | p16-negative/HPV-negative | 67 (14.4) | ||||
| p16-negative/HPV-positive | 16 (3.4) | |||||
| p16-positive/HPV-negative | 7 (1.5) | |||||
| p16-positive/HPV-positive | 377 (80.7) | |||||
| Clinical T Stage, N (%) | T0 - T2 | 286 (76) | 15 (65) | 31 (46) | 332 (71.1) | < 0.001 |
| T3 - T4 | 91 (24) | 8 (35) | 34 (51) | 133 (28.5) | ||
| Morphology, N (%) | Nonkeratinizing | 253 (67) | 7 (30) | 4 (6) | 264 (56.5) | < 0.001 |
| Nonkeratinizing with Maturation | 93 (25) | 11 (48) | 10 (15) | 114 (24.4) | ||
| Keratinizing | 10 (3) | 4 (17) | 52 (78) | 66 (14.1) | ||
| Other | 21 (6) | 1 (4) | 1 (1) | 23 (4.9) | ||
| Treatment, N (%) | Definitive Radiation | 122 (32) | 10 (43) | 27 (40) | 159 (34.1) | < 0.001 |
| Postoperative Radiation | 216 (57) | 11 (48) | 23 (34) | 250 (53.5) | ||
| Surgery Alone | 35 (9) | 0 (0) | 9 (13) | 44 (9.4) | ||
| Other | 4 (1) | 2 (9) | 8 (12) | 14 (3.0) | ||
| Chemotherapy, N (%) | 252 (67) | 18 (78) | 35 (52) | 305 (65.3) | 0.04 | |
| p16/HPV Reclassification, N (%) | Negative | 0 (0) | 3 (13) | 67 (100) | 70 (15.0) | < 0.001 |
| Positive | 376 (100) | 20 (87) | 0 (0) | 396 (84.8) | ||
The 23 discordant patients were demographically and clinically more like the double positive patients (Table 1), being primarily Caucasian men with a high fraction of never smokers, with lower T stage tumors, and with a predominance of nonkeratinizing morphology. Examining anatomic subsites, 11 had tonsil primaries (48%), 9 had base of tongue primaries (39%), and 3 (13%) had soft palate or lateral pharyngeal wall primaries. Compared to double positive patients, discordant patients were less likely to have nonkeratinizing morphology (78% vs 92%) and more likely to be treated with primary chemoradiation than surgery compared to the double positive patients (Table 1). The discordant group and the double negative group differed in several ways as well, including being more likely to be male (91% vs 72%), to be Caucasian (91% vs 67%), and to have nonkeratinizing morphology (78% vs 21%). They were also more likely to receive chemotherapy (78% vs 52%), less likely to be a former or current smoker (70% vs 91%), less likely to suffer disease recurrence/treatment failure (39% vs 64%), and less likely to die in the follow up period (39% vs 61%). In examining the separate discrepant groups, the p16 positive/HPV negative patients consisted of 4 (57%) nonkeratinizing SCC, 2 (29%) nonkeratinizing SCC with maturation, and 1 (14%) keratinizing SCC, compared to the p16 negative/HPV positive patients, who consisted of 3 (19%) nonkeratinizing SCC, 9 (56%) nonkeratinizing SCC with maturation, 3 (19%) keratinizing SCC, and 1 (6%) lymphoepithelial/undifferentiated carcinoma. In short, the discrepant group fell between double positive and double negative patients but was closer overall to the double positive ones.
Survival Analyses
A total of 112 patients (24%) died in the follow up period, with a mean survival of 3.1 years (SD 3.1) and mean time to recurrence of 3.0 years (SD 3.0).
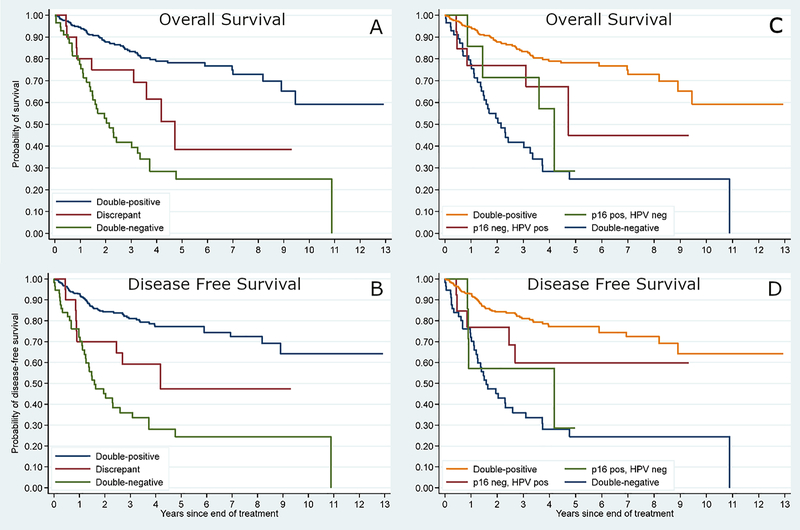
In univariate survival analysis, the discordant patients had statistically significantly greater rates of death (39% vs 16%; p = 0.01) and disease recurrence/treatment failure (39% vs 19% p = 0.02) than the double positive patients. Their survival rates along the Kaplan-Meier curves fell squarely between the two concordant groups (Figure 3).
Figure 3 –
Kaplan-Meier survival plots showing the various cohorts by p16 and HPV testing. Overall survival (A) and disease-free survival (B) for the three cohorts: double positive, double negative, and discrepant. Overall survival (C) and disease-free survival (D) showing with the discrepant patients separated into p16 positive/HPV mRNA negative or p16 negative/HPV mRNA positive.
In multivariate hazards regression analysis for both overall and disease-free survival, discordant patients were not found to have statistically significant differences in survival compared to either double positive or double negative patients (Table 2). For overall survival, p16 and HPV mRNA double negativity (HR 4.13 95% CI 2.67 – 6.37), increasing age (HR 1.03, 95% CI 1.01 – 1.05), ever smoking status (HR 2.11, 95% CI 1.27 – 3.5), T3/T4 disease (HR 1.69, 95% CI 1.11 – 2.56), and whether the patient received definitive radiation therapy with or without chemotherapy (HR 1.65, 95% CI 1.09 – 2.49) were all associated with significantly increased hazard of death (Table 2). For disease free survival, p16 and HPV mRNA double negativity (HR 3.64, 95% CI 2.40 – 5.53), increasing age (HR 1.03, 95% CI 1.01 – 1.04), smoking (HR 2.28, 95% CI 1.40 – 3.73), and T3/T4 disease (HR 1.56, 95% CI 1.05 – 2.33) were all also associated with significantly increased hazard of death or disease recurrence.
Table 2:
Multivariable survival analysis: p16/HPV test combination groups.
| Disease Free Survival | HRR* | 95% CI | Significance (P) |
| Double Positive (Reference category) | 1.00 | ||
| Discordant | 1.91 | 0.93 – 3.95 | 0.08 |
| Double Negative | 3.64 | 2.40 – 5.53 | <0.001 |
| Age (HRR associated with an additional year of life) | 1.03 | 1.01 – 1.05 | 0.004 |
| Former or Current Smoker | 2.28 | 1.40 – 3.73 | 0.001 |
| T3/T4 | 1.56 | 1.05 – 2.33 | 0.03 |
| Definitive / Chemo only | 1.46 | 0.97 – 2.15 | 0.07 |
| Overall Survival | HRR* | 95% CI | Significance (P) |
| Double Positive (Reference category) | 1.00 | ||
| Discordant | 1.91 | 0.91 – 3.99 | 0.09 |
| Double Negative | 4.13 | 2.67 – 6.37 | <0.001 |
| Age (HRR associated with an additional year of life) | 1.03 | 1.01 – 1.05 | 0.01 |
| Former or Current Smoker | 2.11 | 1.27 – 3.50 | 0.004 |
| T3/T4 | 1.69 | 1.11 – 2.56 | 0.01 |
| Definitive / Chemo Only | 1.65 | 1.09 – 2.49 | 0.02 |
| Double Positive (Reference category) | 1.91 | 0.91 – 3.99 | 0.09 |
Hazard Rate Ratio
For the discordant patients, there was a borderline significant trend towards worse overall and disease-free survival compared to double positive patients (HR 1.91, 95% CI 0.91–4.00, 95% CI, p=0.09). Survival for discrepant patients was closer to that of double negative ones, but again not statistically significant (HR 1.91, 95% CI 0.93–3.95, p=0.08).
Reclassification
Our reclassification would have resulted in the use of high-risk HPV mRNA testing in less than 9% of the overall patients, yet would have reclassified 16 of the 23 originally discrepant patients into clear HPV positive or HPV negative classes (Figure 2).
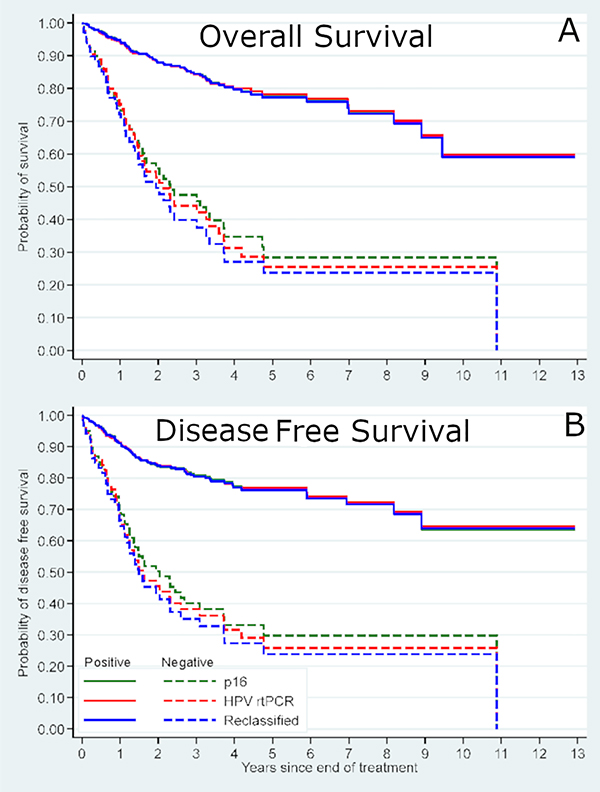
Survival was then examined by p16 status alone, high-risk HPV mRNA status alone, and by our HPV reclassification status (Table 4). In multivariate survival analysis controlling for age, smoking, treatment type, and T classification, hazard ratios for overall and disease-free survival were very comparable for all three approaches with overlapping 95% confidence intervals, but with progressively lower hazard ratios (i.e. progressively better risk stratification between positive and negative results) from p16 alone to HPV mRNA alone to HPV reclassification status alone (Tables 3 and 4, Figure 4). Selective application of HPV mRNA testing to reclassify patients with discrepant p16 and HPV status based on morphology and equivocal p16 status would have resulted in the application of high-risk HPV mRNA testing in only 9% of all patients. This approach would have reclassified 16 of the 23 originally discrepant patients into “HPV positive” or “HPV negative” (two patients moving from p16 positive to final HPV negative and 14 patients moving from p16 negative to final HPV positive). Thus, had it been applied in routine clinical practice for our patient cohorts, this reclassification approach would have resulted in very limited improvement in risk stratification.
Table 4:
Multivariable Survival Analysis: HPV Reclassification Groups
| Disease-Free Survival | HRR* | 95% CI | Significance (P) |
|---|---|---|---|
| Reclassification Negative (Reference category) | 1.00 | ||
| Reclassification Positive | 0.28 | 0.19 – 0.42 | <0.001 |
| Age (HRR associated with an additional year of life) | 1.03 | 1.01 – 1.05 | 0.002 |
| Former or current smoker | 2.28 | 1.39 – 3.72 | 0.001 |
| T3/T4 | 1.54 | 1.03 – 2.29 | 0.03 |
| Definitive / Chemo only | 1.50 | 1.01 – 2.23 | 0.04 |
| Overall Survival | |||
| Reclassification Negative (Reference category) | 1.00 | ||
| Reclassification Positive | 0.25 | 0.16 – 0.37 | <0.001 |
| Age (HRR associated with an additional year of life) | 1.04 | 1.01 – 1.06 | 0.002 |
| Former or current smoker | 2.12 | 1.28 – 3.51 | 0.004 |
| T3/T4 | 1.66 | 1.10 – 2.52 | 0.02 |
| Definitive / Chemo only | 1.71 | 1.14 – 2.57 | 0.01 |
Hazard Rate Ratio
Table 3:
Multivariable Survival Analysis: p16 and HPV mRNA testing (alone) groups
| Disease-Free Survival | HRR* | 95% CI | P |
|---|---|---|---|
| p16-negative (Reference category) | 1.00 | ||
| p16-positive | 0.33 | 0.22 – 0.49 | <0.001 |
| Age (HRR associated with an additional year of life) | 1.03 | 1.01 – 1.05 | 0.008 |
| Former or Current Smoker | 2.40 | 1.47 – 3.92 | <0.001 |
| T3/T4 | 1.54 | 1.03 – 2.31 | 0.04 |
| Definitive / Chemo only | 1.40 | 0.94 – 2.09 | 0.10 |
| Overall Survival | |||
| p16-negative (Reference category) | 1.00 | ||
| p16-positive | 0.30 | 0.20 – 0.46 | <0.001 |
| Age (HRR associated with an additional year of life) | 1.03 | 1.01 – 1.05 | 0.01 |
| Former or Current Smoker | 2.23 | 1.35 – 3.69 | 0.002 |
| T3/T4 | 1.66 | 1.09 – 2.53 | 0.02 |
| Definitive / Chemo Only | 1.56 | 1.03 – 2.36 | 0.04 |
| Disease-Free Survival | |||
| HPV mRNA Negative (Reference category) | 1.00 | ||
| HPV mRNA Positive | 0.29 | 0.20 – 0.43 | <0.001 |
| Age (HRR associated with an additional year of life) | 1.03 | 1.01 – 1.05 | 0.002 |
| Former or current smoker | 2.29 | 1.41 – 3.74 | 0.001 |
| T3/T4 | 1.66 | 1.12 – 2.46 | 0.012 |
| Definitive / Chemo only | 1.48 | 1.00 – 2.19 | 0.05 |
| Overall Survival | |||
| HPV mRNA Negative (Reference category) | 1.00 | ||
| HPV mRNA Positive | 0.26 | 0.17 – 0.39 | <0.001 |
| Age (HRR associated with an additional year of life) | 1.04 | 1.01 – 1.06 | 0.002 |
| Former or current smoker | 2.14 | 1.29 – 3.54 | 0.003 |
| T3/T4 | 1.84 | 1.22 – 2.77 | 0.003 |
| Definitive / Chemo only | 1.69 | 1.12 – 2.54 | 0.01 |
Hazard Rate Ratio
Figure 4 –
Kaplan-Meier survival plots for showing p16 IHC alone, HPV mRNA alone, and reclassification status overlaid. A) Overall survival and B) Disease free survival.
Discussion
HPV-related OPSCC is one of the fastest rising cancers in head and neck oncology, and has been recognized as an entity that is distinct from conventional OPSCC with a more favorable prognosis.1 Treatments specific to these patients are rapidly being developed and evaluated in clinical trials. Given its impact on disease staging, management, and prognosis, it is imperative that HPV status be accurately assigned in patients with OPSCC. While no consensus for testing exists yet, the CAP Guidelines and subsequently the ASCO Panel Guidelines have helped to create some standardization.7,24 The most widely utilized test, p16 IHC, is a surrogate marker for HPV and has been found to disagree with HPV RNA and DNA specific testing in anywhere from 5% to 20% of all OPSCC patients – correlation with HPV RNA being more consistent.15,19,25,26
Most of the reported discrepant cases to date have been p16 positive and HPV DNA PCR or DNA in situ hybridization (ISH) negative (undetectable), and most of these have been reported in relatively lower overall HPV incidence populations.13,14,18,27 Because of concern for these p16 “false positives” and what they might mean for individual patients, some institutions perform HPV specific testing as confirmation of tumor HPV status for all p16 positive OPSCC patients. In fact, the CAP guidelines specifically state that following p16 testing, “additional HPV-specific testing may be performed at the discretion of the pathologist and/or treating clinician or in the context of a clinical trial.”7,24 Despite these recommendations, relatively little data exists on the correlation between p16 and HPV mRNA in high incidence populations.
In this study, we sought to evaluate the rate of discordance between p16 IHC and HPV mRNA testing with the most recognized testing for transcriptionally active high-risk HPV (RT-PCR), and in a population with a high prevalence of HPV-related OPSCC. Viral mRNA expression has widely been recognized as the reference standard for the detection of truly oncogenic, and clinically significant, transcriptionally-active high-risk HPV infection.23,27 The historically high cost, technical complexity, and lack of availability of preserved RNA in tissue blocks have all limited its use in routine clinical practice, leading to the acceptance of p16 IHC as a practical alternative. Indeed, p16 is a particularly useful marker because it is aberrantly overexpressed in transcriptionally-active HPV-related tumors, is independent of HPV type, is widely available in clinical practice, and is easily assessed by practicing pathologists.8 However, its status as a surrogate marker for transcriptionally-active HPV opens the door for situations in which p16 expression levels are discordant with HPV-specific testing results. It has been suggested that patients falling into this category have a prognosis worse than double positive patients and which may be no better than double negative patients.8 To date, no one has thoroughly characterized the frequency and significance of discordance between p16 and high-risk HPV mRNA testing results in populations with high prevalence of HPV-related OPSCC, such as the United States.
Several interesting and informative findings emerge from our study. First, the discordance rate (p16+/HPV mRNA- or p16-/HPV mRNA+) in our cohort is only 5%, reflective of the high positive predictive value of p16 IHC testing for transcriptionally-active HPV in a population where the prevalence of HPV-related OPSCC approaches 85%.4 Even more interesting is that the p16-positive/HPV-negative fraction constitutes only 1.5% of the entire cohort. This contrasts sharply with several European studies using DNA PCR as an HPV-specific confirmatory test, where p16 positive and HPV DNA negative discordant patients are the sole fraction reported, and which constitute up to 20% of all of their OPSCC patients.19,25,26 Our findings are more in keeping with some other studies in United States-based cohorts testing specifically for high risk HPV, where discordant rates have been found in 3.6 – 30% of patients but overall, average approximately 7% (Table 5).13,16–18,27–29
Table 5:
Summary of studies comparing p16 and high risk HPV mRNA testing.
| Study | Testing Type | Year | Oropharyngeal SCC Cases | Discrepant Cases | Discrepancy Rate |
|---|---|---|---|---|---|
| Ukpo | RNA ISH | 2011 | 196 | 7 | 3.6% |
| Bishop | RNA ISH | 2013 | 77 | 6 | 8.0% |
| Kerr | RNA ISH | 2015 | 38 | 0 | 0% |
| Augustin | RNA ISH | 2018 | 65 | 20 | 30.8% |
| Randen-Brady | RNA ISH | 2019 | 357 | 25 | 7.0% |
| Current study | RT-PCR | 2020 | 467 | 23 | 4.90% |
| Total | 1,200 | 81 | 6.8% |
ISH = In Situ Hybridization
SCC = Squamous Cell Carcinoma
RT-PCR = Reverse Transcription Polymerase Chain Reaction
The p16 and HPV mRNA discrepant patients in our study were a somewhat heterogeneous group with characteristics that tended to match more closely with double positive patients than with those who were double negative. They experienced disease-free and overall survival rates intermediate between double-positive or double-negative patients, falling squarely between the two along a Kaplan-Meier curves. While double negative patients demonstrated significantly worse survival than those who were double positive, we found no statistically significant difference in survival between the discordant and double positive or double negative patients. However, a borderline significant trend towards poorer survival for discordant patients when compared to double positive patients was observed (HR 1.91, p = 0.08 in multivariate analysis for disease free survival). When evaluated independently (albeit in very small numbers), as one might have surmised, those patients who were p16 negative but HPV mRNA positive had prognoses somewhat closer to double-positive OPSCC patients, while those who were p16 positive but HPV mRNA negative had prognoses slightly closer to those of double-negative OPSCC patients.
Overexpression of p16 protein with greater than 70% nuclear and cytoplasmic staining on IHC continues to be considered a reliable surrogate marker of transcriptionally active HPV related OPSCC, at least in the United States. While most cases of HPV-related OPSCC show p16 expression well above this threshold (typically approaching 100%), occasionally p16 testing is reported as “equivocal” (Figure 1). This has previously been defined as an observed 50 to 70% of tumor cells showing nuclear and cytoplasmic staining for p16 expression.23,30 At many institutions this is simply treated as a negative result, while others, including a footnote in the CAP guidelines for HPV testing in head and neck carcinomas, have suggested use of HPV specific testing to arbitrate these patients.24 If HPV specific testing is positive, the tumor is considered HPV-related and a favorable prognosis is granted. In the literature, approximately 60% of these patients are reported to be HPV mRNA positive.31,32 Our cohort included four patients that fell into this equivocal category, three (75%) of whom were found to be HPV mRNA positive. While this does confirm the presence of transcriptionally active high-risk HPV, it is not entirely clear if these rare cases carry the same favorable prognosis associated with diffuse p16 overexpression. Our data does support that these “equivocal” patients do carry a more favorable prognosis, which in the absence of better data, seems to represent a reasonable approach.
The logical question that arises in response to the results of our study is whether clinical practice should shift towards routine use of high-risk HPV specific mRNA testing. RNA in situ hybridization is much more commonly available now both in routine practice and in large reference laboratories, and it has been shown to correlate very well with HPV RT-PCR.12 Likewise, precedence for such practices already exists: in geographic regions with lower overall prevalence of HPV-related tumors, the use of p16 testing alone is considered by many to be inadequate, with some organizations recommending additional HPV-specific testing in all cases where p16 is positive.9 Interestingly, there may be significant predictive value in the presence of p16 overexpression itself, which appears to convey some positive prognostic benefit independent of HPV mRNA expression. This is likely due to its role as a tumor suppressor protein in normal cells, such that marked overexpression may have some inhibitory effect on tumor growth.33,34
Previous studies have suggested that comorbidity, smoking status, age, and race significantly influence prognosis, which are similar to the trends we observed in our study.1,4,35 When combined with p16 status and tumor morphology, a comprehensive profile of the patient population in which HPV-driven OPSCC might be expected comes to light. Prototypical HPV-related OPSCC patients are p16 positive with nonkeratinizing morphology and occur in somewhat younger Caucasian men. These tumors are frequently thought of as arising in non-smokers, however, like in most large OPSCC cohorts, we found that more than 50% of patients were either current smokers or had a significant history of smoking. Alternatively, typical HPV-negative OPSCC patients are p16 negative with keratinizing morphology and occur in somewhat older men and women. These patients are almost always current or former smokers, and they frequently have a history of heavy alcohol consumption.33,36,37
While one might consider that the two separate discrepant groups represent different diseases from each other, with p16 positive/HPV negative being “conventional SCC” where p16 is aberrantly overexpressed and p16 negative/HPV positive being HPV-related SCC where p16 is lost by other means, we did not find strong evidence for this. The discrepant groups had similar rates of nonkeratinizing (86% vs 75%) and keratinizing morphology (14% vs 19%). Our data is more suggestive that most of them are likely HPV-related (i.e. like double positive) SCC, but which lack one or more of the otherwise defining features. This probably should not be surprising given the high prevalence of HPV-related SCC in our U.S. population overall.
Non-tonsil, non-base of tongue primary tumors have been speculated to be the source of many patients with discrepant testing, particularly p16+/HPV-, and studies in Europe seem to support this notion. In our study population, among the 23 discordant patients, 11 had tonsil primaries (48%), 9 had base of tongue primaries (39%), and 3 (13%) had other (soft palate and lateral pharyngeal wall) primaries. The latter is a number higher than Jordan et al.15 found for a 235 patient U.S. cohort for the HPV mRNA (RTPCR tested) positive patients (0.6%) and closer to the rate for HPV RNA negative patients (15%). Interestingly, though, all three non-tonsil non-base of tongue patients in our study were p16 negative/HPV positive. There may be some enrichment among the discrepant patients for these non-tonsillar subsites, but the data are too small for any meaningful conclusions.
Discordance between p16 and HPV specific testing tends to arise in patients who deviate from the typical profiles. The most useful feature for identifying patients at risk of discordance is morphology, specifically where p16 is overexpressed and the tumor cells are keratinizing or where p16 testing is negative and the tumor itself is nonkeratinizing. The latter group accounted for 13 out of 23 discordant patients in our cohort yet constituted just 28 patients in total. When combined with the former group (1 out of 11 patients) and those with equivocal p16 staining (3 out of 4), 16 of the 23 (70%) discordant patients could be identified while only applying HPV specific testing to less than 9% of the entire cohort (41 out of 467 total patients). Prognostically, the survival curves shifted slightly for these patients post reclassification (Figure 3), but the improved stratification was not statistically significantly better than what would be predicted by p16 IHC or HPV mRNA RTPCR alone. We found that p16 IHC alone was slightly inferior to HPV RTPCR alone and both were slightly inferior to our reclassification approach of selected HPV RNA use for patient outcome stratification (Tables 3 and 4). While the survival curves and HR shifts are modest (and not statistically significantly different), for the 16 individual patients whose p16/HPV status was “flipped” from positive to negative (or negative to positive) through our reclassification, there would potentially have been significant change in their prognostic counseling and management.
Our study has several important limitations. First, it has such a high prevalence of HPV positive patients that it may not be generalizable to many other populations around the world. In particular, the combination of p16 and high-risk HPV mRNA RT-PCR together may be critical for patients in lower HPV prevalence populations, which through large epidemiological studies, includes most of the rest of the world.38 Also, our study is a retrospective review in a relatively homogenous population, and RT-PCR is a highly technical test for which the cutoff for calling positivity for high risk HPV is somewhat subjective. As previously described, this cutoff is based on where cycle counts plateau rather than on a clear binary cutoff. It is possible that the chosen cutoff may have incorrectly classified a few patients as positive or negative in this study.
In summary, using the reference standard testing for HPV status on a large cohort of OPSCC patients in a high-HPV incidence population, we found that discordant test results are relatively uncommon (~5%). Prognostically, these patients better matched with those who were double positive but still showed disease-free and overall survival rates intermediate between double positive and double negative patients. Furthermore, we found only a non-statistically significant trend for better risk stratification by high-risk HPV RTPCR testing alone versus p16 IHC alone. This still supports the current “p16 IHC alone” approach in high incidence countries like the U.S.
As an alternative to across the board HPV mRNA testing, we proposed a reclassification scheme where HPV mRNA RTPCR testing would be selectively applied only when p16 status and tumor morphology disagreed or when p16 IHC was equivocal. This strategy captures a majority of p16 and HPV mRNA discordant patients while necessitating HPV mRNA PCR testing in only 9% of all patients. Disease-free and overall survival for these patients were only slightly better than for p16 IHC or HPV RTPCR alone. This finding, albeit with small numbers and lacking statistical significance, is still generally supportive of such a strategy. Whether this truly improves the accuracy of classification, prognostic status, and ultimately treatment for these few patients remains unclear.
Acknowledgments
Conflict of Interest and Source Funding: The authors have no conflicts of interest to report. This work was supported by the NIH grant R01DE026471 (Wang).
References
- 1.Sturgis EM, Cinciripini PM. Trends in head and neck cancer incidence in relation to smoking prevalence: an emerging epidemic of human papillomavirus-associated cancers? Cancer. 2007;110(7):1429–1435. doi: 10.1002/cncr.22963 [DOI] [PubMed] [Google Scholar]
- 2.Näsman A, Du J, Dalianis T. A global epidemic increase of an HPV-induced tonsil and tongue base cancer - potential benefit from a pan-gender use of HPV vaccine. J Intern Med. 2020;287(2):134–152. doi: 10.1111/joim.13010 [DOI] [PubMed] [Google Scholar]
- 3.Rettig EM, Fakhry C, Khararjian A, Westra WH. Age Profile of Patients With Oropharyngeal Squamous Cell Carcinoma. JAMA Otolaryngol Head Neck Surg. 2018;144(6):538–539. doi: 10.1001/jamaoto.2018.0310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaturvedi AK, Engels EA, Anderson WF, Gillison ML. Incidence trends for human papillomavirus-related and -unrelated oral squamous cell carcinomas in the United States. J Clin Oncol. Published online 2008. doi: 10.1200/JCO.2007.14.1713 [DOI] [PubMed] [Google Scholar]
- 5.Lewis JS, Beadle B, Bishop JA, et al. Human Papillomavirus Testing in Head and Neck Carcinomas: Guideline From the College of American Pathologists. Arch Pathol Lab Med. 2018;142(5):559–597. doi: 10.5858/arpa.2017-0286-CP [DOI] [PubMed] [Google Scholar]
- 6.Fakhry C, Lacchetti C, Rooper LM, et al. Human Papillomavirus Testing in Head and Neck Carcinomas: ASCO Clinical Practice Guideline Endorsement of the College of American Pathologists Guideline. J Clin Oncol. 2018;36(31):3152–3161. doi: 10.1200/JCO.18.00684 [DOI] [PubMed] [Google Scholar]
- 7.Lewis JS, Beadle B, Bishop JA, et al. Human papillomavirus testing in head and neck carcinomas guideline from the college of American pathologists. Arch Pathol Lab Med. Published online 2018. doi: 10.5858/arpa.2017-0286-CP [DOI] [PubMed] [Google Scholar]
- 8.Larsen CG, Jensen DH, Carlander A-LF, et al. Novel nomograms for survival and progression in HPV+ and HPV- oropharyngeal cancer: a population-based study of 1,542 consecutive patients. Oncotarget. 2016;7(44):71761–71772. doi: 10.18632/oncotarget.12335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nauta IH, Rietbergen MM, van Bokhoven AAJD, et al. Evaluation of the eighth TNM classification on p16-positive oropharyngeal squamous cell carcinomas in the Netherlands and the importance of additional HPV DNA testing. Ann Oncol Off J Eur Soc Med Oncol. 2018;29(5):1273–1279. doi: 10.1093/annonc/mdy060 [DOI] [PubMed] [Google Scholar]
- 10.Freitag J, Wald T, Kuhnt T, et al. Extracapsular extension of neck nodes and absence of human papillomavirus 16-DNA are predictors of impaired survival in p16-positive oropharyngeal squamous cell carcinoma. Cancer. 2020;126(9):1856–1872. doi: 10.1002/cncr.32667 [DOI] [PubMed] [Google Scholar]
- 11.Rietbergen MM, Brakenhoff RH, Bloemena E, et al. Human papillomavirus detection and comorbidity: critical issues in selection of patients with oropharyngeal cancer for treatment De-escalation trials. Ann Oncol Off J Eur Soc Med Oncol. 2013;24(11):2740–2745. doi: 10.1093/annonc/mdt319 [DOI] [PubMed] [Google Scholar]
- 12.Gao G, Chernock RD, Gay HA, et al. A novel RT-PCR method for quantification of human papillomavirus transcripts in archived tissues and its application in oropharyngeal cancer prognosis. Int J cancer. 2013;132(4):882–890. doi: 10.1002/ijc.27739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schache AG, Liloglou T, Risk JM, et al. Validation of a novel diagnostic standard in HPV-positive oropharyngeal squamous cell carcinoma. Br J Cancer. 2013;108(6):1332–1339. doi: 10.1038/bjc.2013.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rietbergen MM, Leemans CR, Bloemena E, et al. Increasing prevalence rates of HPV attributable oropharyngeal squamous cell carcinomas in the Netherlands as assessed by a validated test algorithm. Int J cancer. 2013;132(7):1565–1571. doi: 10.1002/ijc.27821 [DOI] [PubMed] [Google Scholar]
- 15.Jordan RC, Lingen MW, Perez-Ordonez B, et al. Validation of methods for oropharyngeal cancer HPV status determination in US cooperative group trials. Am J Surg Pathol. 2012;36(7):945–954. doi: 10.1097/PAS.0b013e318253a2d1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bishop JA, Ma X-J, Wang H, et al. Detection of transcriptionally active high-risk HPV in patients with head and neck squamous cell carcinoma as visualized by a novel E6/E7 mRNA in situ hybridization method. Am J Surg Pathol. 2012;36(12):1874–1882. doi: 10.1097/PAS.0b013e318265fb2b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kerr DA, Arora KS, Mahadevan KK, et al. Performance of a Branch Chain RNA In Situ Hybridization Assay for the Detection of High-risk Human Papillomavirus in Head and Neck Squamous Cell Carcinoma. Am J Surg Pathol. 2015;39(12):1643–1652. doi: 10.1097/PAS.0000000000000516 [DOI] [PubMed] [Google Scholar]
- 18.Ukpo OC, Flanagan JJ, Ma X-J, Luo Y, Thorstad WL, Lewis JS. High-risk human papillomavirus E6/E7 mRNA detection by a novel in situ hybridization assay strongly correlates with p16 expression and patient outcomes in oropharyngeal squamous cell carcinoma. Am J Surg Pathol. 2011;35(9):1343–1350. doi: 10.1097/PAS.0b013e318220e59d [DOI] [PubMed] [Google Scholar]
- 19.Holzinger D, Schmitt M, Dyckhoff G, Benner A, Pawlita M, Bosch FX. Viral RNA patterns and high viral load reliably define oropharynx carcinomas with active HPV16 involvement. Cancer Res. 2012;72(19):4993–5003. doi: 10.1158/0008-5472.CAN-11-3934 [DOI] [PubMed] [Google Scholar]
- 20.Gondim DD, Haynes W, Wang X, Chernock RD, El-Mofty SK, Lewis JS. Histologic Typing in Oropharyngeal Squamous Cell Carcinoma: A 4-Year Prospective Practice Study With p16 and High-Risk HPV mRNA Testing Correlation. Am J Surg Pathol. 2016;40(8):1117–1124. doi: 10.1097/PAS.0000000000000650 [DOI] [PubMed] [Google Scholar]
- 21.Chernock RD, El-Mofty SK, Thorstad WL, Parvin CA, Lewis JS. HPV-related nonkeratinizing squamous cell carcinoma of the oropharynx: utility of microscopic features in predicting patient outcome. Head Neck Pathol. 2009;3(3):186–194. doi: 10.1007/s12105-009-0126-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis JS, Khan RA, Masand RP, et al. Recognition of nonkeratinizing morphology in oropharyngeal squamous cell carcinoma - a prospective cohort and interobserver variability study. Histopathology. 2012;60(3):427–436. doi: 10.1111/j.1365-2559.2011.04092.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis JS, Chernock RD, Ma X-J, et al. Partial p16 staining in oropharyngeal squamous cell carcinoma: extent and pattern correlate with human papillomavirus RNA status. Mod Pathol. 2012;25(9):1212–1220. doi: 10.1038/modpathol.2012.79 [DOI] [PubMed] [Google Scholar]
- 24.Fakhry C, Lacchetti C, Rooper LM, et al. Human papillomavirus testing in head and neck carcinomas: ASCO clinical practice guideline endorsement of the college of American pathologists guideline. J Clin Oncol. Published online 2018. doi: 10.1200/JCO.18.00684 [DOI] [PubMed] [Google Scholar]
- 25.Rischin D, Young RJ, Fisher R, et al. Prognostic significance of p16INK4A and human papillomavirus in patients with oropharyngeal cancer treated on TROG 02.02 phase III trial. J Clin Oncol. 2010;28(27):4142–4148. doi: 10.1200/JCO.2010.29.2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rietbergen MM, Snijders PJF, Beekzada D, et al. Molecular characterization of p16-immunopositive but HPV DNA-negative oropharyngeal carcinomas. Int J Cancer. 2014;134(10):2366–2372. doi: 10.1002/ijc.28580 [DOI] [PubMed] [Google Scholar]
- 27.Randén-Brady R, Carpén T, Jouhi L, et al. In situ hybridization for high-risk HPV E6/E7 mRNA is a superior method for detecting transcriptionally active HPV in oropharyngeal cancer. Hum Pathol. 2019;90:97–105. doi: 10.1016/j.humpath.2019.05.006 [DOI] [PubMed] [Google Scholar]
- 28.Augustin J, Outh-Gauer S, Mandavit M, et al. Evaluation of the efficacy of the 4 tests (p16 immunochemistry, polymerase chain reaction, DNA, and RNA in situ hybridization) to evaluate a human papillomavirus infection in head and neck cancers: a cohort of 348 French squamous cell carcinomas. Hum Pathol. 2018;78:63–71. doi: 10.1016/j.humpath.2018.04.006 [DOI] [PubMed] [Google Scholar]
- 29.Rooper LM, Gandhi M, Bishop JA, Westra WH. RNA in-situ hybridization is a practical and effective method for determining HPV status of oropharyngeal squamous cell carcinoma including discordant cases that are p16 positive by immunohistochemistry but HPV negative by DNA in-situ hybridization. Oral Oncol. 2016;55:11–16. doi: 10.1016/j.oraloncology.2016.02.008 [DOI] [PubMed] [Google Scholar]
- 30.Schlecht NF, Brandwein-Gensler M, Nuovo GJ, et al. A comparison of clinically utilized human papillomavirus detection methods in head and neck cancer. Mod Pathol. 2011;24(10):1295–1305. doi: 10.1038/modpathol.2011.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barasch S, Mohindra P, Hennrick K, Hartig GK, Harari PM, Yang DT. Assessing p16 Status of Oropharyngeal Squamous Cell Carcinoma by Combined Assessment of the Number of Cells Stained and the Confluence of p16 Staining: A Validation by Clinical Outcomes. Am J Surg Pathol. 2016;40(9):1261–1269. doi: 10.1097/PAS.0000000000000666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen ZW, Weinreb I, Kamel-Reid S, Perez-Ordoñez B. Equivocal p16 immunostaining in squamous cell carcinoma of the head and neck: staining patterns are suggestive of HPV status. Head Neck Pathol. 2012;6(4):422–429. doi: 10.1007/s12105-012-0382-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cai C, Chernock RD, Pittman ME, El-Mofty SK, Thorstad WL, Lewis JS. Keratinizing-Type Squamous Cell Carcinoma of the Oropharynx. Am J Surg Pathol. 2014;38(6):809–815. doi: 10.1097/PAS.0000000000000183 [DOI] [PubMed] [Google Scholar]
- 34.Lewis JS, Thorstad WL, Chernock RD, et al. p16 Positive Oropharyngeal Squamous Cell Carcinoma:An Entity With a Favorable Prognosis Regardless of Tumor HPV Status. Am J Surg Pathol. 2010;34(8):1088–1096. doi: 10.1097/PAS.0b013e3181e84652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Näsman A, Du J, Dalianis T. A global epidemic increase of an HPV-induced tonsil and tongue base cancer – potential benefit from a pan-gender use of HPV vaccine. J Intern Med. Published online 2020. doi: 10.1111/joim.13010 [DOI] [PubMed] [Google Scholar]
- 36.Cooper T, Biron VL, Adam B, Klimowicz AC, Puttagunta L, Seikaly H. Association of Keratinization With 5-Year Disease-Specific Survival in Oropharyngeal Squamous Cell Carcinoma. JAMA Otolaryngol Neck Surg. 2015;141(3):250. doi: 10.1001/jamaoto.2014.3335 [DOI] [PubMed] [Google Scholar]
- 37.Meng H, Miao S, Chen K, et al. Association of p16 as Prognostic Factors for Oropharyngeal Cancer: Evaluation of p16 in 1470 Patients for a 16 Year Study in Northeast China. Biomed Res Int. 2018;2018:1–8. doi: 10.1155/2018/9594568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castellsagué X, Alemany L, Quer M, et al. HPV Involvement in Head and Neck Cancers: Comprehensive Assessment of Biomarkers in 3680 Patients. J Natl Cancer Inst. 2016;108(6):djv403. doi: 10.1093/jnci/djv403 [DOI] [PubMed] [Google Scholar]