Abstract
Emerging genetic testing delivery models have enabled individuals to receive testing without a medical indication. This article will highlight key considerations for patient care in the setting of adult patients with positive results for monogenic disease identified through genomic screening. Suggestions for how to adapt genetic counseling to a genomic screening population will encompass topics such as phenotyping, risk assessments, and the use of existing guidelines and resources. Case examples will demonstrate principles of genotype-first patient care.
Keywords: Population Screening, Secondary Findings, Predictive Genetic Testing, Genetic Counseling, Genetics Services, Risk Assessment
Introduction
Genetic testing service delivery models such as consumer-driven genetic testing, population screening programs, concierge medicine clinics and research studies that return results enable new ways to access genetic information (eMerge Consortium, 2019; “Genomics Working Group of the All of US Research Program Advisory Panel,” 2020; Grzymski et al., 2018; Hagenkord et al., 2020; Lu et al., 2019; Schwartz et al., 2018). This will lead to more patients with a positive genetic finding in the absence of targeted testing based on medical history. While this has been the norm for certain screening applications such as carrier, prenatal and newborn screening, it was not historically the case for adult disorders. Providing genetic counseling (GC) to patients who present based on genotype is distinct from standard phenotype-driven approaches; it provides an opportunity for genetic counselors to embrace the practice of genome-guided preventive medicine (O’Daniel, 2010). Here, we discuss considerations for providing GC to patients who first present for care with a clinical grade (e.g. validated in a CLIA laboratory), pathogenic/likely pathogenic, monogenic result via a test not selected using a professional, personalized risk assessment. We will refer to this method of receiving test results as genomic screening (GS) for the purposes of this article (East et al., 2019; Lu et al., 2019). An emphasis will be placed on results received without formal pre-test GC, however, the topics discussed remain relevant to any patient and/or family who first presents for care based on genotype. Key principles of patient care for this population will be discussed first, followed by case examples to illustrate these principles.
Condensing the care model
Historically, thorough risk assessments based on a personal and/or family history of disease have been used as a first-line screening tool to identify GC and testing candidates (Riley et al., 2012). Concerns about testing cost and an interest in maximizing positive predictive value (PPV) have contributed to the restriction of genetic testing to those likely to have a genetic finding and the prioritization of indication-targeted tests. However, some patients are now bypassing traditional risk assessments and receiving a general genetic test as a first-line screening tool. A convergence of multiple interrelated factors has enabled this shift including decreased sequencing cost, improved data analysis and interpretation abilities, public interest, innovation in the test delivery space, and a recognition that traditional risk assessments are insufficiently sensitive at identifying at-risk individuals (East et al., 2019; Lu et al., 2019). Unlike in populations where medical history triggers testing, illness in a patient or their relative(s) is not a prerequisite for genomic screening (Hooker, Ormond, Sweet, & Biesecker, 2014).
One study of genetic counselor attitudes about population-based screening identified a concern that patients may have higher anxiety from lack of pretest counseling (De Simone, Arjunan, Vogel Postula, Maga, & Bucheit, 2020). However, care for patients with positive results obtained through GS without formal pre-test counseling is not inherently problematic, particularly if patients consented to genetic testing, results are disclosed responsibly, and appropriate follow-up options are available. One study of individuals consented for a GS study by mail found that most individuals had minimal to no decisional conflict about the pursuit of GS and reached a decision about study participation quickly. Of 5,106 participants invited to participate in this study (59.5% consent rate), only eight individuals elected to have the pre-test GC option offered (Pacyna et al., 2019). This suggests that many individuals are willing to consent for GS without receiving pre-test GC.
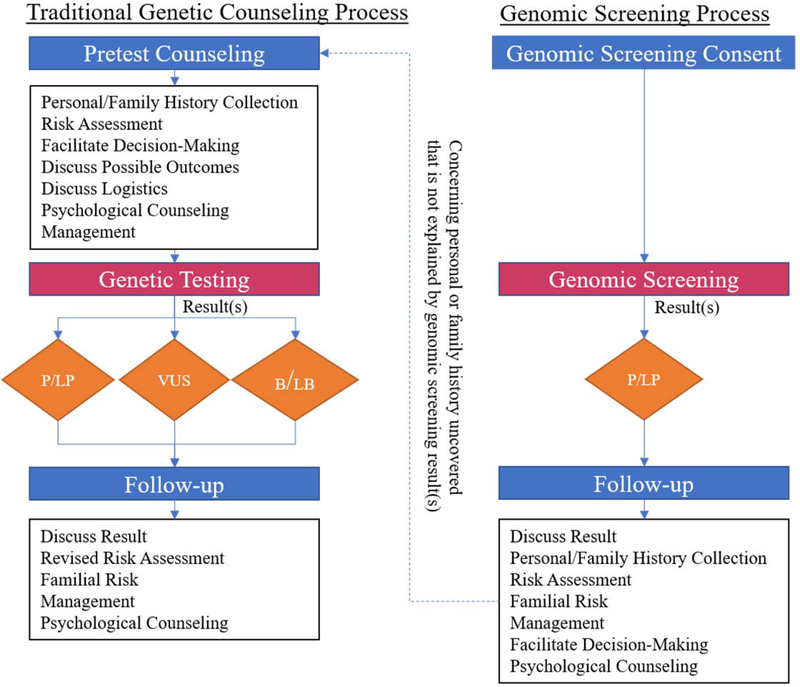
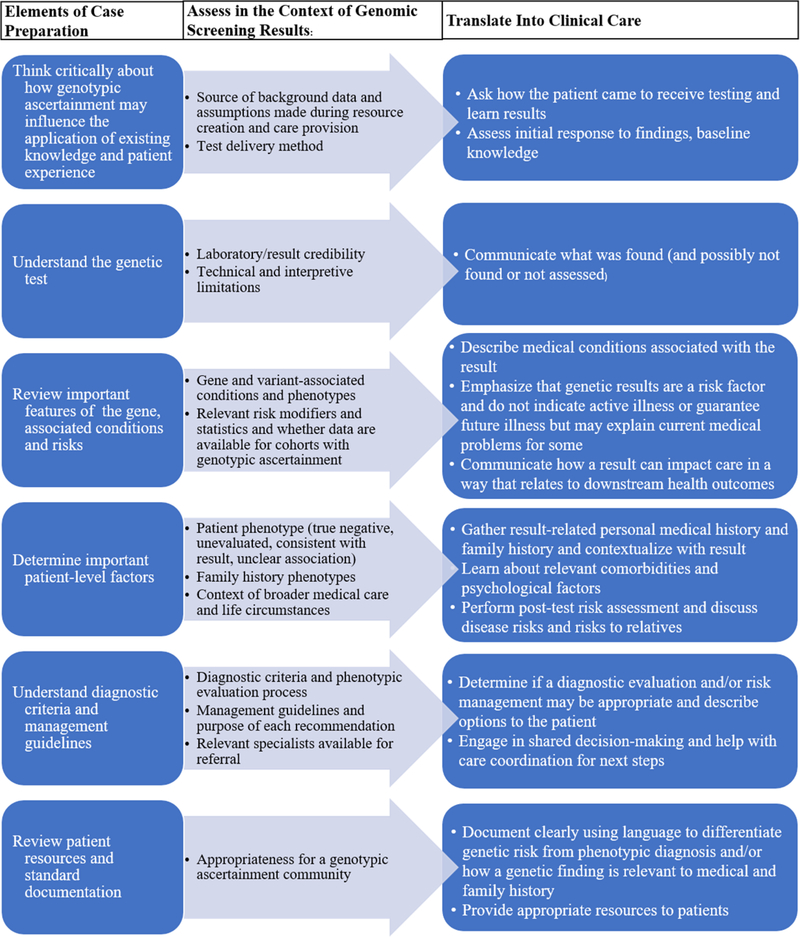
When a person first presents for GC due to a positive genetic test result from GS, the paradigm is a post-test focused GC model rather than the traditional pre/post-test model. Elements from pre- and post-test counseling are combined into a single activity, with some traditional pre-test elements becoming irrelevant in the context of a known result (Figure 1). The session purpose mirrors any positive-result counseling session where the goals are to describe the result and associated genetic risks, contextualize it within the patient’s history, discuss and facilitate management of the identified risk, and provide psychological support (Figure 2). Certain elements, however, require a perspective shift. For example, the fundamental objective of personal and/or family medical history collection changes from determining “Is this history suggestive of a genetic condition and if so, which one(s)?” to “Does this individual’s personal or family history inform the management of this result?” The relevant phenotypes no longer guide the genetic testing choice, but rather contextualize the result by providing information about the potential magnitude of disease risk. In order to provide high quality post-test counseling to GS patients, genetic counselors will need training to understand how ascertainment differences can influence phenotype, disease risk, the utilization of existing clinical resources, and psychological considerations.
Figure 1.
Elements from both pre- and post-test counseling are combined into single genomic screening post-test counseling session. The two pathways are not mutually exclusive as patients with a concerning personal or family history may end up receiving care through both pathways. Abbreviations: Pathogenic/Likely Pathogenic (P/LP), Variant of Uncertain Significance (VUS), Benign/Likely Benign (B/LB)
Figure 2.
Guide for care provision to genomic screening patients
Phenotypic considerations
GS populations have a lower a-priori risk than phenotypically ascertained populations. They may have lower penetrance and a broader phenotypic spectrum which may include milder or atypical phenotypes (Biesecker, 2013; Carruth et al., 2019; Manickam et al., 2018; Murray, 2016; Van Driest et al., 2016). General population penetrance and expressivity data for most genes is unavailable, which complicates counseling about risk, risk management and family communication. Predicting which phenotypes an individual is at risk for can also be challenging, especially for genes with multiple associated phenotypes. For example, pathogenic variants in the LMNA gene have caused diverse phenotypes including dilated cardiomyopathy, muscular dystrophy, neuropathy, and lipodystrophy. While the lab report and literature may note phenotypes from other reported cases with that variant, there may be insufficient data to definitively rule out which phenotypes are not related to the variant.
After a patient receives initial counseling about a result, a diagnostic evaluation to assess for evidence of key gene-associated phenotypes is often appropriate. However, the diagnostic criteria for most genetic conditions assume a phenotype preceded genetic testing and are not optimized for a genomic screening use-case. For example, diagnostic criteria that incorporate genotype may assign significant value to genetic information even in patients with no or limited phenotype such as with criteria that indicate that a positive result is synonymous with a diagnosis of a genetic condition (e.g. Lynch syndrome or PTEN hamartoma tumor syndrome) or incorporate genomic data via a scoring schema (e.g. Arrhythmogenic Right Ventricular Cardiomyopathy (ARVC) Task Force Criteria, Dutch Lipid Clinic Network Criteria for Familial Hypercholesterolemia (FH)(Giardiello et al., 2014; Marcus et al., 2010; Nordestgaard et al., 2013; Pilarski et al., 2013). A key distinction for a GS population is that DNA is not equivalent to a phenotypic diagnosis but is an indicator of risk (Murray, 2016). This is relevant when assigning diagnostic labels since the formal name of some conditions is intended to indicate risk irrespective of phenotype, like Lynch syndrome, whereas for others, a clinical diagnosis implies medical findings distinct from a genetic result, like hypertrophic cardiomyopathy. One schema proposed by Katz et al. is using precise terminology to distinguish between a “clinical finding” (detectable phenotype), a “molecular finding” (genetic result), and a “clinicomolecular diagnosis” (presence of a correlated clinical and molecular finding) (Katz et al., 2020). Documenting which evaluations were completed, which features were present or absent at baseline, the patient’s post-result diagnostic status, and follow-up recommendations that account for baseline phenotype are useful to guide subsequent care. Use of precise terminology to differentiate between risk for a condition and a clinical diagnosis of an associated health problem is important to avoid misunderstanding among patients and healthcare providers. For example, if a patient with a likely pathogenic PTEN variant undergoes a diagnostic evaluation which reveals no PTEN-related features, it may be inappropriate to formally diagnose them with PTEN-hamartoma tumor syndrome based only on a genetic test. In circumstances like this, noting a gene on the electronic health record problem list (e.g. monoallelic mutation of the PTEN gene) rather than the syndrome name may be more appropriate.
Since some patients do not have a relevant personal or family history of disease, the baseline set of assumptions when performing post-test counseling about a result are different. Patients without a known phenotype often lack personal context for the disease. It can be worth explaining the phenotypes they are at risk for, why this information is important for their care, and that a genetic result indicates risk rather than active disease (Figure 2). It is important to convey that identification of future disease is possible, but that some individuals with a genetic risk may never develop related disease. Furthermore, for some patients it may be unclear if a personal or family history of disease is result-related, either because the reported history lacks specificity, or the relationship between a given phenotype and the gene-related disease spectrum is unknown.
It can be valuable to consider and discuss with patients and other medical professionals reasons that the phenotype in a person or family may differ from classic condition descriptions. Underscoring that most gene or condition-specific studies were based on targeted testing for people with certain features, such as a strong history of a specific phenotype, can help patients with a GS result understand why risk data from the literature may over-estimate their risk. Genotypically identified families may have several underlying elements that influence disease risk and contribute to a less striking history such as: (1) genetic modifiers (e.g. protective factors or a lower burden of other risk alleles), (2) different lifestyle and/or environmental risk factors, and (3) identification of the variant earlier in the disease course and/or life-course.
Other factors that impact the sensitivity of history-based risk assessments are also relevant to understanding manifestations in a GS population. For example, limited family structure, inadequate family history information and unrelated premature deaths can mask phenotypes (Riley et al., 2012). Some individuals or their family members may have previously taken preventive measures which would reduce disease risks for reasons unrelated to their genetic result. For example, a hysterectomy for uterine fibroids would also reduce the risk of uterine cancer in an individual with Lynch syndrome. Furthermore, the underlying randomness of who inherits a genetic variant can lead to families in which most members did not inherit the genetic risk due to chance. This can create an illusion of lower penetrance even in families with a larger family structure. Sometimes a new genomic finding can also lead to the proband learning that family members have a relevant phenotype about which the proband was previously unaware.
While some patients may not have a personal and/or family history of disease at the time of result, this is not universal. For those patients with a known phenotype but previously unknown genotype, it is important to emphasize the ways in which this new information may change existing care regimens either for them or family members, as this may not be self-evident. For example, patients with high cholesterol who receive a result for FH may not be aware of how this information may alter estimates of their disease risk and consequently influence their target lipid levels and medication recommendations (Grundy et al., 2019). Additionally, it is important to contextualize a new finding within the overall picture of an individual’s medical history at the time of receiving a positive result from GS. Depending on the circumstances surrounding testing, patients may have different degrees of readiness or willingness to act on a finding. It can be valuable to learn how patients received testing and whether they actively sought a test or if they were recruited for testing through other means. For some individuals, such as the elderly or those with severe co-morbidities, it may be reasonable to acknowledge that some genetic risks may have limited relevance for their personal medical care. In these situations, focusing a discussion on the relevance for relatives may have more value.
Concerns about genetic discrimination may also become salient following a positive GS result, especially absent a related medical history. Genetic counselors should be prepared to counsel about federal and state genetic discrimination laws. Even if a patient consents to genetic testing with a knowledge of discrimination protections and their limitations, pre-emptive steps such as the purchase of life, disability or long-term care insurance may not be prioritized among genomic screening patients with a low probability of a positive test result. It can be helpful to encourage patients who express concerns about genetic discrimination to share their specific fears. This information can then be used to help them contextualize the potential health benefits of genetic information with the risk of genetic discrimination. Additionally, pre-emptively addressing result-specific considerations may be challenging when many genes are screened. For example, GS may evaluate for malignant hyperthermia susceptibility which is considered a disqualifying medical condition by the U.S. Department of Defense (Lee, McGlinch, McGlinch, & Capacchione, 2017). Consequently, for families with current or prospective active duty military members this information may be relevant for post-test discussions about employment choices and family sharing.
Rethinking risk assessments
While general population prevalence, penetrance, and expressivity data remain unavailable for most genetic conditions, emerging data from large unselected cohorts have started to provide data for some genes, particularly those on the American College of Medical Genetics (ACMG) Secondary Findings (SF) V2.0 gene list (Kalia et al., 2017). These data are needed to understand the positive and negative predictive value of current genetic tests and the natural history of disease in GS cohorts. Histories from genomic screening patients can also be used to evaluate the sensitivity of history-based referral and genetic testing criteria had they been appropriately applied to all patients with underlying results. Early work on Geisinger’s Hereditary Breast and Ovarian Cancer Syndrome (HBOC) cohort identified through biobank-based GS found that only 18% had prior positive HBOC testing. Of those without a prior result who had comprehensive personal and family history data available, over half would have met published guidelines for clinical testing (Manickam et al., 2018). Later work on this cohort found that no patients with a result for FH (N=93) had prior knowledge of their variant despite 96% having consistent personal and/or family history features; 87.5% of patients with a Lynch syndrome result were previously unaware of their result despite 61% having a relevant personal and/or family history (Buchanan et al., 2020). Similarly, data from the Healthy Nevada Project found that 90% of carriers of a HBOC, Lynch or FH result were previously undetected despite 26% of carriers having advanced disease (Grzymski et al., 2019). General population penetrance for some genes and conditions may be low, such as ARVC (penetrance estimated at 6%) and some inherited arrhythmias (Carruth et al., 2019; Van Driest et al., 2016).
Similarly, genetics professionals rarely learn about the number of clinically significant genetic findings that their risk assessment practices and test selection preferences fail to identify. It can be professionally challenging to directly observe families with positive results that would have remained undetected if current guidelines and typical care practices were applied. However, observing patients with genetic results obtained through unselected testing practices allows for a more thorough picture of how genetic risks manifest across the full spectrum of positive patients.
Although some GS tests are broader than indication-targeted tests, no contemporary genetic test can detect all possible genetic results. Just as the assessment of post-test residual risk is important in a regular clinical setting, it is particularly relevant when the testing is not indication-based. It is important to understand the analytical completeness (which genes were screened, what types of variants can be detected) and the interpretive completeness (how are variants interpreted and reported) of a GS test as well as whether the laboratory was CLIA/CAP accredited (Lu et al., 2019) (Figure 2). In patients who only receive a post-test risk assessment, attention should be paid to features concerning for other genetic conditions that may warrant additional genetic testing. It is also important for patients to understand that GS is not a substitute for indication-based genetic testing in the setting of a concerning personal and/or family history and that an additional genetic assessment may still be needed.
Facilitating follow-up
In patients without a known phenotype, follow-up care often begins with a diagnostic evaluation (where relevant) followed by subsequent management in the form of continued surveillance or preventive measures as appropriate. Just like diagnostic criteria, most management guidelines and patient-facing materials were developed with the assumption that genetic testing was indication-based and may need modification for GS patients. When applying existing guidelines to this population there is a tension between over- and under- management. Some recommendations may result in overutilization of healthcare resources and unnecessary procedure-related risks, however, the risk of missing a potentially serious phenotype/early diagnosis that could have improved health outcomes is also a concern.
Even when evidence begins to inform management approaches in GS cohorts, it is unlikely that there will be a universal approach to adapting existing recommendations across genes and conditions. Despite these limitations, a shared decision-making approach can be used by patients and condition-specific experts to develop a care plan. Factors that may be incorporated in management discussions include (1) existing management recommendations; (2) relevant personal or family medical history; (3) knowledge about the gene/condition (including data from screening populations); (4) the severity of the medical intervention(s); and (5) other patient-level factors such as age, comorbidities, and psychological factors associated with risk management decisions (e.g. anxiety). Transparency with patients about the limitations of medical knowledge can be useful to guide discussions. When GS penetrance data are unavailable, there may be a preference toward beginning with lower patient burden interventions (e.g., noninvasive cancer screening) over those with a higher patient burden (e.g. prophylactic surgery) if a choice is being made between two or more effective alternatives. For conditions where some data are available on penetrance in a GS cohort, such as FH and ARVC, that information can be useful. For example, in FH the penetrance is high and typical risk management and treatment approaches are low burden (Hunter et al., 2016). In this setting, following the existing management guidelines may be appropriate for most patients. For conditions where penetrance is thought to be lower, like ARVC, that may factor into decisions about the frequency of screenings and at what age to cease follow-up if a patient remains disease free (Haggerty et al., 2018). For example, screening decisions may differ between an elite athlete with an ARVC result (for whom physical activity is thought to amplify risk) and a physically inactive individual with a similar GS result. Additionally, task force diagnostic criteria for ARVC incorporate data from an endomyocardial biopsy. Ordering this invasive test only due to a genetic result may be inappropriate since it has limited utility for management in a phenotypically negative individual. Although genetic counselors do not actively manage patients, they can discuss options with patients and develop a plan for next steps such as facilitating referrals. There is also an opportunity to identify and educate specialist providers with an interest in managing these patients and form multidisciplinary clinics.
Patient-facing materials also make assumptions about the reason for testing and a-priori risks. For example, language use may inaccurately indicate that a person’s experience with a related disease was the indication for genetic testing. Patients may also need guidance to understand how risk estimates or other information they encounter online or through patient materials may not be tailored for their situation. Ideally, materials provided to patients should be reviewed for applicability to GS results prior to distribution. For some conditions, new materials may need to be designed.
As diagnostic criteria, management guidelines, and patient facing materials are generated and revised for common, screened-for genetic conditions, authors of these documents should address genomic screening-use cases. This may include acknowledging the limitations of applying guidelines to GS cohorts when supporting data are unavailable. Precise language to differentiate between recommendations for those with clinical, molecular and clinicomolecular diagnoses would add clarity for genomic and cascade screening situations. Expert panels may benefit from including a member with experience in caring for genomic screening patients for input into consensus guidelines. Large research projects which incorporate genomic screening should also seek to generate data that are lacking in the genomic screening setting to facilitate future evidence-based care. For example, researchers can aim to create evidence frameworks for determining clinical utility in unselected GS populations and collect data on penetrance, risk management behaviors, psychological outcomes, and the impact of genetic risk on morbidity and mortality.
Realizing Genomic Screening in Practice
While GS is not currently standard of care, genetic tests may become sufficiently comprehensive and accessible to allow routine use at scale, without pre-test counseling. A survey of genetic counselors found that most thought that population GS should be implemented in the next 10 years for HBOC (54.5%) and Lynch Syndrome (52.5%) (De Simone et al., 2020). There are already several large-scale studies which have or plan to return GS results such as the All of Us Research Program, the Healthy Nevada Project, and the Geisinger MyCode Community Health Initiative (“Genomics Working Group of the All of US Research Program Advisory Panel,” 2020; Grzymski et al., 2018; Schwartz et al., 2018). Prioritizing provider time and effort on post-test counseling for GS patients with high-impact positive findings would reduce the number of patients without monogenic findings requiring visits and the number of visits per patient relative to a standard pre/post-test counseling approach. This would enable allocation of GC resources to the areas of highest need and could lower barriers to access genetic services (Ormond et al., 2019). Efficiency tools and service delivery innovations such as use of GC assistants and chatbots have been used to support certain tasks in genomic screening programs like testing consent, triaging patient inquiries, and post-result check-ins to enable operation at scale (Hallquist et al., 2020; Hooker et al., 2014; Schmidlen, Schwartz, DiLoreto, Kirchner, & Sturm, 2019).
Case Examples
The following dramatized testing scenarios are based on forms of GS available in the United States. Phenotype information, follow-up decisions and other patient-care circumstances are based on situations that have arisen in real cases from Geisinger’s MyCode Genomic Screening and Counseling Program. This program has returned over 1,800 results for monogenic disease inclusive of the ACMG SF v2.0 and HFE C282Y homozygotes through a biobank-based research initiative that does not include formal pre-test counseling(“MyCode Community Health Initiative,”; Schwartz et al., 2018).
The Absent Phenotype
Sam is a 45-year-old male who agrees to join an institutional genomic screening initiative during a routine visit. Several weeks later he is notified that he has a pathogenic variant in the PKP2 gene. This result is associated with a higher risk of a heart condition called arrhythmogenic cardiomyopathy, or ARVC. Sam has never heard of this heart problem before. Anxious to learn more, he schedules an appointment to discuss this result in more detail with a genetic counselor. He gathers some family history information in advance of the visit and tells the genetic counselor that there is no family history of cardiovascular disease other than in a maternal uncle who had a history of “heart bypass” surgery. He also does not report any family history of sudden death. His brother was a competitive athlete in college without any known concerns. He has never had a cardiac evaluation, so he is referred to cardiology, where he has a cardiac MRI, a 12-lead EKG, and ambulatory rhythm monitoring to look for signs of PKP2-related disease. These tests come back with no evidence of cardiac dysfunction, arrhythmias, or other electrophysiologic abnormalities. His cardiologist expresses that the use of genetic results in this manner is new, so it is unclear exactly what his future risks are and that they can work together to come up with a surveillance plan based on existing disease knowledge and his level of risk tolerance. They discuss a follow-up plan that includes continuation of his current exercise regimen, which does not involve any vigorous activity, and cardiac screening every 5 years until age 60 unless he experiences relevant signs or symptoms of disease. A researcher at Sam’s institution is conducting a research study to learn more about ARVC-related phenotypes in a population identified through GS. He is informed of this project in his clinic visit and agrees to join. Early results from this study suggest that penetrance for ARVC is low in this general population screening cohort (Carruth et al., 2019).
Key points
Some genetic conditions/phenotypes may be unfamiliar to a patient.
Some patients may not have a personal or family history of result-related disease.
The relevance of some existing guidelines to an individual with no known history of related disease remains unclear. While it may make sense to apply existing guidelines, they should be examined critically, and clinical judgment should be exercised. A shared decision-making approach is one patient-centered method that can be used to arrive at a plan with which the patient and provider are comfortable.
Research can be integrated with GS programs to better understand phenotypes and patient outcomes. This may be useful to guide risk assessments and future management recommendations.
The Atypical Phenotype
Jonah is a 38-year-old man who is approached to join a research project that includes GS while he waits for a clinic visit. He agrees to join the study because he is interested in the genetics of type 2 diabetes mellitus due to his recent diagnosis of this condition. Two years later he receives a call from a member of the study team indicating that Jonah has a genetic risk for an adverse reaction to anesthesia. Specifically, he has an pathogenic variant in the RYR1 gene, which can cause a condition called malignant hyperthermia susceptibility. He makes an appointment with the genetics clinic, where he discloses that both he and his daughter had severe prolonged muscle pain following anesthesia for dental surgeries that resolved after 2–3 weeks. He does not report any personal or family history of an acute malignant hyperthermia event. He is counseled that based on what we know about the mechanism of disease it is possible that his muscle pain was related to this finding, but that the reaction would not be considered a classic malignant hyperthermia reaction. Regardless, he and at-risk relatives should avoid triggering agents and consider getting a medical ID. After counseling about his result, he has several questions about the genetics of diabetes. He is counseled about common complex disease and told that the genetic screen did not look for any diabetes-related genetic risk factors.
Key Points
There may be a lag between study consent and result confirmation and disclosure for individuals receiving results in some research settings.
Phenotypes uncovered during an evaluation may seem suspicious of being result-related, even if they are not classic for the condition. For some conditions a judgment of whether a phenotype is related may not impact management recommendations.
Individuals may undergo GS with a specific disease or phenotype in mind. Actual findings may be unrelated to this phenotype, but their original questions can remain.
It can be helpful to understand details of the genetic testing conducted, including which genes were tested and any specific limitations.
The Unmasked Phenotype
Clyde is a 50-year-old man who has a personal interest in genetics and pays for a consumer-initiated elective medical-grade genetic test. He is primarily concerned about his risks for heart disease because several relatives died of heart attacks in their 70’s despite leading an active lifestyle. Through this genetic test he learns that he has a pathogenic variant in the RET gene. He reads about his result and learns that he has up to a 95% risk for medullary thyroid cancer and a lesser risk for pheochromocytomas and hyperparathyroidism. He is not worried because he feels fine and he does not have a family history of any of these conditions, which are unfamiliar to him. Additionally, his lack of a family history of thyroid cancer is not consistent with the very high risk quoted so he believes that his result may be a false positive. At his annual primary care appointment, he shares the result with his doctor who convinces him to follow-up on this result with endocrinology. At his endocrinology appointment he receives surveillance for features associated with this gene. His calcitonin comes back mildly elevated, but thyroid ultrasound and work-up for hyperparathyroidism and pheochromocytoma are negative. His endocrinologist recommends a prophylactic thyroidectomy, which he decides to complete due to his elevated calcitonin. Pathology shows multiple, bilateral, medullary thyroid microcarcinomas with perineural invasion, confined to the thyroid. He does not require additional cancer treatment.
Following his cancer diagnosis, he shares both his new diagnosis and genetic results with his family members. He then learns that one of his father’s cousins had a history of some type of thyroid cancer but specific details about her medical history are unavailable. His parents agree to cascade genetic testing to determine which other family members may be at-risk. His father, who has no known personal history of thyroid cancer, tests positive for the genetic variant. He declines surveillance and prophylactic surgery due to advanced age. A paternal cousin also tests positive for the result and is found on follow-up care to have an early-stage medullary thyroid carcinoma.
Key Points
Personal or family medical history may not be consistent with a result when it is uncovered but further work-up may find subclinical phenotypes or previously unreported family history.
Disease penetrance may be lower and age of onset may trend later for families identified through GS than those reported in the literature from indication-based ascertainment.
GS results provide an opportunity for early disease detection.
Cascade genetic testing in GS families can expand the impact of a positive genetic result by allowing for early detection/prevention in multiple individuals without a known personal or family history of related disease.
Some patients may not be interested in result-related management due to age or other personal reasons.
The Known Phenotype, Dual Risk
Kiara is a 28-year old woman who participates in a biobank-based GS program at her healthcare institution. Several months later, she receives a call from a genetic counselor telling her that she has a positive finding, a pathogenic variant in the LDLR gene, which can cause a type of inherited high cholesterol called FH. She mentions that she is aware that she has high cholesterol and that heart disease runs in her family and she is happy with her current care management. The genetic counselor shares that this result indicates that she has a higher risk for heart attack than previously recognized and that a review of her chart suggests her medications and lipid level goals may not be optimized for FH. She then agrees to follow-up at the lipid multidisciplinary clinic where she meets with a lipidologist, pharmacist, and genetic counselor. A pedigree collected at the visit reveals a family history consistent with FH and early-onset breast and prostate cancers. Her genetic counselor informs her that while she did have genetic testing done, it was a screening test. This means that it is not as comprehensive as a clinical test ordered based on personal and/or family medical history. Some genes related to cancer risk were not included on the test and for those that were, some types of variants are difficult to detect (e.g. large deletions and duplications) with the technology used. She is offered and agrees to further clinical testing for hereditary cancer. This testing uncovers a likely pathogenic CHEK2 variant, which was not a gene covered by her genomic screen. She begins more aggressive lipid-lowering therapy and CHEK2-related cancer surveillance and shares both of her results with family members.
Key points
Some patients may know that a phenotype and/or genotype runs in their family prior to their result.
It may not be clear to patients how learning the genetic underpinning of a known phenotype is relevant to their disease risks or recommended treatment without further explanation.
GS may miss some types of positive genetic results and it is important to recognize the limitations of a genetic test to understand residual risks.
Post-test risk assessment provides an opportunity to uncover other concerning family history and candidates for further genetic testing.
The Missed Result
Jolene has a strong family history of early-onset colon cancer. After her diagnosis of colorectal cancer at age 54 she is referred to cancer genetics. In that appointment she is directed toward a guidelines-based colorectal cancer gene panel which she decides to have. She receives a call a few weeks later informing her that her results were negative. Her oncologist invites her to join a research study investigating the genetics of individuals with a cancer diagnosis and she agrees to participate. This project involves exome sequencing and researchers plan to screen the data for results from the ACMG SF V2.0 list and clinically confirm and return medically actionable results. Six months later, after her treatment is completed, she receives a call that there was a medically significant genetic finding through her participation in that research project. She is told that she tested positive for a result in the BRCA1 gene that puts her at increased risk for additional types of cancer such as breast, ovarian and pancreatic cancers. She is surprised by this information and upset that it was not detected on her clinical genetic test. She is informed that colon cancer is one cancer that is not definitively linked to the BRCA1 gene, and therefore she did not meet genetic testing criteria for this gene which is why it was not evaluated. They have a lengthy discussion about how her personal and family history of colon cancer would typically be thought of as unrelated to this result, with the caveat that we are always learning more about genetics of disease and it is possible that as we expand testing to different groups we may learn of a connection that was previously unrecognized. She expresses that she is afraid of being diagnosed with a second cancer and decides to undergo the recommended BRCA1-related risk management in addition to her routine cancer-related follow-up care.
Key Points
Genetic testing based on a personal/family history-based risk assessment may miss some medically significant positive results.
The constellation of features in a history may be surprising when coupled with a positive result. Sometimes, they may seem concerning for a different genetic predisposition.
It can be important to acknowledge the limitations of both genetic testing and our medical knowledge at any given time.
Conclusions
GS has the potential to expand the reach of genetic information and our knowledge about the genetic underpinnings of disease. Providing care in this setting will require rethinking historical practices and embracing non-traditional approaches to service provision. Genetic counselors will need to become accustomed to a post-test-oriented GC model, and adapt their risk assessments, counseling discussions and materials for GS cohorts. Once data from GS cohorts becomes available, diagnostic criteria, management guidelines, and patient materials developed for those with a personal and/or family medical history may need adaptation. Despite its challenges, providing care for patients who present based on genotype can be particularly valuable because it optimizes the preventive potential of acting on genetic information for both the patient and their family members. Genetic counseling for GS provides an opportunity to deliver preventive care to unique patient populations.
What is known about this topic:
The use of genomic screening without a phenotypic indication is growing. Genotypically ascertained populations have unique characteristics that are relevant for care provision.
What this paper adds to the topic:
This paper provides guidance about how to account for characteristics of genomic screening populations during genetic counseling. Case examples are used to integrate the ideas discussed with clinical scenarios.
Acknowledgements
This publication was supported, in part, by the National Heart, Lung, And Blood Institute of the National Institutes of Health under Award Number R01HL141901. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of Interest
Adam Buchanan has received research grants from the National Institutes of Health and the Marcus Foundation. He has received compensation as a section editor for the Journal of Genetic Counseling. He holds an equity stake in MeTree and You, Inc.
Amy Sturm has received research grants from the National Institutes of Health and salary support from the All of Us Genetic Counseling Resource.
Christopher Haggerty has received research grants from the National Institutes of Health.
Miranda Hallquist and Marci Schwartz declare that they have no conflict of interest.
Footnotes
Human Studies and Informed Consent
No human subject data were collected in the development of this commentary manuscript. The fictional examples used in this manuscript were dramatized from cases identified through the MyCode Community Health Initiative, which has been approved by the Geisinger Institutional Review Board.
Animal Studies
No non-human animal studies were carried out by the authors for this article.
Data Sharing and Data Accessibility
No original research data were used in the development of this commentary manuscript.
Bibliography
- Biesecker LG (2013). Hypothesis-generating research and predictive medicine. Genome Res, 23(7), 1051–1053. doi: 10.1101/gr.157826.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan AH, Lester Kirchner H, Schwartz MLB, Kelly MA, Schmidlen T, Jones LK, … Sturm AC (2020). Clinical outcomes of a genomic screening program for actionable genetic conditions. Genet Med. doi: 10.1038/s41436-020-0876-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carruth ED, Young W, Beer D, James CA, Calkins H, Jing L, … Haggerty CM (2019). Prevalence and Electronic Health Record-Based Phenotype of Loss-of-Function Genetic Variants in Arrhythmogenic Right Ventricular Cardiomyopathy-Associated Genes. Circ Genom Precis Med, 12(11), e002579. doi: 10.1161/CIRCGEN.119.002579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Simone LM, Arjunan A, Vogel Postula KJ, Maga T, & Bucheit LA (2020). Genetic counselors’ perspectives on population-based screening for BRCA-related hereditary breast and ovarian cancer and Lynch syndrome. J Genet Couns. doi: 10.1002/jgc4.1305 [DOI] [PubMed] [Google Scholar]
- East KM, Cochran M, Kelley WV, Greve V, Emmerson K, Raines G, … Bick D (2019). Understanding the present and preparing for the future: Exploring the needs of diagnostic and elective genomic medicine patients. J Genet Couns, 28(2), 438–448. doi: 10.1002/jgc4.1114 [DOI] [PubMed] [Google Scholar]
- eMerge Consortium. (2019). Harmonizing Clinical Sequencing and Interpretation for the eMERGE III Network. Am J Hum Genet, 105(3), 588–605. doi: 10.1016/j.ajhg.2019.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomics Working Group of the All of US Research Program Advisory Panel. (2020). Retrieved from https://allofus.nih.gov/genomics-working-group-all-us-research-program-advisory-panel
- Giardiello FM, Allen JI, Axilbund JE, Boland CR, Burke CA, Burt RW, … Rex DK (2014). Guidelines on genetic evaluation and management of Lynch syndrome: a consensus statement by the US Multi-society Task Force on colorectal cancer. Am J Gastroenterol, 109(8), 1159–1179. doi: 10.1038/ajg.2014.186 [DOI] [PubMed] [Google Scholar]
- Grundy SM, Stone NJ, Bailey AL, Beam C, Birtcher KK, Blumenthal RS, … Yeboah J (2019). 2018 AHA/ACC/AACVPR/AAPA/ABC/ACPM/ADA/AGS/APhA/ASPC/NLA/PCNA Guideline on the Management of Blood Cholesterol: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. J Am Coll Cardiol, 73(24), e285–e350. doi: 10.1016/j.jacc.2018.11.003 [DOI] [PubMed] [Google Scholar]
- Grzymski JJ, Coppes MJ, Metcalf J, Galanopoulos C, Rowan C, Henderson M, … Slonim A (2018). The Healthy Nevada Project: rapid recruitment for population health study. BioRxiv, 250274. doi: 10.1101/250274 [DOI] [Google Scholar]
- Grzymski JJ, Elhanan G, Smith E, Rowan C, Slotnick N, Dabe S, … Lu JT (2019). Population Health Genetic Screening for Tier 1 Inherited Diseases in Northern Nevada: 90% of At-Risk Carriers are Missed. BioRxiv, 650549. doi: 10.1101/650549 [DOI] [Google Scholar]
- Hagenkord J, Funke B, Qian E, Hegde M, Jacobs KB, Ferber M, … Bick D (2020). Design and Reporting Considerations for Genetic Screening Tests. J Mol Diagn, 22(5), 599–609. doi: 10.1016/j.jmoldx.2020.01.014 [DOI] [PubMed] [Google Scholar]
- Haggerty CM, Murray B, Tichnell C, Judge DP, Tandri H, Schwartz M, … James CA (2018). Managing Secondary Genomic Findings Associated With Arrhythmogenic Right Ventricular Cardiomyopathy: Case Studies and Proposal for Clinical Surveillance. Circ Genom Precis Med, 11(7), e002237. doi: 10.1161/CIRCGEN.118.002237 [DOI] [PubMed] [Google Scholar]
- Hallquist MLG, Tricou EP, Hallquist MN, Savatt JM, Rocha H, Evans AE, … Buchanan AH (2020). Positive impact of genetic counseling assistants on genetic counseling efficiency, patient volume, and cost in a cancer genetics clinic. Genet Med. doi: 10.1038/s41436-020-0797-2 [DOI] [PubMed] [Google Scholar]
- Hooker GW, Ormond KE, Sweet K, & Biesecker BB (2014). Teaching genomic counseling: preparing the genetic counseling workforce for the genomic era. J Genet Couns, 23(4), 445–451. doi: 10.1007/s10897-014-9689-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter JE, Irving SA, Biesecker LG, Buchanan A, Jensen B, Lee K, … Goddard KA (2016). A standardized, evidence-based protocol to assess clinical actionability of genetic disorders associated with genomic variation. Genet Med, 18(12), 1258–1268. doi: 10.1038/gim.2016.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia SS, Adelman K, Bale SJ, Chung WK, Eng C, Evans JP, … Miller DT (2017). Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med, 19(2), 249–255. doi: 10.1038/gim.2016.190 [DOI] [PubMed] [Google Scholar]
- Katz AE, Nussbaum RL, Solomon BD, Rehm HL, Williams MS, & Biesecker LG (2020). Management of Secondary Genomic Findings. Am J Hum Genet, 107(1), 3–14. doi: 10.1016/j.ajhg.2020.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MA, McGlinch EB, McGlinch MC, & Capacchione JF (2017). Malignant Hyperthermia Susceptibility and Fitness for Duty. Mil Med, 182(3), e1854–e1857. doi: 10.7205/MILMED-D-16-00186 [DOI] [PubMed] [Google Scholar]
- Lu JT, Ferber M, Hagenkord J, Levin E, South S, Kang HP, … Bick DP (2019). Evaluation for Genetic Disorders in the Absence of a Clinical Indication for Testing: Elective Genomic Testing. J Mol Diagn, 21(1), 3–12. doi: 10.1016/j.jmoldx.2018.09.006 [DOI] [PubMed] [Google Scholar]
- Manickam K, Buchanan AH, Schwartz ML, Hallquist ML, Williams JL, Rahm AK, … Butry LM (2018). Exome Sequencing–Based Screening for BRCA1/2 Expected Pathogenic Variants Among Adult Biobank Participants. JAMA Network Open, 1(5), e182140–e182140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus FI, McKenna WJ, Sherrill D, Basso C, Bauce B, Bluemke DA, … Zareba W (2010). Diagnosis of arrhythmogenic right ventricular cardiomyopathy/dysplasia: proposed modification of the Task Force Criteria. Eur Heart J, 31(7), 806–814. doi: 10.1093/eurheartj/ehq025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MF (2016). Your DNA is not your diagnosis: getting diagnoses right following secondary genomic findings. Genet Med, 18(8), 765–767. doi: 10.1038/gim.2015.134 [DOI] [PubMed] [Google Scholar]
- MyCode Community Health Initiative. Retrieved from https://www.geisinger.org/mycode
- Nordestgaard BG, Chapman MJ, Humphries SE, Ginsberg HN, Masana L, Descamps OS, … European Atherosclerosis Society Consensus, P. (2013). Familial hypercholesterolaemia is underdiagnosed and undertreated in the general population: guidance for clinicians to prevent coronary heart disease: consensus statement of the European Atherosclerosis Society. Eur Heart J, 34(45), 3478–3490a. doi: 10.1093/eurheartj/eht273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Daniel JM (2010). The prospect of genome-guided preventive medicine: a need and opportunity for genetic counselors. J Genet Couns, 19(4), 315–327. doi: 10.1007/s10897-010-9302-4 [DOI] [PubMed] [Google Scholar]
- Ormond KE, Hallquist MLG, Buchanan AH, Dondanville D, Cho MK, Smith M, … Faucett WA (2019). Developing a conceptual, reproducible, rubric-based approach to consent and result disclosure for genetic testing by clinicians with minimal genetics background. Genet Med, 21(3), 727–735. doi: 10.1038/s41436-018-0093-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacyna JE, Radecki Breitkopf C, Jenkins SM, Sutton EJ, Horrow C, Kullo IJ, & Sharp RR (2019). Should pretest genetic counselling be required for patients pursuing genomic sequencing? Results from a survey of participants in a large genomic implementation study. J Med Genet, 56(5), 317–324. doi: 10.1136/jmedgenet-2018-105577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilarski R, Burt R, Kohlman W, Pho L, Shannon KM, & Swisher E (2013). Cowden syndrome and the PTEN hamartoma tumor syndrome: systematic review and revised diagnostic criteria. J Natl Cancer Inst, 105(21), 1607–1616. doi: 10.1093/jnci/djt277 [DOI] [PubMed] [Google Scholar]
- Riley BD, Culver JO, Skrzynia C, Senter LA, Peters JA, Costalas JW, … Trepanier AM (2012). Essential elements of genetic cancer risk assessment, counseling, and testing: updated recommendations of the National Society of Genetic Counselors. J Genet Couns, 21(2), 151–161. doi: 10.1007/s10897-011-9462-x [DOI] [PubMed] [Google Scholar]
- Schmidlen T, Schwartz M, DiLoreto K, Kirchner HL, & Sturm AC (2019). Patient assessment of chatbots for the scalable delivery of genetic counseling. J Genet Couns, 28(6), 1166–1177. doi: 10.1002/jgc4.1169 [DOI] [PubMed] [Google Scholar]
- Schwartz ML, McCormick CZ, Lazzeri AL, D’Andra ML, Hallquist ML, Manickam K, … Frisbie L (2018). A Model for Genome-First Care: Returning Secondary Genomic Findings to Participants and Their Healthcare Providers in a Large Research Cohort. The American Journal of Human Genetics, 103(3), 328–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Driest SL, Wells QS, Stallings S, Bush WS, Gordon A, Nickerson DA, … Roden DM (2016). Association of Arrhythmia-Related Genetic Variants With Phenotypes Documented in Electronic Medical Records. JAMA, 315(1), 47–57. doi: 10.1001/jama.2015.17701 [DOI] [PMC free article] [PubMed] [Google Scholar]