Highlights
-
•
The lncRNA SCHLAP1 predicts poor prognosis in multifocal primary prostate cancer
-
•
Expression of SCHLAP1 is heterogeneous within and between different malignant foci
-
•
Multiple samples per patient is crucial, otherwise high SCHLAP1 may be overlooked
-
•
High SCHLAP1 is associated with reactive stroma and other aggressive features
Keywords: Biomarker, Heterogeneity, lncRNA, Multifocality, Prognosis, Prostate cancer
Abbreviations: BCR, biochemical recurrence; IDCP, intraductal carcinoma of the prostate; lncRNA, long noncoding RNA; PSA, prostate-specific antigen; pT stage, pathological tumor stage
Abstract
In primary prostate cancer, the common multifocality and heterogeneity are major obstacles in finding robust prognostic tissue biomarkers. The long noncoding RNA SCHLAP1 has been suggested, but its prognostic value has not been investigated in the context of tumor heterogeneity. In the present study, expression of SCHLAP1 was investigated using real-time RT-PCR in a multisampled series of 778 tissue samples from radical prostatectomies of 164 prostate cancer patients (median follow-up time 7.4 y). The prognostic value of SCHLAP1 was evaluated with biochemical recurrence as endpoint.
In total, 29% of patients were classified as having high expression of SCHLAP1 in at least one malignant sample. Among these, inter- and intrafocal heterogeneity was detected in 72% and 56%, respectively. High expression of SCHLAP1 was shown to be a predictor of biochemical recurrence in both uni- and multivariable cox regression analyses (P < 0.001 and P = 0.02). High expression of SCHLAP1 was also significantly associated with adverse clinicopathological characteristics, including grade group, high pT stage, invasive cribriform growth/intraductal carcinoma of the prostate, and reactive stroma. In conclusion, high expression of SCHLAP1 in at least one malignant sample is a robust prognostic biomarker in primary prostate cancer. For the first time, high SCHLAP1 expression has been associated with the aggressive histopathologic feature reactive stroma. The expression of SCHLAP1 is highly heterogeneous, and analysis of multiple samples is therefore crucial in determination of the SCHLAP1 status of a patient.
Introduction
Prostate cancer patients have highly variable disease outcomes. Most patients have slow-growing and indolent cancers, while others are diagnosed with either primary metastatic cancers or localized/locally advanced aggressive cancers that relapse after radical treatment and progress to lethal disease. For several cancer types, molecular biomarkers have been incorporated in the clinic to aid in diagnostics, prognostics and prediction of treatment response. However, identification of clinically useful genomic biomarkers for prostate cancer is complex, as the majority of patients have multiple primary tumor foci [1]. In addition, the molecular heterogeneity between the different foci is pronounced [2]. Prostate cancer is known to have a low number of somatic mutations present in the primary setting [3] and few recurrently mutated genes [4]. Long noncoding RNAs (lncRNAs), such as SCHLAP1 and PCA3, have shown promising biomarker potential in prostate cancer [5]. However, studies considering the multifocality and heterogeneity aspects are still required to investigate the prognostic value of SCHLAP1 in prostate cancer.
LncRNA is a class of noncoding RNA more than 200 nucleotides long with no protein-coding capacity [6]. Nonetheless, lncRNAs share many features with protein-coding genes, as they are transcribed and modified in a similar manner. Important functional aspects and cellular mechanisms have been characterized for several lncRNAs [7], and their roles in various diseases have received attention [8]. In addition, many lncRNAs show tissue- and cancer-specific expression patterns [9], and therefore present as potential biomarkers. SCHLAP1 has been found to be highly expressed in approximately 25% of prostate cancers [10], with significantly higher expression in prostate cancer compared to benign prostatic tissue [11]. Knockdown of SCHLAP1 has been found to impair cell invasion, proliferation, and reduced metastatic potential in xenografted nude mice, and expression analyses has shown that SCHLAP1 antagonizes the SWI/SNF-complex [10]. Expression of SCHLAP1 has been associated with gene fusions involving ETS transcription factors, with TMPRSS2-ERG being the most common genetic aberration in primary prostate cancer in Western countries [10,12]. Interestingly, SCHLAP1-UBE2E3 was found to be the most frequent gene fusion in a large Chinese cohort, suggesting geographical or ethnic differences [13].
High expression of SCHLAP1 has been characterized as an independent predictor of metastasis and prostate cancer-specific death [10]. In addition, the lncRNA has been associated with biochemical recurrence (BCR) and adverse histopathological features, including high Gleason score [11], intraductal carcinoma of the prostate (IDCP) and invasive cribriform growth [14,15]. Expression of SCHLAP1 in relation to multifocal prostate cancer has been reported as part of a wider transcriptome analysis [16], but it is unknown to what degree a potentially heterogeneous expression pattern across or within tumor foci will impact its value as a prognostic biomarker. We postulate that expression of SCHLAP1 is dependent on the aggressiveness of the prostate cancer. To investigate this, we have utilized a unique multisampled prostate cancer biobank, consisting of spatially separated fresh-frozen tissue cores collected from prostatectomy specimens, to account for both heterogeneity and multifocality.
Materials and methods
Multisampled patient cohort and follow-up protocol
From a total cohort of 571 prospectively and consecutively included patients who underwent radical prostatectomy between 2010 and 2012 at Oslo University Hospital-Radiumhospitalet, 164 patients were included in this study, none of whom received neoadjuvant treatment. Among these, 121 were consecutively included from the start of the inclusion period, and 43 additional patients were selected where samples were available from clearly separate cancer foci [2,17]. From all 164 patients, multiple tissue samples were collected from the prostatectomy specimens, with a total of 778 fresh-frozen tissue samples (360 from malignant and 418 from benign tissue). Multiple malignant samples were available from 91 patients, and 55 of these had malignant samples from clearly distinct cancer foci. From 20 patients, only benign samples were available. Thus, in total, malignant samples from 144 patients were included.
The clinicopathological and molecular characteristics of all included patients and samples are summarized in Supplementary Tables S1 and S2. Written informed consent was obtained from all included patients and the study was approved by the Regional Ethics Committee South-Eastern Norway (number 2013/595/REK southeast A).
The cohort is monitored by a research nurse. Follow-up data include 2994 measurements of serum concentrations of prostate-specific antigen (PSA; median of 16 measurements per patient), further oncological treatment in case of biochemical relapse, clinical recurrence, and death. The PSA values were obtained from patients returning for regular controls at the hospital, through correspondence with general practitioners, local hospitals, and Fürst Medical Laboratories (Oslo, Norway).
BCR was used as an endpoint in time-to-event analyses and defined as a postoperative PSA ≥ 0.20 ng/ml in two consecutive blood samples. Patients without BCR were censored at the date of the last known PSA measurement. Median follow-up time was 7.4 y for all patients, and 7.8 y for patients without BCR. Clinical recurrence and death (overall survival) were used as other endpoints. Clinical recurrence was defined as radiologic recurrence of prostate cancer in conjunction with PSA relapse, either locally or as distant metastases, most often identified by use of magnetic resonance imaging. Date and cause of death were recorded from the population-based Norwegian Cause of Death Registry with annual updates.
RNA isolation and reverse transcription
Total RNA was isolated from all fresh-frozen tissue samples using the AllPrep DNA/RNA/miRNA Universal kit (Qiagen, Hilden, Germany). cDNA was generated using the High Capacity cDNA Reverse Transcription kit (Thermo Fisher Scientific, Waltham, MA) or SMARTer RACE cDNA Amplification kit (Clontech, Mountain View, CA) according to the manufacturers’ protocols.
Semi-quantitative gene expression analyses
This study is reported according to the Reporting recommendations for tumor marker prognostic studies (REMARK; Supplementary Table S3).
The expression of SCHLAP1 and ERG was investigated using real-time reverse transcription polymerase chain reaction (RT-PCR) using TaqMan Gene Expression Assays (Thermo Fisher Scientific). ABL1 was used as an endogenous control. The applied assays were Hs04968419_m1 (SCHLAP1), Hs01554630_m1 (ERG), and Hs01104728_m1 (ABL1). All samples were run in triplicate on an ABI 7900HT Fast Real-time PCR system (Applied Biosystems, Foster City, CA), with 10 ng cDNA included in each reaction.
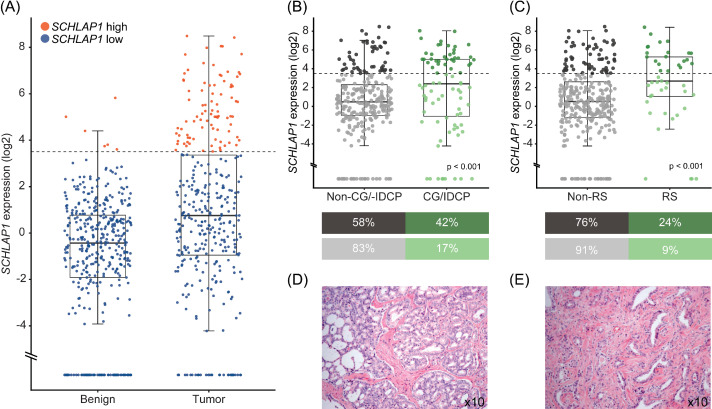
Cycle threshold values were recorded, and the triplicate medians were used. Medians > 35 were considered as no expression of SCHLAP1. Expression was normalized to the endogenous control ABL1 using the standard curve method. The ratio between the median relative quantity of SCHLAP1 and ABL1 in all benign samples was used as calibrator. To separate high and low expression, a threshold was set by evaluation of the distribution of expression levels in all samples, at 3.5 log2 (fold change) (Fig. 1). Samples with no SCHLAP1 expression were placed in the low expression group. ERG expression analyses have been performed (unpublished data).
Fig. 1.
Expression of SCHLAP1 in samples with various histopathological growth patterns. SCHLAP1 expression is often higher in malignant samples, and in tissue with specific histopathological growth patterns associated with poor prognosis. (A) Relative SCHLAP1 expression in benign and malignant tissue samples (n = 418 and 360, respectively). Datapoints at the bottom of the plot indicate samples (68 benign, 34 malignant) where no SCHLAP1 expression was detected. The boxes extend from the first to the third quartiles, and lines in the middle represent the medians. The whiskers extend from the box to the largest value up to 1.5*(interquartile range). (B-C) Expression of SCHLAP1 in malignant samples, separated based on (B) presence or absence of invasive cribriform growth/IDCP or (C) reactive stroma. Dark gray and dark green indicate samples with high expression of SCHLAP1, whereas light colors represent samples with low expression. Associations between SCHLAP1 expression and histopathological features were investigated using χ2 test of independence. (D-E) Representative images of hematoxylin and eosin-stained sections of tumor tissue with (D) invasive cribriform growth and (E) reactive stroma. CG, invasive cribriform growth; IDCP, intraductal carcinoma of the prostate; RS, reactive stroma.
Histopathological assessment
Histopathological parameters (Gleason score, grade group, reactive stroma, invasive cribriform growth/IDCP, pathological tumor (pT) stage, and extraprostatic extension) were re-evaluated blinded to SCHLAP1 status by an experienced uropathologist. Grading was performed according to the 2014 ISUP Modified Gleason system [18]. Reactive stroma was classified as present or absent based on morphological features, as described previously [19,20].
Tumors were classified as distinct foci when separated by at least 2 to 4 mm and showing different tissue morphology (described in [2]). The index tumor was defined as the focus having the highest pT stage [21]. In cases with multiple foci with the same pT stage, the focus with the highest Gleason score was classified as the index. If multiple foci had the same Gleason score, the largest focus in diameter was denoted as the index tumor.
Statistical analyses
Associations between categorical variables were investigated using either χ2 test of independence or Fisher's exact test. Kaplan-Meier plots and log-rank tests were applied to compare time to BCR. Univariable and multivariable Cox regression analyses were performed, and the Schoenfeld test was used to assess whether the proportional hazards assumption was met. An interaction term was included to evaluate interaction between variables. A P value of 0.05 was considered the threshold for statistical significance. Analyses were performed using R (3.5.1) and RStudio (1.1.456), with the survival (2.44-1.1), survminer (0.4.7) and ggplot2 (3.0.0) packages.
Results
Expression of SCHLAP1 in primary prostate cancer
Real-time RT-PCR was used to measure expression of SCHLAP1 in 778 prostate tissue samples from 164 prostate cancer patients treated with radical prostatectomy. A threshold separating high and low expression of SCHLAP1 was set to assign the majority of benign prostate samples in the low group (Fig. 1). Among the malignant samples, 24% (88/360) were classified as having high expression, whereas 1% (6/418) of benign samples were assigned to this category. Using the same threshold, 29% (42/144) of patients were classified as having high expression of SCHLAP1 in at least one malignant sample.
Significant associations were found between SCHLAP1 expression and grade group, pT stage, ERG overexpression (Supplementary Figure S1), invasive cribriform growth/IDCP, and reactive stroma (Table 1, Fig. 1).
Table 1.
Clinicopathological and molecular characteristics for patients stratified by high and low expression of SCHLAP1.
| Expression of SCHLAP1 (Summarized Per Patient) | |||
|---|---|---|---|
| High (n = 42) | Low (n = 102) | P | |
| Age at surgery | 0.1 | ||
| <60 | 13 (31%) | 24 (24%) | |
| 60–70 | 28 (67%) | 64 (63%) | |
| > 70 | 1 (2%) | 14 (14%) | |
| Preoperative PSA | 0.2 | ||
| <10 ng/ml | 19 (45%) | 46 (45%) | |
| 10 ng/ml–20 ng/ml | 12 (29%) | 41 (40%) | |
| > 20 ng/ml | 11 (26%) | 15 (15%) | |
| Grade group | 0.02* | ||
| 1–2 | 12 (29%) | 47 (46%) | |
| 3 | 10 (24%) | 31 (30%) | |
| 4–5 | 20 (48%) | 24 (24%) | |
| pT stage | 0.006* | ||
| pT2 | 6 (14%) | 42 (41%) | |
| pT3a | 26 (62%) | 46 (45%) | |
| pT3b | 10 (24%) | 13 (13%) | |
| NA | 0 (0 %) | 1 (1%) | |
| Lymph node status | 0.1 | ||
| NX | 22 (52%) | 72 (71%) | |
| N0 | 14 (33%) | 27 (26%) | |
| N1 | 6 (14%) | 3 (3%) | |
| Surgical margin | 0.3 | ||
| Positive | 11 (26%) | 17 (17%) | |
| Negative | 31 (74%) | 85 (83%) | |
| ERG overexpression | < 0.001* | ||
| Yes | 36 (86%) | 33 (32%) | |
| No | 6 (14%) | 69 (68%) | |
| Invasive cribriform growth/IDCP | < 0.001* | ||
| Yes | 34 (81%) | 49 (48%) | |
| No | 8 (19%) | 53 (52%) | |
| Reactive stroma | 0.001* | ||
| Yes | 25 (60%) | 30 (29%) | |
| No | 17 (40%) | 72 (71%) | |
| Biochemical recurrence | < 0.001* | ||
| Yes | 23 (55%) | 21 (21%) | |
| No | 19 (45%) | 81 (79%) | |
| Clinical recurrence | 0.1 | ||
| Yes | 6 (14%) | 7 (7%) | |
| No | 36 (86%) | 95 (93%) | |
| Death (overall survival) | 0.01* | ||
| Yes | 10 (24%) | 8 (8%) | |
| No | 32 (76%) | 94 (92%) | |
Patients were classified as having high expression of SCHLAP1 when at least one malignant sample displayed expression levels above the threshold. A χ2 test of independence or Fisher's exact test was applied to investigate associations between categorical variables. Univariable Cox regression was used to assess association with BCR, clinical recurrence and death (overall survival).
IDCP = intraductal carcinoma of the prostate; PSA = prostate-specific antigen; pT stage = pathological tumor stage.
indicate statistical significance, P < 0.05.
High expression of SCHLAP1 is a predictor of biochemical recurrence
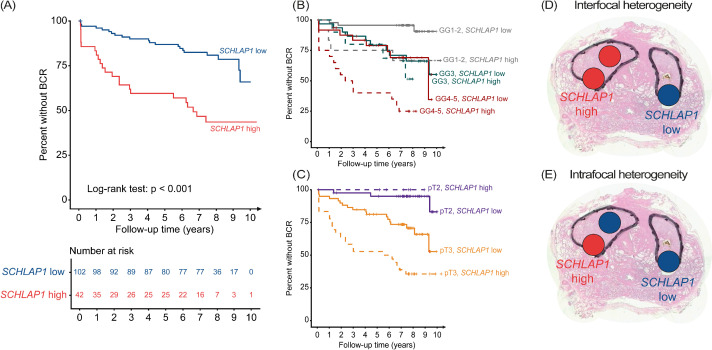
Patients with high expression of SCHLAP1 in at least one malignant sample were found to have a significantly shorter time to BCR compared to patients with low expression (Fig. 2A; log-rank, P < 0.001). Using Cox regression, high expression of SCHLAP1 was found to be a predictor of BCR in univariable analysis, and an independent predictor of BCR in multivariable analysis (Table 2). By testing for association with other endpoints, high expression of SCHLAP1 was not found to predict clinical recurrence (univariable Cox regression, P = 0.1), but predicted a shorter time to death by any cause (overall survival; univariable Cox regression, P = 0.01).
Fig. 2.
High expression of SCHLAP1 is associated with shorter time to BCR, and concepts of inter- and intrafocal heterogeneity in prostate cancer. (A) Kaplan-Meier plot showing the percentage of patients without BCR, stratified by high and low expression of SCHLAP1. (B-C) Percent of patients without BCR, stratified by (B) grade group and expression of SCHLAP1, or (C) stratified by pT stage and expression of SCHLAP1. (D) Interfocal heterogeneity, where SCHLAP1 status varies between tissue samples collected from two different tumor foci. (E) Intrafocal heterogeneity, where the SCHLAP1 status of samples collected from the same focus varies. The outline of each tumor focus is marked with a black line, and the SCHLAP1 status of a sample is indicated by red (high) or blue (low) circles. BCR, biochemical recurrence; GG, grade group; pT, pathological tumor stage.
Table 2.
High expression of SCHLAP1 is a predictor of biochemical relapse.
| Univariable | Multivariable | |||
|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |
| SCHLAP1 | ||||
| Low | Reference | |||
| High | 3.5 (1.9–6.3) | < 0.001* | 2.1 (1.1–4.0) | 0.02* |
| Preoperative PSA | ||||
| < 10 ng/ml | Reference | |||
| 10 ng/ml–≤ 20 ng/mL | 1.9 (0.9–3.9) | 0.07 | 1.3 (0.5–3.0) | 0.6 |
| > 20 ng/ml | 3.3 (1.5–7.1) | 0.002* | 1.0 (0.4–2.9) | 0.9 |
| Grade group | ||||
| 1–2 | Reference | |||
| 3 | 3.3 (1.4–8.0) | 0.007* | 2.0 (0.7–5.8) | 0.2 |
| 4–5 | 6.9 (3.0–15.4) | < 0.001* | 3.0 (1.1–8.2) | 0.03* |
| Surgical margins | ||||
| Negative | Reference | |||
| Positive | 4.1 (2.2–7.3) | < 0.001* | 2.6 (1.3–4.9) | 0.005* |
| pT stage | ||||
| pT2 | Reference | |||
| pT3a | 6.9 (2.4–19.7) | < 0.001* | 4.0 (1.2–13.8) | 0.03* |
| pT3b | 13.2 (4.3–40.6) | < 0.001* | 4.9 (1.2–19.7) | 0.03* |
The multivariable model included clinicopathological characteristics commonly used in risk stratification of prostate cancer patients (preoperative PSA, grade group, surgical margins and pT stage). The assumption of proportional hazards was met in all analyses, except for the univariable analysis of preoperative PSA, and grade group and preoperative PSA in the multivariable model. Nodal stage was excluded from the analysis due to missing data (NX; Table 1).
CI = confidence interval; HR = hazard ratio; PSA = prostate-specific antigen; pT stage = pathological tumor stage.
indicate statistical significance, P < 0.05.
As gene fusions involving ERG, leading to overexpression of the gene, is the most common genetic aberration in prostate cancer, its prognostic value was investigated. Overexpression of ERG in malignant samples was not found to predict a shorter time to BCR (univariable Cox regression, P = 0.1; hazard ratio = 1.6; 95% confidence interval = 0.9−3.0).
When stratifying on both grade group and SCHLAP1 status, patients with grade group 1-2 or 4-5 tumors and high SCHLAP1 expression were found to have a shorter time to BCR than patients within the same grade groups and low SCHLAP1 (Fig. 2B). Interestingly, high SCHLAP1 did not predict a shorter time to BCR in patients with grade group 3 tumors. Patients with grade group 4-5 prostate cancer and low SCHLAP1 expression, or grade group 1-2 and high SCHLAP1, were found to have a similar BCR-free survival as patients within grade group 3. Patients with both pT3 and high SCHLAP1 expression were found to have a shorter time to BCR compared to patients with pT3 and low SCHLAP1 (Fig. 2C). No statistically significant interaction was identified between SCHLAP1 and grade group (P = 0.2 for grade group 3, P = 0.5 for grade group 4-5), or SCHLAP1 and pT stage (P = 1 for both pT3a and pT3b) in Cox regression analyses.
Expression of SCHLAP1 is highly heterogeneous
Interfocal heterogeneity (Fig. 2D) was discovered in 8/11 (73%) of patients with high expression of SCHLAP1 in at least one tumor focus and with samples from more than one focus. Intrafocal heterogeneity (Fig. 2E) was detected in 56% (20/36) of patients with high SCHLAP1 expression in at least one malignant sample, and from whom more than one sample was collected from the same tumor focus. Among patients with high SCHLAP1 and available samples from the index tumor focus, at least one sample was scored as having high expression in 93% (39/42). However, for 54% (19/35) of these patients, the expression of SCHLAP1 was heterogeneous within the index tumor. Among patients with high SCHLAP1 and samples from both the index tumor and at least one other focus, a high expression of SCHLAP1 was detected exclusively in a non-index focus for 27% (3/11) of patients.
Due to the observed degree of heterogeneity, it was deemed necessary to investigate associations between histopathological and molecular characteristics on a “per sample” basis. Significant associations were still found between high SCHLAP1 expression and grade group, ERG overexpression, presence of invasive cribriform growth/IDCP, and reactive stroma (Supplementary Table S4).
Discussion
The present study demonstrates that expression of SCHLAP1 has important prognostic value in primary prostate cancer, despite a high level of heterogeneity between and within tumor foci. Analysis of only 1 tissue biopsy from a prostate cancer patient will not be sufficient in determining SCHLAP1 status, i.e., high or low expression, as more than half of the patients show discordant results among samples from the same tumor.
A high degree of both inter- and intrafocal heterogeneity in SCHLAP1 expression was detected. Samples from clearly separated foci were available from 55 patients, of which 11 patients had high SCHLAP1 expression in at least one tumor focus. To further evaluate the interfocal heterogeneity of SCHLAP1 expression, it would be of relevance to evaluate this biomarker in a larger material. However, as intrafocal heterogeneity was substantial (56%), it is evident that the heterogeneity aspect should not be overlooked in following prostate cancer biomarker research.
We assigned patients to the high SCHLAP1 group when at least 1 malignant sample displayed high expression and found that the high and low groups have clearly distinct times to BCR. This indicates that the identification of any tumor area with a high expression of SCHLAP1 is sufficient to predict poor prognosis for a patient, independently of other adverse clinicopathological features. Previous studies have found differences in time to BCR between patients with high vs. low expression of SCHLAP1, although with less separation [10], best explained by the fact that only one sample per patient was analyzed. The current analysis of multiple malignant samples per patient result in a greater separation of the two groups. However, our analyses include fewer patients than previous reports [10,11].
For prostate cancer patients treated with radical prostatectomy to present with metastatic disease, follow-up for 10 to 15 y is necessary to accurately evaluate clinical recurrence as an endpoint in time-to-event analyses [22]. Our findings using this endpoint were not significant, most likely as a result of few patients (13/164) experiencing clinical recurrence at this time point. The time perspective is also a limitation for evaluating the impact of SCHLAP1 on prostate cancer-specific death and overall survival, with as few as 21 patients in this cohort being registered with death (of which 4 of prostate cancer-specific death). Interestingly, we still find that high SCHLAP1 predicts death (overall survival). SCHLAP1 has also previously been found to predict metastasis and death [23]. Thus, studies with longer follow-up time are required to further evaluate the effect of SCHLAP1 on clinical recurrence, prostate cancer-specific death and overall survival in a multisampled material.
Intrapatient multisample-based SCHLAP1 analysis revealed strong associations between high expression and known adverse histopathological features both on the sample- and patient levels. To the best of our knowledge, the present study is the first to discover the relationship between high SCHLAP1 expression and reactive stroma (P < 0.001 on the sample level). Reactive stroma has previously been found to predict BCR and prostate cancer-specific death [20], and shown to be the strongest prognostic predictor in patients with Gleason 3 + 3 and 3 + 4 (grade groups 1 and 2) [24]. Additionally, reactive stroma is considered to be included in future modifications of the 2014 ISUP grading system [25]. Therefore, further studies evaluating the mechanistic interplay between reactive stroma and SCHLAP1 expression are warranted. We also find associations between high SCHLAP1 and the presence of invasive cribriform growth/IDCP, in agreement with previously published results [14]. Despite such associations, we show that SCHLAP1 expression is a robust biomarker that can be used independently of other adverse clinicopathological markers.
Over the past years, an increasing number of prostate cancer patients in the lower risk groups have chosen active surveillance as a treatment option [26]. However, the present data suggest that low-grade prostate cancers, i.e., grade group 1-2, with high SCHLAP1 expression may benefit from radical treatment, adjuvant oncologic treatment after surgery and/or more intensive follow-up, despite their cancer being classified as a histopathological low grade. In prostate cancer diagnostics, multiple spatially separated needle biopsies are routinely collected, and evaluation of SCHLAP1 expression in this tissue may be a useful tool for making treatment decisions for the patient, also prior to radical treatment. Studies investigating expression of SCHLAP1 in needle biopsies may therefore be a next step toward clinical implementation of this biomarker.
The TMPRSS2-ERG gene fusion is the most common genetic aberration in prostate cancer and leads to overexpression of ERG; however, its prognostic potential is still debated [27,28]. The significant association between high expression of SCHLAP1 and ERG in this sample material may explain why time-to-event analyses often have similar results for the two genes, suggesting that both genes predict poor patient outcomes. However, overexpression of ERG was not found to predict BCR in this study, calling for further investigation.
In light of the discovered heterogeneity in SCHLAP1 expression, diagnostic tools that detect high expression without the requirement for sampling particular tissue areas are highly warranted. To overcome inter- and intrafocal heterogeneity, liquid biopsies, e.g., urine and blood, are emerging as cost effective and minimally invasive tests for investigating molecular aberrations in prostate cancer [29,30]. Expression of SCHLAP1 is highly prostate-specific and has been found to be detectable in urine samples [5]. However, there are several technical limitations that must be overcome to make robust measurements and analysis of RNA in liquid biopsies with sufficient sensitivity [31].
Conclusions
There is substantial inter- and intrafocal heterogeneity in expression of SCHLAP1 in prostate cancer, underscoring the need for multiple, spatially separated samples to accurately determine the SCHLAP1 status of a patient. High expression of SCHLAP1 in at least one malignant sample was sufficient to classify a patient to the high SCHLAP1 group. Patients with high SCHLAP1 expression have a significantly reduced time to BCR. We here, for the first time, report a significant association between high SCHLAP1 expression and reactive stroma. Despite high SCHLAP1 expression being associated with adverse histopathological features, we conclude that high SCHLAP1 expression is an independent predictor of BCR. These results suggest that patients with high expression of SCHLAP1 should be considered for radical treatment independently of histopathological grade group and receive closer follow-up and/or additional oncologic treatment after surgery.
Author contributions
Susanne G. Kidd: Formal analysis, Data curation, Investigation, Writing – Original draft, Visualization.
Kristina T. Carm: Formal analysis, Data curation, Investigation, Writing – Original draft, Visualization.
Mari Bogaard: Investigation, Writing – Review & Editing.
Linn Guro Olsen: Investigation, Writing – Review & Editing.
Anne Cathrine Bakken: Investigation, Writing – Review & Editing.
Marthe Løvf: Conceptualization, Supervision, Writing – Review & Editing.
Ragnhild A. Lothe: Conceptualization, Supervision, Project administration, Funding acquisition, Writing – Review & Editing.
Karol Axcrona: Conceptualization, Supervision, Project administration, Funding acquisition, Resources, Writing – Review & Editing.
Ulrika Axcrona: Conceptualization, Supervision, Project administration, Funding acquisition, Resources, Writing – Review & Editing.
Rolf I. Skotheim: Conceptualization, Supervision, Project administration, Funding acquisition, Writing – Review & Editing
Data availability statement
The data that support the findings of this study are available from the corresponding author, RIS, upon reasonable request.
Acknowledgments
We are grateful for valuable contributions from research nurse Aase Margrethe Victoria Maltau. We also would like to thank the individuals with prostate cancer and their families for contributing to this study.
Footnotes
Funding: The study was supported by the South-Eastern Norway Regional Health Authority (SGK, KTC and MB were financed as PhD students from grants with project numbers 2020063, 2017045, and 2019016), the Research Council of Norway through its FRIPRO funding scheme (262529/F20 and Toppforsk-250993), and the Norwegian Cancer Society (grant number 208197). The study was granted secure storage of computer files and high-performance computation resources from NorStore and University of Oslo's Services for Sensitive Data (NS9013S and p19, respectively).
Conflicts of interest: The authors declare no potential conflicts of interest.
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2021.05.012.
Appendix. Supplementary materials
References
- 1.Andreoiu M., Cheng L. Multifocal prostate cancer: biologic, prognostic, and therapeutic implications. Human pathology. 2010;41(6):781–793. doi: 10.1016/j.humpath.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 2.Løvf M., Zhao S., Axcrona U., Johannessen B., Bakken A.C., Carm K.T., Hoff A.M., Myklebost O., Meza-Zepeda L.A., Lie A.K. Multifocal Primary Prostate Cancer Exhibits High Degree of Genomic Heterogeneity. European urology. 2019;75(3):498–505. doi: 10.1016/j.eururo.2018.08.009. [DOI] [PubMed] [Google Scholar]
- 3.Vogelstein B., Papadopoulos N., Velculescu V.E., Zhou S., Diaz L.A., Kinzler K.W. Cancer Genome Landscapes. Science. 2013;339(6127):1546. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armenia J., Wankowicz S.A.M., Liu D., Gao J., Kundra R., Reznik E., Chatila W.K., Chakravarty D., Han G.C., Coleman I. The long tail of oncogenic drivers in prostate cancer. Nature genetics. 2018;50(5):645–651. doi: 10.1038/s41588-018-0078-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prensner J.R., Zhao S., Erho N., Schipper M., Iyer M.K., Dhanasekaran S.M., Magi-Galluzzi C., Mehra R., Sahu A., Siddiqui J. RNA biomarkers associated with metastatic progression in prostate cancer: a multi-institutional high-throughput analysis of SChLAP1. The Lancet. Oncology. 2014;15(13):1469–1480. doi: 10.1016/S1470-2045(14)71113-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yao R.-W., Wang Y., Chen L.-L. Cellular functions of long noncoding RNAs. Nature Cell Biology. 2019;21(5):542–551. doi: 10.1038/s41556-019-0311-8. [DOI] [PubMed] [Google Scholar]
- 8.DiStefano J.K. The Emerging Role of Long Noncoding RNAs in Human Disease. In: DiStefano J.K., editor. Disease Gene Identification: Methods and Protocols. Springer New York; New York, NY: 2018. pp. 91–110. in. in. Editor. [DOI] [PubMed] [Google Scholar]
- 9.Prensner J.R., Chinnaiyan A.M. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1(5):391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prensner J.R., Iyer M.K., Sahu A., Asangani I.A., Cao Q., Patel L., Vergara I.A., Davicioni E., Erho N., Ghadessi M. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nature Genetics. 2013;45(11):1392–1398. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mehra R., Shi Y., Udager A.M., Prensner J.R., Sahu A., Iyer M.K., Siddiqui J., Cao X., Wei J., Jiang H. A novel RNA in situ hybridization assay for the long loncoding RNA SChLAP1 predicts poor clinical outcome after radical prostatectomy in clinically localized prostate cancer. Neoplasia. 2014;16(12):1121–1127. doi: 10.1016/j.neo.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abeshouse A., Ahn J., Akbani R., Ally A., Amin S., Andry Christopher D., Annala M., Aprikian A., Armenia J., Arora A. The Molecular Taxonomy of Primary Prostate Cancer. Cell. 2015;163(4):1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J., Xu C., Lee H.J., Ren S., Zi X., Zhang Z., Wang H., Yu Y., Yang C., Gao X. A genomic and epigenomic atlas of prostate cancer in Asian populations. Nature. 2020;580(7801):93–99. doi: 10.1038/s41586-020-2135-x. [DOI] [PubMed] [Google Scholar]
- 14.Chua M.L.K., Lo W., Pintilie M., Murgic J., Lalonde E., Bhandari V., Mahamud O., Gopalan A., Kweldam C.F., van Leenders G. A Prostate Cancer "Nimbosus": Genomic Instability and SChLAP1 Dysregulation Underpin Aggression of Intraductal and Cribriform Subpathologies. Eur Urol. 2017;72(5):665–674. doi: 10.1016/j.eururo.2017.04.034. [DOI] [PubMed] [Google Scholar]
- 15.Chen S., Huang V., Xu X., Livingstone J., Soares F., Jeon J., Zeng Y., Hua J.T., Petricca J., Guo H. Widespread and functional RNA circularization in localized prostate cancer. Cell. 2019;176(4) doi: 10.1016/j.cell.2019.01.025. p. 831-843.e22. [DOI] [PubMed] [Google Scholar]
- 16.Salami S.S., Hovelson D.H., Kaplan J.B., Mathieu R., Udager A.M., Curci N.E., Lee M., Plouffe K.R., de la Vega L.L., Susani M. Transcriptomic heterogeneity in multifocal prostate cancer. JCI Insight. 2018;3(21) doi: 10.1172/jci.insight.123468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carm K.T., Hoff A.M., Bakken A.C., Axcrona U., Axcrona K., Lothe R.A., Skotheim R.I., Løvf M. Interfocal heterogeneity challenges the clinical usefulness of molecular classification of primary prostate cancer. Scientific Reports. 2019;9(1):13579. doi: 10.1038/s41598-019-49964-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Epstein J.I., Egevad L., Amin M.B., Delahunt B., Srigley J.R., Humphrey P.A., Committee t.G. Grading of Prostatic Carcinoma: Definition of Grading Patterns and Proposal for a New Grading System. The American journal of surgical pathology. The 2014 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason. 2016;40(2):244–252. doi: 10.1097/PAS.0000000000000530. [DOI] [PubMed] [Google Scholar]
- 19.De Vivar A.D., Sayeeduddin M., Rowley D., Cubilla A., Miles B., Kadmon D., Ayala G. Histologic features of stromogenic carcinoma of the prostate (carcinomas with reactive stroma grade 3) Human pathology. 2017;63:202–211. doi: 10.1016/j.humpath.2017.02.019. [DOI] [PubMed] [Google Scholar]
- 20.Sæter T., Vlatkovic L., Waaler G., Servoll E., Nesland J.M., Axcrona K., Axcrona U. The prognostic value of reactive stroma on prostate needle biopsy: A population-based study. The Prostate. 2015;75(6):662–671. doi: 10.1002/pros.22957. [DOI] [PubMed] [Google Scholar]
- 21.van Leenders G.J.L.H., van der Kwast T.H., Grignon D.J., Evans A.J., Kristiansen G., Kweldam C.F., Litjens G., McKenney J.K., Melamed J., Mottet N. The 2019 International Society of Urological Pathology (ISUP) Consensus Conference on Grading of Prostatic Carcinoma. Am J Surg Pathol, 2020;44(8) doi: 10.1097/PAS.0000000000001497. p. e87-e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han M., Partin A.W., Pound C.R., Epstein J.I., Walsh P.C. Long-term biochemical disease-free and cancer-specific survival following anatomic radical retropubic prostatectomy. The 15-year Johns Hopkins experience. Urol Clin North Am. 2001;28(3):555–565. doi: 10.1016/s0094-0143(05)70163-4. [DOI] [PubMed] [Google Scholar]
- 23.Prensner J.R., Zhao S., Erho N., Schipper M., Iyer M.K., Dhanasekaran S.M., Magi-Galluzzi C., Mehra R., Sahu A., Siddiqui J. RNA biomarkers associated with metastatic progression in prostate cancer: a multi-institutional high-throughput analysis of SChLAP1. The Lancet. Oncology. 2014;15(13):1469–1480. doi: 10.1016/S1470-2045(14)71113-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKenney J.K., Wei W., Hawley S., Auman H., Newcomb L.F., Boyer H.D., Fazli L., Simko J., Hurtado-Coll A., Troyer D.A. Histologic grading of prostatic adenocarcinoma can be further optimized: Analysis of the relative prognostic strength of individual architectural patterns in 1275 patients from the canary retrospective cohort. Am J Surg Pathol. 2016;40(11):1439–1456. doi: 10.1097/PAS.0000000000000736. [DOI] [PubMed] [Google Scholar]
- 25.Epstein J.I., Amin M.B., Fine S.W., Algaba F., Aron M., Baydar D.E., Beltran A.L., Brimo F., Cheville J.C., Colecchia M. The 2019 Genitourinary Pathology Society (GUPS) White Paper on Contemporary Grading of Prostate Cancer. Arch Pathol Lab Med. 2021;145(4):461–493. doi: 10.5858/arpa.2020-0015-RA. [DOI] [PubMed] [Google Scholar]
- 26.Loeb S., Berglund A., Stattin P. Population based study of use and determinants of active surveillance and watchful waiting for low and intermediate risk prostate cancer. The Journal of urology. 2013;190(5):1742–1749. doi: 10.1016/j.juro.2013.05.054. [DOI] [PubMed] [Google Scholar]
- 27.Tomlins S.A., Rhodes D.R., Perner S., Dhanasekaran S.M., Mehra R., Sun X.W., Varambally S., Cao X., Tchinda J., Kuefer R. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310(5748):644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 28.Pettersson A., Graff R.E., Bauer S.R., Pitt M.J., Lis R.T., Stack E.C., Martin N.E., Kunz L., Penney K.L., Ligon A.H. The TMPRSS2:ERG Rearrangement, ERG Expression, and Prostate Cancer Outcomes: A Cohort Study and Meta-analysis. Cancer Epidemiology Biomarkers &amp. Prevention. 2012;21(9):1497. doi: 10.1158/1055-9965.EPI-12-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crea F., Watahiki A., Quagliata L., Xue H., Pikor L., Parolia A., Wang Y., Lin D., Lam W.L., Farrar W.L. Identification of a long non-coding RNA as a novel biomarker and potential therapeutic target for metastatic prostate cancer. Oncotarget. 2014;5(3):764–774. doi: 10.18632/oncotarget.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang F., Ren S., Chen R., Lu J., Shi X., Zhu Y., Zhang W., Jing T., Zhang C., Shen J. Development and prospective multicenter evaluation of the long noncoding RNA MALAT-1 as a diagnostic urinary biomarker for prostate cancer. Oncotarget. 2014;5(22):11091–11102. doi: 10.18632/oncotarget.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Helsmoortel H., Everaert C., Lumen N., Ost P., Vandesompele J. Detecting long non-coding RNA biomarkers in prostate cancer liquid biopsies: Hype or hope? Non-coding RNA Research. 2018;3(2):64–74. doi: 10.1016/j.ncrna.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, RIS, upon reasonable request.